Abstract
Cotton (Gossypium spp) plants produce seed trichomes (cotton fibers) that are an important commodity worldwide; however, genes controlling cotton fiber development have not been characterized. In Arabidopsis thaliana the MYB gene GLABRA1 (GL1) is a central regulator of trichome development. Here, we show that promoter of a cotton fiber gene, RD22-like1 (RDL1), contains a homeodomain binding L1 box and a MYB binding motif that confer trichome-specific expression in Arabidopsis. A cotton MYB protein GaMYB2/Fiber Factor 1 transactivated the RDL1 promoter both in yeast and in planta. Real-time PCR and in situ analysis showed that GaMYB2 is predominantly expressed early in developing cotton fibers. After transferring into Arabidopsis, GL1∷GaMYB2 rescued trichome formation of a gl1 mutant, and interestingly, 35S∷GaMYB2 induced seed-trichome production. We further demonstrate that the first intron of both GL1 and GaMYB2 plays a role in patterning trichomes: it acts as an enhancer in trichome and a repressor in nontrichome cells, generating a trichome-specific pattern of MYB gene expression. Disruption of a MYB motif conserved in intron 1 of GL1, WEREWOLF, and GaMYB2 genes affected trichome production. These results suggest that cotton and Arabidopsis use similar transcription factors for regulating trichomes and that GaMYB2 may be a key regulator of cotton fiber development.
INTRODUCTION
Aerial surfaces of most land plants have epidermal hairs (trichomes), and some plants of systematically unrelated groups, such as cotton (Gossypium spp) and Salix, produce seed trichomes, which are among the longest plant cells. Trichomes are believed to protect plants against insects, microbes, herbivores, and abiotic damages and to assist seed dispersal. Furthermore, seed trichomes of Gossypium plants, namely cotton fibers, are the most widely used natural fibers in the textile industry (Wilkins et al., 2000; Larkin et al., 2003).
Trichomes provide an excellent system for investigation of cell fate determination. In the epidermis, only some cells are committed to becoming a trichome, and the location and timing of trichome differentiation present an example of cell patterning. Once initiated, trichomes often undergo endoduplication and rapid elongation. In addition, trichome production can be genetically manipulated; alterations and even depletion of trichomes often do not affect plant viability and are readily observable (Larkin et al., 2003).
For the model plant Arabidopsis thaliana, trichome development and root epidermal patterning have been studied in depth, and both processes use a common mechanism involving closely related transcription factors and a similar lateral inhibition signaling pathway (Schneider et al., 1997; Schnittger et al., 1999; Larkin et al., 2003). Two transcription factors, GLABRA1 (GL1) and GL3, function as positive regulators of trichome development, and they encode an R2-R3 MYB protein and a basic helix-loop-helix–related protein, respectively (Oppenheimer et al., 1991; Payne et al., 2000). TRANSPARENT TESTA GLABRA1 (TTG1), a WD40 domain–containing protein that is thought to mediate protein–protein interactions, also acts as a positive regulator (Walker et al., 1999). TRIPTYCHON, CAPRICE, and ENHANCER OF TRY AND CPC have been shown to negatively regulate trichome formation. All three genes encode single-repeat MYB proteins lacking a transcriptional activation domain, and they act in a partially redundant manner to mediate lateral inhibition (Wada et al., 1997, 2002; Schnittger et al., 1999; Schellmann et al., 2002; Kirik et al., 2004). It is proposed that the combinatorial action of these patterning genes regulates trichome-specific expression of GL2, which encodes a homeobox (HOX) transcription factor that promotes trichome cell differentiation and growth (Rerie et al., 1994). Thus, GL2 may be considered a regulator of downstream genes of the trichome pathway. However, it has been unclear up to now how these upstream patterning genes, such as GL1, are regulated and whether this regulation also involves lateral inhibition.
Although both are unicellular epidermal hairs, cotton fibers are distinct from Arabidopsis trichomes in that they are produced in a specific organ (seed), their initiation and development are highly synchronous, and they are unbranched and extremely elongated (Kim and Triplett, 2001). Analyses of gene expression patterns have identified numerous genes whose transcripts are enriched in fiber cells (John and Crow, 1992; Li et al., 2002; Ji et al., 2003), including those encoding metabolism enzymes that are essential to cell growth (Pear et al., 1996; Ruan et al., 2003). For transcription factors, a noteworthy work is the isolation of six MYB genes from G. hirsutum (Loguerico et al., 1999; Cedroni et al., 2003). Another MYB gene (GhMYB109) was recently reported and it is mainly expressed in developing fibers (Suo et al., 2003). Functions of all these cotton MYBs, however, remain for investigation.
In this report, we show that the promoter of a cotton fiber gene, RD22-like1 (RDL1), conducts trichome-specific expression in Arabidopsis plants. By dissecting the promoter, we identified two cotton transcription factors, GaMYB2/Fiber Factor 1 (FIF1) and GhHOX3, which are able to activate the RDL1 promoter. GaMYB2 encodes a GL1-like MYB protein and is expressed early in developing fiber cells. In Arabidopsis, GL1∷GaMYB2 fully rescued trichome formation of a gl1 mutant. We found that regulation of trichome formation by GL1-like MYB genes requires an intron (intron 1), which contains a conserved MYB binding site. Interestingly, when expressed under the control of the 35S promoter, the cotton fiber protein GaMYB2 induced Arabidopsis to produce seed trichomes.
RESULTS
Expression Pattern of Cotton RDL1
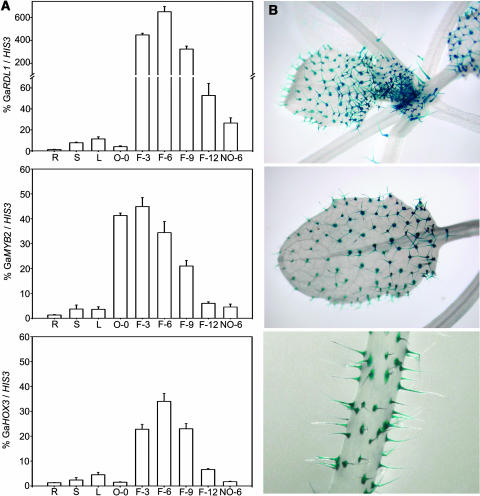
Our previous study showed that transcripts of a cotton RDL gene were enriched in developing fiber cells (Li et al., 2002). DNA gel blot hybridization revealed two RDL genes, namely RDL1 and RDL2, in the genome of G. arboreum (see Supplemental Figure 1 online), an A-genome diploid cotton cultivar (Rong et al., 2004). RT-PCR analysis showed that whereas RDL2 was expressed in various cotton tissues except roots (data not shown), RDL1 was expressed mainly in developing fiber cells (Figure 1A). In ovules, the RDL1 transcripts were undetectable at the day postanthesis (DPA), abundant in rapidly elongating fibers (3 to 9 DPA), and dramatically decreased after 12 DPA. When fibers were separated from the ovule, RDL1 transcripts were mostly present in the fiber portion (Figure 1A). These results indicate that RDL1 is largely a fiber-specific gene.
Figure 1.
Expression Patterns of G. arboreum RDL1, MYB2/FIF1, and HOX3 Genes.
(A) Transcript profile of GaRDL1, GaMYB2, and GaHOX3 in plants of G. arboreum. Relative transcript amounts of each gene were normalized with respect to cotton Histone3 transcript levels (100%). Mean values were obtained from three independent PCR amplifications, and the error bars indicate the standard error of the mean. A break in the scale (=) has been incorporated to show the higher amount of GaRDL1 in fibers. R, roots; S, stems; L, leaves; O-0, 0-DPA ovules; F-3, 3-DPA fibers; F-6, 6-DPA fibers; F-9, 9-DPA fibers; F-12, 12-DPA fibers; NO-6, 6-DPA naked ovules (fibers stripped off).
(B) GUS staining of Arabidopsis plants expressing RDL1-P3∷GUS. Young leaf (top), mature leaf (middle), and stem (bottom) are shown.
To investigate the regulatory mechanism of cotton fiber gene expression, a 7-kb upstream segment of G. arboreum RDL1 gene was isolated by genome walking. According to rapid amplification of cDNA ends (5′-RACE), RDL1 gene transcription starts at the A within the sequence of CCATTCC, which is located at 77 bp upstream to the translation initiation codon ATG (data not shown). A promoter fragment of ∼1200 bp (RDL1-P1) was then fused with a β-glucuronidase (GUS) reporter gene. After transferring into Arabidopsis, GUS activity was present exclusively in trichomes (Figure 1B). Clearly, in Arabidopsis the cotton RDL1-P1 is a trichome-specific promoter.
cis-Elements Conducting Trichome Expression
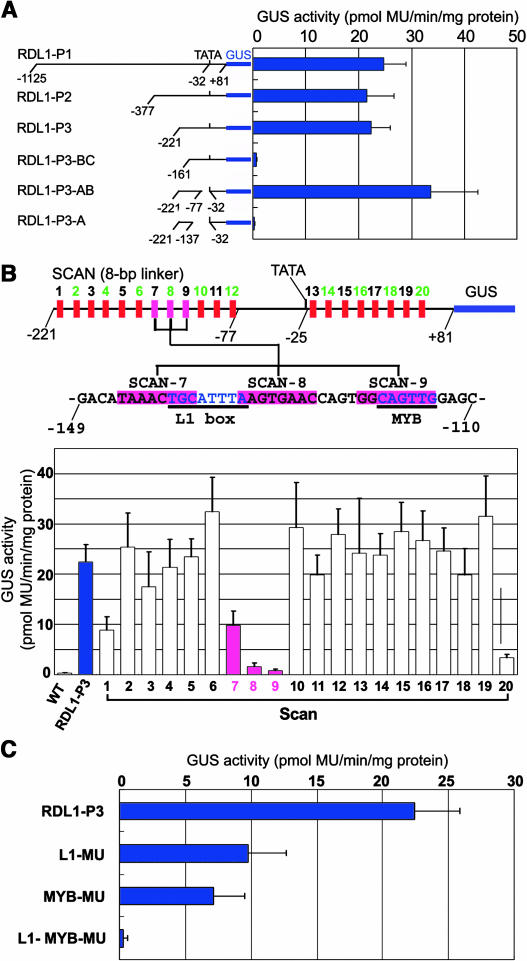
Successive deletions of RDL1-P1 from the 5′-end revealed a 302-bp fragment (RDL1-P3) proximal to the coding region that was already sufficient to conduct trichome-specific expression. Further deletion showed that two DNA segments (−221 ∼ −77 and −32 ∼ +80 bp) contained all the sequence information necessary for the promoter activity (Figure 2A).
Figure 2.
Identification of cis-Elements in the RDL1 Promoter.
(A) GUS activities driven by different upstream fragments of the cotton RDL1 gene. Nucleotides are numbered from the transcription initiation site.
(B) Linker scanning of RDL1-P3. The 8-bp linkers (GGAATTCC or CCTGCAGG) were used. Red and pink boxes represent linkers of SCAN-1 through -20. The L1 box and MYB motif are underlined.
(C) Effects of disruption of L1 box (L1-MU) and MYB motif (MYB-MU) on promoter activity. L1-MYB-MU indicates that both L1 box and MYB motif were mutated. The L1 box was mutated from TGCATTTA to TGCcgTTA and the MYB motif from CAGTTG to CAGTgG.
To identify cis-acting elements in RDL1-P3, 20 DNA replacements were made by PCR-directed linker scanning. We found that changing of a region by three replacements (SCAN-7, -8, and -9) strongly reduced RDL1-P3∷GUS expression (Figure 2B). Sequence analysis by PLACE (http://www.dna.affrc.go.jp/htdocs/PLACE/) showed that this region contains an L1 box (TGCATTTA) and a MYB motif (CAGTTG), which are separated by 13 nucleotides only (Figure 2B). The L1 box is the binding site of HOX proteins, including the L1 layer-specific ATML1 (Abe et al., 2001) and the trichome regulator GL2 (Ohashi et al., 2003). The MYB motif is recognized by MYB transcription factors; Arabidopsis MYB2, for example, binds to the MYB motif of RD22 promoter (Urao et al., 1993). The Arabidopsis GL1, which controls trichome formation, and WEREWOLF (WER), which controls development of non-hair cells of root, are both MYB transcription factors (Oppenheimer et al., 1991; Lee and Schiefelbein, 1999).
To see if these two boxes were responsible for trichome expression, we performed site-directed mutations to change the L1 box from TGCATTTA to TGCcgTTA and the MYB motif from CAGTTG to CAGTgG. Either mutation reduced the promoter activity greatly, though weak GUS staining in trichomes was still observable. Mutation of both cis-elements, however, completely inactivated the promoter (Figure 2C). These results indicate that both L1 box and MYB motif are involved in activating the RDL1-P3 promoter in Arabidopsis trichomes.
Transcription Factors Binding to RDL1-P3
Because both L1 box and MYB motif contribute to the trichome expression of RDL1-P3∷GUS, it is possible that the trichome factors of GL1 and GL2 transactivated RDL1-P3 in transgenic Arabidopsis, and their cotton homologs regulate RDL1 expression in cotton fiber cells.
Searching the database revealed three cotton MYBs that might be related to trichome development. GhMYB1 (Loguerico et al., 1999), also called CotMYBA (Payne et al., 1999), is 33% identical to Antirrhinum majus MIXTA, a positive regulator of floral papillae development (Noda et al., 1994). GhMYB2 (Loguerico et al., 1999) and GhMYB109 (Suo et al., 2003) are 47% identical to each other, and they show 44 and 46% sequence identities to GL1, respectively (see Supplemental Figure 2 online). To avoid confusion with Arabidopsis MYB2, which is not related to trichome development, the cotton MYB2 was also named FIF1 during our investigation. For homeobox proteins, GhHOX1 (AF5309130) and GhHOX2 (AF530914) show 66 and 34% sequence identities to GL2, respectively. We then isolated all these genes or cDNAs from G. arboreum, in addition to a cDNA encoding a novel HOX protein, GhHOX3, which is 37% identical to GL2. These cotton transcription factors were then investigated further.
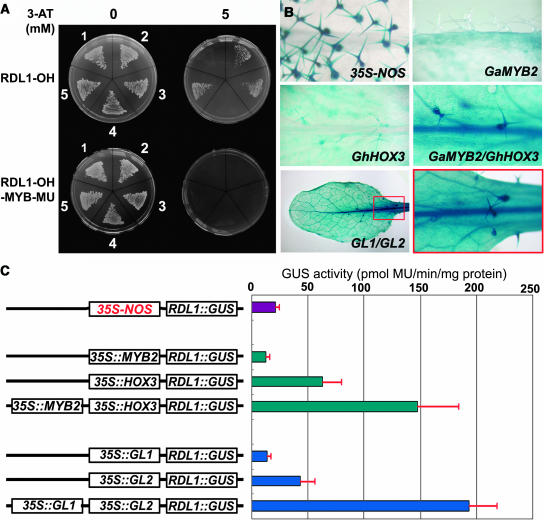
In yeast one-hybrid system, the G. arboreum MYB2 and MYB109 as well as Arabidopsis GL1 and WER exhibited a clear binding activity to a RDL1 promoter fragment (−221 ∼ −33) that contained both the L1 box and MYB motif, whereas another cotton MYB protein, GaMYB1, did not, though its MYB motif showed 65% sequence identity to that of GaMYB2. As mentioned above, GaMYB1 is more similar to Antirrhinum MIXTA than to GaMYB2 and Arabidopsis GL1. When the MYB motif was mutated from CAGTTG to CAGTgG, the MYB proteins were no longer able to bind the DNA (Figure 3A). All the cotton and Arabidopsis HOX proteins tested, however, did not bind to this fragment in yeast, probably because the HOX proteins fused with GAL4 activation domain were too large to be properly expressed or the binding would require other plant proteins to form a complex.
Figure 3.
Transcriptional Activation of RDL1-P3 by GaMYB2 and HOX3.
(A) DNA–protein interactions in yeast one-hybrid system. Yeast strain YM4271 integrating RDL1-OH (−221 to −33 bp) or RDL-OH-MYB-MU was transformed with GAL4 transcriptional activation domain (AD) (1), AD-GL1 (2), AD-WER (3), AD-GaMYB1 (4), and AD-GaMYB2 (5).
(B) Effects of ectopic expression of MYB (GaMYB2 or GL1) and HOX (HOX3 or GL2) genes on the RDL1-P3∷GUS expression pattern in Arabidopsis. In GL1/GL2, the box indicates the magnified region shown at right. 35S-NOS, the 35S promoter and nopaline synthase (NOS) terminator cassette; GaMYB2, 35S∷GaMYB2/RDL1∷GUS; HOX3, 35S∷HOX3/RDL1∷GUS; GaMYB2/HOX3, 35S∷GaMYB2/35S∷HOX3/RDL1∷GUS; GL1/GL2, 35S∷GL1/35S∷GL2/RDL1∷GUS.
(C) GUS activities in Arabidopsis plants expressing both RDL1-P3∷GUS and the transcription factors as indicated. MYB2, GaMYB2.
To provide in planta evidence of RDL1-P3 regulation, related MYB and HOX genes were expressed in wild-type plants of Arabidopsis under the control of the 35S promoter, and their effects on RDL1-P3∷GUS expression were examined. We found that ectopic expression of the MYBs, including GaMYB2, GL1, and WER, induced a weak expression of RDL1-P3∷GUS in nontrichome cells but repressed its expression in trichomes. In plants expressing 35S∷GL2 or 35S∷GhHOX3, the GUS activity was clearly present in trichomes, in addition to a relatively low activity in nontrichome cells. When the MYB and the HOX genes (GL1 and GL2 or GaMYB2 and GaHOX3, respectively) driven by the 35S promoter were cotransferred into Arabidopsis, a strong GUS staining was observed in both trichome and nontrichome cells (Figure 3B), resembling the ubiquitous pattern of the 35S promoter. For each pair of transcription factors, the effects seemed to be synergistic rather than additive (Figure 3C). These results suggest that GL1-like MYB and GL2-like HOX proteins operate together to activate the RDL1-P3 promoter in Arabidopsis plants. In addition, we also transformed the Arabidopsis plants with 35S∷GaMYB2/35S∷HOX1 and 35S∷GaMYB2/35S∷HOX2. We found that, unlike 35S∷GaMYB2/35S∷HOX3, these two combinations did not activate the RDL1 promoter efficiently (data not shown), suggesting that there might be a specific cooperation between GaMYB2 and HOX3 in regulating the RDL1 promoter.
Complementation of Cotton MYB2/FIF1 to gl1 Mutant
To see if GaMYB2 is functionally similar to GL1, complementation was performed on an Arabidopsis gl1 mutant (SALK_039478), which has a T-DNA inserted in the third exon, resulting in a glabrous phenotype (Figure 4A). It is thus a loss-of-function mutation. The coding region of GaMYB2 gene was then placed between the upstream and downstream segments of the GL1 gene (GL1∷GaMYB2); after transferring into gl1 plants, the transgene fully rescued trichome production in >90% of the transformants, similar to the efficiency obtained with Gl1∷GL1 and GL1∷WER (Figure 4B, Table 1). When the three MYB genes driven by the 35S promoter were used for complementation, normal trichome production was also observed in plants of T1 generation. In the wild-type background, these MYB genes driven by either the GL1 sequences or the 35S promoter caused reduced production of leaf trichomes in 30 ∼ 50% of the transgenic plants (Figure 4B, Table 1). Previous studies also showed two opposite effects of 35S∷GL1 expression: suppressing trichome production in the wild-type background and promoting trichome formation in gl1 (Oppenheimer et al., 1991; Larkin et al., 1994), but such effect has not been reported before for MYB genes driven by the GL1 regulatory sequences. Reciprocal complementation using GL1 and WER found that the two genes encode functionally equivalent MYB proteins, though they specify distinct cell types because of nonoverlapping expression patterns (Lee and Schiefelbein, 2001). Our complementation data presented here indicate that the cotton protein GaMYB2 has the same function as Arabidopsis GL1 and WER in regulating trichome development.
Figure 4.
GaMYB2 Regulates Arabidopsis Trichome Development.
(A) The Arabidopsis wild-type and a gl1 (SALK_039478) mutant seedling.
(B) and (C) Trichome phenotypes of wild-type or gl1 mutant plants transformed with various chimerical genes as indicated. For intronless cDNA, a “c” was added as a suffix.
Table 1.
Regulation of Arabidopsis Trichome Development by GL1-Like Genes
| Wild-Type Background
|
gl1 Background
|
||||||
|---|---|---|---|---|---|---|---|
| Construct | T1 Plants Examined | + (%) | ↓ (%) | T1 Plants Examined | + (%) | +/− (%) | − (%) |
| GL1∷GaMYB2 | 47 | 57.5 | 42.5 | 54 | 92.6 | 0.0 | 7.4 |
| GL1∷GaMYB2c | 46 | 100.0 | 0.0 | 52 | 0.0 | 0.0 | 100.0 |
| GL1∷GaMYB109 | 48 | 100.0 | 0.0 | 51 | 11.8 | 64.7 | 23.5 |
| GL1∷GaMYB109c | 48 | 100.0 | 0.0 | 50 | 0.0 | 0.0 | 100.0 |
| GL1∷GL1 | 48 | 56.2 | 43.8 | 54 | 96.3 | 0.0 | 3.7 |
| GL1∷GL1c | 46 | 100.0 | 0.0 | 54 | 0.0 | 0.0 | 100.0 |
| GL1∷WER | 48 | 52.1 | 47.9 | 53 | 100.0 | 0.0 | 0.0 |
| GL1∷WERc | 48 | 100.0 | 0.0 | 54 | 0.0 | 0.0 | 100.0 |
| 35S∷GaMYB2 | 54 | 68.5 | 31.5 | 54 | 92.6 | 0.0 | 7.4 |
| 35S∷GaMYB2c | 52 | 100.0 | 0.0 | 54 | 0.0 | 0.0 | 100.0 |
| 35S∷GaMYB109 | 54 | 100.0 | 0.0 | 48 | 0.0 | 0.0 | 100.0 |
| 35S∷GaMYB109c | 50 | 100.0 | 0.0 | 48 | 0.0 | 0.0 | 100.0 |
| 35S∷GL1 | 54 | 66.7 | 33.3 | 53 | 98.1 | 0.0 | 1.9 |
| 35S∷GL1c | 54 | 100.0 | 0.0 | 50 | 0.0 | 0.0 | 100.0 |
| 35S∷WER | 53 | 66.0 | 34.0 | 54 | 100 | 0.0 | 0.0 |
| 35S∷WERc | 53 | 100.0 | 0.0 | 52 | 0.0 | 0.0 | 100.0 |
| RDL1∷GaMYB2 | 32 | 72.9 | 28.1 | 30 | 96.7 | 0.0 | 3.3 |
| RDL1∷GaMYB2c | 31 | 74.2 | 25.8 | 32 | 100.0 | 0.0 | 0.0 |
| RDL1∷GL1 | 31 | 77.4 | 22.6 | 32 | 100.0 | 0.0 | 0.0 |
| RDL1∷GL1c | 32 | 78.1 | 21.9 | 31 | 100.0 | 0.0 | 0.0 |
| GL1∷GaMYB2-IN-MU | 36 | 69.4 | 30.6 | 36 | 11.1 | 33.3 | 55.6 |
| GL1∷GL1-IN-MU | 36 | 63.9 | 36.1 | 36 | 58.3 | 41.7 | 0.0 |
| GL1∷WER-IN-MU | 36 | 61.1 | 38.9 | 36 | 11.1 | 0.0 | 88.9 |
+, Trichomes normal; ↓, trichome amount reduced (less than one-third of wild-type trichomes); +/−, partial complementation to gl1 (less than half of wild-type trichomes); −, glabrous.
Intron 1 Is Required for Patterning Gene Expression
The Arabidopsis GL1 and WER genes contain two introns, whereas the cotton genes of GaMYB2 and MYB109 contain only one. The cotton intron is equivalent to the first intron of Arabidopsis genes regarding their positions (131 to 146 bp downstream of the translation start codon ATG). During our complementation tests, we also used GaMYB2 cDNA (GaMYB2c). Surprisingly, neither GL1∷GaMYB2c nor 35S∷GaMYB2c rescued trichome formation of the gl1 mutant. In the wild-type background, the two constructs did not exhibit any effect on trichome production. We then extended this test to Arabidopsis GL1 and WER. Under the control of either GL1 sequences or the 35S promoter, the intronless cDNAs did not work (Figure 4B, Table 1). This intron requirement is therefore common to these trichome-related MYB genes, rather than specific to GaMYB2.
When the trichome-specific promoter RDL1-P3 was used to drive GaMYB2, both the intron 1–containing genomic coding region and the cDNA functioned in regulating trichome development in plants of T1 generation (Figure 4C). Similar results were obtained with GL1 (Table 1). In a previous study, ATML1∷GL1c also rescued the gl1 mutant defects (Esch et al., 2003). The ATML1 promoter is expressed at a low level in all the epidermal cells of the developing leaf, and in situ analyses have shown its enhanced expression in young developing trichomes (Sessions et al., 1999). Therefore, cDNAs of the MYB genes could work normally, at least in T1, if expressed properly. These results imply that the intron is involved in regulating the gene expression pattern.
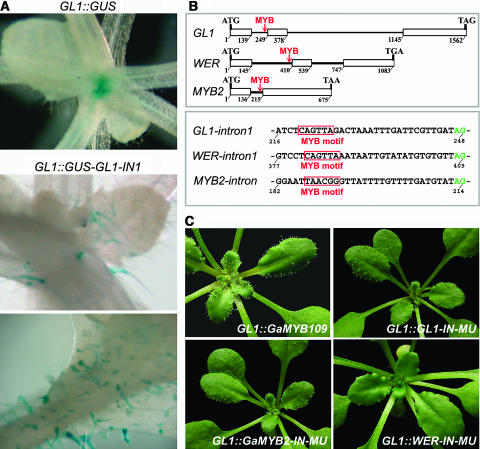
To confirm this assumption, we constructed two types of chimerical GL1∷GUS reporter genes containing different introns in the coding region: one contained the GUS intron in pCAMBIA1301, and the other employed instead the GaMYB2 intron or GL1 intron 1. We found that, when the MYB intron was present, GUS activity was restricted to trichomes and the signal was clear. In plants expressing the original GUS gene, the GUS staining pattern was far from specific (Figure 5A).
Figure 5.
Effects of Intron 1 and the Intronic MYB Motif on Expression of Trichome-Related Genes.
(A) GUS staining of Arabidopsis seedlings expressing GL1∷GUS containing the GUS intron (top) and the first intron of GL1 (middle and bottom).
(B) Intron positions in GL1, WER, and GaMYB2 genes and the MYB motif in the first intron of each gene. MYB2, GaMYB2.
(C) Reduced production of trichomes in gl1 plants expressing GL1∷GaMYB109, whose intron contains no MYB motif, or the MYB genes with disrupted intronic MYB motif. See also Table 1.
Identification of an Intronic MYB Motif
PLACE analysis identified a MYB binding motif (CA/CGTTA) that is present in the GaMYB2 intron and in the first intron of GL1 and WER as well. Furthermore, position of this motif is conserved (22 or 23 bp upstream of the second exon) (Figure 5B). This intronic MYB motif is not found in MYB109 of G. arboreum. We then mutated this MYB motif into CA/CacTA. We found that the activity of promoting trichome formation in gl1 plants was considerably weakened after mutation, although the degree was different depending on the individual genes (Figure 5C, Table 1). For GL1∷GL1-IN-MU, though the complementation rate was still high, >40% of transgenic plants showed abnormal trichome production: the amount of leaf trichomes was decreased and trichomes were mainly distributed on the leaf margin and stem. For GL1∷GaMYB2-IN-MU, this mutation cost >50% of the transgenic plants without complementation. And for GL1∷WER-IN-MU, most of the trichome-regulating activity was lost. When GL1∷GaMYB109 was introduced into gl1 plants, only a partial complementation was achieved (Figure 5C, Table 1), consistent with the fact that the intronic MYB motif is not present. These results indicate that this intronic MYB motif of GL1-related MYB genes plays a role in regulating trichome production.
Expression Patterns of GaMYB2
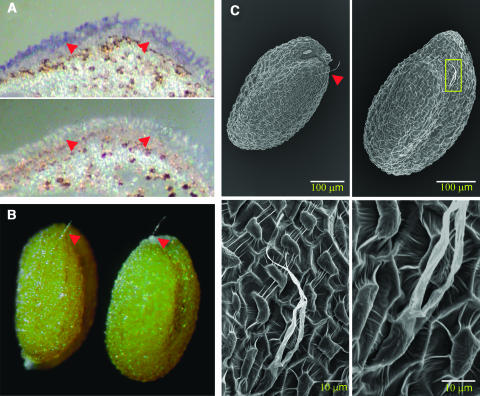
In Arabidopsis, the cotton GaMYB2 gene showed a high activity of controlling trichome formation. If this MYB factor, which is encoded by a single gene in G. arboreum (see Supplemental Figure 1 online), controls cotton fiber development, it should have a higher level of expression in fiber cells at an early stage of ovule development. Real-time PCR showed that GaMYB2 transcripts were indeed highly enriched in developing fiber cells, and the expression level was either rather low or undetectable in all other tissues examined (Figure 1A). A similar expression pattern was observed for the HOX3 and RDL1 genes; however, at the very early stage of ovule development (0 DAP), only GaMYB2 exhibited a substantial level of expression (Figure 1A). In situ mRNA hybridization of 0-DPA ovules localized the GaMYB2 transcripts in fiber initials (Figure 6A), confirming its fiber-specific expression in ovules.
Figure 6.
In Situ Hybridization of Cotton Ovule and the Arabidopsis Seed Trichome.
(A) In situ hybridization of GaMYB2 transcripts in the G. arboreum ovule (0 DPA) with antisense (top) and sense (middle) probes. Arrowheads indicate fiber cells; the mRNA was localized in fiber cells.
(B) Trichome(s) on seed of wild-type Arabidopsis expressing 35S∷GaMYB2 observed under binoculars. Arrowheads indicate the seed-trichome.
(C) Scanning electron micrographs depicting the trichome on seed of transgenic Arabidopsis expressing 35S∷GaMYB2. The box indicates the magnified region shown in the bottom panels. Arrowhead indicates the seed-trichome.
Promoting Arabidopsis Seed-Trichome Production by GaMYB2
Seeds of Arabidopsis are completely glabrous. Our extensive observation of a large number of samples under binoculars did not find any trichomes on seed coat of wild-type plants. We then examined seeds of transgenic 35S∷GaMYB2 plants. Interestingly, the transgene promoted seed-trichome formation in both wild-type and gl1 backgrounds. In the wild-type background, we found that ∼20% (11/54) of T1 plants showed seed-trichome phenotype, among these plants ∼20% (82/416) of the seeds produced epidermal trichomes. The trichomes were often unbranched or occasionally branched, 0.1 to 0.2 mm long (Figures 6B and 6C), and the number of trichomes on each seed was small: frequently one and at most four. Other aspects of seed development appeared normal. In plants transformed with 35S∷GL1, 35S∷WER, and 35S∷GaMYB109, seed trichomes were not observed.
DISCUSSION
Dissection of the cotton RDL1 promoter indicated that both L1 box and MYB motif contribute to its activity of trichome-specific expression in Arabidopsis, and both elements confer a positive regulation. It is interesting that the 302-bp DNA fragment of RDL1-P3 is already sufficient to drive trichome-specific expression efficiently. Previously, a 334-bp promoter of a tobacco (Nicotiana tabacum) P450 gene (CYP71D16) was shown to drive gene expression in distal cells of multicellular trichomes (Wang et al., 2002). These short fragments, particularly the two cis-elements identified here, can be used to design artificial promoters for engineering trichome-based molecular farming, as well as fiber trait improvement.
GL1 is the first transcription factor cloned that controls Arabidopsis trichome development (Herman and Marks, 1989; Marks and Feldmann, 1989). The prevalent model is that GL1, GL3, and probably TTG1 form a transcription complex that regulates expression of GL2, and this homeobox protein regulates downstream target genes to control epidermal cell differentiation (Szymanski et al., 1998; Ohashi et al., 2003). Besides GL2, no other target genes have been identified for GL1, although they were suspected to exist (Larkin et al., 2003). We demonstrate in this paper that GL1-like MYB proteins (GL1, WER, and GaMYB2) are able to bind to the MYB motif of the cotton RDL1 promoter, and both the MYB and HOX proteins are involved in transcriptional regulation of this promoter. Considering that the L1 box and MYB motif are located close to each other in RDL1-P3, the MYB and HOX proteins may form a complex that transactivates this promoter.
This type of trichome-gene regulation is not an isolated case, for the cotton LTP3 gene, which encodes a putative lipid transfer protein, also has L1 box and MYB motifs in its upstream sequence; in transgenic tobacco plants, the LTP3 promoter directed trichome-specific expression of GUS as well (Liu et al., 2000). In cotton ovules, transcripts of both RDL1 and LTP3 genes are distributed mainly in developing fiber cells. It is thus highly possible that the two elements are recruited by cotton transcription factors to confer fiber cell expression of a group of cotton genes. Because both RDL1 and LTP3 are cotton genes, we searched the Arabidopsis genome database to find endogenous genes that have a similar promoter. We then identified several candidates, including a putative membrane bound O-acyl transferase gene (At5g55330). Promoter of this gene indeed directs trichome-specific expression, and its expression requires GL1 (our unpublished results). Based on these findings, we propose that similar molecular machinery is shared by cotton fibers and Arabidopsis trichomes in regulating a subset of downstream target genes, which involves both the GL1-like MYB and GL2-like HOX transcription factors.
Because function of a cell fate–determining gene is closely related to its expression pattern, regulatory elements are of central importance. Previous investigations of GL1 expression provided a vague picture. A promoter from the GL1 5′ noncoding region directed GUS expression in stipules only (Oppenheimer et al., 1991), whereas in situ hybridization showed a predominant distribution of the transcripts in developing trichomes and protoderm cells; the downstream sequence was then suggested to be required for normal expression (Larkin et al., 1993). In this study, we show that the trichome-specific expression can be achieved as long as intron 1 is present. This intron can be considered a repressor in nontrichome cells because it prevented ectopic expression of the MYB genes in nontrichome cells. On the other hand, despite the restricted expression domain because of the presence of this intron, the overall transcript level still increased (data not shown), and the GUS signal in trichomes was comparatively strong (Figure 5A); these data suggest that the intron also acts as an enhancer in trichome cells. This cell-specific bidirectional regulation mediated by the intron identifies with the lateral inhibition model proposed for trichome formation.
That a MYB binding motif is found in the intron (intron 1) of GL1, WER, and GaMYB2 is unlikely a coincidence because this motif is required for normal trichome production. However, unlike deletion of the intron, disruption of this motif reduced, rather than eliminated, the trichome-regulating potential (Table 1). This suggests that the intron may contain other cis-elements. Alternatively, intron removal by a spliceosome may play an additional role.
35S∷GL1 but not 35S∷GL1c rescued the trichome formation of the gl1 mutant. Elaboration of trichomes by 35S∷GL1 or related transgenes is likely owing to the intron (intron 1) because the 35S promoter is active in practically all tissues. However, unlike GL1∷GL1, 35S∷GL1 could not maintain a normal trichome production in plants of T2 and subsequent generations. This indicates that the 5′ and 3′ noncoding sequences of GL1 are also involved in patterning trichome formation. Further dissection is required to clarify the regulatory role for each of these DNA segments.
Most eukaryotic genes are interrupted by one or more introns, which can influence many aspects of gene expression. In plants, a well-characterized example is Arabidopsis AGAMOUS (AG), a MADS box gene required for the development of the internal two whorls (stamens and carpels) of flower. Proper expression of AG involves sequences located in the second intron (Sieburth and Meyerowitz, 1997; Deyholos and Sieburth, 2000; Lohmann et al., 2001; Hong et al., 2003). Besides intronic cis-acting elements, presence of the first intron may enhance plant gene expression (Callis et al., 1987; Fu et al., 1995; Cai et al., 1998). It is now clear that proper expression of GL1 also requires the first intron. This intron-involved regulation of trichome formation is likely conserved in flowering plants because Arabidopsis and cotton represent two distantly related families.
Arabidopsis trichomes and cotton fibers are both unicellular hairs of epidermis, and they may share similar molecular machinery of regulation. Since the cloning of the Arabidopsis GL1 gene, scientists have been searching for MYB transcription factors that control cotton fiber initiation and development. GhMYB1 shows certain homology to MIXTA; when expressed in tobacco under the 35S promoter, it caused phenotypic changes that were global rather than specific, though supernumerary trichomes were observed. In Arabidopsis, however, its expression did not alter trichome production (Payne et al., 1999); and in our yeast one-hybrid assay, GaMYB1 failed to bind to the RDL1 promoter (Figure 3A). The cotton MYB109 shows a relatively high homology to GL1. However, GL1∷MYB109 complemented gl1 mutation of Arabidopsis only partially (Table 1), and its transcripts were barely detectable in ovules at 0 DPA (Suo et al., 2003), when fiber initiation already started (Kim and Triplett, 2001). It is unlikely involved in early initiation of cotton fibers.
Three lines of evidence presented here suggest the cotton MYB2 to be a key regulator of cotton fiber development. First, it showed a clear binding activity to the cotton fiber-specific promoter RDL1 in yeast and, together with cotton HOX3, it regulated RDL1∷GUS expression in Arabidopsis plants. Second, it is highly expressed in fiber cells at an early developmental stage, preceding the expression of other fiber genes, such as RDL1, HOX3 (Figure 1A), and MYB109 (Suo et al., 2003). Clearly, the spatial and temporal expression pattern of GaMYB2 fits its proposed role of controlling fiber cell formation. Third, GaMYB2 protein not only rescued trichome formation of an Arabidopsis gl1 mutant but also promoted the model plant to produce seed trichomes. Besides influencing trichome formation, GaMYB2 did not exhibit other observable effects on Arabidopsis development. Currently, we are transferring sense and antisense constructs of GaMYB2 into cotton plants to obtain more direct evidence of its role in regulating cotton fiber development.
It has been indicated that similar MYBs may play distinct roles depending on cell types in which they are expressed (Glover et al., 1998; Lee and Schiefelbein, 2001). It is therefore not totally unexpected that 35S∷GaMYB2 triggered seed-trichome production in Arabidopsis because seed coat should be included in the expression domain of the 35S promoter. Previous investigations reported that Arabidopsis plants expressing 35S∷GL1 produced one to several ectopic trichomes on the cotyledon, but trichomes on seed were not mentioned (Larkin et al., 1994). In this investigation, we did not find any seed trichomes in plants expressing either 35S∷GL1 or 35S∷WER. Thus, the possibility exists that a specific structure of cotton MYB2 is related to its activity of promoting seed–trichome production. Although leaf trichomes are commonly found in flowering plants, seed-trichome formation is a sporadic event of evolution that happened in several unrelated plant families, including Malvaceae, Salicaceae, and Bombacaceae. Dissection of functional domains of cotton MYB2 and related MYB proteins holds a potential of probing into the mystery of seed-trichome evolution.
METHODS
Plant Material
Cotton plants (Gossypium arboreum cv Qingyangxiaozi and G. hirsutum cv Xuzhou-142) were grown in a greenhouse. Ovules were harvested at various developing stages; fibers were collected by scraping the ovule in liquid nitrogen. Plants of Arabidopsis thaliana (Columbia-0) were grown indoors at 22°C under continuous illumination. A T-DNA insertion mutant of gl1 (SALK_039478) was obtained from ABRC (Ohio State University, Columbus, OH). Arabidopsis tissues above the hypocotyl of the 2-week-old seedlings (four pairs of visible leaves) were collected for analysis, unless otherwise indicated.
DNA and RNA Analysis
Genomic DNA and total RNA were extracted as described (Zhu et al., 2003). For DNA gel blot analysis, hybridization and washing were performed as described (Sambrook et al., 2001). Probes were generated using the Prime-a-Gene Labeling System (Promega, Madison, WI).
Genomic walking was performed on G. arboreum DNA to isolate RDL1 5′ upstream segment according to the Genome Walker kits (Clontech, Palo Alto, CA). To obtain the 5′-terminal sequence of G. arboreum RDL1 transcript, 5′-RACE was performed using the SMART RACE cDNA amplification kit (Clontech).
The cDNAs used in this investigation were amplified by RT-PCR from the total RNAs, which were isolated from G. arboreum ovules and fiber cells, and from A. thaliana seedlings, respectively. GhHOX3 was isolated from a G. hirsutum cDNA library (Zhu et al., 2003). The genes or gene fragments were PCR amplified from the genomic DNA. Pyrobest DNA polymerase (TaKaRa, Dalian, China) was used for the PCR amplification.
The mRNA was quantitatively analyzed by real-time PCR, with gene-specific TagMan primers designed using Primer Express (Applied Biosystems, Foster City, CA), and the primers were given in Supplemental Table 1 online. The 5′ and 3′ ends of these probes were labeled with fluorescent dyes FAM (6-carboxyfluorescein) and TAMRA (6-carboxy-tetramethyl-rhodamine), respectively. Total RNA of 1 μg from various tissues of G. arboretum was reverse transcribed in a 20-μL reaction using the RNA PCR (AMV) kit (TaKaRa). TaqMan RT-PCR assays were performed on a Rotor-Gene 3000 (Corbett Research, Mortlake, Australia). In a final volume of 25 μL, the reaction contained 25 ng of reverse transcription mixture, 0.2 μM forward and reverse primers, and 0.1 μM probe. All reactions were performed in triplicate. Cotton Histone3 (AF024716) mRNA was used as an internal control, and the relative amounts of the mRNA were calculated using the comparative threshold cycle method. Amplification efficiency (82 to 90%) for the four primer-probe sets was determined by amplification of cDNA reverse transcribed from a dilution series of total RNA using 1, 0.5, 0.25, and 0.125 μg per reaction (data not shown).
DNA Mutations and Linker Scanning
Deletion derivatives were PCR constructed using sets of primers (see Supplemental Table 2 online). Linker scanning and point mutations were performed as described (Gustin and Burk, 2000). After PCR amplification, the DNA fragments were inserted into the HindIII-BamHI sites of pBI121 vector (Clontech) to replace the 35S promoter region. Presence of the expected mutations was confirmed by sequencing.
Yeast One-Hybrid Assay
Yeast one-hybrid assay was performed using the MATCHMAKER one-hybrid system (Clontech). An RDL1 promoter region (−221 to −33 bp), RDL1-OH, was ligated into the EcoRI-XbaI sites of pHISi-1 and HindIII-SalI sites of pLacZi, respectively. These two bait constructs were linearized with XhoI and NcoI, respectively, and integrated into the yeast genome (strain YM4271). The level of background HIS was determined by culturing the yeast strain on the synthetic dropout (SD)/-His medium containing various concentrations of 3-amino-1, 2, 4-triazole. A yeast dual reporter strain was prepared by first integrating pLacZi-RDL1, followed by pHISi-1-RDL1, into the genome of yeast. Similarly, yeast strain YM4271 integrating RDL1-OH-MYB-MU was formed and tested. The dual reporter strain was selected and maintained on SD/-His/-Ura medium. For construction of the GAD-MYB or GAD-HOX fusions, the cDNA (open reading frame) was ligated with GAL4 activation domain in pGAD424. Yeast transformants were tested on SD/-Leu/-His/-Ura medium containing 5 mM 3-amino-1, 2, 4-triazole.
Constructs
RDL1-P3, a 302-bp fragment of RDL1 promoter, was used to substitute the 35S promoter region in the HindIII-NcoI sites of pCAMBIA1301 (CAMBIA, Canberra, Australia), forming pRDL1∷GUS. The 35S promoter and NOS terminator were isolated from pBI121 by PCR and inserted into the EcoRI-SacI and the SphI-HindIII sites of pRDL1∷GUS, respectively. Coding regions of MYB and HOX genes were isolated by PCR with gene-specific primers (see Supplemental Table 2 online) and placed between the 35S promoter and NOS terminator, generating p35S∷MYB/RDL1∷GUS and p35S∷HOX/RDL1∷GUS, respectively. The 35S-GL1-NOS cassette was inserted into the EcoRI site of p35S∷GL2/RDL1∷GUS to generate p35S∷GL1/35S∷GL2/RDL1∷GUS. Similarly, the construct of p35S∷GaMYB2/35S∷HOX3/RDL1∷GUS was formed.
The NOS terminator was inserted into the HindIII and SphI sites of pCAMBIA1301, forming p1301-NOS. A 1400-bp upstream fragment and an 1838-bp downstream fragment of Arabidopsis GL1 gene were amplified by PCR. After sequencing, they were inserted into the SphI-PstI and SalI-XbaI sites of p1301-NOS, respectively, forming pGL1-1. Coding regions of the MYB genes and their cDNAs were inserted into the PstI-SalI sites between the 5′-upstream and 3′-downstream fragments of GL1, generating pGL1∷GaMYB2, pGL1∷GaMYB2c, and related constructs of other MYB genes.
RDL1-P3 was inserted into the HindIII-PstI sites of pCAMBIA1301 to generate p1301-RDL1. Then, NOS terminator was inserted into the HindIII and SacI-EcoRI sites of p1301-RDL1 to generate p1301-RDL1-NOS. The cDNAs or genomic coding regions of MYB genes were inserted into the PstI-SalI sites between the RDL1-P3 and NOS of p1301-RDL1-NOS, generating pRDL1∷GaMYB2 and related constructs.
The pGL1-1 was digested with BstEII to remove GUS, and part of GL1 upstream fragment was also removed because it contained a BstEII site. This GL1 upstream fragment was then obtained by PCR and inserted back into BstEII, forming pGL1-2. The coding region of the GUS gene was isolated from pCAMBIA1301 by PCR and inserted into the Pst1 and SalI sites of pGL1-2. The intron of GUS gene was replaced by GL1 intron 1 or GaMYB2 intron, and modified GUS coding regions were inserted into the Pst1 and SalI sites of pGL1-2, generating pGL1∷GUS-GL1-IN1 and pGL1∷GUS-GaMYB2-IN, respectively.
Plant Transformation and GUS Assay
Binary constructs were introduced into Agrobacterium tumefaciens strain GV3101 and subsequently transferred into Arabidopsis using a floral dip method (Clough and Bent, 1998). Transgenic plants were selected on media containing 50 μg/mL of kanamycin or hygromycin.
Histochemical and quantitative GUS assays were performed as described (Jefferson et al., 1987). Samples were stored in 70% ethanol before microscopic observation. For quantitative assays, fluorescence was measured in a minifluorometer (Hofer, San Francisco, CA).
In Situ Hybridization and Scanning Electron Microscopy
G. arboreum ovules were collected at 0 DPA and embedded in paraffin. GaMYB2 mRNA was detected with a digoxigenin-labeled riboprobe using the method as described (Long and Barton, 2000; http://www.wisc.edu/genetics/CATG/barton/protocols.html). PCR was conducted on GaMYB2 cDNA to generate a template for the probe, which was complementary to the 3′ region of GaMYB2 transcript downstream of the conserved MYB domain.
The seed trichome of transgenic Arabidopsis plants expressing 35S∷GaMYB2 was examined under a JSM-6360LV scanning electron microscope (JEOL, Tokyo, Japan).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY641990, AY626160, AY633559, and AY626159 for the GaRDL1, GaMYB2 (FIF1), GaMYB109, and GhHOX3 cDNA clones, respectively. AY633558 is the accession number for the RDL1-P3 promoter.
Supplementary Material
Acknowledgments
We thank Thea Wilkins, Yongbiao Xue, Jeffrey Chen, and Hong Ma for their help and discussions. This research was supported by the Ministry of Science and Technology of China (2002CB111301, 2001AA222121, and JY03-A-02) and by the National Natural Science Foundation of China (30270846).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Xiao-Ya Chen (xychen@sibs.ac.cn).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.024844.
References
- Abe, M., Takahashi, T., and Komeda, Y. (2001). Identification of a cis-regulatory element for L1 layer-specific gene expression, which is targeted by an L1-specific homeodomain protein. Plant J. 26, 487–494. [DOI] [PubMed] [Google Scholar]
- Cai, X.L., Wang, Z.Y., Xing, Y.Y., Zhang, J.L., and Hong, M.M. (1998). Aberrant splicing of intron 1 leads to the heterogeneous 5′ UTR and decreased expression of waxy gene in rice cultivars of intermediate amylose content. Plant J. 14, 459–465. [DOI] [PubMed] [Google Scholar]
- Callis, J., Fromm, M., and Walbot, V. (1987). Introns increase gene expression in cultured maize cells. Genes Dev. 1, 1183–1200. [DOI] [PubMed] [Google Scholar]
- Cedroni, M.L., Cronn, R.C., Adams, K.L., Wilkins, T.A., and Wendel, J.F. (2003). Evolution and expression of MYB genes in diploid and polyploid cotton. Plant Mol. Biol. 51, 313–325. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Deyholos, M.K., and Sieburth, L.E. (2000). Separable whorl-specific expression and negative regulation by enhancer elements within the AGAMOUS second intron. Plant Cell 12, 1799–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch, J.J., Chen, M., Sanders, M., Hillestad, M., Ndkium, S., Idelkope, B., Neizer, J., and Marks, M.D. (2003). A contradictory GLABRA3 allele helps define gene interactions controlling trichome development in Arabidopsis. Development 130, 5885–5894. [DOI] [PubMed] [Google Scholar]
- Fu, H., Kim, S.Y., and Park, W.D. (1995). A potato Sus3 sucrose synthase gene contains a context-dependent 3′ element and a leader intron with both positive and negative tissue-specific effects. Plant Cell 7, 1395–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover, B.J., Perez-Rodriguez, M., and Martin, C. (1998). Development of several epidermal cell types can be specified by the same MYB-related plant transcription factor. Development 125, 3497–3508. [DOI] [PubMed] [Google Scholar]
- Gustin, K., and Burk, R.D. (2000). PCR-directed linker scanning mutagenesis. Methods Mol. Biol. 130, 85–90. [DOI] [PubMed] [Google Scholar]
- Herman, P.L., and Marks, M.D. (1989). Trichome development in Arabidopsis thaliana. II. Isolation and complementation of the GLABROUS1 gene. Plant Cell 1, 1051–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, R.L., Hamaguchi, L., Busch, M.A., and Weigel, D. (2003). Regulatory elements of the floral homeotic gene AGAMOUS identified by phylogenetic footprinting and shadowing. Plant Cell 15, 1296–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, S.J., Lu, Y.C., Feng, J.X., Wei, G., Li, J., Shi, Y.H., Fu, Q., Liu, D., Luo, J.C., and Zhu, Y.X. (2003). Isolation and analyses of genes preferentially expressed during early cotton fiber development by subtractive PCR and cDNA array. Nucleic Acids Res. 31, 2534–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John, M.E., and Crow, L.J. (1992). Gene expression in cotton (Gossypium hirsutum L.) fiber: Cloning of the mRNAs. Proc. Natl. Acad. Sci. USA 89, 5769–5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.J., and Triplett, B.A. (2001). Cotton fiber growth in planta and in vitro. Models for plant cell elongation and cell wall biogenesis. Plant Physiol. 127, 1361–1366. [PMC free article] [PubMed] [Google Scholar]
- Kirik, V., Simon, M., Hülskamp, M., and Schiefelbein, J. (2004). The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev. Biol. 268, 506–513. [DOI] [PubMed] [Google Scholar]
- Larkin, J.C., Brown, M.L., and Schiefelbein, J. (2003). How do cells know what they want to be when they grow up? Lessons from epidermal patterning in Arabidopsis. Annu. Rev. Plant Biol. 54, 403–430. [DOI] [PubMed] [Google Scholar]
- Larkin, J.C., Oppenheimer, D.G., Lloyd, A., Paparozzi, E.T., and Marks, M.D. (1994). The roles of GLABROUS1 and TRANSPARENT TESTA GLABRA genes in Arabidopsis trichome development. Plant Cell 6, 1065–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, J.C., Oppenheimer, D.G., Pollock, S., and Marks, M.D. (1993). Arabidopsis GLABROUS1 gene requires downstream sequences for function. Plant Cell 5, 1739–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M.M., and Schiefelbein, J. (1999). WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99, 473–483. [DOI] [PubMed] [Google Scholar]
- Lee, M.M., and Schiefelbein, J. (2001). Developmentally distinct MYB genes encode functionally equivalent proteins in Arabidopsis. Development 128, 1539–1546. [DOI] [PubMed] [Google Scholar]
- Li, C.H., Zhu, Y.Q., Meng, Y.L., Wang, J.W., Xu, K.X., Zhang, T.Z., and Chen, X.Y. (2002). Isolation of genes preferentially expressed in cotton fibers by cDNA filter arrays and RT-PCR. Plant Sci. 163, 1113–1120. [Google Scholar]
- Liu, H.C., Creech, R.G., Jenkins, J.N., and Ma, D.P. (2000). Cloning and promoter analysis of the cotton lipid transfer protein gene Ltp3. Biochim. Biophys. Acta 1487, 106–111. [PubMed] [Google Scholar]
- Loguerico, L.L., Zhang, J.Q., and Wilkins, T.A. (1999). Differential regulation of six novel MYB-domain genes defines two distinct expression patterns in allotetraploid cotton (Gossypium hirsutum L.). Mol. Gen. Genet. 261, 660–671. [DOI] [PubMed] [Google Scholar]
- Lohmann, J.U., Hong, R., Hobe, M., Busch, M.A., Parcy, F., Simon, R., and Weigel, D. (2001). A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105, 793–803. [DOI] [PubMed] [Google Scholar]
- Long, J., and Barton, M.K. (2000). Initiation of axillary and floral meristems in Arabidopsis. Dev. Biol. 218, 341–353. [DOI] [PubMed] [Google Scholar]
- Marks, M.D., and Feldmann, K.A. (1989). Trichome development in Arabidopsis thaliana. I. T-DNA tagging of the GLABROUS1 gene. Plant Cell 1, 1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda, K., Glover, B.J., Linstead, P., and Martin, C. (1994). Flower colour intensity depends on specialized cell shape controlled by a Myb-related transcription factor. Nature 369, 661–664. [DOI] [PubMed] [Google Scholar]
- Ohashi, Y., Oka, A., Rodrigues-Pousada, R., Possenti, M., Ruberti, I., Morelli, G., and Aoyama, T. (2003). Modulation of phospholipid signaling by GLABRA2 in root-hair pattern formation. Science 300, 1427–1430. [DOI] [PubMed] [Google Scholar]
- Oppenheimer, D.G., Herman, P.L., Sivakumaran, S., Esch, J., and Marks, M.D. (1991). A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67, 483–493. [DOI] [PubMed] [Google Scholar]
- Payne, C.T., Zhang, F., and Lloyd, A.M. (2000). GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156, 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne, T., Clement, J., Arnold, D., and Lloyd, A. (1999). Heterologous myb genes distinct from GL1 enhance trichome production when overexpressed in Nicotiana tabacum. Development 126, 671–682. [DOI] [PubMed] [Google Scholar]
- Pear, J.R., Kawagoe, Y., Schreckengost, W.E., Delmer, D.P., and Stalker, D.M. (1996). Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc. Natl. Acad. Sci. USA 93, 12637–12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerie, W.G., Feldmann, K.A., and Marks, M.D. (1994). The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 8, 1388–1399. [DOI] [PubMed] [Google Scholar]
- Rong, J., et al. (2004). A 3347-locus genetic recombination map of sequence-tagged sites reveals features of genome organization, transmission and evolution of cotton (Gossypium). Genetics 166, 389–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan, Y.L., Llewellyn, D.J., and Furbank, R.T. (2003). Suppression of sucrose synthase gene expression represses cotton fiber cell initiation, elongation, and seed development. Plant Cell 15, 952–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (2001). Molecular cloning: A Laboratory Manual, 3rd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schellmann, S., Schnittger, A., Kirik, V., Wada, T., Okada, K., Beermann, A., Thumfahrt, J., Jurgens, G., and Hülskamp, M. (2002). TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 21, 5036–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, K., Wells, B., Dolan, L., and Roberts, K. (1997). Structural and genetic analysis of epidermal cell differentiation in Arabidopsis primary roots. Development 124, 1789–1798. [DOI] [PubMed] [Google Scholar]
- Schnittger, A., Folkers, U., Schwab, B., Jürgens, G., and Hülskamp, M. (1999). Generation of a spacing pattern: The role of TRIPTYCHON in trichome patterning in Arabidopsis. Plant Cell 11, 1105–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions, A., Weigel, D., and Yanofsky, M.F. (1999). The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 20, 259–263. [DOI] [PubMed] [Google Scholar]
- Sieburth, L.E., and Meyerowitz, E.M. (1997). Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9, 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo, J.F., Liang, X.E., Pu, L., Zhang, Y.S., and Xue, Y.B. (2003). Identification of GhMYB109 encoding a R2R3 MYB transcription factor that expressed specifically in fiber initials and elongating fibers of cotton (Gossypium hirsutum L.). Biochim. Biophys. Acta 1630, 25–34. [DOI] [PubMed] [Google Scholar]
- Szymanski, D.B., Jilk, R.A., Pollock, S.M., and Marks, M.D. (1998). Control of GL2 expression in Arabidopsis leaves and trichomes. Development 125, 1161–1171. [DOI] [PubMed] [Google Scholar]
- Urao, T., Yamaguchi-Shinozaki, K., Urao, S., and Shinozaki, K. (1993). An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5, 1529–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada, T., Kurata, T., Tominaga, R., Koshino-Kimura, Y., Tachibana, T., Goto, K., Marks, M.D., Shimura, Y., and Okada, K. (2002). Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development 129, 5409–5419. [DOI] [PubMed] [Google Scholar]
- Wada, T., Tachibana, T., Shimura, Y., and Okada, K. (1997). Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277, 1113–1116. [DOI] [PubMed] [Google Scholar]
- Walker, A.R., Davison, P.A., Bolognesi-Winfield, A.C., James, C.M., Srinivasan, N., Blundell, T.L., Esch, J.J., Marks, M.D., and Gray, J.C. (1999). The TTG1 (transparent testa, glabra1) locus which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis encodes a WD40-repeat protein. Plant Cell 11, 1337–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, E., Gan, S., and Wagner, G.J. (2002). Isolation and characterization of the CYP71D16 trichome-specific promoter from Nicotiana tabacum L. J. Exp. Bot. 53, 1891–1897. [DOI] [PubMed] [Google Scholar]
- Wilkins, T.A., Rajasekaran, K., and Anderson, D.M. (2000). Cotton biotechnology. Crit. Rev. Plant Sci. 19, 511–550. [Google Scholar]
- Zhu, Y.Q., Xu, K.X., Luo, B., Wang, J.W., and Chen, X.Y. (2003). An ATP-binding cassette transporter GhWBC1 from elongating cotton fibers. Plant Physiol. 133, 580–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.