Abstract
All members of the AP2/ERF family of plant transcription regulators contain at least one copy of a DNA binding domain called the AP2 domain. The AP2 domain has been considered plant specific. Here, we show that homologs are present in the cyanobacterium Trichodesmium erythraeum, the ciliate Tetrahymena thermophila, and the viruses Enterobacteria phage Rb49 and Bacteriophage Felix 01. We demonstrate that the T. erythraeum AP2 domain selectively binds stretches of poly(dG)/poly(dC) showing functional conservation with plant AP2/ERF proteins. The newly discovered nonplant proteins bearing an AP2 domain are predicted to be HNH endonucleases. Sequence conservation extends outside the AP2 domain to include part of the endonuclease domain for the T. erythraeum protein and some plant AP2/ERF proteins. Our phylogenetic analysis of the broader family of AP2 domains supports the possibility of lateral gene transfer. We hypothesize that a horizontal transfer of an HNH-AP2 endonuclease from bacteria or viruses into plants may have led to the origin of the AP2/ERF family of transcription factors via transposition and homing processes.
INTRODUCTION
The AP2/ERF family of transcription regulators is characterized by the presence of the AP2 DNA binding domain (Riechmann and Meyerowitz, 1998; Sakuma et al., 2002). Sakuma et al. (2002) characterized the large AP2/ERF gene family in Arabidopsis thaliana on the basis of number of repetitions and sequence of the AP2 domain. They divided the 144 members found in Arabidopsis into five subfamilies: DREB, ERF, AP2, RAV, and others. The DREB and ERF subfamilies (120 proteins) have one single AP2 domain and a conserved WLG motif. Fourteen proteins have two AP2 repetitions and belong to the AP2 subgroup. The RAV transcription regulators (six members) have a B3 DNA binding domain following the AP2 domain. The four AP2/ERF members of the other subfamily have a single AP2 repetition but lack the WLG motif characteristic of DREB and ERF proteins. Three of this group share a strong sequence similarity with the AP2 subfamily proteins and will be considered members of that subfamily in this paper. The remaining member is a subgroup by itself and will be referred to as the fifth subfamily.
The three-dimensional structure of the Arabidopsis AtERF1 AP2 domain (PDB ID: 1GCC) was solved by heteronuclear multidimensional NMR (Allen et al., 1998). The domain consists of a three-stranded β-sheet and one α-helix running almost parallel to the β-sheet. It contacts DNA via Arg and Trp residues located in the β-sheet. The secondary structure organization of the AtERF1 AP2 domain shares structural similarities with other DNA binding proteins. The Structural Classification of Proteins database (Murzin et al., 1995) groups the DNA binding domain of the Tn916 integrase (Connolly et al., 1998), the λ integrase N-terminal domain (Wojciak et al., 2002), the human methyl-CpG binding domain MBD (Ohki et al., 1999), and the AP2 domain (Allen et al., 1998) in the same superfamily because of the common three-stranded β-sheet and an α-helix structure. Despite the similar secondary structure and topology, no apparent sequence similarity has been found between the AtERF1 AP2 domain and the other domains.
DNA binding specificity has been shown for members of the ERF, DREB, AP2, and RAV subfamilies. Several ERF proteins bind the GCC box (AGCCGCC) where G2, G5, and C7 are essential for binding (Ohme-Takagi and Shinshi, 1995; Buttner and Singh, 1997; Zhou et al., 1997; Hao et al., 1998; Fujimoto et al., 2000; Hao et al., 2002). The dehydration response element ([DRE], TACCGACAT) is recognized by proteins of the DREB subfamily (Yamaguchi-Shinozaki and Shinozaki, 1994; Stockinger et al., 1997). The sequence CCGAC inside the DRE element is the minimal sequence motif for binding, and C4, G5, and C7 are essential for specific interaction (Hao et al., 2002; Sakuma et al., 2002). DREB factors are known to also bind the C-repeat and the low-temperature-responsive element, which share the CCGAC motif with the DRE element (Baker et al., 1994; Jiang et al., 1996; Thomashow, 1999). The Arabidopsis RAV1 transcription factor can bind a bipartite recognition sequence with the B3 and the AP2 domain recognizing the sequences CACCTG and CAACA, respectively (Kagaya et al., 1999). The only member of the AP2 subfamily with a characterized binding sequence is the Arabidopsis AINTEGUMENTA (ANT) protein. The two AP2 domains in ANT selectively bind the consensus sequence gCAC(A/G)N(A/T)TcCC(a/g)ANG(c/t) (Nole-Wilson and Krizek, 2000; Krizek, 2003).
The AP2 domain has been considered plant specific; domains sharing sequence similarity have not been found outside the plant kingdom (Riechmann and Meyerowitz, 1998; Krizek, 2003). AP2/ERF transcription regulators are found throughout the angiosperms in both monocots and eudicots. They are involved in key developmental steps, such as flower organogenesis or seed development, and in many stress responses (Riechmann and Meyerowitz, 1998). Two members of the AP2 subfamily have also been isolated in gymnosperms (Picea abies), but their function is still unknown (Vahala et al., 2001).
In this study, we identified homologs of the AP2 domain in the cyanobacterium Trichodesmium erythraeum, the ciliate Tetrahymena thermophila, and the viruses Enterobacteria phage RB49 and Bacteriophage Felix 01 via different profile-based approaches. The hypothesis that the AP2 domain exists as a DNA binding domain in nonplant species was supported by demonstrating that the T. erythraeum AP2 domain can selectively bind stretches of poly(dG)/poly(dC).
These newly identified proteins bearing an AP2 domain are predicted HNH endonucleases characterized by conserved His and Asn residues (Shub et al., 1994; Dalgaard et al., 1997). Members of the HNH family of endonucleases are homing endonucleases (reviewed in Gimble, 2000; Chevalier and Stoddard, 2001). Homing is a gene conversion process in which an intervening sequence is copied into a cognate allele that lacks it. The homing process is initiated by a homing endonuclease that makes a double-strand break in the target allele. The cleavage will be repaired by the host using the donor allele as a template, thus copying the intervening sequence. Like transposable elements, homing endonucleases are mobile genetic elements. It is speculated that these endonucleases may trigger a transposition event of their encoding genes. They may excise their own genes and make double-strand breaks elsewhere in the genome, allowing the excised genes to transpose into new sites by illegitimate recombination. Homing endonucleases generally behave like selfish genes with no apparent function for the host and are thought to have a dynamic life cycle consisting of host invasion, replication, and eventual loss. Nevertheless, they sometimes evolve a useful function like the HO endonuclease, which triggers yeast mating type interconversion, and homing endonucleases that catalyze intron self-splicing processes. Interestingly, homing endonuclease genes have spread via lateral gene transfer into all biological kingdoms. In eukaryotic cells, they are also found in mitochondrial and chloroplast DNA (Gimble, 2000; Chevalier and Stoddard, 2001; Koufopanou et al., 2002).
Our phylogenetic analysis of the broader family of AP2 domains suggests the possibility of lateral gene transfer. We hypothesize that a horizontal transfer of an HNH-AP2 homing endonuclease from bacteria or viruses into plants may have originated the AP2/ERF family of transcription factors via transposition and homing processes.
RESULTS
Identifying Remote Homologs
Consistent with results of previous analyses (Riechmann and Meyerowitz, 1998), our BLAST (Altschul et al., 1990) searches using known plant AP2 domain proteins yielded no hits outside of plants. However, because the superior performance of profile-based and iterated search methods of remote homolog detection has been widely documented (Karplus et al., 1997; Park et al., 1998), we tried several methods in this class. These approaches produced a significant number of putative AP2-domain proteins outside of plants.
First, we used the AP2 domain HMM from the PFAM database (Bateman et al., 2004) to score the nonredundant (NR) protein database. This search identified nine proteins outside of plants with scores above the PFAM trusted cutoff for that HMM (three of these had E-values < 1e-05). These nine proteins are from the cyanobacterium T. erythraeum, the unicellular eukaryote T. thermophila, and bacteriophages (Figure 1).
Figure 1.
AP2 Homologs.
Muscle 3.2 alignment of a pool of plant AP2 domains representative of all the AP2/ERF subfamilies, the C. reinhardtii AP2 domains, and the sequences found outside the plant kingdom. The nonplant AP2 domains that align to the entire plant AP2 domain are annotated in red.
We cropped the putative AP2 domains from these novel members of the family and used these as seeds in BLAST and PSI-BLAST (Altschul et al., 1997) searches, using conservative E-value cutoffs of 1e-03. This produced a set of other proteins from bacteria and viruses (Figure 1).
To find additional AP2 domains outside of those mentioned above, we performed a series of advanced searches using custom-built HMMs constructed for different AP2 domain subtypes searching databases of predicted proteins (several proteomes and the NR database). None of these searches revealed any additional homolog outside of plants even with a more permissive E-value cutoff of one.
To help elucidate the evolutionary origin of the AP2 domain in plants, we investigated the presence of this domain in algae. The AP2 HMMs were scored against the proteome of the red algae Cyanidioschyzon merolae whose genome was recently completely sequenced (Matsuzaki et al., 2004). No significant hit was found even with a permissive E-value cutoff of one. A TBLASTN search was also tried against the red algae Porphyra yezoensis and the green algae Chlamydomonas reinhardtii EST database (http://www.kazusa.or.jp/en/plant/porphyra/EST/, http://www.kazusa.or.jp/en/plant/chlamy/EST/) using the T. erythraeum putative AP2 domain and AP2 domains from all plant subfamilies as queries. Significant hits (E-value < 1e-03) were retrieved only in C. reinhardtii (Figure 1).
AP2 Domain Analysis
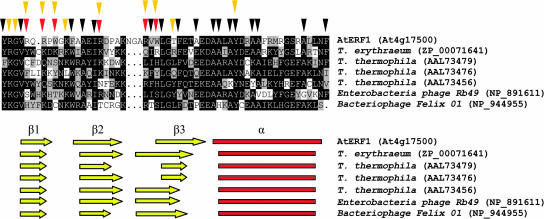
Putative AP2 domains of 29 newly identified nonplant proteins and AP2 domains from members of all the plant subfamilies were aligned with Muscle 3.2 (Edgar, 2004) (Figure 1). Only six of the 29 sequences align to the length of the AP2 domain and were considered further. These six sequences from T. erythraeum, T. thermophila, Enterobacteria phage Rb49, and Bacteriophage Felix 01 align to the plant AP2 domain with an E-value < 5e-04 when used as seeds in BLAST-P. They are also recognized by the Conserved Domain Database (Marchler-Bauer et al., 2003) and the PFAM (Bateman et al., 2004), Phylofacts (http://phylogenomics.berkeley.edu/resources/), and 3D-pssm (Kelley et al., 2000) HMM libraries as AP2 domains. The analysis of their secondary structure in 3D-pssm (Kelley et al., 2000) predicts a three-stranded β-sheet and α-helix conformation similar to the AtERF1 AP2 domain (Allen et al., 1998) (Figure 2). In addition, many DNA-contacting and stabilizing residues of the AtERF1 AP2 domain are conserved (Figure 2). Residues contacting sugar phosphate backbones and stabilizing hydrophobic residues are more conserved than base-contacting amino acids. Thus, the data collected clearly show that the AP2 domain is not unique to plants, and homologs are present in T. erythraeum, T. thermophila, Enterobacteria phage Rb49, and Bacteriophage Felix 01.
Figure 2.
Conservation of Functional Amino Acids and Secondary Structure.
Muscle 3.2 alignment of the AtERF1 AP2 domain and the nonplant AP2 domains. Red and yellow triangles indicate AtERF1 base-contacting and backbone-contacting amino acids, respectively. The hydrophobic residues stabilizing the AtERF1 AP2 domain are marked by black triangles. A schematic representation of the secondary structure as determined in the AtERF1 structure and predicted by 3D-pssm is drawn below the alignment. Yellow arrows indicate β strands, and red rectangles indicate α helices.
DNA Binding Activity of the T. erythraeum AP2 Domain
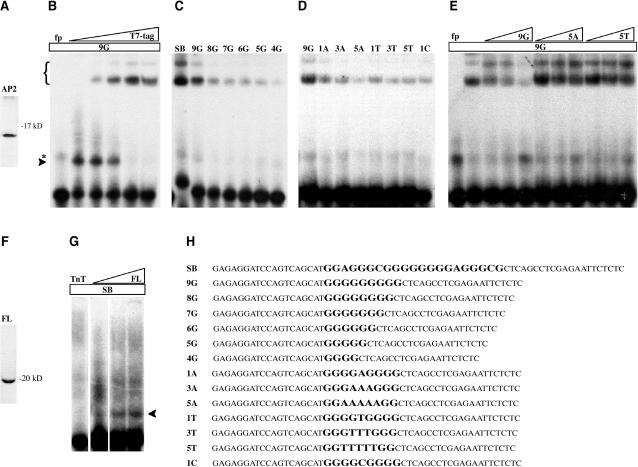
The T. erythraeum AP2 domain that best aligns to plant sequences was tested for DNA binding activity. The coding sequence of the cyanobacterium AP2 domain was cloned by PCR, fused to a His and T7 tag, and overexpressed in Escherichia coli (Figure 3A). Correct translation of the protein was confirmed by protein gel blot analysis (data not shown). A selection and amplification binding assay (SAAB) was performed with this protein to test DNA binding activity against a set of degenerate oligonucleotides. After four cycles of enrichment, 50 putative binding sites were cloned and sequenced (Figure 4). The sequences were submitted to MEME (Bailey and Elkan, 1994) to find common motifs. The software could not identify a significant consensus motif other than homopolymeric stretches of poly(G)/poly(C). These sequences are indeed highly G/C rich with an average G/C content of 65% (Figure 4).
Figure 3.
The T. erythraeum AP2 Domain Selectively Binds DNA.
(A) The T. erythraeum AP2 domain (AP2) was fused to a T7 and His tag, and ∼1 μg of recombinant expressed and purified protein was separated by SDS-PAGE. The molecular mass of the recombinant protein was 12 kD.
(B) EMSA analysis of purified T. erythraeum AP2 domain fused to a T7 and His tag. The protein was incubated with the 9G probe (Figure 3H). The shift is indicated by an arrowhead. In the free probe lane (fp), an artifact was detected at a molecular weight slightly higher than the protein–DNA complexes (indicated by an asterisk). Increasing amounts of anti T7-tag antibody led to a supershift (indicated by brackets).
(C) EMSA analysis of purified T. erythraeum AP2 domain fused to a T7 and His tag. The protein was incubated with seven different probes (Figure 3H): SB, 9G, 8G, 7G, 6G, 5G, and 4G. Anti T7-tag antibodies were added to all samples.
(D) EMSA analysis of purified T. erythraeum AP2 domain fused to a T7 and His tag. The protein was incubated with eight different probes (Figure 3H): 9G, 1A, 3A, 5A, 1T, 3T, 5T, and 1C. Anti T7-tag antibodies were added to all samples.
(E) EMSA analysis of purified T. erythraeum AP2 domain fused to a T7 and His tag. The protein was incubated with the 9G probe (Figure 3H). DNA binding specificity was determined by competing with unlabeled 9G, 5A, or 5T probe (Figure 3H) at 10-, 50-, and 100-fold molar excess over labeled probe. Anti T7-tag antibodies were added to all samples. fp, free probe.
(F) The T. erythraeum full-length protein was translated in the TnT reaction, labeled in vitro with 35S-Met, separated by SDS-PAGE, and analyzed by autoradiography (see Methods). The molecular mass of the protein was 19 kD.
(G) Increasing amounts of the T. erythraeum full-length protein translated in the TnT reaction (triangle named FL) were incubated with the SB probe. The TnT mixture (TnT) was used as a control.
(H) List of the probes used for EMSA analysis. The variable region of the probes is in bold and in a larger font.
Figure 4.
SAAB Assay.
Sequences selected with the T. erythraeum AP2 domain against a pool of random oligonucleotides. G/C and A/T bases are in black and gray, respectively. The diagram at the bottom shows the average percentage of G/C and A/T of the clones sequenced.
To confirm the data obtained with the SAAB assay, electrophoretic mobility shift assays (EMSA) were undertaken. The T. erythraeum AP2 domain strongly binds the G-rich SAAB clone 1 (SB probe, Figures 3C and 3H). A repetition of nine Gs flanked by the SAAB primers (9G probe, Figure 3H) was also bound by the T. erythraeum AP2 domain, and its identity was confirmed by supershifting with an increasing amount of anti-T7 tag antibody (Figure 3B). The 9G probe was used as a standard in further analyses.
A set of five additional probes containing eight to four Gs (8G, 7G, 6G, 5G, and 4G probes, Figure 3H) was used to determine the smallest number of Gs required for binding (Figure 3C). A strong decrease in binding affinity was observed with the 8G probe with respect to the 9G probe. Weaker protein-DNA complexes were detected by reducing the number of Gs, although a signal was still observed with the 4G probe.
To further investigate the DNA binding properties of this new AP2 domain, a battery of mutant oligonucleotides with a disrupted stretch of nine Gs was tested. Six probes were created, mutagenizing one, three, and five Gs in the middle of the nine G stretch into As or Ts (1A, 3A, 5A, 1T, 3T, and 5T probes, Figure 3H). An additional probe with the central G mutagenized into a C was also tested (1C probe, Figure 3H). Substitutions of more than one C were not used to avoid forming a new binding site on the complementary strand. In all cases, the mutagenesis of the 9G probe resulted in a much weaker binding affinity (Figure 3D). In particular, an inverse correlation was observed between the number of As or Ts substituted in the 9G repeat and the extent of protein-DNA complexes. These observations were confirmed by EMSA competition. Figure 3E shows that the cold 9G probe is a much better competitor than the 5A and 5T probes.
Analogous results were displayed by the T. erythraeum full-length protein obtained by in vitro transcription and translation (TnT) (Figure 3F). The full-length protein bound the SB probe in an EMSA experiment (Figure 3G). Increasing amounts of protein resulted in stronger signals. No shift has been detected using the same amount of TnT mixture without the cyanobacterium protein, confirming that the binding is because of the T. erythraeum protein (Figure 3G).
Taken together, these results indicate that the T. erythraeum AP2 domain selectively recognizes stretches of poly(G)/poly(C) but can tolerate some level of nucleotide change in the binding site.
Domain Structure Analysis
To elucidate the possible function of the newly discovered AP2-domain proteins we conducted a domain structure analysis on the full-length proteins. No homologous solved structures could be identified by Phylofacts for the N-terminal region; however, PFAM identifies an HNH endonuclease domain in the three T. thermophila and two viral proteins (Shub et al., 1994; Dalgaard et al., 1997). It also detected a NUMOD4 domain in the T. thermophila proteins (Sitbon and Pietrokovski, 2003).
Figure 5A shows a multiple sequence alignment of all six sequences highlighting the conserved HNH domain. The T. erythraeum putative HNH domain differs from other HNH domains because of two Arg at the C terminus instead of Asn and is not recognized by PFAM. The T. thermophila proteins also contain a NUMOD4 domain, which are often found accompanying HNH domains and are believed to potentially bind DNA (Sitbon and Pietrokovski, 2003). All considered, these six proteins can be predicted to encode HNH endonuclease domains adjacent to AP2 DNA binding domains.
Figure 5.
Domain Analysis.
(A) Muscle 3.2 alignment of the six nonplant proteins containing an AP2 domain. Black boxes indicate the NUMOD4, HNH, and AP2 domains. Arrowheads mark two conserved Asn that are substituted by Arg in the T. erythraeum domain.
(B) Muscle 3.2 alignment of the T. erythraeum predicted HNH domain and the region of the Arabidopsis At4g39780 DREB protein preceding the AP2 domain. A schematic representation of the HNH endonuclease and the At4g39780 protein is drawn above and below the alignment, respectively.
Remarkably, conservation was found between the T. erythraeum protein and plant AP2/ERF factors also outside the AP2 domain. For example, homology between the T. erythraeum protein and the Arabidopsis At4g39780 DREB protein starts well before the AP2 domain (Figure 5B). Extended N-terminal homology of the cyanobacterium protein with plant AP2 domain–containing proteins provides evidence in favor of divergent evolution from a common ancestor and against convergent evolution within the AP2 domain.
Phylogenetic Analysis
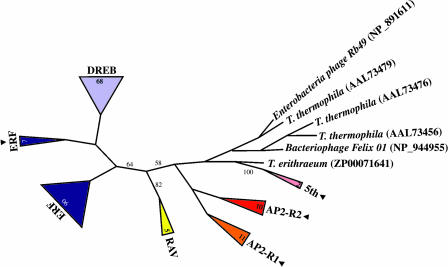
The finding of AP2 domains outside the plant kingdom allowed us to root the phylogenetic tree of the AP2/ERF family of transcription factors. To achieve this goal, the Simple Modular Architecture Research Tool (SMART) (Letunic et al., 2002) was searched for AP2 domains. The AP2 domains of the 434 plant proteins found by SMART were gathered and aligned with Muscle 3.2. The multiple sequence alignment was manually edited to delete partial sequences and was made nonredundant at 90% with BELVU (http://www.cgb.ki.se/cgb/groups/sonnhammer/Belvu.html), narrowing down the number of sequences to 185. We added both the rice (Oryza sativa) and Arabidopsis members of the fifth subfamily and the C. reinhardtii AP2 domains because they were missing. Finally, the AP2 domains selected were aligned with the six newly discovered nonplant AP2 domains (Muscle 3.2). The multiple sequence alignment (see supplemental data online) was used to build a neighbor-joining (NJ) tree with PAUP 4.0 (Figure 6). A maximum likelihood (PHYLIP 3.5 software) and parsimony (PAUP 4.0 software) tree were also constructed from the same alignment (data not shown); intron analysis presented below suggests the NJ tree to be more credible.
Figure 6.
Phylogenetic Analysis.
NJ tree of plant AP2 domains and the newly discovered AP2 domains outside plants. A total of 187 plant AP2 domains belonging to all the subfamilies of AP2/ERF transcription factors, six C. reinhardtii AP2 domains, and the T. erythraeum, T. thermophila, Enterobacteria phage Rb49, and Bacteriophage Felix 01 AP2 domains were aligned using Muscle 3.2 (see supplemental data online), and a NJ consensus tree was generated from 10,000 bootstrap replications, including groups compatible with the 50% majority rule. Monophyletic branches grouping plant AP2 domains of the same subfamily were collapsed in a triangle showing the name of the subfamily. The numbers of taxa collapsed are indicated in the triangles. Numbers on branches indicate bootstrap values. Only values >50% are shown. Branch lengths are not proportional to the distance between sequences. Black arrowheads indicate branches, including AP2 domains coded by Arabidopsis genes bearing introns.
In the NJ tree, the fifth, RAV, and DREB/ERF subfamilies appear to be monophyletic with bootstrap values >50% (10,000 bootstrap repetitions). A monophyletic origin of the first and second AP2 repetitions (AP2-R1 and AP2-R2) is suggested by the NJ tree and is supported by the intron analysis presented below. The AP2 domains of the AP2 proteins with a single repetition are included in the AP2-R1 branch. Seven of the 94 ERFs were grouped as a separate branch. Three of the six C. reinhardtii AP2 domains are grouped in the AP2 subfamily and the others in the ERF subfamily.
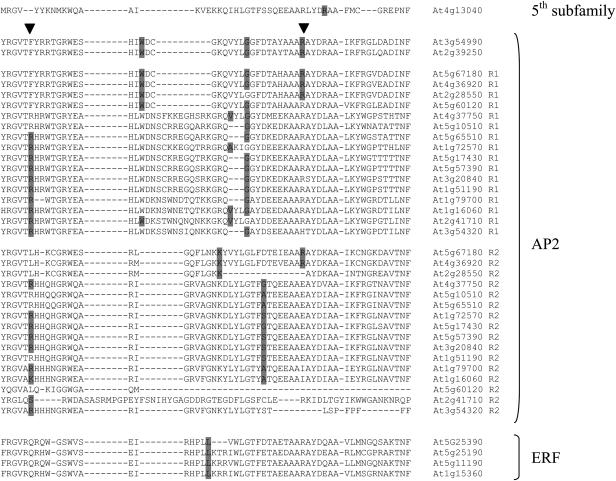
We used intron distribution in the Arabidopsis AP2/ERF genes as an evolutionary marker in the analysis of this gene family. Conserved intronic sites provide evidence of intron evolution before gene duplication and can be used to trace the evolutionary story of genes bearing them. The majority of the AP2/ERF genes are intronless. The 23 Arabidopsis ERF/AP2 genes bearing introns in the coding sequence were analyzed as annotated by The Arabidopsis Information Resource (http://www.Arabidopsis.org). This test includes all members of the AP2 subfamily, the fifth subfamily, and four ERF genes. We focused our analysis on introns breaking the AP2 domain coding sequence to exploit the high sequence conservation of this region. Figure 7 shows highly conserved intronic sites present in the AP2 subfamily genes. Two identical sites are shared by the AP2-R1 and AP2-R2 domains. One of these two intronic markers is also conserved in the AP2 subfamily genes with a single AP2 domain. No conservation was detected among members of different subfamilies. These findings suggest that the evolution of introns in the AP2 subfamily genes occurred before their duplication. It also supports the hypothesis of a common ancestor of both repetitions. All the Arabidopsis ERFs bearing introns have a conserved site and are grouped by the NJ tree in the monophyletic branch that includes seven ERF proteins (Figure 6). The lack of intronic conservation among different subfamilies supports the hypothesis of an early evolution of each subfamily as depicted by NJ. All the Arabidopsis intronic markers in the AP2 coding sequence are also conserved in rice (data not shown), thereby strengthening our hypothesis.
Figure 7.
Intron Analysis.
Analysis of the introns in the AP2 coding sequence of the Arabidopsis AP2/ERF genes. The AP2 domains coded by the Arabidopsis AP2/ERF genes bearing introns were aligned using Muscle 3.2. The proteins At5g60120, At2g41710, and At3g54320 show degenerated second AP2 repetitions that are not annotated as AP2 domains. The C terminus of the At5g60120 repetition was omitted because no similarity was detected with any AP2 domain. One new member of the AP2 subfamily with respect to the characterization made by Sakuma et al. (2002) is included. The position of intronic sites is highlighted in gray in the protein sequence. Arrowheads indicate conserved sites among first and second AP2 repetitions of the AP2 subfamily.
Another important point that emerged from our analysis is the apparent absence of the AP2 domain in many branches of the tree of life. MacClade 4.06 was used to infer a maximum parsimony trace of the AP2 domain character along the tree of life. We considered the AP2 domain absent from a branch if at least one organism with a completely sequenced genome was present in that branch and no AP2 domain was found. Groups with partially sequenced genomes and no AP2 domain identified were considered equivocal. According to the most parsimonious point of view, the AP2 domain character either evolved independently in bacteria, plants, and ciliates or moved horizontally among these organisms (data not shown).
DISCUSSION
The AP2 DNA binding domain characterizes the large AP2/ERF family of transcription factors in plants. In contrast with other important DNA binding domains, such as basic/helix-loop-helix, MYB, and homeodomain, which are conserved in many branches of the tree of life, the AP2 domain has been considered plant specific. Our exhaustive search for AP2 domains outside plants revealed homologs in the cyanobacterium T. erythraeum, the ciliate T. thermophila, and the viruses Enterobacteria phage Rb49 and Bacteriophage Felix 01. The six newly discovered AP2 domains show a striking sequence similarity with plant AP2 domains; they share >40% identity along the entire domain with plant AP2 domains and have the same predicted secondary structure of three-stranded β-sheets and an α-helix. Furthermore, the cyanobacterium AP2 domain shows functional conservation by selectively binding DNA. These newly discovered AP2 domain proteins share an HNH endonuclease domain and are therefore predicted endonucleases. Interestingly, no AP2 domains were detected in eukaryotes except for plants and T. thermophila.
AP2 Domain DNA Binding Affinity
The cyanobacterium AP2 domain binds DNA in vitro showing both amino acid and functional conservation with plant AP2 domains. This domain preferentially binds homopolymeric stretches of poly(G)/poly(C). Mutagenesis of just one base in the stretch drastically decreases the binding affinity, demonstrating that this domain selectively recognizes DNA. A large decrease in DNA–protein complexes was detected by EMSA analysis using the 8G probe compared with the 9G probe. Nevertheless, weak binding was still detectable, decreasing the amount of Gs to four. The domain also tolerates some level of sequence change in the binding site. Even a substitution of five As or Ts does not completely abolish the binding. The substitutions are more easily tolerated when the poly(G)/poly(C) stretches are longer.
Similarities can easily be found between the cyanobacterium and plant AP2 domain DNA binding sites. The plant GCC box bound by ERFs and the DRE, C repeat, and low-temperature-responsive elements recognized by DREBs are G/C rich. The bases essential for binding in these elements are either Cs or Gs, and they share the common motif CCGNC (Baker et al., 1994; Yamaguchi-Shinozaki and Shinozaki, 1994; Ohme-Takagi and Shinshi, 1995; Jiang et al., 1996; Buttner and Singh, 1997; Stockinger et al., 1997; Zhou et al., 1997; Hao et al., 1998, 2002; Thomashow, 1999; Fujimoto et al., 2000; Sakuma et al., 2002). The AP2 transcription factor ANT recognizes a motif that contains an important stretch of three Cs (Nole-Wilson and Krizek, 2000; Krizek, 2003). Interestingly, some AP2/ERF proteins show cross-affinity in vitro for these binding motifs. The ERF Tsi1 and DREB1B and DREB2A can bind both the GCC box and DRE element, and ANT can bind the DRE minimal motif CCGAC in the COR78 and COR15 promoters (Nole-Wilson and Krizek, 2000; Park et al., 2001; Hao et al., 2002; Sakuma et al., 2002). Plant AP2 domains seem to have evolved different DNA binding specificities while conserving a strong affinity for G/C rich motifs. The T. erythraeum AP2 domain fits perfectly well into this scenario, showing selectivity for G/C rich sequences.
HNH Endonucleases Bearing the AP2 Domain
These newly discovered AP2 domain–containing proteins are predicted HNH endonucleases sharing a conserved HNH domain (Shub et al., 1994; Dalgaard et al., 1997). Members of the HNH family of endonucleases are homing endonucleases. Three important features characterize these genes: (1) they transpose from one site into another, (2) they duplicate themselves, exploiting a homing process performed by host cell repair mechanisms, and (3) they move extensively via lateral gene transfer (Gimble, 2000; Chevalier and Stoddard, 2001; Koufopanou et al., 2002). Members of the large family of homing endonucleases were shown to specifically cut long target sequences (14 to 40 bp) but tolerate changes in the cutting sites, thereby guaranteeing their survival despite evolutionary drift of the target sequences (Chevalier and Stoddard, 2001). In accordance with these data, we showed that the AP2 domain in T. erythraeum selectively binds long stretches of Gs/Cs but tolerates a certain level of sequence change in the binding site. Homing endonucleases generally colonize intergenic or intronic regions that have a low impact on host fitness (Gimble, 2000; Chevalier and Stoddard, 2001; Koufopanou et al., 2002). The DNA selectivity of the cyanobacterium AP2 domain may have evolved to recognize noncoding G/C-rich genomic sequences that better tolerate their invasion.
Evolution of the AP2 Domain in Plants
The finding of AP2 domains outside plants opens the possibility of a new scenario to explain the evolution of this domain and the transcription factors that contain it. The presence of AP2 domains in viruses and bacteria and the absence of homologs among eukaryotes other than T. thermophila were unexpected. One possible explanation is an inadequate number of sequenced genomes from which to construct a reliable evolutionary history of this domain. Nevertheless, the ever-increasing number of completely or partially sequenced genomes available today makes this hypothesis less likely. According to the distribution of the AP2 domains among living organisms, three possible hypotheses can be drawn: convergent evolution, divergent evolution followed by multiple loss of the character, or lateral gene transfer.
The hypothesis of lateral gene transfer is supported by many considerations. The strong protein sequence similarity calls for a common evolutionary origin. In contradiction to a convergent evolution hypothesis, this group of putative HNH endonucleases aligns with members of the plant AP2 subfamily outside the AP2 domain. Some AP2/ERF transcription factors show conservation with the cyanobacterium putative HNH motif in the region preceding the AP2 domain. Furthermore, functional conservation was demonstrated for the cyanobacterium AP2 domain, which shows similarity in DNA binding specificity with plant AP2 domains. The most parsimonious explanation for the evolution of the AP2 domain in the tree of life is compatible with the hypothesis of horizontal transfer. The nature of these predicted endonucleases fits well with the hypothesis of lateral transfer; they are known to have spread horizontally into all branches of life and were identified in nuclear and organelle DNA of eukaryotes (Gimble, 2000; Chevalier and Stoddard, 2001; Koufopanou et al., 2002). In accordance with the hypothesis of lateral gene transfer, the three T. thermophila endonuclease genes are located in a new family of mobile genetic elements together with other supposed viral sequences (Wuitschick et al., 2002). The lack of introns in the majority of plant AP2/ERF transcription factors also supports the hypothesis of lateral gene transfer from bacterial or viral origin. In Arabidopsis, only 23 genes out of 145 AP2/ERF genes have introns.
Although the hypotheses of convergent evolution or divergent evolution followed by multiple loss of the character cannot be completely rejected, these data support a direct evolution of plant AP2/ERF transcription factors from bacterial or viral HNH endonucleases. One possible scenario is the evolution of the plant AP2 domain after the endosymbiosis of the ancestral cyanobacterium that gave rise to the chloroplast. Alternatively, the AP2 domain may have moved into plants after viral infections or other lateral gene transfer events. The finding of AP2 domains in the green algae C. reinhardtii implies an early evolution of the AP2 domain in plants. The lack of AP2 domains in the red algae P. yezoensis may be attributable to a later evolution of the AP2 domain or to the loss of the domain in this organism. Like transposons, HNH homing endonucleases are genetic mobile elements that can replicate and move in the genome (Gimble, 2000; Chevalier and Stoddard, 2001; Koufopanou et al., 2002). The ancestral AP2-endonuclease may have colonized the genome, transposing and replicating via a homing process. These endonucleases may have diverged under selection into the AP2/ERF family of transcription factors.
The Evolution of the Plant AP2/ERF Subfamilies
Important clues about the evolution of this large family of transcription factors come from the analysis of intron distribution. In Arabidopsis, all members of the AP2 and fifth subfamily and four ERF genes have introns. The finding of conserved intronic sites suggests the evolution of introns before gene duplication. Nevertheless, the lack of conservation of intron–exon boundaries across subfamilies calls for three independent events. The NJ tree splits the ERF subfamily into two branches, grouping the Arabidopsis ERFs bearing introns together. The tree suggests an early divergence of some ERF proteins as a consequence of intron evolution. The members of the early branching ERF subfamily have a Val and a His or a Val and an Asp at positions 13 and 18 instead of the Val and Glu or Ala and Asp characterizing DREBs and ERFs, respectively (Sakuma et al., 2002). Two intronic markers were found conserved in both the AP2-R1 and AP2-R2 of the AP2 proteins. One site is also present in members with a single AP2 domain. Based on this analysis, we hypothesized a monophyletic evolution of the AP2 subfamily. The ancestor of this subfamily could have evolved introns and then duplicated and diverged. Our hypothesis was confirmed by finding the Arabidopsis intronic markers also conserved in rice.
In Arabidopsis, the DREB and ERF subfamilies are represented many more times than other subfamilies. The presence of introns or the addition of a second DNA binding domain (B3 domain in RAVs and a second AP2 domain in AP2s) correlates with the smaller number of AP2/ERF factors in the other subfamilies. The reduced number of ERFs bearing introns confirms this trend. It is easy to speculate that an early addition of introns or a second DNA binding domain may have impaired the duplicative ability of the hypothesized ancestral HNH endonuclease. A longer piece of DNA would have made a transposition and duplication event less likely.
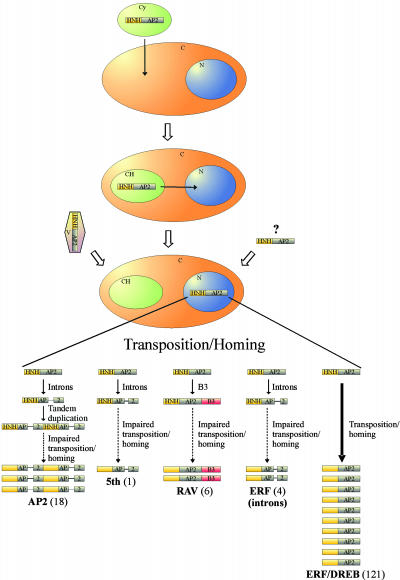
Model
We propose a model for the evolution of the AP2 domain (Figure 8). An HNH endonuclease bearing the AP2 domain may have moved horizontally into plants through the endosymbiosis of a cyanobacterium, viral infections, or other lateral gene transfer events. The endonuclease may have then spread in the genome via transposition and homing processes. Some of the endonucleases may have diverged, losing the HNH domain but retaining the AP2 domain, potentially acquiring new functions. The transposition and duplication of these diverged proteins may still have been triggered by one or a few active HNH endonucleases, eventually resulting in the AP2/ERF family of transcription factors. Three independent events of intron evolution probably affected the ancestors of the AP2, fifth, and part of the ERF subfamilies. The evolution of introns or a new DNA binding domain might have impaired the transposition and homing processes, resulting in a lower rate of duplication.
Figure 8.
Model for the Evolution of the AP2 DNA Binding Domain in Plants.
An HNH-AP2 homing endonuclease may have been transported into plants (C) via the endosymbiosis of an ancestral cyanobacterium (Cy) and may have then moved from the newly formed chloroplast (Ch) into the nucleus (N). Alternatively, it may have been horizontally transferred from viruses (V) or through other lateral gene transfer events. The HNH-AP2 endonuclease may have spread in the genome via transposition and homing processes. An early evolution of introns or B3 domain in the ancestral genes of some of the AP2/ERF subfamilies could have impaired the transposition and homing processes resulting in a reduced number of genes belonging to these subfamilies. Numbers in parentheses indicate the number of members belonging to the specified subfamily in Arabidopsis.
Such lateral gene transfer event would have had a serious impact on the evolution of plant-specific developmental mechanisms because many members of this family are key factors in flower and seed development (Riechmann and Meyerowitz, 1998).
METHODS
Bioinformatics Techniques
The NR database of protein sequences (National Center for Biotechnology Information, National Institutes of Health, Bethesda, MD) was searched using BLAST (Altschul et al., 1990) and PSI-BLAST (Altschul et al., 1997) with an E-value cutoff of 1e-03. The AP2 domain HMM from the PFAM database (Bateman et al., 2004) was scored against the NR protein database and the Saccharomyces cerevisiae, Drosophila melanogaster, Caenorhabditis elegans, and Homo sapiens proteomes. Hits within the PFAM trusted cutoff were considered significant.
Multiple sequence alignments were constructed using Muscle 3.2 (Edgar, 2004) and displayed with Bioedit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) using the BLOSUM62 matrix and a 50% threshold for shading.
Protein secondary structure was predicted using the 3D-PSSM Web server version 2.6.0 (Kelley et al., 2000).
Sequences were submitted to the Conserved Domain Database (Marchler-Bauer et al., 2003), PFAM (Bateman et al., 2004), and the Phylofacts protein structure prediction HMM library (http://phylogenomics.berkeley.edu/resources/) for domain analysis.
We constructed several phylogenetic trees using different methods: neighbor joining and parsimony from the PAUP software suite (http://www.paup.csit.fsu.edu) and maximum likelihood from the PHYLIP software suite (http://evolution.genetics.washington.edu/phylip.html). The NJ tree was constructed using mean character difference, among-site rate variation, and random seed initiation; 10,000 bootstrap replicates were performed followed by identification of the consensus tree including groups compatible with the 50% majority rule. The parsimony tree was constructed from a consensus of 500 trees derived using a heuristic search and the 50% majority rule. The other parameters used were random seed initiation and stepwise addition. Gaps were treated as missing data. The maximum likelihood tree was constructed using the Jones-Taylor-Thornton model.
MacClade software 4.06 (http://macclade.org/macclade.html) was used to trace the AP2 domain character along the tree of life.
Molecular Biology
The Trichodesmium erythraeum AP2 domain coding sequence (NZ_AABK02000006) was amplified using the cyanoF (5′-AAAGGATCCTAAAACATTCATCTAAATACC-3′) and cyanoR (5′-AAAGAATTCATTTCTTTTACCTCTATATTA-3′) primers on a T. erythraeum colony. The amplified product was digested with BamHI and EcoRI and cloned in the pET-21 a-d(+) vector (Novagen, Madison, WI) digested with BamHI and EcoRI in frame with the T7 tag and His tag.
The T. erythraeum gene coding the entire protein containing an AP2 domain (NZ_AABK02000006) was amplified using the cyanoFTnT (5′-ATGTCTAAAGAAATTTTGATTACT-3′) and cyanoRTnT (5′-TTATTATTCTTTTACCTCTATATTATTTA-3′) primers on a T. erythraeum colony. The amplified product was cloned in the pGEMTeasy vector (Promega, Madison, WI). The plasmid was then cut with EcoRI and cloned in the pTnT vector (Promega) digested with EcoRI.
The plasmid was transformed in BL21(DE3)RIL Escherichia coli strain (Stratagene, La Jolla, CA), and overexpression and purification of the recombinant proteins were performed as described in the manufacturer's protocols.
In vitro transcription and translation were performed with the TnT T7-coupled wheat germ extract system (Promega) as described in the manufacturer's protocols.
The SAAB was used to identify the T. erythraeum AP2 domain DNA binding motif as described by Smith et al. (2002). Fifty oligonucleotides identified in the screen were cloned in the pGEMTeasy vector (Promega) and sequenced (Figure 4).
In EMSA, recombinant expressed and purified proteins were mixed with 50,000 cpm of wild-type or mutant DNA probes, 2 μg of poly [d(I·C)] in 1× EMSA buffer (25 mM Tris-HCl, pH 7.5, 40 mM KCl, 0.5 mM DTT, 0.5 mM EDTA, 5% glycerol, 0.5% Nonidet P-40) for 30 min at 4°C in a 20-μL reaction volume. After binding, samples were analyzed by EMSA using 0.5× Tris-borate-EDTA 5% polyacrylamide gels (37.5:1 ratio of acrylamide to bis-acrylamide). After electrophoresis, the gels were dried, developed for 1 to 3 h, and analyzed by autoradiography.
The accession numbers of all proteins mentioned in this article are indicated in the relative figures. Arabidopsis proteins are annotated with Atg numbers and all other proteins with PID numbers. The protein accession numbers displayed in the multiple sequence alignment provided in the supplemental data online are SMART numbers except for the nonplant proteins (PID number), the rice member of the fifth subfamily (PID number), the C. reinhardtii proteins (PID number), and the Arabidopsis member of the fifth subfamily (Atg number).
Supplementary Material
Acknowledgments
We thank Douglas Capone for providing the T. erythraeum colony. We are grateful to Jeremy Dettman, Gabriela Toledo-Ortiz, and members of the Hake lab for helpful discussions.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Enrico Magnani (emagnani@berkeley.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.023135.
References
- Allen, M.D., Yamasaki, K., Ohme-Takagi, M., Tateno, M., and Suzuki, M. (1998). A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 17, 5484–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, T.L., and Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. (Menlo Park, CA: AAAI Press), pp. 28–36. [PubMed]
- Baker, S.S., Wilhelm, K.S., and Thomashow, M.F. (1994). The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 24, 701–713. [DOI] [PubMed] [Google Scholar]
- Bateman, A., et al. (2004). The Pfam protein families database. Nucleic Acids Res. 32, D138–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner, M., and Singh, K.B. (1997). Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc. Natl. Acad. Sci. USA 94, 5961–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier, B.S., and Stoddard, B.L. (2001). Homing endonucleases: Structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 29, 3757–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly, K.M., Wojciak, J.M., and Clubb, R.T. (1998). Site-specific DNA binding using a variation of the double stranded RNA binding motif. Nat. Struct. Biol. 5, 546–550. [DOI] [PubMed] [Google Scholar]
- Dalgaard, J.Z., Klar, A.J., Moser, M.J., Holley, W.R., Chatterjee, A., and Mian, I.S. (1997). Statistical modeling and analysis of the LAGLIDADG family of site-specific endonucleases and identification of an intein that encodes a site-specific endonuclease of the HNH family. Nucleic Acids Res. 25, 4626–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto, S.Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12, 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimble, F.S. (2000). Invasion of a multitude of genetic niches by mobile endonuclease genes. FEMS Microbiol. Lett. 185, 99–107. [DOI] [PubMed] [Google Scholar]
- Hao, D., Ohme-Takagi, M., and Sarai, A. (1998). Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plant. J. Biol. Chem. 273, 26857–26861. [DOI] [PubMed] [Google Scholar]
- Hao, D., Yamasaki, K., Sarai, A., and Ohme-Takagi, M. (2002). Determinants in the sequence specific binding of two plant transcription factors, CBF1 and NtERF2, to the DRE and GCC motifs. Biochemistry 41, 4202–4208. [DOI] [PubMed] [Google Scholar]
- Jiang, C., Iu, B., and Singh, J. (1996). Requirement of a CCGAC cis-acting element for cold induction of the BN115 gene from winter Brassica napus. Plant Mol. Biol. 30, 679–684. [DOI] [PubMed] [Google Scholar]
- Kagaya, Y., Ohmiya, K., and Hattori, T. (1999). RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 27, 470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus, K., Sjolander, K., Barrett, C., Cline, M., Haussler, D., Hughey, R., Holm, L., and Sander, C. (1997). Predicting protein structure using hidden Markov models. Proteins 1 (suppl.), 134–139. [DOI] [PubMed] [Google Scholar]
- Kelley, L.A., MacCallum, R.M., and Sternberg, M.J. (2000). Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299, 499–520. [DOI] [PubMed] [Google Scholar]
- Koufopanou, V., Goddard, M.R., and Burt, A. (2002). Adaptation for horizontal transfer in a homing endonuclease. Mol. Biol. Evol. 19, 239–246. [DOI] [PubMed] [Google Scholar]
- Krizek, B.A. (2003). AINTEGUMENTA utilizes a mode of DNA recognition distinct from that used by proteins containing a single AP2 domain. Nucleic Acids Res. 31, 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic, I., Goodstadt, L., Dickens, N.J., Doerks, T., Schultz, J., Mott, R., Ciccarelli, F., Copley, R.R., Ponting, C.P., and Bork, P. (2002). Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 30, 242–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer, A., et al. (2003). CDD: A curated Entrez database of conserved domain alignments. Nucleic Acids Res. 31, 383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki, M., et al. (2004). Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428, 653–657. [DOI] [PubMed] [Google Scholar]
- Murzin, A.G., Brenner, S.E., Hubbard, T., and Chothia, C. (1995). SCOP: A structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247, 536–540. [DOI] [PubMed] [Google Scholar]
- Nole-Wilson, S., and Krizek, B.A. (2000). DNA binding properties of the Arabidopsis floral development protein AINTEGUMENTA. Nucleic Acids Res. 28, 4076–4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohki, I., Shimotake, N., Fujita, N., Nakao, M., and Shirakawa, M. (1999). Solution structure of the methyl-CpG-binding domain of the methylation-dependent transcriptional repressor MBD1. EMBO J. 18, 6653–6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi, M., and Shinshi, H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J., Karplus, K., Barrett, C., Hughey, R., Haussler, D., Hubbard, T., and Chothia, C. (1998). Sequence comparisons using multiple sequences detect three times as many remote homologues as pairwise methods. J. Mol. Biol. 284, 1201–1210. [DOI] [PubMed] [Google Scholar]
- Park, J.M., Park, C.J., Lee, S.B., Ham, B.K., Shin, R., and Paek, K.H. (2001). Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell 13, 1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann, J.L., and Meyerowitz, E.M. (1998). The AP2/EREBP family of plant transcription factors. Biol. Chem. 379, 633–646. [DOI] [PubMed] [Google Scholar]
- Sakuma, Y., Liu, Q., Dubouzet, J.G., Abe, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009. [DOI] [PubMed] [Google Scholar]
- Shub, D.A., Goodrich-Blair, H., and Eddy, S.R. (1994). Amino acid sequence motif of group I intron endonucleases is conserved in open reading frames of group II introns. Trends Biochem. Sci. 19, 402–404. [DOI] [PubMed] [Google Scholar]
- Sitbon, E., and Pietrokovski, S. (2003). New types of conserved sequence domains in DNA-binding regions of homing endonucleases. Trends Biochem. Sci. 28, 473–477. [DOI] [PubMed] [Google Scholar]
- Smith, H.M., Boschke, I., and Hake, S. (2002). Selective interaction of plant homeodomain proteins mediates high DNA-binding affinity. Proc. Natl. Acad. Sci. USA 99, 9579–9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger, E.J., Gilmour, S.J., and Thomashow, M.F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 94, 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow, M.F. (1999). Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571–599. [DOI] [PubMed] [Google Scholar]
- Vahala, T., Oxelman, B., and von Arnold, S. (2001). Two APETALA2-like genes of Picea abies are differentially expressed during development. J. Exp. Bot. 52, 1111–1115. [DOI] [PubMed] [Google Scholar]
- Wojciak, J.M., Sarkar, D., Landy, A., and Clubb, R.T. (2002). Arm-site binding by lambda-integrase: Solution structure and functional characterization of its amino-terminal domain. Proc. Natl. Acad. Sci. USA 99, 3434–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuitschick, J.D., Gershan, J.A., Lochowicz, A.J., Li, S., and Karrer, K.M. (2002). A novel family of mobile genetic elements is limited to the germline genome in Tetrahymena thermophila. Nucleic Acids Res. 30, 2524–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6, 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J., Tang, X., and Martin, G.B. (1997). The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis-element of pathogenesis-related genes. EMBO J. 16, 3207–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.