Abstract
The Arabidopsis thaliana proteins suppressor of phytochrome A-105 1 (SPA1), SPA3, and SPA4 of the four-member SPA1 protein family have been shown to repress photomorphogenesis in light-grown seedlings. Here, we demonstrate that spa quadruple mutant seedlings with defects in SPA1, SPA2, SPA3, and SPA4 undergo strong constitutive photomorphogenesis in the dark. Consistent with this finding, adult spa quadruple mutants are extremely small and dwarfed. These extreme phenotypes are only observed when all SPA genes are mutated, indicating functional redundancy among SPA genes. Differential contributions of individual SPA genes were revealed by analysis of spa double and triple mutant genotypes. SPA1 and SPA2 predominate in dark-grown seedlings, whereas SPA3 and SPA4 prevalently regulate the elongation growth in adult plants. Further analysis of SPA2 function indicated that SPA2 is a potent repressor of photomorphogenesis only in the dark but not in the light. The SPA2 protein is constitutively nuclear localized in planta and can physically interact with the repressor COP1. Epistasis analysis between spa2 and cop1 mutations provides strong genetic support for a biological significance of a COP1–SPA2 interaction in the plant. Taken together, our results have identified a new family of proteins that is essential for suppression of photomorphogenesis in darkness.
INTRODUCTION
Plants have evolved a variety of mechanisms to adapt growth and development to the ambient light environment. To monitor the light, plants use several classes of photoreceptors. Among these, the red light (R) and far-red light (FR)–sensing phytochromes and the blue light (B)/UV-A–perceiving cryptochromes regulate many aspects of plant development, including seed germination, seedling deetiolation, anthocyanin accumulation, shade avoidance behavior, and the induction of flowering (Neff et al., 2000). Arabidopsis thaliana phytochromes are encoded by a small family comprising five genes (PHYA to PHYE), of which PHYA and PHYB have the most important functions in the plant. Phytochrome A (phyA) is a light-labile photoreceptor that mediates seedling deetiolation responses to very low fluences of light as well as to continuous FR (FRc). The light-stable phytochrome B controls FR-reversible responses to R, whereas the cryptochromes cry1 and cry2 mediate deetiolation in B (Briggs and Olney, 2001; Sullivan and Deng, 2003; Franklin and Whitelam, 2004).
Research efforts have identified positively and negatively acting components involved in the light signaling pathway (Fankhauser and Staiger, 2002; Quail, 2002; Gyula et al., 2003). In darkness, photomorphogenesis is suppressed by the action of 10 repressors (COP/DET/FUS) (Kim et al., 2002; Serino and Deng, 2003). Therefore, mutants deficient in any of these repressors undergo constitutive photomorphogenesis and exhibit features of light-grown seedlings even in complete darkness. One of these repressors, COP1, has E3 ubiquitin ligase activity and promotes the ubiquitination and degradation of the transcription factors HY5 and LAF1, which are positively acting light signaling intermediates (Osterlund et al., 2000; Saijo et al., 2003; Seo et al., 2003). Hence, repression of photomorphogenesis in darkness involves the degradation of positively acting signaling intermediates.
The COP1 protein contains a WD-repeat domain, a coiled-coil domain, and a RING finger typical of a subclass of E3 ubiquitin ligases (Deng et al., 1992; Osterlund et al., 1999). COP1 is primarily active in the dark and is inactivated by light through the functions of phytochromes and cryptochromes (Deng and Quail, 1999). One, but likely not the only, mechanism by which light inactivates COP1 function is a change in the subcellular localization of COP1. COP1 is primarily present and functional in the nucleus in dark-grown seedlings, whereas it is excluded from the nucleus in the light (Von Arnim and Deng, 1994; Subramanian et al., 2004). Recent evidence for a direct physical interaction between COP1 and cryptochromes phyA and phytochrome B suggests that photoreceptors may directly inactivate COP1 (Wang et al., 2001; Yang et al., 2001; Seo et al., 2004). Furthermore, as a possible feedback regulation, COP1 can ubiquitinate phyA and may thereby desensitize phyA-mediated signaling (Seo et al., 2004).
Negative regulators that function in the light have been identified in screens for mutants that exhibit increased responsiveness to light (Hoecker et al., 1998; Büche et al., 2000). Thus, isolated suppressor of phytochrome A-105 1 (spa1) mutants show exaggerated photomorphogenesis in R and FR but not in darkness. The phenotypic effects of spa1 mutations are dependent on a functional PHYA gene, suggesting that SPA1 is important for normal phyA signaling (Hoecker et al., 1998). Moreover, spa1 mutations enhance both modes of phyA action, the response to FRc and the very low fluence response (Baumgardt et al., 2002; Zhou et al., 2002). It was also suggested that growth promotion by SPA1 counteracts phytochrome-mediated growth inhibition in hypocotyls (Parks et al., 2001). SPA1 encodes a nuclear-localized WD-repeat protein that also contains a putative coiled-coil domain and a region related to protein kinases (Hoecker et al., 1999). Its WD-repeat domain shows close sequence similarity to the WD-repeat of COP1, whereas outside the WD-repeat regions the two proteins are unrelated by sequence. SPA1 can physically interact with COP1 via their respective coiled-coil domains, suggesting that SPA1 may function in concert with COP1 to target proteins for degradation and thereby to repress photomorphogenesis in the light (Hoecker and Quail, 2001; Saijo et al., 2003). Indeed, SPA1, like COP1, interacts with HY5 and reduces HY5 protein levels in the light (Saijo et al., 2003). Consistent with this finding, it was recently shown that SPA1 can modulate the E3 ligase activity of COP1. In these studies, SPA1 inhibited in vitro ubiquitination of HY5 by COP1 (Saijo et al., 2003), whereas the coiled-coil domain of SPA1 was able to enhance COP1-mediated in vitro ubiquitination of LAF1 (Seo et al., 2003).
SPA1 is a member of a small family that includes three more proteins (SPA2, SPA3, and SPA4). All members have a similar domain structure, including a WD-repeat domain, a coiled-coil domain, and a kinase-like region, though the sequence similarity among members is highest within the WD-repeat domain (Laubinger and Hoecker, 2003). Mutations in SPA3 and SPA4 cause increased photoresponsiveness in seedlings, indicating that SPA3 and SPA4, like SPA1, function as repressors of photomorphogenesis in the light (Laubinger and Hoecker, 2003). In this study, we have investigated the function of the last member of the SPA1 gene family, SPA2. Furthermore, the availability of mutants in all four SPA genes allowed us to construct and investigate a quadruple mutant that is defective in all four SPA proteins. We consider this of particular importance because the SPA proteins exhibit a very similar domain structure and, moreover, are capable of interacting with COP1 (Laubinger and Hoecker, 2003; this report). Hence, SPA proteins may have redundant functions in the plant.
RESULTS
spa2 Mutant Seedlings Do Not Show Altered Responses to Light
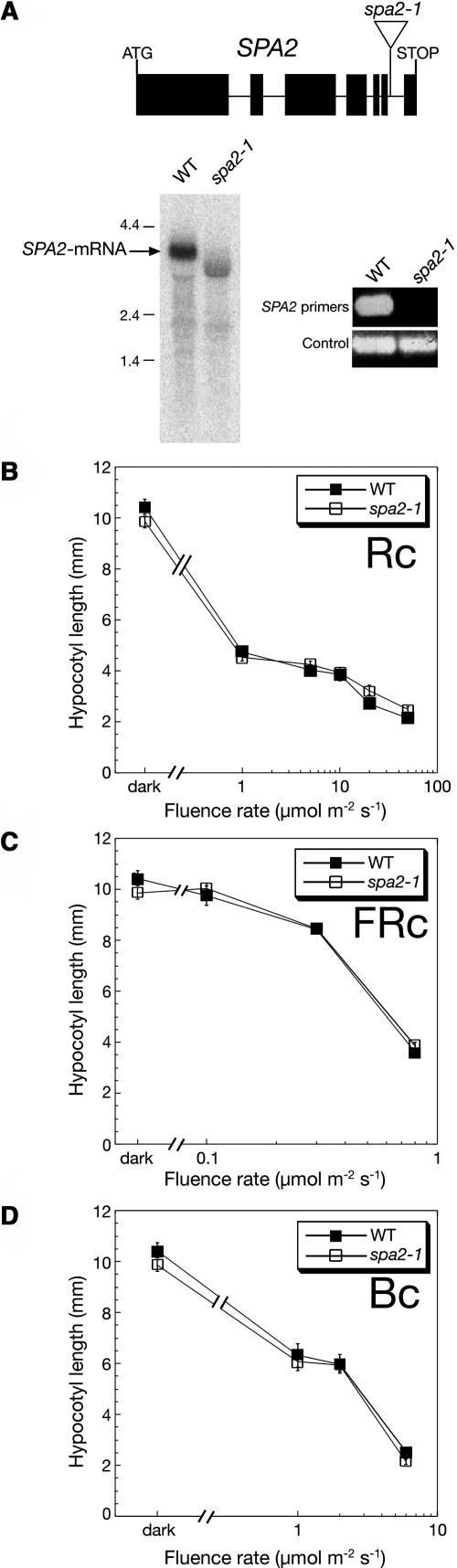
To investigate the function of SPA2, we searched for spa2 mutant alleles in T-DNA insertion populations. In the GABI-Kat population, we identified a line that carries a T-DNA insertion in the last intron of SPA2 at position 4008 bp after the presumed start codon (Figure 1A). In a population segregating for this T-DNA insertion, we identified homozygous spa2-1 mutant plants and homozygous wild-type siblings using PCR-based markers that can discriminate between the mutant and the wild-type SPA2 allele. RNA gel blot analysis revealed that the T-DNA insertion in the spa2-1 mutant affected normal wild-type transcript size (Figure 1A). RT-PCR analysis confirmed that the spa2-1 mutant accumulated no wild-type SPA2 transcript (Figure 1A). These results indicate that spa2-1 mutant plants are disrupted in normal SPA2 gene function. Because the T-DNA in spa2-1 interrupts the WD-repeat–encoding sequence, which is essential for the functions of the related proteins SPA1, SPA3, SPA4, and COP1 (McNellis et al., 1994b; Hoecker et al., 1999; Laubinger and Hoecker, 2003), it is very likely that possibly produced SPA2 protein is nonfunctional.
Figure 1.
Dark- and Light-Grown spa2 Mutant Seedlings Are Indistinguishable from the Wild Type.
(A) Molecular analysis of the spa2-1 mutant. The top panel shows the SPA2 transcript splicing model and the location of the T-DNA insertion site in spa2-1. The bottom left panel shows the RNA gel blot analysis of RNA isolated from Rc-grown wild-type and spa2-1 mutant seedlings. The blot was hybridized with a SPA2-specific probe. The bottom right panel shows an RT-PCR analysis of RNA isolated from wild-type and spa2-1 mutant seedlings. RNA was reverse transcribed and subsequently amplified by PCR using primers flanking the T-DNA insertion site in spa2-1 or, as a control, SPA3-specific primers.
(B) to (D) Hypocotyl length of wild-type and spa2-1 mutant seedlings grown in Rc (B), FRc (C), or Bc (D) of various fluence rates. Error bars denote one standard error of the mean.
To investigate whether the spa2 mutation alters light responses, we compared the phenotypes of continuous R (Rc)-, FRc-, continuous B (Bc)-, and dark-grown spa2 mutant seedlings with those of wild-type seedlings. Under all light conditions tested, the spa2 mutant was indistinguishable from the wild type (Figures 1B to 1D). No morphological changes in hypocotyl elongation, cotelydon opening, or expansion were observed in the spa2 mutant.
Dark-Grown spa1 spa2 Double Mutants Show Features of Light-Grown Seedlings
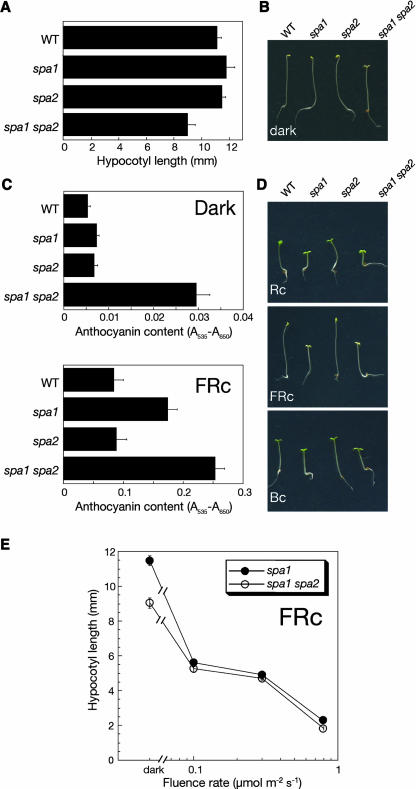
Because the spa2 mutant was indistinguishable from the wild type, we considered the possibility that SPA2 might function redundantly with other SPA genes. The sequence of SPA2 is most closely related to that of SPA1 (Laubinger and Hoecker, 2003). We therefore generated the spa1 spa2 double mutant and investigated its phenotype. When grown in the dark, spa1 spa2 double mutant seedlings exhibited some features of light-grown seedlings: the apical hook was open, cotelydons were unfolded, and the hypocotyl was shorter than that of the wild type or either single mutant seedling (Figures 2A and 2B). From a segregating population, we generated progeny of several homozygous spa1 spa2 mutant plants and observed similar weak constitutive photomorphogenesis in all spa1 spa2 double mutant progenies but in none of the examined progenies of spa1 single mutant siblings. Hence, weak constitutive photomorphogenesis was only observed when both genes, SPA1 and SPA2, were defective.
Figure 2.
spa1 spa2 Double Mutant Seedlings Exhibit Weak Deetiolation in the Dark.
(A) and (B) Hypocotyl length (A) and visual phenotype (B) of dark-grown wild-type, spa1 mutant, spa2 mutant, and spa1 spa2 double mutant seedlings.
(C) Anthocyanin content of seedlings grown in darkness or FRc (3 μmol m−2 s−1).
(D) Visual phenotype of seedlings grown in Rc (8 μmol m−2 s−1), FRc (0.08 μmol m−2 s−1), or Bc (1 μmol m−2 s−1).
(E) Hypocotyl length of spa1 mutant and spa1 spa2 double mutant seedlings grown in FRc of various fluence rates. In all figures, error bars denote one standard error of the mean.
We also examined another normally light-dependent response, the accumulation of anthocyanin. Dark-grown wild-type and spa1 and spa2 single mutant seedlings accumulated very low levels of anthocyanin. spa1 spa2 double mutants, by contrast, accumulated approximately threefold higher levels of anthocyanin in the dark (Figure 2C). Thus, the phenotype of dark-grown spa1 spa2 double mutant seedlings indicates that a loss of SPA1 or SPA2 function has no influence on skotomorphogenesis, whereas a loss of SPA1 and SPA2 function results in partially constitutive photomorphogenesis. Hence, SPA1 and SPA2 act redundantly in the suppression of photomorphogenesis in the dark.
In light-grown seedlings, a lack of SPA2 function, in a spa1 mutant background, had little effect. Under all light conditions tested, the spa1 spa2 double mutant was morphologically indistinguishable from the spa1 single mutant (Figures 2D and 2E). Anthocyanin levels in FRc, however, were slightly higher in the spa1 spa2 double mutant when compared with the spa1 single mutant (Figure 2C). Hence, taken together, these results suggest that SPA2 functions primarily in darkness and has only a limited function in the light. spa1 mutations, by contrast, strongly increase seedling responses to light (Hoecker et al., 1998; Figures 2C and 2D). Thus, in the light, SPA1 appears to be more important than SPA2 in the inhibition of photomorphogenesis.
Complementation of the spa1 spa2 Mutant Phenotype
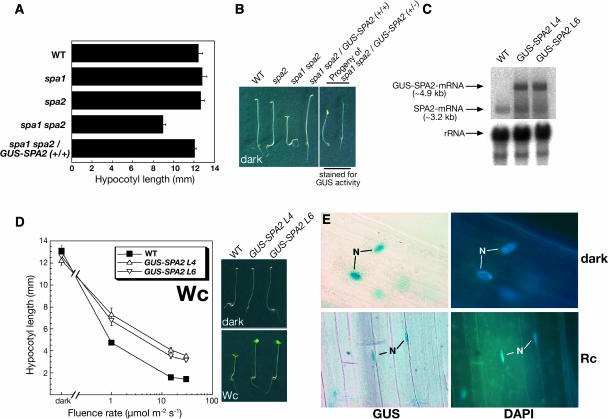
To confirm that the loss of SPA2 function is responsible for the observed deetiolation of dark-grown spa1 spa2 double mutant seedlings and, moreover, to be able to determine the subcellular localization of the SPA2 protein, we introduced a 35S:β-glucuronidase (GUS)-SPA2 transgene into spa1 spa2 double mutant and wild-type plants. Dark-grown spa1 spa2 mutant seedlings that were homozygous for the GUS-SPA2 transgene displayed a wild-type phenotype with long hypocotyls, closed apical hooks, and folded cotyledons (Figures 3A and 3B). This indicates that the GUS-SPA2 transgene fully complemented the spa1 spa2 mutant phenotype.
Figure 3.
Analysis of Seedlings Expressing a GUS-SPA2 Fusion Protein.
(A) and (B) Expression of GUS-SPA2 complements the mutant phenotype of dark-grown spa1 spa2 double mutant seedlings.
(A) Hypocotyl length of dark-grown wild-type, spa1 mutants, spa2 mutants, spa1 spa2 double mutants, and transgenic spa1 spa2 double mutants homozygous for a 35S:GUS-SPA2 transgene (+/+). Error bars denote one standard error of the mean.
(B) Visual phenotype of dark-grown seedlings. Genotypes in the left photograph were as in (A). The right photograph shows representative seedlings of the segregating phenotypic classes found in dark-grown progeny of a homozygous spa1 spa2 double mutant that was hemizygous for the 35S:GUS-SPA2 transgene (+/−). Seedlings were stained for GUS activity.
(C) RNA gel blot analysis of the wild type and two independent 35S:GUS-SPA2 transgenic lines in a wild-type background (L4 and L6). Seedlings were grown in 30 μmol m−2 s−1 Rc. The membrane was hybridized with a SPA2-specific probe. Equal loading was confirmed by rehybridization with an 18S-rRNA–specific probe.
(D) Hypocotyl length of the wild type and two independent 35S:GUS-SPA2 transgenic lines (GUS-SPA2 L4 and GUS-SPA2 L6) grown in white light (Wc) of various fluence rates. The photographs at the right show the visual phenotype of seedlings grown in darkness or Wc (1 μmol m−2 s−1). Error bars denote one standard error of the mean.
(E) Representative cellular GUS staining patterns in the hypocotyl of transgenic GUS-SPA2 L4 seedlings. GUS-SPA2 L4 seedlings were grown in the dark or in Rc (50 μmol m−2 s−1) and were stained for GUS activity and for the positions of the nuclei using 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI).
We also analyzed progeny of a spa1 spa2 double mutant plant that was hemizygous for the GUS-SPA2 transgene. In this population, approximately three-quarters of the dark-grown seedlings displayed a wild-type phenotype, whereas one-quarter of the seedlings showed a weak constitutive photomorphogenesis phenotype, as expected for a segregation of the dominant GUS-SPA2 transgene (Figure 3B). To confirm that the GUS-SPA2 transgene was responsible for the rescue of the spa1 spa2 double mutant phenotype, we stained seedlings for GUS activity. All seedlings with long hypocotyls exhibited GUS activity, whereas all short seedlings lacked the activity of GUS (Figure 3B). Thus, complementation of the spa1 spa2 mutant phenotype cosegregated with the expression of GUS-SPA2. We therefore conclude that the spa2-1 mutation is responsible for the constitutive photomorphogenesis phenotype observed in the spa1 spa2 double mutant.
In a wild-type background, GUS-SPA2 overexpression caused reduced responses to light (Figures 3C and 3D), whereas no change in phenotype was observed in control seedlings expressing GUS (data not shown). The hyposensitivity of GUS-SPA2 overexpressing lines confirms that SPA2 is a repressor in the light signaling pathway. A similar phenotype was also observed when the repressor COP1 was overexpressed (McNellis et al., 1994a).
The SPA2 Protein Is Constitutively Localized to the Nucleus in Planta
Nucleocytoplasmic partitioning has been implicated in the regulation of the activity of several light signaling intermediates, including phytochromes and COP1 (Von Arnim and Deng, 1994; Kircher et al., 1999, 2002; Subramanian et al., 2004). To investigate the subcellular localization of the SPA2 protein, we examined transgenic seedlings overexpressing GUS-SPA2. When grown in the dark, uniform GUS staining was observed in the nucleus (Figure 3E). This GUS staining pattern was not different in Rc-, FRc-, Bc-, or continuous white light (Wc)-grown transgenic seedlings (Figure 3E; data not shown). Transgenic seedlings that expressed GUS alone exhibited GUS activity throughout the cytoplasm (data not shown). Taken together, these results indicate that SPA2 is a constitutively nuclear-localized protein. The nuclear localization of SPA2 is consistent with the finding that SPA2 contains a putative bipartite nuclear localization sequence (386-RRRLGDTSSLSIPAKKQK-403).
The Constitutive Photomorphogenesis Phenotype of spa1 spa2 Double Mutants Is Strongly Enhanced by Additional Loss of SPA3 and/or SPA4 Function
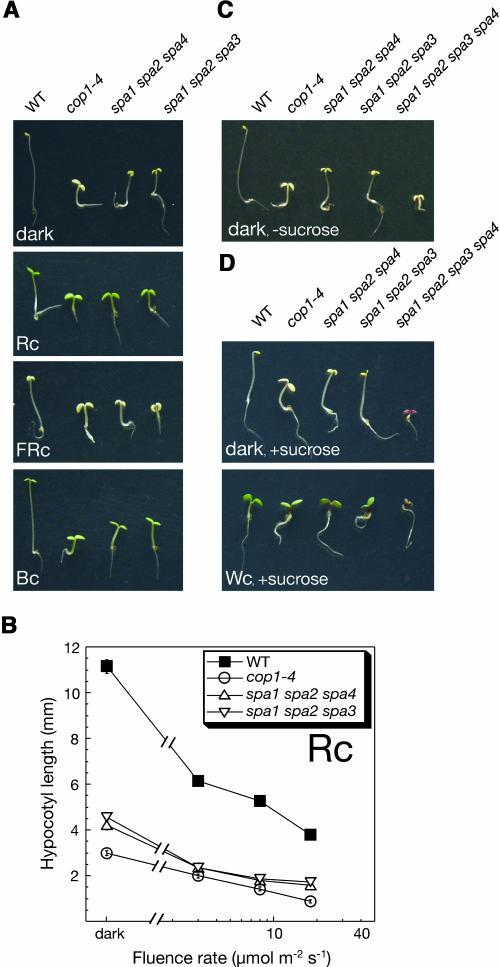
Based on the finding that the spa1 spa2 double mutant exhibited weak deetiolation in the dark, we asked whether this effect is enhanced by additional loss of SPA3 or SPA4 function. Therefore, we constructed spa1 spa2 spa4 and spa1 spa2 spa3 triple mutants and analyzed their deetiolation behavior in the dark. Both triple mutants exhibited more pronounced constitutive photomorphogenesis than the spa1 spa2 double mutant. Dark-grown triple mutant seedlings displayed a short hypocotyl and fully unfolded cotyledons and had an appearance very similar to that of the cop1-4 mutant (Figure 4A). In the light, triple mutant seedlings were also considerably shorter than wild-type seedlings (Figure 4A). Because triple mutants retained some light responsiveness (Figure 4B), they were not fully constitutively photomorphogenic.
Figure 4.
spa Triple Mutant and spa1 spa2 spa3 spa4 Quadruple Mutant Seedlings Exhibit Strong Constitutive Photomorphogenesis.
(A) Visual phenotype of wild-type, cop1-4 mutant, and spa1 spa2 spa4 and spa1 spa2 spa3 triple mutant seedlings grown in darkness, Rc (8 μmol m−2 s−1), FRc (0.08 μmol m−2 s−1), or Bc (1 μmol m−2 s−1).
(B) Hypocotyl length of seedlings grown in Rc of various fluence rates. Genotypes are as in (A). Error bars denote one standard error of the mean.
(C) Visual phenotype of wild-type, cop1-4, two spa triple mutants, and spa1 spa2 spa3 spa4 quadruple mutant seedlings grown in the dark in the absence of sucrose.
(D) Visual phenotype of seedlings grown in the dark or in continuous white light (Wc, 30 μmol m−2 s−1) in the presence of sucrose. Genotypes are as in (C).
To investigate the phenotype of the spa1 spa2 spa3 spa4 quadruple mutant that is defective in the functions of all SPA genes, we analyzed progeny of homozygous triple mutants (spa1 spa2 spa4 or spa1 spa2 spa3) that were heterozygous at the fourth SPA locus (spa3/+ or spa4/+, respectively). Hence, a quarter of the seedlings in these progenies were expected to be spa quadruple mutants. When these populations were grown in the dark, ∼25% of the seedlings underwent very strong deetiolation, showing an extremely short hypocotyl and fully opened cotyledons (Figure 4C). PCR-based analysis of the genotype of these seedlings confirmed that these seedlings were homozygous mutant at all four SPA loci. When these populations were grown on growth medium supplemented with sucrose, spa1 spa2 spa3 spa4 quadruple mutant seedlings exhibited a dark purple coloration as a result of high anthocyanin accumulation (Figure 4D). Dark-grown and light-grown quadruple mutant seedlings had a very similar appearance (Figure 4D), indicating that photomorphogenesis in these mutants was fully constitutive. Hence, the phenotype of the spa quadruple mutant is reminiscent of that of fusca mutants that are defective in COP/DET/FUS genes (Schwechheimer and Deng, 2000).
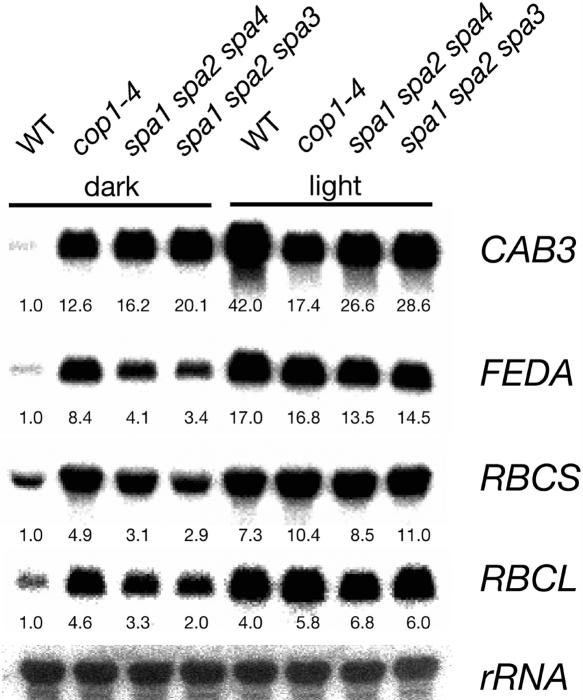
Defects in Multiple SPA Genes Cause Derepression of Light-Regulated Genes in Darkness
To investigate whether defects in multiple SPA genes also cause changes on the molecular level, we examined steady state transcript abundance of several light-induced genes (CAB3, FEDA, RBCS, and RBCL) in homozygous spa triple mutants that show constitutive deetiolation (Figure 5). When grown in darkness, the spa triple mutants spa1 spa2 spa4 and spa1 spa2 spa3 accumulated considerably more of these transcripts than the wild type. Thus, transcript accumulation was derepressed in the spa triple mutants and, moreover, very similar to that observed in the cop1-4 mutant. In conclusion, these spa triple mutants underwent constitutive photomorphogenesis also on the level of gene expression.
Figure 5.
Dark-Grown spa Triple Mutant Seedlings Exhibit Increased Transcript Levels of Light-Induced Genes.
RNA gel blot analysis of total RNA isolated from wild-type, cop1-4 mutant, and spa1 spa2 spa4 and spa1 spa2 spa3 triple mutant seedlings grown in darkness or continuous white light (50 μmol m−2 s−1) for 4 d. Blots were hybridized with CAB3, FEDA, RBCS, RBCL, and 18S rRNA probes. All signals were quantified by phosphor imager analysis. Signals corresponding to the light-regulated genes were normalized with the signal of the 18S rRNA (loading control). Numbers indicate normalized transcript levels relative to the respective signal of dark-grown wild-type seedlings (= 1.0).
SPA2 Is Sufficient to Allow Normal Seedling Development in the Dark, but Not in the Light
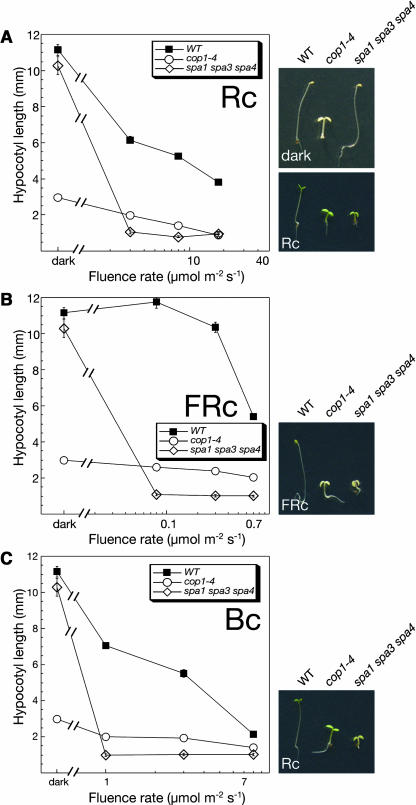
The analyses of spa multiple mutants indicates that SPA genes have redundant functions in repressing photomorphogenesis in dark-grown seedlings. Moreover, the above characterization of spa triple mutants (spa1 spa2 spa3 and spa1 spa2 spa4) that only have one functional SPA gene, SPA3 or SPA4, demonstrates that these SPA gene functions are not sufficient to allow normal seedling development in the dark. To investigate the contribution of SPA2, we examined spa1 spa3 spa4 mutants that only have a normal SPA2 gene. When grown in the dark, wild-type and spa1 spa3 spa4 triple mutant seedlings underwent normal skotomorphogenesis, showing an elongated hypocotyl, a closed apical hook, and folded cotyledons (Figure 6A). Hence, SPA2 function appears to be sufficient to allow normal seedling development in darkness.
Figure 6.
SPA2 Is Sufficient to Repress Photomorphogenesis in the Dark.
Hypocotyl length of wild-type, cop1-4 mutant, and spa1 spa3 spa4 triple mutant seedlings grown in Rc (A), FRc (B), or Bc (C) of various fluence rates. Error bars denote one standard error of the mean. The photographs to the right show the visual phenotype of seedlings grown in darkness or Rc (2 μmol m−2 s−1) (A), FRc (0.08 μmol m−2 s−1) (B), or Bc (1 μmol m−2 s−1) (C).
In the light (Rc, FRc, or Bc), spa1 spa3 spa4 triple mutant seedlings exhibited extremely short hypocotyls. Already rather low fluence rates of light were sufficient to saturate the light response in this spa triple mutant (Figures 6A to 6C). Hence, this mutant was extremely hypersensitive to light. Taken together, these results indicate that SPA2 function is sufficient to allow normal seedling development only in darkness and not in the light.
Because SPA1 is most closely related to SPA2, we investigated whether SPA1, like SPA2, is sufficient to support normal skotomorphogenesis. Indeed, a population that was homozygous SPA1(+/+) spa2 spa2 and segregating for spa3-1 and spa4-1 did not show any deetiolating seedlings when grown in the dark (data not shown), suggesting that spa2 spa3 spa4 mutants undergo etiolated growth. Thus, SPA1 appears to be sufficient for normal dark development of seedlings.
Adult Development of spa Multiple Mutants
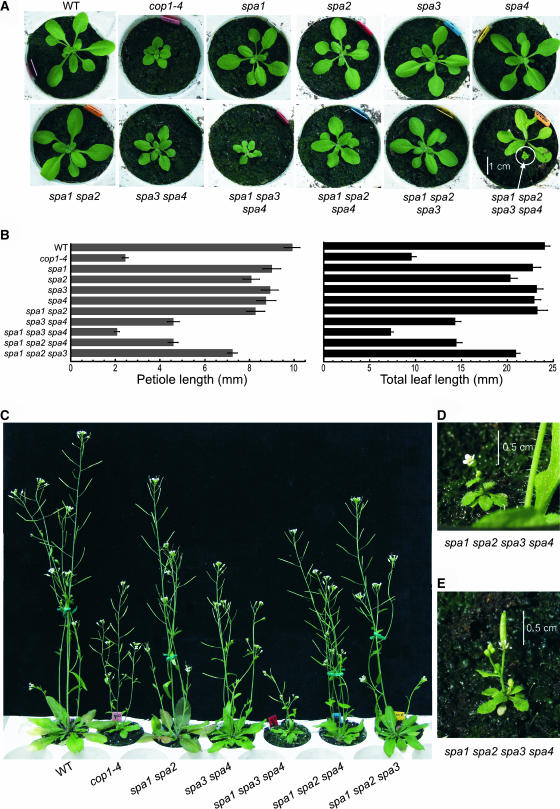
Light also controls growth and development of adult plants. When grown under low fluence rates of light or a low R/FR ratio, plants exhibit long petioles and small leaf blades, whereas high fluence rates of light or a high R/FR ratio induce the opposite response (Casal et al., 2003). Consistent with these responses to light, it was observed that viable constitutively photomorphogenic mutants, like cop1-1 or cop1-4, are very dwarfed (Deng and Quail, 1992). We therefore examined the growth behavior of adult spa mutant plants.
When only one SPA gene was mutated, plants did not show any apparent changes in morphology: rosette size, petiole length, and leaf and inflorescence size were not significantly different in single spa mutants than in the wild type under the growth conditions used (Figures 7A and 7B; data not shown). However, when a population that segregated spa quadruple mutants was sown directly on soil, we observed a fraction of extremely small and dwarfed plants. Rosette diameter of these plants was <1 cm (Figure 7A, bottom right). PCR analysis of the genotype of these plants confirmed that they were mutated at all four SPA loci. These tiny quadruple spa mutant plants developed an inflorescence that produced normal flowers (Figure 7D). Flowers were fertile and produced small siliques containing a few seeds (Figure 7E). Thus, SPA gene function is clearly very important for normal growth of adult plants. Moreover, these results show that there is functional redundancy among SPA genes in the adult stage.
Figure 7.
Phenotypes of Adult Plants with Defects in Individual or Multiple SPA Genes.
(A) Visual phenotype of rosette-stage wild-type, cop1-4 mutant, and spa single, double, triple, and quadruple mutant plants. All plants except for the spa1 spa2 spa3 spa4 quadruple mutant were 19 d old. The bottom right panel shows a 22-d-old spa1 spa2 spa3 spa4/+ plant next to a homozygous spa1 spa2 spa3 spa4 quadruple mutant plant of the same age.
(B) Petiole length and total leaf length of 19-d-old plants. Genotypes are the same as in (A).
(C) Visual phenotype of 33-d-old flowering plants. Genotypes are the same as in (A).
(D) and (E) Visual phenotype of a 26-d-old (D) or a 32-d-old (E) spa1 spa2 spa3 spa4 quadruple mutant plant.
To determine the contributions of the four SPA genes to regulating adult growth, we investigated the phenotypes of spa double and triple mutants. spa1 spa2 double mutant plants appeared normal (Figures 7A to 7C), indicating that SPA1 and SPA2 are not necessary to support normal elongation growth of the adult plant. spa3 spa4 double mutants, by contrast, were smaller and slightly dwarfed, especially early in development, suggesting that SPA3 and SPA4 have a function in the adult plant that cannot be fully replaced by SPA1 and SPA2 (Figures 7A to 7C). When examining spa triple mutants, we found that triple mutants carrying only a functional SPA3 or SPA4 gene, respectively, showed an almost or fully wild-type growth behavior. In particular, spa1 spa2 spa3 triple mutants appeared very similar to the wild type under the growth conditions used (Figures 7A to 7C). Hence, a single functional SPA gene, SPA3 or SPA4, appears to be sufficient to support a rather normal adult morphology under the growth conditions used.
The SPA2 gene, by contrast, was not sufficient for normal adult growth. spa1 spa3 spa4 triple mutant plants carrying only a functional SPA2 gene were very small and dwarfed, exhibiting short petioles and small leaves (Figures 7A and 7B). Also, inflorescences of these triple mutants were very short (Figure 7C). To confirm that this phenotype was associated with the spa1 spa3 spa4 genotype, we also examined plants of a segregating population. This population was homozygous for spa1, spa4, and SPA2(+/+), but segregating at the SPA3 locus (spa3/+). As expected, it segregated dwarfed plants that were confirmed by molecular genotyping to be homozygous mutant for spa3, whereas segregating normal appearing plants carried at least one wild-type allele of SPA3 (data not shown). The dwarfed phenotype of spa1 spa3 spa4 mutants indicates that SPA2 is not sufficient to complement the lack of the other three SPA genes.
Taken together, these results demonstrate that SPA3 and SPA4 make a larger contribution to controlling adult growth than SPA2. Moreover, these analyses demonstrate developmental differences in the functions of the four SPA genes. Although SPA2 is sufficient to allow normal seedling development in the dark, it is not sufficient for normal growth of seedlings in the light or of adult plants. On the contrary, SPA3 and SPA4 are not sufficient for normal seedling development in darkness or in the light, but do support close to normal growth of the adult plant.
SPA2 Physically Interacts with COP1
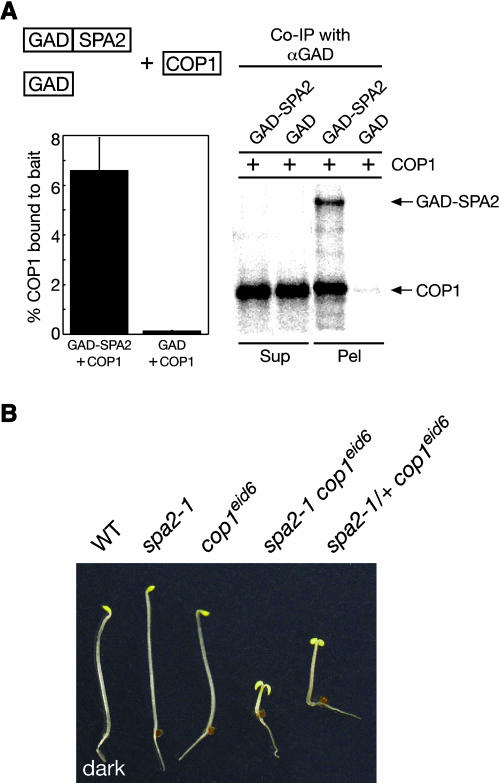
The proteins SPA1, SPA3, and SPA4 can physically interact with COP1, the constitutive negative regulator of photomorphogenesis (Hoecker and Quail, 2001; Laubinger and Hoecker, 2003; Saijo et al., 2003). We therefore tested whether SPA2 is also capable of binding COP1. Indeed, SPA2 interacted with COP1 in an in vitro interaction assay using recombinant proteins (Figure 8A). Approximately 7% of the added COP1 protein was coimmunoprecipitated with the bait GAD-SPA2, whereas <0.2% of the added COP1 bound to the control bait GAD. These results indicate that SPA2 can physically interact with COP1.
Figure 8.
SPA2 Interacts with COP1.
(A) In vitro coimmunoprecipitation of COP1 by SPA2. Recombinant 35S-labeled COP1 was incubated with partially 35S-labeled GAD-SPA2 or GAD, respectively, and coimmunoprecipitated with anti-GAD antibodies. Supernatant fractions (2.15%) (Sup) and 33.3% of the pellet fractions (Pel) were resolved by SDS-PAGE and visualized by autoradiography using a phosphor imager. Quantification of the fractions of COP1 that were coimmunoprecipitated by GAD or GAD-SPA2 are shown in the graph to the left. Error bars denote one standard error of the mean of two replicate experiments.
(B) Genetic interaction of spa2 and cop1 mutations. The photograph shows the phenotypes of wild-type, spa2-1 mutant, cop1eid6 mutant, spa2-1 cop1eid6 double mutant, and spa2-1/+ cop1eid6 seedlings grown in the dark.
spa2 and cop1 Mutations Interact Synergistically
To test whether the observed physical interaction between SPA2 and COP1 is of biological relevance in Arabidopsis, we investigated the epistatic relationship between spa2 and cop1 mutations. We generated a double mutant between spa2-1 and the weak cop1 mutant allele cop1eid6. This recently described nonconstitutive photomorphogenic cop1 allele causes a single amino acid exchange in the RING finger of COP1. The produced mutant COP1 protein is fully functional in the dark but not in the light. Thus, cop1eid6 mutant seedlings undergo normal seedling development in darkness (Dieterle et al., 2003). Because spa2-1 single mutant seedlings also develop normally in darkness (Figure 1), we could examine whether the spa2 mutation genetically interacts with the cop1eid6 allele in dark-grown seedlings.
A segregating F2 population derived from a cross of spa2-1 with cop1eid6 as well as the parents were analyzed for their etiolation behavior in the dark (Figure 8B). As expected, spa2 and cop1eid6 single mutants were fully etiolated and indistinguishable from the wild type. The F2 population derived from these parents segregated three phenotypic classes: fully etiolated seedlings, seedlings undergoing pronounced constitutive photomorphogenesis, and seedlings undergoing weaker constitutive photomorphogenesis. The frequency of the strongly deetiolated seedlings was consistent with the 15:1 segregation ratio expected for spa2 cop1eid6 double mutants (22 short seedlings out of 384 seedlings). Indeed, this genotype was confirmed by molecular analysis using allele-specific PCR-based markers: of 15 short seedlings examined, all were found to be homozygous mutant for spa2-1 and cop1eid6. Tall siblings, by contrast, segregated for these mutations. All seedlings showing weaker deetiolation were found to be spa2-1/+, cop1eid6/cop1eid6 (Figure 8B). They segregated close to the expected ratio of 14:2 (46 weakly consititutively photomorphogenic seedlings out of 384 seedlings). Hence, these results clearly demonstrate a synergistic interaction between the mutations spa2-1 and cop1eid6. They provide strong genetic support for a functional interaction of SPA2 and COP1 in Arabidopsis seedlings. We propose that the mutant COP1EID6 protein requires bound SPA2 protein to be fully functional in dark-grown seedlings. In the absence of SPA2 or in a spa2-1/+ heterozygous situation, COP1EID6 is considerably less active, thereby causing constitutive photomorphogenesis.
We tried to investigate whether spa2-1 and cop1eid6 interacted synergistically also in light-grown seedlings. spa2-1 cop1eid6 and cop1eid6 seedlings were extremely short, even at low fluence rates such as 0.01 μmol m−2 s−1 FRc (data not shown). Thus, because cop1eid6 mutant seedlings are very hypersensitive to light, as described by Dieterle et al. (2003), we were not able to study an additional effect of the spa2 mutation.
DISCUSSION
Negative regulators are important intermediates in the light signaling pathway (Kim et al., 2002). The Arabidopsis repressor COP1 is essential for suppression of photomorphogenesis in dark-grown seedlings, and its activity is inhibited in the light through the functions of several photoreceptors (Osterlund et al., 1999). The COP1-interacting proteins SPA1, SPA3, and SPA4, which are part of a four-member family, have been shown to prevent hyperphotomorphogenesis in light-grown seedlings (Hoecker et al., 1998; Laubinger and Hoecker, 2003). Here, we have analyzed the function of SPA2 and, moreover, have demonstrated that the four SPA proteins act redundantly in repressing photomorphogenesis in the dark. Taken together, we have provided evidence that SPA proteins may be important for normal function of COP1-containing complex(es).
Plants with Defects in Multiple SPA Genes Undergo Constitutive Photomorphogenesis
Because SPA proteins are related by sequence, we considered the possibility that they might act redundantly in regulating plant growth and development. To assess this possibility, we constructed and examined mutants that were defective in the functions of multiple SPA genes. Dark-grown spa quadruple mutant seedlings that were defective in all four members of the SPA1 gene family exhibited features that are normally observed only in light-grown seedlings. These quadruple mutants displayed very short hypocotyls and fully opened cotyledons in darkness. Moreover, they were of dark purple coloration because of high accumulation of anthocyanin. On the molecular level, expression of light-inducible genes was derepressed in dark-grown spa triple mutants that we examined. These results clearly demonstrate that a lack of SPA function causes constitutive photomorphogenesis in the dark. Hence, SPA genes are essential for suppression of photomorphogenesis in dark-grown seedlings.
Adult spa quadruple mutant plants were extremely small and dwarfed. They produced very small inflorescences with one to three flowers that developed a few seeds. Thus, taken together, the phenotype of the spa quadruple mutant is reminiscent of that of constitutively photomorphogenic mutants with defects at the COP/DET/FUS loci. Viable mutants at these loci also show seedling deetiolation in the dark and dwarfed growth as adult plants (Schwechheimer and Deng, 2000).
SPA Proteins May Act in Concert with COP1
Our observation that spa quadruple mutants undergo constitutive photomorphogenesis suggests that the functions of SPA proteins may be related to those of other COP/DET/FUS proteins. In this regard, it is notable that all SPA proteins contain a WD-repeat domain that shows very close sequence similarity with the WD-repeat of COP1 (Hoecker et al., 1999; Laubinger and Hoecker, 2003). Moreover, it was shown previously that SPA1, SPA3, and SPA4 can interact with the coiled-coil domain of COP1 (Hoecker and Quail, 2001; Laubinger and Hoecker, 2003; Saijo et al., 2003) and also that SPA1 modifies the E3 ubiquitin ligase activity of COP1 on the transcription factors HY5 and LAF1 (Saijo et al., 2003; Seo et al., 2003). Here, we have demonstrated that SPA2 is also capable of interacting with COP1 in vitro. Furthermore, epistasis analysis indicated that the spa2-1 mutation genetically interacts with the weak cop1eid6 mutation in dark-grown seedlings: whereas neither the spa2 nor the cop1eid6 mutant showed altered skotomorphogenesis, the double mutant underwent strong constitutive photomorphogenesis in the dark. This is consistent with a previous finding that the spa1 mutation enhances the effect of the weak cop1-6 mutation in dark-grown seedlings (Saijo et al., 2003). Thus, taken together, these results suggest that SPA proteins act in concert with COP1 as parts of nuclear-localized COP1 complex(es). So far, we can only speculate on the biochemical functions of SPA proteins. They may be required for COP1 accumulation in the nucleus, for COP1 ubiquitin ligase activity, or for the interaction of COP1 complex(es) with substrates. Alternatively, they may be essential for stability or assembly of COP1 complex(es).
If SPA proteins are essential for all functions of the postulated COP1 complex(es), we would expect that spa quadruple mutants and COP1-deficient mutants exhibit identical phenotypes. However, cop1-null mutants die early in development after the production of at the most three tiny leaves (McNellis et al., 1994b), whereas spa quadruple mutants are viable and capable of reproducing (this report). This strongly suggests that there may be at least some residual COP1 activities even in the absence of SPA function. However, we cannot fully exclude the possibility that spa quadruple mutants exhibit some SPA function because they may accumulate truncated SPA proteins. The spa1-3 and spa3-1 mutations reside in the first third of the predicted proteins before the coiled-coil domains and are therefore unlikely to produce functional proteins. The T-DNAs in spa2-1 and spa4-1 are inserted toward the end of the respective genes within the WD-repeat–encoding sequence. All previous evidence, however, suggests that a disruption of the WD-repeat domain in SPA proteins or COP1 results in a complete loss of function (McNellis et al., 1994b; Hoecker et al., 1999; Laubinger and Hoecker, 2003). It is therefore not very likely that any of the possibly accumulating truncated SPA proteins have residual function. A quadruple mutant that is truly null for all members of the SPA1 gene family is necessary to unequivocally answer this question.
SPA Genes Have Overlapping but Distinct Functions in Regulating Photomorphogenesis
Our results have demonstrated that SPA genes have redundant functions in regulating photomorphogenesis. This is particularly obvious in dark-grown seedlings and in adult plants. Mutations in any single SPA gene did not cause an apparent change in skotomorphogenesis or adult growth behavior. A change in phenotype was only observed when at least two SPA genes were defective. For example, spa1 spa2 double mutant seedlings exhibited weak deetiolation in the dark. Additional loss of SPA3 or SPA4 function enhanced the constitutive photomorphogenesis, and spa quadruple mutants underwent most pronounced seedling deetiolation in the dark. Similarly, extremely small and dwarfed adult plants were only observed when all four SPA genes were defective. This indicates that SPA genes have overlapping functions throughout plant development.
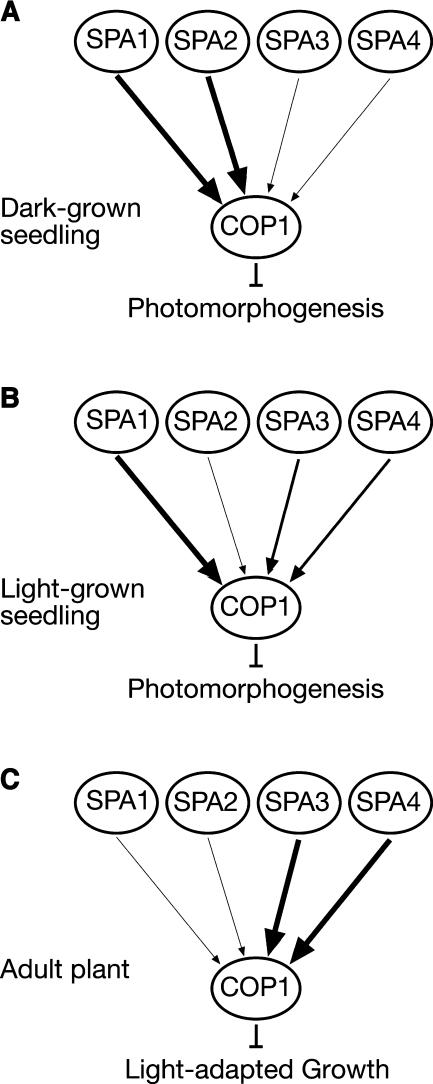
Nevertheless, the phenotypic analyses of spa single, double, triple, and quadruple mutants have also uncovered developmental differences in the functions of the four SPA genes. In dark-grown seedlings, the genes SPA1 and SPA2 dominate in repression of photomorphogenesis (Figure 9A). A single functional SPA gene, SPA1 or SPA2, is sufficient for normal skotomorphogenesis. Consistent with this finding, defects in SPA1 and SPA2 caused weak constitutive photomorphogenesis. On the contrary, a lack of SPA3 and SPA4 function did not affect dark development of seedlings (Laubinger and Hoecker, 2003). Also, neither SPA3 nor SPA4 were sufficient to support normal etiolation in the dark because the respective triple mutants underwent strong constitutive photomorphogenesis (this report). Hence, these results demonstrate differential contributions of the four SPA genes: a member of the SPA1/SPA2 class is necessary and sufficient for normal dark development of seedlings, whereas SPA3 and SPA4 are neither necessary nor sufficient.
Figure 9.
Relative Contributions of the SPA1 Gene Family Members at Different Developmental Stages.
The relative functions of the four SPA genes in dark-grown seedlings (A), light-grown seedlings (B), and adult plants (C) are derived from the phenotypes of mutants with defects in individual or multiple SPA genes.
The contrary was observed when examining elongation growth of adult plants. At this developmental stage, SPA3 and SPA4 were the predominant regulators among the SPA genes. A single functional SPA gene, SPA3 or SPA4, was sufficient to support close to normal adult growth under the growth conditions used. Consistent with this finding, a lack of SPA3 and SPA4 function affected adult growth, producing partially dwarfed plants. By contrast, a lack of SPA1 and SPA2 function had no discernible effect on rosette or inflorescence size, indicating that SPA1 and SPA2 are not essential for normal adult growth. Also, SPA2 was not sufficient to support normal elongation growth of the adult plant because the respective triple mutant was very dwarfed. Whether SPA1, like SPA2, is also not sufficient for normal adult growth remains to be investigated. Thus, a member of the SPA3/SPA4 functional subclass is necessary and close to sufficient for normal adult growth, whereas SPA1 and SPA2 make a smaller contribution to this response (Figure 9C). In conclusion, our results show that SPA3 and SPA4 contribute predominantly to normal elongation growth of the adult plant, whereas SPA1 and SPA2 support especially normal etiolation of dark-grown seedlings (Figures 9A and 9C). The causes for these developmental specificities among SPA genes remain to be tested. However, it is notable that, based on protein sequence, SPA1/SPA2 and SPA3/SPA4 form two subclasses within the SPA1 gene family (Laubinger and Hoecker, 2003).
The Function of SPA2 in Seedling Deetiolation
Plants with a defect only in the SPA2 gene did not show any apparent change in phenotype at any of the developmental stages examined. Hence, SPA2 is the only SPA gene that apparently is not essential for normal plant growth and development. Mutations in SPA1, SPA3, or SPA4, by contrast, cause hypersensitivity of seedlings to light (Hoecker et al., 1998; Laubinger and Hoecker, 2003). Thus, in light-grown seedlings, SPA1, SPA3, and SPA4 contribute more predominantly than SPA2 to the inhibition of photomorphogenesis (Figure 9B).
To uncover the function of SPA2, an analysis of mutants with defects in multiple SPA genes was necessary. The phenotype of a triple mutant that only has a functional SPA2 gene indicates that SPA2 is sufficient to allow normal seedling development only in the dark and not in the light. Consistent with this conclusion, a lack of SPA2 function, in a spa1 mutant background, affected seedling growth only in darkness and not in the light. Hence, the predominant function of SPA2 appears to be limited to suppression of photomorphogenesis in the dark (Figures 9A and 9B). Transgenic lines that overexpressed a GUS-SPA2 fusion protein, however, showed reduced light responses at seedling and adult stages. This indicates that the SPA2 protein, when ectopically overexpressed, is capable of functioning as a repressor of photomorphogenesis also in the light. Hence, SPA2 protein abundance is likely to be an important determinant of SPA2 function.
Interestingly, the phenotype of the spa1 spa3 spa4 triple mutant is very similar to that of the cop1eid6 mutant, which produces a COP1 protein with a missense mutation in the RING-finger domain (Dieterle et al., 2003). Both mutants show normal dark development, extreme hypersensitivity to light, and dwarfed growth as adult plants. Because SPA2 and COP1 are interacting proteins, it is tempting to speculate that nuclear SPA2•COP1 containing complex(es) have similar signaling activities as the COP1EID6 protein in a background carrying four functional SPA genes.
The Functions of SPA1, SPA3, and SPA4 in phyA-Mediated Seedling Deetiolation
Single mutant seedlings that are defective in SPA1, SPA3, or SPA4 exhibit enhanced responses to light in a fashion that is dependent on a functional PHYA gene. Dark-grown seedlings with a mutation in any one of these SPA genes, by contrast, have a wild-type appearance (Hoecker et al., 1998; Laubinger and Hoecker, 2003). Thus, SPA1, SPA3, and SPA4 are required for normal phyA-mediated light signal transduction in seedlings, whereas either one of these proteins is dispensable for normal growth of dark-grown seedlings or light-grown phyA-deficient seedlings. Recently, it was described that COP1 ubiquitinates phyA and thereby may cause desensitization of nuclear-localized phyA through degradation (Seo et al., 2004). Hence, one possibility is that SPA1, SPA3, and SPA4 are important regulators of phyA signaling because they, like COP1, may control phyA abundance. However, an analysis of phyA degradation kinetics in Rc-grown seedlings indicated that spa1 mutations did not cause a delay in phyA degradation (Hoecker et al., 1998). Similar analyses need to be conducted with spa3 and spa4 mutants to unequivocally answer this question. Two alternative possibilities are that phyA is important for accumulation or activity of these SPA proteins in light-grown seedlings or that SPA1, SPA3, and SPA4 function to protect COP1 against phyA-induced inactivation.
In conclusion, our studies have identified a new family of proteins with a central function in suppression of photomorphogenesis. Because members of this SPA protein family likely function in concert with COP1, a biochemical analysis of the COP1 complex(es) in mutants deficient for SPA function will shed light on the mechanisms involved in the regulation of COP1 activity.
METHODS
Plant Material, Growth Conditions, Light Sources, Determination of Hypocotyl Length, and Anthocyanin Content
The spa2-1 mutant allele was selected from the GABI-Kat T-DNA collection (Rosso et al., 2003). The spa1-3 mutant and the spa3-1, spa4-1, and spa3-1 spa4-1 mutants were described previously (Hoecker et al., 1998; Laubinger and Hoecker, 2003). The cop1-4 and cop1eid6 alleles were described in Deng and Quail (1992) and Dieterle et al. (2003), respectively.
Arabidopsis thaliana seeds were surface sterilized (20% Klorix [Colgate Palmolive, Hamburg, Germany] and 0.03% [v/v] Tween-20), rinsed at least four times with sterile water, and plated on agar-solidified medium containing 1× MS without sucrose. For the determination of anthocyanin levels and where indicated, the medium was supplemented with 2% sucrose. After 2 to 4 d of cold treatment (4°C), germination was induced by 3 h of white light at 21°C. Subsequently, plates were kept at 21°C in the dark for 21 h and were then exposed to darkness, Rc, FRc, or Bc for 3 d. Specific light conditions were generated using LED light sources (Quantum Devices, Barneveld, WI). To quantify light responses, seedlings were photographed using a digital camera (Camedia E-10; Olympus, Hamburg, Germany), and hypocotyl length was determined using National Institute of Health IMAGE software (Bethesda, MD). Anthocyanin levels were determined as described in Hoecker et al. (1998).
For determination of the adult phenotype, seeds were planted on soil in a randomized fashion. Plants were grown in a growth chamber under 16-h-light/8-h-dark cycles of 21°C (day) and 18°C (night). Light intensity at 110 μmol m−2 s−1 was produced with FLUORA L58W/77 fluorescent lights (Osram, Munich, Germany). The lengths of the longest petiole and the longest leaf (petiole and leaf blade) were determined for each 3-week-old plant.
Determination of the T-DNA Insertion Site in spa2-1 and Construction of Mutants Defective in Multiple SPA Genes
The T-DNA insertion in the spa2-1 mutant was confirmed by PCR using the gene-specific primer 5′-GCAGTTAGCTATGCGAAGTTC-3′ and the T-DNA–specific primer 5′-CCCATTTGGACGTGAATGTAGACAC-3′. In a population segregating for spa2-1, homzygous spa2-1 mutant and homozygous wild-type plants were identified using PCR-based markers specific for the mutant or wild-type allele, respectively. Progeny of these plants were used for further experiments.
spa1 spa2 double mutants were generated by crossing spa1-3 with spa2-1. Resulting F2 seedlings were screened in weak FRc, and presumed spa1 mutant seedlings (short seedlings) were transferred to soil. The genotype of these plants at the SPA1 and SPA2 loci was verified using PCR-based markers. All of the selected plants were confirmed to be homozygous spa1-3 mutant. In further experiments, progeny of plants homozygous mutant also for spa2 (spa1-3 spa2-1) was compared with progeny of siblings that are homozygous wild-type for SPA2 [spa1-3 SPA2(+)]. For each genotype, several lines were analyzed and showed essentially the same phenotype.
All other multiple mutants (spa1 spa2 spa3, spa1 spa2 spa4, and spa1 spa3 spa4) were derived from a cross of spa1-3 spa2-1 with spa3-1 spa4-1. Resulting F2 seedlings were screened in the dark and in weak Rc (1 μmol m−2 s−1). Dark-grown seedlings that exhibited constitutive photomorphogenesis were selected and transferred to soil. PCR-based genotyping revealed that most of these plants were spa1 spa2 spa3 or spa1 spa2 spa4 triple mutants. A few plants were confirmed to be spa1 spa2 double mutants. To determine the phenotype of spa1 spa2 spa3 spa4 quadruple mutants, F3 progeny of spa1 spa2 spa3 or spa1 spa2 spa4 triple mutants that were heterozygous for spa3 or spa4, respectively, were used.
spa1 spa3 spa4 triple mutants were selected from the segregating F2 population grown in weak Rc. Short seedlings were selected and transferred to soil, and the genotype at all four SPA loci was determined. F3 progeny of thus identified spa1 spa3 spa4 triple mutants was used for further analysis. For each multiple mutant, at least three plants were selected from a segregating population. Each progeny was examined independently and revealed essentially the same phenotype. The sequence of all primers used to determine the genotype at SPA loci will be provided upon request.
To generate spa2-1 cop1eid6 double mutants, spa2-1 was crossed with cop1eid6. Resulting F2 seedlings were grown in darkness, and the number of seedlings showing strong constitutive photomorphogenesis, weaker constitutive photomorphogenesis, and normal skotomorphogenesis was determined. Eight to 10 seedlings from each phenotypic class were harvested individually and used for preparation of genomic DNA. This DNA was used as a template to determine the genotype at the loci SPA2 and COP1.
RNA Analysis
Total RNA was isolated using the RNeasy plant mini kit (Qiagen, Hilden, Germany), separated by standard denaturing gel electrophoresis, and blottet onto nylon membranes. RNA gel blot analysis of spa2-1 mutants and GUS-SPA2 overexpression lines was performed using a radioactively labeled SPA2-specific probe that was comprised of the cDNA sequence of SPA2 from start to stop codon. The FEDA, CAB3, RBCS, and RBCL probes were described in Deng et al. (1992). After overnight hybridization at 65°C, membranes were washed at 65°C once each with 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate), 0.1% (w/v) SDS, 1× SSC, 0.1% (w/v) SDS, 0.5× SSC, 0.1% (w/v) SDS, 0.1× SSC, and 0.1% (w/v) SDS and were then exposed to phosphor imager plates (Fuji, Tokyo, Japan) for 3 d.
For RT-PCR analysis, 1 μg of total RNA was reverse transcribed using an oligo(dT) primer and Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. One micorliter of the RT reaction was used as template to amplify wild-type SPA2 sequence using specific primers flanking the T-DNA insertion site. As a control, SPA3-specific primers were used.
Generation of Transgenic Plants Expressing GUS-SPA2 and Complementation of the Constitutive Deetiolation Phenotype of spa1 spa2 Mutant Seedlings
The open reading frame of SPA2 was cloned into the ClaI site of the vector pRTL2-GUS containing an extended multiple cloning site (Hoecker et al., 1999). The 35S:GUS-SPA2 cassette and, as a control, 35S:GUS were subcloned into the PstI site of the binary vector pPZP211 (Hajdukiewicz et al., 1994). These constructs were introduced into the Agrobacterium strain GV3101, which was subsequently used to transform wild-type plants of the Columbia ecotype. Several independent homozygous lines expressing GUS-SPA2 or GUS from single insertion sites were examined phenotypically, of which 35S:GUS-SPA2 lines L4 and L6 were investigated in detail.
To complement the spa2-dependent constitutive deetiolation phenotype, the homozygous spa1 spa2 double mutant was crossed with the 35S:GUS-SPA2 overexpression line L4. Seventy-seven kanamycin-resistent F2 plants were grown to maturity to obtain F3 generations. Using PCR-based markers and genomic DNA isolated from 10 pooled seedlings, F3 lines were identified that were homozygous spa1 spa2 mutant. To determine whether these lines were homozygous or hemizygous for the 35S:GUS-SPA2 transgene, F3 seeds were germinated on growth medium containing kanamycin, and segregation ratios for kanamycin resistance were scored. Also, seedlings were stained for GUS activity by overnight incubation in a solution containing 100 mM Na-PO4 buffer, pH 7.0, and 1 mM 5-bromo-4-chloro-3-indoyl-β-d-glucuronide at 37°C.
Analysis of GUS Subcellular Localization in Transgenic Arabidopsis Seedlings
Seedlings were transferred into a prestaining solution (100 mM NaPO4, pH 7.0, 2% [v/v] formaldehyde, and 1 mM EDTA), treated with a brief vacuum infiltration, and subsequently incubated for 10 min. Seedlings were washed in 100 mM Na-PO4 buffer, pH 7.0, and then transferred into a GUS-staining solution (100 mM Na-PO4, pH 7.0, 1% (v/v) Triton X-100, 5 mM potassium ferrocyanid, 5 mM potassium ferricyanid, 1 mM EDTA, and 1 mM 5-bromo-4-chloro-3-indoyl-β-d-glucuronide). After brief vacuum infiltration, seedlings were incubated at 37°C for 16 h. Seedlings were fixed in 100 mM Na-PO4, pH 7.0, and 3.7% (v/v) formaldehyde at room temperature for 30 min and subsequently washed twice with 100 mM Na-PO4 buffer, pH 7.0. Chlorophyll was removed by 70% ethanol. For DAPI staining, seedlings were incubated in 100 mM Na-PO4 buffer, pH 7.0, with 0.001% (w/v) DAPI for 1 h at room temperature. Seedlings were analyzed by light microscopy (Axiophot; Zeiss, Jena, Germany), and photos were taken with an Olympus DP50-CO camera (Olympus Optical, Tokyo, Japan).
In Vitro Interaction Assay
For expression of SPA2 protein in vitro, the open reading frame of SPA2 was amplified by PCR and cloned into the NcoI/XhoI sites of the vector pET15b (Novagen, Madison, WI) to create the construct SPA2-pET15b. For the expression of GAD-SPA2, an NcoI fragment of GAD (Hoecker and Quail, 2001) was cloned into the NcoI site of SPA2-pET15b. Synthesis of SPA2, GAD-SPA2, and GAD-COP1 protein by coupled transcription/translation and subsequent coimmunoprecipitations were performed as described in Laubinger and Hoecker (2003).
Acknowledgments
We thank Thomas Kretsch for providing cop1eid6 seed. We acknowledge GABI-Kat (Max-Planck-Institut für Züchtungsforschung, Köln, Germany) for spa2-1 seed. We thank Lena Gebel, Michael Lübeck, and Jan Teune for excellent technical assistance, Wilhelm Rogmann and his greenhouse staff for expert care of our plants, and Udo Gowik, Karin Ernst, Peter Westhoff, and members of the laboratory for helpful discussions. We are grateful to Peter Quail, Patricia Müller-Moulé, and Peter Westhoff for critical reading of the manuscript. This research was supported by the Deutsche Forschungsgemeinschaft (SFB590) to U.H.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Ute Hoecker (hoeckeru@uni-duesseldorf.de).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.024216.
References
- Baumgardt, R.L., Oliverio, K.A., Casal, J.J., and Hoecker, U. (2002). SPA1, a component of phytochrome A signal transduction, regulates the light signaling current. Planta 215, 745–753. [DOI] [PubMed] [Google Scholar]
- Briggs, W.R., and Olney, M.A. (2001). Photoreceptors in plant photomorphogenesis to date. Five phytochromes, two cryptochromes, one phototropin, and one superchrome. Plant Physiol. 125, 85–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büche, C., Poppe, C., Schäfer, E., and Kretsch, T. (2000). eid1: A new Arabidopsis mutant hypersensitive in phytochrome A-dependent high-irradiance responses. Plant Cell 12, 547–558. [PMC free article] [PubMed] [Google Scholar]
- Casal, J.J., Luccioni, L.G., Oliverio, K.A., and Boccalandro, H.E. (2003). Light, phytochrome signalling and photomorphogenesis in Arabidopsis. Photochem. Photobiol. Sci. 2, 625–636. [DOI] [PubMed] [Google Scholar]
- Deng, X.W., Matsui, M., Wei, N., Wagner, D., Chu, A.M., Feldmann, K.A., and Quail, P.H. (1992). COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 71, 791–801. [DOI] [PubMed] [Google Scholar]
- Deng, X.W., and Quail, P.H. (1992). Genetic and phenotypic characterization of cop1 mutants of Arabidopsis thaliana. Plant J. 2, 83–95. [Google Scholar]
- Deng, X.W., and Quail, P.H. (1999). Signalling in light-controlled development. Semin. Cell Dev. Biol. 10, 121–129. [DOI] [PubMed] [Google Scholar]
- Dieterle, M., Buche, C., Schafer, E., and Kretsch, T. (2003). Characterization of a novel non-constitutive photomorphogenic cop1 allele. Plant Physiol. 133, 1557–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser, C., and Staiger, D. (2002). Photoreceptors in Arabidopsis thaliana: Light perception, signal transduction and entrainment of the endogenous clock. Planta 216, 1–16. [DOI] [PubMed] [Google Scholar]
- Franklin, K.A., and Whitelam, G.C. (2004). Light signals, phytochromes and cross-talk with other environmental cues. J. Exp. Bot. 55, 271–276. [DOI] [PubMed] [Google Scholar]
- Gyula, P., Schafer, E., and Nagy, F. (2003). Light perception and signalling in higher plants. Curr. Opin. Plant Biol. 6, 446–452. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. [DOI] [PubMed] [Google Scholar]
- Hoecker, U., and Quail, P.H. (2001). The phytochrome A-specific signaling intermediate SPA1 interacts directly with COP1, a constitutive repressor of light signaling in Arabidopsis. J. Biol. Chem. 276, 38173–38178. [DOI] [PubMed] [Google Scholar]
- Hoecker, U., Tepperman, J.M., and Quail, P.H. (1999). SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science 284, 496–499. [DOI] [PubMed] [Google Scholar]
- Hoecker, U., Xu, Y., and Quail, P.H. (1998). SPA1: A new genetic locus involved in phytochrome A-specific signal transduction. Plant Cell 10, 19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T.H., Kim, B.H., and von Arnim, A.G. (2002). Repressors of photomorphogenesis. Int. Rev. Cytol. 220, 185–223. [DOI] [PubMed] [Google Scholar]
- Kircher, S., Kozma-Bognar, L., Kim, L., Adam, E., Harter, K., Schäfer, E., and Nagy, F. (1999). Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11, 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher, S., Gil, P., Kozma-Bognar, L., Fejes, E., Speth, V., Husselstein-Muller, T., Bauer, D., Adam, E., Schäfer, E., and Nagy, F. (2002). Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell 14, 1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger, S., and Hoecker, U. (2003). The SPA1-like proteins SPA3 and SPA4 repress photomorphogenesis in the light. Plant J. 35, 373–385. [DOI] [PubMed] [Google Scholar]
- McNellis, T.W., Von Arnim, A.G., Araki, T., Komeda, Y., Miséra, S., and Deng, X.-W. (1994. b). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6, 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis, T.W., Von Arnim, A.G., and Deng, X.-W. (1994. a). Overexpression of Arabidopsis COP1 results in partial suppression of light-mediated development: Evidence for a light-inactivable repressor of photomorphogenesis. Plant Cell 6, 1391–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, M.M., Fankhauser, C., and Chory, J. (2000). Light: An indicator of time and place. Genes Dev. 14, 257–271. [PubMed] [Google Scholar]
- Osterlund, M.T., Ang, L.H., and Deng, X.W. (1999). The role of COP1 in repression of Arabidopsis photomorphogenic development. Trends Cell Biol. 9, 113–118. [DOI] [PubMed] [Google Scholar]
- Osterlund, M.T., Hardtke, C.S., Wei, N., and Deng, X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466. [DOI] [PubMed] [Google Scholar]
- Parks, B.M., Hoecker, U., and Spalding, E.P. (2001). Light-induced growth promotion by SPA1 counteracts phytochrome-mediated growth inhibition during de-etiolation. Plant Physiol. 126, 1291–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail, P.H. (2002). Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 3, 85–93. [DOI] [PubMed] [Google Scholar]
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53, 247–259. [DOI] [PubMed] [Google Scholar]
- Saijo, Y., Sullivan, J.A., Wang, H., Yang, J., Shen, Y., Rubio, V., Ma, L., Hoecker, U., and Deng, X.W. (2003). The COP1–SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 17, 2642–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer, C., and Deng, X.W. (2000). The COP/DET/FUS proteins-regulators of eukaryotic growth and development. Semin. Cell Dev. Biol. 11, 495–503. [DOI] [PubMed] [Google Scholar]
- Seo, H.S., Watanabe, E., Tokutomi, S., Nagatani, A., and Chua, N.H. (2004). Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 18, 617–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, H.S., Yang, J.Y., Ishikawa, M., Bolle, C., Ballesteros, M.L., and Chua, N.H. (2003). LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 424, 995–999. [DOI] [PubMed] [Google Scholar]
- Serino, G., and Deng, X.W. (2003). THE COP9 SIGNALOSOME: Regulating plant development through the control of proteolysis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 54, 165–182. [DOI] [PubMed] [Google Scholar]
- Subramanian, C., Kim, B.H., Lyssenko, N.N., Xu, X., Johnson, C.H., and Von Arnim, A.G. (2004). The Arabidopsis repressor of light signaling, COP1, is regulated by nuclear exclusion: Mutational analysis by bioluminescence resonance energy transfer. Proc. Natl. Acad. Sci. USA 101, 6798–6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, J.A., and Deng, X.W. (2003). From seed to seed: The role of photoreceptors in Arabidopsis development. Dev. Biol. 260, 289–297. [DOI] [PubMed] [Google Scholar]
- Von Arnim, A.G., and Deng, X.-W. (1994). Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79, 1035–1045. [DOI] [PubMed] [Google Scholar]
- Wang, H.Y., Ma, L.G., Li, J.M., Zhao, H.Y., and Deng, X.W. (2001). Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294, 154–158. [DOI] [PubMed] [Google Scholar]
- Yang, H.Q., Tang, R.H., and Cashmore, A.R. (2001). The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13, 2573–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y.C., Dieterle, M., Buche, C., and Kretsch, T. (2002). The negatively acting factors EID1 and SPA1 have distinct functions in phytochrome A-specific light signaling. Plant Physiol. 128, 1098–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]