Abstract
MitoNEET, a primary target of type II diabetes drug pioglitazone, has an essential role in regulating energy metabolism, iron homeostasis, and production of reactive oxygen species in mitochondria. Structurally, mitoNEET is anchored to the mitochondrial outer membrane via its N-terminal transmembrane α-helix. The C-terminal cytosolic domain of mitoNEET hosts a redox active [2Fe-2S] cluster via three cysteine and one histidine residues. Here we report that the reduced flavin nucleotides can rapidly reduce the mitoNEET [2Fe-2S] clusters under anaerobic or aerobic conditions. In the presence of NADH and flavin reductase, about 1 molecule of flavin nucleotide is sufficient to reduce 100 molecules of the mitoNEET [2Fe-2S] clusters in 4 minutes under aerobic conditions. The electron paramagnetic resonance (EPR) measurements show that flavin mononucleotide (FMN), but not flavin adenine dinucleotide (FAD), has a specific interaction with mitoNEET. Molecular docking models further reveal that flavin mononucleotide binds mitoNEET at the region between the N-terminal transmembrane α-helix and the [2Fe-2S] cluster binding domain. The closest distance between the [2Fe-2S] cluster and the bound flavin mononucleotide in mitoNEET is about 10 Å, which may facilitate rapid electron transfer from the reduced flavin nucleotide to the [2Fe-2S] cluster in mitoNEET. The results suggest that flavin nucleotides may act as electron shuttles to reduce the mitoNEET [2Fe-2S] clusters and regulate mitochondrial functions in human cells.
Keywords: mitoNEET, type II diabetes, flavin nucleotides, iron-sulfur cluster, redox regulation
INTRODUCTION
Human mitochondrial outer membrane protein mitoNEET was initially identified as a target of type II diabetes drug pioglitazone [1]. While deletion of mitoNEET in mice significantly decreases oxidative phosphorylation activity in mitochondria [2], increased expression of mitoNEET in adipocytes enhances lipid uptake and storage of the cells and inhibits mitochondrial iron transport into matrix [3]. In beta cells, increased expression of mitoNEET leads to hyperglycemia and glucose intolerance [4]. Misregulation of mitoNEET expression has also been attributed to the development of neurodegenerative disease [5, 6] and cardiovascular disease [7], breast cancer proliferation [8–10], browning of white adipose tissue [11], among other pathological conditions [12]. It has thus been proposed that mitoNEET is a key regulator for energy metabolism, iron homeostasis, and production of reactive oxygen species in mitochondria [13].
MitoNEET is a homodimer with each monomer containing an N-terminal transmembrane α-helix (residues 14 to 32) anchored to the mitochondrial outer membrane [1]. The C-terminal cytosolic domain (residues 33–108) of mitoNEET hosts a redox active [2Fe-2S] cluster via a unique ligand arrangement of three cysteine and one histidine residues [14–16]. As mitochondria are the primary sites for iron-sulfur cluster biogenesis [17], several studies have suggested that mitoNEET may mediate transfer of iron-sulfur clusters assembled in mitochondria to target proteins in cytoplasm [18–21]. However, the cluster transfer occurs only when the mitoNEET [2Fe-2S] clusters are in oxidized state [18, 21], indicating the transfer process is controlled by the redox state of the [2Fe-2S] clusters [22, 23]. Alternatively, mitoNEET may directly regulate mitochondrial functions via the redox transition of the [2Fe-2S] clusters [4, 22–24], as mitoNEET forms complexes with multiple mitochondrial proteins including the Parkinson's disease associated protein Parkin [4, 25, 26] and mitochondrial outer membrane import complex protein 1 (MTX1) [27].
Intracellular redox co-enzymes including nicotinamide adenine dinucleotide (NAD+), nicotinamide adenine dinucleotide phosphate (NADP+), flavin mononucleotide (FMN), and flavin adenine dinucleotide (FAD) are considered as natural biomarkers of metabolic activities and mitochondrial anomalies [28]. If mitoNEET is a key regulator of energy metabolism in mitochondria [13], it is expected that mitoNEET will have direct interactions with the redox co-enzymes in cells. Indeed, previous studies have shown that mitoNEET is able to bind NADP(H) (but not NAD(H)) [29], and that binding of NADPH in mitoNEET destabilizes the [2Fe-2S] clusters [29] and inhibits the [2Fe-2S] cluster transfer from mitoNEET to target proteins [30]. However, the interaction between mitoNEET and NADP(H) is fairly weak (with a dissociation constant of about 2.0 mM) [29]. Furthermore, both NADPH and NADH fail to reduce the mitoNEET [2Fe-2S] clusters under anaerobic or aerobic conditions [22, 23, 30]. Here, we report that the reduced flavin nucleotides can rapidly reduce the mitoNEET [2Fe-2S] clusters under anaerobic or aerobic conditions. The electron paramagnetic resonance (EPR) measurements show that flavin mononucleotide, but not flavin adenine dinucleotide, has a specific interaction with mitoNEET. Furthermore, molecular docking models reveal that flavin mononucleotide binds mitoNEET at the region between the N-terminal transmembrane α-helix and the [2Fe-2S] cluster binding domain. Such an arrangement may facilitate rapid electron transfer from the reduced flavin nucleotide to the [2Fe-2S] clusters in mitoNEET. Our results suggest that mitoNEET may sense redox state of intracellular redox co-enzymes to modulate energy metabolism, iron homeostasis, and production of reactive oxygen species in mitochondria.
MATERIALS AND METHODS
1. Protein preparation
Escherichia coli flavin reductase (Fre) was prepared using an E. coli strain hosting an expression plasmid encoding Fre from the ASKA library [31]. Human mitochondrial outer membrane protein mitoNEET33–108 (containing residues 33–108) was purified as described in [23]. MitoNEET with deletion of the segment between Arg-33 and Ile-45 was prepared using the site-directed mutagenesis (Agilent co.), and confirmed by direct sequencing. The purity of purified proteins was greater than 95% as judged by electrophoresis analysis on a 15% polyacrylamide gel containing SDS followed by staining with Coomassie Blue. The protein concentrations of mitoNEET and E. coli Fre were measured at 280 nm using an extinction coefficient of 8.6 and 26.4 mM−1cm−1, respectively. The UV-visible absorption spectra were measured in a Beckman DU640 UV-visible absorption spectrometer equipped with a temperature controller.
2. Iron and sulfide content analyses
The iron content of protein samples was determined using ferroZine [32] as described previously [33]. The sulfide contents of protein samples was determined according to the Siegel’s method [34]. Purified E. coli endonuclease III containing a stable [4Fe-4S] cluster [35] was used as a standard for the iron and sulfide content analyses in protein samples.
3. EPR measurements
The X-band Electron Paramagnetic Resonance (EPR) spectra were recorded using a Bruker model ESR-300 spectrometer equipped with an Oxford Instruments 910 continuous flow cryostat. Routine EPR conditions were: microwave frequency, 9.47 GHz; microwave power, 10.0 mW; modulation frequency, 100 kHz; modulation amplitude, 1.2 mT; temperature, 30 K; receiver gain, 2×105.
4. Chemicals
NADH, NADPH, Isopropyl-β-D-1-thiogalactopyranoside, kanamycin, and ampicillin were purchased from Research Product International co. FMN, FAD, and other chemicals were purchased from Sigma co. Extinction coefficients of 6.2 mM−1cm−1 at 340 nm, 12.5 mM−1cm−1 at 445 nm, and 11.3 mM−1cm−1 at 450 nm were used for NADH/NADPH, FMN and FAD, respectively [28].
5. Molecular docking
The protocol for AutoDockVina-based molecular docking is a modification from Garret M. Morris’ (The Scripps Research Institute, La Jolla, CA) [36]. The validity of this protocol was established during other studies [37]. Partial conformational flexibility was allowed to the residues in direct contacts with targets. Docking is made based on binding energy calculation using Lamarckian Genetic Algorithm implemented in AutoDock3 [36]. The detailed docking parameters per molecule are: the energy evaluations number = 1750000, the population size = 150, the trial number = 20, and the positional rms tolerance = 2.0 Å.
RESULTS
1. Reduced flavin nucleotides rapidly reduce the mitoNEET [2Fe-2S] clusters under aerobic conditions
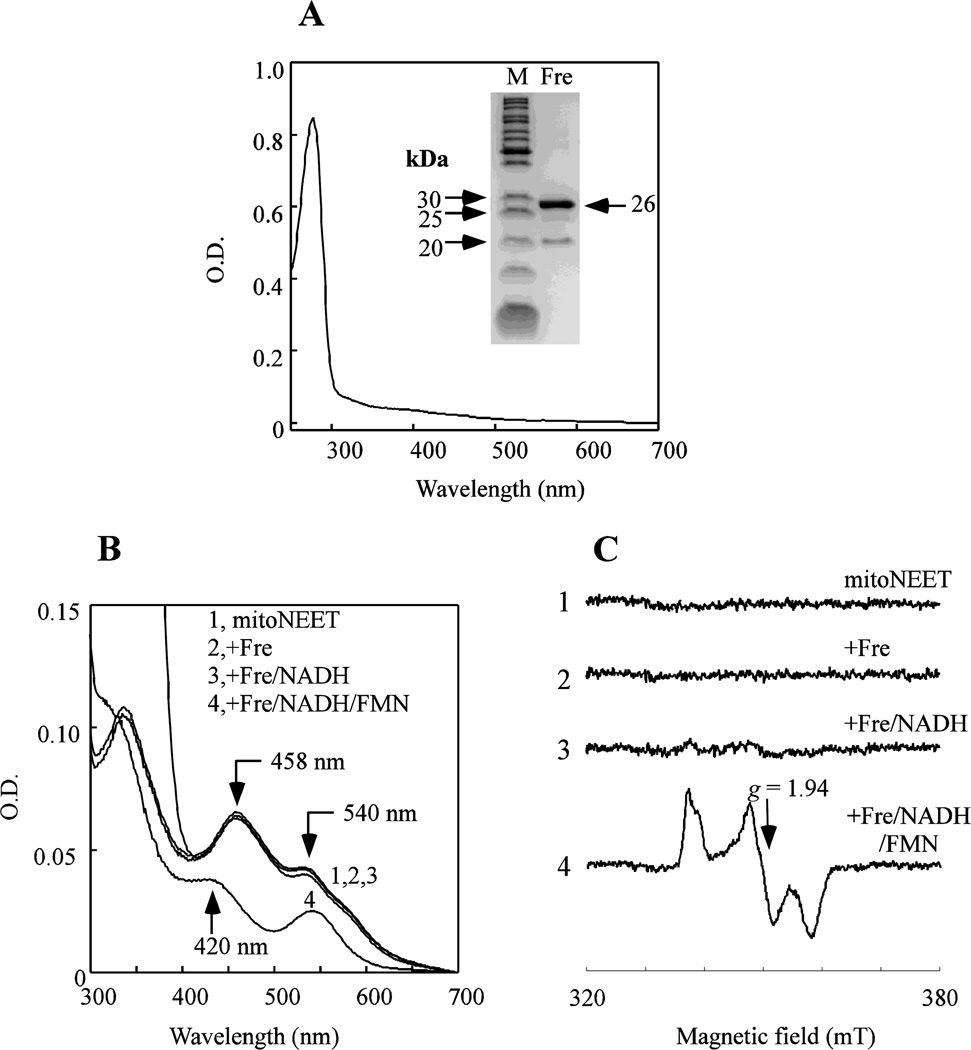
Unlike NADPH or NADH, reduced flavin nucleotides (FMNH2 and FADH2) are not stable under aerobic conditions and not commercially available. To prepare FMNH2 or FADH2, we purified Escherichia coli flavin reductase (Fre) from an E. coli ASKA library strain [31]. Figure 1A shows that purified E. coli Fre does not have any prosthetic groups as reported in [38], and is fully active to reduce FMN/FAD to FMNH2/FADH2 using NADH as electron donor [39].
Figure 1. Reduction of the mitoNEET [2Fe-2S] clusters by reduced flavin nucleotides.
A), UV-visible absorption spectrum of purified E. coli flavin reductase (Fre). Purified Fre (30 µM) was dissolved in buffer containing 500 mM NaCl and 20 mM Tris (pH 8.0). Insert is a photograph of the SDS-PAGE gel of purified E. coli Fre. B), reduction of the mitoNEET [2Fe-2S] clusters by the reduced flavin nucleotides. MitoNEET (containing 10 µM [2Fe-2S] clusters) (spectrum 1) was incubated with E. coli Fre (1 µM) (spectrum 2), Fre and NADH (50 µM) (spectrum 3), Fre, NADH and FMN (1 µM) (spectrum 4) in buffer containing 500 mM NaCl and 20 mM Tris (pH 8.0) at room temperature under aerobic conditions. For each reaction, the UV-visible absorption spectrum was taken after incubation for 2 min. C), EPR spectra of the mitoNEET [2Fe-2S] clusters. The samples were the same as in A). The EPR signal at g = 1.94 indicates the reduced mitoNEET [2Fe-2S] clusters. Data are representative of three independent experiments.
To monitor the redox state of the mitoNEET [2Fe-2S] clusters, we used the UV-visible absorption spectroscopy. Purified human mitoNEET has two major absorption peaks at 458 nm and 540 nm, indicative of the oxidized [2Fe-2S] clusters in the protein [18, 22] (Figure 1B, spectrum 1). When mitoNEET is incubated with a catalytic amount of purified E. coli Fre (Figure 1B, spectrum 2) or Fre and NADH (Figure 1B, spectrum 3) under aerobic conditions, the mitoNEET [2Fe-2S] clusters remain oxidized. However, when mitoNEET is incubated with E. coli Fre, NADH and FMN under aerobic conditions, the mitoNEET [2Fe-2S] clusters are fully reduced, as indicated by a new absorption peak at 420 nm (Figure 1B, spectrum 4). Thus, while Fre or Fre/NADH fails to reduce the mitoNEET [2Fe-2S] clusters, FMN reduced by Fre and NADH can fully reduce the mitoNEET [2Fe-2S] clusters under aerobic conditions. Similar results are observed when FMN is replaced with FAD in the incubation solutions (data not shown).
In parallel experiments, we used the electron paramagnetic resonance (EPR) spectroscopy to evaluate the redox state of the mitoNEET [2Fe-2S] clusters in the above reaction solutions. While the oxidized mitoNEET [2Fe-2S] clusters are EPR silent, the reduced mitoNEET [2Fe-2S] clusters have a distinct EPR signal at g = 1.94 [23, 40]. Figure 1C shows that the mitoNEET [2Fe-2S] clusters are reduced only after mitoNEET is incubated with Fre, NADH and FMN or FAD (not shown). Thus, the mitoNEET [2Fe-2S] clusters can be efficiently reduced by the reduced flavin nucleotides under aerobic conditions.
2. Reversible reduction of the mitoNEET [2Fe-2S] clusters under aerobic conditions
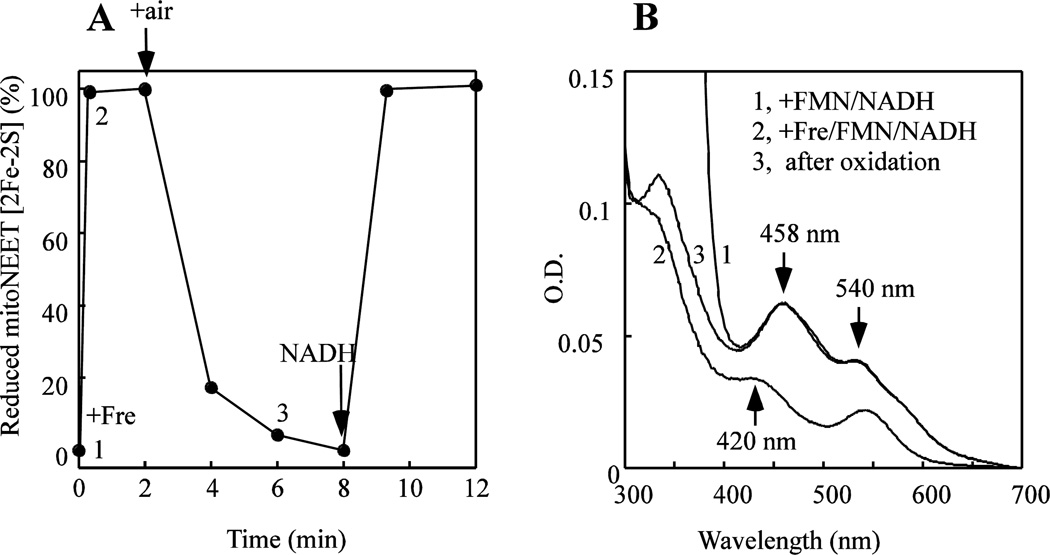
Since the redox midpoint potential (Em7) of the mitoNEET [2Fe-2S] clusters is about 0 mV [24], the reduced [2Fe-2S] clusters in mitoNEET could be readily oxidized by oxygen under aerobic conditions. To test this idea, mitoNEET is pre-incubated with FMN and a limited amount of NADH under anaerobic conditions, followed by addition of a catalytic amount of flavin reductase (Fre). As shown in Figure 2A, addition of Fre leads to a rapid reduction of the mitoNEET [2Fe-2S] clusters under anaerobic conditions. When the reaction solution is exposed to air, the reduced mitoNEET [2Fe-2S] clusters are quickly oxidized (Figure 2B, spectrum 3). However, when additional NADH is added to the reaction solution, the mitoNEET [2Fe-2S] clusters are re-reduced under aerobic conditions (Figure 2A). The results suggest that under aerobic conditions, a continuous supply of NADH is required for maintaining the reduced state of the mitoNEET [2Fe-2S] clusters in the presence of flavin reductase and flavin nucleotide. Thus, the redox state of the mitoNEET [2Fe-2S] clusters may be controlled by the reduced redox co-enzymes in cells under aerobic conditions.
Figure 2. Redox transition of the mitoNEET [2Fe-2S] clusters mediated by flavin nucleotides.
A), redox transition of the mitoNEET [2Fe-2S] clusters. MitoNEET (containing 10 µM [2Fe-2S] clusters) was incubated with FMN (1 µM) and NADH (40 µM) under anaerobic conditions. Addition of Fre (1 µM) rapidly reduced the mitoNEET [2Fe-2S] clusters. Upon exposure to air, the reduced mitoNEET [2Fe-2S] clusters were quickly re-oxidized. Addition of additional NADH (100 µM) to the incubation solution under aerobic conditions re-reduced the mitoNEET [2Fe-2S] clusters. The absorption peak at 458 nm indicates the oxidized [2Fe-2S] clusters of mitoNEET. The peak at 420 nm indicates the reduced [2Fe-2S] clusters of mitoNEET. B), UV-visible absorption spectra of the reaction solution at different time points. Spectrum 1, mitoNEET was incubated with FMN (1 µM) and NADH (40 µM). Spectrum 2, at 15 seconds after addition of Fre (1 µM). Spectrum 3, at 4 min after exposure to air. Data are representative of three independent experiments.
3. Flavin nucleotides act as electron shuttles mediating reduction of the mitoNEET [2Fe-2S] clusters
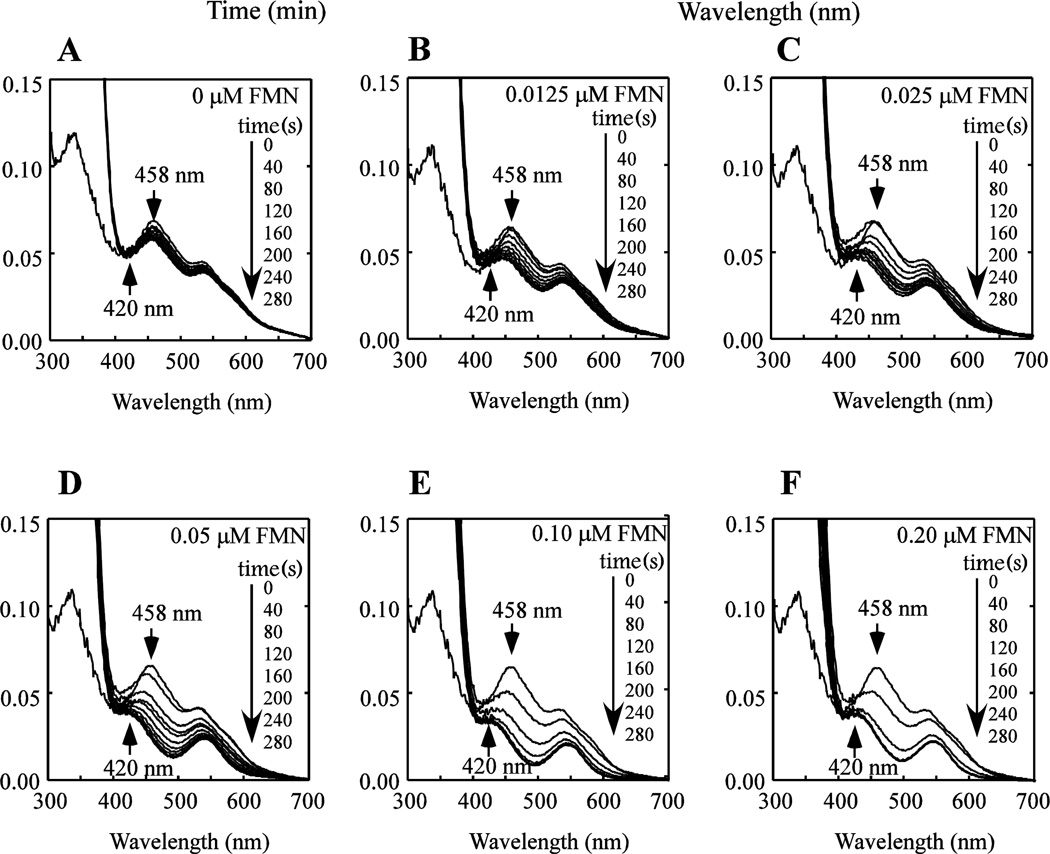
Flavin reductase reduces flavin nucleotides using NADH as electron donor without tightly binding its substrates or products [39]. We assume that flavin nucleotides reduced by flavin reductase will diffuse from the enzyme to reduce the [2Fe-2S] clusters in mitoNEET. To explore this idea, mitoNEET (containing 10 µM [2Fe-2S] cluster) is pre-incubated with Fre (1 µM) and various concentrations of FMN under aerobic conditions. The reduction of the mitoNEET [2Fe-2S] clusters in solutions is initiated by adding NADH (100 µM). Figure 3A shows that without FMN, there is no reduction of the mitoNEET [2Fe-2S] clusters. As the concentration of FMN is increased from 0 to 0.2 µM in the incubation solution (Figure 3B–F), the mitoNEET [2Fe-2S] clusters are quickly reduced under aerobic conditions.
Figure 3. Reduction of the mitoNEET [2Fe-2S] clusters in the presence of different concentrations of FMN.
MitoNEET (containing 10 µM [2Fe-2S] clusters) was pre-incubated with E. coli Fre (1 µM) and different concentrations of FMN under aerobic conditions. NADH (50 µM) was then added to initiate the reaction. UV-Visible absorption spectra were taken every 40 seconds for 280 seconds after addition of Fre. FMN concentration in the reaction solution was: 0 µM (A), 0.0125 µM (B), 0.025 µM (C), 0.05 µM (D), 0.10 µM (E), and 0.20 µM (F). The absorption peak at 458 nm indicates the oxidized [2Fe-2S] clusters of mitoNEET. The peak at 420 nm indicates the reduced [2Fe-2S] clusters of mitoNEET. The results are the representative of three independent experiments.
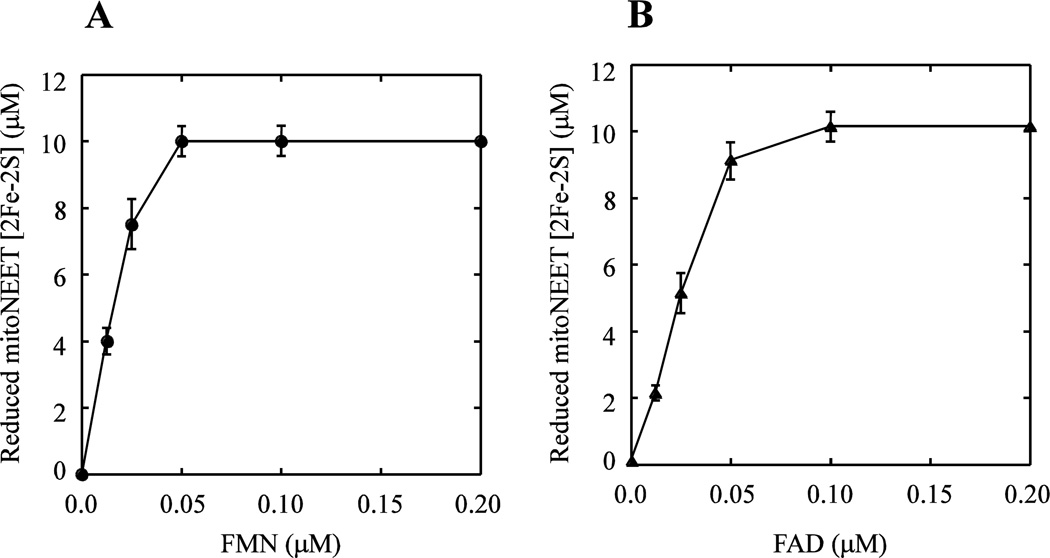
The amounts of the reduced mitoNEET [2Fe-2S] clusters are measured after 4 min incubation with Fre, NADH and various concentrations of FMN or FAD, and plotted as a function of the flavin nucleotide concentrations. Figure 4A shows that about 0.1 µM FMN is sufficient to fully reduce 10 µM mitoNEET [2Fe-2S] clusters after 4 min incubation. Similar results are observed when FMN is replaced with FAD in the reaction solutions (Figure 4B). These results suggest that flavin nucleotides may act as electron shuttles to efficiently reduce the mitoNEET [2Fe-2S] clusters under aerobic conditions.
Figure 4. Titration of flavin nucleotides in reduction of the mitoNEET [2Fe-2S] clusters.
A), titration of FMN. MitoNEET (containing 10 µM [2Fe-2S] clusters) was pre-incubated with E. coli Fre (1 µM) and different concentrations of FMN under aerobic conditions. NADH (50 µM) was then added to initiate the reaction. After 4 min incubation, amounts of the reduced mitoNEET [2Fe-2S] clusters were measured from the UV-visible absorption spectra and plotted as a function of FMN concentrations in the reaction solution. B), titration of FAD. Same as in A), except FMN is replaced with FAD in the reaction solutions. The data represent the averages with standard deviation from three independent experiments.
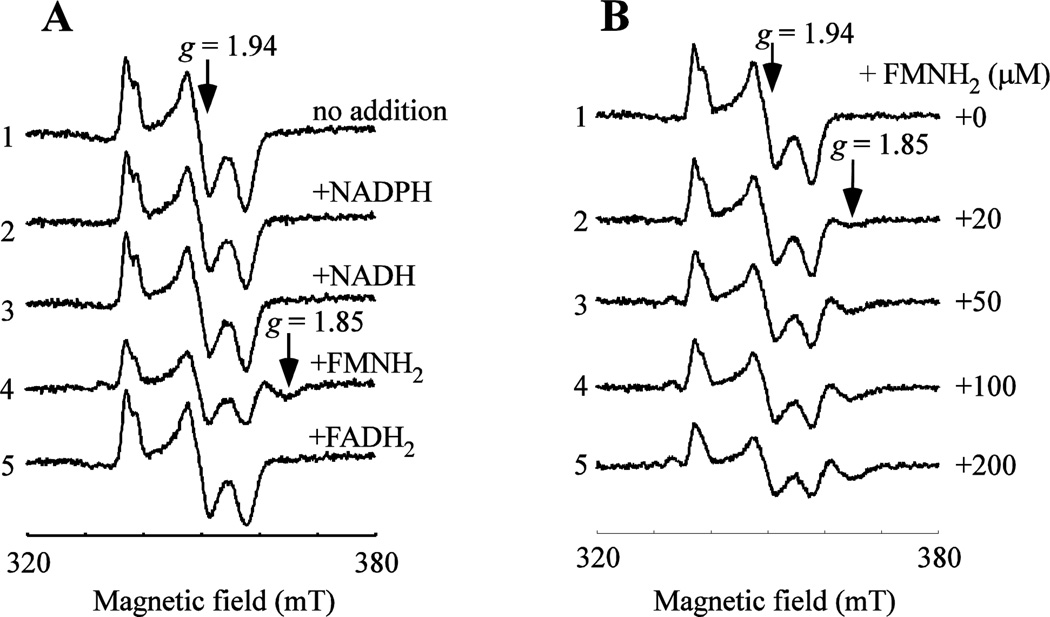
4. EPR evidence for flavin mononucleotide binding in mitoNEET
As a catalytic amount of flavin nucleotides is sufficient for reduction of the mitoNEET [2Fe-2S] clusters (Figure 4), we postulate that mitoNEET may have specific interactions with flavin nucleotides. In the experiments, mitoNEET is pre-incubated with 10-fold excess of NADP+, NAD+, FAD, or FMN in buffer containing 500 mM NaCl and 20 mM Tris (pH 8.0) for 30 min under aerobic conditions. The samples are then reduced with freshly prepared sodium dithionite to fully reduce the redox co-enzyme and the mitoNEET [2Fe-2S] clusters for the EPR measurements, as only the reduced mitoNEET [2Fe-2S] clusters are paramagnetic and EPR visible [23, 40]. Any change of the EPR spectrum would reflect potential interactions between the reduced co-enzyme and the reduced [2Fe-2S] clusters in mitoNEET. Figure 5A shows that NADPH and NADH has very little or no effects on the EPR spectrum of the reduced mitoNEET [2Fe-2S] clusters (spectra 2–3), consistent with the idea that NADPH and NADH have a weak interaction with mitoNEET [29]. In contrast, incubation with FMNH2 produces a new EPR signal at g = 1.85 (spectrum 4), suggesting that FMNH2 may have a specific interaction with the reduced [2Fe-2S] clusters in mitoNEET. Interestingly, incubation with FADH2 does not have any significant effects on the EPR spectrum of the reduced mitoNEET [2Fe-2S] clusters (spectrum 5). Perhaps, the AMP moiety in FAD may obstruct the interaction between FADH2 and the [2Fe-2S] clusters in mitoNEET.
Figure 5. MitoNEET has a specific interaction with FMN.
A), effect of the reduced co-enzymes on the EPR spectrum of the reduced mitoNEET [2Fe-2S] clusters. MitoNEET (containing 10 µM [2Fe-2S] clusters) was pre-incubated with no addition (spectrum 1), NADP+ (spectrum 2), NAD+ (spectrum 3), FMN (spectrum 4), or FAD (spectrum 5) in buffer containing 500 mM NaCl and 20 mM Tris (pH 8.0) at room temperature for 30 min. The concentration of redox co-enzymes in each incubation solution was 100 µM. After incubation, the samples were reduced with freshly prepared sodium dithionite (4 mM) and immediately frozen in liquid nitrogen. The EPR signal at g = 1.94 represents the reduced mitoNEET [2Fe-2S] clusters. Incubation with FMN produced a new EPR signal at g = 1.85 of the reduced mitoNEET [2Fe-2S] clusters. B), EPR spectra of the mitoNEET [2Fe-2S] clusters in the presence of different concentrations of the reduced FMN. MitoNEET (containing 10 µM [2Fe-2S] clusters) was pre-incubated with increasing concentrations of FMN at room temperature for 30 min. After pre-incubation, the samples were reduced with freshly prepared sodium dithionite (4 mM) and immediately frozen in liquid nitrogen for EPR measurements. Data are representative of three independent experiments.
MitoNEET is then incubated with increasing concentrations of FMN under aerobic conditions, followed by reduction with freshly prepared sodium dithionite. Figure 5B shows that as the concentration of FMNH2 increases from 0 to 200 µM, the amplitude of the EPR signal at g = 1.85 gradually increases with concomitant decrease of the EPR signal at g = 1.94. Integration of the new EPR signal at g = 1.85 shows when 10 µM mitoNEET [2Fe-2S] clusters are incubated with 200 µM FMNH2, about 4 µM mitoNEET [2Fe-2S] clusters are converted to the ones with the EPR signal at g = 1.85. The relatively weak interaction could be due to the fact that both FMN and the [2Fe-2S] clusters in mitoNEET are in reduced state. However, attempts to isolate the mitoNEET-FMN complex from the incubation solutions were not successful. It appears that mitoNEET does not form a stable complex with FMN, similar as flavin reductase [38]. Nevertheless, the new EPR signal at g = 1.85 of the reduced mitoNEET [2Fe-2S] clusters at high concentrations of FMNH2 clearly suggests that mitoNEET has a specific interaction with FMN.
5. Molecular docking for the flavin mononucleotide binding in mitoNEET
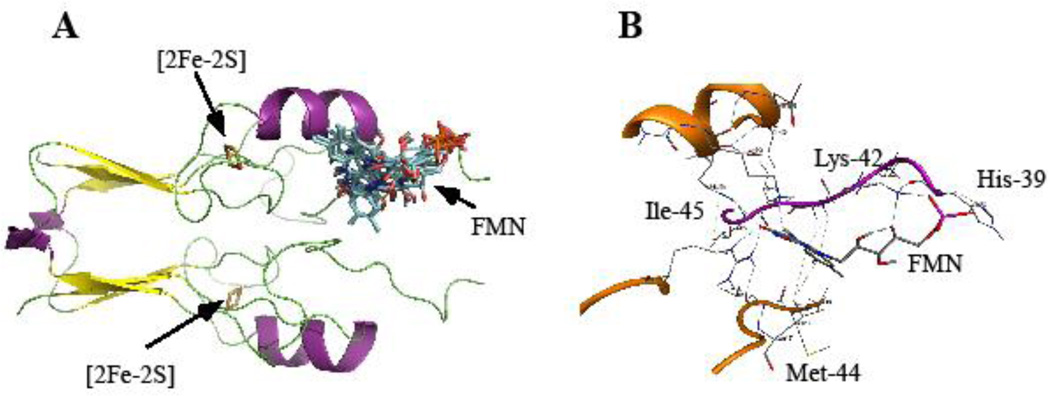
To further explore the FMN binding in mitoNEET, we have used molecular docking approach. Our protocol for AutoDockVina-based Virtual Screening [37] is a modification from Garret M. Morris’ (The Scripps Research Institute, La Jolla, CA) [36]. The best docking models for the FMN binding in mitoNEET are shown in Figure 6A. For clarity, only one monomer in the mitoNEET dimer is shown with the bound FMN molecule. Each conformation of FMN in mitoNEET represents one docking model. In all docking models obtained, FMN always binds mitoNEET at the region between the transmembrane α-helix domain (not shown in Figure 6A) and the [2Fe-2S] cluster domain. The shortest distance between the [2Fe-2S] cluster and the bound FMN in mitoNEET is about 10 Å. A close examination of the FMN binding site in mitoNEET shows that His-39 and Lys-42 from chain 1 form electrostatic interactions with the phosphate group of FMN, while Ile-45 from chain 1 and Met-44 from chain 2 form hydrophobic interactions with the isoalloxazine group of FMN (Figure 6B).
Figure 6. Molecular docking models of FMN binding in mitoNEET.
A), docking models of FMN binding in mitoNEET (PDB: 2QH7, Paddock et al. 2007). For clarity, only one monomer in the mitoNEET dimer was shown with the bound FMN molecule. B), predicted FMN binding site in mitoNEET. The phosphate group of FMN has electrostatic interactions with Lys-42 and His-39, while the isoalloxazine group of FMN forms hydrophobic interactions with Met-44 and Ile-45. Dotted lines represent hydrogen bonds in the mitoNEET-FMN complex.
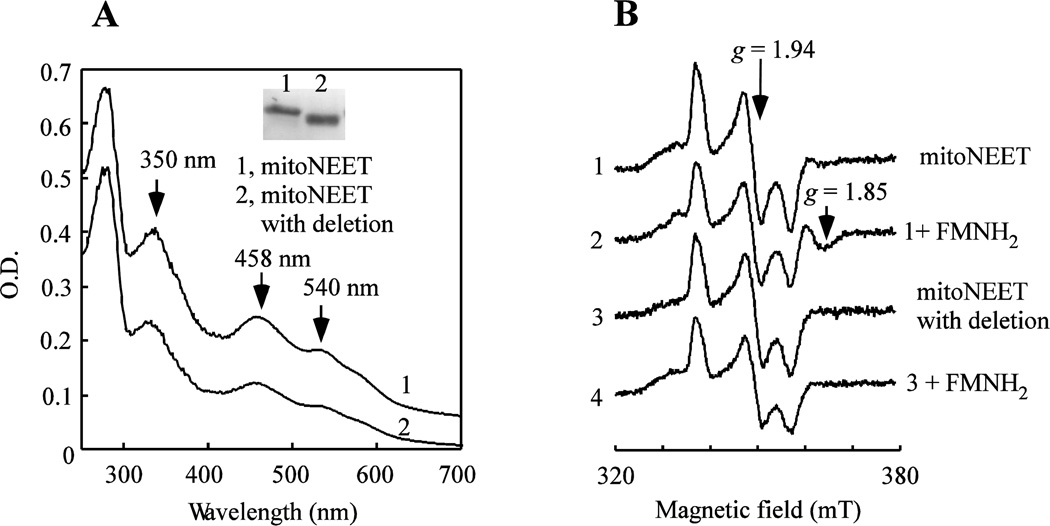
To test potential role of the region between Arg-33 and Ile-45 of mitoNEET in binding FMN, we have constructed a mitoNEET mutant in which the residues between Arg-33 and Ile-45 are in-frame deleted. Deletion of the region between Arg-33 and Ile-45 has only a minor effect on the UV-visible absorption spectrum of the [2Fe-2S] clusters in the protein (Figure 7A). However, the deletion largely abolishes the specific interaction between FMNH2 and the reduced [2Fe-2S] clusters in mitoNEET (Figure 7B), suggesting that the region between Arg-33 and Ile-45 is at least partially responsible for the FMN binding in mitoNEET.
Figure 7. Deletion of the residues between Arg-33 and Ile-45 diminishes the interaction between mitoNEET and the reduced FMN.
MitoNEET with deletion of the region between Arg-33 and Ile-45 was prepared using the site-directed mutagenesis. A), UV-visible absorption spectra of mitoNEET and mitoNEET mutant with the deletion. Spectrum 1, mitoNEET. Spectrum 2, mitoNEET mutant with the deletion. Insert is a photograph of the SDS-PAGE gel of mitoNEET and mitoNEET mutant with the deletion. Lane 1, mitoNEET. Lane 2, mitoNEET mutant with the deletion. B), EPR spectra of mitoNEET and mitoNEET mutant with the deletion after incubation with or without FMNH2. Purified proteins (containing 10 µM [2Fe-2S] clusters) were incubated with FMN (100 µM) for 30 min under aerobic conditions, followed by reduction with freshly prepared sodium dithionite (4 mM). The samples were then subjected to the EPR measurements. Spectrum 1, mitoNEET. Spectrum 2, mitoNEET after incubation with FMNH2. Spectrum 3, mitoNEET mutant. Spectrum 4, mitoNEET mutant with deletion after incubation with FMNH2. Data are representative of three independent experiments.
DISCUSSION
The [2Fe-2S] clusters in mitochondrial outer membrane protein mitoNEET are redox active with a redox midpoint potential (Em7) of 0 mV [21, 24]. However, cellular components responsible for reducing the mitoNEET [2Fe-2S] clusters are not fully understood. In previous studies, we reported that the mitoNEET [2Fe-2S] clusters can be reduced by biological thiols [22] or human glutathione reductase [23]. However, reduction of the mitoNEET [2Fe-2S] clusters mediated by biological thiols [22] or human glutathione reductase [23] is slow, and must be carried out under anaerobic conditions [22, 23]. Here, we find that the reduced flavin nucleotides reduced by flavin reductase and NADH can rapidly reduce the mitoNEET [2Fe-2S] clusters under aerobic conditions (Figure 1). Flavins are ubiquitous redox co-enzymes in cells [41]. While flavin contents vary in different cell lines, riboflavin (3.1–5.7 amol/cell) and FAD (12–26 amol/cell) are found to be predominant flavins with a relatively lower content of FMN (2.7–3.4 atom/cell) in various cell lines [42]. Although riboflavin is considered the precursor of flavin nucleotides [41], we found that the reduced riboflavin, like the reduced FMN and FAD, can also rapidly reduce the mitoNEET [2Fe-2S] clusters under aerobic conditions (data not shown). In this context, we propose that flavin and flavin nucleotides may act as electron shuttles to mediate reduction of the mitoNEET [2Fe-2S] clusters, and that the redox state of the mitoNEET [2Fe-2S] clusters is directly controlled by the reduced flavin and flavin nucleotides.
Unlike the reduced flavin nucleotides, NADH or NADPH fails to reduce the mitoNEET [2Fe-2S] clusters under aerobic or anaerobic conditions [22, 23, 30]. However, we find that NADH can provide electrons to reduce flavin nucleotides via flavin reductase (Figure 1). When NADH is exhausted, the reduced mitoNEET [2Fe-2S] clusters are quickly oxidized by oxygen under aerobic conditions (Figure 2). Thus, the redox state of the mitoNEET [2Fe-2S] clusters may be ultimately controlled by intracellular NADH. In human cells, high glycolytic activity will lead to accumulation of NADH [28]. Excess NADH may be used by flavin reductase [43] to reduce flavin nucleotides which in turn reduce the mitoNEET [2Fe-2S] clusters on mitochondrial outer membrane. As the redox transition of the [2Fe-2S] clusters in mitoNEET is expected to modulate energy metabolism, iron homeostasis and production of reactive oxygen species in mitochondria [13], mitoNEET may act as a crucial link between the redox state of intracellular co-enzymes and mitochondrial functions.
The salient finding of this work is the specific interaction between FMN and mitoNEET. The observation that a catalytic amount of flavin nucleotides is sufficient to reduce the mitoNEET [2Fe-2S] clusters (Figure 4) led us to postulate that the reduced flavin nucleotides may have a specific interaction with mitoNEET in order to rapidly reduce the oxidized [2Fe-2S] clusters in mitoNEET. The EPR measurements show that incubation with the reduced FMN, but not the reduced FAD, indeed produces a new EPR signal at g = 1.85 of the reduced mitoNEET [2Fe-2S] clusters (Figure 5). However, excess amounts of the reduced FMN are required to produce the EPR signal at g = 1.85 of the reduced mitoNEET [2Fe-2S] clusters, indicating that binding of the reduced FMN for mitoNEET is not very strong. It could be that both FMA and the [2Fe-2S] clusters in mitoNEET are reduced. However, attempts to isolate the mitoNEET-FMN complex under various redox states were not successful. Thus, FMN binding in mitoNEET could be transient, similar to that in flavin reductase [38]. We postulate that FMNH2 may rapidly reduce the oxidized [2Fe-2S] clusters in mitoNEET. Upon oxidation, FMN will have much lower binding affinity for mitoNEET and dissociate from mitoNEET. Nevertheless, at high concentrations of FMN, the new EPR signal at g = 1.85 (Figure 5) clearly suggests that mitoNEET has a specific interaction with FMN.
Based on the molecular docking models (Figure 6), FMN most likely binds mitoNEET at the region between the transmembrane α-helix and the [2Fe-2S] cluster binding domain. The shortest distance between the isoalloxazine group of FMN and the [2Fe-2S] cluster in mitoNEET is about 10 Å (Figure 6A). Such an arrangement would allow rapid electron transfer from the reduced FMN to the oxidized [2Fe-2S] cluster in mitoNEET [44]. As reduction of the [2Fe-2S] cluster in mitoNEET is a single-electron event, reduced flavin nucleotide must form a semiquinone intermediate after delivering one electron to the [2Fe-2S] cluster in mitoNEET. However, we were unable to detect any semiquinone signals using EPR (data not shown). A simplest explanation could be that semiquinone intermediates may quickly reduce the other [2Fe-2S] cluster in the mitoNEET dimer (Figure 6A). Evidently, additional experiments are required to illustrate the mechanism by which the mitoNEET [2Fe-2S] clusters are reduced by the reduced flavin nucleotides.
Using molecular docking approaches, Geldenhuys et al. previously identified two major binding sites in mitoNEET for type II diabetes drugs rosiglitazone and pioglitazone [45]. Site 1 consists of Lys-42 and Ala-43 from chain 1 and Met-44, Leu-47, Arg-76, His-90, and Thr-94 from chain 2, while site 2 consists of His-48, Ile-49, Trp-75, Arg-76, Ser-77, and Lys-78 from chain 1 [45]. The FMN binding site in mitoNEET shown in Figure 6A is distinct from site 1 and site 2 for type II diabetes drugs rosiglitazone and pioglitazone [45], but has some significant overlap with site 1. Because of the high insolubility of pioglitazone, the dissociation constant of the interaction between mitoNEET and pioglitazone was not determined (36). A more soluble pioglitazone analog, NL-1, has been shown to bind mitoNEET with a dissociation consistent of 7.4 µM (6). It is expected that pioglitazone or NL-1 will compete with flavin nucleotides for binding in mitoNEET. In previous studies, we reported that pioglitazone inhibits the reduction of the mitoNEET [2Fe-2S] clusters mediated by biological thiols [22] or by human glutathione reductase [23] under anaerobic conditions. Here, we have also found that pioglitazone and NL-1 can effectively inhibit the reduction of the mitoNEET [2Fe-2S] clusters mediated by the reduced flavin nucleotides under aerobic conditions (Wang & Ding, unpublished data). Further investigations on competition between pioglitazone or its analogs and flavin nucleotides for binding in mitoNEET may lead to discovery of new drugs that target mitoNEET to modulate energy metabolism in mitochondria.
Highlights.
A mechanism for redox transition of the mitoNEET [2Fe-2S] clusters is proposed.
Reduced flavin nucleotides can rapidly reduce the mitoNEET [2Fe-2S] clusters.
Flavin mononucleotide appears to have a specific interaction with mitoNEET.
Acknowledgments
Research reported in this publication was supported in part by the National Institutes of Health [Grant number R15GM109399] and by the American Heart Association [Grant number 13GRNT16890014].
List of Abbreviations
- Em7
redox midpoint potential at pH 7.0
- EPR
electron paramagnetic resonance
- FMN
flavin mononucleotide
- FAD
flavin adenine dinucleotide
- Fre
E. coli flavin reductase
- NADH
nicotinamide adenine dinucleotide
- NADPH
nicotinamide adenine dinucleotide phosphate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Colca JR, McDonald WG, Waldon DJ, Leone JW, Lull JM, Bannow CA, Lund ET, Mathews WR. Identification of a novel mitochondrial protein ("mitoNEET") cross-linked specifically by a thiazolidinedione photoprobe. Am J Physiol Endocrinol Metab. 2004;286:E252–E260. doi: 10.1152/ajpendo.00424.2003. [DOI] [PubMed] [Google Scholar]
- 2.Wiley SE, Murphy AN, Ross SA, van der Geer P, Dixon JE. MitoNEET is an iron-containing outer mitochondrial membrane protein that regulates oxidative capacity. Proc Natl Acad Sci U S A. 2007;104:5318–5323. doi: 10.1073/pnas.0701078104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kusminski CM, Holland WL, Sun K, Park J, Spurgin SB, Lin Y, Askew GR, Simcox JA, McClain DA, Li C, Scherer PE. MitoNEET-driven alterations in adipocyte mitochondrial activity reveal a crucial adaptive process that preserves insulin sensitivity in obesity. Nat Med. 2012;18:1539–1549. doi: 10.1038/nm.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kusminski CM, Chen S, Ye R, Sun K, Wang QA, Spurgin SB, Sanders PE, Brozinick JT, Geldenhuys WJ, Li WH, Unger RH, Scherer PE. MitoNEET-Parkin Effects in Pancreatic alpha- and beta-Cells, Cellular Survival, and Intrainsular Cross Talk. Diabetes. 2016;65:1534–1555. doi: 10.2337/db15-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yonutas HM, Sullivan PG. Targeting PPAR isoforms following CNS injury. Current drug targets. 2013;14:733–742. doi: 10.2174/1389450111314070003. [DOI] [PubMed] [Google Scholar]
- 6.Geldenhuys WJ, Leeper TC, Carroll RT. MitoNEET as a novel drug target for mitochondrial dysfunction. Drug discovery today. 2014;19:1601–1606. doi: 10.1016/j.drudis.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Logan SJ, Yin L, Geldenhuys WJ, Enrick MK, Stevanov KM, Carroll RT, Ohanyan VA, Kolz CL, Chilian WM. Novel thiazolidinedione mitoNEET ligand-1 acutely improves cardiac stem cell survival under oxidative stress. Basic Res Cardiol. 2015;110:19. doi: 10.1007/s00395-015-0471-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bai F, Morcos F, Sohn YS, Darash-Yahana M, Rezende CO, Lipper CH, Paddock ML, Song L, Luo Y, Holt SH, Tamir S, Theodorakis EA, Jennings PA, Onuchic JN, Mittler R, Nechushtai R. The Fe-S cluster-containing NEET proteins mitoNEET and NAF-1 as chemotherapeutic targets in breast cancer. Proc Natl Acad Sci U S A. 2015;112:3698–3703. doi: 10.1073/pnas.1502960112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sohn YS, Tamir S, Song L, Michaeli D, Matouk I, Conlan AR, Harir Y, Holt SH, Shulaev V, Paddock ML, Hochberg A, Cabanchick IZ, Onuchic JN, Jennings PA, Nechushtai R, Mittler R. NAF-1 and mitoNEET are central to human breast cancer proliferation by maintaining mitochondrial homeostasis and promoting tumor growth. Proc Natl Acad Sci U S A. 2013;110:14676–14681. doi: 10.1073/pnas.1313198110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salem AF, Whitaker-Menezes D, Howell A, Sotgia F, Lisanti MP. Mitochondrial biogenesis in epithelial cancer cells promotes breast cancer tumor growth and confers autophagy resistance. Cell Cycle. 2012;11:4174–4180. doi: 10.4161/cc.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kusminski CM, Park J, Scherer PE. MitoNEET-mediated effects on browning of white adipose tissue. Nat Commun. 2014;5:3962. doi: 10.1038/ncomms4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wright MB, Bortolini M, Tadayyon M, Bopst M. Minireview: Challenges and opportunities in development of PPAR agonists. Mol Endocrinol. 2014;28:1756–1768. doi: 10.1210/me.2013-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamir S, Paddock ML, Darash-Yahana-Baram M, Holt SH, Sohn YS, Agranat L, Michaeli D, Stofleth JT, Lipper CH, Morcos F, Cabantchik IZ, Onuchic JN, Jennings PA, Mittler R, Nechushtai R. Structure-function analysis of NEET proteins uncovers their role as key regulators of iron and ROS homeostasis in health and disease. Biochim Biophys Acta. 2015;1853:1294–1315. doi: 10.1016/j.bbamcr.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 14.Paddock ML, Wiley SE, Axelrod HL, Cohen AE, Roy M, Abresch EC, Capraro D, Murphy AN, Nechushtai R, Dixon JE, Jennings PA. MitoNEET is a uniquely folded 2Fe 2S outer mitochondrial membrane protein stabilized by pioglitazone. Proc Natl Acad Sci U S A. 2007;104:14342–14347. doi: 10.1073/pnas.0707189104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin J, Zhou T, Ye K, Wang J. Crystal structure of human mitoNEET reveals distinct groups of iron–sulfur proteins. Proceedings of the National Academy of Sciences. 2007;104:14640–14645. doi: 10.1073/pnas.0702426104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou X, Liu R, Ross S, Smart EJ, Zhu H, Gong W. Crystallographic studies of human MitoNEET. J Biol Chem. 2007;282:33242–33246. doi: 10.1074/jbc.C700172200. [DOI] [PubMed] [Google Scholar]
- 17.Paul VD, Lill R. Biogenesis of cytosolic and nuclear iron-sulfur proteins and their role in genome stability. Biochim Biophys Acta. 2015;1853:1528–1539. doi: 10.1016/j.bbamcr.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 18.Zuris JA, Harir Y, Conlan AR, Shvartsman M, Michaeli D, Tamir S, Paddock ML, Onuchic JN, Mittler R, Cabantchik ZI, Jennings PA, Nechushtai R. Facile transfer of [2Fe-2S] clusters from the diabetes drug target mitoNEET to an apo-acceptor protein. Proc Natl Acad Sci U S A. 2011;108:13047–13052. doi: 10.1073/pnas.1109986108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferecatu I, Goncalves S, Golinelli-Cohen MP, Clemancey M, Martelli A, Riquier S, Guittet E, Latour JM, Puccio H, Drapier JC, Lescop E, Bouton C. The Diabetes Drug Target MitoNEET Governs a Novel Trafficking Pathway to Rebuild an Fe-S Cluster into Cytosolic Aconitase/Iron Regulatory Protein 1. J Biol Chem. 2014;289:28070–28086. doi: 10.1074/jbc.M114.548438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baxter EL, Zuris JA, Wang C, Vo PL, Axelrod HL, Cohen AE, Paddock ML, Nechushtai R, Onuchic JN, Jennings PA. Allosteric control in a metalloprotein dramatically alters function. Proc Natl Acad Sci U S A. 2013;110:948–953. doi: 10.1073/pnas.1208286110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golinelli-Cohen MP, Lescop E, Mons C, Goncalves S, Clemancey M, Santolini J, Guittet E, Blondin G, Latour JM, Bouton C. Redox Control of the Human Iron-Sulfur Repair Protein MitoNEET Activity via Its Iron-Sulfur Cluster. J Biol Chem. 2016;291:7583–7593. doi: 10.1074/jbc.M115.711218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landry AP, Ding H. Redox Control of Human Mitochondrial Outer Membrane Protein MitoNEET [2Fe-2S] Clusters by Biological Thiols and Hydrogen Peroxide. J Biol Chem. 2014;289:4307–4315. doi: 10.1074/jbc.M113.542050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landry AP, Cheng Z, Ding H. Reduction of mitochondrial protein mitoNEET [2Fe-2S] clusters by human glutathione reductase. Free Radic Biol Med. 2015;81:119–127. doi: 10.1016/j.freeradbiomed.2015.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bak DW, Zuris JA, Paddock ML, Jennings PA, Elliott SJ. Redox characterization of the FeS protein MitoNEET and impact of thiazolidinedione drug binding. Biochemistry. 2009;48:10193–10195. doi: 10.1021/bi9016445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarraf SA, Raman M, Guarani-Pereira V, Sowa ME, Huttlin EL, Gygi SP, Harper JW. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 2013;496:372–376. doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kusminski CM, Chen S, Ye R, Sun K, Wang QA, Spurgin SB, Sanders PE, Brozinick JT, Li WH, Unger RH, Scherer PE. MitoNEET-Parkin Effects in Pancreatic alpha- and beta-Cells, Cellular Survival and Intra-Insular Crosstalk. Diabetes. 2016 doi: 10.2337/db15-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Havugimana PC, Hart GT, Nepusz T, Yang H, Turinsky AL, Li Z, Wang PI, Boutz DR, Fong V, Phanse S, Babu M, Craig SA, Hu P, Wan C, Vlasblom J, Dar VU, Bezginov A, Clark GW, Wu GC, Wodak SJ, Tillier ER, Paccanaro A, Marcotte EM, Emili A. A census of human soluble protein complexes. Cell. 2012;150:1068–1081. doi: 10.1016/j.cell.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heikal AA. Intracellular coenzymes as natural biomarkers for metabolic activities and mitochondrial anomalies. Biomarkers in medicine. 2010;4:241–263. doi: 10.2217/bmm.10.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou T, Lin J, Feng Y, Wang J. Binding of reduced nicotinamide adenine dinucleotide phosphate destabilizes the iron-sulfur clusters of human mitoNEET. Biochemistry. 2010;49:9604–9612. doi: 10.1021/bi101168c. [DOI] [PubMed] [Google Scholar]
- 30.Zuris JA, Ali SS, Yeh H, Nguyen TA, Nechushtai R, Paddock ML, Jennings PA. NADPH inhibits [2Fe-2S] cluster protein transfer from diabetes drug target MitoNEET to an apo-acceptor protein. J Biol Chem. 2012;287:11649–11655. doi: 10.1074/jbc.M111.319731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. Complete set of ORF clones of Escherichia coli ASKA library (A Complete Set of E. coli K-12 ORF Archive): Unique Resources for Biological Research. DNA Research. 2006;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 32.Cowart RE, Singleton FL, Hind JS. A comparison of bathophenanthrolinedisulfonic acid and ferrozine as chelators of iron(II) in reduction reactions. Anal Biochem. 1993;211:151–155. doi: 10.1006/abio.1993.1246. [DOI] [PubMed] [Google Scholar]
- 33.Ding H, Harrison K, Lu J. Thioredoxin reductase system mediates iron binding in IscA and iron delivery for the iron-sulfur cluster assembly in IscU. Journal of Biological Chemistry. 2005;280:30432–30437. doi: 10.1074/jbc.M504638200. [DOI] [PubMed] [Google Scholar]
- 34.Siegel LM. A Direct Microdetermination of Sulfide. Anal Biochem. 1965;11:126–132. doi: 10.1016/0003-2697(65)90051-5. [DOI] [PubMed] [Google Scholar]
- 35.Rogers PA, Eide L, Klungland A, Ding H. Reversible inactivation of E. coli endonuclease III by nitric oxide via modification of its [4Fe-4S] cluster. DNA Repair. 2003;2:809–817. doi: 10.1016/s1568-7864(03)00065-x. [DOI] [PubMed] [Google Scholar]
- 36.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of computational chemistry. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crochet RB, Cavalier MC, Seo M, Kim JD, Yim YS, Park SJ, Lee YH. Investigating combinatorial approaches in virtual screening on human inducible 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB3): a case study for small molecule kinases. Anal Biochem. 2011;418:143–148. doi: 10.1016/j.ab.2011.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niviere V, Fieschi F, Decout JL, Fontecave M. The NAD(P)H:flavin oxidoreductase from Escherichia coli. Evidence for a new mode of binding for reduced pyridine nucleotides. J Biol Chem. 1999;274:18252–18260. doi: 10.1074/jbc.274.26.18252. [DOI] [PubMed] [Google Scholar]
- 39.Fieschi F, Niviere V, Frier C, Decout JL, Fontecave M. The mechanism and substrate specificity of the NADPH:flavin oxidoreductase from Escherichia coli. J Biol Chem. 1995;270:30392–30400. doi: 10.1074/jbc.270.51.30392. [DOI] [PubMed] [Google Scholar]
- 40.Iwasaki T, Samoilova RI, Kounosu A, Ohmori D, Dikanov SA. Continuous-wave and pulsed EPR characterization of the [2Fe-2S](Cys)3(His)1 cluster in rat MitoNEET. J Am Chem Soc. 2009;131:13659–13667. doi: 10.1021/ja903228w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edwards AM. Structure and general properties of flavins. Methods Mol Biol. 2014;1146:3–13. doi: 10.1007/978-1-4939-0452-5_1. [DOI] [PubMed] [Google Scholar]
- 42.Huhner J, Ingles-Prieto A, Neususs C, Lammerhofer M, Janovjak H. Quantification of riboflavin, flavin mononucleotide, and flavin adenine dinucleotide in mammalian model cells by CE with LED-induced fluorescence detection. Electrophoresis. 2015;36:518–525. doi: 10.1002/elps.201400451. [DOI] [PubMed] [Google Scholar]
- 43.Smith LJ, Browne S, Mulholland AJ, Mantle TJ. Computational and experimental studies on the catalytic mechanism of biliverdin-IXbeta reductase. Biochem J. 2008;411:475–484. doi: 10.1042/BJ20071495. [DOI] [PubMed] [Google Scholar]
- 44.Moser CC, Keske JM, Warncke K, Farid RS, Dutton PL. Nature of biological electron transfer. Nature. 1992;355:796–802. doi: 10.1038/355796a0. [DOI] [PubMed] [Google Scholar]
- 45.Geldenhuys WJ, Funk MO, Barnes KF, Carroll RT. Structure-based design of a thiazolidinedione which targets the mitochondrial protein mitoNEET. Bioorg Med Chem Lett. 2010;20:819–823. doi: 10.1016/j.bmcl.2009.12.088. [DOI] [PubMed] [Google Scholar]