Abstract
Scope
Heat-stabilized rice bran and cooked navy bean powder contain a variety of phytochemicals that are fermented by colonic microbiota and may influence intestinal health. Dietary interventions with these foods should be explored for modulating colorectal cancer risk.
Methods and results
A randomized-controlled pilot clinical trial investigated the effects of eating heat-stabilized rice bran (30g/day) or cooked navy bean powder (35g/day) on gut microbiota and metabolites (NCT01929122). Twenty-nine overweight/obese volunteers with a prior history of colorectal cancer consumed a study-provided meal and snack daily for 28 days. Volunteers receiving rice bran or bean powder showed increased gut bacterial diversity and altered gut microbial composition at 28 days compared to baseline. Supplementation with rice bran or bean powder increased total dietary fiber intake similarly, yet only rice bran intake led to a decreased Firmicutes:Bacteroidetes ratio and increased short chain fatty acids (propionate and acetate) in stool after 14 days but not at 28 days.
Conclusion
These findings support modulation of gut microbiota and fermentation by-products by heat-stabilized rice bran and suggest that foods with similar ability to increase dietary fiber intake may not have equal effects on gut microbiota and microbial metabolism.
Keywords: stool microbiota, short chain fatty acids, bile acids, heat-stabilized rice bran, navy bean
Introduction
Diet is a well-established risk determinant in the development of sporadic (non-hereditary) colorectal cancer (CRC), which is thought to make up as much as 95% of all cases of the disease [1]. Since CRC has the second highest mortality rate of all cancers and is the third most common cancer in the US, nutritional intervention for prevention of CRC has particular value to reduce disease recurrence and mortality [2, 3]. A large body of research associates diets low in whole grains and fiber with increased risk of CRC [1] and diets enriched with legumes and whole grains with reduced risk [4–9]. The underlying protective mechanisms mediated by these foods are thought to involve both fiber and polyphenols. Fiber is composed of polysaccharides and provides structure to the plant while polyphenols defend against UV radiation, herbivores, and infection [10, 11]. Upon consumption, fiber provides the major energy source for microbial fermentation in the colon, which in turn stimulates the catabolism of plant polyphenols. An increase in polyphenol bioavailability results from bacterial enzymes that depolymerize, deglycosylate, and hydrolyze complex polyphenols into smaller phenolic acids capable of crossing the intestinal barrier [12]. Given the importance of these microbial functions it is likely that modulation of the gut microbiota and their metabolic processes is an important mechanism by which whole grains and legumes contribute to CRC prevention.
A growing body of recent research focuses on the role of the gut microbiota in the CRC disease process [13]. Multiple studies report different gut microbial communities in individuals diagnosed with colorectal cancer (CRC) versus healthy individuals [14–17]. In fact, gut microbiota can play a role in either promotion or prevention of CRC through modulation of the inflammatory process due to close contact with host colonic mucosa [18]. Furthermore, microbial-produced metabolites, such as short chain fatty acids (SCFA), modulate host intestinal barrier function [19, 20], immune response [21, 22], and inflammatory state. Tumor formation [23, 24] and proliferation [25] are also directly affected by microbial metabolites of host dietary compounds. Changes in gut commensal microbiota associated with consumption of whole grains and legumes are not well defined, yet are crucial for understanding the contribution to CRC prevention.
Martínez et al. noted altered gut bacterial diversity, and changes in the Bacteroidetes:Firmicutes ratio and abundance of Blautia spp. due to consumption of unidentified non-starch components of whole grain brown rice flakes by healthy adults [26]. We previously demonstrated that dietary supplementation of 30g/day of heat-stabilized rice bran (SRB) selectively stimulated some bacterial groups including Bifidobacterium spp. and Ruminococcus spp. in a pilot human cohort with no history of CRC. In addition, increased stool branched chain fatty acids (BCFA) and polyphenol metabolites were associated with SRB supplementation [27]. Research featuring rice bran fermentation in an in vitro canine microbiome model has also revealed increases in anti-inflammatory short chain fatty acids (SCFA) [28]. Studies investigating effects of dry bean consumption on gut microbiota are limited to animal models and have identified limited effects [29, 30] on gut microbial composition, but functional changes have not previously been investigated to our knowledge. Our pilot study in healthy adults with no history of CRC [31] also failed to identify any changes to gut microbial composition with (35g/day) cooked navy bean powder (NBP) consumption for 28 days (data not published). Taken together, this previous research suggests while SRB and NBP are both high fiber foods, they do not induce similar effects on gut microbiota. Therefore, studying these two foods in conjunction offers a unique opportunity to explore the influence of dietary fiber on gut microbiota and their activities.
The influence of SRB or NBP supplementation on human gut microbial dynamics relevant to risk for CRC remain poorly understood. The present study is an exploratory pilot dietary intervention to investigate the potential for supplementation with SRB or NBP to alter gut microbiota and associated metabolites in CRC survivors. Our previous pilot study in a healthy cohort with no history of CRC [27] suggested that these dietary interventions are well tolerated in healthy adults with no history of CRC. So, the main objectives in the current study were to investigate changes in gut microbial composition, potential genetic functions, and resulting microbial metabolites following a one-month dietary supplementation in a cohort of healthy CRC survivors. Other research has demonstrated an association of CRC with gut microbial dysbiosis [13], suggesting that modulation of gut microbiota could be important in CRC prevention. Therefore, evaluating tolerance and microbial response to SRB and NBP in CRC survivors is an important first step to future studies investigating possible prevention of CRC recurrence by these foods. This research lays important groundwork for possible future investigations into the long-term potential for SRB and NBP supplementation to prevent CRC recurrence.
Material and methods
2.1 Participants, study design and sample collection
A total of 37 CRC survivors, more than four months post cancer treatment (e.g. chemotherapy or radiation), were recruited and 29 completed the dietary invention study (Table 1). The study was a four-week, randomized-controlled single-blinded design with three treatment arms. All participants consumed one study-provided meal and snack daily and were randomly assigned to receive 30g of SRB, 35g of NBP or a macronutrient matched control. The study protocol was approved by the Colorado State University (CSU) Research Integrity and Compliance Review Office and the University of Colorado Health-North (UCH-North) Institutional Review Board (CSU protocol #: 09-1530H, and UCH-North IRB #: 10-1038). All participants provided written informed consent prior to enrollment. Participant recruitment, exclusion criteria, study design, and sample collection is described in detail in Borresen et al. [32].
Table 1.
Study participant characteristics.
| ID | Group | Sex | Age | BMI |
|---|---|---|---|---|
| SRB1m | SRB | FEMALE | 58 | 24.1 |
| SRB2m,s | SRB | MALE | 55 | 33.2 |
| SRB3m,s | SRB | MALE | 70 | 34.5 |
| SRB4m,s | SRB | FEMALE | 68 | 26.1 |
| SRB5m,s,p,b | SRB | MALE | 53 | 31.1 |
| SRB6m,s,p,b | SRB | FEMALE | 63 | 22.4 |
| SRB7m,p,b | SRB | MALE | 51 | 32.5 |
| SRB8s,p,b | SRB | FEMALE | 68 | 20.9 |
| SRB9p,b | SRB | FEMALE | 68 | 32.3 |
| C1m,s | CONTROL | FEMALE | 84 | 25 |
| C2m,s | CONTROL | FEMALE | 61 | 31.3 |
| C3m,s | CONTROL | FEMALE | 53 | 26.3 |
| C4m,s,p | CONTROL | MALE | 75 | 31.4 |
| C5m,s,p | CONTROL | FEMALE | 69 | 24.3 |
| C6m,s,p | CONTROL | MALE | 62 | 28.5 |
| C7m,s,p | CONTROL | MALE | 77 | 31.1 |
| C8m,p | CONTROL | FEMALE | 62 | 22.3 |
| C9s | CONTROL | MALE | 73 | 25.8 |
| C10p | CONTROL | FEMALE | 39 | 25.6 |
| NBP1m | NBP | MALE | 57 | 24.7 |
| NBP2m,s | NBP | FEMALE | 57 | 20.9 |
| NBP3m | NBP | FEMALE | 53 | 35.3 |
| NBP4m | NBP | FEMALE | 48 | 18 |
| NBP5m | NBP | FEMALE | 60 | 25.7 |
| NBP6s,p | NBP | MALE | 67 | 27.5 |
| NBP7s,p | NBP | FEMALE | 70 | 32.8 |
| NBP8s,p | NBP | MALE | 37 | 28.4 |
| NBP9s,p | NBP | FEMALE | 59 | 46.4 |
| NBP10p | NBP | MALE | 80 | 25.4 |
Sample included in
454 pyrosequencing analysis of microbiota,
stool SCFA’s,
stool phenolic acids, and
stool bile acids analysis.
This study analysis includes analysis of stool samples collected at time points 0 (baseline), 14 and 28 days for a total of three samples collected during the intervention. Stool samples were self-collected by participants and transported to the lab within 24 hours of each study visit and immediately stored at −20°C until further processing. Stool samples were homogenized, and three subsamples were collected with sterile cotton swabs. DNA was extracted from all samples using MoBio Powersoil DNA extraction kits (MoBio, Carlsbad, CA) according to the manufacturer’s instructions and stored at −20°C prior to amplification steps. Samples with remaining material were aliquoted and stored at −80°C for metabolite analysis.
2.2 Pyrosequencing of the Bacterial Community
Sampling of human intestinal tissue is invasive and not without risk, therefore we followed the convention of using stool samples from study participants to classify and quantify gut bacteria at day 0 (baseline), day 14 and day 28. Library preparation and pyrosequencing of the V3-V5 region of the bacterial 16S rRNA gene was performed under contract at RTL (Lubbock, Texas). All 16S rRNA gene sequences were filtered for quality and processed with the open source bioinformatics tool mothur, Ver. 1.33 [33].
In order to normalize for differences in sample coverage, stool bacterial communities were randomly subsampled to equal the lowest number in a sample, or 1157 in this case. One sample had less than 800 reads and was excluded from the analysis for this reason. Sub-sampled data was used to calculate stool community diversity, including observed species richness (Sobs), estimated species richness (Chao) and Simpson’s diversity (SD) using the mothur implementation of these calculators. All sequence data are publicly available through the Sequence Read Archive (SRA) under study Accession Number PRJEB14459, which is available at http://www.ebi.ac.uk/ena/data/view/ PRJEB14459.
2.3 Metagenome prediction with PICRUSt and STAMP
A synthetic metagenome was generated based on the observed 16S rRNA sequences for each diet intervention group [34]. The 16S rRNA sequences for each diet intervention group were used to predict the sample metagenome as described previously [35]. First, several scripts distributed with PICRUSt v 1.0.0 were used to prepare the data. OTUs with 99% similarity were clustered together using pick_closed_reference_otus.py and the OTU table was normalized for copies of the 16S rRNA gene using the normalize_by_copy_number.py script. This normalized OTU table was processed with the predict_metagenomes.py script and the predicted metagenome was exported as to .biom format for analysis with the software package STAMP [36]. STAMP includes statistical and visualization tools that were used to identify differences in functional potential for the stool bacterial communities across the three diet intervention groups.
2.4 Stool Metabolite Quantification
2.4.1 Short chain fatty acids (SCFA)
One gram replicates of frozen stool samples were extracted for SCFA using 5mLacidified water (pH 2.5) as previously described [16] and spiked with 1mM of ethylbutyric acid which was used as an internal standard. Samples were separated and analyzed using a TG-WAXMS A column on an Agilent 6890 gas chromatograph equipped with an autosampler and FID detection.. SCFAs were quantified by comparing peak areas to standard curves of commercial standards with acetic acid, propionic acid, isobutyric acid, butyric acid, isovaleric acid, and valeric acid (Sigma, St. Louis, MO, USA). Study participants had significantly different variation in acetic acid at baseline via Wilcoxon rank sum test. Therefore, in order to normalize this difference, stool SCFA levels were presented as fold change (using peak area) at day 14 or 28 relative to day 0.
2.4.2 Bile acids and Phenolic acids
In order to assess changes in stool metabolites associated with the observed changes in microbiota with SRB at 14 and 28 days, quantification of bile acids and phenolic acids was performed on a subset of stool samples from SRB group participants (n=5). Stool samples were thawed to room temperature and 0.4–0.5 g each was mixed with sterile saline solution (0.9 %) at a ratio of 1:10 (w/v). The mixture was vortexed to homogenize and centrifuged for 10 minutes at 1,000 × g at 4°C. Stool solutions were then diluted 1:2 with acetonitrile:water (1:2 v/v) and vacuum filtered using a 0.22 µm nylon filter (Millipore, Billerica, MA, USA). LC-MS grade water was used for dilution. Filtered extract was again centrifuged for 25 minutes at 20,000 × g at 4°C to ensure clarity of the final extract before injection for Ultra-performance liquid chromatography tandem mass-spectrometry (UPLC-MS) analysis. For bile acid analysis, internal standards taurocholic acid-d5, deoxycholic acid-d4, and glycodeoxycholic acid-d4 were added to the extract at a final concentration of 0.67 µg/mL. For phenolic analysis, the extract was concentrated to 1/10 the original volume by drying under nitrogen, then re-suspended in 1:2 acetonitrile:water (v/v). LC-MS for the bile acid analysis was performed on a Waters Acquity UPLC coupled to a Waters Xevo TQ-S triple quadrupole mass spectrometer. Phenolic acid analysis was performed on an Agilent 6220 TOF MS using a dual-ESI source in negative ion polarity. Full details for these analyses have been detailed in the Supporting Information for this research.
2.5 Statistical analyses and data visualization
The student’s t-test was used to compare stool bacterial richness, diversity and phyla composition at each time point to baseline. The SCFA and bile acid data were non-normally distributed. Therefore, the Wilcoxon signed rank sum test, a non-parametric test appropriate for repeated measures, was used to compare stool metabolite concentrations at each time point to baseline. To determine shifts in stool community structure, the mothur implementation of the analysis of molecular variance (AMOVA) test was used. To detect stool bacterial taxa differentiating each 14 and 28 day community from baseline, the online METASTATS [37] tool was used. To assess and visualize predicted functional differences in stool bacterial communities at 14 or 28 days compared to baseline for each supplemented diet we used STAMP [36]. In order to identify associations with potential biological relevance that could be used for hypothesis generation for future testing, a p-value less than 0.05 was considered for determining statistical significance in all analyses.
Results
3.1 Stool bacterial community diversity and composition
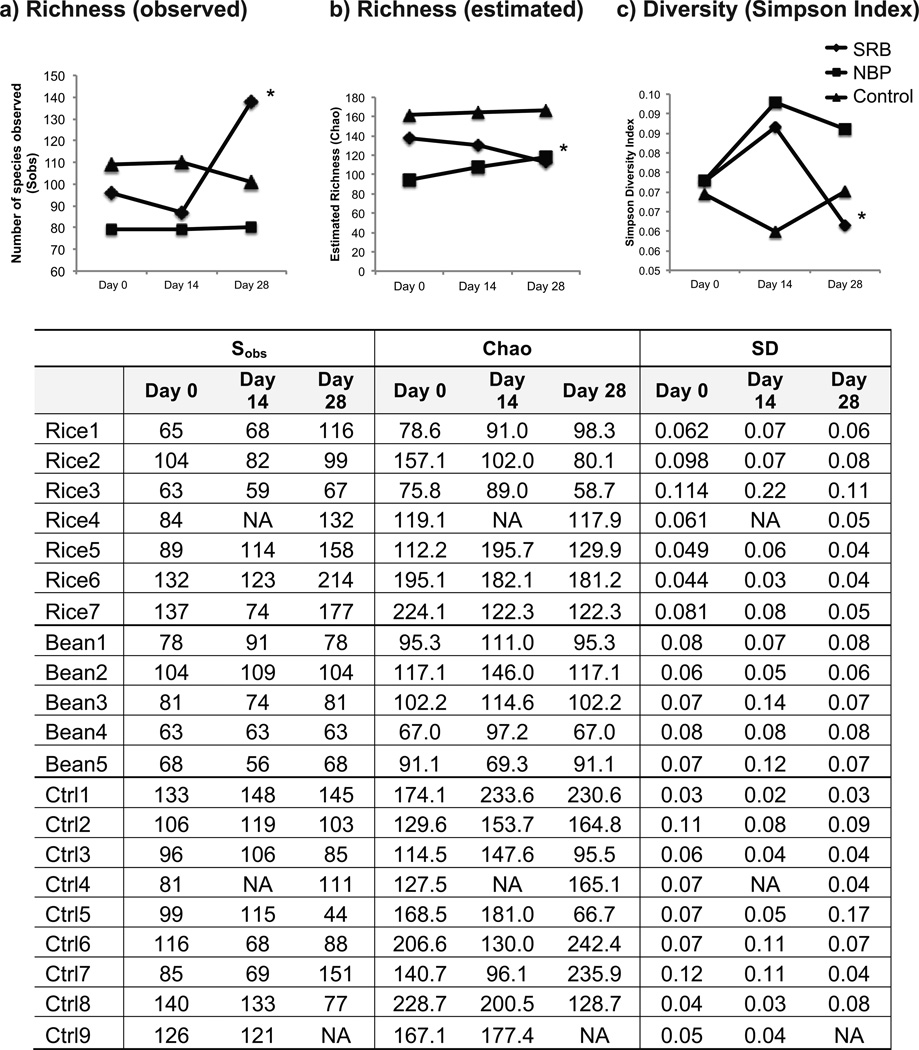
To investigate effects of supplementation with SRB or NBP on stool bacterial communities, richness (total number of OTUs detected), diversity (relative abundance of OTUs present), and composition were measured. Stool bacterial richness and diversity were calculated using Sobs (number of species observed), Chao1, and Simpson diversity indices. The Chao1 richness estimate is used to approximate the number of species potentially not detected due to incomplete sampling using bootstrapping. With SRB supplementation, richness and diversity in stool were not altered at day 14, but were significantly higher than baseline at day 28 (Fig. 1). Stool richness, as estimated by the Chao index, was also increased at day 28 but not day 14 with NBP supplementation (Fig. 1). No changes in richness or diversity were observed in samples from individuals consuming the control diet.
Figure 1.
Stool bacterial diversity at days 0, 14, and 28 for SRB, NBP and control participants. All samples were normalized to 1157 sequences for each participant. Sobs = actual number OTUs detected (richness); Chao = estimated number OTUs (richness); SD = Simpson Diversity Index (lower values reflect higher diversity with this index). The table below the graphs lists data for each participant. The asterisk (*) indicates p < 0.05 with student’s t-test comparing time point to day 0.
To assess potential shifts in stool community structure, principal coordinate analysis (PCoA) including all treatment time points was conducted. Consequently, AMOVA tests were used to compare stool communities at 14 and 28 days to baseline and revealed no significant changes in overall stool community structure in any of the dietary intervention treatment groups (p < 0.05).
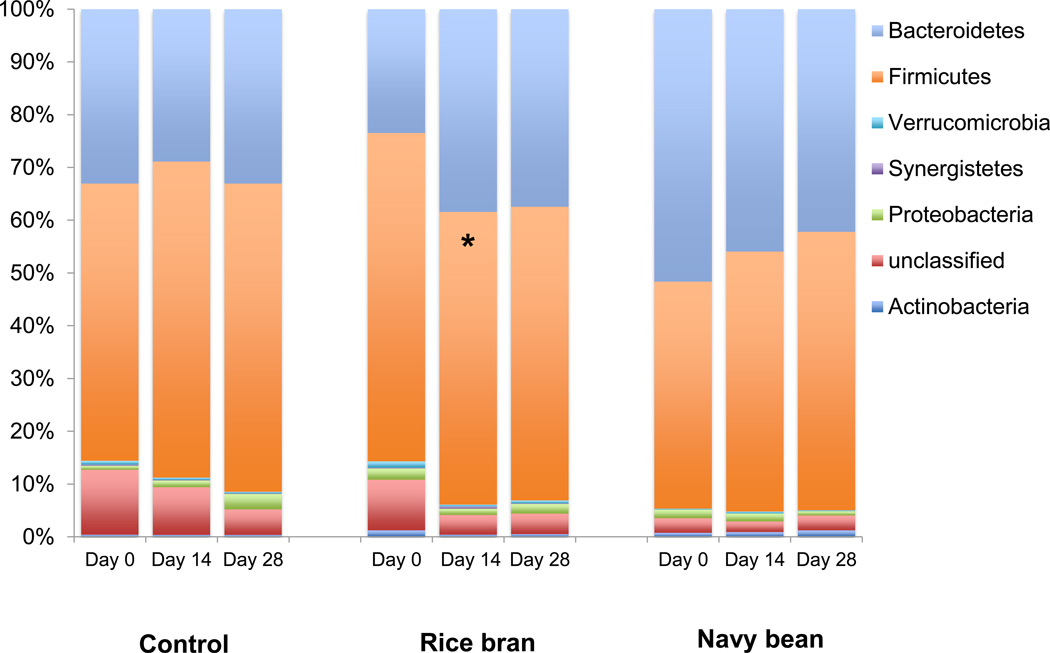
Stool microbial composition on a phyla level showed significant changes with SRB. Overall, SRB supplementation increased abundance of Bacteroidetes and decreased the abundance of Firmicutes resulting in a significantly lower Firmicutes:Bacteroidetes ratio (F:B ratio) at 14 days compared to baseline (Figure 2). Individuals varied in retention of the Firmicutes:Bacteroidetes ratio shift after 28 days of SRB consumption, resulting in no significant difference at 28 days relative to baseline. No changes in microbial composition at the phyla level were detected over time in the NBP or control diet groups.
Figure 2.
SRB is associated with decreased Firmicutes:Bacteroidetes ratio at 14 days relative to baseline, but not 28 days. The asterisk (*) indicates p < 0.05 with student’s t-test comparing time point to day 0.
For each treatment group, differentially abundant OTUs at 14 and 28 days relative to baseline were detected using the software package metastats [38]. No OTUs differed from baseline at either timepoint in the control group. In the NBP group, two OTUs at day 14 and five OTUs at day 28 differentiated the microbial community from baseline (Table S2). At 14 days, Bacteroides fragilis decreased and an unclassified Lachnobacterium increased in mean abundance relative to the NBP baseline microbial community. The reduction in mean abundance of Bacteroides fragilis persisted at 28 days and an unclassified Clostridium and Anaerostipes also had lower mean abundance with NBP. In addition, two OTUs had increased mean abundance at 28 days with NBP including an unclassified Lachnospira and Coprococcus. In the NBP group, four OTUs at day 14 and two OTUs at day 28 differentiated the microbial community from baseline (Table S2).
With SRB at 14 days, an unclassified Ruminococcus and unclassified Ethanoligenens were decreased in mean abundance relative to the baseline SRB microbial community. Conversely, an unclassified Coprococcus and Bacteroides ovatus were increased in mean abundance at 14 days. Bacteroides ovatus showed the second largest increase of any microorganism in this study with a 20-fold increase relative to baseline mean abundance. In addition, eight OTUs were newly detected and eleven OTUs were no longer detected as differential members at14 days compared to the baseline bacterial community (Table S2). The number of OTUs discriminating the bacterial community at 28 days from baseline with SRB was larger than at 14 days (69 versus 23 OTUs). In addition, the largest increase in any single OTU occurred at 28 days with a 24-fold increase in Lachnobacterium relative to baseline mean abundance (Table S2).
3.2 Estimating change in functional potential of stool bacteria
The software package PICRUSt was used as a predictive tool to infer the content of the genome of each bacterium in a community as identified by a 16S rRNA sequencing. This genome was then correlated with a specific functional category as assigned in the Kyoto Encyclopedia of Genes and Genomes (KEGG) for understanding high-level functions of the gut microbial community. Differences in genetic function across treatments and time points in KEGG functional categories can be found using the software package STAMP [36]. Although not a perfect substitute for metagenomic sequencing, this approach allows for developing hypotheses related to the microbial community functions of the gut following dietary intervention.
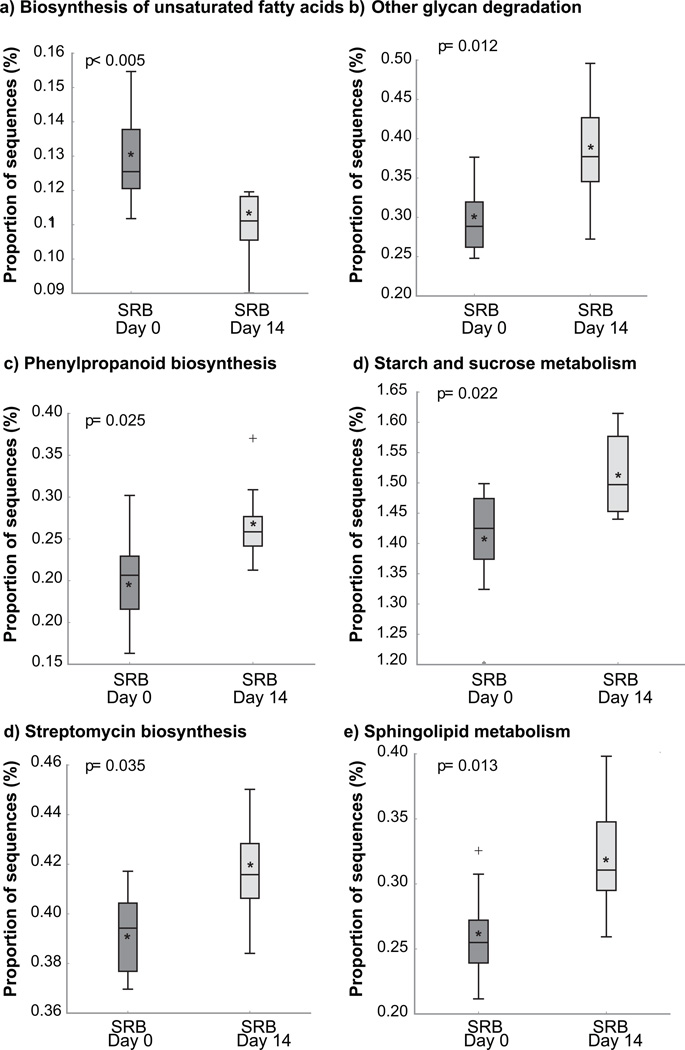
Baseline differences between treatment groups were quite large and ultimately we were looking for responses to specific intervention foods. Therefore, we focused our analysis on a comparison of the 14 and 28-day time points from baseline for each food treatment group, and did not make comparisons between SRB and NBP groups. Several microbial metabolic functions were significantly enriched (p< 0.05) with SRB at 14 days compared to baseline (Figure 3). These functions included “Biosynthesis of unsaturated fatty acids”, “Phenylpropanoid biosynthesis”, “Other glycan degradation”, “Starch and sucrose metabolism”, “Streptomycin biosynthesis”, and “Sphingolipid metabolism” (Figure 3). The most significantly decreasec KEGG functional category was for “Biosynthesis of unsaturated fatty acids”, which represent a significant portion of SRB composition [39]. No significant differences in KEGG functional categories were found at 28 days with SRB, or after 14 or 28 days for the control or NBP interventions when compared to baseline.
Figure 3. Predicted differences in stool bacterial functional categories (PICRUSt).
Only KEGG categories found significant using STAMP analysis are shown, p<0.05.
3.3 Targeted analysis of stool metabolites
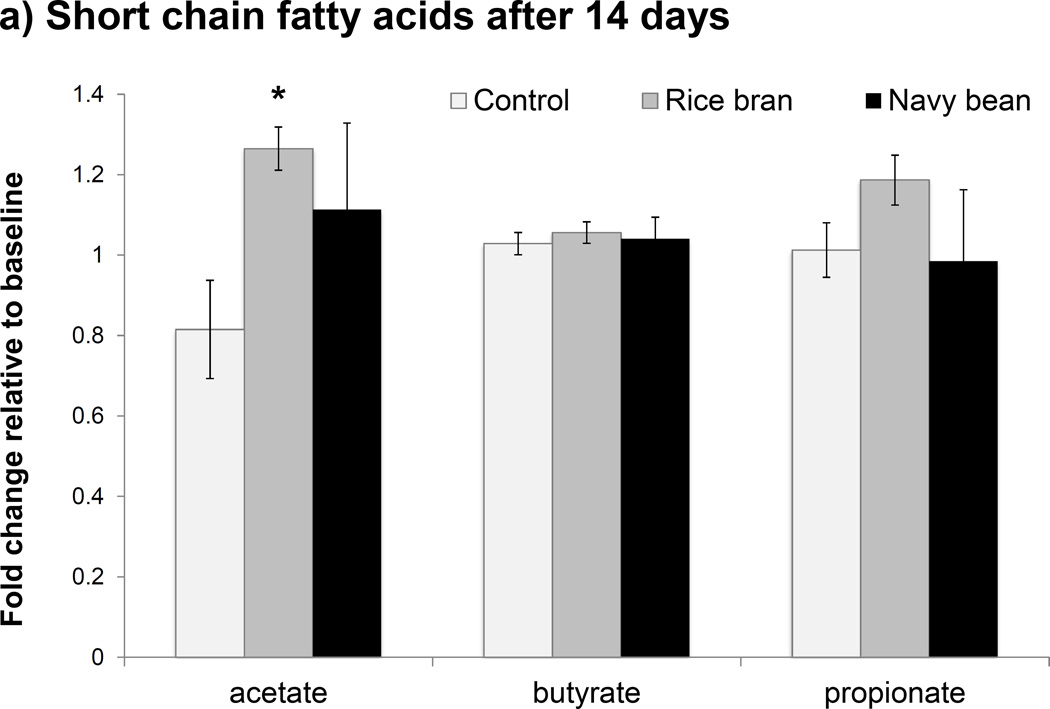
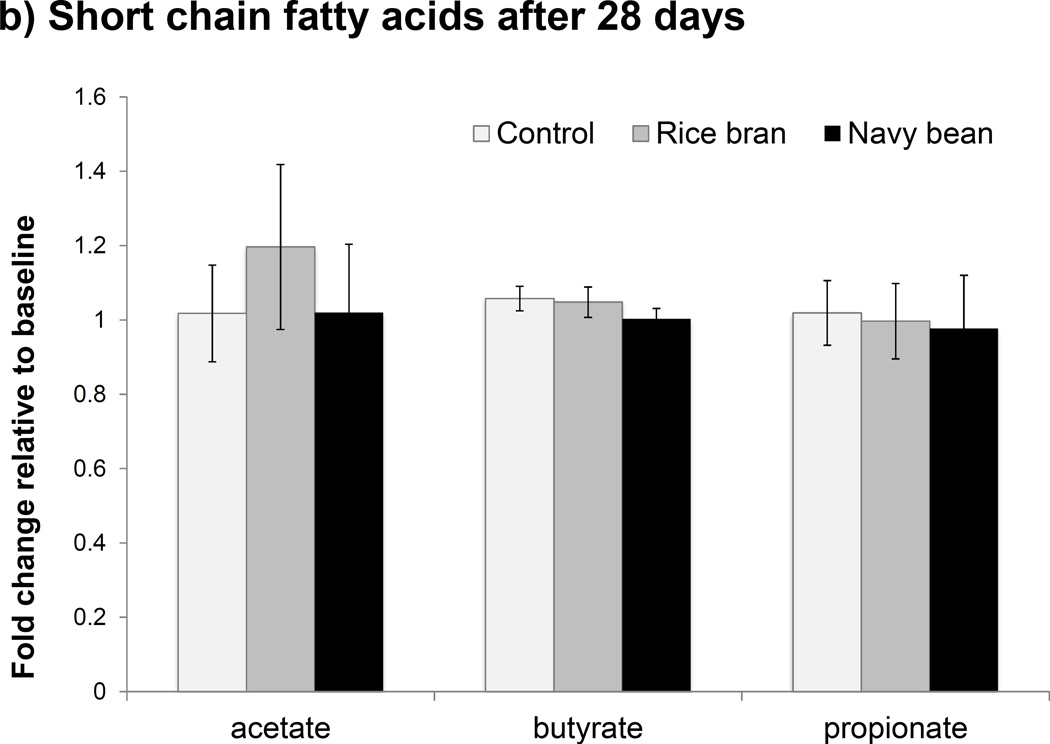
SRB and NBP both contain highly fermentable carbohydrates that can be metabolized by gut bacteria to secondary byproducts such as SCFAs, particularly acetate, propionate and butyrate [40]. Given that functional genes for “Starch and sucrose metabolism” were enriched at 14 days with SRB, SCFAs were quantified to assess potential changes in stool microbial metabolites that resulted with SRB and NBP supplementation. No significant changes in stool SCFAs were observed at 14 or 28 days with the NBP intervention when compared to baseline (Figure 4a); however a large amount of inter-individual variation was observed in this group that could possibly obscure treatment effects. With SRB, no significant changes in stool butyric acid were observed at 14 or 28 days. However, both acetic and propionic acids were significantly increased with SRB at 14 days (p < 0.05); a change that did not persist at 28 days relative to baseline (Figure 4b). Acetic and propionic acids are byproducts of sugar, starch, and glycan fermentation and correspond with the observed enrichment in related metabolic genes at 14 days relative to baseline with SRB. Stool isobutyric and isovaleric acids were also measured based upon increases seen at 4 weeks with SRB consumption in our pilot study [27]. However, these BCFA were below detection limits in all participants using the current study participants and protocols.
Figure 4.
Changes in stool short chain fatty acids (SCFAs) shown in a) Day 14 and b) Day 28. The asterisk (*) indicates p < 0.05 with Wilcoxon rank sum test comparing fold change relative to baseline to control.
Based on differences previously reported for bile acids at 28 days following SRB intake [27], we quantified deoxycholic acid, lithocholic acid, ursodeoxycholic acid, cholic acid, and chenodeoxycholic acid with the current study participants receiving SRB. Stool bile acid concentrations were highly variable across time points with SRB (Figure S1), with primary bile acids cholic acid and chenodeoxycholic trending downward at day 14. Microbial metabolites of polyphenols, including benzoic acid, 3,4-dihydrophenylacetic acid, phenylacetic acid, 3-phenylpropionic acid, 3-hydroxyphenylacetic acid, and 4-hydroxy-3-methoxycinnamic acid were also measured. None of the targeted polyphenol metabolites differed significantly in stool across time points or treatment groups (p < 0.05).
Discussion
Stool samples collected from this pilot human clinical trial supplementing CRC survivors with SRB or NBP revealed changes in gut microbiota, associated metabolites, and predicted functions relative to baseline. None of these changes were observed in stool from participants in the control group. Since previous research has associated CRC with lower gut microbial richness and diversity [41], although somewhat inconsistently [13], we assessed changes to gut microbial richness and diversity with SRB and NBP intervention. In the present study, supplementation with NBP increased gut bacterial richness, while SRB supplementation increased richness and diversity at 28 days. Our results with SRB do not concur with previous studies utilizing fermented rice bran or brown rice flakes, as these foods did not result in significant changes to participants’ gut microbiota richness or composition [26, 42]. However, consuming brown rice flakes (60g/day) did increase stool bacterial diversity after 4 weeks [26]. Since SRB is neither fermented nor refined prior to supplementation, this difference could explain the shift in composition and increased species richness. In addition, our previous research in non-cancer individuals did not show increased stool bacterial diversity with NBP intake, so this effect is likely specific to an unidentified condition present in the CRC survivor cohort.
Changes in gut microbial composition with SRB supplementation included a decrease in F:B ratio after 14 (2.7 down to 1.4), but not 28 days, relative to baseline. Average F:B ratio remained at 1.5 after 28 days, however the change at this time point was not significant due to large variation in individual response. Although an increased F:B ratio is commonly associated with a leaner phenotype in obesity studies, the role of F:B ratio in reference to CRC risk is not well characterized. However, previous studies have shown an increase in Firmicutes in tumor tissue relative to the intestinal lumen with CRC [14]. In addition, Firmicutes are also implicated in increased energy harvest [43], which may promote obesity and influence CRC risk. Therefore, reduction of Firmicutes may provide some protection against CRC. Both NBP and SRB increased total fiber in the participants’ diets [32]; and while NBP increased gut bacterial richness, it did not change the F:B ratio. Furthermore, NBP had 2–5 differentiating bacterial taxa at 14 and 28 days compared to 20+ for SRB (Table S2). It remains unclear whether F:B ratio, or gut bacterial richness or species have more relative importance for CRC risk. So, the relative importance for these findings for CRC chemoprevention cannot be decided.
The effect discrepancies between SRB and NBP suggests that the type of fiber-rich food introduced into the diet may be just as important as total fiber in the diet. Differences in the dominant fiber components of SRB and NBP (i.e. arabinoxylan or xyloglucan) may explain differential effects detected herein on gut microbiota. The other phytochemicals such as polyphenols also influence gut microbial composition [44]. We did not see evidence of changes to polyphenols through measuring phenolic acids in stool with this research, but many forms of these phytochemicals exist beyond what we targeted to measure. Finally, while dry beans and other legumes are typical components of the diet, humans rarely consume SRB and the shift in bacterial community composition may represent a classical ecological response to an environmental disturbance. This is consistent with what appears to be a recovery of some of the baseline-level metabolic activities after 28 days. Mechanistic research identifying which specific components of high-fiber foods alter gut microbial dynamics and metabolic activities will be important for development of dietary therapies targeting the microbiome, including CRC chemoprevention strategies.
In addition to differences in composition of gut microbiota, the study comparing Native Africans (lower CRC risk) to African Americans (higher CRC risk) showed that Native Africans have higher stool SCFAs: acetate, butyrate, and propionate [45]. SCFAs play an important role in gut health by providing a primary energy source, reducing intestinal inflammation and regulating appetite and fat metabolism [46]. Supplementation with SRB increased both acetate and propionate at 14 days relative to baseline, a change that did not persist at 28 days. Acetate and propionate play complex roles in the gut metabolic environment. Propionate is implicated in glucose production and inhibition of cholesterol production from acetate in the liver [47]. While the increase in SCFAs at 14 days is promising for SRB ability to improve overall gut health, more research is necessary to determine what factors influence persistence of increased SCFAs. We did not find changes in SCFA, bile acids or phenolic acids in this study with NBP at 14 or 28 days, which is consistent with findings of other research [48].
Multiple changes in gut microbiota were associated with the increased acetate and propionate with SRB at 14 days. An increase of greater than 20-fold in Bacteroides ovatus in addition to an increase in Lachnospira spp. occurred at 14 days with SRB supplementation. While little is known about Lachnospira spp. function in gut ecology, recent research has revealed that B. ovatus has a specific enzymatic toolkit that makes it uniquely suited to degradation of glycans [49]. The glycan, arabinoxylan, is a dominant constituent of the hemicellulose fraction of rice bran fiber [50]. Primary polysaccharide degraders, like B.ovatus, can release monosaccharides from cellulose and hemicellulose for further metabolism by a wide variety of gut commensals via glycolytic pathways. For example, Lachnobacterium spp., which increased after 28 days, are unable to break down these primary polysaccharides but instead primarily utilize the monosaccharide byproducts of polysaccharide degradation. Pyruvate and oxaloacetate are primary intermediaries of gut fermentation of plant fiber to SCFA metabolites via glycolysis. Blautia spp. utilize the Wood-Ljungdahl pathway to produce acetic acid from pyruvate and were significantly more abundant (13-fold) in stools of the SRB group after 28 days relative to baseline. Propionate is a major fermentation product of Bacteroides spp. and Coprococcus spp. via the succinate and acrylate pathways respectively [51]. Independently of changes in stool SCFA concentrations, genes for “Streptomycin Biosynthesis” were enriched at day 14, which could also play a role in shaping the community at this time point since bacterial species vary in their ability to tolerate this bacteriocide.
Concluding remarks
Previous research has suggested that microbial metabolism of food components during digestion plays a role in potential promotion or chemoprevention of CRC [23–25, 52]. Consumption of high-fiber foods, such as SRB and NBP, has been associated with decreased CRC risk [4–9], which may be a result of changes in gut microbiota and their activities. In this study, both SRB and NBP increased stool bacterial richness or diversity, important stool community characteristics that are reduced in CRC relative to healthy populations. This research with CRC survivors supports the findings of our previous study in a healthy cohort without history of cancer, namely by showing SRB intake of 30 g/day changes stool bacterial populations after 14 days. The current CRC survivor cohort also showed beneficial increases in acetic acid and propionic acid at 14 days that did not persist to 28 days. Further research is needed to determine whether more lasting changes with SRB intervention are possible, perhaps with an increased dose or utilizing other rice cultivars.
Due to the difficulty in recruiting large numbers of CRC survivors to participate in research studies, the number of participants limits interpretation of this study. The outcomes of this research should be used to guide future larger studies designed to control for inter-individual variation. Furthermore, the need for CRC survivor participants did not allow for exclusion and randomization based upon weight status. As such, the number of overweight and obese participants (21 out of 30 total) prevents understanding potential outcomes of SRB and NBP supplementation in a non-overweight CRC survivor cohort. Despite these limitations, these findings support the merit of further investigations of NBP and SRB supplementation as dietary interventions for CRC control and prevention. In addition, these results underscore the importance of tracking microbiota in conjunction with long-term host physiologic responses such as inflammation, tumor recurrence, and intestinal barrier function in future research.
Supplementary Material
Figure S1. Five stoolbile acids were measured with SRB and relative peak intensities were highly variable in participants’ stools. Concentrations at days 14 and 28 did not significantly differ from day 0 via the Wilcoxon Rank Sum Test.
Acknowledgments
NIHR21CA161472 (Multi-PI, Weir and Ryan), the Colorado Agriculture Experiment Station, International Life Sciences Institute and the CSU Proteomics and Metabolomics Facility provided support for this work. We thank Dr. Anna McClung from the USDA-ARS Dale Bumpers Rice Research Unit for supplying the heat-stabilized rice bran, and Gordon Gregory from Archur Daniels Midland Edible Bean Specialties for supplying the cooked navy bean powders. We also appreciate the CSU nutrition science group, Daniel K. Manter for his helpful comments on data interpretation/analysis and Dustin Brown for manuscript comments.
Abbreviations
- BCFA
Branched chain fatty acids
- CRC
Colorectal cancer
- NBP
Cooked navy bean powder
- SCFA
Short chain fatty acids
- SRB
Stabilized rice bran
- UDCA
Ursodeoxycholic acid
- CDCA
Chenodeoxycholic acid
- UPLC-MS
Ultra-performance liquid chromatography tandem mass-spectrometry
Footnotes
Author contributions
Conceived and designed the experiments: TLW, AMS. Implemented trial and human protocol regulatory compliance: EPR, RJB. Coordinated clinical trial and sample collections: ECB. Performed stool extractions: AKW, SL, AMS. Developed stool metabolite quantitation methods: CDB, JSK, CMB. Results interpretations: AMS, TLW. Data analysis and statistics: AMS. Wrote the paper: AMS, TLW, ECB, EPR.
Conflict of Interest
Authors have no conflict of interest to declare for this research.
References
- 1.Gingras D, Béliveau R. Colorectal cancer prevention through dietary and lifestyle modifications. Cancer Microenvironment. 2011;4:133–139. doi: 10.1007/s12307-010-0060-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bingham S. Diet and colorectal cancer prevention. Biochem. Soc. Trans. 2000;28:12–16. doi: 10.1042/bst0280012. [DOI] [PubMed] [Google Scholar]
- 3.Marshall JR. Prevention of colorectal cancer: diet, chemoprevention, and lifestyle. Gastroenterology clinics of North America. 2008;37:73–82. doi: 10.1016/j.gtc.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennink M. Consumption of black beans and navy beans (Phaseolus vulgaris) reduced azoxymethane-induced colon cancer in rats. Nutr. Cancer. 2002;44:60–65. doi: 10.1207/S15327914NC441_8. [DOI] [PubMed] [Google Scholar]
- 5.Bobe G, Barrett KG, Mentor-Marcel RA, Saffiotti U, et al. Dietary cooked navy beans and their fractions attenuate colon carcinogenesis in azoxymethane-induced ob/ob mice. Nutr. Cancer. 2008;60:373–381. doi: 10.1080/01635580701775142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egeberg R, Olsen A, Loft S, Christensen J, et al. Intake of wholegrain products and risk of colorectal cancers in the Diet, Cancer and Health cohort study. Br. J. Cancer. 2010;103:730–734. doi: 10.1038/sj.bjc.6605806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fung TT, Hu FB, Wu K, Chiuve SE, et al. The Mediterranean and Dietary Approaches to Stop Hypertension (DASH) diets and colorectal cancer. The American journal of clinical nutrition. 2010;92:1429–1435. doi: 10.3945/ajcn.2010.29242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lanza E, Hartman TJ, Albert PS, Shields R, et al. High dry bean intake and reduced risk of advanced colorectal adenoma recurrence among participants in the polyp prevention trial. J. Nutr. 2006;136:1896–1903. doi: 10.1093/jn/136.7.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schatzkin A, Mouw T, Park Y, Subar AF, et al. Dietary fiber and whole-grain consumption in relation to colorectal cancer in the NIH-AARP Diet and Health Study. The American journal of clinical nutrition. 2007;85:1353–1360. doi: 10.1093/ajcn/85.5.1353. [DOI] [PubMed] [Google Scholar]
- 10.Huber B, Eberl L, Feucht W, Polster J. Influence of polyphenols on bacterial biofilm formation and quorum-sensing. Zeitschrift für Naturforschung C. 2003;58:879–884. doi: 10.1515/znc-2003-11-1224. [DOI] [PubMed] [Google Scholar]
- 11.Lattanzio V, Lattanzio VM, Cardinali A. Role of phenolics in the resistance mechanisms of plants against fungal pathogens and insects. Phytochemistry: Advances in research. 2006;661:23–67. [Google Scholar]
- 12.Tuohy KM, Conterno L, Gasperotti M, Viola R. Up-regulating the human intestinal microbiome using whole plant foods, polyphenols, and/or fiber. J. Agric. Food Chem. 2012;60:8776–8782. doi: 10.1021/jf2053959. [DOI] [PubMed] [Google Scholar]
- 13.Sheflin AM, Whitney AK, Weir TL. Cancer-Promoting Effects of Microbial Dysbiosis. Curr. Oncol. Rep. 2014;16:1–9. doi: 10.1007/s11912-014-0406-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marchesi JR, Dutilh BE, Hall N, Peters WH, et al. Towards the human colorectal cancer microbiome. PloS one. 2011;6:e20447. doi: 10.1371/journal.pone.0020447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, et al. Microbial dysbiosis in colorectal cancer (CRC) patients. PloS one. 2011;6:e16393. doi: 10.1371/journal.pone.0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weir TL, Manter DK, Sheflin AM, Barnett BA, et al. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PloS One. 2013;8:e70803. doi: 10.1371/journal.pone.0070803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zackular JP, Baxter NT, Iverson KD, Sadler WD, et al. The Gut Microbiome Modulates Colon Tumorigenesis. mBio. 2013;4:e00692–e00613. doi: 10.1128/mBio.00692-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jobin C. Colorectal cancer: looking for answers in the microbiota. Cancer discovery. 2013;3:384–387. doi: 10.1158/2159-8290.CD-13-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelly CJ, Zheng L, Campbell EL, Saeedi B, et al. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell host & microbe. 2015;17:662–671. doi: 10.1016/j.chom.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki T, Yoshida S, Hara H. Physiological concentrations of short-chain fatty acids immediately suppress colonic epithelial permeability. Br. J. Nutr. 2008;100:297–305. doi: 10.1017/S0007114508888733. [DOI] [PubMed] [Google Scholar]
- 21.Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. USA. 2014;111:2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maslowski KM, Vieira AT, Ng A, Kranich J, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vipperla K, O’Keefe SJ. The microbiota and its metabolites in colonic mucosal health and cancer risk. Nutr. Clin. Pract. 2012;27:624–635. doi: 10.1177/0884533612452012. [DOI] [PubMed] [Google Scholar]
- 24.Yoshimoto S, Loo TM, Atarashi K, Kanda H, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- 25.Waldecker M, Kautenburger T, Daumann H, Busch C, Schrenk D. Inhibition of histone-deacetylase activity by short-chain fatty acids and some polyphenol metabolites formed in the colon. J. Nutr. Biochem. 2008;19:587–593. doi: 10.1016/j.jnutbio.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 26.Martínez I, Lattimer JM, Hubach KL, Case JA, et al. Gut microbiome composition is linked to whole grain-induced immunological improvements. ISME J. 2013;7:269–280. doi: 10.1038/ismej.2012.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheflin AM, Borresen EC, Wdowik MJ, Rao S, et al. Pilot dietary intervention with heat-stabilized rice bran modulates stool microbiota and metabolites in healthy adults. Nutrients. 2015;7:1282–1300. doi: 10.3390/nu7021282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogué-Bon E, Khoo C, McCartney AL, Gibson GR, Rastall RA. In vitro effects of synbiotic fermentation on the canine faecal microbiota. FEMS Microbiol. Ecol. 2010;73:587–600. doi: 10.1111/j.1574-6941.2010.00915.x. [DOI] [PubMed] [Google Scholar]
- 29.Queiroz-Monici KdS, Costa GE, da Silva N, Reis SM, de Oliveira AC. Bifidogenic effect of dietary fiber and resistant starch from leguminous on the intestinal microbiota of rats. Nutrition. 2005;21:602–608. doi: 10.1016/j.nut.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 30.Kerr KR, Forster G, Dowd SE, Ryan EP, Swanson KS. Effects of dietary cooked navy bean on the fecal microbiome of healthy companion dogs. PloS one. 2013;8 doi: 10.1371/journal.pone.0074998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borresen EC, Gundlach KA, Wdowik M, Rao S, et al. Feasibility of increased navy bean powder consumption for primary and secondary colorectal cancer prevention. Current nutrition and food science. 2014;10:112. doi: 10.2174/1573401310666140306005934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borresen EC, Brown DG, Harbison G, Taylor L, et al. A randomized-controlled trial to increase navy bean or rice bran consumption in colorectal cancer survivors. Nutr. Cancer. 2016 doi: 10.1080/01635581.2016.1224370. (under review) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schloss PD, Westcott SL, Ryabin T, Hall JR, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langille MG, Zaneveld J, Caporaso JG, McDonald D, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lang JM, Eisen JA, Zivkovic AM. The microbes we eat: abundance and taxonomy of microbes consumed in a day’s worth of meals for three diet types. PeerJ. 2014;2:e659. doi: 10.7717/peerj.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White JR, Nagarajan N, Pop M. Statistical Methods for Detecting Differentially Abundant Features in Clinical Metagenomic Samples. PLoS Comp. Biol. 2009;5:e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol. 2009;5:e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kahlon TS. Rice Bran: Production, Composition, Functionality and Food Applications, Physiological Benefits. Boca Raton: Taylor and Francis Group, LLC; 2009. [Google Scholar]
- 40.Topping DL, Clifton PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol. Rev. 2001;81:1031–1064. doi: 10.1152/physrev.2001.81.3.1031. [DOI] [PubMed] [Google Scholar]
- 41.Ahn J, Sinha R, Pei Z, Dominianni C, et al. Human Gut Microbiome and Risk for Colorectal Cancer. Journal of the National Cancer Institute. 2013;105:1907–1911. doi: 10.1093/jnci/djt300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nemoto H, Ikata K, Arimochi H, Iwasaki T, et al. Effects of fermented brown rice on the intestinal environments in healthy adult. J. Investig. Med. 2011;58:235–245. doi: 10.2152/jmi.58.235. [DOI] [PubMed] [Google Scholar]
- 43.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. nature. 2006;444:1027–1131. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 44.Cardona F, Andrés-Lacueva C, Tulipani S, Tinahones FJ, Queipo-Ortuño MI. Benefits of polyphenols on gut microbiota and implications in human health. J. Nutr. Biochem. 2013;24:1415–1422. doi: 10.1016/j.jnutbio.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 45.Ou J, Carbonero F, Zoetendal EG, DeLany JP, et al. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. The American journal of clinical nutrition. 2013;98:111–120. doi: 10.3945/ajcn.112.056689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sleeth ML, Thompson EL, Ford HE, Zac-Varghese SE, Frost G. Free fatty acid receptor 2 and nutrient sensing: a proposed role for fibre, fermentable carbohydrates and short-chain fatty acids in appetite regulation. Nutrition research reviews. 2010;23:135–145. doi: 10.1017/S0954422410000089. [DOI] [PubMed] [Google Scholar]
- 47.Wolever T, Spadafora P, Eshuis H. Interaction between colonic acetate and propionate in humans. The American journal of clinical nutrition. 1991;53:681–687. doi: 10.1093/ajcn/53.3.681. [DOI] [PubMed] [Google Scholar]
- 48.Perera T, Young MR, Zhang Z, Murphy G, et al. Identification and monitoring of metabolite markers of dry bean consumption in parallel human and mouse studies. Mol. Nutr. Food Res. 2015;59:795–806. doi: 10.1002/mnfr.201400847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Larsbrink J, Rogers TE, Hemsworth GR, McKee LS, et al. A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature. 2014;506:498–502. doi: 10.1038/nature12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shibuya N, Iwasaki T. Structural features of rice bran hemicellulose. Phytochemistry. 1985;24:285–289. [Google Scholar]
- 51.Louis P, Scott K, Duncan S, Flint H. Understanding the effects of diet on bacterial metabolism in the large intestine. Journal of applied microbiology. 2007;102:1197–1208. doi: 10.1111/j.1365-2672.2007.03322.x. [DOI] [PubMed] [Google Scholar]
- 52.David LA, Maurice CF, Carmody RN, Gootenberg DB, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2013 doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Five stoolbile acids were measured with SRB and relative peak intensities were highly variable in participants’ stools. Concentrations at days 14 and 28 did not significantly differ from day 0 via the Wilcoxon Rank Sum Test.