In animal systems, a single signaling network often triggers different developmental goals depending on the cellular context. Several studies, including those related to epidermal patterning, extend such an idea to plant systems (Benfey, 1999; Larkin et al., 2003; Schiefelbein, 2003). Here, I highlight how a network of interacting factors (MYB-BHLH-TTG-GL2) promotes different epidermal cell fates in Arabidopsis depending on their precise localization in the plant body: hairs in the root, stomata in the hypocotyls, and trichomes in the leaf. The emerging picture reveals that the flexibility with which BHLH proteins interact with MYB proteins and the functional redundancy of some components (BHLHs and MYBs) are the secrets that explain how just a few genes can specify the precise patterning of three distinct cell types.
ROOT HAIRS
Root hairs differentiate in a position-dependent manner, with hair-forming cells overlying two cortical cell files and non-hair-forming cells overlying a single cortical file (Dolan et al., 1993, 1994; Galway et al., 1994). A decade ago, Galway et al. (1994) showed that Arabidopsis plants overexpressing the maize BHLH gene R under the control of the Cauliflower mosaic virus 35S (35S) promoter differentiate a reduced number of root hairs because of the production of non-hair cells from epidermal cells located over two cortical cell files. This finding suggested that an R-like BHLH protein in Arabidopsis specifies non-hair cell fate. A recent study based on a qualitative analysis of root hair patterning has shown that GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) encode the long-sought R-like BHLH proteins of Arabidopsis (Zhang et al., 2003). Bernhardt et al. (2003) then confirmed that GL3 and EGL3 together specify non-hair cell fate: mutations of both genes in double mutants give rise to a hairy phenotype caused by the formation of root hairs from epidermal cells overlying a single cortical cell file, and the overexpression of either GL3 or EGL3 under the control of the 35S promoter in wild-type plants produces a reduced number of hairs because epidermal cells above two cortical cell files differentiate as non-hair cells.
Two-hybrid studies in yeast have shown that GL3 contains three protein–protein interaction motifs (Payne et al., 2000; Zhang et al., 2003): (1) an N-terminal domain including the first 97 amino acids interacts with several MYB proteins, among them, GL1, CAPRICE (CPC), and TRIPTYCHON (TRY); (2) a region that encompasses amino acids 212 to 401, required for interaction with a WD repeat–containing protein named TRANSPARENT TESTA GLABRA (TTG); (3) a C-terminal domain containing the BHLH motif of ∼200 amino acids that mediates homodimeric interactions. In addition, a mutation in the 78th codon of GL3 disrupts its interaction with GL1, but it does not affect the interaction with TRY (Esch et al., 2003). A similar but less extensive analysis of EGL3 interactions has revealed that the first 367 amino acids interact with TTG and with several MYB proteins, including GL1, CPC, and TRY (Zhang et al., 2003); its C-terminal domain of 299 amino acids, including the BHLH region, mediates interactions with itself and with GL3 (Zhang et al., 2003). The yeast two-hybrid assay has also shown that both GL3 and EGL3 can interact with the MYB protein WEREWOLF (WER) (Bernhardt et al., 2003). Like GL1, WER is likely to interact with the N-terminal domain of GL3 and EGL3 because WER and GL1 share a high amino acid sequence identity (Lee and Schiefelbein, 1999) and because they are functionally equivalent (Lee and Schiefelbein, 2001). The interaction of the N-terminal region of the BHLH proteins with the MYB proteins extends to the BHLH and MYB proteins that regulate anthocyanin biosynthesis in maize (and probably of other plant species as well), where the N-terminal region of the BHLH proteins R and B interacts with the MYB domain of C1 (Goff et al., 1992; Grotewold et al., 2000).
WER and CPC encode an R2R3 MYB and a single-repeat MYB protein with the highest homology to the R3 region of the R2R3 MYB proteins in their DNA binding domain, respectively (Wada et al., 1997; Lee and Schiefelbein, 1999). The transcripts of both WER and CPC are preferentially restricted to the meristematic and elongation regions of non-hair cell files (Lee and Schiefelbein, 1999; Wada et al., 2002). Like CPC, TRY also encodes a single-repeat MYB protein, which seems to be expressed also in the non-hair-forming cells because mutations or overexpression experiments that produce plants with hairless roots allow TRY expression in all the epidermal cell files (Schellmann et al., 2002). Although WER and CPC/TRY have overlapping expression patterns, they exhibit opposite functions. WER is a negative regulator of hair cell fate as revealed by the hairy phenotype of wer (Lee and Schiefelbein, 1999). By contrast, CPC and TRY redundantly promote root hair cell fate, with cpc exhibiting a few hairs and cpc try being hairless (Wada et al., 1997, 2002; Schellmann et al., 2002). A newly discovered single-MYB repeat gene, ENHANCER OF TRY AND CPC1 (ETC1), also acts in hair patterning (Kirik et al., 2004). Although etc1 on its own does not exhibit an apparent phenotype, the etc1 cpc double mutant reduces the number of root hairs over cpc alone (Kirik et al., 2004). This indicates that ETC1, whose transcripts are preferentially restricted to the non-hair-forming cells (like CPC), acts in a largely redundant fashion with CPC (Kirik et al., 2004). In addition, components with opposite roles regulate one another's transcription (Bernhardt et al., 2003; Larkin et al., 2003; Schiefelbein, 2003; Kirik et al., 2004): in non-hair-forming cells, WER, GL3, and EGL3 positively regulate CPC expression, whereas CPC negatively regulates WER and GL2 expression in hair-forming cells (see below). In hair-forming cells, CPC represses its own expression and, in a redundant manner with TRY, negatively regulates TRY expression (Schellmann et al., 2002; Wada et al., 2002).
But how is root hair patterning controlled? Analysis of transgenic plants expressing a fusion construct of CPC with green fluorescent protein (GFP) under the control of the CPC promoter (CPCP:CPC:GFP) has shown that the CPC protein locates to the hair-forming cell files (Wada et al., 2002). Because CPC is preferentially expressed in non-hair-forming files, Wada et al. (2002) inferred that CPC moves from non-hair-forming cell files to hair-forming ones. It is then logical to propose that CPC (and perhaps TRY and ETC1) might physically interact with GL3 and/or EGL3 in differentiating hair cells and that such interaction would trigger hair cell fate (Bernhardt et al., 2003; Larkin et al., 2003; Schiefelbein, 2003; Figure 1A). WER might interact with the BHLH proteins to prevent hair formation in non-hair-forming cells (Bernhardt et al., 2003; Larkin et al., 2003; Schiefelbein, 2003; Figure 1A). Thus, the spatial compartmentalization between single-repeat MYB proteins and WER might be essential to prevent (or reduce) competition between them for the BHLH binding. Interestingly, plants overexpressing the WER gene resemble wild-type plants in root hair patterning (Lee and Schiefelbein, 1999), suggesting that WER does not interact with the BHLH proteins in epidermal cells overlying two cortical cell files, perhaps because the BHLH proteins are limiting in these cells (Lee and Schiefelbein, 1999; Bernhardt et al., 2003). If this is true, overexpression of either GL3 or EGL3 should produce hairless roots by allowing an accumulation of WER-BHLH complexes in cells that would normally produce root hairs (Bernhardt et al., 2003). Accordingly, expression of GL3 under the control of the 35S promoter (35SP:GL3) and expression of 35SP:EGL3 leads to production of only a few root hairs (Bernhardt et al., 2003). In addition, because 35SP:CPC, 35SP:TRY, and 35SP:ETC1 transgenic plants exhibit a hairy phenotype (Wada et al., 1997; Schellmann et al., 2002; Kirik et al., 2004), an excess of single-repeat MYB proteins might competitively reduce the binding of WER to endogenous BHLH proteins, thus allowing root hair formation in files that normally follow a non-hair cell fate.
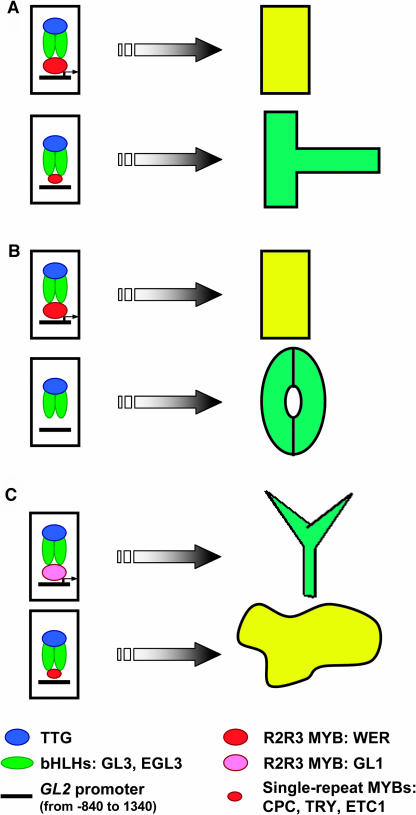
Figure 1.
Interactions among Transcription Factors Involved in Root Hair, Stoma, and Trichome Pattern.
(A) In the root, a complex consisting of WER, GL3 and/or EGL3, and TTG triggers non-hair cell fate determination via GL2 expression. In differentiating hair cells, the complex among a single MYB protein (CPC, TRY, and/or ETC1), GL3 and/or EGL3, and TTG represses GL2 expression.
(B) In the hypocotyl, the BHLH proteins that participate in the multimeric complex that triggers non-stomatal cell fate have not been identified, but it is assumed that they (perhaps GL3 and/or EGL3) interact with TTG and WER to induce GL2 expression. The mechanism that instructs stoma cell fate by repressing GL2 expression is unknown.
(C) In the leaf, similar multimeric complexes to those that operate in the root regulate GL2 expression, but they produce an opposite effect in the hair-forming cells in the two organs: the GL1-GL3/EGL3-TTG complex promotes GL2 expression and trichome cell fate, and the single MYB-GL3/EGL3-TTG complex triggers non-trichome cell fate determination by repressing GL2 expression. The single-MYB redundancy among CPC, TRY, and ETC1 that patterns hairs in the root also operates in trichome pattern formation.
In the three systems, the same 500-bp fragment of the GL2 promoter (−840 to 1340), containing a MYB binding site, is required for a proper regulation of GL2 expression.
The hairy phenotype of ttg indicates that TTG negatively regulates hair cell fate in epidermal cells overlying a single cortical cell file (Galway et al., 1994). Because the overexpression of R in ttg completely represses hair cell fate, it was thought that TTG either encoded or activated an R homolog of Arabidopsis (Galway et al., 1994). An activation function became more likely when TTG was found to encode a WD repeat–containing protein (Walker et al., 1999), indicating a possible role in protein–protein interactions (Neer et al., 1994). The TTG protein physically interacts with GL3 and EGL3 in yeast (Payne et al., 2000; Zhang et al., 2003), and it is assumed that TTG activates these BHLH proteins perhaps by stabilizating their interaction with the MYB proteins (Larkin et al., 2003). TTG is required for the full functioning of GL3 and EGL3 (Bernhardt et al., 2003).
The GL2 gene encodes a homeodomain-leucine zipper protein (Rerie et al., 1994; Di Cristina et al., 1996), and it is also a negative regulator of hair cell fate in files overlying a single cortical cell file as revealed by the hairy gl2 phenotype (Di Cristina et al., 1996; Masucci et al., 1996). GL2 transcripts preferentially locate to the non-hair-forming cells (Masucci et al., 1996; Hung et al., 1998), and in these cells, a basal level of GL2 expression is enhanced by TTG (Hung et al., 1998) and also, redundantly, by GL3 and EGL3 (Bernhardt et al., 2003). WER not only increases the GL2 expression level, it also restricts GL2 transcripts to the non-hair-forming cells as demonstrated by the ectopic expression of GL2 in the wer mutant (Lee and Schiefelbein, 1999). In addition, the cpc mutant exhibits GL2 expression in both non-hair-forming cells and hair-forming ones, revealing that CPC negatively regulates GL2 expression in the hair-forming cells (Wada et al., 2002). The possible effect of TRY and ETC1 on GL2 transcription has not been demonstrated. Taken together, these results suggest that GL2 functions as a downstream effector of the WER-BHLH-TTG complex, repressing hair cell fate in those cells where its expression is positively regulated (Figure 1A). The transcriptional repression of GL2 expression by single MYB-BHLH-TTG complexes, leading to hair cell fate specification by default, is perhaps likely because the single MYB proteins CPC, TRY, and ETC1 lack typical transactivation domains (Wada et al., 1997; Schellmann et al., 2002).
STOMATA
Similar to root hairs, stomata in the hypocotyl develop only from epidermal cells that overlie two cortical cell files (Berger et al., 1998; Hung et al., 1998). Interestingly, mutations in either TTG, WER, or GL2 increase the number of stomata by inducing the formation of stomata in epidermal cells overlying a single cortical cell file (Berger et al., 1998; Hung et al., 1998; Lee and Schiefelbein, 1999). In addition, the overexpression of R under the control of the 35S promoter in wild-type plants produces a reduced number of stomata overlying two cortical cell files (Berger et al., 1998). These findings indicate that TTG, WER, GL2, and an unidentified BHLH protein (R-like) negatively regulate stomatal formation in the epidermal cell files overlying single cortical cell files. WER and GL2 expression is restricted to the upper portion of non-stomatal-forming cell files in the hypocotyl (Hung et al., 1998; Lee and Schiefelbein, 1999), the region where stomatal cell fate determination takes place. Assuming a developmental similarity between root hair and stomatal patterning, GL3 and/or EGL3 might be the BHLH proteins that trigger non-stomatal cell fate and furthermore might provide the physical link with WER and TTG in epidermal cells overlying a single cortical cell file (Figure 1B). Consistent with this idea, the same 500-bp fragment essential for GL2-promoter activity in the root is required for the expression of GL2 in the hypocotyl (Hung et al., 1998; Figure 1B).
Although there is no evidence for a lateral inhibition mechanism guiding stomatal cell fate, the similarity in the expression patterns of CPC, TRY, and ETC1 in the roots and hypocotyls strongly suggests that a similar genetic mechanism guides root hair and stomatal patterning (Kirik et al., 2004). However, because wild-type plants do not develop trichomes in the hypocotyls, whereas etc1 try cpc triple mutants have abundant trichomes in their hypocotyls, the expression patterns of the single-repeat MYB genes may only reflect their roles in repressing trichome cell fate. In any case, some differences in the general mechanism of patterning probably exist between root hairs and the stomata of the hypocotyl because only a few cells in a given epidermal file enter into the stomatal pathway in the hypocotyl (Berger et al., 1998; Hung et al., 1998), whereas all cells in the equivalent file differentiate as hairs in the root (Dolan et al., 1993, 1994; Galway et al., 1994).
TRICHOMES
The spacing of root hairs and trichomes differs significantly. Unlike root hairs, trichomes develop at a minimum distance from their neighbors and never make direct contact with other trichomes (Hülskamp et al., 1994). However, a related molecular mechanism seems to be employed to pattern the trichomes of the leaf. The role of an R-like BHLH protein in trichome patterning was suggested 12 years ago when Lloyd and coworkers (1992) showed that plants overexpressing the maize R gene produce extra trichomes, which strongly suggested that an R homolog in Arabidopsis positively regulates trichome cell fate determination. It is now known that GL3 and EGL3 encode R homologs of Arabidopsis required for trichome determination (Payne et al., 2000; Zhang et al., 2003). Loss-of-function gl3 mutants produce a weak reduction in the number of trichomes (Hülskamp et al., 1994; Payne et al., 2000), and although egl3 exhibits a very subtle phenotype (Zhang et al., 2003), gl3 egl3 double mutants are totally glabrous (Zhang et al., 2003). These findings indicate that GL3 and EGL3 positively regulate trichome cell fate in a redundant fashion (Zhang et al., 2003). These BHLH genes are expressed in young leaves before trichome determination as well as in initiating and developing trichomes (Zhang et al., 2003). This pattern of expression overlaps with that of the GL1 gene (Larkin et al., 1993), which encodes an R2R3 MYB protein required for trichome initiation as indicated by the glabrous phenotype in null alleles (Marks and Feldmann, 1989; Oppenheimer et al., 1991). The TTG gene is also needed for trichome formation as revealed by the absence of trichomes in the null alleles (Koornneef, 1981), and as in the root, TTG activity is also necessary for the full function of both GL3 and EGL3 in trichome initiation (Payne et al., 2000; Zhang et al., 2003).
The proposed protein–protein interactions are supported by genetic interactions, and it has been suggested that a complex comprised of GL3/EGL3, TTG, and GL1 initiates trichome cell fate determination (Larkin et al., 2003; Schiefelbein, 2003; Zhang et al., 2003; Figure 1C). Overexpression of GL3 or EGL3 increases the number of trichomes (Payne et al., 2000; Zhang et al., 2003), suggesting that, as for root hairs, these BHLH proteins are the limiting factors. If this is true, 35SP:GL1 should not trigger the development of extra trichomes. Interestingly, excess trichomes (and trichomes clusters) are generated only when GL1 is co-overexpressed with GL3 (Payne et al., 2000). This coexpression bypasses the need for TTG function (Payne et al., 2000).
Studies using CPC, TRY, and ETC1 extend the molecular parallels between non-root hair and trichome patterning. The try mutant produces trichome clusters (Hülskamp et al., 1994; Schnittger et al., 1998, 1999; Schellmann et al., 2002; Kirik et al., 2004). The frequency of trichome clusters increases in both try cpc double mutants and etc try cpc triple mutants, revealing that these MYB proteins act redundantly in trichome patterning (Schellmann et al., 2002; Kirik et al., 2004). Because TRY, CPC, and ETC1 are preferentially expressed in developing trichomes (Schellmann et al., 2002; Kirik et al., 2004), it has been proposed that the proteins move from trichome precursors to neighboring cells, where they prevent (or reduce) the formation of the multimeric complex among GL1, GL3/EGL3, and TTG, by competing with GL1 for BHLH binding (Schellmann et al., 2002; Larkin et al., 2003; Schiefelbein, 2003; Kirik et al., 2004; Figure 1C). Interestingly, three-hybrid analysis in yeast has shown that TRY physically interacts with GL3 and that this interaction prevents the interaction between GL3 and GL1 (Esch et al., 2003).
GL2 is involved in trichome morphogenesis (Hülskamp et al., 1994; Rerie et al., 1994), and reverse-genetic experiments have revealed that it is also involved in trichome determination because entopically additive expression of GL2 increases the number of trichomes and induces the development of trichome clusters (Ohashi et al., 2002). The expression pattern of GL2 overlaps with that of GL1, except that GL2 transcripts persist in mature trichomes, whereas GL1 transcripts do not (Szymanski et al., 1998). As might be expected, the appropriate level and cell-specific pattern of GL2 expression depend upon TTG, GL1, and GL3/EGL3 (Szymanski et al., 1998; Zhang et al., 2003). In addition, deletion analysis of the GL2 promoter has shown that the same 500-bp fragment required for proper GL2 expression in the root and in the hypocotyl epidermis is also important for regulating GL2 expression in the leaf epidermis (Hung et al., 1998; Szymanski et al., 1998; Figure 1C). Interestingly, this region of the GL2 promoter contains a putative MYB binding site (Hung et al., 1998; Szymanski et al., 1998). Although TRY does not regulate GL2 expression (Szymanski and Marks, 1998), CPC and/or ETC1 may do so. It is important to note that in roots and hypocotyls, the complex consisting of WER, BHLH proteins, and TTG triggers a non-hair cell fate and a non-stomatal fate, respectively. However, a similar complex containing GL1 instead of WER promotes trichome formation in the leaf. The underlying biochemical mechanism might consist of both types of complexes controlling GL2 expression with different outputs in the root and hypocotyl compared with leaves.
The study of epidermal patterning helps not only in understanding how a comparable network triggers different epidermal cell fates depending on the cellular context but also establishes a model that may inform other studies of plant cell specification. The first general principle to emerge from these studies concerns the ability of the BHLH proteins to mediate multiple MYB protein interactions in Arabidopsis, generating a spectrum of multimeric complexes that regulate not only epidermal cell patterning but also anthocyanin production, accumulation of condensed tanins in the seed coat, and probably seed mucilage production (Zhang et al., 2003). Although similar MYB-BHLH complexes regulate anthocyanin synthesis in Arabidopsis and other plant species (for example, Goff et al., 1992; Grotewold et al., 2000), there is currently no evidence that they play roles in the specification of epidermal cell patterns outside the Brassicaceae. Indeed, it is known that (1) multicellular trichome formation in Antirrhinum and Solanaceous species requires R2R3 MYB proteins that are distinct from GL1 (Glover et al., 1998, 2004; Payne et al., 1999; Martin et al., 2002) and (2) R-like BHLH factors seem to play no role in trichome formation in plants other than Arabidopsis, such as tobacco (Lloyd et al., 1992; Mooney et al., 1995; Payne et al., 1999), petunia (Quattrocchio et al., 1993), and tomato (Mooney et al., 1995). The second principle is the functional redundancy of both the BHLH proteins and the single-repeat MYB proteins in epidermal cell patterning in Arabidopsis. This redundancy might ensure proper epidermal cell patterning even if some of these genes are disrupted and more generally probably reflects the importance of epidermal patterning and the development of root hairs, stomata, and trichomes for plant growth and development in natural environments.
Acknowledgments
This work was supported by grants from the Universidad de Castilla-La Mancha, the Ministerio de Ciencia y Tecnología (BMC-2001-1568C), and the Junta de Comunidades de Castilla-La Mancha (GC 02-011). I would like to thank Fred Sack and Lien Lai for the critical reading of the manuscript and for many helpful comments.
References
- Benfey, P.N. (1999). Is the shoot a root with a view? Curr. Opin. Plant Biol. 2, 39–43. [DOI] [PubMed] [Google Scholar]
- Berger, F., Linstead, P., Dolan, L., and Haseloff, J. (1998). Stomata patterning on the hypocotyl of Arabidopsis thaliana is controlled by genes involved in the control of root epidermis patterning. Dev. Biol. 194, 226–234. [DOI] [PubMed] [Google Scholar]
- Bernhardt, C., Lee, M.M., Gonzalez, A., Zhang, F., Lloyd, A., and Schiefelbein, J. (2003). The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130, 6431–6439. [DOI] [PubMed] [Google Scholar]
- Di Cristina, M.D., Sessa, G., Dolan, L., Linstead, P., Baima, S., Ruberti, I., and Morelli, G. (1996). The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant J. 10, 393–402. [DOI] [PubMed] [Google Scholar]
- Dolan, L., Duckett, C.M., Grierson, C., Linstead, P., Schneider, K., Lawson, E., Dean, C., Poethig, S., and Roberts, K. (1994). Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development 120, 2465–2474. [Google Scholar]
- Dolan, L., Janmaat, K., Willemsen, V., Linstead, P., Poethig, S., Roberts, K., and Scheres, B. (1993). Cellular organisation of the Arabidopsis thaliana root. Development 119, 71–84. [DOI] [PubMed] [Google Scholar]
- Esch, J.J., Chen, M., Sanders, M., Hillestad, M., Ndkium, S., Idelkope, B., Neizer, J., and Marks, M.D. (2003). A contradictory GLABRA3 allele helps define gene interactions controlling trichome development in Arabidopsis. Development 130, 5885–5894. [DOI] [PubMed] [Google Scholar]
- Galway, M.E., Masucci, J.D., Lloyd, A.M., Walbot, V., Davis, R.W., and Schiefelbein, J.W. (1994). The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 166, 740–754. [DOI] [PubMed] [Google Scholar]
- Glover, B.J., Bunnewell, S., and Martin, C. (2004). Convergent evolution within the genus Solanum: The specialised anther cone develops through alternative pathways. Gene 331, 1–7. [DOI] [PubMed] [Google Scholar]
- Glover, B.J., Perez-Rodriguez, M., and Martin, C. (1998). Development of several epidermal cell types can be specified by the same MYB-related plant transcriptional factor. Development 125, 3497–3508. [DOI] [PubMed] [Google Scholar]
- Goff, S.A., Cone, K.C., and Chandler, V.L. (1992). Functional analysis of the transcriptional activator encoded by the maize B gene: Evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev. 6, 864–875. [DOI] [PubMed] [Google Scholar]
- Grotewold, E., Sainz, M.B., Tagliani, L., Hernandez, J.M., Bowen, B., and Chandler, V.L. (2000). Identification of the residues in the Myb domain of maize C1 that specify the interaction with the bHLH cofactor R. Proc. Natl. Acad. Sci. USA 25, 13579–13584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung, C.-Y., Lin, Y., Zhang, M., Pollock, S., Marks, M.D., and Schiefelbein, J. (1998). A common position-dependent mechanism controls cell-type patterning and GLABRA2 regulation in the root and hypocotyl epidermis of Arabidopsis. Plant Physiol. 117, 73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hülskamp, M., Misera, S., and Jurgens, G. (1994). Genetic dissection of trichome cell development in Arabidopsis. Cell 76, 555–556. [DOI] [PubMed] [Google Scholar]
- Kirik, V., Simon, M., Huelskamp, M., and Schiefelbein, J. (2004). The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev. Biol. 268, 506–513. [DOI] [PubMed] [Google Scholar]
- Koornneef, M. (1981). The complex syndrome of ttg mutants. Arab. Inf. Serv. 18, 45–51. [Google Scholar]
- Larkin, J., Oppenheimer, D., Pollock, S., and Marks, M. (1993). Arabidopsis GLABROUS1 gene requires downstream sequences for function. Plant Cell 5, 1739–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, J.C., Brown, M.L., and Schiefelbein, J. (2003). How cells know what they want to be when they grow up? Lessons from epidermal patterning in Arabidopsis. Annu. Rev. Plant Biol. 54, 403–430. [DOI] [PubMed] [Google Scholar]
- Lee, M.M., and Schiefelbein, J. (1999). WEREWOLF, a MYB-related protein in Arabidopis, is a position-dependent regulator of epidermal cell patterning. Cell 99, 473–483. [DOI] [PubMed] [Google Scholar]
- Lee, M.M., and Schiefelbein, J. (2001). Developmentally distinct MYB genes encode functionally equivalent proteins in Arabidopsis. Development 128, 1539–1546. [DOI] [PubMed] [Google Scholar]
- Lloyd, A.M., Walbot, V., and Davis, R.W. (1992). Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science 258, 1773–1775. [DOI] [PubMed] [Google Scholar]
- Marks, M.D., and Feldmann, K.A. (1989). Trichome development in Arabidopsis thaliana. I. T-DNA tagging of the GLABROUS1 gene. Plant Cell 1, 1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, C., Bhatt, K., Baumann, K., Jin, H., Zachgo, S., Roberts, K., Schwarz-Sommer, Z., Glover, B., and Perez-Rodrigues, M. (2002). The mechanics of cell fate determination in petals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357, 809–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci, J.D., Rerie, W.G., Foreman, D.R., Zhang, M., Galway, M.E., Marks, M.D., and Schiefelbein, J.W. (1996). The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122, 1253–1260. [DOI] [PubMed] [Google Scholar]
- Mooney, M., Desnos, T., Harrison, K., Jones, J., Carpenter, R., and Coen, E. (1995). Altered regulation of tomato and tobacco pigmentation genes caused by delila gene of Antirrhinum. Plant J. 7, 333–339. [Google Scholar]
- Neer, E.J., Schmidt, C.J., Nambudripad, R., and Smith, T.F. (1994). The ancient regulatory-protein family of WD-repeat proteins. Nature 371, 297–300. [DOI] [PubMed] [Google Scholar]
- Ohashi, Y., Oka, A., Ruberti, I., Morelli, G., and Aoyama, T. (2002). Entopically additive expression of GLABRA2 alters the frequency and spacing of trichome initiation. Plant J. 29, 359–369. [DOI] [PubMed] [Google Scholar]
- Oppenheimer, D.G., Herman, P.L., Sivakumaran, S., Esch, J., and Marks, M.D. (1991). A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67, 483–493. [DOI] [PubMed] [Google Scholar]
- Payne, C.T., Zhang, F., and Lloyd, A.M. (2000). GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156, 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne, T., Clement, J., Arnold, D., and Lloyd, A. (1999). Heterologous myb genes distinct from GL1 enhance trichome production when overexpressed in Nicotiana tabacum. Development 126, 671–682. [DOI] [PubMed] [Google Scholar]
- Quattrocchio, F., Wing, J.F., Leppen, H.T.C., Mol, J.N.M., and Koes, R.E. (1993). Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. Plant Cell 5, 1497–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerie, W.G., Feldmann, K.A., and Marks, M.D. (1994). The GLABRA2 gene encodes a homeodomain protein required for normal trichome development in Arabidopsis. Genes Dev. 8, 1388–1399. [DOI] [PubMed] [Google Scholar]
- Schellmann, S., Schnittger, A., Kirik, V., Wada, T., Okada, K., Beermann, A., Thumfahrt, J., Jürgens, G., and Hülskamp, M. (2002). TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 21, 5036–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein, J. (2003). Cell-fate specification in the epidermis: A common patterning mechanism in the root and shoot. Curr. Opin. Plant Biol. 6, 74–78. [DOI] [PubMed] [Google Scholar]
- Schnittger, A., Folkers, U., Schwab, B., Jürgens, G., and Hülskamp, M. (1999). Generation of a spacing pattern: The role of TRIPTYCHON in trichome patterning in Arabidopsis. Plant Cell 11, 1105–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger, A., Jürgens, G., and Hülskamp, M. (1998). Tissue layer and organ specificity of trichome formation are regulated by GLABRA1 and TRIPTYCHON in Arabidopsis. Development 125, 2283–2289. [DOI] [PubMed] [Google Scholar]
- Szymanski, D.B., Jilk, R.A., Pollock, S.M., and Marks, M.D. (1998). Control of GL2 expression in Arabidopsis leaves and trichomes. Development 125, 1161–1171. [DOI] [PubMed] [Google Scholar]
- Szymanski, D.B., and Marks, M.D. (1998). GLABROUS1 overexpression and TRIPTYCHON alter the cell cycle and trichome cell fate in Arabidopsis. Plant Cell 10, 2047–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada, T., Kurata, T., Tominaga, R., Koshino-Kimura, Y., Tachibana, T., Goto, K., Marks, M.D., Shimura, Y., and Okada, K. (2002). Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development 129, 5409–5419. [DOI] [PubMed] [Google Scholar]
- Wada, T., Tachibana, T., Shimura, Y., and Okada, K. (1997). Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277, 1113–1116. [DOI] [PubMed] [Google Scholar]
- Walker, A.R., Davison, P.A., Bolognesi-Winfield, A.C., James, C.M., Srinivasan, N., Blundell, T.L., Esch, J.J., Marks, M.D., and Gray, J.C. (1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11, 1337–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F., Gonzalez, A., Zhao, M., Payne, C.T., and Lloyd, A. (2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130, 4859–4869. [DOI] [PubMed] [Google Scholar]