Abstract
Biochemical characterization of the purified bile salt hydrolase (BSH) from Bifidobacterium bifidum ATCC 11863 revealed some distinct characteristics not observed in other species of Bifidobacterium. The bsh gene was cloned from B. bifidum, and the DNA flanking the bsh gene was sequenced. Comparison of the deduced amino acid sequence of the cloned gene with previously known sequences revealed high homology with BSH enzymes from several microorganisms and penicillin V amidase (PVA) of Bacillus sphaericus. The proposed active sites of PVA were highly conserved, including that of the Cys-1 residue. The importance of the SH group in the N-terminal cysteine was confirmed by substitution of Cys with chemically and structurally similar residues, Ser or Thr, both of which resulted in an inactive enzyme. The transcriptional start point of the bsh gene has been determined by primer extension analysis. Unlike Bifidobacterium longum bsh, B. bifidum bsh was transcribed as a monocistronic unit, which was confirmed by Northern blot analysis. PCR amplification with the type-specific primer set revealed the high level of sequence homology in their bsh genes within the species of B. bifidum.
Bifidobacteria are one of the major constituents of the human gastrointestinal (GI) microflora (15, 41, 43), and they have attracted particular attention due to their potential health-promoting properties (3). In the GI tracts of humans and animals, most intestinal bacteria encounter significant amounts of bile salts, which are continuously present via enterohepatic circulation (2). Bile salts are synthesized mainly from cholesterol, conjugated with glycine or taurine in the liver, stored in the gall bladder, and released into the duodenum in response to the ingestion of fatty food. In addition to their function in the intestine as natural emulsifiers, bile salts possess some detergent-like antimicrobial properties. Some bacterial species have developed mechanisms to resist the detergent action of bile salts and have evolved to transform bile salts biochemically. Among the biochemical modifications of bile salts that are exhibited by many GI microorganisms, hydrolysis of the conjugated bile salts is considered the primary metabolic activity because bile salts need to be deconjugated before further sterol transformations take place (4). The enzyme responsible, bile salt hydrolase (BSH) (EC 3.5.1.24), has been widely studied in Bacteroides fragilis subsp. fragilis (44), Clostridium perfringens (16), Enterococcus spp. (23), Xanthomonas maltophilia (9), Listeria monocytogenes (11), Lactobacillus spp. (26, 36, 47, 50), and Bifidobacterium spp. (17, 47). In particular, the genus Bifidobacterium has been reported to possess higher BSH activity than other bacterial groups. In previous work (22), BSH enzymes were purified from five strains of bifidobacteria, and three different types of enzyme were identified based on the biochemical characteristics. To date, the bsh genes of C. perfringens (7), Lactobacillus plantarum (5), Lactobacillus johnsonii (12), Bifidobacterium longum (48), and Listeria monocytogenes (11) have been cloned and characterized. With the advent of the genomics era, many microbial genome-sequencing projects are providing several homologous bsh gene sequences.
Bifidobacteria are among the most common genera in the human colon and have been considered as key commensals in promoting host health, but very little is known about the genetics of the genus Bifidobacterium. Among the 32 species of bifidobacteria (21), B. longum strains have been the most studied. Bifidobacterium bifidum is one of the major bifidobacterial species commonly detected in human feces (29), and it was proposed that a high level of B. bifidum was an indication of the typical Bifidobacterium flora in healthy infants as opposed to low levels in allergic infants (19). Attempts have been made in recent years to define some beneficial effects of B. bifidum strains, including antibacterial activities (1, 39), immunostimulating activity (24, 35), antioxidative properties (20), production of bacteriocins (53), improvement of the microbial balance (6), and reduction of inflammation in broiler chickens (14).
For the identification and detection of the bifidobacterial species, genus- and species-specific primers have been developed, mostly on the basis of 16S rRNA sequences (29). Since BSH activity is commonly detected in almost all species of Bifidobacterium (47), further investigation of the conserved and variable regions of the bsh genes from various species could be useful for the development of alternative phylogenetic markers for the genus Bifidobacterium.
In this report, we describe the molecular cloning, sequencing, and characterization of a BSH enzyme from B. bifidum ATCC 11863. The putative bsh promoter sequence was analyzed by primer extension to determine the transcriptional start point (TSP). The bsh gene sequences were obtained by PCR cloning of B. bifidum ATCC 15696 and ATCC 29521, and the sequence comparison revealed the high level of homology (more than 98% nucleotide sequence identity) within the species of B. bifidum.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. All Bifidobacterium strains were verified for genus and species by 16S rRNA gene sequence typing according to the method of Kaufmann et al. (21). Bifidobacteria were propagated anaerobically at 37°C in MRS medium (Difco, Detroit, Mich.) supplemented with 0.05% (wt/vol) cysteine HCl. Escherichia coli cells were propagated at 37°C in Luria-Bertani (LB) broth with vigorous shaking or on LB medium solidified with 1.5% agar. When appropriate, ampicillin (200 μg/ml) and kanamycin (40 μg/ml) were added.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Origin or relevant characteristics | Source or reference |
|---|---|---|
| Bacterial strains | ||
| B. bifidum ATCC 11863 | Wild type for cloning | ATCC |
| B. bifidum ATCC 35914 | Human feces | ATCC |
| B. bifidum ATCC 15696 | Infant intestine | ATCC |
| B. bifidum ATCC 29521 | Type strain; infant feces | ATCC |
| B. bifidum KL 301 | Dairy isolate | This study |
| B. bifidum KL 306 | Dairy isolate | This study |
| B. adolescentis ATCC 15703 | Type strain; adult intestine | ATCC |
| B. longum ATCC 15707 | Type strain; adult intestine | ATCC |
| B. infantis ATCC 15697 | Type strain; infant intestine | ATCC |
| B. breve ATCC 15707 | Type strain; infant intestine | ATCC |
| E. coli DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17 (rK− mK+) deoR | GIBCO-BRL |
| E. coli BL21(DE3) | F−ompT hsdSB (rB− mB−) gal dcm lacY1 (DE3) | Novagen |
| Plasmids | ||
| pBR322 | Apr; cloning vector | Invitrogen |
| pUC19 | Apr; cloning vector | Invitrogen |
| pDrive | Apr Kanr; U-overhang PCR cloning vector | QIAGEN |
| pET36b(+) | Kanr; E. coli expression vector | Novagen |
| pBSH14 | pBR322 with 3.5-kb B. bifidum ATCC 11863 chromosomal insert; BSH+ | This study |
| pBSH27 | pBR322 with 7.5-kb B. bifidum ATCC 11863 chromosomal insert; BSH+ | This study |
| pBSH274 | pUC19 with 4.0-kb PstI insert from pBSH27; BSH+ | This study |
| pDB150 | pDrive with 1.5-kb PCR product; BSH+ | This study |
| pUCB150 | pUC19 with 1.5-kb HindIII & KpnI insert from pDB150; BSH+ | This study |
| pDB095 | pDrive with 950-bp PCR product; BSH+ | This study |
| pUCB095 | pUC19 with 950-bp HindIII & KpnI insert from pDB095; BSH− | This study |
| pBSH36b | pET36b(+) with 950-bp NdeI & HindIII insert from pDB095; BSH+ | This study |
| pBCS | pDB095 with Cys-1-Ser mutation; BSH− | This study |
| pBCT | pDB095 with Cys-1-Thr mutation; BSH− | This study |
Enzyme purification.
Anion-exchange chromatography (Mono Q HR 5/5; Amersham Pharmacia Biotech, Inc., Baie d'Urfé, Québec, Canada) was performed as described by Kim et al. (22). Cell extracts were applied to a column that was equilibrated with buffer A (50 mM bis-Tris propane buffer [pH 6.5]). Proteins were eluted by a linear gradient of 1 M sodium chloride in buffer A at a flow rate of 0.5 ml/min. Fractions (each, 1 ml) were collected and assayed for BSH activity. Hydrophobic interaction chromatography (HIC) was carried out with a HiTrap Butyl FF column (Amersham Pharmacia Biotech, Inc.). The active BSH fractions from the Mono Q column were pooled, desalted, and further concentrated with the Ultrafree-15 centrifugal filter unit (30 molecular weight cutoff; Millipore, Bedford, Mass.). Portions (each, 2 ml) of concentrated sample were applied to the HIC columns and eluted at a flow rate of 0.5 ml/min by a decreasing linear ammonium sulfate gradient (0.8 to 0 M) in 50 mM sodium phosphate buffer (pH 7.0). Fractions exhibiting BSH activity were pooled and assayed for protein and enzymatic activity as described below.
Determination of N-terminal amino acid sequence.
The resulting active fractions were pooled, concentrated, and applied to a mini-electrophoresis unit (Bio-Rad, Hercules, Calif). The purity of the pooled sample was evaluated after the sample was stained with Coomassie brilliant blue R-250. N-terminal amino acid sequencing of the purified BSH was carried out with a Procise protein sequencing system (Applied Biosystems, Foster City, Calif.).
Enzyme assays.
For bifidobacteria, BSH activity in cell extracts was measured by the hydrolysis of sodium taurocholate and/or sodium glycocholate (Sigma, St. Louis, Mo.) at 37°C in sodium phosphate buffer (0.1 M; pH 6.5). The amounts of the amino acids released from conjugated bile salts were measured by the ninhydrin assay (22). One unit of BSH activity was defined as the amount of enzyme that liberated 1 μmol of amino acids from the substrate per min. Specific activity was defined as the number of units per milligram of protein. Protein concentrations were determined by the Bio-Rad (Mississauga, Ontario, Canada) protein assay with bovine serum albumin as a standard. For the measurement of BSH activity in E. coli cells, assays were performed similarly, except that whole cells were disrupted with B-PER reagent (Pierce, Rockford, Ill.) according to the manufacturer's protocol.
Activity staining of polyacrylamide gel.
The BSH activity staining was carried out with a nondenaturing 10% (wt/vol) acrylamide gel by the method of Kim et al. (22). BSH activity band in the gel was visualized by the formation of a white precipitate of deoxycholic acid at the position of the enzyme after the gel was incubated in the reaction mixture containing 10 mM sodium taurodeoxycholate as a substrate for the enzyme.
DNA isolation, manipulation, and transformation.
Bifidobacterium genomic DNA was isolated according to the method of Meile et al. (31), with some modifications. Briefly, cells were harvested by centrifugation from 100 ml of an early-stationary-phase culture in MRS, washed with GTE (50 mM glucose, 25 mM Tris [pH 8.0], 10 mM EDTA) and incubated in lysis buffer (GTE containing 15 mg of lysozyme, 1 kU of mutanolysin, and 50 μg of RNase per ml) at 37°C for 1 h. Cells were lysed with 5% sodium dodecyl sulfate (SDS) at 65°C for 15 min. The cell lysates were extracted first with phenol and then with phenol-chloroform (1:1 [vol/vol]) and chloroform. DNA was precipitated by isopropanol, washed with 70% ethanol, and dissolved in TE buffer (10 mM Tris-1 mM EDTA [pH 8]). Small-scale E. coli plasmid preparations were performed with the QIAprep Spin Miniprep kit (QIAGEN). All of the DNA manipulations in this study were performed according to standard procedures as described previously (40). T4 DNA ligase and other DNA modifying enzymes were purchased from New England Biolabs, Inc., Invitrogen Life Technologies, or Amersham Pharmacia Biotech, Inc., and used according to the manufacturers' specifications. Electroporation was performed with a Gene-Pulser II electroporation apparatus (Bio-Rad) according to the manufacturer's specifications.
Construction and screening of B. bifidum BSH genomic library.
The isolated chromosomal DNA from B. bifidum ATCC 11863 was partially digested with the restriction enzyme Sau3A and analyzed by DNA electrophoresis. DNA fragments ranging from 2.0 to 8.0 kb were isolated with the QIAquick gel extraction kit (QIAGEN) and then ligated to the pBR322 vector that had been restricted with BamHI and dephosphorylated. The ligation mixture was transformed into E. coli DH5α-competent cells to create a plasmid library. The transformed E. coli was plated on LBGCT medium (LB agar plates containing 1% glucose, 0.035% calcium chloride, and 3 mM taurodeoxycholic acid), and BSH-positive clones were detected based on the formation of deoxycholate precipitate around the colony (7).
PCR.
The PCRs were performed with genomic DNA or plasmid DNA from BSH-positive clones as templates for amplifying the target genes. When appropriate, restriction sites were designed at the 5′ end of the primers to facilitate future cloning steps. Template DNA and primers were added to 50 μl of PCR mixture containing 200 μM each deoxynucleoside triphosphate, PCR buffer, and 2.5 U of HotStarTaq DNA polymerase (QIAGEN). The PCR was conducted in a Perkin Elmer GeneAmp system with an initial activation step at 95°C for 15 min, followed by 35 cycles each consisting of a denaturation step at 94°C for 1 min, an annealing step at 55 to 65°C for 30 s, and an extension step at 72°C for 1 min. A final extension step of 10 min at 72°C was performed to ensure complete amplification of all DNA fragments. PCR products were analyzed by electrophoresis in agarose gels containing ethidium bromide (1 μg/ml) and purified with the QIAquick PCR purification kit (QIAGEN).
DNA sequencing and sequence analysis.
The nucleotide sequences were determined by AmpliTaq FS DNA polymerase fluorescent dye terminator reactions with an Applied Biosystems 373 stretch automated sequencer. Assembly and analysis of DNA sequences were performed with DNASIS for Windows (Hitachi Software). Protein homology searches were performed with the Basic Local Alignment Search Tool (BLAST) available at the website of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/).
RNA manipulation.
RNA was isolated from B. bifidum with the RNeasy kit (QIAGEN), starting with digestion of the bacterial cell wall in diethyl pyrocarbonate-treated TE buffer containing 15 mg of lysozyme and 1 kU of mutanolysin per ml for 15 min at 37°C. Purified RNA was treated with RNase-free DNase according to the manufacturer's instructions (QIAGEN) and resuspended in RNase-free water. The absence of any significant DNA contamination of the RNA was confirmed by PCR. The RNA was quantified by spectrophotometric measurement at an optical density of 260 nm and stored at −70°C.
Identification of bsh promoter sequence.
Promoter sequences were predicted by using the Neural Network Promoter Prediction program (NNPP), version 2.2 (http://www.fruitfly.org/seq_tools/promoter.html). To verify the predicted promoter sequence and to determine the TSP of the mRNA, primer extension analysis was performed under conditions described by Swartzman et al. (46). The reverse primer PEA-1 (Table 2), corresponding to nucleotide positions 11 to 30 of the bsh gene, was 5′-end labeled with [γ-32P]ATP by using T4 polynucleotide kinase (Boehringer Mannheim). The purified labeled primer was sealed in a glass microcapillary tube with total RNA (20 μg) in a total volume of 10 μl containing piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) at pH 6.6 and 0.4 M NaCl. Hybridization was carried out at 55°C for 12 to 16 h. The primer was extended using Moloney murine leukemia virus reverse transcriptase (Invitrogen). At the end of the reaction, 2 μl of EDTA (0.5 M) and 1 μl of RNase (10 mg/ml) were added. The labeled cDNAs were purified, heat denatured in 50% formamide and 5 mM EDTA (pH 7.5), and electrophoresed on a sequencing gel (8% [wt/vol] acrylamide-7 M urea). The same primer and a PCR product corresponding to the region from the bsh gene, containing the putative TSP, were used to generate a sequencing ladder by the dideoxy-chain termination method with a T7 sequencing kit (USB Corp., Cleveland, Ohio). The resultant gel was dried and exposed to BioMax MS film (Eastman Kodak, Rochester, N.Y.).
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Nucleotide sequence (5′-3′)a | Locationb | Use |
|---|---|---|---|
| BSH-F | AGTCCATATGTGCACTGGTGTCCGTTTCTCC | 1-24 | Cloning |
| BSH-R | AGTCAAGCTTCAATCGGCGGTGATCAGCTCG | 951-931 | Cloning |
| BF-SER | AGTCCATATGTCAACTGGTGTCCGTTTCTCC | 1-24 | Cys-1-Ser mutation primer |
| BF-THR | AGTCCATATGACTACTGGTGTCCGTTTCTCC | 1-24 | Cys-1-Thr mutation primer |
| BBI-F | CTATGAGTATGGGGCCGAAG | 120-139 | Screening |
| BBI-R | GTTCCGCCTTGCCCCAAGTG | 613-594 | Screening |
| PEA-F | GCGGCATGTACTACGAGGAG | −555 to −536 | Primer extension sequencing |
| PEA-1 | ATCGTCGGAGAAACGGACAC | 30-11 | Primer extension |
Engineered NdeI and HindIII sites are underlined. Changed nucleotide sequences for the site-directed mutagenesis are in boldface.
Based on the ATG start codon as a starting point (position 1).
Northern blotting.
RNA (20 μg) was separated on formaldehyde agarose gels prepared as described previously (40). After the transfer, RNAs were cross-linked to the nylon membrane by UV irradiation. PCR amplicons obtained with primers BBI-F and BBI-R were radiolabeled with [α-32P]dCTP (Amersham) in a Klenow reaction mixture (NEB) according to the manufacturer's specifications. DNA-RNA hybridization was performed as described by Miyamoto et al. (32). The sizes of the transcripts were estimated by direct comparison to a molecular RNA ladder (Invitrogen).
Expression of bsh in E. coli.
To create the plasmid pBSH36b, the bsh gene was amplified with the primers BSH-F and BSH-R (Table 2). An NdeI site was designed in primer BSH-F and a HindIII site was created in primer BSH-R to include the start codon sequence and the stop codon (TGA) sequence, respectively. Cloning into the NdeI-HindIII sites of pET36b resulted in the translational fusion of the bsh gene to the T7 promoter and E. coli ribosome binding site (RBS) of the plasmid. Plasmid pBSH36b was created in E. coli DH5α and transformed into E. coli BL21(DE3) to perform the overexpression studies. For the overexpression, cells (optical density at 600 nm, 0.6) were induced with 1.0 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 37°C for 3 h. Samples were taken at 1-h intervals to measure growth and BSH activity. For the construction of the mutant by PCR-based site-directed mutagenesis, two forward primers (BF-SER and BF-THR) (Table 2) were designed to replace Cys-1 with Ser and Thr, respectively.
Nucleotide sequence accession numbers.
The bsh nucleotide sequences for B. bifidum strains have been deposited in the GenBank database under accession numbers AY506536, AY604516, and AY604517.
RESULTS
Strain selection and BSH purification.
Among many strains of bifidobacteria (Table 1), B. bifidum ATCC 11863 was selected on the basis of the high level of BSH activity as well as the distinct biochemical characteristics that are different from those of B. longum and Bifidobacterium infantis strains (22). Three different types of BSH enzyme (types A, B, and C) from the genus Bifidobacterium have previously been proposed (22), in which B. bifidum BSH belongs to type B.
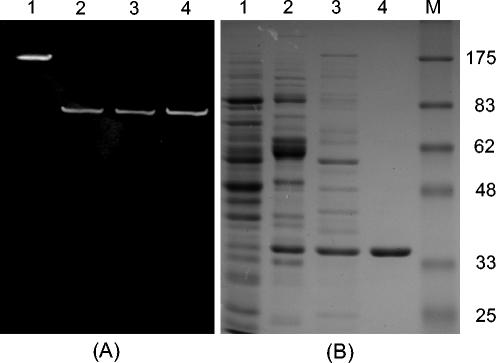
For the purification of the BSH enzyme, the conditions were optimized with anion-exchange and HIC columns. Although BSH activity was eluted from a Mono Q column at an NaCl concentration of between 0.35 and 0.38 M linear gradients, conditions were further optimized by a stepwise gradient using the same molarity to improve selectivity of this column for the BSH enzyme. The established conditions with a Mono Q column were fairly effective in purifying the active BSH from the cell extracts (Fig. 1B, lane 3). When the cell extracts were applied to a Hi-Trap Butyl-FF HIC column, the selectivity profile on the column was very different from that of the Mono Q column. The BSH enzyme could thus be purified to electrophoretic homogeneity by combining these two chromatographic steps (Fig. 1B, lane 4). The enzyme was purified 33-fold over the crude extract, and the yield was about 29%. The subunit molecular mass was estimated to be 35 kDa by SDS-polyacrylamide gel electrophoresis (PAGE), and the native mass was estimated to be about 150 kDa by gel filtration chromatography.
FIG. 1.
(A) Activity staining on a nondenaturing polyacrylamide gel. Lane 1, BSH enzyme from L. acidophilus ATCC 53546 (as a positive control); lane 2, soluble fraction of B. bifidum ATCC 11863 from sonication treatment; lane 3, purified native BSH; lane 4, soluble fraction of IPTG-induced E. coli BL21(DE3) containing pBSH36b from B-PER treatment. (B) SDS-PAGE analysis of cell extracts at each purification step. Lanes 1 through 4: active fractions from B. bifidum ATCC 11863 (lane 1, cell extract; lane 2, HIC column; lane 3, anion-exchange column; lane 4, anion-exchange column plus HIC); lane M, marker proteins (molecular masses in kilodaltons are indicated on the right).
The BSH activity staining on a native PAGE gel revealed that the cell extract showed only one active band with the same Rf value as that of the purified and recombinant BSH enzyme (Fig. 1A).
The N-terminal amino acid sequence of the purified enzyme is XTGVRFSDDEGNMYFGRNLD. A protein sequence comparison showed that this sequence was homologous to the N-terminal amino acid sequences of the BSH enzymes from many enteric bacteria, with the highest identity to that of the B. longum BSH enzyme (42, 48). Only one difference between the amino acids, Thr13 of B. longum BSH and Met13 from B. bifidum BSH, was found.
Cloning and sequence analysis.
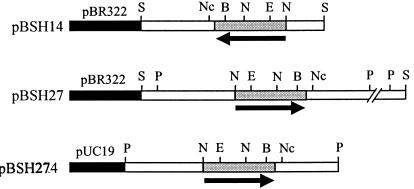
To identify the gene(s) encoding BSH activity in B. bifidum ATCC 11863, an Sau3AI-digested genomic library was created in pBR322 and screened in E. coli DH5α cells. Two positive clones were identified by the formation of white precipitate around the colonies on the selective medium (LBGCT) supplemented with 200 μg of ampicillin/ml and were designated pBSH14 and pBSH27. Preliminary restriction enzyme analysis revealed that pBSH14 contained a 3.5-kb insert and that pBSH27 contained a 7.5-kb insert. With the PstI site of the insert in pBSH27, two subclones, pBSH272 (2.0-kb insert) and pBSH274 (4.0-kb insert), were constructed in pUC19 (Fig. 2), with pBSH274 showing BSH activity. Sequence analysis of the 4.0-kb insert revealed that it included the overlapping fragment of pBSH14. In this sequence, one complete open reading frame (ORF) was detected, and this ORF encoded a 316-amino-acid protein (Fig. 3). The N-terminal amino acid sequence of the deduced protein was identical to that of the purified BSH enzyme. The deduced protein had a theoretical molecular mass of 35,144 Da and a pI of 4.48. The theoretical data derived from the deduced protein were in good agreement with the measured biochemical data obtained from the purified enzyme.
FIG. 2.
Map of plasmids pBSH14, pBSH27, and pBSH274. Black boxes show cloning vectors pBR322 and pUC19, grey boxes indicate the position of the bsh gene, and white boxes are the cloned sequences outside the bsh gene. The arrows indicate the direction of the bsh gene. Restriction enzyme sites used for mapping the clones are shown above the maps (S, Sau3A; Nc, NcoI; B, BamHI; N, NdeI; E, EcoRI; and P, PstI).
FIG. 3.
Nucleotide sequence of the bsh gene. The N-terminal amino acid sequence determined from the purified BSH is underlined. The amino acid sequences of the proposed active site residues (C-1, D-20, N-81, N-172, and R-225) are in double-underlined boldface type. Variable regions used for the B. bifidum-specific primer set are shaded.
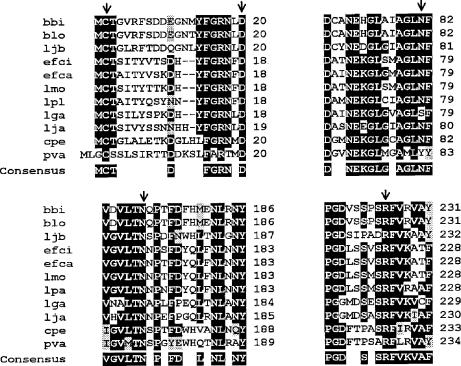
Comparison of the predicted amino acid sequence to the database by BLAST analysis revealed highest similarities to the BSH of B. longum strains (91% identity and 95% similarity) (42, 48). Significant similarities were also found to the BSH enzymes of several Lactobacillus strains (35 to 43% identity) (GenBank accession numbers AF054971, AF305888, AF091248, AF297873, and M96175), as well as to BSH enzymes of Enterococcus strains (36 to 37%) (GenBank accession numbers AY032999 and AY260046), L. monocytogenes (37%) (11), C. perfringens (36%) (7), and penicillin V amidase (PVA) of Bacillus sphaericus (30%) (GenBank accession number M15660) (Fig. 4).
FIG. 4.
Multiple alignment of BSHs of various bacteria and PVAs of B. sphaericus based on the sequences around the proposed active sites of B. sphaericus PVA. Abbreviations for BSHs: bbi, B. bifidum; blo, B. longum; ljb, L. johnsonii BSH-β; efci, Enterococcus faecium; efca, Enterococcus faecalis; lmo, L. monocytogenes; lpa, L. plantarum; lga, Lactobacillus gasseri; lja, L. johnsonii BSH-α; cpe, C. perfringens; pva, B. sphaericus PVA. Conserved amino acids are highlighted in black boxes, and similar amino acids are in grey boxes. Arrows indicate the amino acids proposed to be the key residues in the active sites of B. sphaericus PVA, including C-1, D-20, Y-82, N-175, and R-228. Positions are based on the PVA of B. sphaericus, starting with the Cys residues at the N-terminal end of mature proteins (BSH and PVA).
Characterization of bsh transcript and bsh promoter.
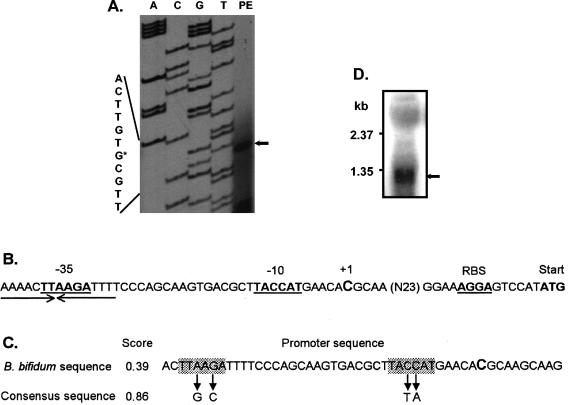
The bsh TSP was identified by primer extension with total RNA isolated from log-phase B. bifidum ATCC 11863 cells. The first base of the transcript was identified as the cytosine located 42 bp upstream of the proposed bsh ATG start codon (Fig. 5). The experimentally verified TSP of +1 was in accordance with the position predicted by NNPP software, version 2.2. The relatively low score value (0.39) (Fig. 5C) of the predicted promoter sequence was partly due to the potential −35 (TTAAGA) and −10 (TACCAT) sequences that are different from those of the consensus hexamer counterparts. A putative rho-independent type transcription terminator sequence (ΔG = −75.5 kcal/mol at 25°C) could be recognized 75 nucleotides downstream of the stop codon, suggesting that the bsh gene is transcribed monocistronically. Furthermore, hybridizations of total RNA from B. bifidum cells with the internal probe revealed a distinct signal of an approximately 1.2-kb mRNA transcript (Fig. 5), indicating that the bsh gene is not cotranscribed with other adjacent genes.
FIG. 5.
Characterization of the bsh transcript and bsh promoter. (A) Identification of the 5′ terminus of the bsh transcript by primer extension. The TSP (+1; indicated by an arrow) is shown in the right lane. The DNA sequence is shown on the left and +1 is indicated by an asterisk. (B) Schematic organization of the bsh promoter. The putative −10 and −35 motifs of the bsh promoter are underlined, and the proposed RBS and start codon (ATG) of bsh are indicated in boldface. The TSP (+1) is enlarged. An inverted repeat sequence in the −35 region is indicated by arrows. (C) A promoter sequence predicted with NNPP software, version 2.2. The consensus −35 (TTGACA) and −10 (TATAAT) sequences are presented under the arrows, and the proposed TSP is shown in enlarged type. Score, the fitness value to the consensus promoter sequences. (D) Northern hybridization of total RNA with the probe prepared with the primers BBI-F and BBI-R. The 1.2-kb transcript corresponding to the bsh transcript is indicated by an arrow.
In the region preceding the ATG start codon of B. bifidum bsh, a putative RBS, a purine-rich region (AAAGGA), was found; this sequence is complementary to the 3′ end of B. bifidum 16S rRNA (3′-UCUUUCCUCC-5′) (Table 3). This potential base-pairing region between mRNA and 16S rRNA is also observed in many other genes of bifidobacteria (33), and the sequence of the 3′ end of 16S rRNA is well conserved in other species of bifidobacteria (Table 3).
TABLE 3.
Alignments of the region immediately upstream of the ATG start colon in Bifidobacterium genes and B. bifidum bsh with the 3′-terminal consensus sequence of 16S RNA from the genus Bifidobacterium
| Organism or sequence | Gene or gene encoding: | Sequence upstream from start codona | ΔG (kcal/mol)b | Distance (nt)c |
|---|---|---|---|---|
| B. breve D88311d | β-Glucosidase | ACTAGAAAGGAATCACCG ATG | −16.2 | 7 |
| B. adolescentisAF124596 | α-Galactosidase | CAAGAAAAGGATGCTGCA ATG | −11.8 | 7 |
| B. adolescentisAF213175 | β-Galactosidase | AAAACAAAGGAGTGGAT ATG | −14.0 | 6 |
| B. bifidumAJ224435 | β-Galactosidase | ATGAAGAAGGAACGTTT ATG | −10.6 | 6 |
| B. infantisAF192265 | β-Galactosidase | GACAGAAAGCAGGAGAAC ATG | −16.2 | 7 |
| B. animalisAJ293946 | F6PPK | GGAGTACAGGAGCACAC ATG | −11.6 | 6 |
| B. longumAE014718 | F6PPK | GGAGTACAGGAGTACAC ATG | −11.6 | 6 |
| B. longumAF148138 | bsh | GATGGAAGGGAGTCCGTT ATG | −12.8 | 7 |
| B. bifidum | bsh | CATGGAAAGGAGTCCAT ATG | −16.2 | 6 |
| 3′-End sequence of bifido- bacterial 16S rRNAe | 3′-TCTTTCCTCCACTAGG-5′ |
Potential base pairing between the purine-rich region of the genes and the complementary region near the 3′ end of 16S rRNA sequence is in boldface italic type. Double underlining indicates a highly conserved area from listed sequences. The central AGGA motif of the SD region in bifidobacteria is indicated in single-underlined boldface type and the ATG start codon is in boldface.
Free energy of complementarity of the SD and the 3′ end of 16S rRNA.
Distance in nucleotides (nt) between ATG and the core AGGA sequence in the SD region.
GenBank nucleotide sequence accession numbers are shown.
This sequence is highly conserved in many strains of the genus Bifidobacterium, including B. animalis DSM10140T (X89513), B. bifidum DSM20456T (S83624), B. infantis ATCC 15697T (U09792), B. breve ATCC 15698 (U09518), and B. adolescentis C1P6461 (U09514) (numbers in parentheses are GenBank accession numbers).
Heterologous expression of B. bifidum BSH in E. coli.
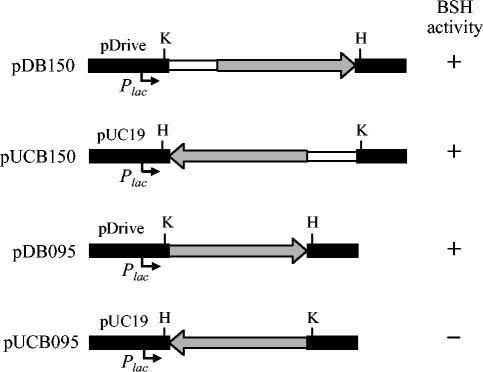
The Bifidobacterium BSH genes on plasmids pBSH14, pBSH27, and pBSH274 were constitutively expressed in E. coli, which was the case in the Bifidobacterium host. No significant difference in their expression levels among the cells containing those plasmids was observed, regardless of the different orientation of the inserts and the different vectors. This finding suggested that the bsh gene on these plasmids could be transcribed by its own promoter sequence and that the initiation of translation might be facilitated by the putative E. coli RBS (AGGA) located directly upstream of the Bifidobacterium bsh gene (Fig. 5). To confirm this hypothesis, PCR products with and without the 550-bp upstream region of the bsh gene were cloned into a PCR cloning vector (pDrive) and screened for BSH activity in E. coli DH5α cells. Colonies from those cells harboring pDB150 and pDB095 (Fig. 6) showed BSH activity when grown on the selective medium, LBGCT. However, when they were cloned in the direction opposite that of the lac promoter in pUC19, BSH activity was observed only from the clone containing the upstream region of the bsh gene in pUC150 (Fig. 6), suggesting that the putative Bifidobacterium bsh promoter and RBS are recognized by E. coli cells.
FIG. 6.
Map of plasmids pDB150, pUCB150, pDB095, and pUCB095. Black boxes indicate the vectors pDrive and pUC19, grey arrows indicate the position and orientation of the bsh gene (950 bp), and white boxes are for the upstream region (550 bp) of the bsh gene. Plac, lac promoter in the vectors. Restriction enzyme sites used for the cloning in the opposite direction are HindIII (H) and KpnI (K).
To facilitate purification and characterization of the enzyme, the BSH was overexpressed in E. coli by using the overexpression vector pET36b(+). The recombinant enzyme showed characteristics similar to those of the native enzyme during the chromatographic steps, and the same migration pattern was observed with native PAGE followed by activity staining (Fig. 1A). Using PCR-based site-directed mutagenesis, we constructed two B. bifidum bsh mutants in which Cys-1 was replaced by other nucleophilic amino acids, Ser (pBCS) or Thr (pBST), which have an OH group instead of an SH group at this position in the original BSH. The resulting mutations did not exhibit BSH activity, while the protein band was still present when they were overexpressed in E. coli with pET36(b) (data not shown). The bsh gene in the mutant plasmids was sequenced to examine unexpected mutations during the PCR cloning experiments, but no such changes were detected.
Substrate specificity of BSH.
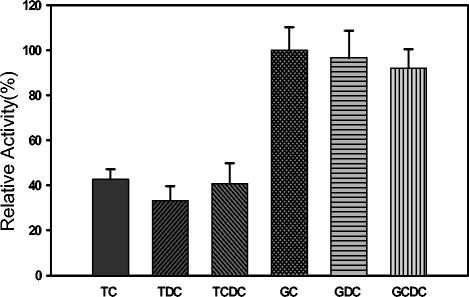
The substrate specificity of the BSH was determined in enzyme assays with the six major human bile salts (22). The BSH enzyme from B. bifidum ATCC 11863 showed the highest enzyme activity with glycocholic acid (defined as 100% activity). The enzymes exhibited a preference for glycine-conjugated bile salts over taurine-conjugated forms, and no difference was observed with di- or trihydroxyconjugated bile salts. (Fig. 7). Recombinant and native enzymes showed similar characteristics in their preference for bile salt substrates (data not shown). The substrate specificity of BSH from B. bifidum was more similar to that of type A than type C (22).
FIG. 7.
Substrate specificity of B. bifidum ATCC 11863 BSH. Six major human bile salts are shown: taurocholic acid (TC), taurodeoxycholic acid (TDC), taurochenodeoxycholic acid (TCDC) glycocholic acid (GC), glycodeoxycholic acid (GDC), and glycochenodeoxycholic acid (GCDC). The relative activity was calculated using GC as a standard at 100%.
PCR screening for B. bifidum strains.
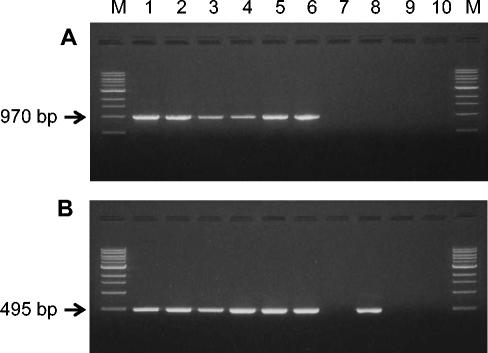
All of the B. bifidum strains screened in this study showed BSH activity towards the conjugated bile salts. To examine the distribution of the bsh locus, genomic DNA from each strain was screened by standard PCR with two primers, BSH-F and BSH-R. All the B. bifidum strains produced the same size (970 bp) of amplicon (Fig. 8). From the PCR cloning of this amplicon into the pDrive vector and screening in E. coli DH5α cells, BSH activity was detected with the amplicons from all the strains tested, suggesting that the bsh gene was selectively amplified from total genomic DNA with the primer set used. Another set of internal primers (BBI-F and BBI-R) was designed to produce a 495-bp amplicon targeting the variable region of the bsh sequence that is different from those of B. longum strains (42, 48). All of the B. bifidum strains and Bifidobacterium adolescentis ATCC 15703 produced the expected size of amplicon. However, the other BSH positive strains, including Bifidobacterium breve, B. infantis, and B. longum, were all negative in the PCR screening with the primer set developed in this study (Fig. 8), implying that these three species might have bsh gene sequences different from those of B. bifidum strains.
FIG. 8.
PCR products of B. bifidum strains with primers BSH-F and BSH-R (A) and BIF-F and BIF-R (B). Lanes: 1, B. bifidum ATCC 11863; 2, B. bifidum ATCC 35914; 3, B. bifidum ATCC 15696; 4, B. bifidum ATCC 29521; 5, B. bifidum KL 301; 6, B. bifidum KL 306; 7, B. longum ATCC 15707; 8, B. adolescentis ATCC 15703; 9, B. infantis ATCC 15697; and 10, B. breve ATCC 15700; M, 1-kb DNA ladder (New England Biolabs). PCR amplicons of 970 bp (A) were obtained with DNA extracted from B. bifidum strains, and amplicons of 495 bp (B) were obtained with DNA extracted from B. bifidum and B. adolescentis strains.
DISCUSSION
In this paper, we present the cloning and transcriptional analysis of a new BSH gene from B. bifidum ATCC 11863. In a previous report, Kim et al. (22) described the purification of three different types of BSH that are found in the genus Bifidobacterium, and their study reported some minor differences in their biochemical characteristics. In this study, supporting evidence for such differences is provided by molecular characterization; to our knowledge, this is the first report of a transcriptional analysis of a bifidobacterial bsh gene.
Purified B. bifidum BSH enzyme exhibited extensive similarities to B. longum BSH (48) with respect to the major characteristics such as subunit size (35 kDa) and homotetrameric structure (4 × 35 kDa = 140 kDa). The N-terminal amino acid sequence of the purified enzyme was the same as that of B. longum BSH, with only one difference. As observed in previous reports (13, 22, 48), the first amino acid was not resolved under experimental conditions. However, the cysteine can be deduced from the genetic data, and this residue is highly conserved in all BSH enzymes. Based on its striking structural similarity to PVA, the BSH enzyme has been proposed as a new member of the N-terminal nucleophile (designated Ntn) hydrolase superfamily with a Cys at the N-terminal end. This Cys residue serves as the nucleophile and proton donor in the catalytic process (45). The absence of a Met residue at the N-terminal end could be explained by the fact that the Cys-1 residue becomes a catalytic center after removal of the initiation formyl methionine by an autoproteolytic process, which is one of the common features of the Ntn hydrolase superfamily (34). The importance of the SH group in the N-terminal cysteine was confirmed by the fact that replacing Cys with other potential nucleophilic residues, Ser or Thr, results in an inactive enzyme.
From the homology comparison of the deduced protein to previously known sequences, B. bifidum BSH shares the highest amino acid sequence similarity with the BSH from B. longum strains (42, 48). The predicted ORF of 951 bp displayed 83% DNA sequence identity and 95% deduced amino acid sequence similarity to BSH from B. longum NCC 2705 (42). Such a discrepancy is due to different codon usage, mostly at the third position. However, DNA sequences flanking the bsh gene were not conserved in these two species. Partial ORFs flanking the bsh gene that have been identified by BLAST do not display significant sequence similarity to any known genes, indicating that they are either specific to B. bifidum or yet to be determined in other genomes.
Although the TSP of the bsh gene has been identified in L. monocytogenes (11), that of Bifidobacterium spp. has only now been determined by primer extension analysis. The putative bsh promoter sequence was different from the consensus promoter sequences in the consensus −35 (TTGACA) and −10 (TATAAT) hexamer sequences and in the absence of a TG motif upstream of the −10 hexamer. In addition, the spacer (20 nucleotides) between the region spanning −35 to −10 was longer than that of the consensus promoter sequences, which were reported to be 17 ± 1 nucleotides (18, 30). Interestingly, a palindromic sequence in the −35 region was identified (Fig. 5B), which was also found in the bsh promoter of L. monocytogenes (11). The spacer between the palindromic sequence and the −10 position, which was 16 nucleotides in B. bifidum and 18 nucleotides in L. monocytogenes, is close to the spacer of the consensus promoter sequences. Further experiments will be required to establish how this palindromic sequence works as an RNA polymerase recognition site in B. bifidum. Since little is known about the the vegetative RNA polymerase in Bifidobacterium species compared to those of other bacterial species, we could not exclude the possibility that the sequence investigated in this study is not representative of typical bifidobacterial −35 and −10 hexamers. Until now, there have been only a few reports of primer extension analysis being used to determine a TSP of genes from the genus Bifidobacterium (27, 38).
The putative rho-independent transcription terminator sequence located 75 nucleotides downstream of the stop codon of the bsh gene and the 1.2-kb transcript size shown by Northern hydridization indicates that the bsh gene of B. bifidum is transcribed as a monocistronic unit. Monocistronic bsh transcripts in L. plantarum (5) and L. monocytogenes (11) have been previously reported. On the contrary, Tanaka et al. (48) proposed that the bsh gene of B. longum SBT2928 is part of an operon in which the bsh gene is transcribed with at least one more gene, the glutamine synthetase gene (glnE). The same genetic organization was revealed from the recently completed genomic projects with B. longum NCC 2705 (42) and the ongoing project with B. longum DJO10A (www.jgi.doe.gov/JGI_microbial/html). Directly upstream of the bsh gene in both strains, a gene with high homology to the 5,10-methylenetetrahydrofolate reductase gene (metF) was found. If these three genes, metF, bsh, and glnE, were cotranscribed in one operon as reported by Tanaka et al. (48) it would be interesting to find out the functional relationships between the three enzymes they encode. To date, the extent of the operon in B. longum strains has not been revealed. Another polycistronic bsh transcript has been reported by Elkins et al. (13). They identified a complete BSH operon from L. johnsonii 100-100 and L. acidophilus KS-13, in which the bsh gene is transcribed with another gene for a putative conjugated bile salt transporter.
It has been proposed that acquisition of the bsh gene occurs by horizontal (lateral) gene transfer in lactobacilli (13) and in L. monocytogenes (11). Whether the bsh gene in the genus Bifidobacterium is duplicated or acquired by horizontal transfer is not known at this time. However, the following four facts contradict the hypothesis that the bsh gene in B. bifidum is acquired by horizontal gene transfer from other enteric bacteria. (i) The G+C content (58%) of the bsh gene reflects the overall G+C content (57 to 64%) of the Bifidobacterium genus chromosome. (ii) The bsh gene sequence was highly conserved within all the B. bifidum strains tested in this study. For the similarity comparison, two more bsh genes obtained by PCR amplification from B. bifidum ATCC 15696 and ATCC 29521 were sequenced, and they showed more than 98% nucleotide sequence similarity to the genes of B. bifidum ATCC 11863. Furthermore, three B. longum bsh genes available from GenBank showed a high level of similarity. (iii) There are no reports of BSH activity or the bsh gene from any G+C-rich gram-positive bacteria other than the Bifidobacterium species. (iv) In the genus Bifidobacterium, the BSH phenotype is not strain specific and is rather widespread among almost all the strains within the genus. This indicates that the bsh gene in the bifidobacterial genome could be a paralogous gene, not an orthologous gene acquired by lateral gene transfer. Given these facts, it appears that BSH activity is important at some level for bifidobacteria to respond efficiently to bile salts and to colonize at the lower part of the large intestine of human and animal.
Finally, it remains to be determined whether the BSH activity of the probiotics, including many commercially available Bifidobacterium strains, is beneficial or detrimental to the host. While the potential positive aspects of BSH activity of the probiotics have been previously discussed (10, 36, 37, 49), other possible negative concerns about BSH activity have also been raised (11, 28, 51). Recently, Kurdi et al. (25) proposed one possible beneficial consequence of BSH activity in bifidobacteria upon investigation of the cholic acid transport and accumulation in some intestinal Bifidobacterium strains, including two B. bifidum strains. They observed that cholic acid, the main free bile acid produced by BSH activity in the intestine, could accumulate inside the bacterial cell in the intestine so long as the bacteria were energized. The entrapment of free bile acids by bifidobacteria can contribute to the decreased production of secondary bile acids, which are considered cytotoxic and precarcinogenic. The enzyme responsible for this undesirable reaction, 7α-dehydroxylase, has been found in Clostridium and Eubacterium species (8, 52) but not in lactic acid bacteria or bifidobacteria (25). Since the hydrolytic products of BSH have properties toxic to cells and can damage the membrane of mammalian cells (51), the accumulation of free bile acids within BSH-producing bacteria could counteract the toxic nature of deconjugated bile salts. On the other hand, it has yet to be determined what advantage BSH activity provides Bifidobacterium strains. So far, there is no report of the ability of bifidobacteria to produce energy from bile salts or of any additional catabolic pathways of the steroid ring structure of bile salts in bifidobacteria. Further experiments will be needed to unveil the physiological significance of BSH activity for the enzyme-producing bacterial cells as well as for the mammalian hosts.
Acknowledgments
This research was supported by a research operating grant from Natural Sciences and Engineering Research Council of Canada (NSERC-222026) and by Medical Research Grant MT-7672. G.-B.K. is the recipient of a Fonds pour la Formation de Chercheurs et l'Aide a la Recherche (FCAR) scholarship from the Government of Quebec.
REFERENCES
- 1.Asahara, T., K. Shimizu, K. Nomoto, M. Watanuki, and R. Tanaka. 2001. Antibacterial effect of fermented milk containing Bifidobacterium breve, Bifidobacterium bifidum, and Lactobacillus acidophilus against indigenous Escherichia coli infection in mice. Microb. Ecol. Health Dis. 13:16-24. [Google Scholar]
- 2.Bahar, R. J., and S. Andrew. 1999. Bile acid transport. Gastroenterol. Clin. N. Am. 28:27-58. [DOI] [PubMed] [Google Scholar]
- 3.Ballongue, J. 1998. Bifidobacteria and probiotic action, p. 519-587. In S. Salminen and A. Von Wright (ed.), Lactic acid bacteria. Microbiology and functional aspects. Marcel Dekker, Inc., New York, N.Y.
- 4.Batta, A. K., G. Salen, R. Arora, S. Shefer, M. Batta, and A. Person. 1990. Side chain conjugation prevents bacterial 7-dehydroxylation of bile acids. J. Biol. Chem. 265:10925-10928. [PubMed] [Google Scholar]
- 5.Christiaens, H., R. J. Leer, P. H. Pouwels, and W. Verstraete. 1992. Cloning and expression of a conjugated bile acid hydrolase gene from Lactobacillus plantarum by using a direct plate assay. Appl. Environ. Microbiol. 58:3792-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung, Y. J., S. K. Kim, and E. C. Choi. 1997. The effects of Bifidobacterium bifidum OFR9, a strain resistant to antituberculosis and antileprosy agents, on fecal flora in mice. J. Gen. Appl. Microbiol. 43:61-66. [DOI] [PubMed] [Google Scholar]
- 7.Coleman, J. P., and L. L. Hudson. 1995. Cloning and characterization of a conjugated bile acid hydrolase gene from Clostridium perfringens. Appl. Environ. Microbiol. 61:2514-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coleman, J. P., W. B. White, M. Lijewski, and P. B. Hylemon. 1988. Nucleotide sequence and regulation of a gene involved in bile acid 7-dehydroxylation by Eubacterium sp. strain VPI 12708. J. Bacteriol. 170:2070-2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dean, M., C. Cervellati, E. Casanova, M. Squerzanti, V. Lanzara, A. Medici, P. P. de Laureto, and C. M. Bergamini. 2002. Characterization of cholylglycine hydrolase from a bile-adapted strain of Xanthomonas maltophilia and its application for quantitative hydrolysis of conjugated bile salts. Appl. Environ. Microbiol. 68:3126-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Smet, I., P. De Boever, and W. Verstraete. 1998. Cholesterol lowering in pigs through enhanced bacterial bile salt hydrolase activity. Br. J. Nutr. 79:185-194. [DOI] [PubMed] [Google Scholar]
- 11.Dussurget, O., D. Cabanes, P. Dehoux, M. Lecuit, C. Buchrieser, P. Glaser, and P. Cossart. 2002. Listeria monocytogenes bile salt hydrolase is a virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol. Microbiol. 45:1095-1106. [DOI] [PubMed] [Google Scholar]
- 12.Elkins, C. A., and D. C. Savage. 1998. Identification of genes encoding conjugated bile salt hydrolase and transport in Lactobacillus johnsonii 100-100. J. Bacteriol. 180:4344-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elkins, C. A., S. A. Moser, and D. C. Savage. 2001. Genes encoding bile salt hydrolases and conjugated bile salt transporters in Lactobacillus johnsonii 100-100 and other Lactobacillus species. Microbiology 147:3404-3412. [DOI] [PubMed] [Google Scholar]
- 14.Estrada, A., D. C. Wilkins, and M. Drew. 2001. Administration of Bifidobacterium bifidum to chicken broilers reduces the number of carcass condemnations for cellulitis at the abattoir. J. Appl. Poult. Res. 10:329-334. [Google Scholar]
- 15.Franks, A. H., H. J. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gopal-Srivastava, R., and P. B. Hylemon. 1988. Purification and characterization of conjugated bile salt hydrolase from Clostridium perfringens. J. Lipid Res. 29:1079-1085. [PubMed] [Google Scholar]
- 17.Grill, J. P., F. Schneider, J. Crociani, and J. Ballongue. 1995. Purification and characterization of conjugated bile salt hydrolase from Bifidobacterium longum BB536. Appl. Environ. Microbiol. 61:2577-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harley, C. B., and R. P. Reynolds. 1987. Analysis of E. coli promoter sequences. Nucleic Acids Res. 15:2343-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He, F., A. C. Ouwehand, E. Isolauri, H. Hashimoto, Y. Benno, and S. Salminen. 2001. Comparison of mucosal adhesion and species identification of bifidobacteria isolated from healthy and allergic infants. FEMS Immunol. Med. Microbiol. 30:43-47. [DOI] [PubMed] [Google Scholar]
- 20.Ito, M., H. Sawada, K. Ohishi, Y. Yoshida, W. Yokoi, T. Watanabe, and T. Yokokura. 2001. Suppressive effects of bifidobacteria on lipid peroxidation in the colonic mucosa of iron-overloaded mice. J. Dairy Sci. 84:1583-1589. [DOI] [PubMed] [Google Scholar]
- 21.Kaufmann, P., A. Pfefferkorn, M. Teuber, and L. Meile. 1997. Identification and quantification of Bifidobacterium species isolated from food with genus-specific 16S rRNA-targeted probes by colony hybridization and PCR. Appl. Environ. Microbiol. 63:1268-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, G.-B., S. H. Yi, and B. Lee. 2004. Purification and characterization of three different types of bile salt hydrolase from Bifidobacterium strains. J. Dairy Sci. 87:258-266. [DOI] [PubMed] [Google Scholar]
- 23.Knarreborg, A., R. M. Engberg, S. K. Jensen, and B. B. Jensen. 2002. Quantitative determination of bile salt hydrolase activity in bacteria isolated from the small intestine of chickens. Appl. Environ. Microbiol. 68:6425-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ko, E. J., J. S. Goh, B. J. Lee, S. H. Choi, and P. H. Kim. 1999. Bifidobacterium bifidum exhibits a lipopolysaccharide-like mitogenic activity for murine B lymphocytes. J. Dairy Sci. 82:1869-1876. [DOI] [PubMed] [Google Scholar]
- 25.Kurdi, P., H. Tanaka, H. W. van Veen, K. Asano, F. Tomita, and A. Yokota. 2003. Cholic acid accumulation and its diminution by short-chain fatty acids in bifidobacteria. Microbiology 149:2031-2037. [DOI] [PubMed] [Google Scholar]
- 26.Lundeen, S. G., and D. C. Savage. 1990. Characterization and purification of bile salt hydrolase from Lactobacillus sp. strain 100-100. J. Bacteriol. 172:4171-4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacConaill, L. E., D. Butler, M. O'Connell-Motherway, G. F. Fitzgerald, and D. van Sinderen. 2003. Identification of two-component regulatory systems in Bifidobacterium infantis by functional complementation and degenerate PCR approaches. Appl. Environ. Microbiol. 69:4219-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marteau, P., and J. C. Rambaud. 1993. Potential of using lactic acid bacteria for therapy and immunomodulation in man. FEMS Microbiol. Rev. 12:207-220. [DOI] [PubMed] [Google Scholar]
- 29.Matsuki, T., K. Watanabe, and R. Tanaka. 2003. Genus- and species-specific PCR primers for the detection and identification of Bifidobacteria. Curr. Issues Intest. Microbiol. 4:61-69. [PubMed] [Google Scholar]
- 30.McCracken, A., and P. Timms. 1999. Efficiency of transcription from promoter sequence variants in Lactobacillus is both strain and context dependent. J. Bacteriol. 181:6569-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meile, L., L. M. Rohr, T. A. Geissmann, M. Herensperger, and M. Teuber. 2001. Characterization of the d-xylulose 5-phosphate/d-fructose 6-phosphate phosphoketolase gene (xfp) from Bifidobacterium lactis. J. Bacteriol. 183:2929-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyamoto, C. M., A. D. Graham, M. Boylan, J. F. Evans, K. W. Hasel, E. A. Meighen, and A. F. Graham. 1985. Polycistronic mRNAs code for polypeptides of the Vibrio harveyi luminescence system. J. Bacteriol. 161:995-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Møller, P. L., F. Jørgensen, O. C. Hansen, S. M. Madsen, and P. Stougaard. 2001. Intra- and extracellular β-galactosidases from Bifidobacterium bifidum and B. infantis: molecular cloning, heterologous expression, and comparative characterization. Appl. Environ. Microbiol. 67:2276-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oinonen, C., and J. Rouvinen. 2000. Structural comparison of Ntn-hydrolases. Protein Sci. 9:2329-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park, J. H., J. I. Um, B. J. Lee, J. S. Goh, S. Y. Park, W. S. Kim, and P. H. Kim. 2002. Encapsulated Bifidobacterium bifidum potentiates intestinal IgA production. Cell. Immunol. 219:22-27. [DOI] [PubMed] [Google Scholar]
- 36.Pereira, D. I. A., A. L. McCartney, and G. R. Gibson. 2003. An in vitro study of the probiotic potential of a bile-salt-hydrolyzing Lactobacillus fermentum strain, and determination of its cholesterol-lowering properties. Appl. Environ. Microbiol. 69:4743-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pereira, D. I. A., and G. R. Gibson. 2002. Cholesterol assimilation by lactic acid bacteria and bifidobacteria isolated from the human gut. Appl. Environ. Microbiol. 68:4689-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rossi, M., L. Altomare, A. Gonzalez Vara y Rodriguez, P. Bridigi, and D. Matteuzzi. 2000. Nucleotide sequence, expression and transcriptional analysis of the Bifidobacterium longum MB219 lacZ gene. Arch. Microbiol. 174:74-80. [DOI] [PubMed] [Google Scholar]
- 39.Sabikhi, L., and B. N. Mathur. 2000. Studies on the antibacterial activity of Bifidobacterium bifidum against selected enteropathogens and spoilage organisms. Indian J. Dairy Sci. 53:227-230. [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Scardovi, V. 1984. Genus Bifidobacterium Orla-Jensen 1924, 472, p. 1418-1434. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. The Williams & Wilkins Co., Baltimore, Md.
- 42.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sghir, A., G. Gramet, A. Suau, V. Rochet, P. Pochart, and J. Dore. 2000. Quantification of bacterial groups within the human fecal flora by oligonucleotide probe hybridization. Appl. Environ. Microbiol. 66:2263-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stellwag, E. J., and P. B. Hylemon. 1976. Purification and characterization of bile salt hydrolase from Bacteroides fragilis subsp. fragilis. Biochim. Biophys. Acta 452:165-176. [DOI] [PubMed] [Google Scholar]
- 45.Suresh, C. G., A. V. Pundle, H. SivaRaman, K. N. Rao, J. A. Brannigan, C. E. McVey, C. S. Verma, Z. Dauter, E. J. Dodson, and G. G. Dodson. 1999. Penicillin V acylase crystal structure reveals new Ntn-hydrolase family members. Nat. Struct. Biol. 6:414-416. [DOI] [PubMed] [Google Scholar]
- 46.Swartzman, E., C. Miyamoto, A. Graham, and E. Meighen. 1990. Delineation of the transcriptional boundaries of the lux operon of Vibrio harveyi demonstrates the presence of two new lux genes. J. Biol. Chem. 265:3513-3517. [PubMed] [Google Scholar]
- 47.Tanaka, H., K. Doesburg, T. Iwasaki, and I. Mierau. 1999. Screening of lactic acid bacteria for bile salt hydrolase activity. J. Dairy Sci. 82:2530-2535. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka, H., H. Hashiba, J. Kok, and I. Mierau. 2000. Bile salt hydrolase of Bifidobacterium longum-—biochemical and genetic characterization. Appl. Environ. Microbiol. 66:2502-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tannock, G. W., M. P. Dashkevicz, and S. D. Feighner. 1989. Effect of sodium taurocholate on the in vitro growth of Lactobacilli. Microb. Ecol. 33:163-167. [DOI] [PubMed] [Google Scholar]
- 50.Taranto, M. P., F. Sesma, and G. F. Valdez. 1999. Localization and primary characterization of bile salt hydrolase activity from Lactobacillus reuteri. Biotechnol. Lett. 21:935-938. [Google Scholar]
- 51.Thomas, L. A., M. J. Veysey, T. Bathgate, A. King, G. French, N. C. Smeeton, G. M. Murphy, and R. H. Dowling. 2000. Mechanism for the transit-induced increase in colonic deoxycholic acid formation in cholesterol cholelithiasis. Gastroenterology 119:806-815. [DOI] [PubMed] [Google Scholar]
- 52.Wells, J. E., and P. B. Hylemon. 2000. Identification and characterization of a bile acid 7α-dehydroxylation operon in Clostridium sp. strain TO-931, a highly active 7α-dehydroxylating strain isolated from human feces. Appl. Environ. Microbiol. 66:1107-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yildirim, Z., D. K. Winters, and M. G. Johnson. 1999. Purification, amino acid sequence and mode of action of bifidocin B produced by Bifidobacterium bifidum NCFB 1454. J. Appl. Microbiol. 86:45-54. [DOI] [PubMed] [Google Scholar]