Abstract
In profiling studies, the analysis of a single dataset often leads to unsatisfactory results because of the small sample size. Multi-dataset analysis utilizes information of multiple independent datasets and outperforms single-dataset analysis. Among the available multi-dataset analysis methods, integrative analysis methods aggregate and analyze raw data and outperform meta-analysis methods, which analyze multiple datasets separately and then pool summary statistics. In this study, we conduct integrative analysis and marker selection under the heterogeneity structure, which allows different datasets to have overlapping but not necessarily identical sets of markers. Under certain scenarios, it is reasonable to expect some similarity of identified marker sets – or equivalently, similarity of model sparsity structures – across multiple datasets. However, the existing methods do not have a mechanism to explicitly promote such similarity. To tackle this problem, we develop a sparse boosting method. This method uses a BIC/HDBIC criterion to select weak learners in boosting and encourages sparsity. A new penalty is introduced to promote the similarity of model sparsity structures across datasets. The proposed method has an intuitive formulation and is broadly applicable and computationally affordable. In numerical studies, we analyze right censored survival data under the AFT (accelerated failure time) model. Simulation shows that the proposed method outperforms alternative boosting and penalization methods with more accurate marker identification. The analysis of three breast cancer prognosis datasets shows that the proposed method can identify marker sets with increased similarity across datasets and improved prediction performance.
Keywords: Integrative analysis, Model sparsity structure, Heterogeneity structure, Sparse boosting, Marker identification
1. Introduction
Profiling studies have been extensively conducted in the search for genetic markers associated with disease outcomes and phenotypes such as risk, progression, and response to treatment. Data generated in such studies have the “large d, small n” characteristic, with the number of covariates (for example gene expressions profiled) d much larger than the sample size n. Results generated from the analysis of a single dataset are often unsatisfactory [1]. Many factors contribute to the unsatisfactory results, with the most important one likely being the small sample sizes. Fortunately, for many diseases there are multiple datasets from independent studies with comparable designs. Multi-dataset analysis combines information across datasets, increases sample size, and can outperform single-dataset analysis. Multi-dataset analysis methods include meta-analysis [2, 3] and integrative analysis methods. In “classic” meta-analysis, multiple datasets are initially analyzed separately, and then summary statistics are pooled across datasets. In contrast, in integrative analysis, the raw data from multiple datasets are pooled and analyzed. Recent studies have shown that integrative analysis outperforms meta-analysis with more accurate marker identification [4, 5].
Consider the integrative analysis of M independent datasets with the same type of response variable. In dataset m(= 1, …, M), denote Ym as the response variable, and Xm as the length-d vector of covariates (for example, gene expressions or SNPs). For simplicity of notation, it is assumed that the same set of covariates is measured in all M datasets. In whole-genome studies, the sets of covariates measured in different datasets are usually very similar. The rescaling approach [5] can easily accommodate covariates measured in some but not all datasets. As one of the main goals of multi-dataset analysis is to find the similarity/difference across datasets, integrative analysis may not be sensible if different datasets measure significantly different sets of covariates. In dataset m, assume nm i.i.d. observations. Assume that , where the form of model ϕ is known, and βm is the length-d vector of unknown regression coefficients. Denote the jth component of βm as βm,j. Our goal is to identify markers associated with the response variables or, equivalently, to determine which βm,j’s are nonzero.
The genetic basis of the M datasets, as measured by the identified marker sets, can be described using the homogeneity structure or the heterogeneity structure [5]. Under the homogeneity structure, the same set of markers is identified for all datasets, and so the M models have the same sparsity structure. That is, I(βm,j = 0) = I(βk,j = 0) for all m, k = 1, …, M and j = 1, …, d. The heterogeneity structure differs from the homogeneity one by allowing the M models to have possibly different sparsity structures. Here, it is possible that I(βm,j = 0) ≠ I(βk,j = 0) for some (j, m, k)’s. The heterogeneity structure includes the homogeneity structure as a special case and is more flexible [5, 6].
In this study, we conduct integrative analysis under the heterogeneity structure. Although multiple datasets are allowed to have different sets of markers, as the basis of integrating multiple datasets, it is reasonable to expect that they share some common markers. Further, under certain scenarios, it is of interest to promote the similarity of model sparsity structures across datasets. As the first example, consider multiple independent datasets generated under similar protocols [7]. Because of the experimental differences, the homogeneity structure, which requires the same model sparsity structure across datasets, can be too restrictive. However, as multiple datasets measure the same set of response variable and covariates, it is reasonable to expect and hence to encourage multiple datasets to have similar marker sets. The second example is the analysis of data on different response variables. For example in the study conducted by Liu and others [5], each dataset is on the risk of a different cancer type. Despite great differences across cancer types, multiple genes and pathways have been identified as associated with a large number of cancers [8]. Compared with cancer type-specific markers, those shared by multiple cancers are more likely to define the fundamental characteristics of cancer. Thus, in multi-cancer analysis it is also of interest to promote markers to be identified in multiple datasets. The existing methods do not have a mechanism that explicitly promotes the similarity of model sparsity structures across datasets.
In integrative analysis under the heterogeneity model, we adopt sparse boosting for marker selection and estimation. Sparse boosting, first developed by Buhlmann and Yu [9] and others, is a family of methods especially suitable for high-dimensional data and sparse models. This study differs from the existing sparse boosting studies [4, 9, 10] by conducting the integrative analysis of multiple datasets and by assuming the heterogeneity structure. The most significant advancement is the introduction of a new penalty in the boosting algorithm, which explicitly promotes the similarity of model sparsity structures across datasets. This penalty has a simple form and an intuitive interpretation.
2. Integrative Analysis and Marker Selection using Sparse Boosting
For dataset m(= 1, …, M), denote Rm(βm) as the loss function. The most fundamental requirement on the loss is that it leads to a consistent estimate under the “classic” condition with nm ≫ d. The most common choice is the negative likelihood function. For models such as the logistic, an intercept term is needed beyond βm. We omit the intercept term as it will not be subject to selection and be very easy to deal with.
2.1. Sparse boosting a single dataset
As described above, with high-dimensional covariates, we focus on linear covariate effects. Under this setting, boosting assembles a set of individual covariates (weak learners) into a comprehensive model (a strong learner, e.g., an effective linear combination of covariates). Advantages of boosting include its simple and intuitive form, broad applicability, affordable computational cost, and satisfactory numerical performance. We refer to Buhlmann and Hothorn [11] and others for comprehensive reviews.
With ordinary boosting, marker selection is achieved with an early stopping. However, Buhlmann and Yu [9] and several other studies find that the ordinary boosting results may not be “sparse enough”. That is, too many covariates may be identified as associated with response. Sparse boosting is developed to tackle this problem. For the integrity of this article, we first present a version of sparse boosting based on Buhlmann and Yu [9] for the analysis of a single dataset, say dataset m.
Algorithm 0: Sparse boosting a single dataset
Step 1: Initialization. k = 0. Denote as the estimate of βm in the kth iteration, and its jth component as . Initialize for j = 1, …, d. With each component of Xm being a weak learner, the strong learner is .
-
Step 2: Fit and update. k = k + 1.
Compute , where 1s is the length-d vector with the sth component equal to 1 and all others equal to 0. pen(·) is the penalty function on model complexity (more details are provided below).
Update and , where ν is the step size. It has been suggested that the choice of ν is not critical as long as it is small [9]. We set ν = 0.1 following published studies.
Step 3: Iteration. Repeat Step 2 for K times. K is a large number.
Step 4: Selection of optimal stopping. At iteration k(= 1, …, K), compute . Select the optimal number of iterations as k̂ = argmin1≤k≤KFm(k). The final strong learner is . Covariates corresponding to the nonzero components of are identified as associated with the response.
Different from the ordinary boosting which uses Rm only, the sparse boosting introduces the penalty function pen(·) in selecting the weak learners and optimal stopping. By penalizing model complexity, it can lead to sparser models. Choices of pen(·) include BIC, AIC, MDL (minimum description length), and others [9]. In the literature there is still a lack of investigation on when a penalty is preferred over the others. To be more flexible, the model complexity penalties in weak learner selection and stopping can be different.
2.2. Integrative sparse boosting multiple datasets
Consider extending sparse boosting to the integrative analysis of M independent datasets. In the integrative analysis under the heterogeneity structure, two-level marker selection is needed [5]. Use similar notations as for Algorithm 0. We propose the following algorithm.
Algorithm 1: Sparse boosting for integrative analysis
Step 1: Initialization. k = 0. For m = 1, …, M, initialize for j = 1, …, d. The strong learner for dataset m is .
-
Step 2: Fit and update. k = k + 1. For m = 1, …, M:
Compute .
Update and .
Step 3: Iteration. Repeat Step 2 for K times. K is a large number.
Step 4: Selection of optimal stopping. At iteration k(= 1, …, K), compute . Select the optimal number of iterations as k̂ = argmin1≤k≤KF(k). For dataset m, the final strong learner is . Covariates corresponding to the nonzero components of are identified as associated with the response in dataset m.
In integrative analysis, we need to jointly analyze M datasets. In each iteration, we consecutively apply sparse boosting to each dataset. When selecting the weak learners and updating the strong learners, multiple datasets are considered separately. However, the stopping rule is selected by jointly considering the M datasets. Loosely speaking, this amounts to applying a comparable amount of regularization to all datasets, which has been suggested in integrative analysis using the penalization technique [5, 6, 7].
2.3. Promoting the similarity of model sparsity structures
Algorithm 1 fully relies on data to determine how similar the sparsity structures are. However, there is no mechanism to encourage the similarity. Denote β·,j = (β1,j, …, βM,j) as the jth column of β. We propose the following algorithm.
Algorithm 2: Sparse boosting for integrative analysis that promotes the similarity
Step 1: Initialization. The same as in Algorithm 1.
-
Step 2: Fit and update. k = k + 1. For m = 1, …, M:
- Compute
Here , |u|0 = 1 if u ≠ 0 and = 0 otherwise, λ ≥ 0 is a data-dependent tuning parameter, ||β·,j||2 is the ℓ2-norm of β·,j, and 1m,s is a d × M matrix with (s,m)th element set to 1.
Update and .
Step 3: Iteration. Repeat Step 2 for K times. K is a large number.
Step 4: Selection of optimal stopping. At iteration k(= 1, …, K), compute . Select the optimal number of iterations as k̂ = argmin1≤k≤K F(k).
Advancing from the existing studies, we propose the pens penalty to encourage similarity. In , the numerator Σm,j |βm,j|0 counts how many covariates are selected across the M datasets, and the denominator Σj |||β·,j ||2|0 counts how many unique covariates are selected. pens is closely related to the Jaccard index of similarity [12] and takes value in . It is minimized if the M datasets identify the same set of covariates, and is maximized if there is no covariate identified in more than one dataset. Thus, it has the capability of promoting similarity. λ determines the degree of regularization. When λ = 0, the proposed method goes back to Algorithm 1. On the other hand, when λ = ∞, the proposed method reinforces that the same set of covariates is selected in all datasets, i.e, the homogeneity structure, which is an extreme case of the heterogeneity structure. We encounter Σj |||β·,j ||2|0 = 0 in the first step of boosting. To ensure that the boosting is not “trapped”, we take 0/0=1.
With the proposed method, λ needs to be determined data-dependently. In addition, we need to specify a proper pen function for selecting weak learners and a possibly different pen function for stopping. In the sparse boosting literature [9], multiple pen functions have been suggested. However, there is a lack of study showing when one may be better than the others [13]. In this study, we adopt the BIC-based approaches because of their simple forms, broad applicability, and satisfactory numerical performance.
As a specific example, consider a dataset with sample size n and under a linear regression model. With a specific strong learner, denote the residual sum of squares as RSS and degree of freedom as df. We adopt the duo
where the BIC criterion is log(RSS) + df × log(n)/n, and the HDBIC criterion is log(RSS) + df × log(n) log(d)/n [14]. Adopting the BIC criterion for selecting weak learners has been motivated by published studies [13]. The HDBIC criterion imposes more penalty than BIC and can generate sparser models. In the literature, there are other BIC-type criteria [15]. We adopt the proposed combination because of its satisfactory performance. It is beyond the scope of this article to comprehensively compare different model complexity criteria.
We have developed an R program implementing the proposed approach. To illustrate the usage, we have also provided demo with sample survival datasets. The code and demo are publicly available at https://github.com/shuanggema/IntSpBoost. The computer time can be potentially reduced by adopting parallel computing.
To further examine the working characteristics of proposed method, we simulate one replicate with three independent datasets. More details on the simulation settings are provided in Table 1 and the next section. We analyze the simulated data using four different methods. The first two do not have pens and serve as a reference. More specifically, the first method (Alt.1) is the ordinary boosting and uses Rm(·)’s as the criterion for selection and HDBIC for stopping. The second (Alt.2) is a sparse boosting method and uses BIC for selection and HDBIC for stopping. There are two versions of the proposed method. One uses BIC+pens for selection and HDBIC for stopping (New), and the other uses BIC+pens for selection and HDBIC+pens for stopping (New+). Table 1 shows that for this specific replicate, the methods New and New+ yield identical estimates (which is not always true). They outperform Alt.1 and Alt.2 by identifying fewer false positives. Alt.2 outperforms Alt.1 by using BIC in selecting weak learners.
Table 1.
Analysis of one replicate with nm = 100, d = 100, ρ = 0.2, and half-overlapping marker sets. All nonzero regression coefficients=1. For the (selection, stopping) duo: Alt.1=(L2, HDBIC), Alt.2=(BIC, HDBIC), New=(BIC+, HDBIC), and New+=(BIC+, HDBIC+). Estimated coefficient in each cell.
| cov | Dataset 1 | Dataset 2 | Dataset 3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alt.1 | Alt.2 | New | New+ | Alt.1 | Alt.2 | New | New+ | Alt.1 | Alt.2 | New | New+ | |
| 1 | 0.898 | 0.992 | 0.961 | 0.961 | 0.667 | 0.692 | 0.658 | 0.658 | 1.144 | 1.206 | 1.192 | 1.192 |
| 2 | 0.968 | 1.061 | 1.031 | 1.031 | 1.011 | 1.082 | 1.069 | 1.069 | 0.681 | 0.751 | 0.736 | 0.736 |
| 3 | 0.798 | 0.774 | 0.774 | 0.774 | 0.635 | 0.758 | 0.717 | 0.717 | 0.858 | 0.938 | 0.903 | 0.903 |
| 4 | 0.885 | 0.984 | 0.958 | 0.958 | ||||||||
| 5 | 0.880 | 0.937 | 0.921 | 0.921 | ||||||||
| 6 | 0.875 | 1.028 | 0.984 | 0.984 | ||||||||
| 7 | 0.760 | 0.883 | 0.857 | 0.857 | ||||||||
| 8 | 1.075 | 1.072 | 1.072 | 1.072 | ||||||||
| 9 | 1.018 | 1.145 | 1.103 | 1.103 | ||||||||
| 10 | 0.821 | 0.937 | 0.906 | 0.906 | ||||||||
| 11 | 0.637 | 0.691 | 0.656 | 0.656 | ||||||||
| 12 | 0.741 | 0.852 | 0.808 | 0.808 | ||||||||
| 19 | −0.053 | |||||||||||
| 31 | −0.058 | −0.024 | ||||||||||
| 40 | −0.030 | |||||||||||
| 58 | −0.024 | |||||||||||
| 77 | −0.050 | −0.023 | ||||||||||
| 92 | −0.022 | −0.022 | ||||||||||
2.4. Potential extensions
Carefully examining the proposed algorithm suggests that the newly added penalty (for promoting similarity of model sparsity structure) is relatively independent of the boosting loss function. Boosting analysis of censored survival data is definitely not limited to the AFT model. In Appendix, we also describe applying the proposed strategy to the Cox model, which is more popular than the AFT model. With high-dimensional data, the disadvantage of the Cox model is its higher computational cost (compared to the AFT model). Another possible extension is to accommodate nonlinear covariate effects using trees as weak learners. In the literature, boosting survival trees has been investigated in multiple studies. In Appendix, we describe “coupling” boosting survival trees with the new penalty for promoting similarity in sparsity structure. In the following numerical study, we mostly focus on the AFT model. In addition, in simulation, we also consider using trees as weak learners (results in Appendix); and in data analysis, we also consider analyzing under the Cox model (results in Appendix).
3. Numerical Study
The proposed method is potentially applicable to a large number of data and model settings and R(β) functions. As a specific example, we consider right censored survival data under the AFT (accelerated failure time) model. Details on the data settings and estimation procedure are described in Appendix.
3.1. Simulation
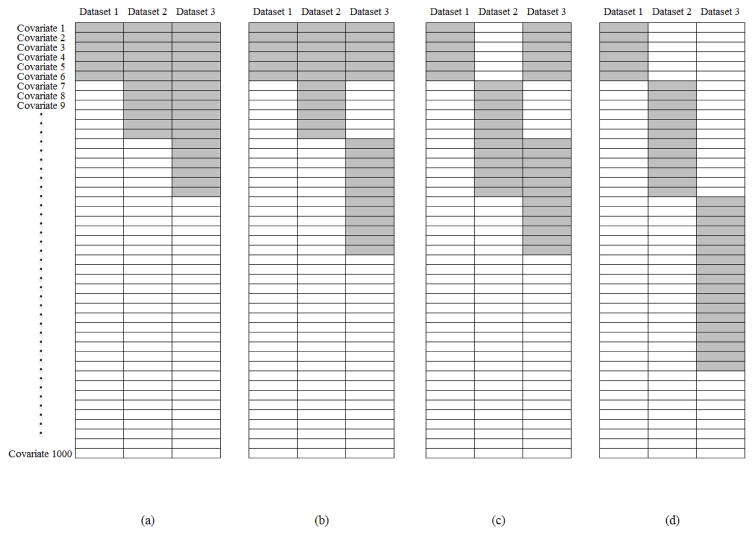
We simulate M = 3 independent datasets. In each dataset, the sample size is nm = 100. Mimicking gene expression data, we simulate d = 100, 500, and 1,000 continuously distributed covariates with a multivariate normal distribution. The marginal means are equal to 0, and the marginal variances are equal to 1. We consider an auto-regressive correlation structure where covariates j and k have correlation coefficient ρ|j−k| with ρ = 0.2, 0.5, and 0.8, corresponding to weak, moderate, and strong correlations. In each dataset, there are 6 covariates with nonzero regression coefficients. Thus, the total number of truly important covariates is 18. The nonzero regression coefficients are generated from Unif[0.2, 1] or all equal to 1, representing two levels of signals. Regarding the overlapping of important covariates, we consider three scenarios: (a) Complete overlapping: all three datasets have the same set of six important covariates. (b) Half overlapping: the three datasets share three common important covariates. In addition, each dataset has three dataset-specific important covariates. For each dataset, the percentage of shared important covariates is 50%. (c) None overlapping. There is no important covariate shared by any two datasets. In the AFT models, the intercepts are set as 0.5. The random errors are simulated from N(0, σ2) with σ2 = 1 and 3, representing two noise levels. We generate the log censoring times from normal distributions. The censoring distributions are adjusted so that the overall censoring rate is about 33%. To better gauge the proposed method, we also apply alternative analysis methods. We first consider three alternative sparse boosting methods: (a) indiv-SB. This method applies sparse boosting (Algorithm 0) to each dataset separately, and evaluation (as described below) is first separately conducted and then combined. This method does not account for potential shared information across datasets. (b) pool-SB. This method pools the three datasets together and then applies sparse boosting (Algorithm 0). When the datasets are highly similar, this is expected to be the most effective method. (c) alg1-SB, which applies Algorithm 1. This method is the closest to the proposed. Note that it is also referred to as “Alt.2” in Table 1. For the analysis of high-dimensional data, a large number of regularization methods have been developed. Here we compare with penalization, which is one of the most popular regularization methods. We consider four penalization methods built on the MCP [16], which has been shown to have superior theoretical and empirical properties. The applied penalization methods include (a) indiv-MCP, which takes a similar approach as indiv-SB and applies MCP to each dataset separately; (b) pool-MCP, which takes a similar approach as pool-SB and applies MCP to the pooled dataset; and (c) sgroup-MCP, which applies the sparse group MCP method [6]. This method has been designed to conduct two-level selection and is suitable for the heterogeneity structure; and (d) group-MCPT. This method first applies the group MCP [5]. As the group MCP conducts one-level selection and may not be appropriate for the heterogeneity structure, a thresholding is conducted and sets small estimates as zero. Here the cutoffs are selected in a way that the numbers of identified true positives are similar to the proposed method.
As expected, the proposed method is computationally more expensive. However, simulation suggests that it is still affordable. For example, under the setting described in Table 2, indiv-SB and alg1-SB have similar cost, with the analysis of one replicate taking about 40 seconds on a regular desktop PC. The pool-SB analysis takes about 160 seconds. In comparison, the proposed method takes about 309 seconds.
For each method and each replicate, we evaluate marker identification performance using TP (number of true positives) and FP (number of false positives). In addition, we evaluate prediction and estimation performance. Specifically, prediction is quantified using PMSE (prediction MSE) which is defined as Σm(β̂m − βm)′Var(Xm)(β̂m − βm)/σ2. Estimation is evaluated using EMSE (estimation MSE) which is defined as Σm(β̂m − βm)′(β̂m − βm).
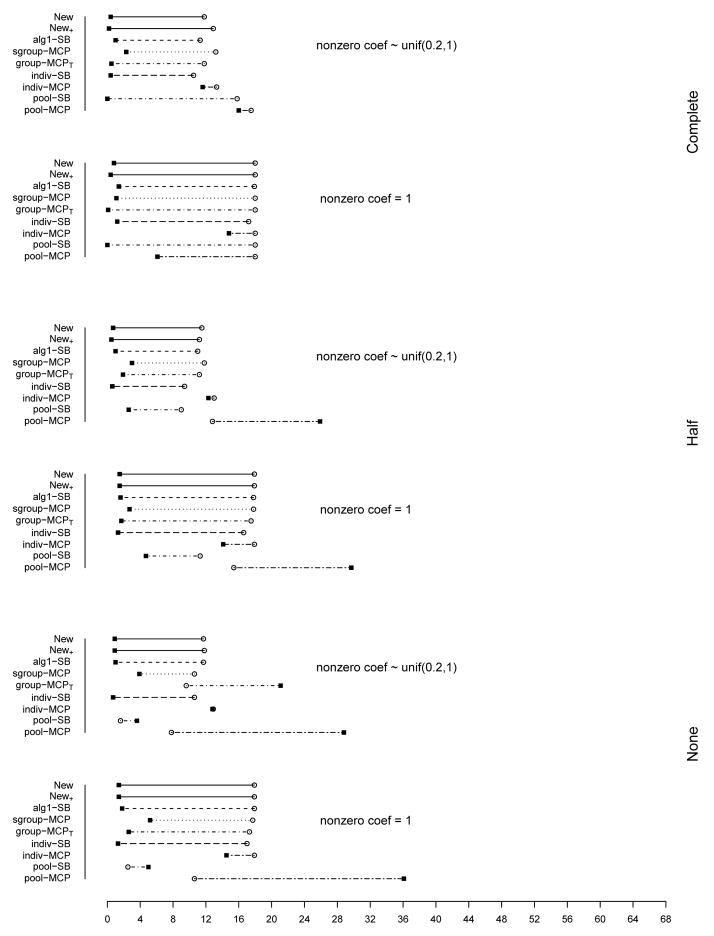
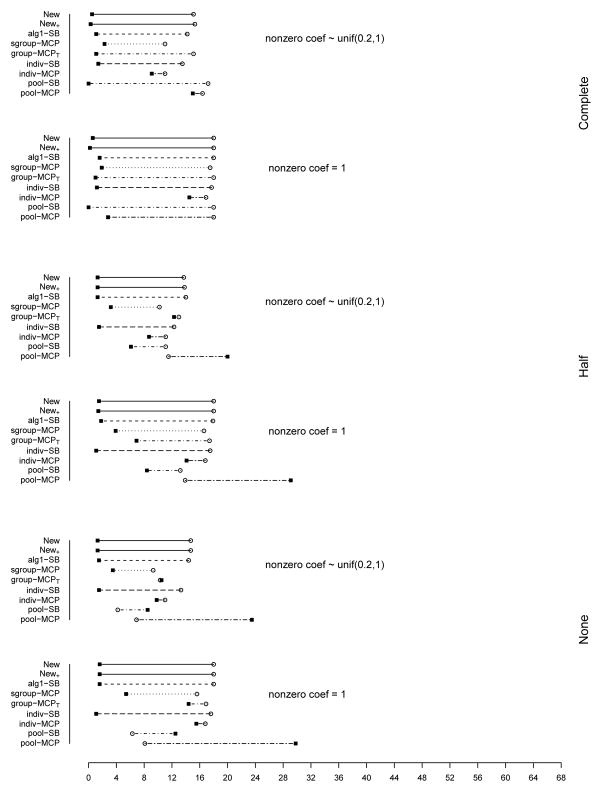
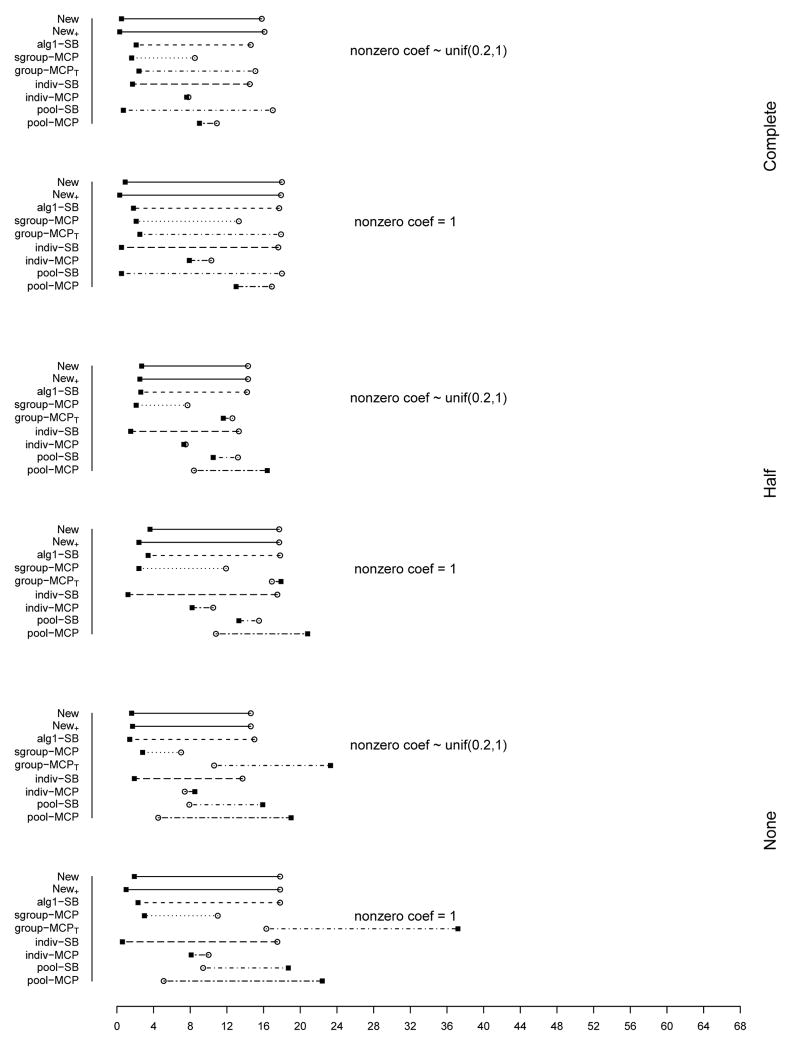
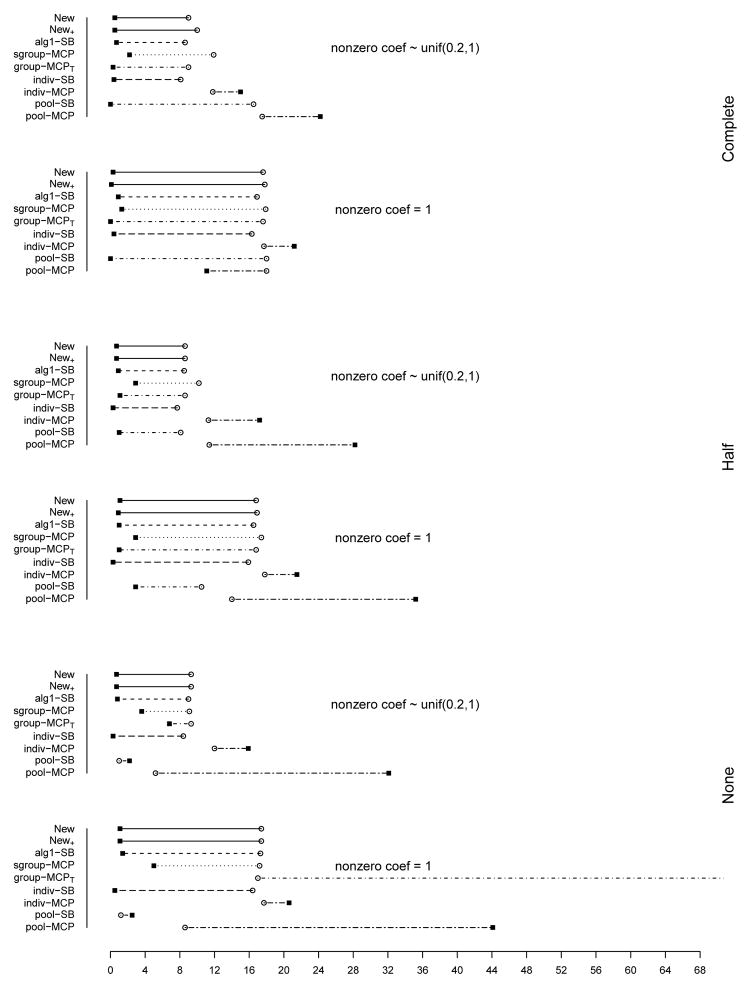
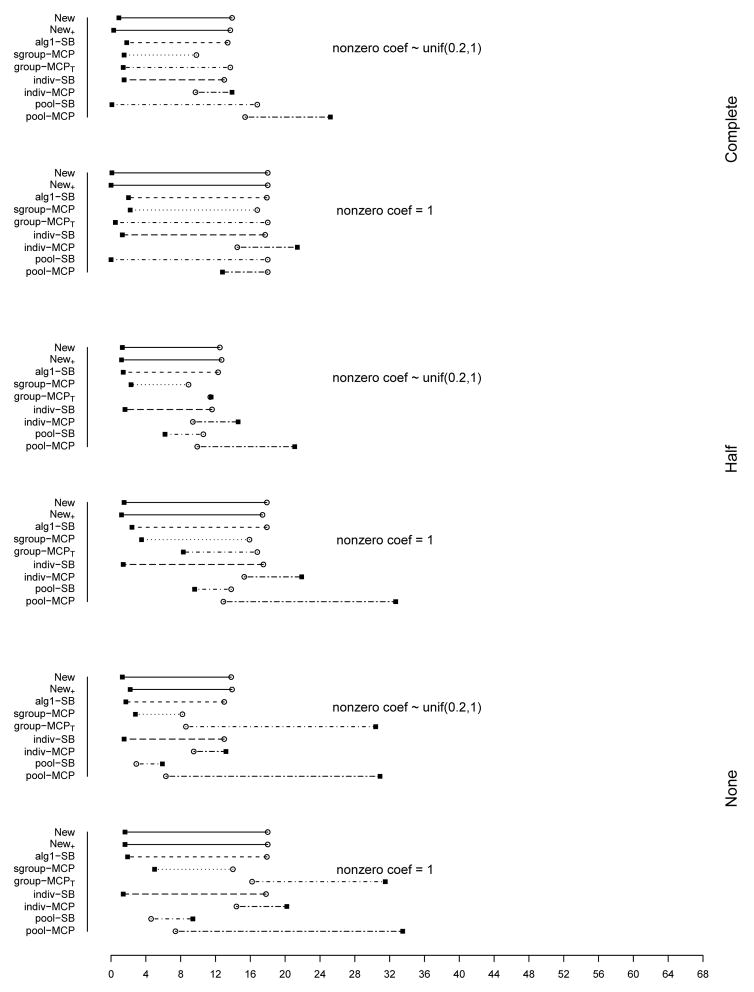
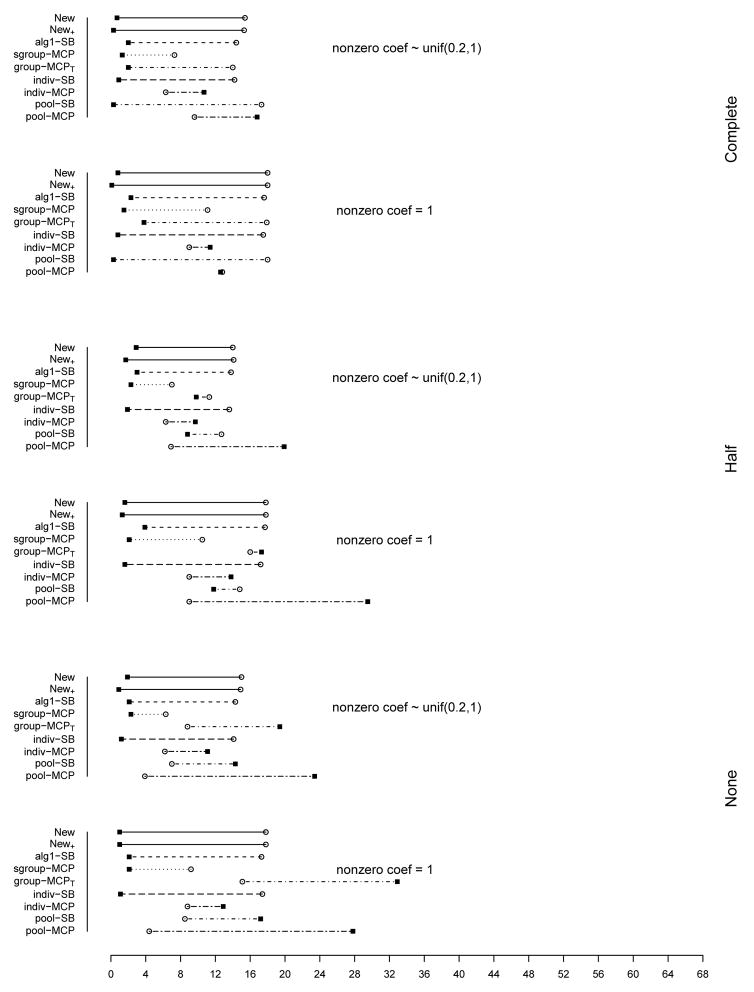
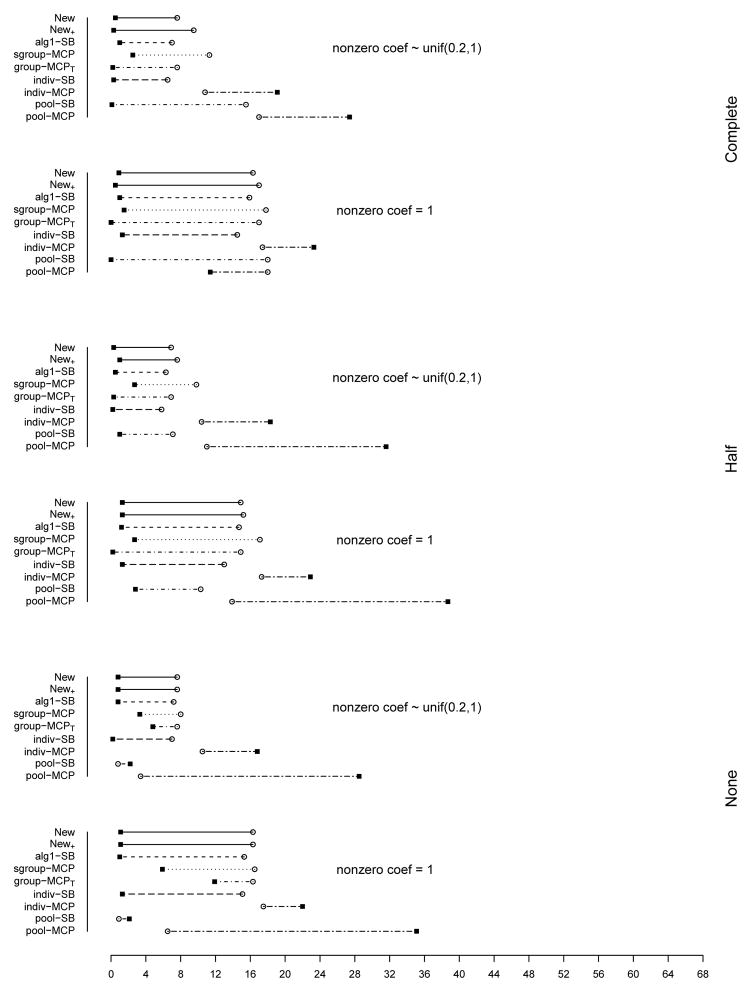
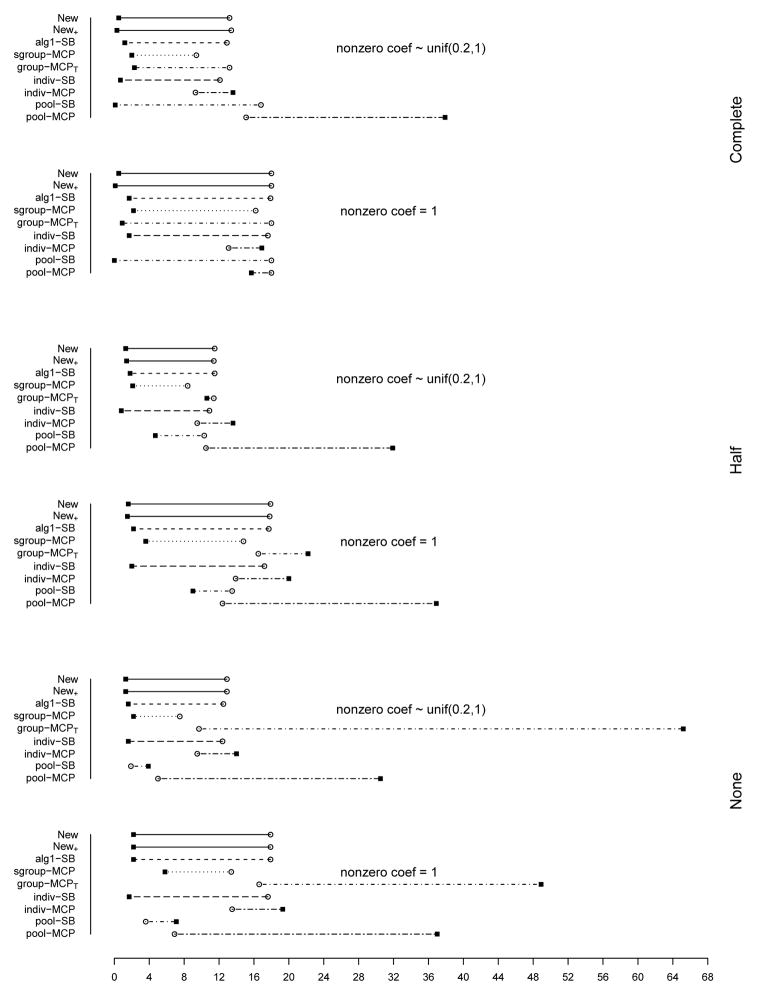
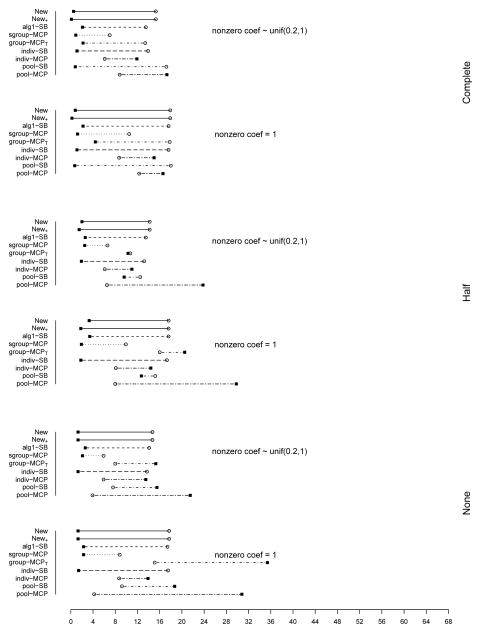
The summary of identification results for the setting with d = 1, 000 and ρ = 0.2 is presented in Figure 1 (σ2 = 1) and Table 2 (σ2 = 1 and 3). The rest of the figures and tables for identification are presented in Appendix. The estimation and prediction results are separately summarized and also presented in Appendix. Simulation suggests competitive performance of the proposed method. For example in Table 2 with σ2 = 1 and the nonzero regression coefficients generated from Unif[0.2, 1], the proposed New+ identifies 15.7 (complete overlapping), 13.9 (half overlapping), and 14.9 (non overlapping) true positives, with a very small number of false positives. Under most of the simulation settings, New+ outperforms the method New with more true positives and fewer false positives. The indiv-SB method has good performance, however, inferior to the proposed under most of the simulation settings. Its performance is not strongly affected by the overlapping structure, as it analyzes each dataset separately. As expected, under the complete overlapping scenario, the proposed method has advantages. For example, under the scenario described above and complete overlapping, indiv-SB identifies 13.8 true positives (compared to 15.7 of the proposed). The pool-SB method reinforces that all datasets identify the same set of important covariates. It has superior performance under the complete overlapping scenario but poor performance under the half and none overlapping scenarios. The alg1-SB method, which is similar to the proposed but does not promote the similarity across datasets, performs inferior to the proposed method. For example when σ2 = 1 and the nonzero coefficients are all equal to 1, it identifies 4.9, 5.2, and 4.8 false positives, compared to 0.4, 1.4, and 1.2 with the proposed method. The penalization methods have reasonable but in general inferior performance. Under quite a few simulation scenarios, indiv-MCP and group-MCPT identify a relatively large number of false positives (although the performance with true positives is satisfactory). Similar to pool-SB, pool-MCP has good performance under the complete overlapping scenario but behaves poorly under the other two scenarios. The sgroup-MCP method has better performance than the other three penalization methods but is slightly inferior to the proposed method. In general, it is observed that the proposed method is favored over the alternatives in terms of identification. The proposed method also has favorable estimation and prediction results. However, it is also noted that, when factoring in variation, the proposed method and some alternatives may have comparable estimation and prediction performance.
Figure 1.
Plots of mean TP and FP for simulations with d = 1, 000, ρ = 0.2, and σ2 = 1. The circles stand for TP and black squares stand for FP.
Table 2.
Simulation: summary statistics on identification. In each cell, mean (sd). d = 1, 000 and ρ = 0.2.
| σ2 = 1 | σ2 = 3 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Complete | Half | None | Complete | Half | None | |||||||
|
| ||||||||||||
| TP | FP | TP | FP | TP | FP | TP | FP | TP | FP | TP | FP | |
| nonzero coef ~ unif(0.2,1) | ||||||||||||
| New | 15.2 (2.7) | 0.1 (0.3) | 14.0 (1.8) | 1.8 (1.3) | 14.8 (1.7) | 1.9 (1.7) | 7.6 (2.8) | 0.5 (0.7) | 6.9 (3.3) | 0.3 (0.5) | 7.6 (2.4) | 0.8 (1.0) |
| New+ | 15.7 (3.2) | 0.0 (0) | 13.9 (2.1) | 1.6 (1.4) | 14.9 (1.7) | 2.0 (1.8) | 9.5 (3.2) | 0.3 (0.6) | 7.6 (2.5) | 1.0 (1.4) | 7.6 (2.4) | 0.8 (1.0) |
| alg1-SB | 13.8 (1.8) | 1.8 (1.2) | 14.1 (1.9) | 2.0 (1.5) | 14.1 (2.1) | 2.0 (1.7) | 7.0 (2.4) | 1.0 (1.1) | 6.3 (2.5) | 0.5 (0.8) | 7.2 (2.7) | 0.8 (0.9) |
| sgroup-MCP | 15.6 (1.8) | 1.1 (1.7) | 14.5 (1.9) | 1.5 (1.6) | 13.4 (2.1) | 2.5 (2.6) | 11.3 (2.3) | 2.5 (2.8) | 9.8 (2.5) | 2.7 (3.0) | 8.0 (2.4) | 3.3 (4.2) |
| group-MCPT | 14.9 (1.8) | 0.2 (0.7) | 13.9 (1.8) | 1.0 (1.8) | 13.6 (2.7) | 57.5 (94.8) | 7.6 (2.1) | 0.2 (0.4) | 6.9 (2.4) | 0.3 (0.7) | 7.6 (3.2) | 4.8 (8.8) |
| indiv-SB | 13.2 (1.8) | 1.6 (0.6) | 12.5 (1.9) | 1.5 (0.6) | 13.3 (1.9) | 1.4 (0.6) | 6.5 (1.7) | 0.3 (0.5) | 5.8 (1.6) | 0.2 (0.4) | 7.0 (1.5) | 0.2 (0.4) |
| indiv-MCP | 14.8 (1.7) | 18.7 (7.6) | 14.5 (1.8) | 18.3 (8.2) | 14.9 (1.6) | 19.2 (8.6) | 10.8 (2.3) | 19.1 (7.3) | 10.4 (2.1) | 18.3 (7.0) | 10.5 (2.3) | 16.8 (6.1) |
| pool-SB | 17.9 (0.6) | 0.0 (0) | 9.7 (1.4) | 3.0 (1.8) | 1.1 (0.5) | 2.3 (0.9) | 15.5 (2.5) | 0.1 (0.6) | 7.1 (2.5) | 1.0 (1.4) | 0.8 (0.4) | 2.2 (0.4) |
| pool-MCP | 18.0 (0) | 20.3 (21.4) | 13.2 (1.5) | 35.9 (20.8) | 6.6 (2.0) | 38.7 (21.2) | 17.0 (1.8) | 27.4 (26.5) | 11.0 (2.5) | 31.6 (26.6) | 3.4 (2.3) | 28.5 (25.1) |
|
| ||||||||||||
| nonzero coef = 1 | ||||||||||||
| New | 18.0 (0) | 1.4 (1.4) | 18.0 (0) | 4.8 (2.1) | 18.0 (0) | 3.8 (1.9) | 16.3 (2.0) | 0.9 (0.7) | 14.9 (3.1) | 1.3 (0.9) | 16.3 (1.7) | 1.1 (1.1) |
| New+ | 18.0 (0) | 0.4 (0.8) | 18.0 (0) | 1.4 (2.0) | 18.0 (0) | 1.2 (2.1) | 17.0 (1.5) | 0.5 (0.6) | 15.2 (3.0) | 1.3 (0.9) | 16.3 (1.7) | 1.1 (1.1) |
| alg1-SB | 18.0 (0) | 4.9 (1.7) | 18.0 (0) | 5.2 (1.8) | 18.0 (0) | 4.8 (2.1) | 15.9 (2.1) | 1.0 (1.1) | 14.7 (2.4) | 1.2 (1.1) | 15.3 (2.5) | 1.0 (1.1) |
| sgroup-MCP | 18.0 (0) | 0.1 (0.4) | 18.0 (0) | 0.4 (0.8) | 18.0 (0.1) | 0.8 (1.2) | 17.8 (0.6) | 1.5 (1.5) | 17.1 (1.1) | 2.7 (2.5) | 16.5 (1.7) | 5.9 (4.6) |
| group-MCPT | 18.0 (0) | 0.0 (0) | 18.0 (0.1) | 0.2 (0.4) | 18.0 (0) | 0.0 (0) | 17.0 (1.0) | 0.0 (0) | 14.9 (1.7) | 0.2 (0.5) | 16.3 (1.7) | 11.9 (10.1) |
| indiv-SB | 18.0 (0) | 1.0 (0.2) | 18.0 (0) | 1.1 (0.3) | 18.0 (0) | 1.1 (0.3) | 14.5 (2.0) | 1.3 (0.5) | 13.0 (1.9) | 1.3 (0.5) | 15.1 (1.8) | 1.3 (0.5) |
| indiv-MCP | 18.0 (0) | 11.4 (8.6) | 18.0 (0) | 11.9 (7.9) | 18.0 (0) | 11.2 (8.1) | 17.4 (0.9) | 23.3 (8.0) | 17.3 (1.0) | 22.9 (7.3) | 17.5 (0.7) | 22.0 (7.1) |
| pool-SB | 18.0 (0) | 0.0 (0) | 11.7 (1.3) | 5.4 (2.5) | 1.0 (0.3) | 2.2 (0.5) | 18.0 (0) | 0.0 (0) | 10.3 (0.9) | 2.8 (1.9) | 0.9 (0.4) | 2.1 (0.4) |
| pool-MCP | 18.0 (0) | 3.5 (6.9) | 15.7 (1.5) | 43.7 (22.8) | 9.2 (2.5) | 51.2 (24.7) | 18.0 (0) | 11.4 (13.1) | 13.9 (1.7) | 38.7 (25.1) | 6.5 (3.1) | 35.1 (22.1) |
In the above simulation, different datasets have the same level of complexity (the same number of truly important covariates). We also examine the scenario where different datasets have different complexity structures. Consider the setting with d = 1, 000 and ρ = 0.2, 0.5, and 0.8. As shown in Figure 19 (Appendix), the three datasets have 6, 12, and 18 truly important covariates respectively. We simulate four different overlapping scenarios. Based on the observations described above, we analyze data using the proposed method as well as indiv-SB and alg1-SB. The summary identification results are shown in Table 3. When ρ = 0.5 and 0.8, the proposed method outperforms alg1-SB. Compared to indiv-SB, it has comparable performance in terms of true positives but slightly more false positives. When ρ = 0.2, the proposed method significantly outperforms indiv-SB.
Table 3.
Simulation: summary statistics on identification when the datasets have different complexity structures. In each cell, mean (sd). d = 1, 000.
| (a) | (b) | (c) | (d) | |||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| TP | FP | TP | FP | TP | FP | TP | FP | |
| ρ = 0.2 | ||||||||
| New | 26.5(4.3) | 2.0(1.6) | 26.1(4.7) | 2.9(1.9) | 23.4(5.6) | 2.3(2.0) | 24.8(5.8) | 3.0(2.2) |
| New+ | 25.7(7.6) | 1.6(1.6) | 26.1(4.7) | 2.9(2.0) | 24.3(4.9) | 2.3(2.0) | 24.4(6.7) | 2.9(2.2) |
| alg1-SB | 21.3(7.1) | 2.2(2.3) | 22.9(6.3) | 1.9(1.8) | 23.3(6.7) | 1.9(1.6) | 24.3(7.2) | 2.6(2.4) |
| indiv-SB | 15.3(5.2) | 2.9(1.9) | 13.8(4.6) | 2.5(1.6) | 15.1(4.5) | 1.8(1.5) | 16.1(4.3) | 1.9(1.7) |
|
| ||||||||
| ρ = 0.5 | ||||||||
| New | 35.2(0.9) | 4.7(2.3) | 34.9(1.2) | 6.9(2.5) | 34.4(1.7) | 6.8(3.1) | 35.1(1.1) | 8.3(3.3) |
| New+ | 33.8(1.4) | 1.6(1.8) | 34.5(1.7) | 5.2(2.6) | 34.4(1.7) | 4.8(2.8) | 35.1(1.1) | 4.3(3.3) |
| alg1-SB | 35.2(0.9) | 8.3(2.6) | 34.6(1.5) | 8.2(3.1) | 33.8(1.9) | 7.1(3.0) | 35.0(1.0) | 8.2(3.1) |
| indiv-SB | 35.8(0.7) | 3.7(1.6) | 34.1(3.9) | 3.7(2.2) | 33.3(5.1) | 2.9(2.0) | 35.6(1.3) | 4.3(1.7) |
|
| ||||||||
| ρ = 0.8 | ||||||||
| New | 35.4(0.7) | 6.3(2.0) | 35.2(0.8) | 9.5(2.2) | 35.0(0.8) | 10.1(2.7) | 35.2(1.1) | 9.5(2.1) |
| New+ | 35.0(1.0) | 4.5(1.8) | 35.1(1.1) | 4.0(2.6) | 35.2(0.9) | 4.1(1.9) | 35.2(1.1) | 4.7(2.3) |
| alg1-SB | 35.3(0.9) | 10.4(2.0) | 35.0(0.8) | 11.2(2.7) | 35.1(0.8) | 10.3(2.7) | 35.2(0.8) | 10.6(2.9) |
| indiv-SB | 35.5(0.6) | 2.6(1.3) | 35.6(0.5) | 3.2(1.3) | 35.7(0.6) | 2.8(1.8) | 35.5(0.5) | 2.2(1.4) |
Overall, simulation suggests competitive performance of the proposed method. It is interesting to note that it has satisfactory performance even under the none overlapping scenario. Thus, it provides a “safe” choice for practical data analysis where the degree of overlapping is unknown. The observed improvement over the alternative integrative analysis methods is not dramatic, which is reasonable. The newly added component (which promotes similarity across datasets) accommodates finer or secondary structure of data. The “first order” selection still depends on the sparse boosting component descried in Algorithm 1.
We have also conducted simulation using trees as weak learners to accommodate nonlinear covariate effects. More details and results are provided in Appendix. This set of simulation demonstrates the broad applicability of proposed strategy and again the merit of promoting similarity of sparsity structure in integrative analysis.
3.2. Analysis of breast cancer prognosis studies
Worldwide, breast cancer is the commonest cancer death among women. An estimated 226,870 new cases of invasive breast cancer were expected to occur among women in the U.S. in 2012. An estimated 39,510 breast cancer deaths were expected. Multiple profiling studies have been conducted, showing that genomic markers may be independently associated with prognosis beyond clinical risk factors and environmental exposures. Notable findings include the 97-gene signature [17], which includes genes UBE2C, PKNA2, TPX2, FOXM1, STK6, CCNA2, BIRC5, MYBL2, and others, and the 70-gene signature [18], which involves the hallmarks of cancer including cell cycle, metastasis, angiogenesis, and invasion.
We collect three gene expression datasets on breast cancer prognosis. The first dataset was initially described in Huang et al. [19]. Affymetrix chips were used to profile 12,625 genes on 71 samples. The second dataset was first described in Sotiriou et al. [20]. cDNA chips were used to profile 7,650 genes on 98 samples. The third dataset was first described in van’t Veer et al. [18]. Oligonucleotide chips were used to profile 24,481 genes on 78 samples. We refer to the original publications for more details.
The proposed method has been described for the scenario where the same set of covariates is measured in all datasets. If a covariate is not measured in a dataset, its corresponding coefficient can be set as zero [5], and the proposed method is then directly applicable. Most multi-dataset analyses target finding the similarity/difference across datasets. If a covariate is only measured in one or a few datasets but not others, it can be difficult or impossible to draw across-dataset conclusion. In the pangenomic era, the standard platforms conduct whole-genome profiling. In terms of methodology, the similarity of sparsity structures gets less meaningful when the overlaps of measured covariates get smaller. In our data analysis, we focus on the 2,555 genes measured in all three datasets. With practical data, preprocessing is needed. With gene expression data, we first conduct normalization. With Affymetrix data, a floor and a ceiling are added, and then measurements are log2 transformed. For both Affymetrix and cDNA data, there are a small number of missing values. We fill in using means across samples. We then standardize each gene expression to have zero mean and unit variance. As with some existing integrative analysis methods [4], the proposed method does not require direct comparability of measurements in different datasets. Thus, cross-dataset processing is not needed.
As simulation suggests inferior performance of the penalization methods, we focus on data analysis using the boosting methods. The estimation results using the six boosting methods are shown in Table 4. Different methods identify overlapping but different sets of markers. Even for genes identified by multiple methods (for example gene BCKDHB in dataset 1), the estimates can be different. We find that introducing the overlapping penalty improves similarity across datasets. Specifically, we calculate to be 0.33 (Alt.1), 0.33 (alg1-SB), 0.39 (New), 0.67 (New+), and 0.33 (indiv-SB), respectively. Note that under pool-SB, this measure is always equal to 1. Quick literature search suggests that some of the identified genes, for example PPP3CB and BAP1, have important implications in breast cancer progression. However, there is no objective way of determining which set of markers is “more meaningful”.
Table 4.
Analysis of the breast cancer datasets: identified genes and estimates.
| Unigene | gene | Alt.1 | alg1-SB | New | New+ | indiv-SB | pool-SB |
|---|---|---|---|---|---|---|---|
| Dataset 1 | |||||||
| Hs.100090 | TSPAN3 | 0.067 | |||||
| Hs.101382 | TNFAIP2 | 0.028 | |||||
| Hs.10247 | ALCAM | 0.063 | |||||
| Hs.153752 | CDC25B | 0.037 | 0.037 | ||||
| Hs.106778 | ATP2C1 | 0.059 | 0.059 | 0.111 | 0.100 | ||
| Hs.111126 | PTTG1IP | 0.041 | 0.035 | ||||
| Hs.115617 | CRHBP | 0.097 | 0.097 | 0.131 | 0.097 | ||
| Hs.124029 | INPP5A | 0.045 | 0.072 | ||||
| Hs.1265 | BCKDHB | −0.109 | −0.109 | −0.158 | −0.111 | −0.109 | |
| Hs.151531 | PPP3CB | −0.051 | −0.096 | −0.043 | −0.191 | ||
| Hs.153687 | INPP4B | 0.042 | 0.079 | 0.189 | |||
|
| |||||||
| Dataset 2 | |||||||
| Hs.100090 | TSPAN3 | 0.067 | |||||
| Hs.101382 | TNFAIP2 | 0.028 | |||||
| Hs.10247 | ALCAM | 0.063 | |||||
| Hs.101813 | SLC9A3R2 | 0.071 | 0.071 | 0.037 | 0.071 | ||
| Hs.102456 | GEMIN2 | 0.126 | 0.126 | 0.128 | 0.157 | ||
| Hs.105806 | GNLY | 0.028 | 0.028 | 0.097 | |||
| Hs.108332 | UBE2D2 | ||||||
| Hs.1265 | BCKDHB | 0.008 | 0.008 | ||||
| Hs.1311 | CD1C | 0.163 | 0.202 | 0.167 | 0.202 | ||
|
| |||||||
| Dataset 3 | |||||||
| Hs.100090 | TSPAN3 | 0.034 | 0.032 | 0.031 | 0.067 | ||
| Hs.101382 | TNFAIP2 | 0.028 | |||||
| Hs.10247 | ALCAM | 0.063 | |||||
| Hs.100030 | TERF2 | 0.026 | 0.044 | 0.022 | 0.026 | ||
| Hs.103081 | RPS6KB2 | 0.026 | 0.025 | ||||
| Hs.106674 | BAP1 | −0.117 | −0.116 | −0.077 | −0.086 | ||
| Hs.108332 | UBE2D2 | −0.063 | −0.063 | −0.096 | −0.035 | −0.063 | |
| Hs.110707 | DCAF8 | −0.026 | −0.026 | −0.025 | |||
| Hs.1265 | BCKDHB | −0.006 | −0.006 | ||||
We examine the prediction performance of different methods which may provide some insights into the analysis results. We randomly split each dataset into a training and a testing set with sizes 3:1. Estimates are generated using the training data and used to make prediction for subjects in the testing sets. We then dichotomize the testing subjects’ risk scores at the median and create two hypothetical risk sets. We calculate the logrank test statistic which measures the survival difference between the two sets. To avoid an extreme split, we repeat the above process 100 times and calculate the average logrank statistics as 2.176 (Alt.1), 2.338 (alg1-SB), 4.632 (New), 3.863 (New+), 2.880 (indiv-SB), and 0.544 (pool-SB), respectively. The proposed method leads to improved prediction performance.
With practical data, the “true” model is unknown. We also analyze data under the Cox model, which is a popular choice for survival data. More details are provided in Appendix.
4. Discussion
In the analysis of high-dimensional profiling data, integrative analysis provides an effective way of combining multiple datasets and increasing effective sample size and outperforms single-dataset analysis and classic meta-analysis. In this study, we consider the heterogeneity structure, which is more flexible and more challenging than the homogeneity structure. Sparse boosting is adopted for marker selection. To the best of our knowledge, this is the first study applying sparse boosting to the heterogeneity structure. As described in Introduction, there are scenarios under which it is desirable to encourage the similarity of sparsity structures across datasets. This study has proposed a new sparse boosting method, which explicitly promotes such similarity. The proposed method has an intuitive interpretation and is computationally feasible. In simulation, it shows competitive performance and can be preferred over the alternative boosting and penalization methods. In the analysis of three breast cancer prognosis datasets, the proposed method identifies markers different from the alternatives. The identified markers have a higher degree of similarity across datasets and better prediction performance.
In this article, we have focused on methodological development for marker selection. Practical data analysis demands extensive additional considerations. Specifically, different studies may have different research goals and designs. Quality control such as inclusion and exclusion has been discussed for meta-analysis [21] and is also needed here. The high dimensionality and other complexities of genomic data make this even more challenging [22]. In addition, it has been observed that prevalence may also affect selection [23]. We refer to the novel framework of Li and Fine [24] and others for more discussions. Prior to analysis, preprocessing, for example matching covariates across datasets and imputing missing measurements, is needed. The sensitivity of the proposed method on quality control, data selection, and data processing demands attention in practice. Potentially, there are multiple ways of conducting marker selection under the heterogeneity structure. In our simulation, we compare against penalization because of its popularity. We also conjecture that it is possible to couple the proposed penalty on similarity with the penalization methods. A limitation of this study is that theoretical properties are not established. For example, the convergence property is unclear, and there is no direct control of the number of selected markers. We do note that in our extensive simulations, convergence is achieved for all datasets, and only a small number of markers are identified. The theoretical properties are extremely difficult even under much simpler settings [9]. The heterogeneity across datasets and the new penalty make theoretical investigation even more challenging. In data analysis, bioinformatics and biological analysis is needed to fully comprehend the results.
Table 5.
Simulation: summary statistics on estimation and prediction. In each cell, mean (sd). d = 100 and ρ = 0.2.
| σ2 = 1 | σ2 = 3 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Complete | Half | None | Complete | Half | None | |||||||
|
| ||||||||||||
| PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | |
| nozero coef ~ unif(0.2,1) | ||||||||||||
| New | 1.12 (1.03) | 0.92 (0.79) | 0.95 (0.24) | 0.82 (0.21) | 0.97 (0.31) | 0.80 (0.25) | 1.21 (0.34) | 2.84 (0.78) | 1.24 (0.27) | 3.00 (0.64) | 1.18 (0.37) | 2.79 (0.81) |
| New+ | 1.08 (1.04) | 0.90 (0.80) | 0.96 (0.26) | 0.83 (0.22) | 0.97 (0.31) | 0.80 (0.25) | 1.14 (0.37) | 2.69 (0.86) | 1.30 (0.31) | 3.14 (0.76) | 1.17 (0.37) | 2.77 (0.81) |
| alg1-SB | 1.07 (0.45) | 0.87 (0.33) | 1.18 (0.33) | 0.99 (0.28) | 0.92 (0.33) | 0.75 (0.25) | 1.18 (0.29) | 2.79 (0.66) | 1.23 (0.36) | 2.96 (0.85) | 1.21 (0.33) | 2.86 (0.74) |
| sgroup-MCP | 0.44 (0.19) | 0.41 (0.18) | 0.59 (0.28) | 0.56 (0.27) | 0.74 (0.31) | 0.69 (0.30) | 0.65 (0.27) | 1.85 (0.81) | 0.77 (0.30) | 2.29 (0.96) | 0.89 (0.31) | 2.66 (1.01) |
| group-MCPT | 0.31 (0.15) | 0.33 (0.16) | 0.73 (0.31) | 0.77 (0.31) | 0.92 (0.34) | 0.97 (0.36) | 1.76 (0.77) | 1.80 (0.76) | 2.43 (1.00) | 2.45 (0.94) | 4.03 (1.07) | 4.12 (1.04) |
| indiv-SB | 1.13 (0.28) | 1.00 (0.24) | 1.30 (0.45) | 1.15 (0.40) | 1.22 (0.43) | 1.05 (0.36) | 1.14 (0.28) | 2.76 (0.65) | 1.24 (0.40) | 3.05 (0.95) | 1.17 (0.34) | 2.81 (0.76) |
| indiv-MCP | 0.62 (0.24) | 0.66 (0.25) | 0.66 (0.24) | 0.69 (0.26) | 0.65 (0.23) | 0.69 (0.25) | 0.78 (0.30) | 2.37 (0.95) | 0.84 (0.31) | 2.50 (0.92) | 0.81 (0.27) | 2.42 (0.82) |
| pool-SB | 1.03 (0.28) | 1.01 (0.29) | 4.86 (1.12) | 4.00 (0.88) | 9.22 (1.64) | 6.89 (1.19) | 0.63 (0.15) | 1.75 (0.45) | 1.88 (0.38) | 4.58 (0.90) | 3.17 (0.56) | 7.09 (1.20) |
| pool-MCP | 0.76 (0.23) | 0.78 (0.23) | 4.02 (0.91) | 3.53 (0.79) | 7.87 (1.51) | 6.24 (1.20) | 0.42 (0.11) | 1.32 (0.38) | 1.52 (0.37) | 4.06 (0.99) | 2.82 (0.53) | 6.68 (1.31) |
|
| ||||||||||||
| nonzero coef = 1 | ||||||||||||
| New | 0.49 (0.12) | 0.45 (0.12) | 0.63 (0.19) | 0.55 (0.16) | 0.62 (0.18) | 0.52 (0.15) | 1.00 (0.30) | 2.48 (0.75) | 1.24 (0.37) | 3.15 (0.92) | 1.10 (0.38) | 2.74 (0.93) |
| New+ | 0.48 (0.11) | 0.45 (0.11) | 0.66 (0.20) | 0.57 (0.17) | 0.63 (0.18) | 0.52 (0.15) | 1.10 (0.48) | 2.68 (1.11) | 1.24 (0.37) | 3.15 (0.92) | 1.10 (0.38) | 2.74 (0.93) |
| alg1-SB | 0.70 (0.22) | 0.60 (0.19) | 0.64 (0.22) | 0.56 (0.18) | 0.68 (0.20) | 0.57 (0.14) | 1.03 (0.31) | 2.55 (0.76) | 1.18 (0.46) | 3.01 (1.14) | 1.11 (0.32) | 2.70 (0.79) |
| sgroup-MCP | 0.25 (0.09) | 0.24 (0.09) | 0.25 (0.10) | 0.24 (0.10) | 0.26 (0.12) | 0.25 (0.12) | 0.31 (0.18) | 0.88 (0.49) | 0.44 (0.25) | 1.24 (0.75) | 0.56 (0.30) | 1.57 (0.84) |
| group-MCPT | 0.23 (0.08) | 0.24 (0.08) | 0.25 (0.16) | 0.27 (0.16) | 0.24 (0.08) | 0.25 (0.08) | 0.68 (0.33) | 0.73 (0.35) | 1.62 (0.99) | 1.71 (1.01) | 2.07 (1.13) | 2.16 (1.17) |
| indiv-SB | 0.83 (0.22) | 0.76 (0.21) | 0.94 (0.38) | 0.86 (0.33) | 0.86 (0.36) | 0.78 (0.32) | 1.38 (0.36) | 3.73 (1.02) | 1.63 (0.68) | 4.34 (1.64) | 1.40 (0.48) | 3.72 (1.20) |
| indiv-MCP | 0.28 (0.12) | 0.30 (0.12) | 0.29 (0.13) | 0.31 (0.13) | 0.28 (0.13) | 0.30 (0.13) | 0.60 (0.26) | 1.93 (0.83) | 0.64 (0.33) | 2.01 (1.03) | 0.64 (0.29) | 2.07 (0.92) |
| pool-SB | 0.22 (0.08) | 0.19 (0.08) | 11.10 (0.70) | 8.82 (0.50) | 23.80 (0.65) | 17.23 (0.37) | 0.30 (0.11) | 0.79 (0.30) | 3.96 (0.28) | 9.39 (0.66) | 8.09 (0.20) | 17.50 (0.33) |
| pool-MCP | 0.08 (0.06) | 0.09 (0.06) | 8.79 (0.47) | 7.49 (0.44) | 20.36 (0.76) | 15.81 (0.63) | 0.09 (0.06) | 0.29 (0.18) | 3.13 (0.22) | 8.04 (0.57) | 7.01 (0.30) | 16.33 (1.00) |
Table 6.
Simulation: summary statistics on estimation and prediction. In each cell, mean (sd). d = 100 and ρ = 0.5.
| σ2 = 1 | σ2 = 3 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Complete | Half | None | Complete | Half | None | |||||||
|
| ||||||||||||
| PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | |
| nonzero coef ~ unif(0.2,1) | ||||||||||||
| New | 0.57 (0.20) | 0.69 (0.35) | 0.78 (0.24) | 0.71 (0.19) | 0.59 (0.19) | 0.58 (0.20) | 0.63 (0.19) | 1.51 (0.50) | 0.76 (0.23) | 1.80 (0.59) | 0.68 (0.21) | 1.68 (0.49) |
| New+ | 0.57 (0.19) | 0.68 (0.34) | 0.81 (0.26) | 0.73 (0.20) | 0.58 (0.19) | 0.57 (0.20) | 0.64 (0.24) | 1.52 (0.53) | 0.75 (0.23) | 1.79 (0.60) | 0.67 (0.21) | 1.68 (0.48) |
| alg1-SB | 0.63 (0.18) | 0.56 (0.16) | 0.71 (0.26) | 0.66 (0.23) | 0.62 (0.15) | 0.60 (0.19) | 0.72 (0.17) | 1.71 (0.49) | 0.76 (0.20) | 1.92 (0.53) | 0.70 (0.18) | 1.70 (0.42) |
| sgroup-MCP | 0.80 (0.42) | 0.48 (0.22) | 0.94 (0.41) | 0.58 (0.23) | 1.38 (0.58) | 0.79 (0.31) | 1.08 (0.42) | 1.92 (0.72) | 1.21 (0.46) | 2.29 (0.85) | 1.47 (0.47) | 2.66 (0.78) |
| group-MCPT | 0.27 (0.18) | 0.39 (0.22) | 0.69 (0.32) | 1.04 (0.43) | 1.04 (0.35) | 1.60 (0.46) | 1.06 (0.55) | 1.60 (0.90) | 2.20 (0.81) | 3.26 (1.14) | 3.18 (0.87) | 4.69 (1.19) |
| indiv-SB | 0.66 (0.17) | 0.75 (0.23) | 0.77 (0.21) | 0.90 (0.27) | 0.64 (0.18) | 0.76 (0.23) | 0.70 (0.22) | 1.96 (0.66) | 0.82 (0.22) | 2.31 (0.67) | 0.69 (0.19) | 2.00 (0.69) |
| indiv-MCP | 0.67 (0.23) | 1.09 (0.41) | 0.65 (0.25) | 1.06 (0.42) | 0.68 (0.29) | 1.18 (0.50) | 0.80 (0.23) | 3.84 (1.40) | 0.76 (0.22) | 3.48 (1.24) | 0.81 (0.26) | 4.08 (1.49) |
| pool-SB | 0.80 (0.27) | 0.86 (0.23) | 5.05 (0.99) | 3.88 (0.85) | 12.92 (2.54) | 6.59 (1.25) | 0.44 (0.11) | 1.43 (0.44) | 1.94 (0.34) | 4.30 (0.95) | 4.58 (0.85) | 6.84 (1.28) |
| pool-MCP | 0.69 (0.27) | 0.78 (0.23) | 4.66 (1.07) | 4.21 (1.04) | 11.92 (2.53) | 7.52 (1.84) | 0.39 (0.14) | 1.67 (0.77) | 1.79 (0.45) | 5.26 (1.78) | 4.12 (0.84) | 7.88 (2.00) |
|
| ||||||||||||
| nonzero coef = 1 | ||||||||||||
| New | 0.46 (0.20) | 0.62 (0.34) | 0.54 (0.13) | 0.49 (0.13) | 0.49 (0.15) | 0.50 (0.17) | 0.58 (0.21) | 1.65 (0.59) | 0.77 (0.24) | 2.08 (0.70) | 0.64 (0.14) | 1.88 (0.46) |
| New+ | 0.46 (0.20) | 0.62 (0.34) | 0.56 (0.14) | 0.50 (0.13) | 0.49 (0.15) | 0.50 (0.17) | 0.61 (0.22) | 1.69 (0.58) | 0.78 (0.24) | 2.10 (0.71) | 0.64 (0.14) | 1.88 (0.46) |
| alg1-SB | 0.50 (0.16) | 0.49 (0.11) | 0.59 (0.17) | 0.57 (0.17) | 0.53 (0.21) | 0.51 (0.21) | 0.66 (0.27) | 1.91 (0.76) | 0.71 (0.21) | 1.99 (0.73) | 0.66 (0.19) | 1.80 (0.61) |
| sgroup-MCP | 0.35 (0.17) | 0.23 (0.12) | 0.36 (0.21) | 0.24 (0.12) | 0.49 (0.43) | 0.31 (0.23) | 0.58 (0.39) | 1.09 (0.69) | 1.03 (0.74) | 1.90 (1.22) | 1.53 (0.86) | 2.62 (1.21) |
| group-MCPT | 0.23 (0.11) | 0.35 (0.16) | 0.25 (0.10) | 0.36 (0.14) | 0.40 (0.23) | 0.56 (0.30) | 0.78 (0.55) | 1.11 (0.64) | 2.03 (0.93) | 2.98 (1.38) | 3.29 (1.19) | 4.93 (2.06) |
| indiv-SB | 0.52 (0.14) | 0.62 (0.21) | 0.60 (0.16) | 0.75 (0.25) | 0.50 (0.16) | 0.62 (0.27) | 0.67 (0.19) | 2.35 (0.85) | 0.85 (0.25) | 3.15 (1.02) | 0.67 (0.18) | 2.45 (0.89) |
| indiv-MCP | 0.27 (0.11) | 0.40 (0.17) | 0.26 (0.09) | 0.39 (0.15) | 0.26 (0.11) | 0.40 (0.19) | 0.77 (0.34) | 3.81 (2.03) | 0.72 (0.32) | 3.63 (1.95) | 0.73 (0.32) | 3.87 (2.06) |
| pool-SB | 0.15 (0.06) | 0.17 (0.09) | 11.51 (0.76) | 8.38 (0.61) | 33.66 (1.72) | 16.48 (0.62) | 0.18 (0.07) | 0.58 (0.34) | 4.13 (0.28) | 9.00 (0.79) | 11.52 (0.60) | 16.69 (0.62) |
| pool-MCP | 0.07 (0.05) | 0.10 (0.08) | 10.59 (0.53) | 8.99 (0.67) | 31.19 (1.24) | 19.40 (2.18) | 0.07 (0.05) | 0.32 (0.24) | 3.75 (0.27) | 9.88 (1.05) | 10.62 (0.58) | 20.17 (2.87) |
Table 7.
Simulation: summary statistics on estimation and prediction. In each cell, mean (sd). d = 100 and ρ = 0.8.
| σ2 = 1 | σ2 = 3 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Complete | Half | None | Complete | Half | None | |||||||
|
| ||||||||||||
| PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | |
| nonzero coef ~ unif(0.2,1) | ||||||||||||
| New | 0.36 (0.10) | 1.00 (0.39) | 0.41 (0.12) | 1.04 (0.35) | 0.38 (0.12) | 1.02 (0.38) | 0.31 (0.09) | 2.23 (0.85) | 0.48 (0.14) | 2.27 (0.67) | 0.35 (0.12) | 2.29 (0.69) |
| New+ | 0.37 (0.11) | 1.00 (0.39) | 0.43 (0.14) | 1.04 (0.35) | 0.38 (0.12) | 1.02 (0.38) | 0.31 (0.09) | 2.23 (0.85) | 0.48 (0.13) | 2.27 (0.67) | 0.35 (0.12) | 2.29 (0.69) |
| alg1-SB | 0.39 (0.11) | 0.97 (0.41) | 0.44 (0.12) | 1.05 (0.40) | 0.40 (0.16) | 1.03 (0.42) | 0.39 (0.10) | 2.41 (0.57) | 0.45 (0.13) | 2.36 (0.76) | 0.34 (0.10) | 2.31 (0.70) |
| sgroup-MCP | 2.44 (1.55) | 0.54 (0.27) | 3.03 (1.39) | 0.72 (0.31) | 3.96 (1.49) | 0.85 (0.27) | 2.36 (0.77) | 1.64 (0.56) | 2.68 (0.75) | 2.07 (0.58) | 3.07 (0.91) | 2.22 (0.68) |
| group-MCPT | 0.31 (0.19) | 0.98 (0.42) | 0.62 (0.26) | 2.20 (0.71) | 0.94 (0.34) | 3.42 (0.90) | 1.05 (0.58) | 3.53 (1.66) | 1.79 (0.64) | 5.77 (1.56) | 2.44 (0.69) | 8.33 (2.17) |
| indiv-SB | 0.38 (0.11) | 1.05 (0.42) | 0.42 (0.13) | 1.17 (0.53) | 0.41 (0.14) | 1.22 (0.50) | 0.37 (0.11) | 2.40 (0.74) | 0.43 (0.11) | 2.73 (0.91) | 0.37 (0.10) | 2.61 (0.88) |
| indiv-MCP | 0.86 (0.22) | 4.51 (1.52) | 0.86 (0.26) | 4.14 (1.73) | 0.93 (0.29) | 4.83 (1.61) | 0.71 (0.17) | 8.81 (2.72) | 0.76 (0.19) 8.65 | (2.16) | 0.76 (0.25) | 9.16 (2.64) |
| pool-SB | 0.68 (0.40) | 0.99 (0.34) | 3.58 (0.87) | 3.88 (1.01) | 13.82 (2.73) | 7.12 (1.44) | 0.33 (0.16) | 1.70 (0.70) | 1.34 (0.33) | 4.40 (1.21) | 4.77 (0.92) | 7.38 (1.60) |
| pool-MCP | 0.83 (0.47) | 2.02 (1.28) | 3.79 (0.87) | 6.08 (1.78) | 14.07 (2.57) | 10.85 (2.09) | 0.47 (0.15) | 5.13 (2.06) | 1.44 (0.32) | 7.51 (1.99) | 4.87 (0.89) | 11.84 (2.48) |
|
| ||||||||||||
| nonzero coef = 1 | ||||||||||||
| New | 0.37 (0.11) | 1.30 (0.49) | 0.47 (0.12) | 1.30 (0.59) | 0.46 (0.13) | 1.30 (0.48) | 0.38 (0.14) | 3.47 (1.46) | 0.47 (0.16) | 3.52 (1.34) | 0.37 (0.14) | 3.08 (1.14) |
| New+ | 0.39 (0.12) | 1.31 (0.49) | 0.47 (0.12) | 1.30 (0.59) | 0.46 (0.13) | 1.30 (0.48) | 0.42 (0.19) | 3.48 (1.49) | 0.48 (0.17) | 3.53 (1.34) | 0.37 (0.14) | 3.07 (1.15) |
| alg1-SB | 0.44 (0.18) | 1.45 (0.68) | 0.49 (0.15) | 1.37 (0.51) | 0.41 (0.11) | 1.34 (0.48) | 0.38 (0.12) | 2.91 (1.04) | 0.45 (0.12) | 3.29 (1.30) | 0.43 (0.10) | 3.26 (1.21) |
| sgroup-MCP | 1.60 (1.43) | 0.40 (0.33) | 2.60 (1.81) | 0.63 (0.39) | 4.13 (2.89) | 0.88 (0.51) | 3.06 (1.62) | 1.98 (0.83) | 3.88 (1.36) | 2.50 (0.75) | 4.59 (1.39) | 2.83 (0.68) |
| group-MCPT | 0.26 (0.14) | 0.96 (0.48) | 0.68 (0.30) | 2.40 (0.98) | 1.12 (0.44) | 3.79 (1.31) | 0.95 (0.57) | 2.96 (1.57) | 1.99 (0.90) | 7.19 (2.14) | 3.01 (0.90) | 11.34 (2.57) |
| indiv-SB | 0.37 (0.12) | 1.23 (0.55) | 0.42 (0.16) | 1.44 (0.76) | 0.41 (0.13) | 1.45 (0.65) | 0.39 (0.11) | 3.29 (1.16) | 0.45 (0.14) | 4.05 (1.74) | 0.42 (0.13) | 4.00 (1.46) |
| indiv-MCP | 1.41 (0.33) | 9.09 (2.51) | 1.08 (0.33) | 6.30 (2.09) | 1.51 (0.36) | 9.71 (2.58) | 0.97 (0.24) | 15.33 (3.45) | 1.01 (0.23) | 14.72 (3.45) | 1.03 (0.25) | 16.31 (3.39) |
| pool-SB | 0.10 (0.06) | 0.29 (0.19) | 7.84 (0.35) | 8.09 (0.55) | 36.74 (1.39) | 18.13 (1.75) | 0.11 (0.06) | 0.90 (0.60) | 2.76 (0.19) | 8.85 (0.98) | 12.43 (0.57) | 18.25 (2.01) |
| pool-MCP | 0.08 (0.09) | 0.27 (0.25) | 8.38 (0.51) | 12.69 (2.94) | 37.40 (1.54) | 27.67 (3.06) | 0.21 (0.16) | 2.57 (2.61) | 3.00 (0.21) | 14.46 (3.00) | 12.64 (0.62) | 28.59 (3.44) |
Table 8.
Simulation: summary statistics on estimation and prediction. In each cell, mean (sd). d = 500 and ρ = 0.2.
| σ2 = 1 | σ2 = 3 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Complete | Half | None | Complete | Half | None | |||||||
|
| ||||||||||||
| PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | |
| nonzero coef ~ unif(0.2,1) | ||||||||||||
| New | 1.69 (1.12) | 1.37 (0.86) | 1.81 (0.53) | 1.48 (0.41) | 1.81 (0.59) | 1.41 (0.44) | 1.78 (0.42) | 4.07 (0.96) | 1.79 (0.41) | 4.25 (0.92) | 1.71 (0.39) | 3.91 (0.90) |
| New+ | 1.58 (1.14) | 1.28 (0.88) | 1.85 (0.56) | 1.51 (0.44) | 1.80 (0.59) | 1.41 (0.44) | 1.68 (0.47) | 3.84 (1.07) | 1.82 (0.53) | 4.32 (1.20) | 1.71 (0.39) | 3.91 (0.90) |
| alg1-SB | 1.73 (0.48) | 1.34 (0.36) | 1.99 (0.65) | 1.61 (0.51) | 1.69 (0.40) | 1.33 (0.30) | 1.87 (0.45) | 4.26 (1.01) | 1.77 (0.36) | 4.21 (0.84) | 1.73 (0.44) | 3.97 (1.00) |
| sgroup-MCP | 0.49 (0.22) | 0.46 (0.22) | 0.69 (0.27) | 0.65 (0.26) | 0.86 (0.33) | 0.80 (0.31) | 0.79 (0.31) | 2.29 (0.97) | 1.01 (0.35) | 3.03 (1.08) | 1.11 (0.31) | 3.38 (0.96) |
| group-MCPT | 0.37 (0.25) | 0.39 (0.26) | 0.74 (0.32) | 0.78 (0.33) | 1.53 (0.74) | 1.60 (0.69) | 2.62 (0.90) | 2.53 (0.79) | 3.48 (1.38) | 3.42 (1.17) | 4.42 (1.37) | 4.18 (1.53) |
| indiv-SB | 1.53 (0.60) | 1.23 (0.44) | 1.61 (0.45) | 1.35 (0.34) | 1.56 (0.49) | 1.26 (0.37) | 1.80 (0.49) | 4.12 (1.10) | 1.77 (0.51) | 4.22 (1.19) | 1.81 (0.46) | 4.13 (1.04) |
| indiv-MCP | 0.88 (0.30) | 0.91 (0.31) | 0.90 (0.24) | 0.94 (0.27) | 0.88 (0.24) | 0.92 (0.27) | 1.04 (0.30) | 2.89 (0.82) | 1.03 (0.32) | 2.93 (0.99) | 1.00 (0.28) | 2.83 (0.90) |
| pool-SB | 1.03 (0.30) | 0.99 (0.27) | 5.07 (1.06) | 4.13 (0.82) | 9.57 (1.77) | 7.13 (1.27) | 0.76 (0.21) | 1.98 (0.49) | 1.98 (0.40) | 4.78 (0.92) | 3.23 (0.60) | 7.21 (1.29) |
| pool-MCP | 0.75 (0.23) | 0.77 (0.22) | 4.21 (0.91) | 3.64 (0.78) | 8.33 (1.48) | 6.50 (1.15) | 0.45 (0.10) | 1.40 (0.31) | 1.63 (0.37) | 4.25 (0.98) | 3.02 (0.54) | 7.02 (1.23) |
|
| ||||||||||||
| nonzero coef = 1 | ||||||||||||
| New | 0.95 (0.31) | 0.77 (0.25) | 1.33 (0.31) | 1.10 (0.25) | 1.19 (0.28) | 0.98 (0.24) | 1.91 (0.85) | 4.53 (1.87) | 2.28 (0.81) | 5.60 (1.92) | 1.99 (0.58) | 4.75 (1.36) |
| New+ | 1.18 (0.43) | 0.94 (0.34) | 1.33 (0.31) | 1.10 (0.25) | 1.19 (0.28) | 0.97 (0.24) | 1.87 (0.68) | 4.45 (1.53) | 2.30 (0.75) | 5.65 (1.79) | 1.99 (0.58) | 4.75 (1.36) |
| alg1-SB | 1.38 (0.34) | 1.11 (0.27) | 1.16 (0.24) | 0.96 (0.19) | 1.27 (0.35) | 1.04 (0.27) | 2.08 (1.01) | 4.92 (2.27) | 2.65 (1.13) | 6.42 (2.66) | 2.08 (0.79) | 4.90 (1.77) |
| sgroup-MCP | 0.23 (0.11) | 0.22 (0.11) | 0.25 (0.11) | 0.23 (0.11) | 0.25 (0.10) | 0.23 (0.09) | 0.34 (0.21) | 0.94 (0.58) | 0.62 (0.43) | 1.72 (1.15) | 0.79 (0.44) | 2.16 (1.18) |
| group-MCPT | 0.20 (0.07) | 0.22 (0.08) | 0.24 (0.14) | 0.26 (0.14) | 0.28 (0.17) | 0.29 (0.17) | 0.89 (0.67) | 0.94 (0.69) | 2.11 (1.27) | 2.25 (1.37) | 7.66 (4.73) | 7.49 (4.23) |
| indiv-SB | 0.90 (0.66) | 0.78 (0.57) | 0.91 (0.55) | 0.80 (0.45) | 0.90 (0.53) | 0.78 (0.44) | 2.14 (0.76) | 5.13 (1.64) | 2.36 (0.74) | 5.85 (1.66) | 2.15 (0.76) | 5.14 (1.65) |
| indiv-MCP | 0.29 (0.12) | 0.30 (0.14) | 0.29 (0.10) | 0.31 (0.10) | 0.33 (0.18) | 0.35 (0.19) | 0.87 (0.31) | 2.70 (0.92) | 0.83 (0.28) | 2.59 (0.85) | 0.87 (0.31) | 2.75 (1.03) |
| pool-SB | 0.20 (0.10) | 0.17 (0.08) | 11.28 (0.61) | 8.89 (0.42) | 24.50 (0.45) | 17.64 (0.25) | 0.29 (0.14) | 0.73 (0.36) | 4.09 (0.26) | 9.57 (0.56) | 8.24 (0.10) | 17.76 (0.14) |
| pool-MCP | 0.11 (0.10) | 0.11 (0.10) | 9.38 (0.61) | 7.90 (0.62) | 21.32 (1.14) | 16.35 (0.93) | 0.11 (0.10) | 0.35 (0.31) | 3.40 (0.29) | 8.60 (0.87) | 7.39 (0.43) | 16.90 (1.11) |
Table 9.
Simulation: summary statistics on estimation and prediction. In each cell, mean (sd). d = 500 and ρ = 0.5.
| σ2 = 1 | σ2 = 3 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Complete | Half | None | Complete | Half | None | |||||||
|
| ||||||||||||
| PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | |
| nonzero coef ~ unif(0.2,1) | ||||||||||||
| New | 0.78 (0.24) | 0.71 (0.30) | 1.11 (0.33) | 0.88 (0.28) | 0.84 (0.20) | 0.65 (0.16) | 0.91 (0.28) | 1.92 (0.56) | 1.20 (0.35) | 2.54 (0.70) | 0.99 (0.24) | 2.00 (0.51) |
| New+ | 0.78 (0.26) | 0.71 (0.30) | 1.36 (1.18) | 1.03 (0.69) | 0.84 (0.20) | 0.65 (0.16) | 1.00 (0.37) | 2.03 (0.67) | 1.20 (0.34) | 2.53 (0.67) | 0.95 (0.26) | 1.95 (0.51) |
| alg1-SB | 1.02 (0.26) | 0.78 (0.21) | 1.19 (0.40) | 0.92 (0.30) | 0.93 (0.26) | 0.73 (0.23) | 0.99 (0.33) | 2.12 (0.71) | 1.23 (0.35) | 2.63 (0.67) | 1.02 (0.24) | 2.14 (0.52) |
| sgroup-MCP | 1.00 (0.42) | 0.57 (0.22) | 1.22 (0.47) | 0.74 (0.28) | 1.68 (0.69) | 0.93 (0.35) | 1.40 (0.49) | 2.44 (0.84) | 1.50 (0.45) | 2.83 (0.76) | 1.69 (0.47) | 3.12 (0.86) |
| group-MCPT | 0.34 (0.22) | 0.47 (0.27) | 0.87 (0.45) | 1.28 (0.56) | 1.33 (0.70) | 1.95 (0.82) | 1.48 (0.83) | 2.37 (1.30) | 2.82 (1.33) | 4.15 (1.60) | 4.41 (1.63) | 6.27 (1.62) |
| indiv-SB | 0.77 (0.20) | 0.73 (0.18) | 0.85 (0.23) | 0.82 (0.20) | 0.77 (0.31) | 0.73 (0.24) | 0.92 (0.21) | 2.12 (0.46) | 1.08 (0.26) | 2.47 (0.49) | 0.94 (0.29) | 2.07 (0.56) |
| indiv-MCP | 0.98 (0.33) | 1.65 (0.52) | 0.96 (0.33) | 1.56 (0.59) | 0.97 (0.28) | 1.70 (0.51) | 1.04 (0.27) | 4.71 (1.47) | 1.07 (0.28) | 4.71 (1.43) | 1.01 (0.27) | 4.48 (1.35) |
| pool-SB | 0.81 (0.28) | 0.86 (0.25) | 5.07 (1.03) | 3.75 (0.75) | 13.66 (2.38) | 6.66 (1.11) | 0.47 (0.13) | 1.38 (0.37) | 2.02 (0.39) | 4.24 (0.85) | 4.81 (0.85) | 6.87 (1.14) |
| pool-MCP | 0.77 (0.33) | 0.92 (0.36) | 4.88 (1.22) | 4.40 (1.17) | 11.96 (2.30) | 7.15 (1.48) | 0.51 (0.15) | 2.29 (0.91) | 1.88 (0.37) | 5.45 (1.16) | 4.18 (0.77) | 7.30 (1.58) |
|
| ||||||||||||
| nonzero coef = 1 | ||||||||||||
| New | 0.60 (0.21) | 0.58 (0.28) | 0.93 (0.28) | 0.75 (0.23) | 0.79 (0.23) | 0.68 (0.22) | 0.89 (0.26) | 2.59 (0.87) | 1.12 (0.40) | 2.71 (1.06) | 0.92 (0.24) | 2.30 (0.71) |
| New+ | 0.61 (0.26) | 0.59 (0.32) | 1.02 (0.31) | 0.80 (0.24) | 0.78 (0.24) | 0.68 (0.23) | 0.91 (0.28) | 2.63 (0.85) | 1.45 (1.35) | 3.28 (2.42) | 0.92 (0.24) | 2.30 (0.71) |
| alg1-SB | 0.82 (0.23) | 0.67 (0.21) | 0.94 (0.26) | 0.75 (0.21) | 0.82 (0.21) | 0.72 (0.22) | 0.98 (0.25) | 2.37 (0.59) | 1.04 (0.35) | 2.63 (0.85) | 1.01 (0.29) | 2.46 (0.73) |
| sgroup-MCP | 0.38 (0.21) | 0.25 (0.14) | 0.42 (0.29) | 0.29 (0.18) | 0.52 | (0.52) 0.34 (0.32) | 0.92 (0.59) | 1.62 (0.93) | 1.40 (0.77) | 2.51 (1.28) | 2.27 (1.15) | 3.70 (1.64) |
| group-MCPT | 0.26 (0.14) | 0.37 (0.16) | 0.29 (0.14) | 0.40 (0.18) | 0.46 (0.32) | 0.59 (0.36) | 0.75 (0.41) | 1.08 (0.47) | 2.86 (1.57) | 4.19 (1.91) | 5.57 (2.61) | 7.79 (3.07) |
| indiv-SB | 0.52 (0.15) | 0.56 (0.15) | 0.60 (0.24) | 0.67 (0.33) | 0.52 (0.20) | 0.55 (0.18) | 0.82 (0.23) | 2.45 (0.74) | 0.95 (0.27) | 2.89 (0.86) | 0.84 (0.37) | 2.36 (0.80) |
| indiv-MCP | 0.29 (0.09) | 0.39 (0.12) | 0.35 (0.40) | 0.50 (0.76) | 0.31 (0.14) | 0.41 (0.17) | 1.48 (0.36) | 8.21 (2.43) | 1.38 (0.37) | 7.18 (1.96) | 1.47 (0.27) | 8.19 (1.79) |
| pool-SB | 0.15 (0.07) | 0.16 (0.08) | 11.41 (0.60) | 8.18 (0.48) | 35.96 (2.15) | 16.57 (0.53) | 0.19 (0.08) | 0.53 (0.28) | 4.19 (0.27) | 8.81 (0.58) | 12.43 (0.72) | 16.88 (0.51) |
| pool-MCP | 0.08 (0.04) | 0.11 (0.06) | 10.86 (0.43) | 9.35 (0.78) | 31.84 (1.32) | 18.90 (1.74) | 0.10 (0.06) | 0.41 (0.23) | 3.94 (0.25) | 10.74 (1.36) | 10.84 (0.55) | 18.74 (1.52) |
Table 10.
Simulation: summary statistics on estimation and prediction. In each cell, mean (sd). d = 500 and ρ = 0.8.
| Σ2 = 1 | σ2 = 3 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Complete | Half | None | Complete | Half | None | |||||||
|
| ||||||||||||
| PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | |
| nonzero coef ~ unif(0.2,1) | ||||||||||||
| New | 0.44 (0.15) | 0.97 (0.35) | 0.65 (0.17) | 1.06 (0.26) | 0.54 (0.17) | 1.09 (0.43) | 0.47 (0.16) | 2.35 (0.95) | 0.59 (0.15) | 2.61 (0.97) | 0.44 (0.12) | 2.17 (0.69) |
| New+ | 0.49 (0.16) | 0.98 (0.35) | 0.65 (0.17) | 1.06 (0.26) | 0.54 (0.17) | 1.09 (0.43) | 0.51 (0.17) | 2.40 (1.02) | 0.62 (0.17) | 2.61 (0.98) | 0.44 (0.12) | 2.17 (0.69) |
| alg1-SB | 0.57 (0.16) | 1.19 (0.38) | 0.58 (0.21) | 1.00 (0.31) | 0.54 (0.15) | 1.01 (0.24) | 0.55 (0.16) | 2.42 (0.76) | 0.60 (0.19) | 2.20 (0.66) | 0.54 (0.15) | 2.22 (0.60) |
| sgroup-MCP | 3.31 (1.65) | 0.71 (0.30) | 4.08 (1.64) | 0.91 (0.35) | 5.11 (1.86) | 1.06 (0.37) | 2.84 (0.79) | 1.93 (0.52) | 2.91 (0.73) | 2.41 (0.71) | 3.38 (0.92) | 2.60 (0.91) |
| group-MCPT | 0.32 (0.26) | 1.04 (0.59) | 0.76 (0.38) | 2.36 (0.74) | 1.11 (0.48) | 3.70 (1.12) | 1.15 (0.73) | 3.83 (1.71) | 2.10 (0.92) | 6.38 (1.95) | 2.84 (0.97) | 8.97 (2.44) |
| indiv-SB | 0.45 (0.18) | 1.15 (0.54) | 0.48 (0.12) | 1.22 (0.47) | 0.45 (0.11) | 1.15 (0.48) | 0.46 (0.19) | 2.65 (1.04) | 0.55 (0.18) | 2.90 (1.11) | 0.47 (0.10) | 2.66 (0.71) |
| indiv-MCP | 1.25 (0.39) | 6.51 (2.16) | 1.32 (0.36) | 6.74 (2.18) | 1.28 (0.35) | 6.64 (1.97) | 0.94 (0.24) | 11.23 (2.48) | 0.95 (0.18) | 10.41 (2.03) | 0.96 (0.20) | 11.35 (2.54) |
| pool-SB | 0.68 (0.35) | 1.05 (0.34) | 3.62 (0.80) | 4.01 (0.81) | 14.08 (2.90) | 7.11 (1.45) | 0.33 (0.12) | 1.80 (0.60) | 1.36 (0.30) | 4.54 (1.11) | 4.94 (1.01) | 7.31 (1.46) |
| pool-MCP | 1.06 (0.48) | 3.94 (1.7) | 4.08 (0.87) | 7.36 (1.98) | 14.47 (2.92) | 11.37 (2.55) | 0.56 (0.18) | 6.48 (2.27) | 1.58 (0.35) | 9.18 (2.69) | 5.04 (1.00) | 11.90 (3.26) |
|
| ||||||||||||
| nonzero coef = 1 | ||||||||||||
| New | 0.52 (0.22) | 1.95 (1.12) | 0.68 (0.16) | 1.42 (0.51) | 0.52 (0.18) | 1.43 (0.72) | 0.48 (0.17) | 3.21 (1.1) | 0.65 (0.23) | 3.38 (1.13) | 0.49 (0.15) | 3.11 (1.37) |
| New+ | 0.52 (0.22) | 1.95 (1.12) | 0.69 (0.16) | 1.43 (0.52) | 0.52 (0.17) | 1.44 (0.72) | 0.53 (0.22) | 3.26 (1.11) | 0.67 (0.25) | 3.42 (1.13) | 0.49 (0.15) | 3.11 (1.37) |
| alg1-SB | 0.62 (0.22) | 1.51 (0.65) | 0.69 (0.25) | 1.51 (0.85) | 0.59 (0.18) | 1.48 (0.74) | 0.54 (0.15) | 3.30 (1.12) | 0.64 (0.17) | 3.65 (1.33) | 0.58 (0.12) | 3.52 (0.99) |
| sgroup-MCP | 2.50 (2.61) | 0.58 (0.51) | 4.25 (3.33) | 0.96 (0.68) | 6.82 (3.64) | 1.33 (0.63) | 4.35 (1.66) | 2.58 (0.84) | 4.89 (1.32) | 2.98 (0.72) | 5.95 (1.29) | 3.45 (0.86) |
| group-MCPT | 0.29 (0.19) | 0.97 (0.44) | 0.79 (0.38) | 2.20 (1.09) | 1.46 (0.78) | 4.49 (1.55) | 1.20 (1.20) | 3.16 (1.77) | 2.25 (0.94) | 7.80 (2.29) | 3.69 (1.68) | 12.60 (3.19) |
| indiv-SB | 0.43 (0.16) | 1.39 (0.56) | 0.44 (0.10) | 1.43 (0.50) | 0.43 (0.12) | 1.45 (0.62) | 0.48 (0.18) | 3.72 (1.26) | 0.51 (0.13) | 4.12 (1.51) | 0.48 (0.11) | 3.84 (1.43) |
| indiv-MCP | 2.11 (0.50) | 13.00 (2.66) | 1.88 (0.55) | 10.45 (3.30) | 1.96 (0.37) | 12.37 (2.29) | 1.32 (0.26) | 20.11 (2.82) | 1.34 (0.40) | 19.59 (4.05) | 1.34 (0.26) | 20.55 (3.11) |
| pool-SB | 0.11 (0.06) | 0.31 (0.17) | 7.99 (0.36) | 8.52 (0.87) | 37.08 (1.53) | 17.60 (1.54) | 0.12 (0.08) | 0.96 (0.65) | 2.84 (0.19) | 9.36 (1.27) | 12.62 (0.62) | 17.97 (1.51) |
| pool-MCP | 0.98 (0.45) | 6.92 (3.67) | 8.96 (0.49) | 15.54 (2.85) | 38.30 (1.36) | 29.74 (2.29) | 0.47 (0.19) | 9.30 (4.57) | 3.29 (0.32) | 18.28 (4.29) | 13.10 (0.58) | 30.88 (3.06) |
Table 11.
Simulation: summary statistics on estimation and prediction. In each cell, mean (sd). d = 1,000 and ρ = 0.2.
| σ2 = 1 | σ2 = 3 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Complete | Half | None | Complete | Half | None | |||||||
|
| ||||||||||||
| PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | |
| nonzero coef ~ unif(0.2,1) | ||||||||||||
| New | 2.00 (0.69) | 1.57 (0.53) | 2.32 (0.71) | 1.85 (0.53) | 2.13 (0.66) | 1.66 (0.49) | 2.09 (0.59) | 4.75 (1.27) | 2.12 (0.55) | 4.98 (1.3) | 1.95 (0.41) | 4.41 (0.93) |
| New+ | 1.83 (0.87) | 1.46 (0.68) | 2.38 (0.81) | 1.90 (0.61) | 2.12 (0.66) | 1.65 (0.50) | 1.82 (0.51) | 4.17 (1.11) | 2.02 (0.60) | 4.77 (1.39) | 1.95 (0.41) | 4.41 (0.93) |
| alg1-SB | 2.66 (0.98) | 2.03 (0.71) | 2.36 (0.74) | 1.90 (0.58) | 2.36 (0.90) | 1.81 (0.68) | 2.16 (0.51) | 4.90 (1.15) | 2.22 (0.44) | 5.22 (1.03) | 2.05 (0.52) | 4.62 (1.16) |
| sgroup-MCP | 0.58 (0.25) | 0.54 (0.23) | 0.81 (0.30) | 0.77 (0.29) | 1.01 (0.39) | 0.97 (0.39) | 0.86 (0.32) | 2.50 (0.96) | 1.05 (0.34) | 3.18 (1.14) | 1.31 (0.44) | 4.09 (1.43) |
| group-MCPT | 0.50 (0.25) | 0.51 (0.25) | 0.83 (0.35) | 0.86 (0.36) | 1.99 (1.11) | 2.03 (1.03) | 3.32 (1.09) | 3.09 (0.85) | 4.02 (1.57) | 3.81 (1.31) | 5.38 (1.66) | 4.83 (1.78) |
| indiv-SB | 2.11 (0.76) | 1.66 (0.56) | 2.26 (0.82) | 1.85 (0.62) | 2.06 (0.70) | 1.64 (0.52) | 2.11 (0.49) | 4.78 (1.13) | 2.13 (0.57) | 5.04 (1.37) | 1.97 (0.49) | 4.50 (1.11) |
| indiv-MCP | 1.05 (0.32) | 1.08 (0.32) | 1.06 (0.32) | 1.08 (0.33) | 0.98 (0.33) | 1.02 (0.34) | 1.22 (0.30) | 3.40 (0.87) | 1.23 (0.33) | 3.47 (0.96) | 1.19 (0.30) | 3.33 (0.82) |
| pool-SB | 1.13 (0.39) | 1.05 (0.37) | 5.41 (1.13) | 4.36 (0.89) | 9.66 (1.72) | 7.18 (1.24) | 0.96 (0.28) | 2.42 (0.65) | 2.20 (0.45) | 5.24 (1.03) | 3.24 (0.58) | 7.23 (1.26) |
| pool-MCP | 0.80 (0.28) | 0.83 (0.28) | 4.27 (0.83) | 3.68 (0.70) | 8.69 (1.67) | 6.70 (1.28) | 0.52 (0.18) | 1.61 (0.54) | 1.72 (0.27) | 4.41 (0.72) | 3.12 (0.56) | 7.17 (1.28) |
|
| ||||||||||||
| nonzero coef = 1 | ||||||||||||
| New | 1.35 (0.39) | 1.07 (0.29) | 1.68 (0.52) | 1.37 (0.42) | 1.57 (0.40) | 1.24 (0.31) | 2.79 (1.12) | 6.49 (2.51) | 3.34 (1.31) | 8.01 (3.01) | 2.97 (1.02) | 6.92 (2.28) |
| New+ | 1.54 (0.50) | 1.22 (0.37) | 1.81 (0.53) | 1.47 (0.44) | 1.55 (0.40) | 1.23 (0.31) | 2.78 (0.99) | 6.44 (2.2) | 3.28 (1.27) | 7.87 (2.91) | 2.97 (1.02) | 6.92 (2.28) |
| alg1-SB | 1.49 (0.39) | 1.19 (0.29) | 1.69 (0.50) | 1.39 (0.41) | 1.58 (0.48) | 1.25 (0.37) | 3.02 (1.14) | 7.00 (2.54) | 3.58 (1.28) | 8.57 (2.93) | 3.55 (1.31) | 8.15 (2.82) |
| sgroup-MCP | 0.23 (0.09) | 0.22 (0.09) | 0.26 (0.11) | 0.25 (0.11) | 0.29 (0.16) | 0.27 (0.14) | 0.40 (0.31) | 1.11 (0.81) | 0.77 (0.47) | 2.13 (1.29) | 1.17 (0.72) | 3.20 (2.00) |
| group-MCPT | 0.22 (0.09) | 0.23 (0.10) | 0.30 (0.25) | 0.32 (0.24) | 0.26 (0.10) | 0.28 (0.11) | 1.40 (1.03) | 1.46 (1.05) | 3.61 (1.83) | 3.72 (1.82) | 6.81 (2.61) | 6.39 (2.35) |
| indiv-SB | 1.02 (0.34) | 0.87 (0.29) | 1.06 (0.32) | 0.95 (0.28) | 0.96 (0.24) | 0.83 (0.21) | 3.18 (0.90) | 7.42 (1.97) | 3.80 (0.84) | 9.19 (1.90) | 2.96 (0.80) | 7.02 (1.83) |
| indiv-MCP | 0.40 (0.28) | 0.42 (0.28) | 0.37 (0.22) | 0.39 (0.24) | 0.36 (0.24) | 0.38 (0.24) | 1.32 (0.51) | 4.17 (1.61) | 1.29 (0.52) | 4.11 (1.68) | 1.20 (0.42) | 3.80 (1.35) |
| pool-SB | 0.24 (0.10) | 0.21 (0.09) | 11.78 (0.75) | 9.24 (0.49) | 24.79 (0.22) | 17.80 (0.12) | 0.38 (0.14) | 0.96 (0.36) | 4.41 (0.41) | 10.24 (0.87) | 8.30 (0.05) | 17.86 (0.08) |
| pool-MCP | 0.08 (0.04) | 0.08 (0.04) | 9.35 (0.54) | 7.85 (0.44) | 21.93 (1.03) | 16.56 (0.61) | 0.11 (0.06) | 0.34 (0.19) | 3.44 (0.22) | 8.63 (0.41) | 7.64 (0.37) | 17.08 (0.66) |
Table 12.
Simulation: summary statistics on estimation and prediction. In each cell, mean (sd). d = 1,000 and ρ = 0.5.
| σ2 = 1 | σ2 = 3 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Complete | Half | None | Complete | Half | None | |||||||
|
| ||||||||||||
| PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | |
| nonzero coef ~ unif(0.2,1) | ||||||||||||
| New | 0.85 (0.26) | 0.78 (0.31) | 1.29 (0.42) | 0.98 (0.31) | 1.09 (0.26) | 0.79 (0.17) | 1.13 (0.34) | 2.25 (0.62) | 1.46 (0.47) | 2.96 (0.82) | 1.12 (0.32) | 2.20 (0.54) |
| New+ | 0.83 (0.26) | 0.77 (0.31) | 1.36 (0.45) | 1.01 (0.34) | 1.09 (0.26) | 0.79 (0.17) | 1.13 (0.36) | 2.24 (0.62) | 1.46 (0.52) | 2.97 (0.87) | 1.12 (0.32) | 2.20 (0.54) |
| alg1-SB | 1.11 (0.24) | 0.85 (0.19) | 1.35 (0.36) | 0.98 (0.24) | 1.09 (0.21) | 0.83 (0.19) | 1.15 (0.27) | 2.25 (0.49) | 1.38 (0.31) | 2.79 (0.63) | 1.25 (0.36) | 2.45 (0.69) |
| sgroup-MCP | 1.20 (0.53) | 0.66 (0.27) | 1.39 (0.53) | 0.82 (0.29) | 1.94 (0.84) | 1.09 (0.46) | 1.51 (0.45) | 2.69 (0.87) | 1.59 (0.52) | 3.05 (0.91) | 1.83 (0.54) | 3.50 (1.03) |
| group-MCPT | 0.43 (0.55) | 0.59 (0.59) | 1.10 (0.67) | 1.52 (0.68) | 2.03 (1.15) | 2.68 (1.12) | 1.79 (1.09) | 2.57 (1.36) | 3.47 (1.73) | 4.67 (1.86) | 5.45 (2.24) | 7.11 (2.00) |
| indiv-SB | 1.02 (0.36) | 0.82 (0.25) | 1.12 (0.43) | 0.96 (0.29) | 0.98 (0.29) | 0.82 (0.26) | 1.28 (0.49) | 2.44 (0.76) | 1.52 (0.62) | 3.03 (1.06) | 1.26 (0.40) | 2.47 (0.76) |
| indiv-MCP | 1.18 (0.36) | 1.97 (0.76) | 1.18 (0.40) | 1.85 (0.67) | 1.23 (0.41) | 2.07 (0.76) | 1.22 (0.31) | 4.78 (1.32) | 1.17 (0.33) | 4.35 (1.26) | 1.15 (0.28) | 4.74 (1.39) |
| pool-SB | 0.89 (0.32) | 0.90 (0.27) | 5.40 (1.12) | 3.79 (0.78) | 14.03 (2.86) | 6.80 (1.25) | 0.56 (0.21) | 1.47 (0.46) | 2.24 (0.43) | 4.33 (0.81) | 4.95 (1.00) | 7.02 (1.28) |
| pool-MCP | 0.84 (0.29) | 1.02 (0.36) | 4.98 (1.04) | 4.44 (1.10) | 12.36 (2.35) | 7.37 (1.39) | 0.59 (0.17) | 2.68 (0.80) | 1.95 (0.35) | 5.26 (1.09) | 4.35 (0.84) | 7.42 (1.40) |
|
| ||||||||||||
| nonzero coef = 1 | ||||||||||||
| New | 0.64 (0.20) | 0.76 (0.39) | 1.07 (0.25) | 0.85 (0.18) | 1.02 (0.23) | 0.81 (0.20) | 0.99 (0.34) | 2.38 (0.92) | 1.40 (0.45) | 3.23 (0.96) | 1.01 (0.21) | 2.36 (0.61) |
| New+ | 0.64 (0.20) | 0.76 (0.39) | 1.18 (0.31) | 0.92 (0.20) | 1.02 (0.23) | 0.81 (0.20) | 1.10 (0.40) | 2.57 (1.06) | 1.41 (0.45) | 3.25 (0.97) | 1.01 (0.21) | 2.36 (0.61) |
| alg1-SB | 0.89 (0.22) | 0.71 (0.20) | 1.04 (0.31) | 0.87 (0.27) | 0.95 (0.23) | 0.72 (0.19) | 1.11 (0.26) | 2.62 (0.63) | 1.33 (0.28) | 3.20 (0.75) | 1.18 (0.30) | 2.67 (0.81) |
| sgroup-MCP | 0.38 (0.22) | 0.24 (0.13) | 0.42 (0.25) | 0.28 (0.17) | 0.53 (0.45) | 0.35 (0.27) | 1.21 (0.84) | 2.01 (1.19) | 1.87 (0.87) | 3.20 (1.32) | 2.63 (1.18) | 4.38 (1.66) |
| group-MCPT | 0.25 (0.13) | 0.39 (0.20) | 0.40 (0.28) | 0.50 (0.29) | 0.51 (0.45) | 0.63 (0.46) | 0.94 (1.00) | 1.31 (1.11) | 4.28 (2.64) | 5.86 (2.56) | 7.49 (3.10) | 9.39 (2.78) |
| indiv-SB | 0.65 (0.23) | 0.59 (0.16) | 0.69 (0.28) | 0.71 (0.26) | 0.63 (0.19) | 0.64 (0.22) | 1.17 (0.45) | 2.91 (0.97) | 1.33 (0.53) | 3.55 (1.08) | 1.16 (0.42) | 3.02 (1.04) |
| indiv-MCP | 0.32 (0.14) | 0.44 (0.20) | 0.36 (0.19) | 0.50 (0.26) | 0.35 (0.14) | 0.49 (0.21) | 1.78 (0.39) | 9.95 (2.28) | 1.79 (0.46) | 9.14 (2.24) | 1.73 (0.33) | 9.63 (1.89) |
| pool-SB | 0.18 (0.09) | 0.17 (0.08) | 11.85 (0.72) | 8.17 (0.58) | 36.72 (1.65) | 16.81 (0.41) | 0.24 (0.13) | 0.62 (0.34) | 4.46 (0.35) | 8.85 (0.59) | 12.69 (0.60) | 17.10 (0.40) |
| pool-MCP | 0.08 (0.07) | 0.12 (0.09) | 11.12 (0.62) | 9.43 (1.07) | 32.46 (1.32) | 18.64 (2.10) | 0.12 (0.09) | 0.50 (0.39) | 4.11 (0.37) | 11.08 (1.94) | 11.06 (0.56) | 18.73 (2.35) |
Table 13.
Simulation: summary statistics on estimation and prediction. In each cell, mean (sd). d = 1,000 and ρ = 0.8.
| σ2 = 1 | σ2 = 3 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Complete | Half | None | Complete | Half | None | |||||||
|
| ||||||||||||
| PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | PMSE | EMSE | |
| nonzero coef ~ unif(0.2,1) | ||||||||||||
| New | 0.50 (0.15) | 1.01 (0.37) | 0.72 (0.14) | 1.08 (0.31) | 0.58 (0.15) | 1.15 (0.40) | 0.50 (0.17) | 2.47 (1.12) | 0.69 (0.21) | 2.57 (0.74) | 0.51 (0.11) | 2.18 (0.79) |
| New+ | 0.57 (0.19) | 1.03 (0.38) | 0.75 (0.17) | 1.09 (0.31) | 0.58 (0.15) | 1.15 (0.40) | 0.53 (0.18) | 2.47 (1.12) | 0.73 (0.20) | 2.60 (0.74) | 0.51 (0.11) | 2.18 (0.79) |
| alg1-SB | 0.57 (0.14) | 1.11 (0.32) | 0.72 (0.17) | 1.14 (0.34) | 0.63 (0.12) | 1.13 (0.33) | 0.65 (0.17) | 2.82 (0.91) | 0.70 (0.23) | 2.65 (0.83) | 0.57 (0.14) | 2.57 (0.87) |
| sgroup-MCP | 3.86 (1.84) | 0.81 (0.35) | 4.65 (1.65) | 1.03 (0.38) | 5.82 (1.96) | 1.19 (0.39) | 3.04 (0.83) | 2.13 (0.65) | 3.13 (0.75) | 2.73 (0.81) | 3.69 (0.97) | 2.85 (0.85) |
| group-MCPT | 0.44 (0.50) | 1.11 (0.73) | 0.85 (0.51) | 2.59 (0.91) | 1.31 (0.58) | 3.98 (1.27) | 1.31 (0.89) | 4.31 (1.89) | 2.27 (1.08) | 6.86 (1.95) | 3.24 (1.21) | 10.07 (2.88) |
| indiv-SB | 0.48 (0.15) | 0.95 (0.41) | 0.53 (0.15) | 1.11 (0.47) | 0.48 (0.16) | 1.07 (0.43) | 0.52 (0.17) | 2.23 (0.89) | 0.62 (0.19) | 2.43 (0.96) | 0.51 (0.16) | 2.39 (0.88) |
| indiv-MCP | 1.41 (0.40) | 6.96 (1.75) | 1.41 (0.38) | 6.76 (2.34) | 1.47 (0.43) | 7.33 (2.42) | 1.03 (0.20) | 11.58 (2.88) | 0.97 (0.19) | 9.98 (2.60) | 1.07 (0.23) | 12.06 (2.97) |
| pool-SB | 0.71 (0.44) | 0.99 (0.39) | 3.66 (0.78) | 3.89 (0.95) | 14.11 (2.67) | 6.79 (1.36) | 0.36 (0.19) | 1.63 (0.73) | 1.40 (0.28) | 4.43 (0.99) | 4.95 (0.89) | 6.98 (1.38) |
| pool-MCP | 1.13 (0.49) | 4.36 (1.48) | 4.27 (0.99) | 7.97 (2.47) | 14.62 (2.78) | 11.91 (2.64) | 0.62 (0.19) | 7.32 (2.25) | 1.65 (0.31) | 9.74 (1.91) | 5.03 (0.95) | 12.10 (2.95) |
|
| ||||||||||||
| nonzero coef = 1 | ||||||||||||
| New | 0.52 (0.20) | 1.63 (0.95) | 0.76 (0.26) | 1.64 (0.91) | 0.64 (0.27) | 1.58 (0.89) | 0.50 (0.14) | 3.15 (0.89) | 0.72 (0.22) | 3.19 (1.01) | 0.58 (0.14) | 3.53 (1.38) |
| New+ | 0.56 (0.28) | 1.66 (0.98) | 0.77 (0.26) | 1.65 (0.91) | 0.64 (0.27) | 1.58 (0.89) | 0.53 (0.17) | 3.19 (0.90) | 0.75 (0.24) | 3.22 (1.03) | 0.58 (0.14) | 3.53 (1.38) |
| alg1-SB | 0.64 (0.20) | 1.35 (0.72) | 0.71 (0.19) | 1.45 (0.60) | 0.63 (0.17) | 1.35 (0.60) | 0.61 (0.22) | 3.72 (1.6) | 0.79 (0.24) | 3.93 (1.66) | 0.64 (0.17) | 3.53 (1.36) |
| sgroup-MCP | 3.05 (2.70) | 0.71 (0.56) | 5.37 (2.97) | 1.15 (0.61) | 8.50 (4.53) | 1.61 (0.79) | 4.74 (1.40) | 2.79 (0.71) | 5.30 (1.41) | 3.42 (1.04) | 6.34 (1.50) | 3.78 (1.10) |
| group-MCPT | 0.32 (0.31) | 0.96 (0.54) | 0.97 (0.76) | 2.75 (1.40) | 1.59 (0.70) | 4.00 (1.48) | 1.39 (1.35) | 3.46 (2.43) | 2.74 (1.49) | 8.46 (3.02) | 4.28 (1.98) | 13.53 (4.13) |
| indiv-SB | 0.44 (0.14) | 1.21 (0.59) | 0.49 (0.17) | 1.40 (0.71) | 0.46 (0.17) | 1.41 (0.67) | 0.50 (0.17) | 3.21 (1.48) | 0.57 (0.20) | 3.86 (1.75) | 0.51 (0.17) | 3.67 (1.45) |
| indiv-MCP | 2.22 (0.55) | 13.55 (3.03) | 2.60 (0.85) | 14.54 (4.41) | 2.28 (0.48) | 14.04 (2.54) | 1.43 (0.27) | 21.05 (2.44) | 1.54 (0.30) | 22.27 (3.79) | 1.45 (0.34) | 21.57 (3.36) |
| pool-SB | 0.12 (0.07) | 0.28 (0.18) | 7.93 (0.30) | 8.12 (1.14) | 37.30 (1.30) | 17.08 (1.10) | 0.14 (0.08) | 0.90(0.62) | 2.82 (0.16) | 8.81 (1.39) | 12.66 (0.48 | 17.28 (1.31) |
| pool-MCP | 1.17 (0.37) | 7.87 (2.84) | 9.18 (0.58) | 16.71 (3.39) | 38.85 (1.38) | 30.73 (2.65) | 0.52 (0.17) | 9.49 (3.52) | 3.36 (0.21) | 20.16 (3.74) | 13.10 (0.45) | 31.13 (2.64) |
Table 14.
Simulation: summary statistics on identification. In each cell, mean (sd). d = 100 and ρ = 0.2.
| σ2 = 1 | σ2 = 3 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Complete | Half | None | Complete | Half | None | |||||||
|
| ||||||||||||
| TP | FP | TP | FP | TP | FP | TP | FP | TP | FP | TP | FP | |
| nonzero coef ~ unif(0.2,1) | ||||||||||||
| New | 16.8 (3.1) | 0.03 (0.2) | 16.6 (1.0) | 1.7 (1.1) | 16.6 (1.2) | 1.6 (1.3) | 11.8 (2.5) | 0.4 (0.6) | 11.5 (2.3) | 0.7 (1.1) | 11.7 (2.7) | 0.9 (0.9) |
| New+ | 17.0 (3.2) | 0.0 (0) | 16.7 (1.0) | 1.6 (1.1) | 16.6 (1.2) | 1.6 (1.3) | 12.9 (2.9) | 0.2 (0.5) | 11.2 (3.1) | 0.5 (0.7) | 11.8 (2.7) | 0.9 (0.9) |
| alg1-SB | 16.4 (1.4) | 2.1 (1.2) | 16.0 (1.3) | 2.1 (1.3) | 16.4 (1.2) | 2.0 (1.4) | 11.3 (2.0) | 1.0 (1.3) | 11.0 (2.1) | 1.0 (1.0) | 11.7 (2.2) | 1.0 (1.3) |
| sgroup-MCP | 16.7 (1.2) | 1.5 (2.0) | 15.6 (1.3) | 2.5 (2.9) | 14.8 (1.6) | 3.7 (3.4) | 13.2 (2.3) | 2.3 (2.4) | 11.8 (2.5) | 3.0 (2.8) | 10.6 (2.5) | 3.9 (3.2) |
| group-MCPT | 16.7 (1.1) | 0.2 (1.0) | 16.1 (1.4) | 15.9 (5.7) | 14.6 (1.9) | 4.6 (2.8) | 11.8 (2.1) | 0.5 (1.0) | 11.2 (2.6) | 1.9 (1.8) | 9.6 (2.7) | 21.1 (7.9) |
| indiv-SB | 14.7 (1.5) | 0.2 (0.4) | 14.2 (1.7) | 0.3 (0.6) | 14.4 (1.5) | 0.3 (0.6) | 10.5 (1.8) | 0.4 (0.6) | 9.4 (2.4) | 0.6 (0.6) | 10.6 (2.2) | 0.7 (0.8) |
| indiv-MCP | 16.3 (1.2) | 10.3 (4.7) | 16.4 (1.3) | 10.9 (4.6) | 16.5 (1.2) | 11.3 (4.8) | 13.3 (2.0) | 11.6 (5.8) | 13.0 (2.0) | 12.3 (5.9) | 12.9 (2.2) | 12.8 (5.9) |
| pool-SB | 17.6 (1.3) | 0.0 (0) | 10.7 (1.8) | 4.5 (2.0) | 3.0 (1.5) | 6.1 (3.0) | 15.8 (2.5) | 0.0 (0) | 9.0 (2.3) | 2.6 (1.6) | 1.6 (1.1) | 3.6 (2.3) |
| pool-MCP | 18.0 (0) | 10.1 (11.2) | 14.6 (1.4) | 27.1 (14.4) | 10.0 (1.8) | 37.0 (12.4) | 17.5 (1.6) | 16.0 (14.1) | 12.8 (2.2) | 25.9 (14.0) | 7.8 (2.5) | 28.8 (14.2) |
|
| ||||||||||||
| nonzero coef = 1 | ||||||||||||
| New | 18.0 (0) | 0.2 (0.5) | 18.0 (0) | 2.5 (1.7) | 18.0 (0) | 3.5 (1.5) | 18.0 (0) | 0.8 (1.1) | 17.9 (0.3) | 1.5 (1.4) | 17.9 (0.4) | 1.4 (1.0) |
| New+ | 18.0 (0) | 0.2 (0.8) | 18.0 (0) | 1.1 (1.7) | 18.0 (0) | 1.5 (1.5) | 18.0 (0) | 0.4 (0.7) | 17.9 (0.3) | 1.5 (1.4) | 17.9 (0.4) | 1.4 (1.0) |
| alg1-SB | 18.0 (0) | 3.4 (1.7) | 18.0 (0) | 3.5 (2.3) | 18.0 (0) | 3.6 (1.5) | 17.9 (0.4) | 1.4 (1.2) | 17.8 (0.5) | 1.6 (1.3) | 17.9 (0.4) | 1.8 (1.1) |
| sgroup-MCP | 18.0 (0) | 0.4 (1.1) | 18.0 (0) | 0.6 (1.0) | 18.0 (0) | 1.0 (1.8) | 18.0 (0.2) | 1.1 (1.6) | 17.8 (0.5) | 2.7 (2.3) | 17.7 (0.6) | 5.2 (3.2) |
| group-MCPT | 18.0 (0) | 0.0 (0) | 18.0 (0.1) | 0.0 (0) | 18.0 (0) | 0.0 (0) | 18.0 (0) | 0.1 (0.6) | 17.5 (0.8) | 1.7 (2.0) | 17.3 (1.0) | 2.6 (2.3) |
| indiv-SB | 18.0 (0) | 0.1 (0.3) | 18.0 (0) | 0.0 (0) | 18.0 (0) | 0.0 (0.2) | 17.2 (0.8) | 1.2 (0.4) | 16.6 (1.4) | 1.3 (0.5) | 17.0 (1.0) | 1.3 (0.5) |
| indiv-MCP | 18.0 (0) | 3.3 (2.8) | 18.0 (0) | 4.3 (3.7) | 18.0 (0) | 4.2 (4.2) | 18.0 (0.2) | 14.8 (5.9) | 17.9 (0.4) | 14.1 (5.9) | 17.9 (0.4) | 14.5 (5.5) |
| pool-SB | 18.0 (0) | 0.0 (0) | 12.1 (1.4) | 6.3 (2.8) | 3.7 (1.7) | 7.4 (3.4) | 18.0 (0) | 0.0 (0) | 11.3 (1.2) | 4.7 (2.4) | 2.5 (1.4) | 5.0 (3.0) |
| pool-MCP | 18.0 (0) | 3.6 (8.0) | 16.4 (1.3) | 32.5 (13.5) | 12.0 (1.9) | 40.7 (14.7) | 18.0 (0) | 6.1 (8.7) | 15.4 (1.3) | 29.7 (11.3) | 10.6 (1.9) | 36.1 (16.3) |
Table 15.
Simulation: summary statistics on identification. In each cell, mean (sd). d = 100 and ρ = 0.5.
| σ2 = 1 | σ2 = 3 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Complete | Half | None | Complete | Half | None | |||||||
|
| ||||||||||||
| TP | FP | TP | FP | TP | FP | TP | FP | TP | FP | TP | FP | |
| nonzero coef ~ unif(0.2,1) | ||||||||||||
| New | 17.7 (0.8) | 0.1 (0.3) | 17.0 (1.0) | 2.2 (1.5) | 17.3 (0.8) | 1.7 (1.3) | 15.1 (1.5) | 0.5 (0.9) | 13.7 (1.5) | 1.3 (1.2) | 14.7 (1.6) | 1.3 (0.8) |
| New+ | 17.8 (0.8) | 0.0 (0) | 16.9 (1.0) | 1.9 (1.3) | 17.3 (0.8) | 1.9 (1.3) | 15.3 (1.6) | 0.3 (0.6) | 13.8 (1.5) | 1.3 (1.2) | 14.7 (1.6) | 1.3 (0.9) |
| alg1-SB | 17.3 (0.7) | 2.0 (1.3) | 17.2 (0.9) | 1.9 (1.2) | 16.8 (0.9) | 1.9 (1.5) | 14.2 (1.8) | 1.1 (1.1) | 14.0 (1.7) | 1.3 (1.1) | 14.4 (1.7) | 1.5 (0.9) |
| sgroup-MCP | 15.0 (1.8) | 1.5 (2.1) | 14.3 (1.7) | 2.5 (2.3) | 13.0 (2.0) | 3.4 (2.9) | 11.0 (2.1) | 2.3 (2.8) | 10.2 (2.0) | 3.2 (3.4) | 9.3 (1.9) | 3.5 (3.5) |
| group-MCPT | 17.7 (0.7) | 1.1 (2.9) | 16.2 (1.2) | 17.8 (7.9) | 15.1 (1.7) | 33.2 (8.0) | 15.1 (2.4) | 1.1 (2.2) | 13.0 (2.4) | 12.3 (5.9) | 10.3 (2.5) | 10.5 (5.2) |
| indiv-SB | 16.1 (1.2) | 0.3 (0.7) | 15.6 (1.2) | 0.2 (0.5) | 16.1 (1.2) | 0.2 (0.6) | 13.5 (1.6) | 1.4 (0.9) | 12.3 (1.4) | 1.5 (0.9) | 13.3 (1.4) | 1.5 (0.7) |
| indiv-MCP | 14.8 (1.5) | 10.7 (5.5) | 14.9 (1.6) | 9.8 (4.8) | 14.6 (1.7) | 11.4 (5.4) | 11.0 (1.5) | 9.1 (5.2) | 11.1 (1.6) | 8.7 (4.8) | 11.0 (1.6) | 9.8 (5.1) |
| pool-SB | 18.0 (0) | 0.0 (0) | 12.7 (1.6) | 8.4 (2.3) | 6.2 (1.5) | 12.5 (3.0) | 17.2 (1.6) | 0.0 (0) | 11.1 (1.5) | 6.1 (2.5) | 4.2 (1.4) | 8.5 (2.8) |
| pool-MCP | 18.0 (0) | 8.2 (7.7) | 13.2 (1.4) | 22.3 (11.2) | 8.2 (1.1) | 29.6 (13.6) | 16.4 (2.3) | 15.0 (9.6) | 11.5 (2.3) | 20.0 (13.0) | 6.9 (1.4) | 23.5 (10.8) |
|
| ||||||||||||
| nonzero coef = 1 | ||||||||||||
| New | 18.0 (0) | 0.0 (0) | 18.0 (0) | 2.4 (1.7) | 18.0 (0) | 2.3 (1.3) | 18.0 (0) | 0.6 (0.9) | 18.0 (0.2) | 1.5 (1.0) | 18.0 (0) | 1.6 (1.3) |
| New+ | 18.0 (0) | 0.0 (0) | 18.0 (0) | 1.1 (1.7) | 18.0 (0) | 1.4 (1.3) | 18.0 (0) | 0.2 (0.5) | 18.0 (0.2) | 1.4 (1.0) | 18.0 (0) | 1.6 (1.3) |
| alg1-SB | 18.0 (0) | 2.3 (1.6) | 18.0 (0) | 2.6 (1.7) | 18.0 (0) | 2.5 (1.3) | 18.0 (0) | 1.6 (1.3) | 17.9 (0.3) | 1.8 (1.1) | 18.0 (0) | 1.6 (1.1) |
| sgroup-MCP | 18.0 (0) | 0.3 (0.9) | 18.0 (0.1) | 0.6 (1.1) | 18.0 (0.2) | 1.4 (1.7) | 17.5 (0.9) | 1.9 (2.4) | 16.6 (1.6) | 3.9 (3.5) | 15.6 (2.0) | 5.4 (3.5) |
| group-MCPT | 18.0 (0) | 0.0 (0) | 18.0 (0) | 0.0 (0.1) | 18.0 (0.1) | 0.6 (0.7) | 18.0 (0) | 1.0 (3.2) | 17.4 (0.9) | 6.9 (4.4) | 16.9 (1.7) | 14.4 (5.4) |
| indiv-SB | 18.0 (0) | 0.0 (0.2) | 18.0 (0) | 0.0 (0) | 18.0 (0) | 0.0 (0.2) | 17.7 (0.5) | 1.2 (0.5) | 17.5 (0.6) | 1.1 (0.3) | 17.6 (0.5) | 1.1 (0.3) |
| indiv-MCP | 18.0 (0) | 4.9 (3.4) | 18.0 (0) | 4.0 (2.9) | 18.0 (0) | 4.9 (3.5) | 16.9 (1.1) | 14.5 (5.2) | 16.8 (1.2) | 14.1 (4.2) | 16.8 (1.1) | 15.5 (4.5) |
| pool-SB | 18.0 (0) | 0.0 (0) | 14.5 (1.1) | 11.1 (2.2) | 7.1 (1.5) | 14.2 (3.1) | 18.0 (0) | 0.0 (0) | 13.2 (1.2) | 8.4 (2.3) | 6.3 (1.5) | 12.5 (2.9) |
| pool-MCP | 18.0 (0) | 1.8 (5.7) | 14.7 (0.8) | 28.9 (14.5) | 8.8 (1.1) | 32.0 (14.0) | 18.0 (0) | 2.8 (6.1) | 13.9 (0.8) | 29.1 (13.6) | 8.1 (1.1) | 29.8 (13.1) |
Table 16.
Simulation: summary statistics on identification. In each cell, mean (sd). d = 100 and ρ = 0.8.
| σ2 = 1 | σ2 = 3 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Complete | Half | None | Complete | Half | None | |||||||
|
| ||||||||||||
| TP | FP | TP | FP | TP | FP | TP | FP | TP | FP | TP | FP | |
| nonzero coef ~ unif(0.2,1) | ||||||||||||
| New | 17.7 (0.7) | 0.7 (0.8) | 16.8 (1.3) | 3.5 (1.5) | 16.8 (0.9) | 2.2 (1.5) | 15.8 (1.9) | 0.5 (0.7) | 14.3 (1.2) | 2.7 (1.2) | 14.6 (1.5) | 1.6 (1.4) |
| New+ | 17.7 (0.6) | 0.5 (0.9) | 16.8 (1.3) | 3.2 (1.5) | 16.8 (0.9) | 2.3 (1.4) | 16.1 (1.8) | 0.3 (0.6) | 14.3 (1.2) | 2.5 (1.2) | 14.6 (1.5) | 1.7 (1.3) |
| alg1-SB | 17.0 (1.0) | 2.2 (1.2) | 16.7 (1.0) | 3.7 (1.7) | 16.9 (0.8) | 2.4 (1.4) | 14.6 (1.8) | 2.1 (1.5) | 14.2 (1.7) | 2.6 (1.3) | 15.0 (1.8) | 1.4 (1.1) |
| sgroup-MCP | 12.7 (2.4) | 1.9 (2.7) | 11.8 (2.0) | 2.5 (2.5) | 10.6 (1.9) | 2.9 (2.8) | 8.5 (1.8) | 1.6 (2.3) | 7.7 (1.6) | 2.1 (1.9) | 7.0 (1.4) | 2.8 (2.9) |
| group-MCPT | 17.7 (0.8) | 2.3 (4.0) | 16.2 (1.4) | 17.5 (6.1) | 15.4 (1.8) | 34.2 (8.6) | 15.1 (2.9) | 2.4 (4.8) | 12.6 (2.6) | 11.6 (7.1) | 10.6 (2.6) | 23.3 (8.0) |
| indiv-SB | 16.6 (1.1) | 1.5 (0.6) | 16.2 (1.4) | 2.2 (1.1) | 16.4 (1.3) | 1.7 (0.9) | 14.5 (1.6) | 1.7 (0.8) | 13.3 (2.0) | 1.5 (1.3) | 13.7 (1.8) | 1.9 (1.0) |
| indiv-MCP | 10.1 (1.0) | 7.3 (3.3) | 10.6 (1.3) | 8.0 (3.9) | 9.9 (1.0) | 7.0 (3.6) | 7.8 (1.1) | 7.6 (3.6) | 7.5 (1.2) | 7.3 (4.1) | 7.4 (1.1) | 8.5 (4.1) |
| pool-SB | 18.0 (0) | 0.6 (1.5) | 14.7 (1.3) | 12.2 (2.1) | 8.8 (1.4) | 17.7 (2.8) | 17.0 (1.4) | 0.7 (1.5) | 13.2 (2.1) | 10.5 (2.4) | 7.9 (1.5) | 15.9 (3.0) |
| pool-MCP | 14.9 (3.0) | 11.9 (10.6) | 9.6 (2.1) | 15.7 (7.4) | 5.0 (1.0) | 22.7 (8.9) | 10.9 (2.2) | 9.0 (7.6) | 8.4 (1.5) | 16.4 (8.6) | 4.5 (1.1) | 19.0 (8.7) |
|
| ||||||||||||
| nonzero coef = 1 | ||||||||||||
| New | 18.0 (0) | 0.6 (1.0) | 18.0 (0) | 3.8 (1.9) | 18.0 (0) | 2.3 (1.5) | 18.0 (0) | 0.9 (1.1) | 17.7 (0.5) | 3.6 (1.7) | 17.8 (0.4) | 1.9 (1.5) |
| New+ | 18.0 (0) | 0.4 (0.9) | 18.0 (0) | 2.8 (2.0) | 18.0 (0) | 1.2 (1.4) | 17.9 (0.6) | 0.3 (0.6) | 17.7 (0.5) | 2.4 (1.6) | 17.8 (0.4) | 1.0 (1.6) |
| alg1-SB | 18.0 (0) | 1.8 (1.2) | 18.0 (0) | 3.5 (1.2) | 18.0 (0) | 2.4 (1.5) | 17.7 (0.5) | 1.8 (1.2) | 17.8 (0.6) | 3.4 (1.9) | 17.8 (0.4) | 2.3 (1.3) |
| sgroup-MCP | 17.5 (0.9) | 1.1 (1.7) | 16.8 (1.2) | 2.1 (2.1) | 15.9 (1.8) | 3.0 (2.4) | 13.3 (2.7) | 2.1 (2.4) | 11.9 (2.0) | 2.4 (2.6) | 11.0 (1.8) | 3.0 (2.9) |
| group-MCPT | 18.0 (0) | 0.2 (0.7) | 18.0 (0.2) | 12.7 (3.2) | 18.0 (0.2) | 19.7 (4.8) | 17.9 (0.7) | 2.5 (4.1) | 16.9 (1.2) | 17.9 (5.8) | 16.3 (1.7) | 37.2 (8.0) |
| indiv-SB | 18.0 (0) | 1.4 (0.6) | 18.0 (0) | 2.0 (1.0) | 18.0 (0) | 1.5 (0.8) | 17.6 (0.6) | 0.5 (0.7) | 17.5 (0.7) | 1.2 (1.0) | 17.5 (0.6) | 0.6 (0.9) |
| indiv-MCP | 12.7 (1.2) | 4.0 (2.4) | 14.3 (1.3) | 5.7 (2.8) | 12.5 (1.4) | 4.3 (2.5) | 10.3 (1.0) | 7.9 (3.5) | 10.5 (1.2) | 8.2 (4.0) | 10.0 (1.3) | 8.1 (4.0) |
| pool-SB | 18.0 (0) | 0.3 (1.2) | 16.2 (1.1) | 14.6 (2.3) | 9.4 (1.4) | 18.8 (2.9) | 18.0 (0) | 0.5 (1.4) | 15.5 (1.0) | 13.3 (2.1) | 9.4 (1.8) | 18.7 (3.6) |
| pool-MCP | 18.0 (0) | 2.3 (5.3) | 11.6 (1.8) | 18.3 (10.3) | 5.4 (1.1) | 22.1 (9.9) | 16.9 (1.7) | 13.0 (9.2) | 10.8 (1.8) | 20.8 (10.9) | 5.1 (1.1) | 22.4 (9.2) |
Table 17.
Simulation: summary statistics on identification. In each cell, mean (sd). d = 500 and ρ = 0.2.
| σ2 = 1 | σ2 = 3 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Complete | Half | None | Complete | Half | None | |||||||
|
| ||||||||||||
| TP | FP | TP | FP | TP | FP | TP | FP | TP | FP | TP | FP | |
| nonzero coef ~ unif(0.2,1) | ||||||||||||
| New | 16.4 (3.2) | 0.0 (0.2) | 15.1 (1.7) | 2.5 (1.5) | 15.6 (1.8) | 1.8 (1.8) | 9.0 (2.4) | 0.5 (0.9) | 8.6 (2.2) | 0.7 (0.7) | 9.3 (2.1) | 0.7 (0.9) |
| New+ | 16.7 (3.2) | 0.0 (0) | 15.0 (1.7) | 1.3 (1.4) | 15.6 (1.8) | 1.9 (1.8) | 10.0 (2.6) | 0.5 (1.0) | 8.6 (2.5) | 0.7 (0.9) | 9.3 (2.1) | 0.7 (0.9) |
| alg1-SB | 14.8 (1.7) | 2.5 (1.3) | 14.9 (1.6) | 2.4 (1.5) | 15.3 (1.6) | 2.2 (1.5) | 8.6 (2.3) | 0.7 (1.2) | 8.5 (2.2) | 0.9 (1.0) | 9.0 (2.5) | 0.8 (1.0) |
| sgroup-MCP | 16.1 (1.5) | 1.2 (1.4) | 15.0 (1.8) | 2.1 (2.3) | 14.0 (2.1) | 3.1 (2.8) | 11.9 (2.5) | 2.2 (2.6) | 10.2 (2.1) | 2.9 (2.8) | 9.1 (2.1) | 3.6 (3.0) |
| group-MCPT | 16.3 (1.5) | 0.5 (2.0) | 15.0 (1.6) | 2.1 (2.5) | 13.8 (2.4) | 27.9 (35.9) | 9.0 (2.0) | 0.3 (0.7) | 8.6 (2.7) | 1.1 (1.9) | 9.3 (4.0) | 6.8 (5.5) |
| indiv-SB | 14.4 (1.8) | 0.6 (0.7) | 13.7 (1.7) | 0.3 (0.6) | 14.0 (1.5) | 0.4 (0.6) | 8.1 (2.4) | 0.4 (0.6) | 7.7 (2.6) | 0.3 (0.5) | 8.4 (2.1) | 0.3 (0.6) |
| indiv-MCP | 15.5 (1.5) | 17.4 (7.0) | 15.3 (1.6) | 16.8 (6.9) | 15.4 (1.2) | 15.3 (7.1) | 11.8 (1.9) | 15.0 (7.3) | 11.3 (1.8) | 17.2 (6.1) | 12.0 (2.0) | 15.9 (6.3) |
| pool-SB | 17.8 (0.8) | 0.0 (0) | 10.4 (1.6) | 3.4 (2.1) | 1.5 (0.6) | 2.9 (1.3) | 16.5 (1.9) | 0.0 (0) | 8.1 (2.0) | 1.0 (1.0) | 1.0 (0.3) | 2.2 (0.5) |
| pool-MCP | 17.9 (0.6) | 14.2 (20.6) | 13.6 (1.5) | 30.9 (15.4) | 8.2 (2.1) | 41.8 (18.8) | 17.5 (1.1) | 24.2 (23.4) | 11.4 (2.1) | 28.2 (20.3) | 5.2 (3.0) | 32.1 (27.1) |
|
| ||||||||||||
| nonzero coef = 1 | ||||||||||||
| New | 18.0 (0) | 1.8 (1.7) | 18.0 (0) | 1.6 (1.8) | 18.0 (0) | 1.9 (1.6) | 17.6 (0.7) | 0.3 (0.6) | 16.8 (1.2) | 1.1 (1.0) | 17.4 (0.9) | 1.1 (1.3) |
| New+ | 18.0 (0) | 0.4 (0.8) | 18.0 (0) | 1.6 (1.8) | 18.0 (0) | 2.1 (1.5) | 17.8 (0.4) | 0.1 (0.3) | 16.9 (1.0) | 0.9 (1.0) | 17.4 (0.9) | 1.1 (1.3) |
| alg1-SB | 18.0 (0) | 4.1 (2.0) | 18.0 (0) | 4.1 (2.0) | 18.0 (0) | 4.6 (1.6) | 16.9 (1.5) | 0.9 (1.2) | 16.5 (1.6) | 1.0 (0.9) | 17.3 (1.0) | 1.4 (1.0) |
| sgroup-MCP | 18.0 (0) | 0.5 (1.9) | 18.0 (0) | 0.5 (1.0) | 18.0 (0) | 0.8 (1.2) | 17.9 (0.3) | 1.3 (1.7) | 17.4 (0.9) | 2.9 (2.7) | 17.2 (1.1) | 5.0 (3.8) |
| group-MCPT | 18.0 (0) | 0.0 (0) | 18.0 (0.1) | 0.0 (0.1) | 18.0 (0.1) | 0.0 (0) | 17.6 (0.7) | 0.0 (0.2) | 16.8 (1.2) | 1.0 (1.4) | 17.0 (1.8) | 110.8 (123.8) |
| indiv-SB | 17.9 (0.4) | 1.1 (0.4) | 17.9 (0.4) | 1.1 (0.3) | 17.9 (0.4) | 1.1 (0.3) | 16.3 (1.5) | 0.4 (0.6) | 15.9 (1.3) | 0.3 (0.5) | 16.4 (1.5) | 0.5 (0.6) |
| indiv-MCP | 18.0 (0) | 8.1 (4.6) | 18.0 (0) | 9.4 (5.0) | 18.0 (0) | 9.2 (6.2) | 17.7 (0.6) | 21.2 (6.3) | 17.8 (0.6) | 21.5 (6.6) | 17.7 (0.7) | 20.6 (6.5) |
| pool-SB | 18.0 (0) | 0.0 (0) | 11.6 (1.1) | 5.3 (2.3) | 1.7 (1.0) | 3.4 (2.0) | 18.0 (0) | 0.0 (0) | 10.5 (0.8) | 2.9 (1.6) | 1.2 (0.4) | 2.5 (0.9) |
| pool-MCP | 18.0 (0) | 9.3 (18.7) | 15.6 (1.3) | 39.0 (23.1) | 10.2 (2.6) | 49.6 (22.3) | 18.0 (0) | 11.1 (18.2) | 14.0 (1.3) | 35.2 (21.2) | 8.6 (3.1) | 44.1 (21.8) |
Table 18.
Simulation: summary statistics on identification. In each cell, mean (sd). d = 500 and ρ = 0.5.
| σ2 = 1 | σ2 = 3 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Complete | Half | None | Complete | Half | None | |||||||
|
| ||||||||||||
| TP | FP | TP | FP | TP | FP | TP | FP | TP | FP | TP | FP | |
| nonzero coef ~ unif(0.2,1) | ||||||||||||
| New | 17.1 (1.3) | 0.0 (0.2) | 16.6 (1.1) | 2.0 (1.4) | 17.0 (1.0) | 2.3 (2.0) | 13.9 (2.0) | 0.9 (1.3) | 12.5 (2.4) | 1.3 (1.1) | 13.8 (1.8) | 1.3 (1.1) |
| New+ | 17.2 (1.3) | 0.0 (0) | 16.2 (2.7) | 1.7 (1.6) | 17.0 (1.0) | 2.3 (1.9) | 13.7 (2.4) | 0.3 (0.6) | 12.7 (2.2) | 1.2 (1.0) | 13.9 (1.8) | 2.2 (2.0) |
| alg1-SB | 17.0 (0.8) | 2.6 (1.7) | 16.1 (1.4) | 2.4 (1.4) | 16.7 (1.2) | 3.0 (1.4) | 13.4 (2.2) | 1.8 (1.1) | 12.3 (2.4) | 1.4 (1.4) | 13.0 (1.5) | 1.7 (1.6) |
| sgroup-MCP | 14.4 (1.7) | 1.3 (1.6) | 13.5 (1.8) | 2.1 (2.0) | 12.2 (2.0) | 2.7 (2.4) | 9.8 (1.9) | 1.5 (2.0) | 8.9 (1.8) | 2.3 (2.3) | 8.2 (1.5) | 2.8 (2.4) |
| group-MCPT | 17.1 (1.1) | 0.9 (2.4) | 15.6 (1.8) | 17.5 (12.6) | 14.3 (2.3) | 20.4 (13.3) | 13.7 (2.7) | 1.4 (3.8) | 11.4 (2.6) | 11.5 (14.3) | 8.6 (3.0) | 30.4 (63.1) |
| indiv-SB | 16.1 (1.2) | 1.4 (0.5) | 16.0 (0.9) | 1.3 (0.6) | 16.0 (1.2) | 1.4 (0.6) | 13.0 (2.0) | 1.5 (0.6) | 11.6 (2.3) | 1.6 (0.9) | 13.0 (1.8) | 1.5 (0.6) |
| indiv-MCP | 13.2 (1.2) | 15.3 (5.8) | 13.7 (1.5) | 16.1 (5.6) | 13.2 (1.5) | 16.7 (6.1) | 9.7 (1.2) | 13.9 (6.9) | 9.4 (1.3) | 14.6 (6.2) | 9.5 (1.7) | 13.2 (6.1) |
| pool-SB | 17.9 (0.6) | 0.1 (0.6) | 12.7 (1.5) | 8.5 (2.7) | 4.4 (1.7) | 9.0 (3.6) | 16.8 (2.0) | 0.1 (0.6) | 10.6 (2.2) | 6.2 (2.3) | 2.9 (1.7) | 5.9 (3.2) |
| pool-MCP | 17.7 (1.2) | 15.0 (12.4) | 12.2 (1.6) | 32.5 (19.6) | 7.2 (1.2) | 33.1 (14.6) | 15.4 (3.0) | 25.2 (17.9) | 9.9 (2.0) | 21.1 (13.8) | 6.3 (1.5) | 30.9 (16.6) |
|
| ||||||||||||
| nonzero coef = 1 | ||||||||||||
| New | 18.0 (0) | 0.2 (0.4) | 18.0 (0) | 3.1 (1.4) | 18.0 (0) | 2.8 (1.6) | 18.0 (0) | 0.1 (0.4) | 17.9 (0.3) | 1.5 (1.2) | 18.0 (0) | 1.6 (1.2) |
| New+ | 18.0 (0) | 0.1 (0.6) | 18.0 (0) | 1.9 (1.3) | 18.0 (0) | 2.9 (1.7) | 18.0 (0) | 0.0 (0) | 17.4 (2.2) | 1.2 (1.3) | 18.0 (0) | 1.6 (1.2) |
| alg1-SB | 18.0 (0) | 2.3 (1.6) | 18.0 (0) | 3.5 (1.5) | 18.0 (0) | 3.2 (1.4) | 17.9 (0.3) | 2.0 (1.3) | 17.9 (0.3) | 2.4 (1.3) | 17.9 (0.3) | 1.9 (1.4) |
| sgroup-MCP | 18.0 (0.1) | 0.3 (0.7) | 18.0 (0.2) | 0.6 (1.1) | 17.9 (0.4) | 1.5 (2.2) | 16.8 (1.3) | 2.2 (1.9) | 15.9 (1.6) | 3.5 (2.7) | 14.0 (2.5) | 5.0 (4.3) |
| group-MCPT | 18.0 (0) | 0.0 (0.2) | 18.0 (0) | 0.0 (0.2) | 18.0 (0) | 0.8 (1.1) | 18.0 (0) | 0.5 (1.5) | 16.8 (1.3) | 8.3 (7.4) | 16.2 (2.5) | 31.5 (21.0) |
| indiv-SB | 18.0 (0) | 1.1 (0.3) | 18.0 (0) | 1.1 (0.4) | 18.0 (0) | 1.1 (0.3) | 17.7 (0.5) | 1.3 (0.6) | 17.5 (0.6) | 1.4 (0.6) | 17.8 (0.5) | 1.4 (0.6) |
| indiv-MCP | 18.0 (0) | 11.2 (4.3) | 17.9 (0.6) | 10.0 (5.2) | 18.0 (0) | 11.7 (5.7) | 14.5 (1.7) | 21.4 (9.1) | 15.3 (1.3) | 21.9 (6.8) | 14.4 (1.0) | 20.2 (7.5) |
| pool-SB | 18.0 (0) | 0.0 (0) | 14.6 (1.3) | 11.3 (2.5) | 5.9 (2.0) | 11.8 (3.9) | 18.0 (0) | 0.0 (0) | 13.8 (1.3) | 9.6 (2.6) | 4.6 (2.0) | 9.4 (3.9) |
| pool-MCP | 18.0 (0) | 4.5 (8.8) | 13.8 (0.9) | 28.5 (14.4) | 7.8 (1.3) | 37.0 (17.6) | 18.0 (0) | 12.8 (16.1) | 12.9 (1.1) | 32.7 (14.5) | 7.4 (1.4) | 33.5 (19.1) |
Table 19.
Simulation: summary statistics on identification. In each cell, mean (sd). d = 500 and ρ = 0.8.
| σ2 = 1 | σ2 = 3 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Complete | Half | None | Complete | Half | None | |||||||
|
| ||||||||||||
| TP | FP | TP | FP | TP | FP | TP | FP | TP | FP | TP | FP | |
| nonzero coef ~ unif(0.2,1) | ||||||||||||
| New | 17.3 (0.8) | 1.2 (1.1) | 16.8 (1.0) | 3.6 (1.3) | 16.8 (1.0) | 2.6 (1.6) | 15.4 (1.9) | 0.7 (0.7) | 14.0 (1.8) | 2.9 (1.5) | 15.0 (1.5) | 1.9 (1.6) |
| New+ | 17.2 (0.8) | 0.5 (0.9) | 16.8 (1.0) | 2.6 (1.4) | 16.8 (1.0) | 1.6 (1.5) | 15.3 (2.4) | 0.3 (0.6) | 14.1 (1.8) | 1.7 (1.4) | 14.9 (1.5) | 0.9 (1.2) |
| alg1-SB | 16.6 (1.0) | 2.4 (1.3) | 17.0 (1.0) | 3.8 (1.9) | 17.1 (0.8) | 2.4 (1.3) | 14.4 (1.3) | 2.0 (1.4) | 13.8 (1.5) | 3.0 (1.7) | 14.3 (2.0) | 2.1 (1.2) |
| sgroup-MCP | 11.1 (1.9) | 1.4 (1.7) | 10.3 (1.7) | 2.0 (1.9) | 9.2 (1.6) | 2.2 (2.0) | 7.3 (1.3) | 1.3 (1.8) | 7.0 (1.3) | 2.3 (2.4) | 6.3 (1.3) | 2.3 (2.8) |
| group-MCPT | 17.2 (1.5) | 1.8 (5.2) | 15.6 (1.6) | 18.2 (9.4) | 14.6 (2.2) | 34.5 (10.8) | 14.0 (3.1) | 2.0 (4.9) | 11.3 (2.6) | 9.8 (7.7) | 8.8 (2.3) | 19.4 (8.7) |
| indiv-SB | 16.5 (1.2) | 0.9 (0.8) | 16.4 (1.0) | 1.7 (1.1) | 16.7 (1.0) | 1.0 (0.9) | 14.2 (1.5) | 0.9 (0.9) | 13.6 (1.6) | 1.9 (1.1) | 14.1 (1.6) | 1.2 (1.0) |
| indiv-MCP | 8.8 (0.9) | 11.0 (6.2) | 8.5 (1.0) | 10.5 (5.8) | 8.7 (0.8) | 12.0 (6.3) | 6.3 (1.2) | 10.7 (5.5) | 6.3 (0.8) | 9.7 (5.5) | 6.2 (1.1) | 11.1 (6.1) |
| pool-SB | 17.9 (0.6) | 0.4 (1.0) | 14.2 (1.5) | 11.3 (2.3) | 8.5 (1.5) | 17.0 (3.1) | 17.3 (1.5) | 0.3 (0.9) | 12.7 (2.0) | 8.8 (2.5) | 7.0 (1.7) | 14.3 (3.3) |
| pool-MCP | 11.5 (1.9) | 9.4 (8.2) | 8.2 (1.8) | 24.9 (14.4) | 4.3 (0.7) | 26.6 (18.8) | 9.6 (1.2) | 16.8 (16.3) | 6.9 (1.9) | 19.9 (13.8) | 3.9 (0.8) | 23.4 (17.8) |
|
| ||||||||||||
| nonzero coef = 1 | ||||||||||||
| New | 18.0 (0) | 0.1 (0.3) | 18.0 (0) | 4.3 (1.8) | 18.0 (0) | 2.5 (1.2) | 18.0 (0.2) | 0.8 (1.0) | 17.8 (0.4) | 1.6 (1.4) | 17.8 (0.5) | 1.0 (1.4) |
| New+ | 18.0 (0) | 0.0 (0) | 18.0 (0) | 2.0 (1.8) | 18.0 (0) | 1.4 (1.1) | 18.0 (0) | 0.1 (0.4) | 17.8 (0.4) | 1.3 (1.4) | 17.8 (0.5) | 1.0 (1.4) |
| alg1-SB | 18.0 (0) | 1.9 (1.2) | 18.0 (0) | 4.4 (1.8) | 18.0 (0) | 3.2 (1.4) | 17.6 (0.6) | 2.3 (1.4) | 17.7 (0.5) | 3.9 (1.8) | 17.3 (0.8) | 2.1 (1.1) |
| sgroup-MCP | 16.8 (1.6) | 1.1 (1.6) | 15.8 (2.0) | 1.9 (2.0) | 14.1 (2.1) | 2.9 (2.6) | 11.1 (2.2) | 1.5 (2.2) | 10.5 (1.5) | 2.1 (2.6) | 9.2 (1.3) | 2.1 (2.2) |
| group-MCPT | 18.0 (0) | 0.4 (1.2) | 17.8 (0.5) | 6.0 (2.9) | 17.8 (0.5) | 37.0 (9.4) | 17.9 (0.4) | 3.8 (9.3) | 16.0 (1.6) | 17.3 (7.4) | 15.1 (2.3) | 32.9 (13.6) |
| indiv-SB | 18.0 (0) | 0.8 (0.7) | 18.0 (0) | 1.5 (1.1) | 18.0 (0) | 0.9 (0.8) | 17.5 (0.7) | 0.8 (0.8) | 17.2 (0.7) | 1.6 (1.1) | 17.4 (0.8) | 1.1 (1.1) |
| indiv-MCP | 11.0 (1.1) | 9.9 (4.7) | 12.3 (1.5) | 13.0 (5.4) | 11.2 (0.9) | 10.2 (4.2) | 9.0 (0.6) | 11.4 (5.7) | 9.0 (1.1) | 13.8 (6.2) | 8.8 (0.8) | 12.9 (6.4) |
| pool-SB | 18.0 (0) | 0.1 (0.6) | 15.7 (1.2) | 13.7 (2.6) | 9.4 (1.7) | 19.0 (3.7) | 18.0 (0) | 0.3 (0.9) | 14.8 (1.4) | 11.8 (2.6) | 8.5 (1.8) | 17.2 (3.7) |
| pool-MCP | 13.8 (2.0) | 0.4 (1.0) | 9.9 (1.6) | 31.0 (17.9) | 4.7 (0.6) | 27.3 (15.7) | 12.8 (2.2) | 12.6 (8.1) | 9.0 (1.9) | 29.5 (21.4) | 4.4 (1.0) | 27.8 (18.7) |
Table 20.
Simulation: summary statistics on identification. In each cell, mean (sd). d = 1, 000 and ρ = 0.5.
| σ2 = 1 | σ2 = 3 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Complete | Half | None | Complete | Half | None | |||||||
|
| ||||||||||||
| TP | FP | TP | FP | TP | FP | TP | FP | TP | FP | TP | FP | |
| nonzero coef ~ unif(0.2,1) | ||||||||||||
| New | 17.3 (1.4) | 0.0 (0.2) | 16.4 (1.3) | 2.4 (1.4) | 16.8 (1.0) | 2.4 (1.4) | 13.2 (1.9) | 0.5 (0.7) | 11.5 (2.1) | 1.3 (1.6) | 12.9 (2.2) | 1.3 (1.0) |
| New+ | 17.5 (1.4) | 0.0 (0) | 16.3 (1.5) | 1.9 (1.3) | 16.8 (1.0) | 2.4 (1.4) | 13.4 (2.4) | 0.3 (0.6) | 11.4 (2.3) | 1.4 (1.7) | 12.9 (2.2) | 1.3 (1.0) |
| alg1-SB | 16.3 (1.3) | 2.5 (1.6) | 15.8 (1.5) | 2.5 (1.4) | 16.8 (1.0) | 3.3 (2.0) | 12.9 (2.3) | 1.2 (1.3) | 11.5 (2.5) | 1.8 (1.2) | 12.5 (1.8) | 1.6 (1.3) |
| sgroup-MCP | 13.9 (1.9) | 1.4 (1.9) | 13.1 (1.7) | 2.3 (2.8) | 11.9 (2.0) | 3.5 (3.4) | 9.4 (2.0) | 2.0 (2.8) | 8.4 (1.8) | 2.1 (3.1) | 7.5 (1.4) | 2.2 (2.5) |
| group-MCPT | 17.3 (1.3) | 3.2 (10.4) | 15.8 (1.7) | 23.6 (18.4) | 14.9 (2.2) | 45.5 (29.1) | 13.2 (2.7) | 2.3 (4.7) | 11.4 (2.8) | 10.6 (14.6) | 9.7 (4.2) | 65.2 (115.0) |
| indiv-SB | 14.8 (1.4) | 0.7 (0.9) | 15.2 (1.6) | 0.9 (0.8) | 15.8 (1.5) | 1.9 (0.9) | 12.1 (2.1) | 0.7 (0.7) | 10.9 (2.4) | 0.8 (0.7) | 12.4 (2.1) | 1.6 (0.7) |
| indiv-MCP | 12.5 (1.4) | 16.5 (7.1) | 12.9 (1.6) | 14.6 (6.4) | 12.8 (1.7) | 16.5 (7.5) | 9.3 (1.6) | 13.6 (8.0) | 9.5 (1.5) | 13.6 (6.8) | 9.5 (1.3) | 14.0 (6.4) |
| pool-SB | 17.8 (0.8) | 0.1 (0.6) | 12.6 (1.7) | 8.3 (2.6) | 3.9 (1.3) | 7.8 (2.6) | 16.8 (1.5) | 0.1 (0.6) | 10.3 (2.0) | 4.7 (2.5) | 1.9 (1.1) | 3.9 (2.1) |
| pool-MCP | 17.4 (1.2) | 24.4 (19.9) | 12.0 (1.7) | 31.8 (21.8) | 6.3 (1.2) | 38.1 (20.7) | 15.1 (2.6) | 37.9 (27.4) | 10.5 (1.9) | 31.9 (21.1) | 5.0 (1.3) | 30.5 (19.0) |
|
| ||||||||||||
| nonzero coef = 1 | ||||||||||||
| New | 18.0 (0) | 0.0 (0) | 18.0 (0) | 3.0 (1.5) | 18.0 (0) | 2.9 (1.4) | 18.0 (0.2) | 0.5 (0.6) | 17.9 (0.4) | 1.6 (1.0) | 17.9 (0.3) | 2.2 (1.2) |
| New+ | 18.0 (0) | 0.0 (0) | 18.0 (0) | 2.0 (1.6) | 18.0 (0) | 2.9 (1.4) | 18.0 (0.2) | 0.1 (0.3) | 17.8 (0.4) | 1.5 (1.1) | 17.9 (0.3) | 2.2 (1.2) |
| alg1-SB | 18.0 (0) | 3.1 (1.5) | 18.0 (0) | 3.7 (1.5) | 18.0 (0) | 3.2 (1.4) | 17.9 (0.3) | 1.7 (1.0) | 17.7 (0.7) | 2.2 (1.5) | 17.9 (0.3) | 2.2 (1.2) |
| sgroup-MCP | 18.0 (0.1) | 0.2 (0.5) | 18.0 (0.2) | 0.8 (1.2) | 17.9 (0.3) | 1.4 (1.7) | 16.2 (1.8) | 2.2 (2.2) | 14.8 (1.9) | 3.6 (3.3) | 13.4 (2.6) | 5.8 (4.6) |
| group-MCPT | 18.0 (0) | 0.0 (0.1) | 18.0 (0) | 0.4 (0.8) | 18.0 (0) | 0.6 (1.1) | 18.0 (0.2) | 0.9 (3.1) | 16.5 (1.5) | 22.2 (22.2) | 16.6 (2.2) | 48.9 (30.0) |
| indiv-SB | 18.0 (0) | 1.3 (0.5) | 18.0 (0) | 1.4 (0.6) | 18.0 (0) | 1.4 (0.6) | 17.6 (0.6) | 1.7 (0.8) | 17.2 (0.9) | 2.0 (1.0) | 17.6 (0.6) | 1.7 (0.8) |
| indiv-MCP | 18.0 (0) | 13.9 (5.4) | 18.0 (0) | 13.7 (6.4) | 18.0 (0) | 14.7 (6.0) | 13.1 (1.6) | 16.9 (7.2) | 13.9 (1.4) | 20.0 (9.7) | 13.5 (1.6) | 19.3 (8.4) |
| pool-SB | 18.0 (0) | 0.0 (0) | 14.6 (1.3) | 11.4 (2.6) | 4.8 (1.4) | 9.7 (2.7) | 18.0 (0) | 0.0 (0) | 13.5 (1.1) | 9.0 (2.3) | 3.6 (1.5) | 7.1 (2.9) |
| pool-MCP | 18.0 (0) | 7.9 (15.3) | 14.0 (0.7) | 35.7 (23.8) | 7.3 (1.4) | 36.3 (20.0) | 18.0 (0) | 15.7 (15.8) | 12.4 (1.7) | 36.9 (23.5) | 6.9 (1.2) | 37.0 (20.6) |
Table 21.
Simulation: summary statistics on identification. In each cell, mean (sd). d = 1, 000 and ρ = 0.8.
| σ2 = 1 | σ2 = 3 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Complete | Half | None | Complete | Half | None | |||||||
|
| ||||||||||||
| TP | FP | TP | FP | TP | FP | TP | FP | TP | FP | TP | FP | |
| nonzero coef ~ unif(0.2,1) | ||||||||||||
| New | 17.5 (0.6) | 1.2 (1.4) | 16.9 (1.0) | 2.5 (2.1) | 16.6 (0.9) | 1.7 (1.9) | 15.3 (2.0) | 0.5 (0.7) | 14.2 (1.7) | 2.0 (1.9) | 14.7 (1.2) | 1.3 (1.1) |
| New+ | 17.5 (0.6) | 0.4 (0.8) | 16.9 (1.0) | 2.8 (2.2) | 16.6 (0.9) | 1.7 (1.9) | 15.3 (2.0) | 0.1 (0.4) | 14.2 (1.7) | 1.5 (1.8) | 14.7 (1.2) | 1.3 (1.1) |
| alg1-SB | 16.6 (1.1) | 2.4 (1.5) | 16.4 (1.1) | 4.2 (1.9) | 16.5 (1.3) | 2.7 (1.3) | 13.5 (1.6) | 2.1 (1.1) | 13.5 (1.8) | 2.6 (1.5) | 14.1 (1.7) | 2.6 (1.6) |
| sgroup-MCP | 10.7 (2.1) | 1.3 (1.8) | 9.9 (1.6) | 2.1 (2.0) | 8.8 (1.5) | 2.3 (2.3) | 7.0 (1.2) | 0.9 (1.3) | 6.6 (1.2) | 2.5 (2.8) | 5.9 (1.0) | 2.1 (2.2) |
| group-MCPT | 17.5 (1.5) | 4.4 (11.1) | 15.4 (1.7) | 19.6 (12.9) | 14.4 (2.5) | 36.7 (13.6) | 13.4 (3.3) | 2.2 (6.2) | 10.7 (2.6) | 10.3 (11.1) | 8.0 (2.4) | 15.3 (9.6) |
| indiv-SB | 16.5 (1.0) | 1.2 (1.2) | 16.0 (1.4) | 2.0 (1.2) | 16.7 (0.9) | 1.6 (1.1) | 13.9 (1.4) | 1.1 (1.2) | 13.2 (1.6) | 1.9 (1.3) | 13.7 (1.2) | 1.3 (1.1) |
| indiv-MCP | 8.4 (1.1) | 12.4 (4.7) | 8.1 (0.9) | 12.4 (7.5) | 8.0 (1.1) | 12.6 (6.3) | 6.1 (1.2) | 11.9 (6.5) | 6.1 (0.8) | 11.0 (6.1) | 5.9 (0.9) | 13.5 (7.5) |
| pool-SB | 17.9 (0.6) | 0.8 (1.6) | 14.8 (1.6) | 12.5 (2.6) | 8.8 (1.7) | 18.0 (3.2) | 17.2 (1.4) | 0.8 (1.6) | 12.5 (2.2) | 9.6 (2.7) | 7.6 (1.4) | 15.5 (2.6) |
| pool-MCP | 10.9 (2.0) | 10.9 (13.0) | 7.8 (1.7) | 26.2 (18.7) | 4.1 (0.7) | 29.8 (23.2) | 8.8 (1.8) | 17.3 (15.2) | 6.5 (1.6) | 23.8 (18.0) | 3.9 (0.7) | 21.5 (16.1) |
|
| ||||||||||||
| nonzero coef = 1 | ||||||||||||
| New | 18.0 (0) | 0.4 (0.6) | 18.0 (0) | 2.4 (1.3) | 18.0 (0) | 1.7 (1.5) | 17.9 (0.3) | 0.8 (1.2) | 17.6 (0.6) | 3.3 (1.8) | 17.7 (0.7) | 1.3 (1.4) |
| New+ | 18.0 (0) | 0.0 (0) | 18.0 (0) | 2.2 (1.2) | 18.0 (0) | 1.7 (1.6) | 17.9 (0.3) | 0.2 (0.5) | 17.6 (0.6) | 1.8 (1.6) | 17.7 (0.7) | 1.3 (1.4) |
| alg1-SB | 18.0 (0) | 2.6 (1.6) | 18.0 (0) | 5.0 (1.7) | 18.0 (0) | 2.5 (1.4) | 17.6 (0.7) | 2.2 (1.1) | 17.6 (0.8) | 3.4 (1.3) | 17.4 (0.9) | 2.3 (1.4) |
| sgroup-MCP | 16.5 (1.7) | 1.2 (1.6) | 15.0 (1.8) | 2.0 (1.7) | 13.3 (2.4) | 3.0 (2.7) | 10.5 (1.7) | 1.2 (1.8) | 9.9 (1.5) | 1.9 (1.9) | 8.8 (1.3) | 2.3 (2.4) |
| group-MCPT | 18.0 (0) | 0.7 (2.1) | 17.9 (0.5) | 21.2 (8.2) | 17.9 (0.4) | 16.8 (5.9) | 17.8 (1.2) | 4.4 (8.3) | 16.0 (1.7) | 20.5 (11.8) | 15.1 (2.6) | 35.4 (14.5) |
| indiv-SB | 18.0 (0) | 1.1 (1.1) | 18.0 (0) | 1.8 (1.4) | 18.0 (0) | 1.4 (1.1) | 17.6 (0.6) | 1.1 (1.0) | 17.3 (0.9) | 1.8 (1.1) | 17.5 (0.8) | 1.4 (1.1) |
| indiv-MCP | 10.8 (1.1) | 11.0 (5.4) | 10.5 (1.6) | 13.4 (6.5) | 10.5 (0.9) | 12.3 (6.4) | 8.7 (0.7) | 15.0 (6.1) | 8.1 (1.1) | 14.4 (5.5) | 8.7 (0.7) | 13.9 (7.2) |
| pool-SB | 18.0 (0) | 0.7 (1.5) | 16.1 (1.0) | 14.6 (2.2) | 9.6 (1.1) | 19.4 (2.2) | 18.0 (0) | 0.7 (1.5) | 15.2 (1.0) | 12.7 (2.0) | 9.2 (1.4) | 18.7 (2.8) |
| pool-MCP | 13.0 (1.6) | 1.6 (2.8) | 9.3 (1.7) | 32.4 (18.4) | 4.3 (0.8) | 32.7 (24.9) | 12.3 (1.8) | 16.6 (16.2) | 8.0 (1.3) | 29.8 (19.5) | 4.2 (0.6) | 30.8 (22.2) |
Acknowledgments
We thank the associate editor and reviewer for their careful review and insightful comments, which have led to a significant improvement of the manuscript. This study was partly supported by CA142774 and CA016359 from NIH; VA Cooperative Studies Program of the Department of Veterans Affairs, Office of Research and Development; Duke-NUS Graduate Medical School WBS: R-913-200-098-263; National Social Science Foundation of China (13&ZD148, 13CTJ001); and National Natural Science Foundation of China (71471152, 71201139, 71301162).
Appendix
A. Estimation under the AFT model
For survival data, we consider the AFT (accelerated failure time) model. We note that some alternative models, especially the Cox model, have been more popular for “classic” low-dimensional data. With high-dimensional data, the simple form, low computational cost, and lucid interpretations of the AFT model make it especially attractive. This model has been adopted in multiple genetic and genomic studies. Denote T as the logarithm of failure time. The AFT model assumes that
α is the unknown intercept, X is the length-d vector of covariates, β is the vector of unknown regression coefficients, and ε is the random error. Under right censoring, denote C as the logarithm of censoring time. We observe (Y = min(T,C), δ = I(T ≤ C),X). Assume n i.i.d. observations.
When the distribution of ε is known, the parametric likelihood function can be easily constructed. Here we consider the more flexible case where this distribution is unknown. The weighted least squares estimator first proposed by Stute [26] is adopted, as it has statistical properties comparable to but computational cost lower than for example the Buckley-James and rank-based approaches.
Let F̂ be the Kaplan-Meier estimator of the distribution function F of T. , where ωi’s can be computed as
Y(1) ≤ · · · ≤ Y(n) are the order statistics of Yi’s, and δ(1), …, δ(n) are the associated censoring indicators. Denote X(i) as the covariate associated with (Y(i), δ(i)).
The weighted least squares estimator . We center X(i) and Y(i) using their ωi-weighted means, respectively. Define
Let and , respectively. With the weighted centered values, the intercept is zero. The weighted least squares objective function can be rewritten as
B. Boosting with tree-based weak learners
In the algorithm described in the main text, linear weak learners are considered. Boosting is a very flexible tool and can also accommodate nonlinear weak learners. As an example, we consider tree-based weak learners. Boosting survival trees has been studied in multiple published studies. Here we consider applying the proposed promoting similarity in sparsity structure to boosting survival trees. Following the literature, we take simple weak learners where each boosting step considers a single variable for growing the tree. A popular choice in the literature is stump, which is a tree learner with two terminal nodes [9]. We allow a user-specified maximum depth for growing a tree based on a single variable. The detailed algorithm is as follows.
Algorithm 2 (tree): Sparse boosting for integrative analysis with tree-based weak learners
Step 1: Initialization.
Initialize k = 0 and S[0] = 0M×d. For m = 1, …, M, initialize and .
-
Step 2: Fit and update. k = k + 1. For m = 1, …, M:
Compute . Here denotes the fitted tree model based on Xm,s with a pre-specified depth. is defined as , where Inm is the nm × nm identity matrix and Hm,s is an nm × nm symmetric matrix whose (i, j)th element equals one over the subjects that are in the same terminal node as the ith subject if the ith and jth subjects are in the same terminal node, and zero otherwise. The matrix S[k −1],s takes the same values as S[k − 1] except that the (m, s)th element is 1. To calculate pen(·), trace( ) is used as the degree of freedom [9]. pens(·) is calculated in the same way as in Algorithm 2.
Update. Set the (m, ŝ)th element of S[k − 1] to 1 and obtain S[k]. Let and .
Step 3: Iteration. Repeat Step 2 for K times. K is a large number.
Step 4: Selection of optimal stopping. At iteration k(= 1, …, K), compute . Select the optimal number of iterations as k̂ = argmin1≤k≤KF(k).
When using the tree-based weak learners, different from that in the main text, there is no well-defined regression coefficient β. To accommodate this difference, modifications are made to Step 2. Specifically, Hm,s is introduced for calculating pen, and S[k] is introduced to record selection for calculating pens.
We conduct simulation to assess performance of the proposed strategy with tree-based weak learners. Specifically, we generate M = 3 independent datasets. In each dataset, the sample size is nm = 100, and the dimension is d = 1, 000. Event times are generated from the model α + β′X2 + ε, where X is the length-d vector of covariates following a multivariate normal distribution, and β is the vector of regression coefficients. Here we consider normal errors with zero mean and unit variance. The specifications for the multivariate normal distribution, β, and other settings are the same as in Section 3.
For comparison, we also consider two alternative methods: (a) alg1-SB, which applies Algorithm 1 with tree-based weak learners, and (b) indiv-SB, which applies sparse boosting with tree-based weak learners to each dataset separately. The summary identification results are shown in Table 22 (Appendix). The overall observed patterns are similar to those with linear weak learners: the proposed strategy has obvious advantages under the complete overlapping scenario and competitive performance under the half and none overlapping scenarios.
C. Boosting under the Cox model
For censored survival data, the most popular model is the Cox model. For high-dimensional data, it has higher computational cost than the AFT model, making it less advantaged. Below we describe applying the proposed strategy to the Cox model.
The Cox model assumes that
where λ0(T) is the baseline hazard function. Consider dataset m with nm iid observations ( ) for i = 1, …, nm. Here denotes the observed time, denotes the event indicator, and denotes the covariates. Denote . The log partial likelihood function is
Consider the loss function Rm(fm) = −log PLm(fm).
Algorithm 2 (Cox): Sparse boosting for integrative analysis under the Cox model
Step 1: Initialization. The same as in Algorithm 1 and 2.
-
Step 2: Fit and update. k = k + 1. For m = 1, …, M:
Obtain the working response with .
For each j = 1, …, d, fit a linear regression of on Xm,j and obtain the coefficients γ̂j. Select ŝ that minimizes . The pen(·) and pens(·) are defined the same as in Algorithm 2.
Update and .
Step 3: Iteration. Repeat Step 2 for K times. K is a large number.
Step 4: Selection of optimal stopping. At iteration k(= 1, …, K), compute . Select the optimal number of iterations as k̂ = argmin1≤k≤KF(k).
The above algorithm is very similar to that for the AFT model. To accommodate the Cox model, we follow Ridgeway [25] and construct a working response variable.
We also analyze the breast cancer data under the Cox model. The identification and estimation results are provided in Table 23 (Appendix). Different sets of genes are identified under the AFT and Cox models. With high-dimensional data, model diagnostics and specifying model forms are very challenging and have not been carefully investigated. We leave the selection between AFT and Cox models to future research. Under the Cox model, the overlapping percentages (which are calculated in the same manner as under the AFT model) are 0.35 (Alt.1), 0.33 (alg1-SB), 0.43 (New), 0.46 (New+), and 0.33 (indiv-SB), respectively. Prediction performance is assessed in the same way as under the AFT model. The average logrank statistics are 3.76 (Alt.1), 3.53 (alg1-SB), 5.03 (New), 5.03 (New+), 3.58 (indiv-SB), and 4.04 (pool-SB), respectively. The proposed approach leads to improved prediction.
D. Additional tables and figures
Figure 2.
Plots of mean TP and FP for simulations with d = 100, ρ = 0.2, and σ2 = 1. The circles stand for TP and black squares stand for FP.
Figure 3.
Plots of mean TP and FP for simulations with d = 100, ρ = 0.5, and σ2 = 1. The circles stand for TP and black squares stand for FP.
Figure 4.
Plots of mean TP and FP for simulations with d = 100, ρ = 0.8, and σ2 = 1. The circles stand for TP and black squares stand for FP.
Figure 5.
Plots of mean TP and FP for simulations with d = 500, ρ = 0.2, and σ2 = 1. The circles stand for TP and black squares stand for FP.
Figure 6.
Plots of mean TP and FP for simulations with d = 500, ρ = 0.5, and σ2 = 1. The circles stand for TP and black squares stand for FP.
Figure 7.
Plots of mean TP and FP for simulations with d = 500, ρ = 0.8, and σ2 = 1. The circles stand for TP and black squares stand for FP.
Figure 8.
Plots of mean TP and FP for simulations with d = 1, 000, ρ = 0.5, and σ2 = 1. The circles stand for TP and black squares stand for FP.
Figure 9.
Plots of mean TP and FP for simulations with d = 1, 000, ρ = 0.8, and σ2 = 1. The circles stand for TP and black squares stand for FP.
Figure 10.
Plots of mean TP and FP for simulations with d = 100, ρ = 0.2, and σ2 = 3. The circles stand for TP and black squares stand for FP.
Figure 11.
Plots of mean TP and FP for simulations with d = 100, ρ = 0.5, and σ2 = 3. The circles stand for TP and black squares stand for FP.
Figure 12.
Plots of mean TP and FP for simulations with d = 100, ρ = 0.8, and σ2 = 3. The circles stand for TP and black squares stand for FP.
Figure 13.
Plots of mean TP and FP for simulations with d = 500, ρ = 0.2, and σ2 = 3. The circles stand for TP and black squares stand for FP.
Figure 14.
Plots of mean TP and FP for simulations with d = 500, ρ = 0.5, and σ2 = 3. The circles stand for TP and black squares stand for FP.
Figure 15.
Plots of mean TP and FP for simulations with d = 500, ρ = 0.8, and σ2 = 3. The circles stand for TP and black squares stand for FP.
Figure 16.
Plots of mean TP and FP for simulations with d = 1, 000, ρ = 0.2, and σ2 = 3. The circles stand for TP and black squares stand for FP.
Figure 17.
Plots of mean TP and FP for simulations with d = 1, 000, ρ = 0.5, and σ2 = 3. The circles stand for TP and black squares stand for FP.
Figure 18.
Plots of mean TP and FP for simulations with d = 1, 000, ρ = 0.8, and σ2 = 3. The circles stand for TP and black squares stand for FP.
Figure 19.
Four simulation scenarios where datasets have different complexity structures. The grey cells correspond to covariates with nonzero coefficients.
Table 22.
Simulation with trees as weak learners: summary statistics on identification. In each cell, mean (sd). d = 1, 000.
| Complete | Half | None | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| TP | FP | TP | FP | TP | FP | |
| nonzero coef ~ unif(0.2,1) | ||||||
| ρ = 0.2 | ||||||
| New | 6.1 (2.3) | 3.1 (3.0) | 5.3 (2.1) | 5.2 (3.4) | 4.8 (2.2) | 5.2 (2.7) |
| New+ | 6.0 (2.0) | 2.5 (2.7) | 5.2 (2.2) | 4.7 (3.5) | 4.8 (2.1) | 5.2 (2.8) |
| alg1-SB | 4.9 (1.5) | 5.6 (2.9) | 5.1 (2.1) | 6.0 (2.8) | 4.9 (2.3) | 5.2 (2.8) |
| indiv-SB | 4.8 (1.4) | 4.3 (1.9) | 5.3 (2.0) | 4.7 (2.1) | 5.2 (1.9) | 4.7 (2.1) |
| ρ = 0.5 | ||||||
| New | 8.4 (2.8) | 1.2 (1.7) | 7.2 (2.0) | 2.8 (2.0) | 7.0 (1.8) | 4.1 (1.7) |
| New+ | 8.8 (3.5) | 0.9 (1.5) | 7.1 (1.7) | 2.6 (2.2) | 7.0 (1.9) | 4.0 (1.7) |
| alg1-SB | 7.3 (1.9) | 3.4 (2.4) | 6.7 (1.8) | 4.3 (2.1) | 7.0 (1.8) | 4.1 (1.6) |
| indiv-SB | 7.2 (1.7) | 2.6 (1.4) | 7.1 (1.9) | 3.6 (1.9) | 6.8 (2.0) | 2.7 (1.6) |
| ρ = 0.8 | ||||||
| New | 12.1 (3.2) | 0.9 (1.6) | 11.2 (2.3) | 1.9 (1.3) | 12.9 (1.6) | 2.3 (1.9) |
| New+ | 12.4 (3.2) | 0.5 (0.8) | 10.8 (3.0) | 1.5 (1.3) | 13.0 (1.8) | 2.4 (2.1) |
| alg1-SB | 13.1 (2.0) | 1.9 (1.6) | 11.3 (2.2) | 2.2 (1.3) | 12.8 (1.8) | 2.2 (1.7) |
| indiv-SB | 12.8 (2.0) | 1.4 (1.5) | 10.8 (1.9) | 1.3 (1.0) | 12.8 (1.8) | 1.5 (1.4) |
|
| ||||||
| nonzero coef = 1 | ||||||
| ρ = 0.2 | ||||||
| New | 7.4 (2.7) | 4.4 (3.5) | 6.5 (1.7) | 5.1 (3.2) | 6.1 (2.4) | 8.2 (2.7) |
| New+ | 6.9 (2.7) | 3.9 (3.4) | 6.3 (1.7) | 4.9 (3.3) | 5.9 (2.4) | 8.2 (2.8) |
| alg1-SB | 6.1 (1.8) | 8.8 (3.5) | 5.8 (1.5) | 8.1 (3.1) | 6.1 (2.4) | 8.3 (2.8) |
| indiv-SB | 6.1 (1.9) | 7.5 (3.0) | 6.0 (1.6) | 7.4 (2.3) | 6.1 (2.3) | 7.5 (3.2) |
| ρ = 0.5 | ||||||
| New | 11.3 (3.0) | 1.7 (1.6) | 9.0 (2.1) | 4.0 (2.1) | 8.7 (2.1) | 4.7 (1.7) |
| New+ | 11.0 (3.6) | 1.3 (1.7) | 9.0 (2.4) | 4.3 (2.9) | 8.7 (2.1) | 4.8 (1.8) |
| alg1-SB | 9.2 (2.0) | 4.8 (2.2) | 8.8 (2.3) | 5.3 (2.1) | 8.7 (2.1) | 4.8 (1.8) |
| indiv-SB | 9.2 (2.1) | 4.4 (2.0) | 8.3 (2.0) | 4.2 (2.1) | 8.6 (2.4) | 3.9 (1.6) |
| ρ = 0.8 | ||||||
| New | 15.4 (2.0) | 1.7 (2.3) | 14.2 (1.6) | 2.7 (1.7) | 15.2 (1.4) | 3.2 (1.9) |
| New+ | 15.1 (3.0) | 0.9 (1.8) | 13.9 (2.7) | 2.3 (1.9) | 15.3 (1.5) | 3.2 (1.8) |
| alg1-SB | 15.4 (1.5) | 3.2 (2.6) | 14.2 (1.5) | 3.1 (2.1) | 15.3 (1.5) | 3.1 (1.8) |
| indiv-SB | 15.0 (1.8) | 2.5 (2.4) | 13.4 (2.0) | 2.0 (1.7) | 15.0 (1.6) | 2.7 (2.5) |
Table 23.
Analysis of the breast cancer datasets: identified genes and estimates under the Cox model.
| UniGene | Alt.1 | alg1-SB | New | New+ | indiv-SB | pool-SB |
|---|---|---|---|---|---|---|
| Dataset 1 | ||||||
| Hs.646 | −0.187 | |||||
| Hs.19413 | 0.227 | 0.292 | ||||
| Hs.19699 | −0.135 | |||||
| Hs.25351 | −0.179 | −0.287 | −0.393 | −0.343 | −0.287 | |
| Hs.75149 | −0.049 | −0.117 | −0.117 | |||
| Hs.78881 | −0.204 | −0.342 | −0.367 | −0.257 | −0.271 | |
| Hs.82548 | 0.073 | 0.305 | 0.392 | 0.308 | 0.305 | |
| Hs.95821 | 0.211 | |||||
| Hs.154443 | 0.057 | |||||
| Hs.154797 | −0.093 | |||||
| Hs.274382 | −0.061 | −0.030 | −0.030 | |||
| Hs.288319 | 0.025 | |||||
| Hs.431584 | −0.160 | −0.518 | −0.560 | −0.419 | −0.428 | |
|
| ||||||
| Dataset 2 | ||||||
| Hs.646 | −0.034 | −0.187 | ||||
| Hs.2421 | −0.032 | |||||
| Hs.15303 | −0.109 | −0.536 | −0.544 | −0.489 | −0.296 | |
| Hs.19699 | −0.135 | |||||
| Hs.25351 | 0.011 | 0.011 | ||||
| Hs.82548 | −0.032 | −0.015 | −0.015 | |||
| Hs.89506 | 0.032 | |||||
| Hs.95821 | 0.053 | 0.255 | 0.278 | 0.211 | ||
| Hs.154443 | 0.088 | 0.057 | ||||
| Hs.154797 | −0.013 | −0.013 | −0.093 | |||
| Hs.407372 | 0.087 | |||||
|
| ||||||
| Dataset 3 | ||||||
| Hs.646 | −0.187 | |||||
| Hs.1578 | 0.047 | 0.070 | 0.070 | |||
| Hs.14541 | 0.067 | |||||
| Hs.19699 | −0.135 | |||||
| Hs.75617 | 0.022 | |||||
| Hs.89399 | −0.049 | |||||
| Hs.95821 | 0.211 | |||||
| Hs.154443 | 0.057 | |||||
| Hs.154797 | −0.185 | −0.556 | −0.635 | −0.448 | −0.565 | −0.093 |
| Hs.177584 | 0.163 | 0.421 | 0.466 | 0.315 | 0.442 | |
| Hs.206770 | 0.085 | 0.509 | 0.598 | 0.401 | 0.528 | |
| Hs.301094 | −0.021 | |||||
References
- 1.Guerra R, Goldstein DR. Meta-Analysis and Combining Information in Genetics and Genomics. Chapman and Hall/CRC; 2009. [Google Scholar]
- 2.Ritz J, Demidenko E, Spiegelman D. Multivariate meta-analysis for data consortia, individual patient meta-analysis, and pooling projects. Journal of Statistical Planning and Inference. 2008;138:1919–1933. [Google Scholar]
- 3.Stukel T, Demidenko E, Dykes J, Karagas M. Two-stage methods for the analysis of pooling data. Statistics in Medicine. 2011;20:2115–2130. doi: 10.1002/sim.852. [DOI] [PubMed] [Google Scholar]
- 4.Huang Y, Huang J, Shia BC, Ma S. Identification of cancer genomic markers via integrative sparse boosting. Biostatistics. 2012;13:509–522. doi: 10.1093/biostatistics/kxr033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J, Huang J, Ma S. Integrative analysis of cancer diagnosis studies with composite penalization. Scandinavian Journal of Statistics. 2014;41(1):87–103. doi: 10.1111/j.1467-9469.2012.00816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, Huang J, Xie Y, Ma S. Sparse group penalized integrative analysis of multiple cancer prognosis datasets. Genetics Research. 2013;95:68–77. doi: 10.1017/S0016672313000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma S, Huang J, Wei F, Xie Y, Fang K. Integrative analysis of multiple cancer prognosis studies with gene expression measurements. Statistics in Medicine. 2011;30:3361–3371. doi: 10.1002/sim.4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. PNAS. 2004;101(25):9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buhlmann P, Yu B. Sparse boosting. Journal of Machine Learning Research. 2006;7:1001–1024. [Google Scholar]
- 10.Zhang J, Ramadge PJ. Sparse boosting. International Conference on Acoustics, Speech and Signal Processing.2009. [Google Scholar]
- 11.Buhlmann P, Hothorn T. Boosting algorithms: regularization, prediction and model fitting (with discussion) Stat Sci. 2007;22:477–505. [Google Scholar]
- 12.Tan PN, Steinbach M, Kumar V. Introduction to Data Mining. Addison-Wesley; 2005. [Google Scholar]
- 13.Ma S, Huang Y, Huang J, Fang K. Gene network-based cancer prognosis analysis with sparse boosting. Genetics Research. 2012;94:205–221. doi: 10.1017/S0016672312000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ing C, Lai T. A stepwise regression method and consistent model selection for high-dimensional sparse linear models. Statistica Sinica. 2011;21:1473–1513. [Google Scholar]
- 15.An H, Huang D, Yao Q, Zhang C. Stepwise searching for feature variables in high-dimensional linear regression. Technical report. 2008 available at http://stats.lse.ac.uk/q.yao/qyao.links/paper/ahyz08.pdf.
- 16.Zhang C. Nearly unbiased variable selection under minimax concave penalty. Annals of Statistics. 2010;38(2):894–942. [Google Scholar]
- 17.Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. JNCI. 2006;98:262–72. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 18.van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–36. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 19.Huang E, Cheng SH, Dressman H, Pittman J, Tsou MH, Horng CF, et al. Gene expression predictors of breast cancer outcomes. Lancet. 2003;361:1590–6. doi: 10.1016/S0140-6736(03)13308-9. [DOI] [PubMed] [Google Scholar]
- 20.Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, et al. Breast cancer classification and prognosis based on gene expression profiles from a population based study. PNAS. 2003;100:10393–8. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang DD, Sibille E, Kaminski N, Tseng GC. MetaQC: objective quality control and inclusion/exclusion criteria for genomic meta-analysis. Nucleic Acids Research. 2011;40:1–14. doi: 10.1093/nar/gkr1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tseng GC, Ghosh D, Feingold E. Comprehensive literature review and statistical considerations for microarray meta-analysis. Nucleic acids research. 2012;40:3785–3799. doi: 10.1093/nar/gkr1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brenner H, Gefeller O. Variation of sensitivity, specificity, likelihood ratios and predictive values with disease prevalence. Stat Med. 1997;16:981–991. doi: 10.1002/(sici)1097-0258(19970515)16:9<981::aid-sim510>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Fine J. Assessing the dependence of sensitivity and specificity on prevalence in meta-analysis. Biostatistics. 2011;12:710–722. doi: 10.1093/biostatistics/kxr008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ridgeway G. Generalized Boosted Models: A guide to the gbm package. 2007 Available at http://www.saedsayad.com/docs/gbm2.pdf.
- 26.Stute W. Distributional convergence under random censorship when covariates are present. Scandinavian Journal of Statistics. 1996;23:461–471. [Google Scholar]