Abstract
The ARP2/3 complex is an important regulator of actin nucleation and branching in eukaryotic organisms. All seven subunits of the ARP2/3 complex have been identified in Arabidopsis thaliana, and mutation of at least three of the subunits results in defects in epidermal cell expansion, including distorted trichomes. However, the mechanisms regulating the activity of the ARP2/3 complex in plants are largely unknown. In mammalian cells, WAVE and WASP proteins are involved in activation of the ARP2/3 complex. WAVE1 activity is regulated by a protein complex containing NAP1/HEM/KETTE/GEX-3 and PIR121/Sra-1/CYFIP/GEX-2. Here, we show that the WAVE1 regulatory protein complex is partly conserved in plants. We have identified Arabidopsis genes encoding homologs of NAP1 (NAPP), PIR121 (PIRP), and HSPC300 (BRK1). T-DNA inactivation of NAPP and PIRP results in distorted trichomes, similar to ARP2/3 complex mutants. The napp-1 mutant is allelic to the distorted mutant gnarled. The actin cytoskeleton in napp-1 and pirp-1 mutants shows orientation defects and increased bundling compared with wild-type plants. The results presented show that activity of the ARP2/3 complex in plants is regulated through an evolutionarily conserved mechanism.
INTRODUCTION
Polarized cell growth in plant cells is believed to take place through either diffuse polar growth or tip growth. Diffuse growth is generally anisotropic, expansion being driven by turgor pressure and occurring across the entire cell surface. The polarity of diffuse growth is considered to be determined by the arrangement of cellulose microfibrils deposited during expansion, restricting polar growth to the perpendicular axis of the cellulose microfibrils. Tip growth is a unidirectional process in which secretory vesicles are directed to the apical growth site, delivering plasma membrane and cell wall material required for cell extension (Martin et al., 2001; Mathur and Hülskamp, 2002; Smith, 2003). Microtubules and actin microfilaments are central players in both diffuse growth and tip growth in plant cells. In the current models, microtubules participate in determination of cell polarity, whereas actin filaments direct secretory vesicles to the growth site (Mathur and Hülskamp, 2002).
Cell structure, morphogenesis, and movement in eukaryotic cells are dependent on a dynamic actin cytoskeleton. The balance between polymerization and depolymerization of actin filaments is governed by a diverse array of actin binding proteins, of which many are conserved in all or most eukaryotes (Vantard and Blanchoin, 2002). A very well conserved activator of actin polymerization is the ARP2/3 complex, first identified in the primitive eukaryote Acanthamoeba castellanii (Machesky et al., 1994). The complex consists of seven subunits: two proteins with similarity to conventional actins (ARP2 and ARP3) and five unique polypeptides (ARPC1-5) (Machesky and Gould, 1999). Upon activation, the ARP2/3 complex binds to a preexisting actin filament and nucleates a new filament at an angle of 70° on the side of the parental filament (Amann and Pollard, 2001). As a result, a branched network of actin filaments is created.
In mammalian cells, the ARP2/3 complex is activated by two distinct protein families: the Wiskott-Aldrich syndrome protein (WASP) family and the cortactin family (Weaver et al., 2003). WASP family proteins activate the ARP2/3 complex through their C-terminal VCA domains, consisting of one or two verprolin homology (V) regions, a hydrophobic central (C) region, and an acidic (A) region. The V region binds actin monomers, whereas the C and A regions bind and activate the ARP2/3 complex (Marchand et al., 2001). The WASP family can be divided into two subfamilies. WASP and N-WASP are autoinhibited through the folding of the N-terminal part of the protein onto the VCA domain (Takenawa and Miki, 2001). These proteins also contain a Cdc42/Rac-interactive binding motif that is bound by activated Rho GTPases, mainly Cdc42. Upon binding of Cdc42, the autoinhibition is relieved and the VCA domain is free to interact with the ARP2/3 complex (Rohatgi et al., 1999). The members of the other subfamily, SCAR/WAVE1-3, do not contain a Cdc42/Rac-interactive binding motif and therefore do not interact directly with Rho GTPases. In addition, WAVE1-3 are not autoinhibited. Purification of WAVE1 from bovine brain indicates that the inactive form exists in a pentameric protein complex, consisting of WAVE1, PIR121, NAP1, HSPC300, and Abelson interactor2 (Abi2) (Eden et al., 2002). Active Rac1 binds and induces dissociation of the PIR121/NAP1/Abi2 subcomplex. Active WAVE1 and HSPC300 stay associated and activate the ARP2/3 complex. Homologs of all seven subunits of the ARP2/3 complex have been identified in Arabidopsis thaliana (Klahre and Chua, 1999; Le et al., 2003; Li et al., 2003; Mathur et al. 2003b). However, the mechanism by which the ARP2/3 complex is activated in plants is still unclear.
Arabidopsis trichomes offer an attractive model system for studying cell morphogenesis in plant cells (Hülskamp et al., 1994). Trichomes develop from precursor cells that usually undergo four rounds of endoreplication. The incipient trichome cell grows out of the leaf surface and initiates two successive branching events. Subsequently, the trichome cell begins rapid cell elongation concomitant with vacuolization. Studies of trichome mutants have shown that the first growth phase is dependent on microtubules and that the second is dependent on actin cytoskeleton (Hülskamp, 2000). A group of eight genes called the DISTORTED genes are believed to act during the second growth phase (Hülskamp et al., 1994). Studies of the distorted (dis) mutants dis1, dis2, gnarled (grl), klunker (klk), wurm (wrm), crooked (crk), and alien uncovered defects in organization of the actin cytoskeleton (Mathur et al., 1999; Szymanski et al., 1999; Schwab et al., 2003).
We report here a functional analysis of two Arabidopsis genes that are similar to components of the mammalian regulatory WAVE protein complex. NAPP and PIRP show a moderate but significant similarity to human NAP1 and PIR121, respectively. Transgenic plants carrying T-DNA knockouts of NAPP and PIRP have a distorted trichome phenotype, indicating that the gene products are involved in organization of the actin cytoskeleton. Indeed, the organization of microfilaments is disturbed in both NAPP and PIRP knockout plants. Complementation experiments and sequencing show that the napp-1 mutant is allelic to grl, one of the dis trichome mutants.
RESULTS
Identification of Potential Arabidopsis Homologs of the WAVE Regulatory Complex Subunits
The finding by Eden et al. (2002) that the mammalian WAVE1, when inactive, exists in a complex with four other proteins prompted us to search the Arabidopsis genome for potential homologs of the subunits of this protein complex. All the subunits of the WAVE1 regulatory protein complex are conserved in several metazoan organisms, such as man, mouse, Drosophila melanogaster, and Caenorhabditis elegans (Baumgartner et al., 1995; Schenck et al., 2001; Soto et al., 2002) as well as the mycetozoan Dictyostelium discoideum (Blagg et al., 2003).
Extensive database searches did not reveal any Arabidopsis gene products with apparent homology to Abi2. The three other components of the protein complex, HSPC300, Nap1, and PIR121, show significant similarity to single copy genes in the Arabidopsis genome. The smallest component of the complex, HSPC300, is very well conserved in Arabidopsis. The gene At2g22640 has two exons and encodes a protein with 34% identical and 57% similar amino acid residues as compared with HSPC300 (Figure 1A). The predicted amino acid sequence of At2g22640 is highly similar (79% identical and 88% similar amino acid residues) to the BRK1 gene product in maize (Zea mays) (Frank and Smith, 2002) and was therefore named BRK1. The two other subunits of the WAVE1 regulatory protein complex that are conserved in Arabidopsis, Nap/HEM/GEX-3 and PIR121/Sra-1/CYFIP/GEX-2, are both relatively large proteins, and no full-length cDNA sequence encoding Arabidopsis homologs of these genes has been published to date. We therefore used RT-PCR to amplify the transcripts of the putative Arabidopsis homologs of NAP and PIR.
Figure 1.
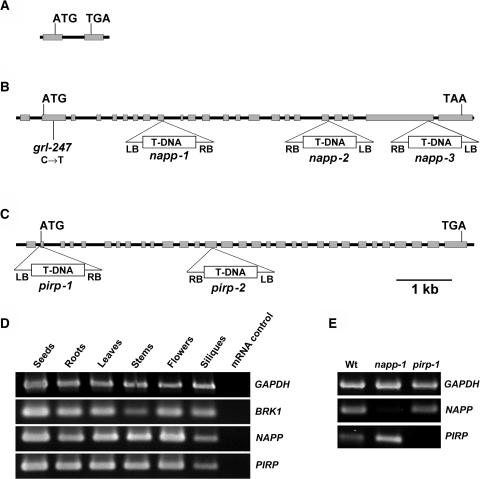
Molecular Organization and Expression Analysis of BRK1, NAPP, and PIRP and Characterization of napp and pirp Mutants.
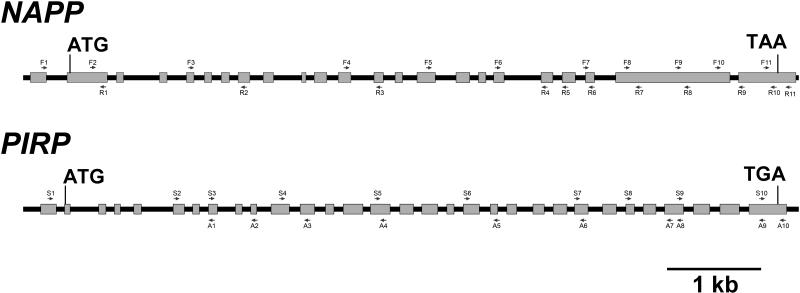
(A), (B), and (C) Gene structure and location of mutations in BRK1 (A), NAPP (B), and PIRP (C). Gray boxes represent exons, and black lines represent introns and untranscribed flanking sequences. The location and orientation of T-DNA insertions in NAPP and PIRP are shown. grl-247 is an ethyl methanesulfonate–generated allele.
(D) RT-PCR analysis of NAPP, PIRP, and BRK1 expression in various tissues. mRNA was extracted from different parts of Arabidopsis plants and used for RT-PCR analysis as described in the text. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a positive control. mRNA not subjected to reverse transcription was used as a negative control for each gene.
(E) RT-PCR analysis of NAPP and PIRP expression in wild-type napp-1 and pirp-1. mRNA was extracted from leaves of soil-grown wild-type plants and homozygous mutant plants and subjected to RT-PCR. GAPDH was used as a positive control.
Metazoan Nap/HEM proteins contain no domains of known function. The gene At2g35110, which we named NAPP (NAP of plants), consists of 23 exons. The 5′ untranslated region (UTR) contains an intron (Figure 1B). The NAPP gene encodes a protein with a predicted molecular mass of 156 kD, showing moderate similarity to metazoan Nap/HEM proteins. NAPP contains 15% identical and 32% similar amino acid residues compared with Nap1, which is the most similar of the two human Nap/HEM proteins (Table 1, Figure 2). Similarity is mainly found in the central part of NAPP, corresponding to the C-terminal 1000 amino acids of human Nap/HEM, C. elegans GEX-3, Drosophila DHEM/KETTE, and Dictyostelium Nap. NAPP also contains additional N- and C-terminal domains of ∼80 and 120 amino acids, respectively (Figure 2).
Table 1.
Comparison of NAPP, PIRP, and BRK1 with Their Homologs in Other Organisms
| Percentage of Amino Acid Identity (Similarity)
|
|||||
|---|---|---|---|---|---|
| Subunit | Arabidopsis Gene Accession No. (MATDB)a | Human | Drosophila | C. elegans | Dictyostelium |
| NAPP | At2g35110 | 15 (32)b | 16 (33) | 14 (31) | 14 (31) |
| PIRP | At5g18410 | 28 (49)c | 26 (47) | 25 (48) | 25 (46) |
| BRK1 | At2g22640 | 34 (57) | 34 (56) | 34 (55) | 33 (54) |
Munich Information Center for Protein Sequences Arabidopsis thaliana database.
Compared with human Nap1/HEM-2.
Compared with human PIR121/CYFIP2.
Figure 2.
Amino Acid Alignment of NAPP.
NAPP aligned with human Nap1 (HsNAP1) and Dictyostelium Nap (DdNAP). Amino acid residues in black indicate identity, and those in gray indicate conserved substitutions.
PIR121/Sra-1/CYFIP/GEX-2 is the largest subunit of the WAVE1 regulatory complex in animal cells. The Arabidopsis gene At5g18410, which we named PIRP (PIR of plants), consists of 31 exons (Figure 1C). Interestingly, PIRP also has a 5′-UTR intron. The PIRP gene product has a putative molecular mass of 146 kD and contains 28% identical and 49% similar amino acid residues compared with human PIR121, distributed along most of the amino acid sequence (Table 1, Figure 3). Similarity was also found in the N-terminal 420 amino acids of PIRP, which corresponds to the Rac-interacting region of Sra-1, the human isogene of PIR121 (Kobayashi et al., 1998). RT-PCR analyses showed that BRK1, NAPP, and PIRP are expressed in all tissues studied (Figure 1D).
Figure 3.
Amino Acid Alignment of PIRP.
PIRP aligned with human PIR121 (HsPIR121) and Dictyostelium PIR121 (DdPIR121). Amino acid residues in black indicate identity, and those in gray indicate conserved substitutions.
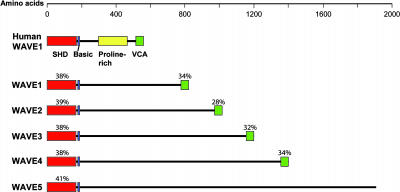
We next wanted to identify putative WAVE homologs in Arabidopsis. As shown in Figure 3, mammalian WAVE proteins contain an N-terminal Scar homology domain (SHD), followed by a short region of basic residues, a Pro-rich region, and a VCA domain at the C-terminal end (Takenawa and Miki, 2001). The Arabidopsis genome contains five genes encoding proteins with an N-terminal domain sharing similarity with the WAVE SHD domain (Figure 4). Four of these putative proteins also carry a C-terminal domain with similarity to the VCA domain. ESTs for all five genes are present in GenBank, indicating that all five genes are expressed. Thus, four of the five subunits constituting the WAVE regulatory complex in mammalian cells are conserved in Arabidopsis.
Figure 4.
Domain Structure of the Putative Arabidopsis WAVE Homologs.
Comparison of the domain structure of human WAVE1 and the putative Arabidopsis WAVE homologs. The SHD domain, the basic region, the Pro-rich region, and the VCA domain are shown. The percentage of conserved amino acid residues between human WAVE1 and the putative Arabidopsis WAVE homologs are indicated for the SHD and the VCA domains. Arabidopsis ID numbers are as follows: WAVE1 (At2g34150), WAVE2 (At1g29170), WAVE3 (At5g01730), WAVE4 (At2g38440), and WAVE5 (At4g18600).
Identification and Verification of NAPP and PIRP T-DNA Knockout Mutants
Having identified potential candidates for proteins involved in Rac-mediated regulation of the ARP2/3 complex in Arabidopsis, we next wanted to study the functional consequences of disrupting these genes. The SALK T-DNA insertion mutant collection (Alonso et al., 2003) contained one potential T-DNA knockout mutant for PIRP and several for NAPP but none for BRK1. The line SALK_014298 (subsequently called napp-1) contains a T-DNA insertion in the ninth exon of NAPP, leading to the truncation of NAPP after 338 amino acids (Figure 1B). SALK_038799 (napp-2) contains a T-DNA insertion in the splice donor site of intron 19, whereas T-DNA in SALK_009695 (napp-3) is inserted in exon 22. The line SALK_106757 (subsequently called pirp-1) has T-DNA inserted in the first intron of PIRP, 15 bp upstream of the second exon containing the start codon of PIRP (Figure 1C). The insertion occurs 10 bp downstream of a putative splice acceptor site and may therefore result in defective splicing of the first intron. The line EXM115 (pirp-2), which was obtained from the T-DNA insertion collection at INRA-Versailles (Bechtold et al., 1993), has a T-DNA insertion in exon 14 of PIRP.
To verify that the expression of NAPP and PIRP was disrupted in napp-1 and pirp-1, mRNA was isolated from the two mutants. RT-PCR showed that expression of NAPP was absent in napp-1, whereas PIRP expression was strongly reduced in pirp-1 (Figure 1E). Sensitive RT-PCR resulted in weak amplification of PIRP in pirp-1 (data not shown). Expression of NAPP in pirp-1 and of PIRP in napp-1 was at levels comparable to wild types.
Disruption of PIRP and NAPP Lead to a Distorted-Like Phenotype of Arabidopsis Trichomes
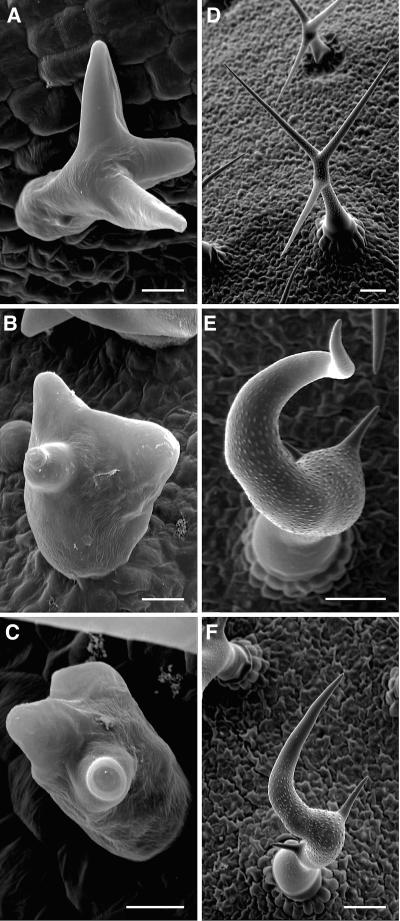
napp and pirp plants displayed similar phenotypes, both when grown on agar and in soil. When grown in soil, the general morphology of mutant plants did not differ much from wild-type plants (Figures 5A to 5C). However, trichome morphology in napp-1, napp-2, napp-3, pirp-1, and pirp-2 plants was notably different compared with wild-type trichomes (Figures 5 and 6). The phenotypes were first evident after trichome branch initiation, when branches started elongating. The stalk diameter of napp-1 and pirp-1 trichomes (Figures 6B and 6C) at this stage was notably increased, and branch elongation appeared reduced compared with wild-type trichomes (Figure 5C). Mature wild-type trichomes on Arabidopsis leaves have a very uniform shape, typically with three branches of similar length (Figures 5A and 6D), whereas stem trichomes are generally unbranched (Figure 5D). The shape of napp-1 and pirp-1 trichomes was highly variable (Figures 5B and 5C). Trichomes of napp-1 and pirp-1 plants had crooked branches and stunted branch length, particularly of secondary and tertiary branches. Stalk length of napp-1 and pirp-1 trichomes was reduced, and stalk diameter increased compared with wild-type trichomes (Figures 6E and 6F). Stem trichomes on napp-1 and pirp-1 plants (Figures 5E and 5F) were shorter and much less regularly shaped compared with wild-type stem trichomes (Figure 5D).
Figure 5.
napp-1 and pirp-1 Plants Exhibit Defects in Trichome Development.
(A) Trichomes on 2-week-old wild-type leaves have a uniform shape, generally with three branches.
(B) and (C) Trichomes on 2-week-old leaves of napp-1 (B) and pirp-1 (C) have highly variable shapes, whereas leaf shape is normal.
(D) Stem trichomes of wild-type plants are straight.
(E) and (F) Stem trichomes of napp-1 (E) and pirp-1 (F) plants are shorter and crooked.
Bars = 0.5 mm in (A), (B), and (C) and 1 mm in (D), (E), and (F).
Figure 6.
Scanning Electron Microscopy Analyses of Wild-Type, napp-1, and pirp-1 Trichomes.
(A) Developing wild-type trichome with elongating branches.
(B) and (C) Developing napp-1 (B) and pirp-1 (C) trichomes (stage 4); branch initiation is apparently normal, but branch elongation is delayed/reduced, and trichome stalk diameter is increased.
(D) Mature wild-type trichome with a thin stalk and branches of similar length.
(E) and (F) Mature napp-1 (E) and pirp-1 (F) trichomes with increased stalk diameter, irregular branch position, and highly reduced secondary and tertiary branch lengths.
Bars = 10 μm in (A), (B), and (C) and 50 μm in (D), (E), and (F).
Trichoblasts expand by tip growth, thereby forming the highly polarized mature root hairs (Carol and Dolan, 2002). Under normal growth conditions, root hair growth and morphology of napp and pirp plants was not affected. Changes in root hair growth and morphology in dis mutants has previously been observed when root hairs were challenged to elongate rapidly by growing the plants at a 20° downward angle from vertical position (Mathur et al., 2003a, 2003b). However, we were not able to notice consistent differences between wild-type and mutant root hairs under these growth conditions (data not shown).
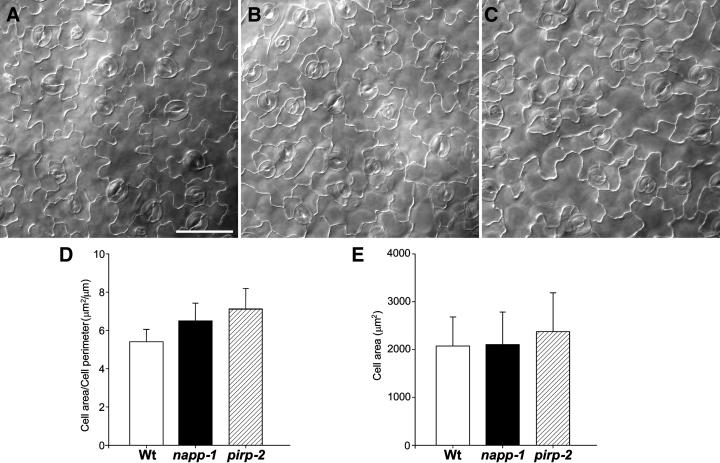
Wild-type leaf epidermal pavement cells in Arabidopsis are thought to expand by both polar tip growth and diffuse growth to produce mature cells with highly wavy cell outlines (Fu et al., 2002) as shown in Figure 7A. Microscopic inspection indicated that the pavement cell lobes in napp-1 and pirp-2 were shorter than those in the wild type, resulting in a less wavy outline (Figures 7B and 7C). Quantification of this phenotype by measuring the ratio between the area and the perimeter of pavement cells (n = 207) confirmed this observation (Figure 7D). The sampled pirp-2 cells were slightly larger than the wild-type and napp-1 cells that were measured (Figure 7E); this is probably the cause of the difference in the area-to-perimeter ratio between napp-1 and pirp-2. pirp-1 pavement cells showed similar defects in lobe elongation (data not shown).
Figure 7.
Pavement Cell Lobe Structure in Wild-Type and napp/pirp Mutants.
(A), (B), and (C) Leaf pavement cell shapes of wild-type (A), napp-1 (B), and pirp-2 (C) plants. The fourth rosette leaf of 2-week-old plants was cleared, and the adaxial pavements cells were observed using phase-contrast light microscopy. Bar = 50 μm.
(D) Comparison of lobe extensions in wild-type and mutant leaf pavement cells. The area and the perimeter were measured on 118 pavement cells in wild-type and mutant plants, and the ratio between area and perimeter was calculated for each cell.
(E) Comparison of cell area of wild-type and mutant pavement cells.
napp-1 Is Allelic to the dis Mutant grl
The trichome phenotype in napp-1 and pirp-1 mutants is reminiscent of the phenotype of the distorted group of trichome mutants. We therefore compared the chromosomal localization of NAPP and PIRP with the mapped positions of the DIS genes (Hülskamp et al., 1994; Schwab et al., 2003). Interestingly, NAPP mapped to the region of the grl locus. grl-247 plants (seeds kindly donated by Martin Hülskamp, University of Köln, Germany) were crossed with napp-1 and pirp-1 plants. None of the progeny from a cross between napp-1 and grl-247 (n = 17) showed a reversion to normal trichome phenotype. By contrast, the progeny of a cross between grl-247 and pirp-1 plants (n = 16) had a normal trichome phenotype. Sequencing of the NAPP transcript in grl-247 revealed a C-T transition in the second exon, generating a stop codon 196 bp downstream of the start codon (Figure 1B). Thus, the napp-1 and grl-247 mutants are allelic, both producing highly truncated NAPP proteins. napp-3 plants, which carry a T-DNA insertion toward the 3′-end of NAPP, display identical phenotypes as napp-1 and grl-247 plants. This observation indicates that the C-terminal, nonconserved part is important for proper NAPP function.
Organization of the Actin Cytoskeleton Is Disrupted in napp-1 and pirp-1 Trichomes
To study the actin cytoskeleton in the napp-1 and pirp-1 mutants, we crossed mutant plants with transgenic plants expressing the actin binding domain of mouse talin (mTalin) fused to yellow fluorescent protein (YFP) under control of the 35S promoter of Cauliflower mosaic virus. Similar protein fusion constructs have earlier been shown to be ideal for noninvasive labeling of microfilaments (Kost et al., 1998; Mathur et al., 1999). In young wild-type trichomes with elongating branches, actin filaments were mostly longitudinally oriented, extending along the entire length of the branch (Figure 8A). The branch tips were dominated by dense actin (Figure 8A, arrows). The branches of young pirp-1 trichomes at a comparable stage of development contained actin filaments with a more random orientation. Many filaments terminated against the sides of the branches; long filaments were rarely observed (Figure 8B). Dense actin was not seen at the branch tips (Figure 8B, arrowhead). At a later stage of development, increased bundling of actin filaments was observed in pirp-1 trichomes (Figure 8C).
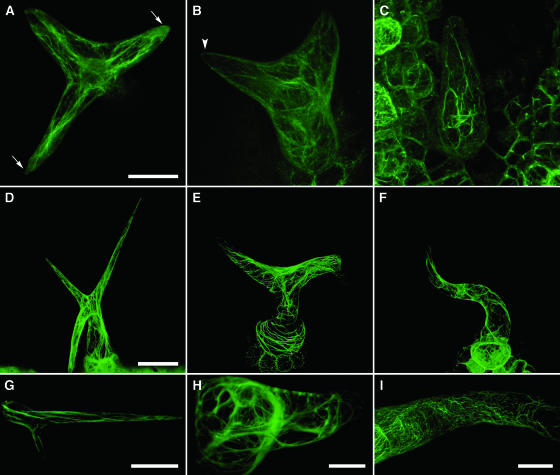
Figure 8.
Organization of F-Actin in Leaf Trichomes of Wild-Type, napp-1, and pirp-1 Plants.
F-actin was visualized by stable expression of YFP-mTalin in 8-d-old plants ([A] to [C]) and 9-d-old plants ([D] to [F]) and by transient expression in 14-d-old plants ([G] to [I]). Young ([A] to [C]) and mature ([D] to [I]) trichomes were imaged using confocal microscopy. Three-dimensional projections were generated from multiple optical sections.
(A) A young wild-type trichome with elongating branches. Actin filaments are longitudinally oriented; two of the branch tips are dominated by diffuse actin.
(B) A pirp-1 trichome at a similar stage of development as in (A). Actin filaments show a more random orientation but not increased bundling. Diffuse actin is not observed in the branch tips.
(C) A pirp-1 trichome at a slightly later stage contains thick actin cables extending both longitudinally and transversely across the branch.
(D) A (mature) wild-type trichome showing a fine network of longitudinally oriented actin filaments.
(E) A pirp-1 trichome with thick actin cables that are mainly transversely oriented, especially in the radially expanded stalk. Note the high density of actin filaments at the branch point between the two branches.
(F) A napp-1 trichome showing transversely oriented filaments.
(G) A branch of a wild-type trichome with a population of diffuse actin at the branch tip.
(H) A transverse cross section through a pirp-1 trichome. Thick actin cables extend across the stalk. The branch tip has small amounts of both actin filaments and diffuse actin.
(I) Portion of the branch of a moderately affected napp-1 trichome. Thick actin cables are not seen, but actin filaments show varying degrees of transverse orientation.
Bars = 20 μm in (A) to (C), (H), and (I), 50 μm in (D) to (F), and 40 μm in (G).
The actin cytoskeleton in mature wild-type trichomes was organized in a fine network of longitudinally oriented filaments (Figure 8D). In mature napp-1 and pirp-1 trichomes, actin was organized in thicker cables, resulting in a less-dense network throughout the trichome (Figures 8E and 8F). The actin cables were generally more transversely orientated in napp-1 and pirp-1 trichomes than in wild-type trichomes. The density of actin cables was often observed to be very high at branch points, especially when branch growth was stunted (Figure 8E). Optical cross sections of mutant trichomes often revealed a loose mesh of transversely interconnecting actin cables (Figure 8H). The tips of mature wild-type trichome branches generally contained dense actin, and filaments were less prominent (Figure 8G). By contrast, the tips of napp-1 and pirp-1 trichome branches were devoid of dense actin (Figures 8E, 8F, and 8H). In the napp-1 and pirp-1 trichomes examined, the severity of the phenotype appeared to correlate with the degree of actin organization defects (cf. Figures 8E and 8I).
The actin cytoskeleton of wild-type pavement cells is generally organized in a dense cortical mesh, and subcortical actin cables often extend along the long axis of the cell (Li et al., 2003; our unpublished observations). By contrast, subcortical actin cables of napp-1 and pirp-1 pavement cells with reduced lobes were more transversely oriented, and long actin cables were rarely observed (data not shown).
DISCUSSION
In this study, we have identified plant homologs of proteins with a role in regulation of actin cytoskeleton organization in animal cells and analyzed the function they might have in plant cell development and morphogenesis. Database searches led to the identification of three Arabidopsis genes encoding products similar to subunits of a protein complex regulating WAVE1 activity in mammalian cells (Eden et al., 2002). The T-DNA knockout mutants of NAPP and PIRP, napp and pirp, displayed abnormal development of epidermal cells as a result of disruption of normal actin filament organization. Our data support a role for NAPP and PIRP in regulation of the ARP2/3 complex in plants. Furthermore, the identification of NAPP, PIRP, and BRK1 suggests that a WAVE-like protein complex is present in plants.
The Mammalian WAVE1 Regulatory Protein Complex Is Partly Conserved in Plants
The five subunits of the inactive WAVE1 protein complex have been identified in several metazoan organisms (Eden et al., 2002; Soto et al., 2002; Bogdan and Klämbt, 2003), as well as the mycetozoan Dictyostelium (Blagg et al., 2003). We have identified three genes in the Arabidopsis genome that encode proteins similar to the mammalian NAP1/2, PIR121/CYFIP, and HSPC300 proteins and five genes that encode putative WAVE homologs. Actins and several protein classes involved in actin dynamics and organization are encoded by multigene families, both in plants and other eukaryotic organisms. By contrast, NAPP, PIRP, and BRK1 as well as six of seven Arabidopsis ARP2/3 complex subunits are single-copy genes. Similarly, these proteins are encoded by only one or two genes in all eukaryotic genomes investigated. Thus, evolutionary pressure inhibiting the production of multiple copies of these genes appears to exist throughout eukaryota. An explanation for this observation could be that higher protein doses have a negative effect on fitness.
An interesting feature of the gene structure of NAPP and PIRP is the presence of an intron in the 5′-UTR of both transcripts. At least 9% of Arabidopsis genes are predicted to have one or several introns in their UTRs (Zhu et al., 2003). 5′-UTR introns have been shown to contain gene regulatory elements necessary for proper expression of several plant genes, such as potato (Solanum tuberosum) sucrose synthase (Fu et al., 1995), tobacco (Nicotiana tabacum) polyubiquitin (Plesse et al., 2001), and the Arabidopsis actin gene ACT1 (Vitale et al., 2003). A similar effect on the expression of NAPP and PIRP could also be expected.
NAPP and PIRP Are Involved in Regulation of the Actin Cytoskeleton
Several lines of evidence point toward a role for PIRP and NAPP in regulation of the actin cytoskeleton. First, the phenotype of napp and pirp epidermal cells, such as trichomes and leaf pavement cells, is highly similar to that of the dis class of trichome mutants, which have previously been shown to have defects in actin organization. napp-1 is allelic to grl, further confirming this connection. Second, the actin cytoskeleton in napp-1 and pirp-1 trichomes shows an abnormal organization, forming thick bundles that are transversely orientated. Third, four other mutants of the dis class, wrm, dis1, crk, and dis2, have been mapped to genes encoding the ARP2/3 complex subunits ARP2, ARP3, ARPC5, and ARPC2, respectively (Le et al., 2003; Li et al., 2003; Mathur et al., 2003a, 2003b; El-Assal et al., 2004). Thus, NAPP and PIRP appear to operate in the same pathway as the ARP2/3 complex. Inactivation of Nap, PIR121, and ARP2/3 complex subunit homologs also produce similar cellular defects in other organisms. RNA interference depletion of the Nap homolog Kette, the PIR121 homolog Sra-1, and the p20 subunit of the ARP2/3 complex in Drosophila S2 cells all resulted in a stellate cell morphology (Kunda et al., 2003; Rogers et al., 2003). During embryogenesis in C. elegans, the PIR121 mutant gex-2 and the Nap mutant gex-3 display defects in ventral enclosure similar to those observed for lines with RNA interference depletion of ARP2/3 complex subunits (Soto et al., 2002; Sawa et al., 2003).
We did not observe any consistent defects in growth and morphology of napp and pirp root hairs challenged to rapid growth, as has been reported for ARP2/3 complex mutants (Mathur et al., 2003a, 2003b). The results could indicate that the ARP2/3 complex acts independently of NAPP and PIRP in root hairs. More likely, subtle differences in growth conditions in these experiments could possibly interfere with appearance of root hair phenotypes.
PIRP expression doesn't appear to be totally abolished in the pirp-1 mutant. A possible explanation for this result is that a small amount of PIRP transcripts are correctly spliced despite the presence of T-DNA. The levels of correctly spliced PIRP transcript are probably too low to maintain proper function of PIRP.
The chromosomal localization of PIRP is very close to the genetic markers surrounding the klk allele. Furthermore, klk is the only reported dis mutant that maps to chromosome 5, and no genes encoding ARP2/3 complex subunits are located on this chromosome. Although no complementation experiments have been performed between pirp-1 and klk, it is tempting to speculate that these two mutants are allelic. A double mutant of klk and the ARP3 mutant dis1 showed an additive phenotype, indicating that ARP3 and KLK act in the same process (Schwab et al., 2003). Evidence that PIRP and KLK are identical would confirm the functional connection between the WAVE regulatory complex and the ARP2/3 complex in Arabidopsis. The putative maize homolog of HSPC300, BRK1, has also been implicated in the regulation of the actin cytoskeleton. The maize brk1 mutant causes defects in the formation of lobes in epidermal cells, similar to what is observed in the napp1, pirp-1, and ARP2/3 complex mutants in Arabidopsis (Frank and Smith, 2002), further supporting a role for the WAVE regulatory protein complex in actin microfilament organization in plants. However, none of the reported dis mutants map to the vicinity of BRK1. The small size of the BRK1 gene makes it a difficult target for conventional mutagenesis; the lack of any dis mutants mapping to BRK1 suggests that the mutant screens may not have been completely saturated.
The WAVE Complex Is Likely Present in Plants
In animal cells, NAP/HEM/GEX-2 and PIR121/CYFIP/GEX-3 proteins function as negative regulators of the ARP2/3 complex through their inhibitory effects on WAVE. However, there have been no reports to date of WAVE/WASP-like proteins in plants. A possible explanation is that plants lack a functional equivalent of WAVE proteins. NAPP and PIRP would then have roles other than the regulation of ARP2/3 complex activity. This hypothesis is contradicted by the phenotypical similarity of napp, pirp, and ARP2/3 complex mutants. What proteins perform the role of WAVE in plants? Four Arabidopsis genes encode proteins with a domain composition similar to mammalian WAVEs, with an N-terminal SHD domain and a C-terminal VCA domain. Although no functional data presently exist regarding these genes, they are likely candidates for being Arabidopsis WAVE homologs, as has also been noted by Deeks and Hussey (2003). A fifth protein, which contains the SHD domain, but lacks the VCA domain, might constitute a novel plant gene. This protein may be regulated by the Arabidopsis WAVE regulatory complex but interact with other effectors than the ARP2/3 complex.
Recent publications place Abi at the core of the mammalian WAVE regulatory complex, interacting with Nap, WAVE, and HSPC300 (Gautreau et al., 2004; Innocenti et al., 2004). It is therefore puzzling that the Arabidopsis genome apparently does not encode Abi homologs. Considering the relatively low similarity of NAPP to Nap homologs from other organisms, it is not unlikely that putative Arabidopsis Abi homologs are highly divergent from animal Abi proteins. Alternatively, plants may have developed a modified WAVE regulatory complex, involving a novel protein performing a role similar to that of Abi.
At first glance, it would seem counterintuitive that the phenotypes of the napp-1 and pirp-1 mutants be identical to that of the ARP2/3 complex mutants. If NAPP and PIRP inhibit the activity of any Arabidopsis WAVE equivalent, one would expect the inactivation of NAPP and PIRP to result in unregulated activation of the ARP2/3 complex, as suggested by Deeks and Hussey (2003). The organization of the actin cytoskeleton in napp-1 and pirp-1 would then be expected to be radically different from the ARP2/3 complex mutants, with excessive branching of actin filaments. However, inactivation of WAVE regulatory complex subunits in animal cells has been shown to lead to the degradation of WAVE itself (Blagg et al., 2003; Rogers et al., 2003). The explanation for this phenomenon seems to be that active, uncomplexed WAVE protein is prone to degradation and that the regulatory complex protects WAVE from this degradation. If this mechanism is conserved in plants, inactivation of NAPP or PIRP would result in rapid proteolysis of the Arabidopsis WAVE homolog, and the ARP2/3 complex would remain inactive.
An alternative model for the role of the WAVE regulatory complex in mammalian cells has been presented (Innocenti et al., 2004; Steffen et al., 2004). According to this model, the WAVE regulatory complex stays associated with WAVE upon activation by Rac1 and is essential for WAVE activity. If a WAVE-like complex is necessary for proper activation and/or localization of the Arabidopsis WAVE homologs, the disruption of NAPP or PIRP would be likely to block WAVE activity. Therefore, both current models for the regulation of WAVE activity can explain the observed phenotypes of the napp and pirp mutants.
The ARP2/3 Complex Might Not Be Essential for Actin Polymerization in Plants
Mutants lacking either functional NAP or PIR are embryonic lethals in C. elegans and Drosophila, showing defects in cell sheet morphogenesis and development of the central nerve system, respectively (Hummel et al., 2000; Soto et al., 2002; Schenck et al., 2003). By comparison, the napp-1 and pirp-1 mutant phenotypes are relatively mild. One explanation could be that inactivation of NAPP or PIRP does not completely abolish ARP2/3 complex activity. However, mutations in ARP2/3 complex subunits result in similar phenotypes, such as napp-1 and pirp-1, and a more likely explanation is that the ARP2/3 complex is not essential for actin polymerization. The formins are another protein class that exhibit actin-nucleating activity (Wallar and Alberts, 2003; Zigmond, 2004). In metazoan cells, formins stimulate de novo polymerization of actin filaments and produce actin cables. The Arabidopsis genome encodes at least 21 putative formins (Deeks et al., 2002). Overexpression of the Arabidopsis formin AFH1 in tobacco pollen tubes induced formation of actin cables throughout the cytoplasm and resulted in growth depolarization and growth arrest of transformed pollen tubes (Cheung and Wu, 2004). The actin-nucleating activity of formins could be sufficient for normal cell function in most cells. There may be an equilibrium between formin activity and ARP2/3 complex activity in plant cells, the former producing actin cables and the latter producing cortical actin networks. Disruption of ARP2/3 complex activity would then be expected to lead to a shift toward formin activity, resulting in the reduction of cortical actin and overproduction of actin cables, as observed in napp-1, pirp-1, and ARP2/3 complex mutants.
Are Plant RAC/ROP GTPases Involved in Activation of the ARP2/3 Complex?
In human cells, the N-terminal domain of the PIRP homolog Sra-1 interacts specifically with GTP-bound Rac1 (Kobayashi et al., 1998). This domain is also partially conserved in PIRP, suggesting that physical interaction might occur between PIRP and one or several of the 11 RAC/ROP proteins in Arabidopsis. Thus, the results presented in this work provide a link between plant RAC/ROP GTPases and regulation of the ARP2/3 complex. Expression studies show that the AtRAC genes have specific but overlapping expression patterns; promoter studies indicate that at least one AtRAC gene is specifically expressed during trichome development (P. Winge, T. Brembu, and A.M. Bones, unpublished results). Redundancy as a consequence of overlapping expression could explain why mutation of AtRAC genes has not been reported as resulting in morphological phenotypes.
In conclusion, we have shown that an evolutionarily conserved protein complex involved in regulation of the actin cytoskeleton through the ARP2/3 complex exists in plants. Disruption of two of the complex subunits, NAPP and PIRP, leads to defects in actin filament organization, resulting in aberrant morphogenesis of several epidermal cell types. The results provide a possible mechanism for RAC/ROP-mediated regulation of actin dynamics through the ARP2/3 complex in plants.
METHODS
Plant Materials and Growth Conditions
All plants were in the Columbia-0 background, except the grl mutant, which was in the Landsberg erecta background, and the pirp-2 mutant, which was in the Wassilewskija background. The grl mutant was isolated in an ethyl methanesulfonate mutagenesis screen (Hülskamp et al., 1994). Arabidopsis thaliana plants were grown in vitro (0.6% agar with 1× MS salts [Sigma, St. Louis, MO] and 3% sucrose) and in soil in growth rooms under a light regime of 8 h of darkness and 16 h of light.
Mutant Identification, Reverse Transcription, and PCR Analysis
Searches of the SALK T-DNA insertion mutant collection (http://signal.salk.edu/cgi-bin/tdnaexpress) led to the discovery of T-DNA insertion lines for At2g35110 (napp-1, SALK_014298; napp-2, SALK_038799; napp-3, SALK_009695) and At5g18410 (pirp-1, SALK_106757). The pirp-2 mutant line (EXM115) was found in the INRA-Versailles T-DNA insertion collection (http://flagdb-genoplante-info.infobiogen.fr/projects/fst/). Seeds were obtained from the Nottingham Arabidopsis Stock Center (http://nasc.nott.ac.uk/), except the pirp-2 mutant, which was obtained from INRA-Versailles (http://dbsgap.versailles.inra.fr/publiclines/informations.php). Genomic DNA was isolated from the mutant lines, and T-DNA insertions were confirmed by PCR reactions using T-DNA left border primers and gene-specific primers, followed by sequencing of the PCR products.
Total RNA was isolated from the rosette leaves of 3-week-old wild-type napp-1, pirp-1, and grl-247 plants using the RNeasy midi kit (Qiagen, Hilden, Germany). mRNA was isolated from ∼50 μg of total RNA using the Micro FastTrack kit (Invitrogen, Breda, The Netherlands). For tissue expression analysis, 100 mg of tissue from different parts of 5-week-old plants was used for mRNA isolation, using Dynabeads mRNA DIRECT kit (Dynal, Oslo, Norway), according to the manufacturer's instructions. For first strand cDNA synthesis, 100 ng (mutant analysis) or 20 ng (tissue expression analysis) of mRNA was reverse transcribed using hexanucleotide primers and M-MuLV (Amersham Biosciences, Buckinghamshire, UK). The PCR profile was 94°C for 30 s, 53°C for 30 s, and 72°C for 1 min. For NAPP, PIRP, and BRK1, 35 cycles (mutant analysis) or 40 cycles (gene expression analysis) of PCR amplification were used. For the positive control GAPDH, 30 cycles (mutant analysis) or 35 cycles (tissue expression analysis) were used. As a negative control, mRNA was used as template in all experiments. Twenty microliters of each PCR product was loaded on a 1.5% (w/v) agarose gel.
Wild-type cDNA transcripts of NAPP and PIRP were sequenced using the cloned genes (see below) as template and gene-specific primers (Table 2, Figure 9). NAPP transcripts in grl were amplified using RT-PCR with the same primers as wild-type NAPP, and the PCR products were purified and sequenced directly. For sequencing of the 5′- and 3′-UTR of NAPP and PIRP, cDNA was prepared with the GeneRacer kit (Invitrogen) following the manufacturer's instructions. Peptide alignments were made with the GeneDoc program (http://www.psc.edu/biomed/genedoc/).
Table 2.
Primers Used in This Study
| Primer Name | Sequence (5′/3′) |
|---|---|
| NAPPF1 (F1) | AGCTTCTCAGAGATTATTGTA |
| NAPPR1 (R1) | TTGCTGAAATGCTCAGGGAACT |
| NAPPF2 (F2) | TTGGAGTATCCTGATCCTGTTG |
| NAPPR2 (R2) | GAATCTAGGATGATATGGGCTT |
| NAPPF3 (F3) | TGTGCATTTGTCCGAGTCAATC |
| NAPPR3 (R3) | GGATCTTGAGGATGCAATACCA |
| NAPPF4 (F4) | TTCACTGATCAACCAAGTTTGC |
| NAPPR4 (R4) | CTCGACGTAAAGCACAGCATCT |
| NAPPF5 (F5) | TTTCAGCGTATTGTGCAACATC |
| NAPPR5 (R5) | TTGTTCTGGAAGAAGCTGAGAT |
| NAPPF6 (F6) | GCATTGTTGTGCATGGCTCAGT |
| NAPPR6 (R6) | CACAAATAGGCTCCATGTCGTT |
| NAPPF7 (F7) | GCTGCAATTCAAAGGTTGACTA |
| NAPPR7 (R7) | TTCGAATGTATGCAAAGATGAC |
| NAPPF8 (F8) | ATGGACCTAACCCAAGGCATCA |
| NAPPR8 (R8) | TTCTTCAGGTATGTGCTCAACG |
| NAPPF9 (F9) | GACAATGCATCCTTGATACACT |
| NAPPR9 (R9) | CACGAGATGCGATCTGTTGGCT |
| NAPPF10 (F10) | GTAAGGTTACAACGTGAATACC |
| NAPPR10 (R10) | GCTCGAGCTGTAATCCAATGGT |
| NAPPF11 (F11) | ATTCAGGCTACTTCAAAGGCTC |
| NAPPR11 (R11) | AACAAGAAGACTAGTGCGAGGGAT |
| PIRPF1 (S1) | CAATTGACTTGAAGTAGTAGCA |
| PIRPR1 (A1) | TCAAGCAACTTTAGCATTGACC |
| PIRPF2 (S2) | CATAGTCAAGCGGATTTGTATC |
| PIRPR2 (A2) | CTTGTTTACACGGAACATCTCA |
| PIRPF3 (S3) | ACACATCTTTGGTCAATGCT |
| PIRPR3 (A3) | TGAAAGTATACCGACAATTC |
| PIRPF4 (S4) | ACACCATCTGAGAAAGATACCG |
| PIRPR4 (A4) | TTGAACTTCGGCATGTATTGTC |
| PIRPF5 (S5) | GATGCTTTATGGGAGACAATAC |
| PIRPR5 (A5) | AAGAAGCTCACTTGCTGACCAA |
| PIRPF6 (S6) | CCATGGATGCTGATAGACTACA |
| PIRPR6 (A6) | TGGTTTCTGAGTAGGAGGAACT |
| PIRPF7 (S7) | ATGCAGAGTGATTTCCTACCAA |
| PIRPR7 (A7) | TAATCCCAACCATGGAGCTGTT |
| PIRPF8 (S8) | ACAACACTTGAGCCAATGATCT |
| PIRPR8 (A8) | TTCTGCCTGTTTGGACATGGTA |
| PIRPF9 (S9) | TCTATACCATGTCCAAACAGGC |
| PIRPR9 (A9) | AAAGCAGACACGGTATTCTCAA |
| PIRPF10 (S10) | TTGAGAATACCGTGTCTGCTTT |
| PIRPR10 (A10) | TGTTCAGAAAGAAAGGTGGATACATGGT |
The position and orientation of each primer is indicated in Figure 9.
Figure 9.
Position of NAPP and PIRP Primers Used in This Study.
The arrows indicate the orientation of the primers.
Microscopy Analysis of Epidermal Cell Phenotypes
Fresh plant tissue was observed with a stereomicroscope (SMZ1500; Nikon, Tokyo), and pictures were taken with a Nikon Coolpix 990 digital camera. For visualization of leaf pavement cells, the fourth rosette leaf from 2-week-old plants grown in vitro was cleared with chloral hydrate as described by Hamada et al. (2000) and observed using differential interference contrast microscopy (Nikon S800). Images were captured with a cooled SPOT CCD camera (Diagnostic Instruments, Sterling Heights, MI), and measurement of pavement cell perimeter and area were done using analySIS 3.1 (Soft Imaging System, Münster, Germany). For scanning electron microscopy studies, developing leaves from 4-week-old soil-grown plants were dehydrated in an ethanol series (20, 40, 50, 60, 70, 80, 90, and 100% [v/v]). After critical point drying and coating, the specimens were observed with a JEOL JSM-840 scanning electron microscope, and images were taken using SemAfore 4.02 software (JEOL, Sollentuna, Sweden).
DNA Manipulation, Plasmid Constructs, and Transformation
The YFP-mTalin construct was generated in pBI121 (Clontech, Palo Alto, CA; GenBank accession number AF485783). Full-length cDNA transcripts of NAPP and PIRP were constructed in the pGreen binary vector (Hellens et al., 2000; GenBank accession number AJ007829). Furthermore, all constructs were cloned downstream of the Cauliflower mosaic virus 35S promoter. Pfu DNA polymerase (Stratagene, La Jolla, CA) was used for all the PCR amplifications. The actin binding domain of mTalin was amplified from mouse cDNA using the primers 5′- GAGTGCATGCACTTTGAGGAACAAATCC and 3′- GCATGAGCTCCACAGGGCTTTTTTAGTGC. The PCR product was digested with SphI and SacI and fused to the C-terminal end of YFP as described by Fu et al. (2001). The full-length cDNA transcripts of NAPP and PIRP were cloned in multiple steps because of the size of the transcripts. cDNA from wild-type Columbia rosette leaves was produced as described previously, and the coding sequence was amplified using RT-PCR. A SalI site was introduced in the 5′-UTR of NAPP, and a BamHI site was introduced in the coding sequence using the PCR primers 5′-CGTTGTCGACTATTACTGGGTTTTTGGA and 3′-CTGCGGATCCAGTGGTTTGCTGTT. The T-C substitution resulting in the generation of the BamHI site was neutral, with both codons encoding Ser. Digestion with SalI and BamHI produced an ∼2.7-kb fragment, which was cloned to the SalI and BamHI sites of pGreen. The 3′ part of NAPP was amplified with the primers 5′-AACAGCAAACCACTGGATCCGCAG and 3′- AACAAGAAGACTAGTGCGAGGGAT, introducing a BamHI site in the same position as in the first cloning step and a SpeI site in the 3′-UTR. Digestion with BamHI and SpeI produced an ∼1.6-kb fragment, which was fused to the BamHI site of the 2.7-kb fragment. PIRP was cloned in three steps. First, the 3′ part of PIRP was amplified using the primers 5′-GGTTGTATGAAGCTTATACGAGAG and 3′-TGTTCAGAAAGAAAGGTGGATCCATGGT, the latter primer introducing a BamHI site in the 3′-UTR. The PCR product was digested with HindIII and BamHI, producing an ∼950-bp fragment, which was cloned to the HindIII and BamHI sites of pGreen. Second, the primers 5′-GCATTGTTGTGCATGGCTCAGT and 3′-TAATCCCAACCATGGAGCTGTT were used to amplify a PCR product, which after treatment with SalI and HindIII, yielded an ∼400-bp fragment. The fragment was cloned to the SalI sites, upstream of the first fragment of PIRP. Third, the 5′ part of PIRP was amplified using the primers 5′-GTTGTTGGTCGACTCAATGGCGGTTC and 3′-CTGCATCTCTGACCAAATCTGTG, the former introducing a SalI site in the 5′-UTR of PIRP. Digestion with SalI resulted in an ∼2.5-kb fragment, which was cloned to the SalI site, upstream of the second fragment of PIRP. Finally, the orientation of the SalI fragment was checked. All constructs were sequenced using BigDye chemistry (Applied Biosystems, Foster City, CA). For stable transformation of Arabidopsis plants, pBI121 plasmids (carrying YFP-mTalin) were transformed into Agrobacterium tumefaciens (strain GV3101), which were used for transformation of Arabidopsis plants with vacuum infiltration (Bechtold et al., 1993).
Particle Bombardment–Mediated Transient Expression in Arabidopsis Leaves
Two-week-old Arabidopsis plants grown in vitro were used for the bombardment experiments. All plasmids were purified from Escherichia coli XL1-blue strain cells using the alkaline lysis method (Sambrook and Russel, 2001) followed by phenol/chloroform extraction. Plants were bombarded with 1-μm-diameter gold particles coated with plasmids using a Bio-Rad PDS-1000/He biolistic particle delivery system (Bio-Rad, Hercules, CA). In all experiments, 1 μg of plasmid was used. Bombarded plants were grown under normal conditions for 20 to 48 h before observation with a confocal microscope.
Visualization of Fluorescent Protein Fusions and Confocal Microscopy
Arabidopsis plants stably expressing YFP-mTalin were produced. The YFP-mTalin transgene was crossed into napp-1 and pirp-1 plants. Alternatively, YFP-mTalin was transiently expressed in wild-type, napp-1, and pirp-1 plants through particle bombardment as described above. Specimens were observed with a confocal microscope (LSM510; Zeiss, Jena, Germany). Optical sections were scanned, and three-dimensional projections were made using Zeiss LSM510 Image Examiner 3.0 software. The resulting images were processed using Adobe Photoshop 7.0 (Mountain View, CA).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under the following accession numbers: AY496700 (NAPP/At2g35110 mRNA), AY496701 (PIRP/At5g18410 mRNA), BK004072 (NAPP gene), BK004072 (PIRP gene), AF370530 (BRK1/At2g22640 mRNA), and AC006340 (BRK1 gene). Accession numbers for BAC clones containing putative WAVE homologs are as follows: AC002341 (WAVE1/At2g34150), AC021043 (WAVE2/At1g29170), AL161946 (WAVE3/At5g01730), AAC28760 (WAVE4/At2g38440), and AL021710 (WAVE5/At4g18600).
Acknowledgments
We thank Martin Hülskamp (University of Köln) for the grl-247 seeds, Kjell Evjen for assistance with the scanning electron microscope, Anne Mortensen for technical support, Ishita Ahuja and Nancy Bazilchuk for critical reading of the manuscript. This work was supported by the functional genomics program of the Norwegian Research Council.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Atle M. Bones (atle.bones@bio.ntnu.no).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.023739.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Amann, K.J., and Pollard, T.D. (2001). The Arp2/3 complex nucleates actin filament branches from the sides of pre-existing filaments. Nat. Cell Biol. 3, 306–310. [DOI] [PubMed] [Google Scholar]
- Baumgartner, S., Martin, D., Chiquet-Ehrismann, R., Sutton, J., Desai, A., Huang, I., Kato, K., and Hromas, R. (1995). The HEM proteins: A novel family of tissue-specific transmembrane proteins expressed from invertebrates through mammals with an essential function in oogenesis. J. Mol. Biol. 251, 41–49. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR Acad. Sci. III–Vie 316, 1194–1199. [Google Scholar]
- Blagg, S.L., Stewart, M., Sambles, C., and Insall, R.H. (2003). PIR121 regulates pseudopod dynamics and SCAR activity in Dictyostelium. Curr. Biol. 13, 1480–1487. [DOI] [PubMed] [Google Scholar]
- Bogdan, S., and Klämbt, C. (2003). Kette regulates actin dynamics and genetically interacts with Wave and Wasp. Development 130, 4427–4437. [DOI] [PubMed] [Google Scholar]
- Carol, R.J., and Dolan, L. (2002). Building a hair: Tip growth in Arabidopsis thaliana root hairs. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357, 815–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, A.Y., and Wu, H.M. (2004). Overexpression of an Arabidopsis formin stimulates supernumerary actin cable formation from pollen tube cell membrane. Plant Cell 16, 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks, M.J., and Hussey, P.J. (2003). Arp2/3 and ‘the shape of things to come’. Curr. Opin. Plant Biol. 6, 561–567. [DOI] [PubMed] [Google Scholar]
- Deeks, M.J., Hussey, P.J., and Davies, B. (2002). Formins: Intermediates in signal-transduction cascades that affect cytoskeletal reorganization. Trends Plant Sci. 7, 492–498. [DOI] [PubMed] [Google Scholar]
- Eden, S., Rohatgi, R., Podtelejnikov, A.V., Mann, M., and Kirschner, M.W. (2002). Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature 418, 790–793. [DOI] [PubMed] [Google Scholar]
- El-Assal, S.E., Le, J., Basu, D., Mallery, E.L., and Szymanski, D.B. (2004). DISTORTED2 encodes an ARPC2 subunit of the putative Arabidopsis ARP2/3 complex. Plant J. 38, 526–538. [DOI] [PubMed] [Google Scholar]
- Frank, M.J., and Smith, L.G. (2002). A small, novel protein highly conserved in plants and animals promotes the polarized growth and division of maize leaf epidermal cells. Curr. Biol. 12, 849–853. [DOI] [PubMed] [Google Scholar]
- Fu, H., Kim, S.Y., and Park, W.D. (1995). High-level tuber expression and sucrose inducibility of a potato Sus4 sucrose synthase gene require 5′ and 3′ flanking sequences and the leader intron. Plant Cell 7, 1387–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y., Li, H., and Yang, Z. (2002). The ROP2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. Plant Cell 14, 777–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y., Wu, G., and Yang, Z. (2001). Rop GTPase-dependent dynamics of tip-localized F-actin controls tip growth in pollen tubes. J. Cell Biol. 152, 1019–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautreau, A., Ho, H.Y., Li, J., Steen, H., Gygi, S.P., and Kirschner, M.W. (2004). Purification and architecture of the ubiquitous Wave complex. Proc. Natl. Acad. Sci. USA 101, 4379–4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada, S., Onouchi, H., Tanaka, H., Kudo, M., Liu, Y.G., Shibata, D., Machida, C., and Machida, Y. (2000). Mutations in the WUSCHEL gene of Arabidopsis thaliana result in the development of shoots without juvenile leaves. Plant J. 24, 91–101. [DOI] [PubMed] [Google Scholar]
- Hellens, R.P., Edwards, E.A., Leyland, N.R., Bean, S., and Mullineaux, P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832. [DOI] [PubMed] [Google Scholar]
- Hummel, T., Leifker, K., and Klämbt, C. (2000). The Drosophila HEM-2/NAP1 homolog KETTE controls axonal pathfinding and cytoskeletal organization. Genes Dev. 14, 863–873. [PMC free article] [PubMed] [Google Scholar]
- Hülskamp, M. (2000). How plants split hairs. Curr. Biol. 10, R308–R310. [DOI] [PubMed] [Google Scholar]
- Hülskamp, M., Misra, S., and Jürgens, G. (1994). Genetic dissection of trichome cell development in Arabidopsis. Cell 76, 555–566. [DOI] [PubMed] [Google Scholar]
- Innocenti, M., Zucconi, A., Disanza, A., Frittoli, E., Areces, L.B., Steffen, A., Stradal, T.E., Fiore, P.P., Carlier, M.F., and Scita, G. (2004). Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nat. Cell Biol. 6, 319–327. [DOI] [PubMed] [Google Scholar]
- Klahre, U., and Chua, N.H. (1999). The Arabidopsis actin-related protein 2 (AtARP2) promoter directs expression in xylem precursor cells and pollen. Plant Mol. Biol. 41, 65–73. [DOI] [PubMed] [Google Scholar]
- Kobayashi, K., Kuroda, S., Fukata, M., Nakamura, T., Nagase, T., Nomura, N., Matsuura, Y., Yoshida-Kubomura, N., Iwamatsu, A., and Kaibuchi, K. (1998). p140Sra-1 (specifically Rac1-associated protein) is a novel specific target for Rac1 small GTPase. J. Biol. Chem. 273, 291–295. [DOI] [PubMed] [Google Scholar]
- Kost, B., Spielhofer, P., and Chua, N.H. (1998). A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16, 393–401. [DOI] [PubMed] [Google Scholar]
- Kunda, P., Craig, G., Dominguez, V., and Baum, B. (2003). Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr. Biol. 13, 1867–1875. [DOI] [PubMed] [Google Scholar]
- Le, J., El Assal, S., Basu, D., Saad, M.E., and Szymanski, D.B. (2003). Requirements for Arabidopsis ATARP2 and ATARP3 during epidermal development. Curr. Biol. 13, 1341–1347. [DOI] [PubMed] [Google Scholar]
- Li, S., Blanchoin, L., Yang, Z., and Lord, E.M. (2003). The putative Arabidopsis arp2/3 complex controls leaf cell morphogenesis. Plant Physiol. 132, 2034–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky, L.M., Atkinson, S.J., Ampe, C., Vandekerckhove, J., and Pollard, T.D. (1994). Purification of a cortical complex containing two unconventional actins from Acanthamoeba by affinity chromatography on profilin-agarose. J. Cell Biol. 127, 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky, L.M., and Gould, K.L. (1999). The Arp2/3 complex: A multifunctional actin organizer. Curr. Opin. Cell Biol. 11, 117–121. [DOI] [PubMed] [Google Scholar]
- Marchand, J.B., Kaiser, D.A., Pollard, T.D., and Higgs, H.N. (2001). Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat. Cell Biol. 3, 76–82. [DOI] [PubMed] [Google Scholar]
- Martin, C., Bhatt, K., and Baumann, K. (2001). Shaping in plant cells. Curr. Opin. Plant Biol. 4, 540–549. [DOI] [PubMed] [Google Scholar]
- Mathur, J., and Hülskamp, M. (2002). Microtubules and microfilaments in cell morphogenesis in higher plants. Curr. Biol. 12, R669–R676. [DOI] [PubMed] [Google Scholar]
- Mathur, J., Mathur, N., Kernebeck, B., and Hülskamp, M. (2003. b). Mutations in actin-related proteins 2 and 3 affect cell shape development in Arabidopsis. Plant Cell 15, 1632–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur, J., Mathur, N., Kirik, V., Kernebeck, B., Srinivas, B.P., and Hülskamp, M. (2003. a). Arabidopsis CROOKED encodes for the smallest subunit of the ARP2/3 complex and controls cell shape by region specific fine F-actin formation. Development 130, 3137–3146. [DOI] [PubMed] [Google Scholar]
- Mathur, J., Spielhofer, P., Kost, B., and Chua, N. (1999). The actin cytoskeleton is required to elaborate and maintain spatial patterning during trichome cell morphogenesis in Arabidopsis thaliana. Development 126, 5559–5568. [DOI] [PubMed] [Google Scholar]
- Plesse, B., Criqui, M.C., Durr, A., Parmentier, Y., Fleck, J., and Genschik, P. (2001). Effects of the polyubiquitin gene Ubi. U4 leader intron and first ubiquitin monomer on reporter gene expression in Nicotiana tabacum. Plant Mol. Biol. 45, 655–667. [DOI] [PubMed] [Google Scholar]
- Rogers, S.L., Wiedemann, U., Stuurman, N., and Vale, R.D. (2003). Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 162, 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi, R., Ma, L., Miki, H., Lopez, M., Kirchhausen, T., Takenawa, T., and Kirschner, M.W. (1999). The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell 97, 221–231. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and Russel, D.W. (2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sawa, M., Suetsugu, S., Sugimoto, A., Miki, H., Yamamoto, M., and Takenawa, T. (2003). Essential role of the C. elegans Arp2/3 complex in cell migration during ventral enclosure. J. Cell Sci. 116, 1505–1518. [DOI] [PubMed] [Google Scholar]
- Schenck, A., Bardoni, B., Langmann, C., Harden, N., Mandel, J.L., and Giangrande, A. (2003). CYFIP/Sra-1 controls neuronal connectivity in Drosophila and links the Rac1 GTPase pathway to the fragile X protein. Neuron 38, 887–898. [DOI] [PubMed] [Google Scholar]
- Schenck, A., Bardoni, B., Moro, A., Bagni, C., and Mandel, J.L. (2001). A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc. Natl. Acad. Sci. USA 98, 8844–8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab, B., Mathur, J., Saedler, R., Schwarz, H., Frey, B., Scheidegger, C., and Hülskamp, M. (2003). Regulation of cell expansion by the DISTORTED genes in Arabidopsis thaliana: Actin controls the spatial organization of microtubules. Mol. Genet. Genomics 269, 350–360. [DOI] [PubMed] [Google Scholar]
- Smith, L.G. (2003). Cytoskeletal control of plant cell shape: Getting the fine points. Curr. Opin. Plant Biol. 6, 63–73. [DOI] [PubMed] [Google Scholar]
- Soto, M.C., Qadota, H., Kasuya, K., Inoue, M., Tsuboi, D., Mello, C.C., and Kaibuchi, K. (2002). The GEX-2 and GEX-3 proteins are required for tissue morphogenesis and cell migrations in C. elegans. Genes Dev. 16, 620–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen, A., Rottner, K., Ehinger, J., Innocenti, M., Scita, G., Wehland, J., and Stradal, T.E. (2004). Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. EMBO J. 23, 749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski, D.B., Marks, M.D., and Wick, S.M. (1999). Organized F-actin is essential for normal trichome morphogenesis in Arabidopsis. Plant Cell 11, 2331–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenawa, T., and Miki, H. (2001). WASP and WAVE family proteins: Key molecules for rapid rearrangement of cortical actin filaments and cell movement. J. Cell Sci. 114, 1801–1809. [DOI] [PubMed] [Google Scholar]
- Vantard, M., and Blanchoin, L. (2002). Actin polymerization processes in plant cells. Curr. Opin. Plant Biol. 5, 502–506. [DOI] [PubMed] [Google Scholar]
- Vitale, A., Wu, R.J., Cheng, Z., and Meagher, R.B. (2003). Multiple conserved 5′ elements are required for high-level pollen expression of the Arabidopsis reproductive actin ACT1. Plant Mol. Biol. 52, 1135–1151. [DOI] [PubMed] [Google Scholar]
- Wallar, B.J., and Alberts, A.S. (2003). The formins: Active scaffolds that remodel the cytoskeleton. Trends Cell Biol. 13, 435–446. [DOI] [PubMed] [Google Scholar]
- Weaver, A.M., Young, M.E., Lee, W.L., and Cooper, J.A. (2003). Integration of signals to the Arp2/3 complex. Curr. Opin. Cell Biol. 15, 23–30. [DOI] [PubMed] [Google Scholar]
- Zhu, W., Schlueter, S.D., and Brendel, V. (2003). Refined annotation of the Arabidopsis genome by complete expressed sequence tag mapping. Plant Physiol. 132, 469–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond, S. (2004). Formin' adherens junctions. Nat. Cell Biol. 6, 12–14. [DOI] [PubMed] [Google Scholar]