Abstract
The balance between cell proliferation and differentiation is crucial in multicellular organisms, where it is regulated by complex gene expression networks. This is particularly relevant in plants because organogenesis is a continuous postembryonic process. Here, we investigate the function of Arabidopsis thaliana E2Ff, an atypical member of the E2F family of transcription factors, which acts independently of a dimerization partner. We have focused our analysis on roots and hypocotyls, organs where (1) cell proliferation and differentiation are spatially and/or temporally separated, (2) growth depends on cell expansion in the longitudinal axis, and (3) the AtE2Ff promoter is active. AtE2Ff overexpression produced a reduction in the size of differentiated cells of these organs. Cells of mutant e2ff-1 plants with reduced levels of AtE2Ff mRNA were larger, especially in the hypocotyl, suggesting a role as a growth regulator. These effects of AtE2Ff are not associated with changes in nuclear ploidy levels or in the expression of cell cycle marker genes. However, expression of a subset of cell wall biogenesis genes is misregulated in an AtE2Ff-dependent manner, and based on chromatin immunoprecipitation experiments, they seem to be direct E2F targets. Our results highlight the complex regulatory function exerted by E2F and suggest a possible role of AtE2Ff in repressing cell wall biosynthesis genes during cell elongation in differentiated cells.
INTRODUCTION
The availability of cellular factors required for multiple physiological processes in living organisms is regulated at different levels, one of them of primary importance being transcriptional regulation. The temporally and spatially coordinated action of transcription factors is crucial for a correct function both at cellular and organismal levels. One of the best studied is the family of E2F/dimerization partner (DP) transcription factors. They participate in controlling cell cycle transitions in multicellular organisms, both animals and plants (Harbour and Dean, 2000a; De Veylder et al., 2002; Gutierrez et al., 2002). In addition, they regulate non-cell-cycle functions by mechanisms still poorly understood (Harbour and Dean, 2000a; Müller et al., 2001; Stevaux and Dyson, 2002; Del Pozo et al., 2002; Dimova et al., 2003; Ramirez-Parra et al., 2003; Vlieghe et al., 2003). Members of the retinoblastoma (RB) family that cooperate in the formation of repressor complexes modulate E2F/DP activity. RB-mediated repression occurs by blocking the activation domain of E2Fs and/or by recruiting chromatin remodeling factors (Harbour and Dean, 2000b; Trimarchi and Lees, 2002; Rossi et al., 2003).
Mammalian E2F members constitute a rather complex family (Trimarchi and Lees, 2002). E2F1, E2F2, and E2F3 regulate cell cycle entry and progression by activating the expression of a set of genes, and their activity is modulated by the RB protein. E2F4 and E2F5 act in differentiated cells where they behave largely as repressors in cooperation with other RB family members, p107 and p130. E2F6, a potent transcriptional repressor, is able to regulate gene expression independently of RB. All of these E2Fs perform their function as heterodimers with the DP1 or DP2 protein partners. The last member to be identified is E2F7 (De Bruin et al., 2003; Di Stefano et al., 2003), a transcriptional repressor whose functional role has not been defined yet. On the other side, in Drosophila, only E2F1 and E2F2 are present, which contribute to a complex regulatory network (Stevaux and Dyson, 2002; Cayirlioglu et al., 2003; Dimova et al., 2003).
After the initial identification of E2F family members in plants (Ramirez-Parra et al., 1999; Sekine et al., 1999; Albani et al., 2000; Ramirez-Parra and Gutierrez, 2000), the availability of the Arabidopsis thaliana genome sequence has facilitated further analysis on E2F/DP transcription factors. These studies are revealing that the situation in Arabidopsis is also complex, and the role of each E2F family member is still far from being fully understood. Three Arabidopsis E2Fs, AtE2Fa, AtE2Fb, and AtE2Fc, share a common domain organization, including domains for DNA binding, dimerization with DP, interaction with the retinoblastoma-related (RBR) protein, and transcriptional regulation (De Jager et al., 2001; Mariconti et al., 2002). AtE2Fa is an activator of gene expression and stimulates cell division and endoreplication (De Veylder et al., 2002; Rossignol et al., 2002; Kosugi and Ohashi, 2003). However, AtE2Fc is an inhibitor of cell division that restricts cell proliferation in the dark (Del Pozo et al., 2002). Postembryonic growth relies entirely on the continuous balance between cell proliferation and differentiation; consequently, these AtE2Fs affect plant morphogenesis. Previous observations show that ectopic expression of AtE2Fa/AtDPa or AtE2Fc in Arabidopsis results in hyperplasia or hypoplasia, respectively. These effects strengthen the important role of the E2F-DP pathway not only in the control of cell proliferation but also in development. E2F in plants also seems to regulate the expression of non-cell-cycle genes, as revealed recently using genomic approaches (Ramirez-Parra et al., 2003; Vlieghe et al., 2003), thus expanding the cellular processes that may be under the control of E2F/DP transcription factors.
The other three Arabidopsis E2F family genes, E2Fd, E2Fe, and E2Ff (Mariconti et al., 2002), also known as DEL2, DEL1, and DEL3 (Vandepoele et al., 2002), E2L1, E2L3, and E2L2 (Kosugi and Ohashi, 2002a), or ELP3, ELP2, and ELP1 (De Jager et al., 2001), respectively, have been identified by data mining. Only a basic molecular characterization of these novel E2F members is available, but it is clear that they are atypical in that, while interacting with E2F consensus sites (Kosugi and Ohashi, 2002a; Mariconti et al., 2002), they have a duplicated DNA binding domain and act independently of a DP. These properties are shared with the recently identified mammalian E2F7 (De Bruin et al., 2003; Di Stefano et al., 2003). So far, nothing is known about the physiological role of these atypical E2F family members in the context of a whole organism.
Here, we have studied AtE2Ff, one of these atypical E2F members, which seems to act as a key regulator of Arabidopsis growth and development through the control of a subset of E2F targets in an organ-specific manner. Our studies have defined a novel role for AtE2F in regulating the expression of key genes involved in plant cell wall biosynthesis, a process coupled to plant cell differentiation and organ growth. This leads us to propose that AtE2Ff is part of a crucial regulatory network required for the differentiation of certain cell types during Arabidopsis postembryonic growth and development.
RESULTS
AtE2Ff, a Unique Member That Does Not Bind to the RB Protein
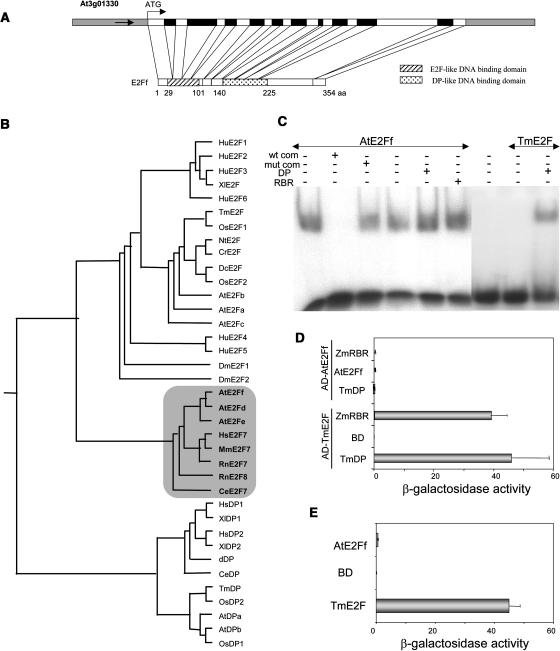
The Arabidopsis E2Ff gene (At3g01330) encodes a 354–amino acid protein (Figure 1A) that differs from the genomic prediction (Arabidopsis Genome Initiative, 2000) in that it contains one extra intron and one extra exon at the C terminus. The atypical E2F members, which have a duplicated DNA binding domain and act independently of a DP, had been only identified in Arabidopsis and were considered to be unique to plants (Kosugi and Ohashi, 2002a; Shen, 2002; Vandepoele et al., 2002). A search in the available databases led us to identify putative homologs of AtE2Ff in human, mouse, rat, and Caenorhabditis elegans. The alignment of these sequences confirmed that the N-terminal DNA binding domain showed the highest identity to the E2F family members, whereas the other is more similar to the DP DNA binding domain, in agreement with previous reports. It also revealed that the three branches (typical E2F, atypical E2F, and DP) can be distinguished (Figure 1B). While this work was in progress, mammalian cDNAs with homology to Arabidopsis AtE2Ff were reported (De Bruin et al., 2003; Di Stefano et al., 2003).
Figure 1.
Structural and Phylogenetic Organization of Atypical E2F Family Members and Molecular Characterization of the AtE2Ff Protein.
(A) Structure of the AtE2Ff gene showing introns (black boxes), exons, and noncoding regions (white boxes). The two DNA binding domains are shaded as indicated.
(B) Dendrogram of E2F and DP superfamilies calculated using the ClustalW 1.81 program. Accession numbers of the atypical branch (shadowed) of E2F factors are as follows: AtE2Ff (AB074532), AtE2Fe (AB074533), AtE2Fd (AB074531), HsE2F7 (XP084871), MmE2F7 (XP196008), RnE2F7 (XP235118), RnE2F8 (XP218601), and CeE2F7 (NP495771).
(C) Electrophoretic mobility shift assay (EMSA) with MBP-AtE2Ff (50 ng) using an oligonucleotide containing the consensus E2F motif (Ramirez-Parra and Gutierrez, 2000) with different additions, as indicated: wild-type (wt com) or mutated (mut com) competitor oligonucleotide and recombinant wheat DP (100 ng) or maize RBR (100 ng). TmE2F protein (50 ng) was used as control.
(D) Interaction of AtE2Ff with the indicated proteins in the yeast two-hybrid system. Saccharomyces cerevisiae HF7c cells were cotransformed with the indicated plasmids expressing AtE2Ff or TmE2F protein fused to the Gal4 activation domain (Gal4AD) and TmDP, ZmRBR, or AtE2Ff protein fused to the Gal4 DNA binding domain (Gal4BD), as indicated. Galactosidase activity was expressed as Miller units. Data correspond to two independent experiments, which were performed in triplicate.
(E) Transactivation assay of AtE2Ff in the yeast two-hybrid system. HF7c yeast cells were transformed with plasmids expressing Gal4BD alone (vector) or fused to AtE2Ff or TmE2F (positive control), as indicated. Galactosidase activity was measured as in (D).
We show that purified AtE2Ff alone is fully able to bind in vitro to a consensus E2F binding site (Figure 1C), in agreement with previous data (Mariconti et al., 2002) and in contrast with others (Egelkrout et al., 2002), likely because of the use of a truncated protein in this case. AtE2Ff binding was specific because it can be competed out with an excess of a DNA containing an E2F binding site (Figure 1C, wt com) but not with the same DNA carrying a point mutation that destroys the binding site (Figure 1C, mut com). AtE2Ff-DNA complex formation was not affected by the presence of either DP or a plant RBR protein (Figure 1C). Interaction of DP with wheat (Triticum aestivum) E2F, which needs it for DNA binding, was included as a control (Figure 1C). We also confirmed that the full-length AtE2Ff protein does not homodimerize or heterodimerize with DP in the yeast two-hybrid system (Figure 1D; Kosugi and Ohashi, 2002a). AtE2Ff did not interact efficiently with the RBR protein (Figure 1D) and did not show any detectable transactivation ability in yeast (Figure 1E). Therefore, we conclude that AtE2Ff encodes a member of the E2F/DP family, related to mammalian E2F7, that interacts with DNA containing consensus E2F binding sites in a DP-independent manner and that DNA binding in vitro is not significantly affected by RBR.
AtE2Ff mRNA Accumulates in S-Phase upon Reentry into the Cell Cycle
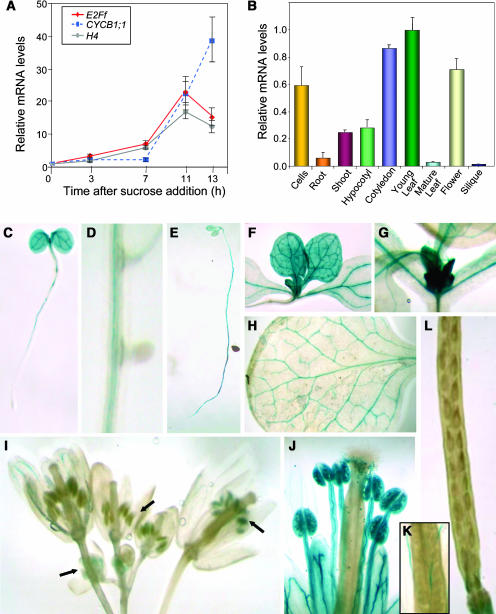
To study whether AtE2Ff expression depends on the proliferative stage, we determined the AtE2Ff mRNA levels in Arabidopsis cell suspension cultures. Sucrose starvation arrests Arabidopsis cell proliferation, which is resumed synchronously upon sucrose addition (Menges and Murray, 2002). Using this protocol (Figure 2A), we found that AtE2Ff gene expression was shut down in arrested cells, and it was stimulated upon reentry in the cell cycle, reaching a maximum in mid-S-phase (Menges and Murray, 2002). This coincided with histone H4 gene expression (Figure 2A) that occurs during most of the S-phase (Reichheld et al., 1998) and preceded that of the cyclinB1;1 gene (Figure 2A), a marker of G2/M (Doerner et al., 1996). Similar results were obtained in aphidicolin-arrested cells (data not shown). These results suggest that AtE2Ff may play a role after S-phase in proliferating cells, although this aspect was not further analyzed in this study.
Figure 2.
Expression Pattern of AtE2Ff.
(A) Cell cycle–dependent expression of the AtE2Ff gene. Expression was determined using real-time RT-PCR on mRNA prepared from cells released after 24 h of sucrose starvation. We used the levels of histone H4 as a marker of S-phase cells, of AtCYCB1;1 as a G2/M marker, and of AtACT2 as a loading control. Values were made relative to the mRNA amount detected at the zero time point for each gene.
(B) Expression pattern of AtE2Ff analyzed by real-time RT-PCR. Measurements were normalized to the amount of AtACT2 and then the AtE2Ff values made relative to the amount present in young leaves. Samples were prepared from asynchronous growing cells, 2-week-old roots, 4-d-old hypocotyls (dark grown) and cotyledons, 10-d-old first leaves, 4-week-old rossette leaves (leaf #1/2), flowers at different stages, and mature siliques.
(C) to (L) Histochemical localization of GUS activity in Arabidopsis pE2Ff:GUS transgenic plants. Four-day-old seedlings grown in the light (C); ten-day-old light-grown seedlings with developing lateral roots (D); four-day-old seedlings grown in the dark (E); young leaves and leaf primordia (F) and (G); mature leaves (H); flowers at different stages of development ([I], arrows); a mature flower showing stamens with pollen (J) and pistils (K) and a mature silique (L).
AtE2Ff Shows an Organ- and Developmental Stage-Specific Expression Pattern
To gain insight into the role of AtE2Ff in planta, AtE2Ff mRNA levels were analyzed by real-time RT-PCR in samples prepared from different organs. A high expression of the AtE2Ff gene occurred in young cotyledons and leaves, hypocotyls, and roots, whereas it is moderate in flowers (see also below) and barely detectable in mature leaves and siliques (Figure 2B).
The spatial regulation of the AtE2Ff promoter was analyzed in detail using transgenic plants expressing the β-glucuronidase (uidA) reporter gene under the AtE2Ff promoter (pE2Ff:GUS plants). In 4-d-old seedlings, we observed high levels of expression in cotyledons, the shoot apical meristem, and the differentiated part of the root (Figure 2C). Interestingly, neither the primary root meristem (Figure 2C) nor the emerging lateral roots showed detectable AtE2Ff gene expression (Figure 2D). Dark-grown seedlings had a similar expression pattern (Figure 2E). These results indicate that AtE2Ff expression occurs in some, but not all, proliferative tissues. In 10-d-old seedlings, AtE2Ff expression has disappeared in cotyledons, whereas leaf primordia and young leaves showed a strong expression (Figures 2F and 2G). Developing trichomes had a moderate AtE2Ff expression level that disappeared in mature leaves (Figure 2H). In older plants, AtE2Ff expression was largely restricted to the vascular tissue and flower buds, whereas AtE2Ff expression was detectable at early stages of developing anthers (Figure 2I, left arrow), likely during active cell proliferation and/or meiosis of pollen mother cells. Then, AtE2Ff expression transiently disappeared at approximately flower stage 10 (Figure 2I, middle arrow); later, it was again very prominent in mature pollen grains (Figures 2I, right arrow, and 2J). By contrast, pistils did not show detectable reporter gene activity at any stage of development analyzed (Figure 2I), as was also the case in developing embryos (Figure 2K) and siliques (Figure 2L).
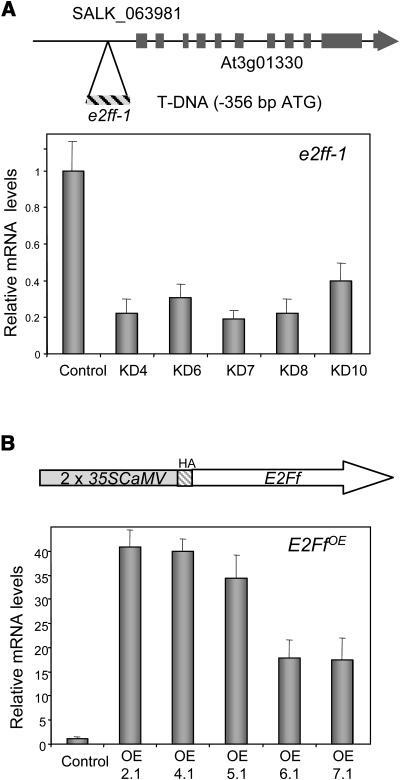
Altered Levels of AtE2Ff mRNA Are Compatible with Plant Growth
To probe the physiological role of AtE2Ff in vivo, we asked whether plants could grow and develop with altered AtE2Ff expression. We identified an Arabidopsis line with a T-DNA insertion (SALK_063981) located 356 bp upstream from the putative initiator ATG (Figure 3A). Homozygous plants (called e2ff-1) showed an approximately threefold to fivefold reduction in AtE2Ff mRNA levels (Figure 3A), indicating that AtE2Ff promoter activity is affected. We also generated several independent lines expressing AtE2Ff under the constitutive 35S promoter of Cauliflower mosaic virus (called E2FfOE) in which AtE2Ff expression was between ∼15- and ∼40-fold higher than in the controls transformed with an empty vector (Figure 3B). Thus, we conclude that plant development is compatible with altered AtE2Ff expression. Both e2ff-1 and E2FfOE plants had a rather normal architecture, including mature leafs and flowers, as well as normal pollen morphology and viability.
Figure 3.
Arabidopsis Plants with Altered Levels of AtE2Ff mRNA.
(A) Scheme of the AtE2Ff locus to show the position of the T-DNA insertion site (e2ff-1, line SALK_063981) in the putative promoter (top panel). Decreased AtE2Ff mRNA levels of several homozygous plants (7-d-old seedlings; e2ff-1 plants; bottom panel). Values are the average of at least three measurements performed on different cDNAs normalized to the amount of AtACT2 mRNA. Control refers to transgenic plants transformed with an empty vector.
(B) Scheme of the construct used to generate AtE2Ff overexpressor plants (E2FfOE) under the control of the constitutive 35S promoter of Cauliflower mosaic virus (CaMV) (top panel). Determination of AtE2Ff mRNA levels in these plants was performed as described in (A). Two lines (OE2.1 and OE4.1) were used for further experiments.
AtE2Ff Is a Regulator of Cell Growth in Roots and Hypocotyls
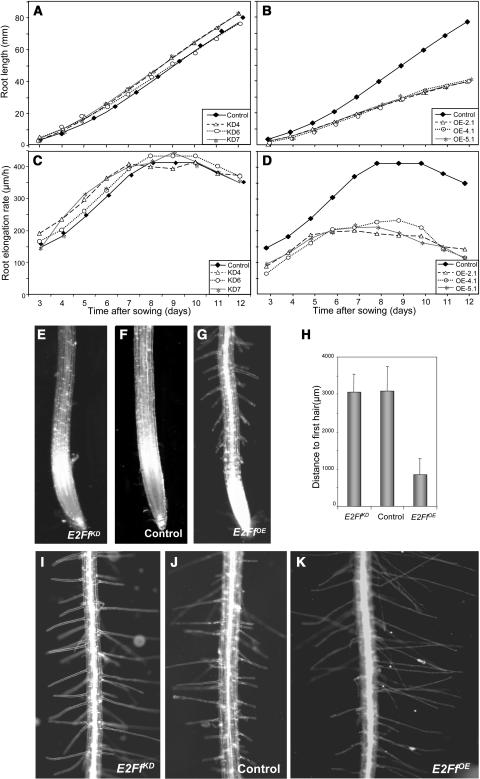
AtE2Ff is highly expressed in dark-grown hypocotyls as well as in the upper region of the roots, locations that contain differentiated cells, but not in the root meristem. Because in these organs cell proliferation and differentiation are temporally and/or spatially separated and organ growth relies largely on cell expansion in one axis, we chose to focus this study on roots and hypocotyls. The final root length and growth rate (Figures 4A and 4C), as well as the size of the transition zone (Table 1), of e2ff-1 plants were indistinguishable from the controls. By contrast, these three parameters were severely reduced (∼50%) in E2FfOE plants (Figures 4B and 4D, Table 1). However, altered levels of E2Ff did not affect meristem size (Table 1). AtE2Ff overexpression also produced a severalfold reduction in the distance from the root tip to the point at which the first root hairs develop (Figures 4E to 4H). At the cellular level, the size of trichoblasts at this point is reduced by 25% in the E2FfOE plants, an effect that was similar in fully developed trichoblasts (Table 1). These data suggest that cell expansion is reduced by an excess of E2Ff once the cells exit the meristem. Hair length was larger in the E2FfOE plants than in the controls (Figures 4I to 4K, Table 1). An increased hair length, concomitant with a decreased trichoblast size, has been reported when root hair development is altered (Lopez-Bucio et al., 2003).
Figure 4.
AtE2Ff Affects Root Growth and Morphology.
(A) and (B) Root growth in vertical plates of control and different lines of e2ff-1 plants (A) and control and different lines of E2FfOE plants (B). Measurements are derived from 30 seedlings for each line indicated in the inset.
(C) and (D) Root elongation rate of control and e2ff-1 plants (C) and control and E2FfOE plants (D).
(E) to (G) Morphology of the root tip in control, e2ff-1, and E2FfOE plants (7 d old) as indicated.
(H) Quantification of the distance from the tip to the first hair in control, e2ff-1, and E2FfOE plants grown as in (E) to (G).
(I) to (K) Root hair morphology of control, e2ff-1, and E2FfOE plants (7 d old) as indicated.
Table 1.
Effect of AtE2Ff Misregulation on Root Growtha
| e2ff-1 | Control | E2FfOE | |
|---|---|---|---|
| Root length (mm) | 32.5 ± 2.0 | 29.8 ± 1.6 | 18.1 ± 2.9* |
| Root growth rate (μm/h) | 341.7 ± 31.8 | 371.4 ± 27.8 | 210.7 ± 36.8* |
| Meristem length (μm) | 673.6 ± 47.3 | 657.5 ± 36.4 | 646.3 ± 75.5 |
| Transition zone length (μm) | 2058.9 ± 202.6 | 2093.9 ± 187.6 | 513.3 ± 268.4* |
| Young trichoblast length (μm) | 62.8 ± 13.2 | 61.2 ± 12.3 | 45.8 ± 12.3* |
| Mature trichoblast length (μm) | 159.5 ± 14.0 | 164.6 ± 31.2 | 122.3 ± 24.3* |
| Hair length (μm) | 179.5 ± 86.2 | 177.2 ± 91.7 | 249.5 ± 101.2* |
| Hair density (number per mm) | 31.2 ± 10.7 | 29.8 ± 10.3 | 36.2 ± 13.6* |
Values indicate the mean ± sd. Asterisks indicate values whose differences with the control measurements are statistically significant (P < 0.01).
Root growth measured in 7-d-old plants.
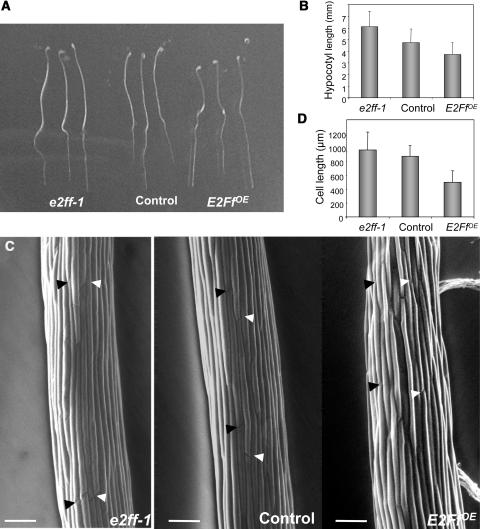
Hypocotyl growth can be more readily analyzed in dark-grown seedlings. We found that e2ff-1 and E2FfOE plants showed ∼30% longer and shorter hypocotyls, respectively, compared with controls (Figures 5A and 5B). This change in organ size was not because of changes in cell number but because of a change in cell size (Figures 5C and 5D). Hypocotyl growth in the light was affected in a similar way, although the final effect on cell size was less pronounced than in the dark (data not shown). These data together indicate that AtE2Ff regulates cell size, an effect particularly evident in hypocotyls.
Figure 5.
AtE2Ff Affects Cell Elongation in Dark-Grown Hypocotyl Cells.
(A) Seedlings germinated in darkness for 4 d.
(B) Length of hypocotyls grown as described in (A).
(C) Scanning electron microscopy of epidermal hypocotyl cells of e2ff-1, control, or E2FfOE transgenic seedlings grown in darkness for 4 d. Bar = 100 μm. Arrowheads mark cellular ends of different cells.
(D) Length of epidermal hypocotyl cells grown as described in (B).
Altered Levels of AtE2Ff Do Not Affect Cell Proliferation in Roots and Hypocotyls
The data shown above indicate that AtE2Ff seems to be a key regulator of root and hypocotyl growth through changes in cell size and growth rate. Root growth rate is influenced by the rate of cell production in the meristems as well as by cell elongation during differentiation in the transition and differentiated regions of the root (reviewed in Beemster et al., 2003). Hypocotyl growth in the dark is largely dependent on cell expansion that occurs in coordination with endocycles (Gendreau et al., 1997; Sugimoto-Shirasu and Roberts, 2003). To determine the mechanism by which AtE2Ff regulates organ growth, we analyzed first several parameters to address the possible contribution of cell proliferation.
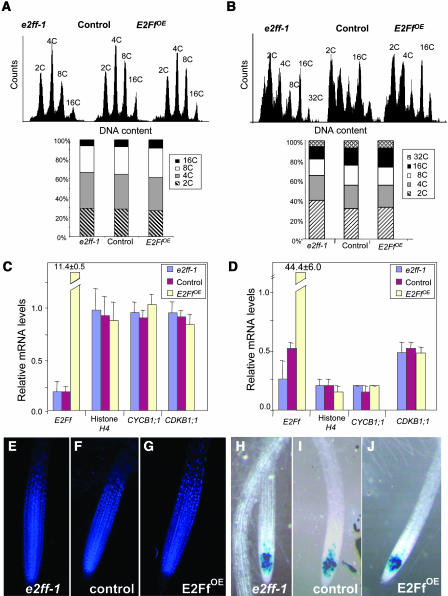
Altered levels of AtE2Ff did not produce significant changes in the nuclear ploidy distribution that was similar to the controls in the roots and hypocotyls of e2ff-1 and E2FfOE plants (Figures 6A and 6B). We also determined by real-time RT-PCR the mRNA levels of three cell cycle marker genes: namely, histone H4 for S-phase (Reichheld et al., 1998), CDKB1;1 for S/G2/M (Fobert et al., 1996; Porceddu et al., 2001), and CYCB1;1 for G2/M (Doerner et al., 1996). We also analyzed other putative E2F targets, such as CDC6a and ORC1b, involved in initiation of DNA replication, containing E2F sites in their promoters and expressed in a cell cycle–regulated manner (Castellano et al., 2001; Ramirez-Parra et al., 2003; S. Diaz-Trivino and C. Gutierrez, unpublished results). However, they are expressed at very low levels in these organs and were not useful for whole-organ analysis (data not shown). The levels of these cell cycle marker genes in whole root and hypocotyl extracts did not seem to be affected in e2ff-1 and E2FfOE plants (Figures 6C and 6D), suggesting that the amount of proliferating cells or their status was not dramatically affected by AtE2Ff.
Figure 6.
Effect of AtE2Ff on Cell Proliferation.
(A) Ploidy distribution of root nuclei in 7-d-old e2ff-1, control, and E2FfOE plants.
(B) Ploidy distribution of hypocotyl nuclei in 7-d-old e2ff-1, control, and E2FfOE plants.
(C) mRNA levels of cell cycle markers in 7-d-old roots. Measurements and normalization were performed as in Figure 3A.
(D) mRNA levels of cell cycle markers in 7-d-old hypocotyls. Measurements and normalization were performed as in Figure 3A.
(E) to (G) Nuclear distribution (DAPI staining) in the root meristem of e2ff-1, control, and E2FfOE plants (7 d old) as indicated.
(H) to (J) Detection of G2 cells (CYCB1;1:GUS positive cells) in 4-d-old e2ff--1, control, and E2FfOE plants as indicated.
We also analyzed by 4′,6-diamidino-2-phenylindole (DAPI) staining the overall morphology of the root meristems, which in the e2ff-1 and E2FfOE plants appeared to be similar to that of controls (Figures 6E to 6G). We also expressed a translational fusion of CYCB1;1 to the uidA (GUS) gene (Colón-Carmona et al., 1999) that allows identification of G2/M cells in e2ff-1, control, and E2FfOE plants. Altered levels of AtE2Ff did not seem to produce dramatic changes in the amount of cycling cells present in the root meristem (Figures 6H to 6J). Similar analysis performed in the hypocotyl, where CYCB1;1 is not detected in the controls, revealed that GUS expression was negative in the e2ff-1 and E2FfOE plants (data not shown). Altogether these data indicate that the phenotypic effects of altered levels of AtE2Ff in roots and hypocotyls were not mediated by changes in cell proliferation markers.
AtE2Ff Binds in Vivo to the Promoters of Cell Wall Biosynthesis Genes and Regulates Their Expression
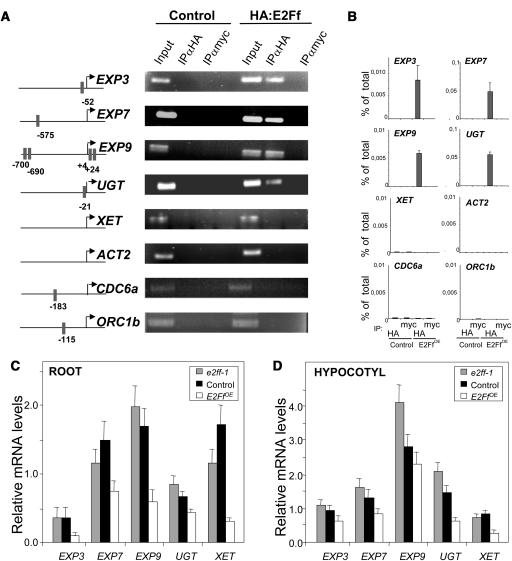
The effect of altering AtE2Ff mRNA on cell growth and size suggests that cellular processes other than those directly controlling cell cycle transitions may be major targets of the AtE2Ff action. Previous studies have revealed the presence of E2F binding sites in promoters of cell cycle and non-cell-cycle functional categories (Ramirez-Parra et al., 2003; Vlieghe et al., 2003). Root and hypocotyl growth relies extensively on individual cell expansion, a process that occurs after cells stop proliferation and differentiate, and depends on increase in cell wall biosynthesis (Sugimoto-Shirasu and Roberts, 2003). Thus, we looked for the presence of consensus E2F binding sites in the promoters of genes involved in cell wall biosynthesis, including expansins, which among others are key players in cell wall growth (Vissenberg et al., 2000; Li et al., 2003).
Although the list of genes searched is not complete, we found that, interestingly, several of them contained consensus E2F binding sites in their putative promoters. These were three expansin genes, EXP3 (5′-CTTCCCGC-3′ at −52 from the putative ATG), EXP7 (5′-TCTCCCGC-3′ at −575), and EXP9 (5′-TATGGCGG-3′ at +24, 5′-TTCGCCGC-3′ at +4, 5′-ATTCGCGG-3′ at −690, and 5′-TTCGCCGC-3′ at −700). In addition, we included in our analysis two genes, the xyloglucan-endo-[1,4,β-d-glucanase] transglycosylase (XET) and the UDP-glucose-glycosyl transferase (UGT), which are upregulated in E2Fa-DPa overexpressing plants (Vlieghe et al., 2003). Whereas UGT contains one E2F binding site in its promoter (5′-TTTCGCGC-3′ at −21 from the putative ATG), XET does not (Figure 7A, left panel).
Figure 7.
AtE2Ff Binds in Vivo to the Promoters of Cell Wall Biosynthesis Genes and Regulates Their Expression.
(A) Scheme of the cell wall biosynthesis gene promoters studied. Gray boxes indicate potential E2F binding sites and their positions relative to the putative ATG (left panel). Chromatin immunoprecipitation (ChIP) analysis of 12-d-old Arabidopsis seedlings using antibodies specific for the HA-epitope with control or HA:E2Ff overexpressing plants (right panel). Anti-myc was used as a negative control IgG and conventional PCR was made using 1 μL of 50 μL of total eluate. Immunoprecipitated genomic DNA was amplified with primers specific for the indicated promoters using between 37 and 45 cycles of amplification, depending of each promoter. The actin-2 (ACT2) promoter was used as a negative control.
(B) ChIP analysis of AtE2Ff binding to promoters in extracts prepared as described in (A) using real-time PCR.
(C) and (D) Expression profile of cell wall biosynthesis genes in 10-d-old roots (C) and dark-growth hypocotyls (D) of e2ff-1, control, and E2FfOE plants. Measurements and normalization were performed as in Figure 6C.
To address whether these genes behave as direct E2F targets in vivo, we used a chromatin immunoprecipitation approach taking advantage of the plants that express an HA-tagged version of AtE2Ff. Nuclei with cross-linked chromatin were purified from E2FfOE seedlings and sonicated, and chromatin was isolated as described in Methods. Specific immunoprecipitation was conducted with an anti-HA antibody, and an anti-Myc antibody was used as an unspecific control IgG. AtACT2 was used as a control for a non-E2F regulated gene. As shown in Figure 7A (right panel), promoter sequences that do not contain E2F consensus binding sites, such as those of XET and ACT2 genes, were not recovered from the immunoprecipitates with either anti-HA or anti-Myc antibodies. However, promoter fragments of the EXP3, EXP7, EXP9, and UGT genes, which contain E2F binding sites, were specifically amplified from the anti-HA immunoprecipitates of E2FfOE extracts. AtE2Ff binding was also quantitatively determined using real-time PCR of immunoprecipitates with either anti-HA or anti-myc antibodies. The results fully corroborated the specific binding of AtE2Ff to these promoters in vivo (Figure 7B) and strongly suggested that it regulates their expression. Both AtCDC6a and AtORC1a genes, also included in this analysis, did not show detectable binding to AtE2Ff (Figures 7A and 7B).We next determined mRNA levels of this set of putative E2F target genes using real-time RT-PCR in root and hypocotyl extracts of e2ff-1, control, and E2FfOE plants (Figures 7C and 7D). The expression level of all these genes was reduced significantly in root (Figure 7C) and hypocotyl (Figure 7D) extracts of E2FfOE plants. These effects were particularly evident for EXP3, EXP9, and XET in roots (Figure 7C) and for UGT and XET in hypocotyls (Figure 7D). Significant changes were also found in e2ff-1 plants where an increase in mRNA levels (e.g., EXP9) was clearly observed in hypocotyls (Figure 7D). It should be noted that mRNA levels of AtE2Ff in e2ff-1 plants do not change significantly in roots (Figure 7C). As already described above, AtCDC6a and AtORC1b expression in differentiated organs (e.g., roots and hypocotyls) was below detectable levels (Castellano et al., 2001; data not shown). Consequently, a correlation between expression of these genes and binding of AtE2Ff to their promoters is not straightforward. These data together demonstrate that a set of cell wall biogenesis genes are likely direct E2F targets in vivo and uncover a role of AtE2Ff in their transcriptional regulation. This role of AtE2Ff may have a significant impact on cell wall growth and may explain, at least in part, the root and hypocotyl phenotypes of e2ff-1 and E2FfOE plants.
DISCUSSION
The mechanisms controlling the balance among cell proliferation, growth, and differentiation are important for development in multicellular organisms. This is crucial in plants because postembryonic growth relies exclusively on continuous proliferation and differentiation throughout the entire life of the organism. The gene networks that operate during cell division and differentiation are complex and poorly understood (Gutierrez et al., 2002; Dewitte and Murray, 2003). In this study, we define an unforeseen role of an Arabidopsis E2F in regulating the expression of a subset of genes involved in plant cell wall growth. This role is particularly striking in organs, such as hypocotyls and roots, where growth relies largely on cell enlargement in one axis. Our results also suggest that the E2F network, or part of it, may function as a checkpoint that coordinates cell cycle control and cell wall growth.
A Unique Subfamily of E2F/DP Transcription Factors in Plants and Animals
Completion of the Arabidopsis genome sequencing led to the identification of three novel members of the E2F/DP family of transcription factors that have received different names (De Jager et al., 2001; Kosugi and Ohashi, 2002a; Mariconti et al., 2002; Vandepoele et al., 2002). We favor the acronyms E2Fd, E2Fe, and E2Ff to highlight the ability of these proteins to interact with consensus E2F sequences, to regulate the expression of genes with E2F sites in their promoters, and to reinforce its structural similarity with animal counterparts (e.g., mammalian E2F7) (De Bruin et al., 2003; Di Stefano et al., 2003).
These atypical E2Fs, which have two DNA binding domains (Egelkrout et al., 2002; Kosugi and Ohashi, 2002a; Mariconti et al., 2002; Stevens et al., 2002), bind to E2F consensus sequences in the absence of DP. This suggests that they have evolved to fulfill in a single molecule the requirements of contacts with DNA. Computer modeling indicates that the two binding domains fit into the human E2F4-DP2 crystal structure (Zheng et al., 1999; R. Campos-Oliva, unpublished data). Another striking feature is that these atypical E2Fs do not need to dimerize with DP for DNA binding and transient gene expression in cultured cells (Egelkrout et al., 2002; Kosugi and Ohashi, 2002a, 2002b; Mariconti et al., 2002) and do not interact with plant RBR (this work).
While this work was in progress, mouse and human cDNAs encoding E2F7, whose domain organization is similar to the Arabidopsis E2Fd-f, were identified (De Bruin et al., 2003; Di Stefano et al., 2003). Homologs in other organisms, such as C. elegans and rat, are also available in the databases. These findings expand the complexity of the E2F/DP family of transcription factors.
Role of AtE2Ff in Cell Growth Control
In cultured plant cells, overexpression of AtE2Fd-f represses the E2F responsive promoters of tobacco (Nicotiana tabacum) and rice (Oryza sativa) PCNA genes (Kosugi and Ohashi, 2002b). Likewise, mammalian E2F7 represses E2F target genes (Di Stefano et al., 2003). However, the effects of overexpressing human E2F7 in cultured cells on the cell cycle are relatively small (De Bruin et al., 2003; Di Stefano et al., 2003). We have found that altered levels of AtE2Ff mRNAs do not lead to changes in the nuclear ploidy distribution in roots and hypocotyls. We do not detect significant changes in the expression of cell cycle markers (histone H4, cyclin B1;1, and CDKB1;1) or binding of AtE2Ff in vivo to the promoters of AtCDC6a and AtORC1b, which contain E2F binding sites. In proliferating human cells, association of E2F7 to promoters increases in S-phase, but only in a subset of them, whereby repressing genes required for cell cycle progression (Di Stefano et al., 2003). If a similar situation occurs in Arabidopsis cultured cells, AtE2Ff may cooperate in repressing E2F target genes that may have been activated by another E2F family member (e.g., AtE2Fa) earlier during the cell cycle. Although the peak of AtE2Ff expression in mid-S-phase would be consistent with that idea, this is an aspect open for future studies. It should be kept in mind that a large number of cell cycle genes are highly expressed in suspension cultured cells (Menges and Murray, 2002), and this may not necessarily represent the situation in planta. The responsiveness of E2F targets to AtE2Ff may also depend on the organ. Thus, it is tempting to speculate that AtE2Ff can compete for the occupancy of E2F site responsive promoters, depending on the temporal and spatial availability of different E2Fs.
AtE2Ff is expressed in dark-growing hypocotyls and elongating roots. Growth of these organs is severely reduced in E2FfOE plants. In e2ff-1 plants, significant phenotypic effects were not observed in roots, where AtE2Ff mRNA remains at almost normal levels, whereas hypocotyl growth is enhanced as a consequence of an increase in cell size. In this context, in etiolated seedlings, reduced expression of CDKB1;1 (formerly Cdc2b) produces short hypocotyls, by inhibition of cell elongation, without changes in ploidy levels (Yoshizumi et al., 1999). CDKB1;1 contains E2F binding sites in its promoter (De Jager et al., 2001) and is upregulated in AtE2Fa/AtDPa overexpressing seedlings (Vlieghe et al., 2003). However, we have not found changes in CDKB1;1 expression in either E2FfOE or e2ff-1 plants. Based on the lack of changes in cell proliferation markers, our results strongly suggest that the phenotypic effects of altering AtE2Ff levels are not primarily mediated by modifications of cell proliferation. Furthermore, we observed no significant changes in the amount of cycling cells in the root meristem as deduced from the analysis of CYCB1;1:GUS expression in either a E2FfOE or e2ff-1 background. In any case, the possible effects on cell proliferation may be indirect and organ specific. For example, a reduction in cell production in the root meristem may contribute to a reduced root growth rate (Beemster et al., 2003). AtE2Fa induces ectopic divisions and inhibits cell differentiation (De Veylder et al., 2002). Thus, detailed future analysis would be needed to address possible combinatorial effects of different E2Fs on cell differentiation and cell cycle transitions in planta.
In proliferating and differentiating cells, cell growth involves an increase in nuclear, cytoplasmic, and cell wall components. Cell wall growth relies on the remodeling of the preexisting cell wall, a process that requires integration of structural elements, cell wall loosening, and polymerization of new components (for review, see Showalter, 1993; Cosgrove, 1997). We found that a set of genes encoding enzymatic activities key for cell wall growth contain E2F binding sites in their promoters and bind AtE2Ff in vivo. Thus, particularly in roots and hypocotyls, the expression of XET and UGT genes as well as that of three expansin genes (EXP3, EXP7, and EXP9) is affected by AtE2Ff levels. Interestingly, in AtE2Fa/DPa overexpressor plants, a large proportion (∼16%) of the upregulated genes belong to the cell wall biosynthesis category, some of which contain E2F sites in their promoters (Vlieghe et al., 2003). This provides further support to a possible role of AtE2Ff in roots and hypocotyls by regulating genes involved in cell wall biosynthesis. AtE2Ff does not act as an activator in reporter gene analysis (Kosugi and Ohashi, 2002b; Mariconti et al., 2002), and it reduces the mRNA levels of a set of genes (this study). Thus, it is conceivable that it acts as a repressor in differentiated cells. This is supported by the reduced cell size of young trichoblasts in the E2FfOE plants, consistent with an inhibition of cell expansion once the cells exit the meristem. However, at present the role of AtE2Ff seems to be organ specific and cannot be extrapolated to other organs as indicated by the macroscopic normal phenotype of mature leaves and flowers.
The emerging picture for the E2F regulatory network is much more complex than previously anticipated. This is in agreement with data from other systems where an increasing list of E2F targets are also expressed in a temporally and spatially concerted manner (Ren et al., 2002; Stevaux and Dyson, 2002; Cam and Dynlacht, 2003; Dimova et al., 2003). Genomic approaches in Arabidopsis have revealed that genes belonging not only to the cell cycle but also to other functional categories are likely regulated by E2F/DP (Ramirez-Parra et al., 2003; Vlieghe et al., 2003). The temporal and tissue/organ-specific expression pattern of AtE2F genes (Del Pozo et al., 2002; De Veylder et al., 2002; this work) may be crucial for the transcriptional response of E2F targets. Consistent with this, differences in the regulation of putative E2F target genes in different organs are observed. Our studies provide support for a link between AtE2Ff and cell expansion, which we have revealed here for hypocotyls and roots, through the transcriptional regulation of cell wall biosynthesis genes. However, this is likely only part of the organ- and developmental stage-specific role played by AtE2Ff. A detailed understanding of the complex network of transcriptional regulation dependent on different combinations of E2F/DP factors awaits future studies as well as detailed analysis of profiling data and phenotypic effects in plants with altered levels of E2F activities.
METHODS
Protein Interaction and EMSA
The full-length AtE2Ff (At3g01330) cDNA was obtained by PCR of Arabidopsis thaliana cultured cells. Yeast two-hybrid assays were performed as described (Ramirez-Parra and Gutierrez, 2000). Plasmid pGBT-AtE2Ff and pGAD-AtE2Ff were generated by cloning the full-length AtE2Ff coding sequence in frame into the pGBT8 and pACT2 vectors (Clontech, Palo Alto, CA). Quantification of β-galactosidase assays was done in liquid culture using o-nitrophenyl-β-d-galactopyranoside (Sigma, St. Louis, MO) as substrate (Miller, 1972).
EMSA was performed using purified MBP-AtE2Ff, MBP-TmDP, GST-TmE2F, or His-RBR as described (Ramirez-Parra and Gutierrez, 2000). Plasmid pMBP-AtE2Ff was constructed by cloning the AtE2Ff coding sequence in frame into the pMal-c2 vector (New England Biolabs, Beverly, MA), transferred to Escherichia coli BL21(DE3), and the recombinant protein purified using amylose beads (New England Biolabs).
Cell Culture Synchronization
Arabidopsis MM2d cultured cells were used (Menges and Murray, 2002). Cell cycle arrest by sucrose starvation and synchronization with aphidicolin were performed as described by Menges and Murray (2002) and Ramirez-Parra et al. (2003), respectively.
Transgenic Plants
For expression analysis, 850 bp of the genomic region containing the AtE2Ff promoter was fused to the GUS coding sequence in a pBI101.1 vector (Jefferson et al., 1987) and used for transformation of Arabidopsis (pAtE2Ff:GUS) plants. For ectopic expression studies, the AtE2Ff cDNA was cloned in frame with the HA epitope using pPily vector (Ferrando et al., 2000), and subsequently into the pROK2 binary vector (Baulcombe et al., 1986), under the control of the 35S promoter of Cauliflower mosaic virus. In all cases, Arabidopsis (Columbia-0 ecotype) was transformed with Agrobacterium tumefaciens C58CRifR by the floral dip method (Clough and Bent, 1998). Transformed seedlings (T0 generation) were selected on MS agar plates containing 50 μg mL−1 of kanamycin and transferred to soil. T2 homozygous plants were selected for further analysis. Histochemical detection of GUS activity was done using 5-bromo-4-chloro-3-indolyl-β-d-glucuronide with slight modifications (Jefferson et al., 1987). The Arabidopsis T-DNA insertion line SALK-063981 was obtained from the ABRC.
RNA Extraction, RT-PCR, and Real-Time PCR
Total RNA was extracted using the Trizol reagent (Invitrogen, Carlsbad, CA), and RT-PCR was performed with the ThermoScript RT system (Invitrogen). The LightCycler system with the FastStart DNA Master SYBR Green I (Roche, Indianapolis, IN) was used for real-time quantitative RT-PCR. The concentration of ubiquitin10 (AtUBQ10) or actin (AtACT2) mRNAs in each sample was determined to normalize for differences of total RNA amount. The data were derived from two independent experiments performed in duplicate, and in the case of the analysis of transgenic plants, at least two independent lines were used. To avoid amplification of contaminating genomic DNA, primers were designed for scanning exon–exon junctions. The primer sequences used are available upon request.
Optical and Scanning Electron Microscopy
For scanning electron microscopy, a FEI QUANTA 200 microscope (FEI, Philips, The Netherlands) was used in ambient mode or low vacuum conditions with unfixed material. The images of dark-grown hypocotyls were processed for cell length and cell area measurements using calibrated ImageJ software (NIH version 1.27). At least 200 cells taken from eight different cotyledons, leaves, or hypocotyls were measured. The length of root hairs was measured and the number of root hair cells counted in a 1-mm section of the root where trichoblasts had fully expanded. To measure trichoblast cell length, 160 mature trichoblasts from 10 different roots were examined, photographed with a digital Coolsnap FX camera (Roper Scientific, Trenton, NJ) mounted in an Axioskop2 plus microscope (Zeiss, Jena, Germany), and processed with the ImageJ software. Nuclear visualization was done by staining with DAPI (0.1 μg/mL) for 2 h. Samples were washed and analyzed by fluorescence microscopy using an Axioskop2 Plus microscope (Zeiss) and the images captured with a digital Coolsnap FX camera (Roper Scientific).
Root Growth Measurement
For analysis of root growth, root length was measured every 24 h after germination for a period of 13 d, during which seedlings were grown in a vertical position. Length was basically determined as described (De Veylder et al., 2001). Digitized images from scanning were processed using the ImageJ software. Average growth rate was calculated by expressing daily growth as a function of time.
Flow Cytometry Measurements
Roots and hypocotyls were chopped and resuspended in cold nuclear isolation buffer (Galbraith et al., 1983). This crude preparation of isolated nuclei was filtered through 60-μm nylon mesh and stained with propidium iodide (50 μg/mL; Sigma). DNA histograms corresponding to 104 isolated nuclei were made with a FACScalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ).
ChIP Assays
ChIP assays and data analysis were performed basically as previously described (Gendrel et al., 2002). Briefly, whole 12-d-old Arabidopsis seedlings were treated with 1% formaldehyde under vacuum and then cross-linking reaction was stopped with 0.125 M Gly. Arabidopsis nuclei were extracted, lysed in SDS buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, and 1% SDS), and sonicated to shear DNA to an average size of 700 to 1500 bp. Crude chromatin lysates were precleared with protein G agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA), blocked with salmon sperm DNA, and then incubated overnight at 4°C with anti-HA (Roche) or anti-Myc 9E10 (Santa Cruz Biotechnology) antibodies. Immunocomplexes were recovered using protein G agarose, extensively washed, and eluted from beads. Cross-links were reversed, the samples treated with proteinase K, and the DNA was recovered after phenol/chloroform extraction by ethanol precipitation. DNA was resuspended in 50 μL of water and 1-μL aliquots were used for real-time or conventional PCR as indicated. The primer sequences used are available upon request.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AB074532.
Acknowledgments
Authors are indebted to S. Llorens-Berzosa for technical assistance. We also thank P. Doerner for CYCB1;1:uidA reporter Arabidopsis seeds, the ABRC stock center for seeds, Bayer CropScience for the Arabidopsis MMd2 cell suspension, P. Lastres (Centro Investigaciones Biologicas-CSIC) and L. Tormo (Museo Nacional Ciencias Naturales-CSIC) for their help with flow cytometry and scanning electron microscopy, respectively, and E. Martinez-Salas for comments on the manuscript. This work has been partially supported by grants BMC2000-1004 and BMC2003-2131 (Ministerio de Ciencia y Tecnologia), QLRT/1999/00454 (ECCO-EU), 07G/0033/00, and 07B/053/2002 (Comunidad de Madrid) and by an institutional grant from Fundación Ramón Areces.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Crisanto Gutierrez (cgutierrez@cbm.uam.es).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.023978.
References
- Albani, D., Mariconti, L., Ricagno, S., Pitto, L., Moroni, C., Helin, K., and Cella, R. (2000). DcE2F, a functional plant E2F-like transcriptional activator from Daucus carota. J. Biol. Chem. 275, 19258–19267. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D.C., Saunders, G.R., Bevan, M.W., Mayo, M.A., and Harrison, B.D. (1986). Expression of biologically-active viral satellite RNA from the nuclear genome of transformed plants. Nature 321, 446–449. [Google Scholar]
- Beemster, G.T.S., Fiorani, F., and Inzé, D. (2003). Cell cycle: The key to plant growth control? Trends Plant Sci. 8, 154–158. [DOI] [PubMed] [Google Scholar]
- Cam, H., and Dynlacht, B.D. (2003). Emerging roles for E2F: Beyond the G1/S transition and DNA replication. Cancer Cell 3, 311–316. [DOI] [PubMed] [Google Scholar]
- Castellano, M.M., del Pozo, J.C., Ramirez-Parra, E., Brown, S., and Gutierrez, C. (2001). Expression and stability of Arabidopsis CDC6 are associated with endoreplication. Plant Cell 13, 2671–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayirlioglu, P., Ward, W.O., Silver-Key, S.C., and Duronio, R.J. (2003). Transcriptional repressor functions of Drosophila E2F1 and E2F2 cooperate to inhibit genomic DNA synthesis in ovarian follicle cells. Mol. Cell. Biol. 23, 2123–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Colón-Carmona, A., You, R., Haimovitch-Gal, T., and Doerner, P. (1999). Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20, 503–508. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J. (1997). Assembly and enlargement of the primary cell wall in plants. Annu. Rev. Cell Dev. Biol. 13, 171–201. [DOI] [PubMed] [Google Scholar]
- De Bruin, A., Maiti, B., Jakoi, L., Timmers, C., Buerki, R., and Leone, G. (2003). Identification and characterization of E2F7, a novel mammalian E2F family member capable of blocking cellular proliferation. J. Biol. Chem. 278, 42041–42049. [DOI] [PubMed] [Google Scholar]
- De Jager, S.M., Menges, M., Bauer, U.M., and Murray, J.A.H. (2001). Arabidopsis E2F1 binds a sequence present in the promoter of S-phase-regulated gene AtCDC6 and is a member of a multigene family with differential activities. Plant Mol. Biol. 47, 555–568. [DOI] [PubMed] [Google Scholar]
- De Veylder, L., Beeckman, T., Beemster, G.T.S., de Almeida-Engler, J., Ormenese, S., Maes, S., Naudts, M., van der Schueren, E., Jacqmard, A., Engler, G., and Inzé, D. (2002). Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J. 21, 1360–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder, L., Beemster, G.T., Beeckman, T., and Inze, D. (2001). CKS1At overexpression in Arabidopsis thaliana inhibits growth by reducing meristem size and inhibiting cell-cycle progression. Plant J. 25, 617–626. [DOI] [PubMed] [Google Scholar]
- Del Pozo, J.C., Boniotti, M.B., and Gutierrez, C. (2002). Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCF(AtSKP2) pathway in response to light. Plant Cell 14, 3057–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte, W., and Murray, J.A. (2003). The plant cell cycle. Annu. Rev. Plant Biol. 54, 235–264. [DOI] [PubMed] [Google Scholar]
- Di Stefano, L., Jensen, M.R., and Helin, K. (2003). E2F7 a novel E2F featuring DP-independent repression of a subset of E2F-regulated genes. EMBO J. 22, 6289–6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova, D.K., Stevaux, O., Frolov, M.V., and Dyson, N.J. (2003). Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev. 17, 2308–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerner, P., Jorgensen, J.E., You, R., Steppuhn, J., and Lamb, C. (1996). Control of root growth and development by cyclin expression. Nature 380, 520–523. [DOI] [PubMed] [Google Scholar]
- Egelkrout, E.M., Mariconti, L., Settlage, S.B., Cella, R., Robertson, D., and Hanley-Bowdoin, L. (2002). Two E2F elements regulate the proliferating cell nuclear antigen promoter differently during leaf development. Plant Cell 14, 3225–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando, A., Farras, R., Jasik, J., Schell, J., and Koncz, C. (2000). Intron-tagged epitope: A tool for facile detection and purification of proteins expressed in Agrobacterium transformed plant cells. Plant J. 22, 553–560. [DOI] [PubMed] [Google Scholar]
- Fobert, P.R., Gaudin, V., Lunness, P., Coen, E.S., and Doonan, J.H. (1996). Distinct classes of cdc2-related genes are differentially expressed during the cell division cycle in plants. Plant Cell 8, 1465–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith, D.W.H.K., Maddox, J.M., Ayres, N.M., Sharma, D.P., and Firoozabadi, E. (1983). Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220, 1049–1051. [DOI] [PubMed] [Google Scholar]
- Gendreau, E., Traas, J., Desnos, T., Grandjean, O., Caboche, M., and Hofte, H. (1997). Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 114, 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel, A., Lippman, Z., Yordan, C., Colot, V., and Martienssen, R.A. (2002). Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297, 1871–1873. [DOI] [PubMed] [Google Scholar]
- Gutierrez, C., Ramirez-Parra, E., Castellano, M.M., and del Pozo, J.C. (2002). G(1) to S transition: More than a cell cycle engine switch. Curr. Opin. Plant Biol. 5, 480–486. [DOI] [PubMed] [Google Scholar]
- Harbour, J.W., and Dean, D.C. (2000. a). The Rb/E2F pathway: Expanding roles and emerging paradigms. Genes Dev. 14, 2393–2409. [DOI] [PubMed] [Google Scholar]
- Harbour, J.W., and Dean, D.C. (2000. b). Chromatin remodeling and Rb activity. Curr. Opin. Cell Biol. 12, 685–689. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanaugh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi, S., and Ohashi, Y. (2002. a). E2Ls, E2F-like repressors of Arabidopsis that bind to E2F sites in a monomeric form. J. Biol. Chem. 277, 16553–16558. [DOI] [PubMed] [Google Scholar]
- Kosugi, S., and Ohashi, Y. (2002. b). E2F sites that can interact with E2F proteins cloned from rice are required for meristematic tissue-specific expression of rice and tobacco proliferating cell nuclear antigen promoters. Plant J. 29, 45–59. [DOI] [PubMed] [Google Scholar]
- Kosugi, S., and Ohashi, Y. (2003). Constitutive E2F expression in tobacco plants exhibits altered cell cycle control and morphological change in a cell type-specific manner. Plant Physiol. 132, 2012–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L.C., Bedinger, P.A., Volk, C., Jones, A., and Cosgrove, D.J. (2003). Purification and characterization of four beta-expansins (Zea m 1 isoforms) from maize pollen. Plant Physiol. 132, 2073–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bucio, J., Cruz-Ramirez, A., and Herrera-Estrella, L. (2003). The role of nutrient availability in regulating root architecture. Curr. Opin. Plant Biol. 6, 280–287. [DOI] [PubMed] [Google Scholar]
- Mariconti, L., Pellegrini, B., Cantoni, R., Stevens, R., Bergonioux, C., Cella, R., and Albani, D. (2002). The E2F family of transcription factors from Arabidopsis thaliana. Novel and conserved components of the retinoblastoma pathway in plants. J. Biol. Chem. 277, 9911–9919. [DOI] [PubMed] [Google Scholar]
- Menges, M., and Murray, J.A. (2002). Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J. 30, 203–212. [DOI] [PubMed] [Google Scholar]
- Miller, J.H. (1972). Experiments in Molecular Genetics. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Müller, H., Bracken, A.P., Vernell, R., Moroni, M.C., Christians, F., Grassilli, E., Prosperini, E., Vigo, E., Oliner, J.D., and Helin, K. (2001). E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 15, 267–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porceddu, A., Stals, H., Reichheld, J.P., Segers, G., De Veylder, L., Barroco, R.P., Casteels, P., Van Montagu, M., Inze, D., and Mironov, V. (2001). A plant-specific cyclin-dependent kinase is involved in the control of G2/M progression in plants. J. Biol. Chem. 276, 36354–36360. [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra, E., Frundt, C., and Gutierrez, C. (2003). A genome-wide identification of E2F-regulated genes in Arabidopsis. Plant J. 33, 801–811. [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra, E., and Gutierrez, C. (2000). Characterization of wheat DP, a heterodimerization partner of the plant E2F transcription factor which stimulates E2F-DNA binding. FEBS Lett. 486, 73–78. [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra, E., Xie, Q., Boniotti, M.B., and Gutierrez, C. (1999). The cloning of plant E2F, a retinoblastoma-binding protein, reveals unique and conserved features with animal G(1)/S regulators. Nucleic Acids Res. 27, 3527–3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichheld, J.P., Gigot, C., and Chaubet-Gigot, N. (1998). Multilevel regulation of histone gene expression during the cell cycle in tobacco. Nucleic Acids Res. 26, 3255–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, B., Cam, H., Takahashi, Y., Volkert, T., Terragni, J., Young, R.A., and Dynlacht, B.D. (2002). E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Genes Dev. 16, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi, V., Locatelli, S., Lanzanova, C., Boniotti, M.B., Varotto, S., Pipal, A., Goralik-Schramel, M., Lusser, A., Gatz, C., Gutierrez, C., and Motto, M. (2003). A maize histone deacetylase and retinoblastoma-related protein physically interact and cooperate in repressing gene transcription. Plant Mol. Biol. 51, 401–413. [DOI] [PubMed] [Google Scholar]
- Rossignol, P., Stevens, R., Perennes, C., Jasinski, S., Cella, R., Tremosaygue, D., and Bergounioux, C. (2002). AtE2F-a and AtDP-a, members of the E2F family of transcription factors, induce Arabidopsis leaf cells to re-enter S-phase. Mol. Genet. Genomics 266, 995–1003. [DOI] [PubMed] [Google Scholar]
- Sekine, M., Ito, M., Uemukai, K., Maeda, Y., Nakagami, H., and Shinmyo, A. (1999). Isolation and characterization of the E2F-like gene in plants. FEBS Lett. 460, 117–122. [DOI] [PubMed] [Google Scholar]
- Shen, W.H. (2002). The plant E2F-Rb pathway and epigenetic control. Trends Plant Sci. 7, 505–511. [DOI] [PubMed] [Google Scholar]
- Showalter, A.M. (1993). Structure and function of plant cell wall proteins. Cell 5, 9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevaux, O., and Dyson, N.J. (2002). A revised picture of the E2F transcriptional network and RB function. Curr. Opin. Cell Biol. 14, 684–691. [DOI] [PubMed] [Google Scholar]
- Stevens, R., Mariconti, L., Rossignol, P., Perennes, C., Cella, R., and Bergounioux, C. (2002). Two E2F sites in the Arabidopsis MCM3 promoter have different roles in cell cycle activation and meristematic expression. J. Biol. Chem. 277, 32978–32984. [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu, K., and Roberts, K. (2003). “Big it up”: Endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 6, 544–553. [DOI] [PubMed] [Google Scholar]
- Trimarchi, J.M., and Lees, J.A. (2002). Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell. Biol. 3, 11–20. [DOI] [PubMed] [Google Scholar]
- Vandepoele, K., Raes, J., De Veylder, L., Rouzé, P., Rombauts, S., and Inzé, D. (2002). Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14, 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissenberg, K., Martinez-Vilchez, I.M., Verbelen, J.P., Miller, J.G., and Fry, S.C. (2000). In vivo colocalization of xyloglucan endotransglycosylase activity and its donor substrate in the elongation zone of Arabidopsis roots. Plant Cell 12, 1229–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieghe, K., Vuylsteke, M., Florquin, K., Rombauts, S., Maes, S., Ormenese, S., Van Hummelen, P., Van de Peer, Y., Inze, D., and De Veylder, L. (2003). Microarray analysis of E2Fa-DPa-overexpressing plants uncovers a cross-talking genetic network between DNA replication and nitrogen assimilation. J. Cell Sci. 116, 4249–4259. [DOI] [PubMed] [Google Scholar]
- Yoshizumi, T., Nagata, N., Shimada, H., and Matsui, M. (1999). An Arabidopsis cell cycle-dependent kinase-related gene, CDC2b, plays a role in regulating seedling growth in darkness. Plant Cell 11, 1883–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, N., Fraenkel, E., Pabo, C.O., and Pavletich, N.P. (1999). Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F-DP. Genes Dev. 13, 666–674. [DOI] [PMC free article] [PubMed] [Google Scholar]