Abstract
Recently, we have provided evidence that the polymorphic self-incompatibility (S) locus-encoded F-box (SLF) protein AhSLF-S2 plays a role in mediating a selective S-RNase destruction during the self-incompatible response in Antirrhinum hispanicum. To investigate its role further, we first transformed a transformation-competent artificial chromosome clone (TAC26) containing both AhSLF-S2 and AhS2-RNase into a self-incompatible (SI) line of Petunia hybrida. Molecular analyses showed that both genes are correctly expressed in pollen and pistil in four independent transgenic lines of petunia. Pollination tests indicated that all four lines became self-compatible because of the specific loss of the pollen function of SI. This alteration was transmitted stably into the T1 progeny. We then transformed AhSLF-S2 cDNA under the control of a tomato (Lycopersicon esculentum) pollen-specific promoter LAT52 into the self-incompatible petunia line. Molecular studies revealed that AhSLF-S2 is specifically expressed in pollen of five independent transgenic plants. Pollination tests showed that they also had lost the pollen function of SI. Importantly, expression of endogenous SLF or SLF-like genes was not altered in these transgenic plants. These results phenocopy a well-known phenomenon called competitive interaction whereby the presence of two different pollen S alleles within pollen leads to the breakdown of the pollen function of SI in several solanaceaous species. Furthermore, we demonstrated that AhSLF-S2 physically interacts with PhS3-RNase from the P. hybrida line used for transformation. Together with the recent demonstration of PiSLF as the pollen determinant in P. inflata, these results provide direct evidence that the polymorphic SLF including AhSLF-S2 controls the pollen function of S-RNase–based self-incompatibility.
INTRODUCTION
Self-incompatibility (SI) in the Solanaceae, Scrophulariaceae, and Rosaceae is controlled by S-RNases that have been thought to act as S-allele–specific cytotoxins that inhibit the growth of pollen bearing an S-allele matching to either of the style S-alleles (McClure et al., 1990; Gray et al., 1991; McCubbin and Kao, 2000). However, a distinct gene controlling the pollen specificity known as pollen S is postulated to interact with S-RNases to accomplish the pollen rejection or acceptance (Golz et al., 1999, 2001).
Recently, S-locus F-box (SLF) proteins have been shown to be potential candidates for pollen S in Antirrhinum hispanicum, a member of the Scrophulariaceae (Lai et al., 2002; Zhou et al., 2003), several species of the Rosaceae (Entani et al., 2003; Ushijima et al., 2003; Yamane et al., 2003), and also the Solanaceae (Kao and Tsukamoto, 2004; Sijacic et al., 2004). As predicted for pollen S, these S-linked genes are polymorphic with a pollen-specific expression pattern. Moreover, we have found that AhSLF-S2 physically interacts with S-RNases in a nonallele-specific fashion probably through a proposed SCFAhSLF-S2 (Skp1/Cullin or CDC53/F-box) complex that targets S-RNase destruction during compatible rather than incompatible response (Qiao et al., 2004).
In certain species of Nicotiana and petunia, heteroallelic S-allele duplications have been found to be associated with pollen-part self-compatible mutants (PPM) (Brewbaker and Natarajan, 1960; Pandy, 1965; Golz et al., 1999; reviewed in de Nettancourt, 2001). The genetic behavior of these mutants could be explained if pollen S products act as an inhibitor of all S-RNases except for its cognate S-RNase (Golz et al., 1999). It is known that tetraploids derived from self-incompatible diploid plants often lose the pollen function of SI through a competitive interaction between two heteroallelic S-alleles within the diploid pollen but remain cross-incompatible as females with normal self-related haploid pollen (Crane and Lewis, 1942; Stout and Chandler, 1942; Lewis, 1947; reviewed in de Nettancourt, 2001). In Nicotiana alata, Golz et al. (1999) have isolated PPMs with γ-ray radiation that specifically affects the SI phenotype of pollen but not the SI phenotype of the style. These PPMs are predominantly associated with centric fragments that carry a duplicated copy of an S-allele. Furthermore, S-allele duplications lacking duplicated S-RNases still produce PPMs (Golz et al., 2001), showing that the pollen S duplication alone is capable of generating pollen compatibility. These observations provide a test for the validity of a pollen S candidate in an S-RNase–based SI system through genetic transformation (Kao and Tsukamoto, 2004). In fact, Sijacic et al. (2004) have recently used this approach to demonstrate that PiSLF determines the pollen–self-incompatibility in P. inflata.
In this study, we transformed an Antirrhinum transformation-competent artificial chromosome (TAC) clone containing both AhS2-RNase and AhSLF-S2 into a self-incompatible line of Petunia hybrida. In a separate experiment, we transformed the AhSLF-S2 cDNA driven by a pollen-specific promoter into the same line. In both cases, the introduced genes were correctly expressed in the reproductive tissues of transgenic petunia plants that significantly all became self-compatible. Pollination tests showed that they had specifically lost the pollen function of SI and phenocopied the competitive interaction because endogenous SLF or SLF-like genes maintained normal expression in these transgenic plants. Moreover, we detected a physical interaction between AhSLF-S2 and PhS3-RNase. Taken together, these results provide direct evidence that AhSLF-S2 mediates the pollen function of SI in the S-RNase–based SI species.
RESULTS
Generation and Analysis of Petunia Plants Transformed with TAC26
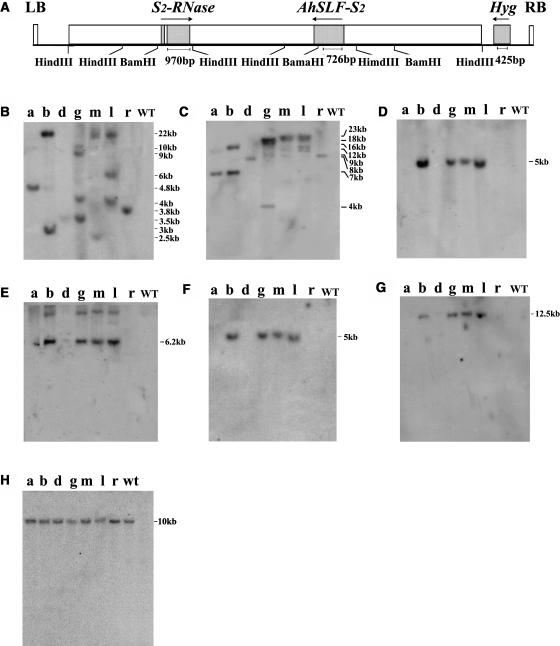
To test the validity of the pollen S candidate AhSLF-S2, we created the S-allele duplication by genetic transformation of a self-incompatible line of P. hybrida, a member of the Solanaceae. In the initial experiment, a TAC26 clone containing both AhSLF-S2 and AhS2-RNase genes (Figure 1A) was transformed into an S3S3 self-incompatible line of P. hybrida (Robbins et al., 2000) via an Agrobacterium tumefaciens–mediated approach. Sequence analysis of the TAC insert of ∼55 kb revealed that this genomic region mainly contains repetitive sequences except for two genes, AhSLF-S2 and AhS2-RNase (Lai et al., 2002; Zhou et al., 2003). The TAC clone was used to transform petunia via an Agrobacterium-mediated approach, resulting in seven hygromycin-resistant T0 lines. DNA gel blot analysis of their leaf genomic DNA digested with HindIII or BamHI, using a Hyg fragment as probe (Figures 1B and 1C), showed that lines a, d, and r had a single copy insertion of the hygromycin transgene, lines b and m two copies, line l three copies, and line g four copies. As a negative control, no hybridization signal was detected in the untransformed wild-type line.
Figure 1.
Molecular Analysis of TAC26 Transgenic Petunia Plants.
(A) A schematic structure of TAC26. Probes for AhS2-RNase and AhSLF-S2 used are also indicated and HindIII and BamHI sites with the genomic region covered by TAC26. LB, left border; RB, right border of the T-DNA; Hyg, hygromycin resistance gene.
(B) to (G) DNA gel blot analysis of the T0 transgenic lines a, b, d, g, m, l, r, and the untransformed control (WT). Leaf DNA (5 μg) was digested with HindIII ([B], [D], and [F]) or BamHI ([C], [E], and [G]) and was blotted and probed with the Hyg fragment ([B] and [C]), AhSLF-S2 ([D] and [E]) and S2-RNase ([F] and [G]).
(H) DNA gel blot of HindIII-digested leaf DNA was probed with a PhS3-RNase cDNA fragment.
Sizes of the markers ([B] and [C]) and the hybridizing bands ([D] to [H]) are indicated in kilobase pairs.
To determine whether an intact insert was transferred into the genome, further DNA gel blot analysis was performed using both AhSLF-S2 (Figures 1D and 1E) and AhS2-RNase (Figures 1F and 1G) cDNA fragments as probes. Out of the seven lines, four lines (b, g, l, and m) had both complete genes, indicated by the two predicted hybridizing bands of 5-kb HindIII and 6.2-kb BamHI fragments using AhSLF-S2 as a probe and 5-kb HindIII and 12.5-kb BamHI fragments using AhS2-RNase as a probe (Xue et al., 1996; Lai et al., 2002), suggesting that these four lines contain an intact copy(s) of the TAC26 insert. These results indicated that the genomic region encompassing both AhSLF-S2 and AhS2-RNase had been successfully integrated into the petunia genome in the four independent transformants. The S-haplotype of the primary transformants was confirmed using PhS3-RNase fragment as a probe (Figure 1H). Notably, none of the single hygromycin copy insertion lines (a, d, and r) had intact target genes (Figures 1B to 1G). This might be caused by an incomplete transfer of the T-DNA as occasionally observed for Agrobacterium-mediated transformation (Liu et al., 2000).
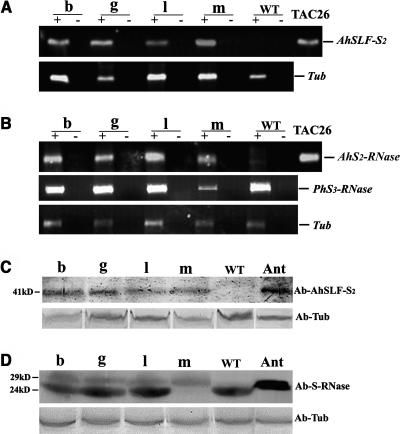
We examined the expression of AhSLF-S2 and AhS2-RNase in the four transgenic lines with both target genes in detail (b, g, l, and m). RNA transcripts from AhSLF-S2 (Figure 2A) and AhS2-RNase (Figure 2B) were detected by RT-PCR in mature anther and pistil, respectively. PCR products were exclusively detected in lanes with reverse transcriptase added (+ lanes) and the positive control of the TAC26 plasmid (lane 11 in Figures 2A and 2B) containing both AhSLF-S2 and AhS2-RNase. No product was detected in lanes without reverse transcriptase (− lanes) and the negative control of cDNA from the untransformed S3S3 petunia plant, suggesting no genomic DNA contamination in the cDNA used. An RT-PCR reaction with tubulin primers was used as a control for RNA loading.
Figure 2.
Expression Analysis of AhSLF-S2 and AhS2-RNase in the Transgenic Petunia Lines.
(A) and (B) RT-PCR analysis of RNA isolated from pollens or styles, with (+) or without (–) reverse transcriptase in the synthesis of cDNA.
(A) Top panel, RT-PCR analysis of RNA isolated from pollen of the transgenic lines b, g, l, m, and S3S3 wild-type petunia plant using specific primers of AhSLF-S2. TAC26 plasmid (TAC26) was used as a positive control. Bottom panel, RT-PCR analysis of tubulin for loading control.
(B) Top panel, RT-PCR analysis of RNA isolated from styles of transgenic lines b, g, l, m, and the wild type using specific primers of AhS2-RNase, and the TAC26 plasmid (TAC26) was used as a positive control. Middle panel, RT-PCR analysis of RNA isolated from styles of the trangenic lines using specific primers of PhS3-RNase. Bottom panel, RT-PCR analysis of tubulin for loading control.
(C) and (D) Immunoblot detection of AhSLF-S2 or S-RNases.
(C) Top panel, detection of AhSLF-S2 from total pollen proteins of transgenic lines b, g, l, m, and wild-type S3S3 petunia plant and Antirrhinum of S2S4 (Ant) by polyclonal antibody against AhSLF-S2. Bottom panel, detection of tubulin for loading control.
(D) Top panel, detection of AhS2-RNase (∼29 kD) and endogenous S3-RNase (∼24 kD) from total style proteins of transgenics lines b, g, l, m, wild-type S3S3 petunia plant and Antirrhinum (Ant) by polyclonal antibody against AhS-RNases. Bottom panel, detection of tubulin for loading control. Sizes of the detected protein bands are indicated in kilodaltons.
To further confirm normal expression of the transgenes, AhSLF-S2 expression was examined in anther by immunoblot analysis using an antibody against the C-terminal region (Qiao et al., 2004) and was only detected in the transgenic lines at a similar level to that in Antirrhinum (Figure 2C). We also examined S-RNase expression in pistil using an antibody against AhS-RNase (Qiao et al., 2004) (Figure 2D). The immunoblot showed that AhS2-RNase (∼29 kD) was also detected in each transgenic line, proving that it is expressed and processed normally in styles. The endogenous PhS3-RNase (∼24 kD) present in the untransformed petunia was detected in three lines (lines b, g, and l) except one (line m). Why PhS3-RNase was not detected in line m was not clear, but it could be because of gene silencing of the transgenes (Fagard and Vaucheret, 2000). In fact, two copies of the transgenes were detected in line m (see above). However, it might not be because of cosuppression because AhS2-RNase and PhS3-RNase show a low degree of nucleotide sequence identities (48.9%; Xue et al., 1996; Robbins et al., 2000). Furthermore, lower levels of RNA transcripts of PhS3-RNase were detected in this line (Figure 2B, middle panel). No expression of the two Antirrhinum genes was detected in vegetative tissues in the transgenic plants (data not shown). Taken together, these results clearly showed that both AhSLF-S2 and AhS2-RNase are expressed in the majority of the transgenic plants in the same tissues as that observed in Antirrhinum.
Phenotypic Analysis of TAC26 Primary Transformants
We did not observe any obvious morphological change of the transgenic plants compared with that of the untransformed plants. To test the pollination behavior of the transformants, they were self- or cross-pollinated. All of the pollinations were performed at the open flower stage, and the results are shown in Table 1. After self-pollination, all four transgenic lines lost their SI behavior and became self-compatible (SC). They set variable-sized capsules with an average seed number from 75.2 to 241 per capsule, comparable to a fully compatible pollination in petunia. To further test whether the conversion of SI into SC is because of the complete loss of the S-locus function or the loss of male or female component function, the transgenic plants were reciprocally crossed with an untransformed petunia line either as pollen donor or accepter (Table 1). First, we pollinated the untransformed S3S3 plants using the four transgenic lines as pollen donors. As a control, we self-pollinated the untransformed S3S3 plants, and no single seed set was obtained, suggesting that they maintain normal SI function in their pollen and pistil. After being pollinated by each transgenic line, they set middle-sized capsules, and the average seed number per capsule they set ranged from 98.4 to 135.2, also comparable to a fully compatible pollination. We also pollinated the four transgenic lines using S3S3 wild-type plants as pollen donor. The results showed that three lines (b, g, and l) set no seed, except line m, which set full-size capsules with an average seed number of 213.5. The abnormal SI behavior of line m is consistent with our observation that it expressed very low levels of the endogenous PhS3-RNase (Figure 2C). Thus, it should accept S3 pollen. The above data showed that the transformation of TAC26 into petunia leads to the breakdown of the pollen function of self-incompatibility, a phenocopy of competitive interaction, suggesting that the TAC26 clone contains the pollen S.
Table 1.
Pollination Behaviors of the TAC26 T0 Transgenic Petunia Plants
| Cross-Pollination
|
|||
|---|---|---|---|
| Planta | Self-Pollinationb | WT Femalec | WT Maled |
| b | 15/16 (88.4 ± 9.7) | 10/10 (105.7 ± 23.5) | 0/12 (0) |
| g | 15/15 (241.0 ± 39.5) | 11/12 (105.2 ± 21.9) | 0/18 (0) |
| l | 15/17 (75.2 ± 10.2) | 9/10 (135.2 ± 35.2) | 0/16 (0) |
| me | 11/12 (170.0 ± 15.5) | 7/7 (98.4 ± 32.2) | 13/13 (213.5 ± 32.4) |
| WT | 0/35 (0) | –f | – |
b, g, l, and m are four transgenic lines, and WT is the wild-type S3S3 plant.
Data are represented as mature capsules/total pollination sets (average seeds per capsule ± sd).
The pollen from the transgenic plants were used to pollinate mature styles from the wild-type S3S3 plants.
The pollen from the wild-type plants were used to pollinate mature styles from the transgenic plants.
Plants with low expression of endogenous PhS3-RNase.
Not applicable.
Molecular and Phenotypic Analysis of the Transgenic Progeny
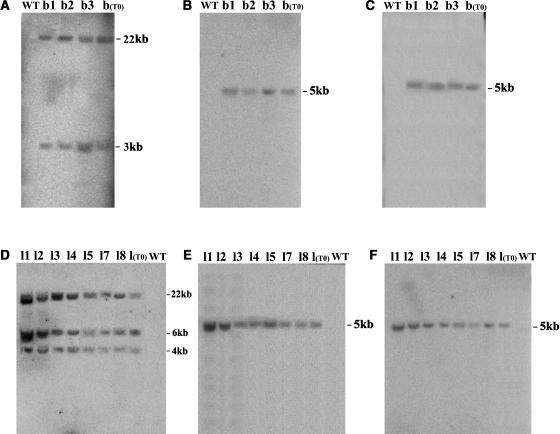
To determine whether the alteration of self-incompatibility induced by expressing TAC26 is heritable, the transgenic T1 progeny were raised from the seeds obtained from self-pollinations of two lines, b and l. Three T1 plants were obtained from line b. The TAC26 insert was inherited stably, as detected by DNA gel blot hybridization with hyg, AhSLF-S2, and AhS2-RNase fragments as probes (Figures 3A to 3C). Compared with the T0 primary transgenic line, no apparent recombination or independent segregation occurred for the transgenes. Seven T1 plants were obtained from line l and were also found to inherit the transgenes stably (Figures 3D to 3F).
Figure 3.
Molecular Analysis of DNA from T1 Progeny of the TAC26 Transgenic Petunia Lines.
(A) to (C) Genomic DNA gel blot analysis of the T1 progeny (b1 to b3), T0 transformant b, and the untransformed control. Leaf DNA digested by HindIII was blotted and probed with the Hyg fragment (A), AhSLF-S2 (B), and AhS2-RNase (C), respectively.
(D) to (F) DNA gel blot analysis of the T1 progeny (l1 to 5, 7, and 8), T0 transformant, and the untransformed control. Leaf DNA digested by HindIII was blotted and probed with the Hyg fragment (D), AhSLF-S2 (E), and AhS2-RNase (F), respectively. Sizes of the hybridizing fragments are indicated in kilobase pairs.
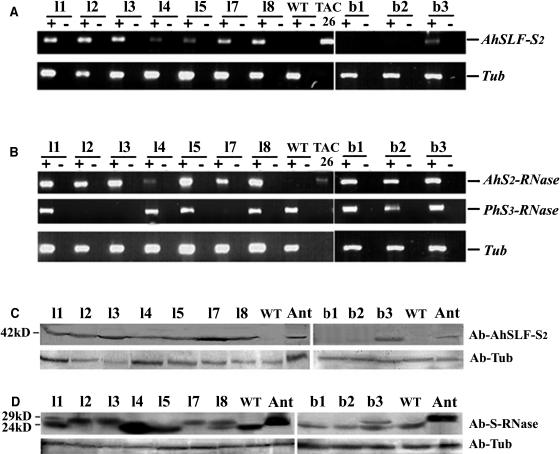
To examine the transgenes expression in the progeny, RT-PCR analysis was performed. As shown in Figures 4A and 4B, the seven progeny from line l all expressed AhSLF-S2 and AhS2-RNase in pollen and style, respectively. As for the three progeny from line b, only one (b3) expressed AhSLF-S2 normally (Figures 4A and 4C). No or very low expression of AhS2-RNase was detected in the T1 siblings b1 and b2 despite of the presence of their RNA transcripts (Figures 4B and 4D), compared with that in b3, which had a normal AhS2-RNase expression. Furthermore, protein gel blot analysis showed that AhSLF-S2 was detected in all the seven line l progeny (Figure 4C). AhS2-RNase was detected normally in five plants from line l (Figure 4D, progeny 1, 2, 3, 7, and 8) consistent with the lower levels of expression observed in the RT-PCR (Figure 4B). It was also found that endogenous PhS3-RNase was not expressed in the three l progeny (l2, 3, and 7) consistent with the absence of RNA transcripts detected in these lines (Figure 4B). In addition, genomic PCR analysis showed that these progeny contained no Sv allele used initially to develop S3S3 line from S3Sv plant (Robbins et al., 2000; data not shown). The reason why AhSLF-S2, AhS2-RNase, or endogenous PhS3-RNase could not be detected in some progeny is not clear, but some posttranscriptional gene silencing could lead to this. It has been known that gene silencing triggered by a transgene can lead to the mRNA degradation of the transgene and/or endogenous gene with no or low expression of the protein, rendering the protein undetectable by antibody (Fagard and Vaucheret, 2000).
Figure 4.
Expression Analysis of AhSLF-S2 and AhS2-RNase in the Transgenic Petunia T1 Progeny.
(A) and (B) RT-PCR analysis of RNA isolated from pollen or style, with (+) or without (−) reverse transcriptase in the synthesis of cDNA.
(A) Top panel, RT-PCR analysis of RNA isolated from pollen of the transgenic T1 lines l1, 2, 3, 4, 5, 7, and 8, lines b1 to 3, and wild-type plant using AhSLF-S2–specific primers with TAC26 plasmid as a positive control. Bottom panel, RT-PCR analysis of tubulin for loading control.
(B) Top panel, RT-PCR analysis of RNA isolated from styles of the transgenic progeny using AhS2-RNase–specific primers. Middle panel, RT-PCR analysis of RNA isolated from styles of the transgenic progeny using specific primers of PhS3-RNase. Bottom panel, RT-PCR analysis of tubulin for loading control.
(C) and (D) Immunoblot detection of AhSLF-S2 or S-RNases.
(C) Top panel, detection of AhSLF-S2 from total pollen proteins of the transgenic lines l1 to 5, 7, and 8, b1 to 3, wild-type plant, and Antirrhinum (Ant) by polyclonal antibody against AhSLF-S2. The bottom panel is tubulin for loading control.
(D) Top panel, detection of AhS2-RNase (∼29 kD) and endogenous S3-RNase (∼24 kD) from total style proteins of the transgenic lines l1 to 5, 7, and 8, b1 to 3, wild-type plant, and Antirrhinum (Ant) by polyclonal antibody against S-RNases. Bottom panel, detection of tubulin for loading control.
To investigate whether the alteration of SI to SC displayed by the primary transgenic lines was transmitted into in the progeny, pollination tests were performed on the progeny plants of lines b and l (Table 2). After self-pollination, all seven progeny from line l (l1 to 5, 7, and 8) and one progeny b3 from line b still maintained their self-compatibility and set variable-sized capsules. However, the two progeny b1 and b2, in which AhSLF-S2 was undetectable using an AhSLF-S2 antibody (Figure 4C), set no capsule. The self-incompatibility may be restored as a result of the loss of AhSLF-S2 expression in pollen. Furthermore, the wild-type plants pollinated by the seven l progeny and the b3 set large capsules, demonstrating that the alteration of SI to SC is stably transmitted into the progeny. By contrast, the wild-type plants set no seed after being pollinated by two b1 and b2 progeny. This was consistent with the results of self-pollination test of these two plants described above.
Table 2.
Pollination Behaviors of the TAC26 Transgenic Petunia T1 Progeny
| Cross-Pollination
|
|||
|---|---|---|---|
| Planta | Self-Pollinationb | WT Femalec | WT Maled |
| l1 | 9/11 (89.1 ± 19.6) | 10/10 (102.0 ± 12.3) | 0/12 (0) |
| l2e | 10/10 (222.7 ± 42.5) | 11/12 (117.5 ± 20.8) | 13/13 (201.5 ± 23.7) |
| l3e | 10/10 (79.9 ± 10.98) | 9/10 (109.7 ± 24.4) | 11/11 (176.5 ± 18.3) |
| l4f | 7/8 (186.43 ± 29.44) | 7/7 (91.4 ± 8.6) | 13/13 (0) |
| l5f | 8/9 (104.0 ± 16.1) | 11/12 (196.5 ± 18.2) | 0/16 (0) |
| l7e | 11/11 (225.5 ± 40.8) | 10/10 (230.7 ± 21.2) | 8/8 (207.9 ± 20.0) |
| l8 | 10/10 (202.1 ± 35.3) | 14/14 (230.6 ± 21.2) | 0/18 (0) |
| b1fg | 0/17 (0) | 0/18 (0) | 0/12 (0) |
| b2fg | 0/15 (0) | 0/21 (0) | 0/14 (0) |
| b3 | 10/12 (70.9 ± 14.0) | 12/12 (260 ± 22.7) | 0/16 (0) |
| WT | 0/25 (0) | –h | – |
b, g, l, and m are four transgenic lines, and WT is the wild-type S3S3 plant.
Data are represented as mature capsules/total pollination sets (average seeds per capsule ± sd).
The pollen from the transgenic plants were used to pollinate mature styles from the wild-type S3S3 plants.
The pollen from the wild-type plants were used to pollinate mature styles from the transgenic plants.
Plants with low or undetectable expression of PhS3-RNase.
Plants with low or undetectable expression of introduced AhS2-RNase.
Plants with low or undetectable expression of introduced AhSLF-S2.
Not applicable.
Finally, the T1 progeny were pollinated using wild-type S3S3 plants as pollen donor. The result showed that three b progeny maintain normal SI function in styles, consistent with the normal expression of endogenous PhS3-RNase, and out of seven l progeny, four plants (l1, 4, 5, and 8) maintain normal style SI function, whereas three plants (l2, 3, and 7) lost their normal style function, consistent with the result that no endogenous PhS3-RNase transcript or protein could be detected (Figures 4B and 4D). These results also suggest that expression of AhS2-RNase is not related to the conversion of SI to SC in the transgenic lines because l4 and l5 progeny lost the expression of AhS2-RNase in their styles but maintained the SC trait as with those still having AhS2-RNase in styles, such as l1, 8, and b3. Thus, these results further indicate that the TAC26 encodes the pollen component of self-incompatible response.
Expression of AhSLF-S2 in Pollen Causes the Breakdown of Pollen Self-Incompatibility in Transgenic Petunia
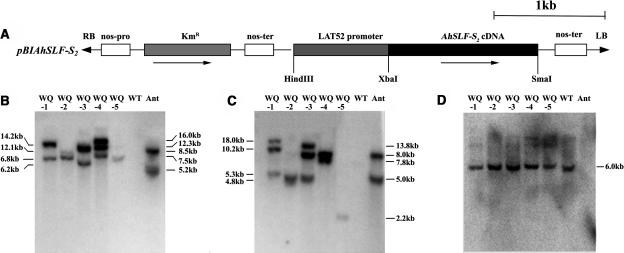
Because no predicted genes except AhSLF-S2 and AhS2-RNase are found in the genomic region covered by the TAC26 (Lai et al., 2002; Zhou et al., 2003) and S-RNases do not function in pollen (Kao and McCubbin, 1996), the alteration of the pollen function of SI in these transgenic lines is very likely because of the presence of AhSLF-S2. To confirm this and further exclude the possibility that the repetitive or unknown sequences flanking AhSLF-S2 could somehow influence the phenotype, we constructed a transformation vector, pBIAhSLF-S2 (Figure 5A). To express AhSLF-S2 in pollen, we used a tomato (Lycopersicon esculentum) pollen-specific promoter LAT52 (Twell et al., 1991) to drive the expression of AhSLF-S2 and introduced the construct into the S3S3 homozygotes of P. hybrida by Agrobacterium-mediated transformation. Five kanamycin-resistant T0 lines were generated. DNA gel blot analysis of leaf genomic DNA digested with HindIII or EcoRI was performed on these plants using AhSLF-S2 cDNA as a probe (Figures 5B and 5C). The results showed that two lines (WQ-2 and WQ-5) had one copy and the other there lines contained three copies of the AhSLF-S2 transgene and that all five plants were independently transformed. The S3S3 genotype of the transgenic lines was also confirmed using PhS3-RNase cDNA as a probe (Figure 5D).
Figure 5.
Molecular Analysis of AhSLF-S2 Transgenic Petunia Plants.
(A) A schematic diagram of pBIAhSLF-S2. RB and LB, right and left borders of the T-DNA; Kmr, kanamycin resistance (neomycin phosphortransferase) gene. Several restriction sites also are indicated.
(B) and (C) DNA gel blot analysis of the transgenic lines WQ-1, WQ-2, WQ-3, WQ-4, and WQ-5 and the untransformed control (WT). Leaf genomic DNA was digested with HindIII (B) or EcoRI (C) was blotted and probed with AhSLF-S2. Leaf DNA of an S2S4 Antirrhinum line (Ant) was as a positive control.
(D) Leaf DNA digested by EcoRI from the transgenic lines was blotted and probed with PhS3-RNase.
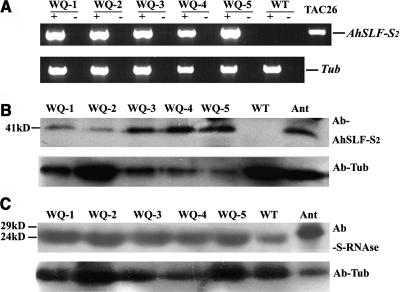
To examine the expression of AhSLF-S2, RT-PCR analysis was conducted. Pollen cDNA from the five transgenic lines and the wild-type S3S3 plants with or without reverse transcriptase were used. As shown in Figure 6A, the PCR products of ∼700 bp were exclusively detected in the five transgenic lines and Antirrhinum, whereas the wild-type S3S3 petunia showed no product using AhSLF-S2 specific primers. To further confirm the normal expression of the AhSLF-S2 transgene, total protein extracts from the pollen of the five transgenic lines were subjected to immunoblot analysis using the antibody against the AhSLF-S2 C-terminal region (Qiao et al., 2004). The result showed that its expression could be detected in the transgenic lines (Figure 6B) at a similar size (∼41 kD) to that observed in Antirrhinum, whereas no similar protein could be detected in the pollen of wild-type petunia plant. To exclude the possibility of abnormal expression of the endogenous PhS3-RNase in pistil, total protein extracts from pistil were examined using the polyclonal antibody against Antirrhinum S-RNases (Qiao et al., 2004). The result showed that the expression level of PhS3-RNase in transformed lines was similar to that of the wild-type line (Figure 6C). These results showed that the AhSLF-S2 transgene is expressed in pollen in the five independent transgenic lines.
Figure 6.
Expression Analysis of AhSLF-S2 Transgenic Petunia Plants.
(A) RT-PCR analysis of RNA isolated from pollen of the AhSLF-S2 transgenic lines WQ-1, WQ-2, WQ-3, WQ-4, and WQ-5 and wild-type plant, using specific primers of AhSLF-S2 with (+) or without (−) reverse transcriptase in the synthesis of cDNA. The TAC26 plasmid (lane 13) was used as a positive control (top panel). RT-PCR analysis of tubulin for loading control (bottom panel).
(B) Top panel, immunoblot detection of AhSLF-S2 from total pollen protein of the transgenic plants WQ-1, WQ-2, WQ-3, WQ-4, and WQ-5 and wild-type S3S3 petunia by polyclonal antibody against AhSLF-S2. Bottom panel, detection of tubulin for loading control. Antirrhinum pollen protein (Ant) was used as a positive control.
(C) Immunoblot detection of PhS3-RNase from total style proteins of the transgenic lines of WQ-1, WQ-2, WQ-3, WQ-4, and WQ-5 and wild-type plant by polyclonal antibody against AhS-RNase (top panel). Detection of tubulin for loading control (bottom panel). Antirrhinum style protein (Ant) was used as a positive control.
As with the lines transformed by TAC26, no obvious morphological change of the AhSLF-S2 transgenic plants was observed compared with the untransformed plant. The self-incompatibility phenotype of all AhSLF-S2 transgenic plants was examined by a series of self- and cross-pollinations (Table 3). To check whether the expression of AhSLF-S2 could change the SI behavior as observed in the TAC26 transgenic plants, the five transgenic lines were self-pollinated, and all of them lost their SI and became SC. They also set variable-sized capsules with an average seed number of 49.0 to 98.7, slightly lower than a normal compatible pollination. The reason why these lines set lower numbers of seeds than that of the TAC26 lines is not clear, and perhaps it is related to the different promoters used or physiological conditions of the plants. The fruit size was not related to the insert copy number. To observe how many seeds were set after pollination by compatible S-genotype pollen, we pollinated these plants using homozygous Sv pollen and found they also set a similar number of seeds (∼60 to 80 seeds per capsule). To exclude the possibility that the breakdown of SI was because of the change in style function, we pollinated mature style of the wild-type S3S3 plant using the transgenic lines as pollen donor. As shown in Table 3, they also set fruits, and the average seed number per capsule set ranged from 177.2 to 213.2. The wild-type plants maintain their SI rigorously because 34 flowers were self-pollinated and none could set seed. We also pollinated the five transgenic lines using the wild-type plant as pollen donor and found that no fruit was set. These results clearly demonstrate that the styles of the transgenic lines maintain the normal SI function, but they had lost the pollen SI function. Taken together, these results directly show that the expression of AhSLF-S2 in pollen leads to the loss of the S-locus pollen function, and the AhSLF-S2 gene alone could control the pollen function of S-RNase–based self-incompatibility.
Table 3.
Pollination Behaviors of the AhSLF-S2 Transgenic Petunia Plants
| Cross-Pollination
|
|||
|---|---|---|---|
| Planta | Self-Pollinationb | WT Femalec | WT Maled |
| WQ-1 | 12/14 (56.6 ± 23.4) | 8/8 (174.0 ± 30.6) | 0/12 (0) |
| WQ-2 | 11/11 (69.8 ± 13.5) | 11/11 (190.9 ± 26.3) | 0/18 (0) |
| WQ-3 | 20/24 (49.0 ± 16.2) | 9/10 (177.2 ± 16.7) | 0/16 (0) |
| WQ-4 | 34/37 (50.3 ± 27.1) | 7/7 (213.2 ± 36.4) | 0/12 (0) |
| WQ-5 | 30/33 (98.7 ± 21.1) | 13/13 (200.0 ± 24.3) | 0/14 (0) |
| WT | 0/34 (0) | –e | – |
WQ-1 to -5 are five transgenic lines, and WT is the wild-type S3S3 plant.
Data are represented as mature capsules/total pollination sets (average seeds per capsule ± sd).
The pollen from the transgenic plants were used to pollinate mature styles from the wild-type S3S3 plants.
The pollen from the wild-type plants were used to pollinate mature styles from the transgenic plants.
Not applicable.
The Physical Interaction between AhSLF-S2 and PhS3-RNase
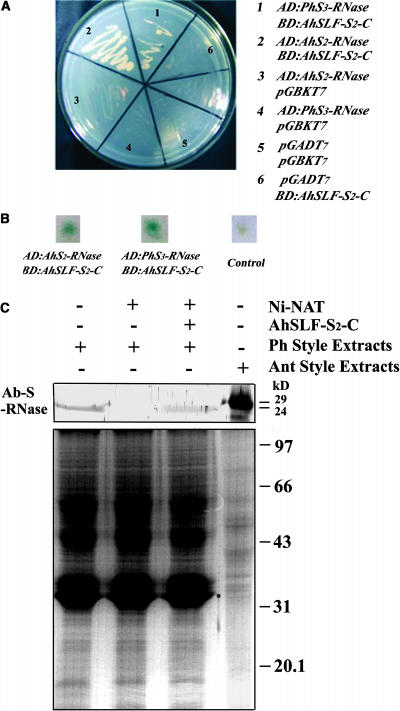
The above results indicated that P. hybrida contains an AhSLF-S2 ortholog and that AhSLF-S2 is capable of physically interacting with P. hybrida S3-RNase. To examine the possibility of the physical interaction between AhSLF-S2 and PhS3-RNase, we used a yeast two-hybrid screening procedure. Our previous work showed that only C-terminal AhSLF-S2 interacts physically with AhS-RNases in yeast (Qiao et al., 2004). Thus, we also used the C-terminal AhSLF-S2 to test whether it is able to interact physically with PhS3-RNase in yeast. The C-terminal AhSLF-S2 was introduced into pGBKT7 vector and expressed as a fusion to GAL4 DNA binding domain (BD), whereas AhS2-RNase and PhS3-RNase were introduced into pGADT7 vector and expressed as a fusion to transcriptional activating domain (AD) as described previously (Qiao et al., 2004). The AD: AhS2-RNase or AD: PhS3-RNase was transformed into yeast AH109 in combination with BD: AhSLF-S2-C, respectively. As shown in Figure 7A, transformed yeast cells by AD: AhS2-RNase/BD: AhSLF-S2-C and AD: PhS3-RNase/BD: AhSLF-S2-C grew well on both -Leu/-Trp and -Leu/-Trp/-His/-Ade media, indicating that a physical interaction had occurred between AhSLF-S2-C and PhS3-RNase. Yeast transformed with the control plasmids AD: PhS3-RNase and pGBKT7 or BD: AhSLF-S2-C and pGADT7 did not grow (Figure 7A). Furthermore, the β-galactosidase reporter gene activity was detected and appeared to be positive in yeast cells cotransformed with AD: PhS3-RNase and BD: AhSLF-S2-C as with the positive controls AD: AhS2-RNase and BD: AhSLF-S2-C (Figure 7B), showing that AhSLF-S2 and PhS3-RNase physically interact in yeast cells.
Figure 7.
Physical Interaction between AhSLF-S2 and PhS3-RNase.
(A) Yeast cells containing various combinations of BD and AD fusions were tested for their growth on -Leu/-Trp/-His/-Ade dropout media. Plasmid pGBKT7 with various AD:constructs and plasmid pGADT7 with various BD:constructs were used as negative controls.
(B) The strains were grown further to test for expression of the β-galactosidase reporter gene.
(C) A pull-down assay for the physical interaction between AhSLF-S2 and PhS3-RNase. Ni-NTA resin and the purified fusion proteins of His-AhSLF-S2-C were incubated with the style extract of the S3S3 line of P. hybrida (Ph Style Extracts). Bound proteins were pulled down with Ni-NTA resin, eluted with the lysis buffer, separated by 12% SDS-PAGE, transferred to membranes, and analyzed by immunoblotting using the anti-S-RNase antibody (top panel). Style extracts from Antirrhinum (S2S5) (Ant Style Extracts) were also included as a control. Input style total protein was used as a positive control (bottom panel). Molecular mass markers are indicated in kilodaltons.
To further confirm this interaction, a pull-down assay was performed using a recombinant AhSLF-S2 protein fused with an N-terminal His-tag (Figure 7C). The purified His-AhSLF-S2-C fusion proteins were incubated with the style extracts from wild-type S3S3 P. hybrida plant. After washing with buffer, the nickel-nitrilotriacetic acid agarose (Ni-NTA) resin–bound proteins were assayed by SDS-PAGE and examined by immunoblot analysis with the polyclonal AhS-RNase antibody (Qiao et al., 2004). As shown in Figure 7C, a specific protein of ∼24 kD, similar to that detected in the wild-type S3S3 P. hybrida style, was also detected by the antibody when using the His-AhSLF-S2-C fusion protein with the style extracts. The ∼24-kD protein was not detected when using the style extract only with the resin, indicating that the C-terminal part of AhSLF-S2 physically interacts with PhS3-RNase in vitro, similar to the interaction observed previously for Antirrhinum AhSLF-S2 and S-RNases (Qiao et al., 2004).
Expression of AhSLF-S2 Does Not Interfere with the Expression of Endogenous SLF or SLF-Like Genes in Pollen
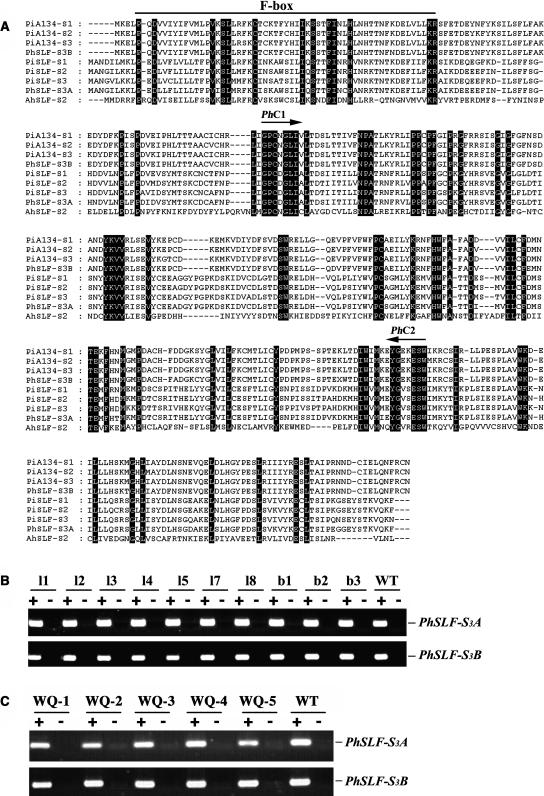
To test for the normal expression of an endogenous SLF gene in these transgenic plants, we designed two degenerate primers based on two conserved regions of the SLF proteins (Figure 8A) and subsequently cloned two SLF genes from mature pollen of S3S3 P. hybrida. Their full-length cDNA were obtained by rapid amplification of cDNA ends (RACE). Amino acid comparison revealed that the first gene named PhSLF-S3A has ∼90% amino acid identity to PiSLF-S1, S2, and S3 (Sijacic et al., 2004) (Figure 8A), similar to the identity observed for the three P. inflata alleles, which have been demonstrated as the pollen determinant genes in P. inflata (Sijacic et al., 2004). The second gene, named PhSLF-S3B, has ∼96% amino acid identity to PiA134-S1, S2, and S3, which are tightly linked to the S-locus in P. inflata (Wang et al., 2003). Sequence alignment showed that AhSLF-S2 shares ∼30% amino acid identity to PhSLF-S3A and PhSLF-S3B. RT-PCR analysis showed that these two genes also are specifically expressed in pollen (data not shown). Although further work is required to formally demonstrate that PhSLF-S3A and PhSLF-S3B are orthologs of PiSLF and PiA134, respectively, their sequence similarities strongly suggest that they are functionally related.
Figure 8.
Amino Acid Sequence Alignment of Predicted SLF Polypeptides from Antirrhinum and Petunia and Expression Analysis of Endogenous SLF Genes in Transgenic Petunia Plants.
(A) Alignment of the predicted polypeptide sequences from the SLF family. Antirrhinum, AhSLF-S2 (CAC33010); P. inflata, PiA134-S1 (AAR15914), PiA134-S2 (AAR15915), PiA134-S3 (AAR15916), PiSLF-S1 (AY500390), PiSLF-S2 (AY500391), and PiSLF-S3 (AY500392); P. hybrida, PhSLF-S3A (AY639403) and PhSLF-S3B (AY639402). PhC1 and PhC2 indicate the conserved regions that were used to design degenerate primers for cloning PhSLF-S3A and PhSLF-S3B. The F-box domain is also indicated.
(B) and (C) RT-PCR analysis of RNA isolated from pollen of the TAC26 and AhSLF-S2 transgenic lines and wild-type plant, using specific primers of PhSLF-S3A and PhSLF-S3B with (+) or without (−) reverse transcriptase in the synthesis of cDNA. Tubulin control was the same as in Figures 4A and 6A.
To examine whether these two endogenous SLF genes are expressed normally in pollen of the transgenic plants, RT-PCR were conducted using specific primers of these two genes. Ten T1 TAC26 plants and five AhSLF-S2 cDNA transgenic plants all expressed PhSLF-S3A and PhSLF-S3B normally in pollen (Figures 8B and 8C), showing that the expression of AhSLF-S2 does not interfere with the normal expression of the endogenous SLF or their related genes in transgenic plants and the compatibility phenotype observed in the transgenic plants could not be caused by the loss of expression of the endogenous SLF genes.
DISCUSSION
AhSLF-S2 Controls the Pollen Function of S-RNase–Based Self-Incompatibility
The finding that the allelic SLF genes in several species from three families with S-RNase–based self-incompatibility are expressed specifically in pollen suggests that they encode the elusive pollen S function (Kao and Tsukamoto, 2004). Consistent with this view, we have shown that AhSLF-S2 physically interacts with S-RNases albeit without allelic specificity (Qiao et al., 2004). In this study, the breakdown of the pollen component of self-incompatibility by expressing AhSLF-S2 in self-incompatible petunia provides direct evidence that it controls the pollen function of the S-RNase–based self-incompatible reaction. In fact, Sijacic et al. (2004) have demonstrated that PiSLF-S2 also controls the pollen self-incompatibility in P. inflata. Together, these results show that these polymorphic SLF proteins determine the pollen self-incompatibility in both the Solanaceae and Scrophulariaceae and possibly in the Rosaceae as well.
Two different models, the gatekeeper model and the inhibitor model, have been proposed to explain how S-RNases function inside the pollen tube (Thompson and Kirch, 1992; Kao and McCubbin, 1996). The inhibitor model predicts that the pollen S products are cytosolic RNase inhibitors, with each allelic product specifically inhibiting the RNase activity of all nonself S-RNases but not that of its cognate self S-RNase. In the gatekeeper model, the pollen S presumably encodes an allele-specific receptor for S-RNases whereby only self S-RNases would be taken up into the cytoplasm of a pollen tube. Several lines of evidence support the inhibitor model (reviewed in Kao and Tsukamoto, 2004). Luu et al. (2000) used immunocytochemistry to clearly show that there is unspecific uptake of S-RNases by the pollen tube, thus providing a strong support for the inhibitor model. This model can explain a well-known phenomenon termed competitive interaction, which refers to a breakdown of pollen function in SI caused by the presence of two heteroallelic S-haplotypes in pollen (Crane and Lewis, 1942; Stout and Chandler, 1942; Lewis, 1947; reviewed in de Nettancourt, 2001). Based on recent results (Qiao et al., 2004; Sijacic et al., 2004; this study), we propose a modified inhibitor model to explain how AhSLF-S2 recognizes self- or nonself-S-RNase and leads to self-incompatibility or compatibility in the S-RNase–based self-incompatible reaction. After self-S-RNase and nonself-S-RNase are taken up into the growing pollen tube, AhSLF-S2 interacts with both S1- and S2-RNases. The interaction of AhSLF-S2 with self S2-RNase is somehow not effective in forming a functional SCF complex to ubiquitinate S-RNase, thus allowing its action in inhibiting the pollen tube growth. By contrast, the interaction of AhSLF-S2 with nonself S1-RNases produces a functional SCF complex that in turn leads to its subsequent ubiquitination and destruction by the 26S proteasome.
Thus, in our transformation experiments, AhSLF-S2 or PhSLF- S2 recognizes endogenous PhS3-RNase or introduced AhS2-RNase as a nonself S-RNase and inhibits its RNase activity, which leads to the successful growth of the pollen tube in pistil, a phenocopy of the well-known phenomenon of competitive interaction. This model also provides a possible explanation to an apparently contradictory observation of the domain-swapping experiments in petunia (Kao and McCubbin, 1996), Nicotiana (Zurek et al., 1997), and Solanum (Matton et al., 1997). The swapping of hypervariable regions and other domains between alleles results in the production of a chimeric ribonuclease. In petunia and Nicotiana, the chimeric S-RNases generated between two significantly diverged alleles were unable to reject self-pollen. It is likely that the alteration of their structure compared with the native S-RNases are so gross that it renders them to behave as nonself S-RNases, and the pollen could grow through the styles because the chimeric S-RNases were degraded as nonself S-RNases. In Solanum, a so-called dual-specificity chimeric S-RNase, named S11/13-RNase, was generated between two highly similar alleles (Matton et al., 1997; Luu et al., 2001), and this would produce two very similar S-RNases apart from the S11- and S13-allele specificity domains. Plants that produce the S11/13-RNase reject both S11 and S13 pollen, showing that the chimeric S-RNase is not degraded in the pollen tube. In addition, the pistil of transgenic plants that produce the S11/13-RNase also is able to reject the pollen from tetraploid plants of S11S11S13S13 genotype (Luu et al., 2001), suggesting that the S11/13-RNase is still functional within the diploid pollen. The hybrid S11/13-RNase would be recognized as a self RNase by either of the pollen S products present in the pollen because of its little alteration in three-dimensional conformation; subsequently, it would not be degraded, thus preventing the pollen tube growth. This possibility could be tested after cloning the SLF-S11 and -S13 genes in Solanum chacoense and using a genetic transformation approach.
AhSLF-S2 Could Determine Both Allelic Specificity and S-RNase Inhibitory Activity
The breakdown of the pollen self-incompatibility by AhSLF-S2 in petunia and the fact that AhSLF-S2 is an F-box protein suggests that it could control both S-allele specificity and inhibition of S-RNase activity. Kao and Tsukamoto (2004) consider that the putative pollen S contains two domains, an S-allele–specific domain and an inhibitor domain. The S-RNase also contains two separated functional domains, an S-allele specificity domain and a catalytic domain. In the case of self-interaction, the S-allele–specific domain of pollen S interacts with S-allele–specific domain of self S-RNase by virtue of the match between the paired domains, leaving the catalytic domain of S-RNase active. However, in the case of nonself interaction, the inhibitor domain of pollen S interacts with the catalytic domain of the S-RNase in the absence of a match between their S-allele specificity domains, thus inhibiting the RNase activity of S-RNase.
It is known that S-RNases show a high degree of allelic sequence diversity and domain-swapping experiments have demonstrated that S-allele specificity is scattered throughout the molecule and not restricted to specific sites in both petunia and Nicotiana (Kao and McCubbin, 1996; Zurek et al., 1997). Nevertheless, in some cases, S-RNase alleles share very high similarity, for example, S1- and Sr1-RNase in Solanum tuberosum (Kaufmann et al., 1991) and S11- and S13-RNases in S. chacoense (Despres et al., 1994; Saba-el-Leil et al., 1994) all show >95% homology and yet they are phenotypically distinct. Despite the low degree of polymorphism of four AhSLF alleles (∼95% identity) (Zhou et al., 2003) and three PiSLF alleles (∼90% identity) (Sijacic et al., 2004) compared with PdSFB and PmSLF alleles (∼68 to 80% similarity) (Entani et al., 2003; Ushijima et al., 2003; Yamane et al., 2003), the alteration of the pollen function by AhSLF-S2 clearly shows that this Antirrhinum F-box gene is capable of determining male specificity.
Although two hypervariable regions have been described in the SFB/SLF proteins from the Rosaceae as in the case of S-RNase alleles (Ushijima et al., 2003; Ikeda et al., 2004; Kao and Tsukamoto, 2004), amino acid differences among four AhSLF-S alleles are scattered throughout the proteins, including the F-box domain (Lai et al., 2002; Zhou et al., 2003), which is similar to the three PiSLF alleles in P. inflata (Sijacic et al., 2004). It is likely that these minor allelic differences could possibly lead to the change of the higher-order conformation of the protein and eventually affect the outcome of their interaction with S-RNases. The amino acid similarity between AhS2-RNase and PhS3-RNase is as low as 36.9%, but AhSLF-S2 does appear to physically interact with PhS3-RNase (Figure 7), indicating the structural and functional conservation between AhSLF-S2 and the endogenous ortholog of P. hybrida.
Recently, we have shown that AhSLF-S2 interacts with Ask-1 and Cul-1 like proteins and the 26S proteasome pathway is required for the inhibition of S-RNase during a compatible pollination (Qiao et al., 2004). These findings indicated that, as a possible substrate of AhSLF-S2, nonself S-RNase is ubiquinated by SCFAhSLF-S2 complex and degraded through the 26S proteasome pathway. It remains to be determined what differences lead to self S-RNase stability in contrast with nonself S-RNase being degraded. In addition, most known substrates of the SCF complexes in animals and plants need to be phosphorylated before being recognized by the F-box protein (Hershko and Ciechanover, 1998). Kunz et al. (1996) found that S-RNases could be phosphorylated by a calcium-dependent protein kinase from pollen tube without allelic specificity. Thus, it needs to be tested whether the phosphorylation condition will affect the degradation of S-RNases in vivo. The ubiquitin/26S proteasome has been found to participate in other mating systems, such as in yeast and sporophytic SI (Hicke and Riezman, 1996; Stone et al., 2003), and its possible participation in S-RNase–based SI systems suggests the conservation and importance of the ubiquitin/26S proteasome pathway in the operation of a diverse range of breeding systems.
Functional and Evolutionary Implications for the SLF Proteins in S-RNase–Based SI Families
The SLF sequences are extensively diverged among the three families Solanaceae, Scrophulariaceae, and Rosaceae (Kao and Tsukamoto, 2004), but they possibly maintain a similar three-dimensional structure because of the presence of several conserved domains. Our results and that obtained in P. inflata (Sijacic et al., 2004) suggest that the SLF sequences closest to S-RNases are functionally conserved. However, it has been found that clusters of F-box genes are present in the S-locus of Antirrhinum (Zhou et al., 2003) and several other solanaceous and rosaceous species (Entani et al., 2003; Ushijima et al., 2003; Wang et al., 2003). Although the parologous copies of AhSLF-S2 have been found not to physically interact with S-RNases (Qiao et al., 2004), it remains unclear if they have any roles in other yet-defined processes. As a recognition locus, the disease resistance R locus in plants also is characterized by gene clusters. For example, the tomato Pto gene belongs to a complex locus consisting of a tightly linked cluster of five to seven genes. Pto confers resistance to Pseudomonas syringae (Martin et al., 1993), whereas the tightly linked paralog Fen confers sensitivity to an organophosphate insecticide (Martin et al., 1994; Loh and Martin, 1995). In other cases, potato Gpa2 and Rx1 share 88.7% homology as paralogs, but they confer resistance to the potato cryst nematode Globodera pallida and potato virus X, respectively (van der Vossen et al., 2000). Further functional analysis of these S-locus paralogs will provide a better understanding of their functions in plant development as well as the origin and evolution of the S-RNase–based self-incompatibility.
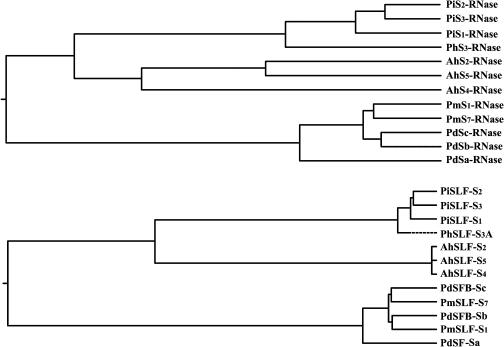
The functional conservation of AhSLF-S2 in P. hybrida has important implication for the evolution of S-RNase–based self-incompatibility. Previous analyses have found that S-RNases from the Scrophulariaceae, Solanaceae, and Roasaceae are most likely monophyletic (Xue et al., 1996; Igic and Kohn, 2001). The Solanaceae and Scrophulariaceae belong to the subclass Asteridea, whereas the Rosaceae are in the subclass Rosidea. Together they comprise roughly three-quarters of dicot families (Cronquist, 1981). That the expression of AhSLF-S2 from the Scrophulariaceae is able to change the pollen behavior of P. hybrida from the Solanaceae further supports a common origin of gametophytic SI system in these two distantly related families. Figure 9 represents the schematic diagrams of the phylogenetic relationships for several pollen determinant SLF and possible pollen determinant SLF proteins and S-RNases in the three families. A similar topology was observed for them, supporting the notion that they are coevolved. Nevertheless, it is unclear how this coevolution occurs. Further comparative analyses of the S-locus structures from members of the three families will reveal their origin and evolution.
Figure 9.
Schematic Representations of the Phylogenetic Relationships for Predicted SLF and S-RNase Polypeptides.
The phylogentic relationships are shown for the S-RNases (top) and SLFs (bottom), respectively. Antirrhinum, AhSLF-S2, AhSLF-S4 (CAD56661), AhSLF-S5 (CAD56664), AhS2-RNase (CAC33020), AhS4-RNase (Q38717), and AhS5-RNase (CAA65318); P. inflata, PiSLF-S1, PiSLF-S2, PiSLF-S3, PiS1-RNase (S20989), PiS2-RNase (AAG21384), and PiS3-RNase (AAA33727); P. hybrida, PhSLF-S3A and PhS3-RNase (AJ271065); Prunus dulcis, PdSFB-Sa (BAC65206), PdSFB-Sb (BAC65207), PdSFB-Sc (BAC65201), PdSa-RNase (BAA95317), PdSb-RNase (T12078), and PdSc-RNase (T12076); P. mume, PmSLF-S1 (BAC66622), PmSLF-S7 (BAC66623), PmS1-RNase (BAC56115), and PmS7-RNase (BAC56116).
METHODS
Plant Materials and Transformation
The self-incompatible line of S3S3 Petunia hybrida and SI lines of Antirrhinum hispanicum have been described previously (Xue et al., 1996; Robbins et al., 2000). Antirrhinum TAC26 was obtained from a TAC genomic DNA library constructed using pTAC as described previously (Liu et al., 2000; Zhou et al., 2003). A pollen-specific promoter LAT52 from tomato (Lycopersicon esculentum) was used to drive the expression of AhSLF-S2 (Twell et al., 1991). HindIII and XbaI double digested LAT52 fragment was ligated to pBI101.1 where the β-glucuronidase gene has been removed previously and then XbaI and SmaI double digested AhSLF-S2 full-length coding region was ligated to pBILAT52. The plasmid TAC26 and pBIAhSLF-S2 were electroporated into Agrobacterium tumefaciens C58 and LBA4404, respectively. Transformation of leaf strips of P. hybrida of S3S3 genotype with Agrabacterium and regeneration of transgenic plants were performed as previously described (Lee et al., 1994; Harbord et al., 2000). For TAC26 transformation, leaf disks of P. hybrida with S3S3 genotype were infected with Agrobacterium by the cocultivation method on MS medium supplemented with 6-benzylaminopurine (2.0 mg L−1) and naphthalene acetic acid (0.2 mg L−1). Shoots were regenerated on fresh MS medium supplemented with hygromycin (50 μg mL−1) and carbenicillin (500 μg mL−1). Regenerated shoots were transferred to hormone-free MS medium containing the same concentrations of antibiotics to induce root formation. For pBIAhSLF-S2 transformation, shoots were regenerated on fresh MS medium supplemented with kanamycin (100 μg mL−1) and carbenicillin (500 μg mL−1).
DNA Gel Blotting Analyses
Genomic DNA isolation was performed as described previously (Xue et al., 1996). DNA (5 μg) was digested, separated on 0.8% agarose gel, and transferred onto Hybond N+ (Amersham, Buckinghamshire, UK) membrane. Prehybridization, hybridization, and washing of the blot were performed as recommended by the manufacturers. Probes were labeled with 32P by random priming using the Prime-a-Gene labeling system (Promega, Madison, WI).
RT-PCR Analysis
Total RNA was prepared as previously described (Lai et al., 2002) and was digested with DNase I (TaKaRa, Dalian, China). Reverse transcriptase (Invitrogen, Carlsbad, CA) was used to synthesis the first strand cDNA. The RT-PCR was performed using specific primers of AhSLF-S2, AhS2-RNase, and P. hybrida Sv- and S3-RNase as previously described (Robbins et al., 2000; Zhou et al., 2003). Specific primers of PhSLF-S3A used for RT-PCR were as follows: 5′-CTGATGGTTATCCTGGTCC-3¢ (forward) and 5′-CAGCTCGTGCGTAAT ACGAC-3′ (reverse). Specific primers of PhSLF-S3B used for RT-PCR were as follows: 5′-CAAGGAACCTTGTGACAAAG-3′ (forward) and 5′-GTTGGACTACTAGGCATTGG-3′ reverse).
Immunoblot Analysis
For protein blot analysis, style or pollen was homogenized in SDS-loading buffer (0.2 M Tris-HCl, pH 6.8, 0.5 M DTT, 4% SDS, and 25% glycerol;), boiled, and centrifuged. Proteins were separated in 12% Tris-Tricine gels (Schägger and von Jagow, 1987) and blotted onto Nitrobind (Micron Separations, Westborough, MA) using a Bio-Rad Transblot SD wet electroblotting apparatus (Hercules, CA). Blots were treated with the rabbit AhS-RNase (1:1000) or AhSLF-S2 antiserum (1:1000) (Qiao et al., 2004) and a mouse monoclonal anti-tubulin Ab (Sigma, St. Louis, MO), and immune complexes were detected using alkaline phosphatase–conjugated secondary antibodies and nitroblue tetrazolumy 5-bromo-4-chloro-3-indolyl phosphate (Harlow and Lane, 1988) or a chemiluminescent immunodetection system (Invitrogen).
Yeast Two-Hybrid Cotransformation Assay
P. hybrida S3-RNase open reading frame lacking signal peptide was cloned into pGADT7 (AD) (Clontech, Palo Alto, CA), and the construction of C-terminal regions of AhSLF-S2 and AhS2-RNase was described before. The BD and AD vectors were cotransformed into AH109 and grown on -Leu/-Trp medium containing 2% agar at 30°C for 4 to 5 d. The clones were further grown on -Leu/-Trp/-His/-Ade medium containing 2% agar at 30°C for 3 to 4 d to test interaction.
For liquid β-galactosidase assay, yeast clones grown for 48 h at 30°C were transferred onto filter paper, and the clones were lysed in liquid nitrogen for 1 min, and then 5 mL of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, and 1 mM MgSO4, pH 7.0) containing β-mercaptoethanol (27 μL/10 mL) was added and incubated at 30°C (for <8 h) and checked periodically for the appearance of blue color.
Pull-Down Assay
Recombinant fusion proteins His-AhSLF-S2C were expressed in Escherichia coli BL21 (DE3) strain (Promega) after induction by 0.1 mM isopropyl-d-thio-galactopyranoside (Sigma) for 3 h at 37°C and purified on Ni-NTA (Pharmacia Biotech, Piscataway, NJ) following the manufacturer's recommendation (Sigma, P6611). Ten micrograms of recombinant protein was coupled to 100 μL of a 50% suspension (v/v) of beads in equilibration buffer for 20 min at 4°C. After prewashing, style extracts were transferred to a clean tube containing His-AhSLF-S2C coupled to the Ni-NTA resin and incubated overnight with constant rotation at 4°C. Subsequently, the beads were washed more than five times with ice-cold 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 1 mM EDTA buffer, and then 50 μL of SDS-PAGE sample buffer was added to each sample. Bound proteins were dissociated by heating at 37°C for 1 h and resolved by SDS-PAGE in 12% gels as described above.
Pollination
All of the pollinations were performed using open flowers. Except for self-pollination, anthers were removed from the flower of the plant serving as female recipient before dehiscence to prevent self-pollination. Pollination was done by collecting pollen from the male donor of the cross and applying it directly to the stigma of the female recipient. Pollinated flowers were covered with paper bags, and a positive result was scored as the formation of a mature capsule with seeds.
Cloning and Sequence Analysis of PhSLF Genes
Degenerate primers PhC1 and PhC2 were used to clone partial cDNA sequences of PhSLF-S3A and -S3B from S3S3 P. hybrida pollen cDNA. PhC1 was 5′-(T/C)TIATIGGICCITG(T/C)(A/G)A(T/C)GG-3′, and PhC2 was 5′-CCAI(C/G)A(T/C)TCII(T/A)I(T/A)CICC-3′. The first strand pollen cDNA was synthesized using forward primer 5′-AAGCAGTGGTATCAACGCAGAGTGGCCATTATGGCCGGG-3′ and reverse primer oligo(dT)17 following the manufacturer's recommendation (Invitrogen). For PhSLF-S3A, 3′RACE was conducted using the primer 5′-CGCACGAGCTGTATTATGGC-3′ and the primer 5′-GTACGGTGTAAGCGAGTCTTGG-3′; 5′RACE was conducted using the primer 5′-GGACCAGGATAACCATCAG-3′ and the primer 5′-GAGGACTCTGAAGTTTCTGG-3′. For PhSLF-S3B, 3′RACE was conducted using the primer 5′-CCAATGCCTAGTAGTCCAAC-3′ and the primer 5′-CGGTGAAAAGGAGTCTTGG-3′; 5′RACE was conducted using the primer 5′-CCGCTGATGGAACGTCTGAAAC-3′ and the primer 5′-GGCATGGTGGGATTAGTCTG-3′.
BLASTp and BLAST2 (http://www.ncbi.nlm.nih.gov/BLAST/) and ClustalW (http://www.ebi.ac.uk) were used for DNA sequence analysis and the alignment. The phylogenetic tree was generated with ClustalW using a neighbor-joining feature from the DNASTAR package. The GenBank accession numbers for and are, respectively.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY639403 (PhSLF-S3A) and AY639402 (PhSLF-S3B).
Acknowledgments
We thank E.S. Coen and R. Carpenter for providing Antirrhinum plants and constant support. We also thank S. McCormick for providing LAT52 promoter and Weicai Yang for critically reading the manuscript. The work was supported by the Chinese Academy of Sciences and the National Natural Science Foundation of China (39825103 and 30221002).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Yongbiao Xue (ybxue@genetics.ac.cn).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.024919.
References
- Brewbaker, J.L., and Natarajan, A.T. (1960). Centric fragments and pollen-part mutation of self-incompatibility alleles in Petunia. Genetics 45, 699–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane, M.B., and Lewis, D. (1942). Genetical studies in pears. III. Incompatibility and sterility. J. Genet. 43, 31–42. [Google Scholar]
- Cronquist, A. (1981). An Integrated System of Classification of Flowering Plants. (New York: Columbia University Press).
- Despres, C., Saba-el-Leil, M., Rivard, S., Morse, D., and Cappadocia, M. (1994). Molecular cloning of two Solanum chacoense S-alleles and a hypothesis concerning their evolution. Sex. Plant Reprod. 7, 169–176. [Google Scholar]
- de Nettancourt, D. (2001). Incompatibility and Incongruity in Wild and Cultivated Plants. (Berlin: Springer-Verlag).
- Entani, T., Iwano, M., Shiba, H., Che, F.S., Isogai, A., and Takayama, S. (2003). Comparative analysis of the self-incompatibility (S-) locus region of Prunus mume: Identification of a pollen-expressed F-box gene with allelic diversity. Genes Cells 8, 203–213. [DOI] [PubMed] [Google Scholar]
- Fagard, M., and Vaucheret, H. (2000). (Trans) gene silencing in plants: How many mechanisms? Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 167–194. [DOI] [PubMed] [Google Scholar]
- Golz, J.F., Oh, H.-Y., Su, V., Kusaba, M., and Newbigin, E. (2001). Genetic analysis of Nicotiana pollen-part mutants is consistent with the presence of an S-ribonuclease inhibitor at the S-locus. Proc. Natl. Acad. Sci. USA 98, 15372–15376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golz, J.F., Su, V., Clarke, A.E., and Newbigin, E. (1999). A molecular description of mutations affecting the pollen component of Nicotiana alata S locus. Genetics 152, 1123–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, J.E., McClure, B.A., Bonig, I., Anderson, M.A., and Clarke, A.E. (1991). Action of the style product of the self-incompatibility gene of Nicotiana alata (S-RNase) on in vitro–grown pollen tubes. Plant Cell 3, 271–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbord, R.M., Napoli, C.A., and Robbins, T.P. (2000). Segregation distortion of T-DNA markers linked to the self-incompatibility (S) locus in Petunia hybrida. Genetics 154, 1323–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D. (1988). Antibodies: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), p. 349.
- Hershko, A., and Ciechanover, A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Hicke, L., and Riezman, H. (1996). Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell 84, 277–287. [DOI] [PubMed] [Google Scholar]
- Igic, B., and Kohn, J.R. (2001). Evolutionary relationships among self-incompatibility RNases. Proc. Natl. Acad. Sci. USA 98, 13167–13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda, K., Igic, B., Ushijima, K., Yamane, H., Hauck, N.R., Nakano, R., Sassa, H., Iezzoni, A.F., Kohn, J.R., and Tao, R. (2004). Primary structural features of the S haplotype-specific F-box protein, SFB, in Prunus. Sex. Plant Reprod. 16, 235–243. [Google Scholar]
- Kao, T.H., and McCubbin, A.G. (1996). How flowering plants discriminate between self and non-self pollen to prevent inbreeding. Proc. Natl. Acad. Sci. USA 93, 12059–12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao, T.H., and Tsukamoto, T. (2004). The molecular and genetic bases of S-RNase-based self-incompatibility. Plant Cell 16 (suppl.), S72–S83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann, H., Salamini, F., and Thompson, R. (1991). Sequence variability and gene structure at the self-incompaibility locus of Solanum tuberosum. Mol. Gen. Genet. 226, 457–466. [DOI] [PubMed] [Google Scholar]
- Kunz, C., Chang, A., Faure, J.D., Clarke, A.E., Polya, G.M., and Anderson, M.A. (1996). Phosphorylation of style S-RNases by Ca-[2+]-dependent protein kinases from pollen tubes. Sex. Plant Reprod. 9, 25–34. [Google Scholar]
- Lai, Z., Ma, W., Han, B., Liang, L., Zhang, Y., Hong, G., and Xue, Y. (2002). An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Mol. Biol. 50, 29–42. [DOI] [PubMed] [Google Scholar]
- Lee, H.S., Huang, S., and Kao, T.H. (1994). S proteins control rejection of incompatible pollen in Petunia inflata. Nature 367, 560–563. [DOI] [PubMed] [Google Scholar]
- Lewis, D. (1947). Competition and dominance of incompatibility alleles in diploid pollen. Heredity 1, 85–108. [Google Scholar]
- Liu, Y.G., Nagaki, K., Fujita, M., Kawaura, K., Uozumi, M., and Ogihara, Y. (2000). Development of an efficient maintenance and screening system for large-insert genomic DNA libraries of hexaploid wheat in transformation-competent artificial chromosome (TAC) vector. Plant J. 23, 687–695. [DOI] [PubMed] [Google Scholar]
- Loh, Y.T., and Martin, G.B. (1995). The disease-resistance gene Pto and the fenthion-sensitivity gene Fen encode closely related functional protein kinases. Proc. Natl. Acad. Sci. USA 92, 4181–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu, D.T., Qin, X., Laublin, G., Yang, Q., Morse, D., and Cappadocia, M. (2001). Rejection of S-heteroallelic pollen by a dual-specific S-RNase in Solanum chacoense predicts a multimeric SI pollen component. Genetics 159, 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu, D.T., Qin, X., Morse, D., and Cappadocia, M. (2000). S-RNase uptake by compatible pollen tubes in gametophytic self-incompatibilty. Nature 407, 649–651. [DOI] [PubMed] [Google Scholar]
- Martin, G.B., Brommonschenkel, S.H., Chunwongse, J., Frary, A., Ganal, M.W., Spivey, R., Wu, T., Earle, E.D., and Tanksley, S.D. (1993). Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262, 1432–1436. [DOI] [PubMed] [Google Scholar]
- Martin, G.B., Frary, A., Wu, T., Brommonschenkel, S., Chunwongse, J., Earle, E.D., and Tanksley, S.D. (1994). A member of the tomato Pto gene family confers sensitivity to fenthion resulting in rapid cell death. Plant Cell 6, 1543–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matton, D.P., Maes, O., Laublin, G., Xike, Q., Bertrand, C., Morse, D., and Cappadocia, M. (1997). Hypervariable domains of self-incompatibility RNases mediate allele-specific pollen recognition. Plant Cell 9, 1757–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure, B.A., Gray, J.E., Anderson, M.A., and Clarke, A.E. (1990). Self-incompatibility in Nicotiana alata involves degradation of pollen rRNA. Nature 250, 937–941. [Google Scholar]
- McCubbin, A.G., and Kao, T.H. (2000). Molecular recognition and response in pollen and pistil interactions. Annu. Rev. Cell Dev. Biol. 16, 333–364. [DOI] [PubMed] [Google Scholar]
- Pandy, K.K. (1965). Centric chromosome fragments and pollen-part mutation of the incompatibility gene in Nicotiana alata. Nature 205, 792–795. [Google Scholar]
- Qiao, H., Wang, H., Zhao, L., Zhou, J., Huang, J., Zhang, Y., and Xue, Y. (2004). The F-Box protein AhSLF-S2 physically interacts with S-RNases that may be inhibited by the ubiquitin/26S proteasome pathway of protein degradation during compatible pollination in Antirrhinum. Plant Cell 16, 582–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins, T.P., Harbord, R.M., Sonneveld, T., and Clark, K. (2000). The molecular genetics of self-incompatibility in Petunia hybrida. Ann. Bot. 85 (suppl. A), 105–112. [Google Scholar]
- Saba-el-Leil, M.K., Rivard, S., Morse, D., and Cappadocia, M. (1994). The S11 and S13 self-incompatibility alleles in solanum chacoense Bitt. are remarkably similar. Plant Mol. Biol. 24, 571–583. [DOI] [PubMed] [Google Scholar]
- Schägger, H., and von Jagow, G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379. [DOI] [PubMed] [Google Scholar]
- Sijacic, P., Wang, X., Skirpan, A.L., Wang, Y., Dowd, P.E., McCubbin, A.G., Huang, S., and Kao, T.-h. (2004). Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature 429, 302–305. [DOI] [PubMed] [Google Scholar]
- Stone, S.L., Anderson, E.M., Mullen, R.T., and Goring, D.R. (2003). ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell 15, 885–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout, A.B., and Chandler, C. (1942). Hereditary transmission of induced tetraploidy and compatibility in fertilization. Science 96, 257–258. [DOI] [PubMed] [Google Scholar]
- Thompson, R., and Kirch, H. (1992). The S-locus of flowering plants: When self-rejection is self-interest. Trends Genet. 9, 381–387. [DOI] [PubMed] [Google Scholar]
- Twell, D., Yamaguchi, J., Wing, R.A., Ushiba, J., and McCormick, S. (1991). Promoter analysis of genes that are coordinately expressed during pollen development reveals pollen-specific enhancer sequences and shared regulatory elements. Genes Dev. 5, 496–507. [DOI] [PubMed] [Google Scholar]
- Ushijima, K., Sassa, H., Dandekar, A.M., Gradziel, T.M., Tao, R., and Hirano, H. (2003). Structural and transcriptional analysis of the self-incompatibility locus of almond: Identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. Plant Cell 15, 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vossen, E., Voort, J., Kanyuka, K., Bendahmane, A., Sandbrink, H., Baulcombe, D.C., Bakker, J., Stiekema, W.J., and Klein-Lankhorst, R.M. (2000). Homologues of a single resistance-gene cluster in potato confer resistance to distinct pathogens: A virus and a nematode. Plant J. 23, 567–576. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Wang, X., McCubbin, A.G., and Kao, T.-h. (2003). Genetic mapping and molecular characterization of the self-incompatibility (S-) locus in Petunia inflata. Plant Mol. Biol. 53, 565–580. [DOI] [PubMed] [Google Scholar]
- Xue, Y., Carpenter, R., Dickinson, H.G., and Coen, E.S. (1996). Origin of allelic diversity in Antirrhinum S locus RNases. Plant Cell 8, 805–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane, H., Ikeda, K., Ushijima, K., Sassa, H., and Tao, R. (2003). A pollen-expressed gene for a novel protein with an F-box motif that is very tightly linked to a gene for S-RNase in two species of cherry, Prunus cerasus and P. avium. Plant Cell Physiol. 44, 764–769. [DOI] [PubMed] [Google Scholar]
- Zhou, J., Wang, F., Ma, W., Zhang, Y., Han, B., and Xue, Y. (2003). Structural and transcriptional analysis of S-Locus F-box genes in Antirrhinum. Sex. Plant Reprod. 16, 165–177. [Google Scholar]
- Zurek, D.M., Mou, B., Beecher, B., and McClure, B. (1997). Exchanging sequence domains between S-RNases from Nicotiana alata disrupts pollen recognition. Plant J. 11, 797–808. [DOI] [PubMed] [Google Scholar]