Abstract
BRASSINOSTEROID-INSENSITIVE 1 (BRI1) is a Leu-rich-repeat (LRR) receptor kinase that functions as a critical component of a transmembrane brassinosteroid (BR) receptor. It is believed that BRI1 becomes activated through heterodimerization with BAK1, a similar LRR receptor kinase, in response to BR signal. A yeast two-hybrid screen using the kinase domain of BRI1 identified an Arabidopsis thaliana Transthyretin-Like protein (TTL) as a potential BRI1 substrate. TTL interacts with BRI1 in a kinase-dependent manner in yeast and is phosphorylated by BRI1 in vitro. TTL displays a similar expression pattern with BRI1 and is associated with the plasma membrane. Overexpression of the TTL gene results in a phenotype that was observed in weak bri1 mutants and null bak1 mutants. By contrast, two T-DNA insertional mutations in the TTL gene promote plant growth and enhance BR sensitivity. We hypothesized that TTL might directly regulate certain biochemical activities near the plasma membrane to control plant growth.
INTRODUCTION
Brassinosteroids (BRs) are a special class of plant polyhydroxysteroids that are structurally similar to the well-studied animal steroid hormones and play important roles throughout the plant development. Like animal steroids, BR can elicit both genomic responses that depend on gene expression and nongenomic effects that are independent of DNA transcription and protein synthesis. In animals, steroids use intracellular steroid receptors that are ligand dependent transcriptional factors to control gene expression in the nucleus (Aranda and Pascual, 2001) but rely on membrane steroid receptors to induce rapid nongenomic effects at the cell membrane or in the cytosol (Cato et al., 2002). By contrast, the plant steroids are mainly, if not exclusively, perceived by transmembrane steroid receptors to initiate signaling pathways that could control both genomic and nongenomic processes (Clouse 2002a, 2002b; Thummel and Chory, 2002; Peng and Li, 2003).
For the past several years, three different approaches have been successfully used to identify potential BR signaling components that transduce the plant steroid signals from the cell surface into the nucleus. The first approach, direct screening for BR-insensitive mutants, led to the identification of two BR-insensitive genes in Arabidopsis thaliana, BRASSINOSTEROID-INSENSITIVE 1 (BRI1) encoding a Leu-rich-repeat (LRR) receptor-like kinase that functions as a key component of a membrane BR receptor (Clouse et al., 1996; Kauschmann et al., 1996; Li and Chory, 1997; Noguchi et al., 1999; He et al., 2000; Wang et al., 2001), and BRASSINOSTEROID-INSENSITIVE 2 (BIN2), encoding a highly conserved GSK3 kinase that negatively regulates BR signaling (Choe et al., 2002; Li and Nam, 2002; Perez-Perez et al., 2002). The second approach, screening for extragenic mutations that suppress weak bri1 phenotypes or are resistant to a specific BR-biosynthesis inhibitor, resulted in the identification of a putative Ser carboxypeptidase (BRS1) that was hypothesized to regulate an early BR signaling event upstream of BR perception (Li et al., 2001a), a BRI1-associated receptor kinase (BAK1) thought to heterodimerize with BRI1 in initiating BR signaling at the cell surface (Li et al., 2002), two putative transcriptional factors (BES1 and BZR1) critical for regulating the expression of many BR-responsive genes (Wang et al., 2002; Yin et al., 2002), and a nuclear phosphatase (BSU1) that counteracts the BIN2 action (Mora-Garcia et al., 2004). The third approach, yeast two-hybrid screening, was successfully used in our laboratory to independently identify BAK1 as a potential coreceptor for BRI1 (Nam and Li, 2002) and to discover BES1 and BZR1 as two potential substrates for the BIN2 GSK3 kinase (Zhao et al., 2002).
A working model for BR signaling emerges from the biochemical and genetic studies of the discovered BR signaling proteins (Peng and Li, 2003). In the absence of BR, both BRI1 and BAK1 are inactive, but BIN2 is a constitutively active kinase that phosphorylates BES1 and BZR1 to target them for a proteasome-mediated protein degradation process (He et al., 2002), thus blocking the transduction of BR signals into the nucleus. In the presence of BR, BRI1 and BAK1 are activated, most likely through receptor heterodimerization and the resulting transphosphorylation, which can then inhibit BIN2 via a yet unknown mechanism, resulting in increased protein stability and subsequent accumulation of both BES1 and BZR1 in the nucleus where they regulate gene activities to influence many cellular processes.
In contrast with the rapid progress in understanding how the plant steroid signals reach the nucleus to regulate gene activities, little is known about how BRs induce nongenomic responses at the cell membrane or in the cytosol. Two competing models were proposed to explain how the plant steroid signaling is initiated at the cell surface and transmitted into the plant cell. Based on studies using the Escherichia coli–expressed recombinant proteins of the cytoplasmic domains of BRI1 and BAK1, Li et al. (2002) proposed a two-step activation model involving sequential phosphorylation of BRI1 and BAK1. BR binds and activates BRI1, which can then phosphorylate and activate BAK1, and the activated BAK1 is responsible for transmitting BR signal into the cytosol by phosphorylating cytoplasmic BR signaling components. We proposed a transphosphorylation model based on our biochemical studies of the two full-length receptor kinases expressed in yeast cells (Nam and Li, 2002). BRI1 and BAK1, existing mainly as inactive monomers, are in equilibrium with a small pool of BRI1/BAK1 heterodimers. BR binding stabilizes the BRI1-BAK1 heterodimer, shifting the equilibrium toward the BRI1-BAK1 dimer formation. Such a BR-stimulated BRI1-BAK1 dimerization would activate both receptor kinases through transphosphorylation, which can then phosphorylate a multitude of cytoplasmic substrates to transduce the BR signal into the cytosol or directly regulate the membrane activities that are important for plant growth. In this article, we report the identification of a novel Arabidopsis protein, TTL (for Transthyretin-Like), by a yeast two-hybrid library screening as a potential substrate of BRI1. Our genetic studies suggest that TTL is a negative regulator in BR-mediated plant growth.
RESULTS
Identification of an Arabidopsis TTL Protein as a BRI1-Specific Interactor
To identify potential targets of the activated BRI1 receptor kinase in response to BR signal, we conducted a yeast two-hybrid screening using the kinase domain of BRI1 (BRI1CK) as the bait. Out of 96 positive BRI1-interacting clones, 60 were found to be derived from the same Arabidopsis gene (At5g58220) that encodes a hypothetical protein of 324 amino acids. To eliminate the possibility that this hypothetical protein is promiscuous in protein interaction, we performed a specificity test in which its interaction with BRI1 was compared with its interactions with some nonrelevant proteins, including a C-terminal segment of the Arabidopsis phytochrome A (Fankhauser et al., 1999), an N-terminal fragment of an Arabidopsis NADPH oxidase (Keller et al., 1998), and the kinase domain of a similar LRR receptor kinase, CLV1, that regulates shoot meristem development (Clark et al., 1997). As shown in Figure 1A, the newly identified BRI1-interacting protein only interacted with BRI1, but failed to interact with any of the control proteins, suggesting that the observed interaction is BRI1 specific.
Figure 1.
Identification of TTL as a BRI1-Interacting Protein by Yeast Two-Hybrid Screening.
(A) TTL showed specific interaction with BRI1 kinase domain (BRI1CK), examined by the growth on His-lacking synthetic media and the production of blue colonies on the 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal)–containing media, compared with its interactions with nonrelevant baits, including CLV1 kinase domain (CLV1CK), the N-terminal portion of an Arabidopsis NADPH oxidase (N-gp91), the C-terminal segment of the Arabidopsis phytochrome A (C-PHYA), and the Gal4 binding domain (GAL4BD) alone.
(B) Amino acid sequence alignment of TTL and its homologs from various organisms, including LeTTL from Lycopersicon esculentum, StTTL from Solanum tuberosum, GmTTL from Glycine max, TaTTL from Triticum aestivum, HvTTL from Hordeum vulgare, OsTTL from Oriza sativa, MmTRP from Mus musculus, DmTRP from Drosophila melanogaster, CeTRP from Caenorhabditis elegans, SpTRP from Schizosaccharomyces pombe, and EcTRP from Escherichia coli. Except TTL, other full-length plant TTL sequences were deduced from overlapping EST sequences as follows: LeTTL (AW649495, BE432987, NM535270, and BF114284), StTTL (BE920936, BM405599, and BE919514), GmTTL (AW100149, BE022630, BI468680, and BE824466), TaTTL (BE400327, BQ245413, and BE444381), HvTTL (BE602694, BU989926, and AL503126), and OsTTL (AC092075, AU067993, and CB214315). Sequence alignment was conducted using ClustalW software.
A database search revealed that the BRI1-interacting protein displays ∼42% sequence identity within its C-terminal half (198 to 324 amino acids) with the vertebrate transthyretin, a thyroid binding protein that is thought to mediate thyroid hormone transfer into target tissues in animals (Palha, 2002) and indirectly implicated in the transport of vitamin A via association with retinol binding protein (Monaco, 2000). In human, transthyretin is often associated with amyloidosis (Benson and Uemichi, 1996). Proteins similar to the vertebrate transthyretin, collectively called transthyretin-related proteins (TRPs), were found in a wide variety of species, including bacteria, plants, and animals (Eneqvist et al., 2003). We named this new BRI1-interacting protein TTL. Whereas all known transthyretins and most TRPs are ∼120 to 130 amino acids long, TTL contains a long N-terminal extension of ∼200 amino acids that is conserved among all known plant TTLs (Figure 1B). A BLAST search against the entire Arabidopsis genome revealed that TTL is a single-copy gene.
The N Terminus of TTL Is Crucial for TTL–BRI1 Interaction
Because all the TTL clones that were recovered from the yeast two-hybrid screen contained the complete TTL open reading frame (ORF) or lacked no more than three amino acids at the N terminus, we suspected that the N terminus of TTL is important for its interaction with BRI1. To test our hypothesis, we deleted the N-terminal 29 amino acids to create a truncated TTL protein, TTLΔN29, and tested its interaction with the BRI1 kinase domain by the yeast two-hybrid assay. As shown in Table 1, TTLΔN29 failed to interact with BRI1CK, confirming that these 29 amino acids are crucial for the TTL–BRI1 interaction. To determine if such a 29–amino acid peptide is sufficient for binding to BRI1, we made a chimeric protein, TTLN29:BES1, by fusing the 29–amino acid peptide to BES1, a nuclear BR signaling protein that does not interact with BRI1 (Yin et al., 2002; Zhao et al., 2002). The resulting chimeric protein was then tested by the yeast two-hybrid assay for its interaction with BRI1CK. As shown in Table 1, the TTLN29:BES1 fusion protein did not interact with BRI1CK in yeast cells. These results indicated that whereas the N-terminal 29 amino acids are essential for the TTL–BRI1 interaction, they are not sufficient to bind BRI1.
Table 1.
The N-Terminal 29 Amino Acids Are Critical for TTL-BRI1 Interaction
| Constructs | Relative β-Galactosidase Activity (%) | |
|---|---|---|
| GAL4-DB Fusion | GAL4-AD Fusion | |
| pAS2 | pACT2-TTL | NDa |
| pAS2-BRI1CK | pACT2-TTL | 100.00 |
| pAS2-BRI1CK | pACT2-TTLΔN29 | 0.35 |
| pAS2-BRI1CK | pACT2-TTLN29:BES1 | 0.35 |
Nondetectable.
TTL Is a Potential Substrate of BRI1
Using the yeast two-hybrid assay, we also tested the interaction between TTL with three mutated BRI1CK, each containing a unique single amino acid change that was identified in three mutant bri1 alleles (bri1-101 changes Glu1078 to Lys, bri1-104 mutates Ala1031 to Thr, and bri1-115 substitutes Gly1048 with Asp). Whereas all three bri1 mutations were known to destroy the in vivo function of BRI1 (Li and Chory, 1997), they have different effects on the BRI1 kinase activity. As shown in Figure 2A, bri1-101 completely eliminates and bri1-115 greatly reduces the autophosphorylation activity of an E. coli–expressed glutathione S-transferase (GST)-BRI1CK fusion protein, whereas bri1-104 mutation reduces the kinase activity of GST-BRI1CK by only ∼50%. Results shown in Figure 2B revealed that an active kinase activity was required for the BRI1–TTL interaction. Both bri1-101 and bri1-115 mutations completely abrogate the BRI1–TTL interaction. By contrast, the BRI1CK containing the bri1-104 mutation can still interact with TTL to the same extent as the wild-type BRI1CK.
Figure 2.
TTL Is a Putative Substrate for BRI1.
(A) Different bri1 mutations have different effects on the kinase activity of BRI1. In vitro autophosphorylation activity was measured using the E. coli expressed GST fusion proteins of the wild-type BRI1 cytoplasmic kinase domain and its three mutated counterparts containing bri1-101, bri1-104, and bri1-114, respectively. The top panel shows the amount of purified recombinant proteins visualized by Coomassie blue staining, and the bottom panel reveals the autophosphorylation level by autoradiography.
(B) Yeast two-hybrid assay of the interaction between TTL and the three mutated BRI1 cytoplasmic kinases.
(C) In vitro transphosphorylation assay of TTL by BRI1. The wild type and mutated BRI1 cytoplasmic kinase domains were fused to GST, whereas TTL was fused to MBP for recombinant protein expression in E. coli cells. Transphosphorylation assay was performed by mixing MBP-TTL with GST-fused wild type or mutated BRI1CK. Lane 1, GST-BRI1CK alone; lane 2, GST-BRI1 (one-fifth of the amount loaded on lane 1) mixed with MBP-TTL; lane 3, GST-BRI1CK (bri1-101) alone; lane 4, GST-BRI1CK (bri1-101) (one-fifth of the amount loaded on lane 3) mixed with MBP-TTL; lane 5, MBP-TTL alone; lane 6, MBP alone; lane 7, GST-BRI1CK mixed with MBP. The top panel displays the purified recombinant protein by Coomassie blue staining, and the bottom panel shows the level of protein phosphorylation by autoradiography.
The fact that only the kinase active BRI1CKs were capable of interacting with TTL prompted us to investigate whether TTL might be a substrate for the BRI1 kinase. We fused the full-length TTL with the maltose binding protein (MBP) and expressed the resulting fusion protein in E. coli. After purification, the MBP-TTL fusion protein was mixed with the wild type or the bri1-101–mutated GST-BRI1CK fusion protein and assayed protein phosphorylation. As shown in Figure 2C, the wild-type GST-BRI1CK fusion protein exhibited strong autophosphorylation activity in vitro, whereas no autophosphorylation was detected for the MBP-TTL fusion protein. However, TTL was phosphorylated when the two fusion proteins were mixed together in the kinase assay solution. Because of the similar electrophoretic mobility of the two fusion proteins, only one-fifth of the GST-BRI1CK of the autophosphorylation assay was used in the transphosphorylation assay to clearly reveal the phosphorylated MBP-TTL band. When incubated with the kinase-dead GST-BRI1CK fusion protein, no phosphorylation of TTL was detected, eliminating the possibility that the observed TTL phosphorylation was catalyzed by an unknown E. coli kinase that might be copurified with the GST-BRI1CK fusion protein. No transphosphorylation was detected when the GST-BRI1CK fusion protein was incubated with the MBP alone. Taken together, these in vitro phosphorylation results indicated that BRI1 could phosphorylate TTL in vitro.
TTL Exhibits a Similar Gene Expression Pattern with BRI1
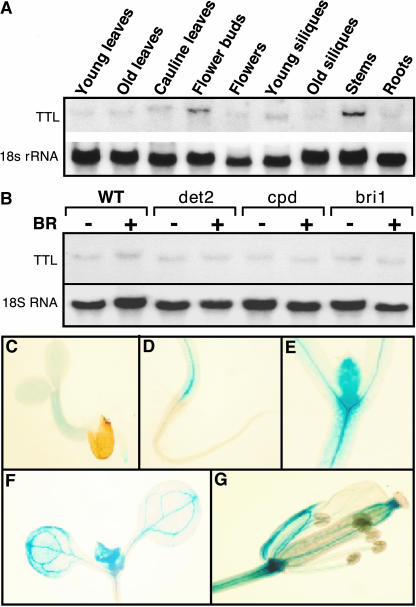
If BRI1 and TTL interact with each other in vivo, we would expect that their gene expression domains should overlap. RNA gel blot analysis was conducted with total RNAs isolated from various tissues of the wild-type Arabidopsis plants using the full-length TTL cDNA as a probe. As shown in Figure 3A, TTL was expressed in all the tissues that we analyzed, with the flower buds and elongating inflorescence stems accumulating the highest levels of the TTL transcripts. Such a ubiquitous expression pattern was previously observed for both BRI1 and BAK1 genes (Li and Chory, 1997; Li et al., 2002; Nam and Li, 2002). The expression of the TTL gene is not regulated by the plant steroids. RNA gel blot analysis of total RNAs, isolated from mock- or brassinolide-treated seedlings of wild-type, two BR-deficient mutants, det2 and cpd, and a strong BR-insensitive bri1 mutant, revealed no detectable difference in the abundance of the TTL transcript between mock- and BR-treated seedlings or between wild-type plants and BR mutants (Figure 3B). To obtain a better picture of tissue specificity of TTL gene expression, we constructed a TTL-β-glucuronidase (GUS) reporter gene by fusing a 1.6-kb genomic fragment containing the native TTL promoter to a GUS reporter gene. As shown in Figures 3C to 3F, the GUS activity was detected mainly in root tips, hypocotyls, leaf primordia, newly expanding young leaves, and vascular tissues of flower organs, which were known to express BRI1 (Friedrichsen et al., 2000; Caño-Delgado et al., 2004).
Figure 3.
The Expression Pattern of the TTL Gene.
(A) RNA gel blot analysis of the TTL gene expression. Total RNAs were isolated from various tissues of 5-week-old Arabidopsis plants, including young rosette leaves, old leaves, cauline leaves, floral buds, flowers, young siliques, old siliques, stems, and roots.
(B) The expression of TTL is not regulated by BR. Total RNAs were isolated from 10-d-old seedlings of wild-type, det2, cpd, and bri1 mutants treated with or without 1 μM brassinolide.
(C) to (G) Histochemical analysis of the GUS reporter gene expression driven by the TTL promoter. GUS signal was detected in root tips of 3-d-old seedling (C), hypocotyls (D), leaf primordia (E), expanding young leaves (F) of 10-d-old seedlings, and flowers (G) of mature plants.
TTL Is a Membrane-Associated Protein
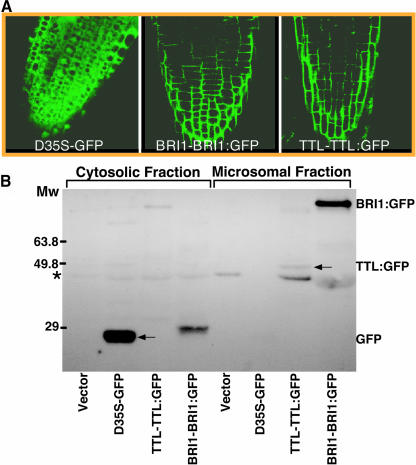
Because BRI1 is a transmembrane receptor kinase and TTL is predicted to be a cytosolic protein, a BRI1–TTL interaction would imply that TTL should be a membrane-associated protein. To test our prediction, we translationally fused the full-length TTL at its C terminus with green fluorescent protein (GFP) and used the native TTL promoter to drive the expression of the TTL:GFP fusion protein in Arabidopsis plants. Confocal microscopic analysis of the resulting TTL:GFP transgenic plants revealed a similar green fluorescent pattern between TTL:GFP and BRI1:GFP, indicating that TTL was indeed associated with the plasma membrane (Figure 4A). The association of TTL with the plasma membrane was not influenced by BRI1 activity because no detectable change in the membrane localization pattern of the TTL:GFP fusion protein was observed when the TTL:GFP transgenic seedlings were treated with brassinazole (data not shown), a specific BR biosynthesis inhibitor that prevents the activation of BRI1 (Wang et al., 2001). The membrane association of TTL was also confirmed by a cell fractionation assay. Membrane proteins of a transgenic TTL:GFP plant were separated from cytosolic proteins by differential centrifugation, these proteins were separated by SDS-PAGE, and the presence of TTL:GFP in the two fractions was detected by immunoblotting using a commercial anti-GFP antiserum. As shown in Figure 4B, both BRI1:GFP and TTL:GFP were mainly in the membrane fraction. By contrast, the GFP itself was exclusively in the cytosolic fraction. Both the gene expression analysis and the protein localization studies strongly suggested that BRI1 could interact with TTL near the plasma membrane.
Figure 4.
TTL Is a Membrane-Associated Protein.
(A) Confocal microscopic analysis of GFP fusion proteins in the root tips of 4-d-old transgenic plants containing the BRI1-BRI1:GFP or TTL-TTL:GFP transgene. The green fluorescent signal of the GFP itself driven by two copies of the strong 35S promoter of Cauliflower mosaic virus was used as a control for cytosolic and nuclear localization.
(B) Subcellular fractionation analysis of the TTL localization. Cytosolic (lanes 1 to 4) and membrane fractions (lanes 5 to 8) were prepared from the 7-d-old seedlings of transgenic plants containing the pPZP212 vector alone, D35S-GFP, TTL-TTL:GFP, or BRI1-BRI1:GFP transgene. The similar amount of total proteins of each fractionation was analyzed for the presence of GFP or GFP-fusion proteins by immunostaining with anti-GFP antibodies (Molecular Probes, Eugene, OR). The asterisk indicates a nonspecific cross-reacting band, and the arrow indicates the TTL:GFP fusion protein.
Overexpression of the TTL Gene Inhibits Plant Growth
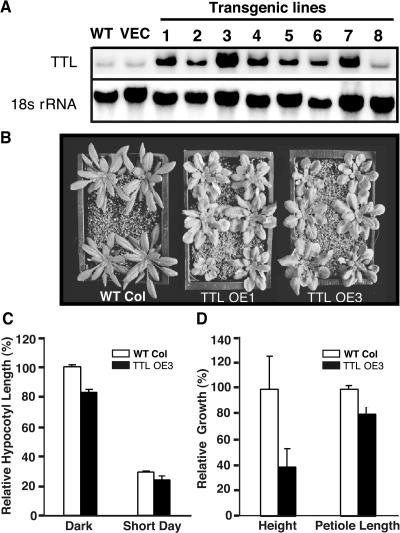
To investigate the physiological function of TTL in plant growth, we generated transgenic Arabidopsis plants expressing the TTL gene using its native promoter. The resulting transgenic plants were screened by RNA gel blot analysis for lines that accumulated high levels of the TTL transcript (Figure 5A), which were chosen for phenotypic comparison with the wild-type plants. Interestingly, the TTL-TTL–overexpressing transgenic plants displayed rounder rosette leaves with shorter petioles (Figures 5B and 5D), a phenotype that was previously observed in the bak1 mutant (Li et al., 2002; Nam and Li, 2002) and a weak bri1-301 mutant (Li and Nam, 2002), suggesting that TTL might function as a negative regulator to modulate BR-mediated plant growth. As shown in Figure 5C, overexpression of the TTL gene also led to an ∼20% reduction in hypocotyl length when grown in complete darkness or under a short-day (8-h-light/16-h-light) growth condition. The most striking phenotype was the height of the adult plants. As indicated in Figure 5D, the height of TTL overexpression transgenic plants of 4 weeks old (TTL OE3) was only ∼40% of that of the wild-type plants of the same developmental age.
Figure 5.
ttl Overexpression Leads to Growth Inhibition.
(A) RNA gel blot analysis of ttl overexpression in eight independent transgenic lines with the 32P-labeled full-length ttl cDNA probe.
(B) Phenotypic comparison of two ttl-overexpressing transgenic lines with the corresponding wild-type (Col-0) plants. Plants shown here were 5 weeks old grown under long-day growth condition (16 h light/8 h dark).
(C) Quantitative analysis of the hypocotyl growth of the two ttl-overexpressing transgenic lines grown for 5 d in the dark or under a short-day (8-h-light/16-h-dark) growth condition. Hypocotyl growth is expressed as a percentage of the hypocotyl length of the 5-d-old wild-type seedling grown in the dark, and each data point represents the average of ∼50 seedlings.
(D) Quantitative analysis of the height and petiole length of the ttl transgenic plants relative to those of the wild-type plants. The wild-type control and the two ttl-overexpressing transgenic lines were grown in soil for 7 weeks under a long-day (16-h-light/8-h-dark) growth condition. The length of the main inflorescence stem was used as the height of an adult plant, and the average petiole length of the three longest rosette leaves of each analyzed plant was taken as the petiole length. Each data point represents the average of 14 plants, and the error bar denotes standard error.
The T-DNA Insertional ttl Mutations Promote Plant Growth
Further support for a negative role of TTL in modulating plant growth came from the analysis of two independent ttl T-DNA insertional mutants, ttl-1 and ttl-2, which were identified from a population of ∼140,000 transgenic Arabidopsis plants (in the Wassilewskija [Ws] ecotype) using a PCR-based reverse-genetic approach (Winkler et al., 1998). The ttl-1 mutant contains a single T-DNA insertion in the 117th codon downstream from the presumed start codon, whereas the ttl-2 has a single T-DNA insertion at the 15th base pair upstream of the suspected initiation codon (Figure 6A). RNA gel blot analysis with RNAs isolated from the two insertional mutants and the wild-type control plants revealed that both insertions led to a complete disappearance of the TTL transcripts, suggesting that the two T-DNA insertional mutations are null (Figure 6B). The two ttl knockout mutants were carefully examined for any phenotypic difference from the corresponding Ws wild-type controls. Consistent with a negative role for regulating plant growth, the two ttl mutants were bigger than the wild-type plants of the same developmental age with larger rosette leaves and longer petioles. However, they were different than the transgenic plants overproducing BR or BRI1 (Choe et al., 2001; Wang et al., 2001) that are characterized by narrow leaves with elongated petioles, suggesting that TTL is not a major signaling component to transduce the plant steroid signal into the nucleus but plays a role in regulating a subset of BR responses. The hypocotyls of dark-grown ttl mutants were longer than those of the corresponding wild-type controls (Figure 6D). Consistent with the observed size difference between the ttl-1 mutants and the wild-type plants, the mature ttl mutants were much taller than the wild-type plants of the same developmental age and their leaves have longer petioles (Figures 6E and 6F).
Figure 6.
Two T-DNA Insertional Mutations Promote Plant Growth.
(A) Schematic representation of two T-DNA insertion sites in the TTL gene. ttl-1 contains the T-DNA insertion in the 117th codon of the second exon, whereas ttl-2 has the T-DNA inserted 15 bp upstream of the presumed ATG start codon.
(B) RNA gel blot analysis of the TTL gene expression in the two ttl mutants.
(C) Phenotypic comparison between the two ttl mutants and the corresponding wild-type plant (Ws-0) grown for 5 weeks under the 16-h-light/8-h-dark growth condition.
(D) Quantitative analysis of the hypocotyl growth of the ttl-1 mutants grown for 5 d in the dark or under a short-day (8-h-light/16-h-dark) growth condition. Hypocotyl growth is expressed as a percentage of the hypocotyl length of the 5-d-old wild-type seedlings grown in the dark, and each data point represents the average of ∼50 seedlings.
(E) Quantitative analysis of the height and petiole length of the ttl-1 mutants relative to those of the wild-type control plants. Both the wild-type control and the ttl-1 mutants were grown in soil for 7 weeks under the 16-h-light/8-h-dark growth condition. The length of the main inflorescence stem was used as the height of an adult plant, and the average petiole length of the three longest rosette leaves of each analyzed plant was taken as the petiole length. For (D) and (E), each data point represents the average of 16 plants, and the error bar denotes standard error.
(F) Shown are a 5-week-old ttl-1 mutant and a 5-week-old wild-type (Ws-0) plant.
TTL Is Involved in BR-Mediated Growth Responses
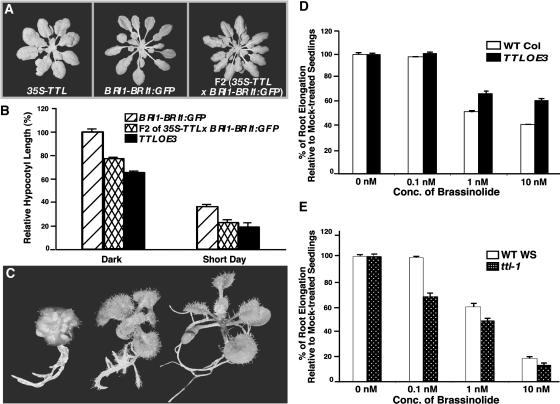
To examine a potential role of TTL in modulating BR-mediated plant growth, we crossed a 35S-TTL transgenic line, which is phenotypically similar to TTL-OE1 or TTL-OE3 transgenic plants, with a BRI1-overexpressing transgenic line that displays longer and narrower rosette leaves compared with the wild-type plants, a characteristic phenotype for increased BR production or enhanced BR signaling (Choe et al., 2001; Wang et al., 2001). If TTL functions as a negative regulator in BR-mediated cell growth, its overexpression could suppress the BRI1-overexpressing phenotype. Indeed, the resulting F2 transgenic plants homozygous for both transgenes displayed wider and shorter rosette leaves than the BRI1-overexpressing transgenic plants (Figure 7A). Similar to the previous hypocotyl measurement of the TTL-TTL transgenic plants, the hypocotyl lengths of the double transgenic line were 77 and 62% of the BRI1-overexpression line when grown in the dark and under short-day growth conditions, respectively (Figure 7B). Consistent with these overexpression results, the ttl-1 mutants are partially resistant to brassinazole, which was known to inhibit BR biosynthesis (Asami et al., 2000) and prevents BRI1 activation (Wang et al., 2001). As shown in Figure 7C, the brassinazole treatment resulted in a strong dwarf morphology of wild-type seedlings but induced a weak BR-deficient phenotype on the ttl-1 mutants. As a control, transgenic plants overexpressing the BRI1 gene were quite resistant to the dwarfing effect of the BR inhibitor. This result suggested that the elimination of TTL could partially compensate for the reduced BRI1 activity caused by the inhibition of BR biosynthesis.
Figure 7.
TTL Modulates BR-Mediated Growth Response.
(A) Overexpression of the TTL gene suppresses the BRI1 overexpression phenotype. Shown (from left to right) are a 35S-TTL transgenic plant, a BRI1-BRI1:GFP transgenic line, and a F2 plant homozygous for both the 35S-TTL and BRI1-BRI1:GFP transgenes.
(B) Quantitative analysis of the hypocotyl growth of the 5-d-old transgenic Arabidopsis seedlings grown in the dark or under the 8-h-light/16-h-dark growth condition. Hypocotyl growth is expressed as a percentage of the hypocotyl length of the dark-grown seedlings of the BRI1-overexpressing transgenic line, and each data point represents the average of ∼30 seedlings.
(C) The ttl mutants display a brassinazole-resistant phenotype. Shown (from left to right) are a wild-type seedling, a ttl-1 mutant, and a BRI1-BRI1:GFP transgenic plant grown on 1 μM brassinazole-containing medium.
(D) and (E) Quantitative analysis of BR sensitivity of the TTL-overexpressing plants (D) and the ttl-1 mutants (E). Seedlings were germinated and grown on medium containing increasing concentrations of brassinolide. Root elongation was measured 7 d after germination. Each data point represents the average root elongation of ∼50 seedlings. Inhibition of root growth by brassinolide is expressed as a percentage of the root elongation of the wild-type controls grown on medium containing the same volume of 80% (v/v) ethanol used to dilute brassinolide from a 2 mM stock solution. Error bar denotes standard error.
Further support for a role of ttl in BR-mediated growth response came from two root-growth inhibition assays. Seeds of a TTL overexpression transgenic line, the ttl-1 mutant, and their corresponding wild-type controls were germinated and grown on synthetic medium containing different concentrations of brassinolide for 7 d, and their root lengths were measured. As shown in Figure 7C, the root growth of the TTL-OE3 plants was less inhibited by 1 and 10 nM concentrations of brassinolide than that of the wild-type controls, indicating a reduced BR sensitivity for the TTL-TTL transgenic plants. By contrast, the ttl-1 mutant was more sensitive to brassinolide compared with its wild-type control, especially at low concentrations. As shown in Figure 7D, the most dramatic difference between ttl-1 mutants and the corresponding Ws wild-type seedlings was seen at 0.1 nM brassinolide, which had virtually no effect on the root growth of the wild-type seedlings but inhibited the root elongation of the ttl-1 mutants by ∼30%.
DISCUSSION
Despite the rapid progress in recent years in identifying several BR signaling components and BR-regulated genes, no substrate has been identified for BRI1, a key component of a membrane BR receptor. In this report, we describe the identification of an Arabidopsis TTL protein as a potential BRI1 substrate and provide both genetic and transgenic results to implicate a negative role of TTL in BR-mediated plant growth.
In yeast, TTL interacts specifically with BRI1 but lacks interaction with three nonrelevant proteins, including CLV1, a similar LRR receptor-like kinase involved in meristem development, indicating a specific interaction between TTL and BRI1. The dependence of the BRI1-TTL interaction on the BRI1 kinase activity in yeast and the demonstration of TTL phosphorylation by BRI1 in vitro strongly suggest that TTL is a potential substrate for BRI1. BRI1 and TTL share similar gene expression patterns as revealed by both RNA gel blot and histochemical analyses of a GUS reporter gene. Confocal microscopic examination and cellular fractionation analysis of TTL:GFP transgenic plants suggested that TTL is loosely associated with membrane where BRI1 is localized, suggesting that TTL can potentially interact with BRI1. However, repeated coimmunoprecipitation experiments failed to show a direct physical interaction between the two proteins, most likely because of the transient nature of the kinase/substrate interaction. The biological function of TTL in modulating plant growth was revealed by both gain-of-function and loss-of-function studies. Overexpression of the TTL gene inhibited plant growth and resulted in a reduced BR sensitivity as revealed by the root growth inhibition assay. By contrast, the T-DNA insertional mutations of the TTL gene promoted plant growth, enhanced BR sensitivity, and increased brassinazole resistance, suggesting that TTL functions as a negative regulator in BR-mediated plant growth.
A database search indicated that TTL is a member of a growing family of TRPs, which share significant sequence identity with the vertebrate transthyretin, an extracellular thyroid hormone carrier protein, whose mutations in human are often associated with two clinical forms of amyloidosis (Benson and Uemichi, 1996). So far, at least 49 TRP sequences have been identified from 47 species, including E. coli, yeast, Drosophila, C. elegans, human, and many plant species (Eneqvist et al., 2003). Almost all of them contain a characteristic four amino acid motif Tyr-Arg-Gly-Ser at their C-terminal ends, which distinguishes the TRPs from vertebrate transthyretins and other transthyretin-like sequences in databases. Sequence comparison and phylogenetic analysis of all 49 TRP sequences and the known vertebrate transthyretins indicated that TRPs might represent the ancestor of the vertebrate thyroid hormone carriers and have evolved independently to perform one or more different biochemical functions (Eneqvist et al., 2003). A recent study has shown that the E. coli TRP, which exhibits 35% sequence identity and displays a similar three-dimensional structure to the human transthyretin, does not bind thyroid hormones but contains a distinct binding motif shared by all known TRPs for unknown ligands (Eneqvist et al., 2003).
Sequence comparison among the known TRP sequences also reveals a unique feature for the plant TTLs. Whereas most TRPs of bacteria and animals are 120 to 130 amino acids long, the plant TTLs contain 320 to 340 amino acids with a long N-terminal extension of 200 to 210 amino acids, which is found in a variety of hypothetical bacterial proteins (Eneqvist et al., 2003) and a hypothetic mouse protein (encoded by a mouse EST sequence BI328404). Interestingly, in some bacteria genomes, the ORF encoding an ∼200–amino acid polypeptide similar to the TTL N-terminal domain overlaps with the TRP ORF, suggesting that plant TTLs are created by an ancient gene fusion event. For example, in the Streptomyces coelicolor A3(2) genome (NC_003888), the TGA stop codon of the SC2G5.30 ORF encoding a hypothetic protein similar to the N-terminal domain of TTL overlaps with the ATG start codon of the downstream TRP ORF. The occurrence of this domain in many unrelated proteins and the fact that this domain seems to be critical for interacting with BRI1 suggest that it might have a regulatory function to control the activity of TTLs in the plants.
A potential role for TRPs in purine catabolism was suggested by the demonstration that the Bacillus subtilis TRP is essential for a uricase activity (Ashultz et al., 2001), which catalyzes the oxidation of uric acid to allantoin by reduction of O2 to H2O2. However, TTL seems unlikely to participate in this process for the following two reasons. First, uricases were identified in many different plant species, including Arabidopsis, and many were known to be involved in purine catabolism; however, none of them show any detectable sequence similarity to TTL, although weak homology was detected between the N-terminal half of TTL and the N-terminal 170 amino acids of a uricase precursor from Bacillus sp TB-90 (Yamamoto et al., 1996). Second, it is well known that purine catabolism takes place inside peroxisomes that contain high uricase activity, and the only alleged uricase of Arabidopsis (At2g26230) does have a conserved Ser-Lys-Leu peroxisomal targeting signal at its C-terminal end. By contrast, TTL lacks such a peroxisomal targeting signal and is associated instead with the plasma membrane, as revealed by our confocal microscopic analysis of TTL:GFP transgenic plants. Our conclusion that TTL is not involved in purine catabolism is consistent with the existence of an expressed TRP gene (BC044232) in human known to lack uricase (Wu et al., 1992). Despite the facts that TRPs are evolutionarily conserved proteins and that TTL is a single copy gene in the Arabidopsis genome, the two T-DNA insertional ttl mutants exhibit a relatively weak morphological phenotype under normal growth conditions, suggesting that TRP is not an essential component of a fundamental biological process. However, the possibility that TTL might function redundantly with certain proteins of dissimilar sequences in a critical cellular process could not be ruled out.
Although the exact function of TTL awaits further investigation, its membrane association and its effects on BR-mediated growth responses suggest that this protein might interact with membrane proteins or activities previously implicated in BR-stimulated cell elongation, such as cell wall biosynthesis/reorganization of cortical microtubule (Mayumi and Shibaoka, 1995; Yamamuro et al., 2000; Catterou et al., 2001), the plasma membrane ATPases (Katsumi, 1991), and aquaporins (Morillon et al., 2001). Plant growth and morphogenesis are largely contributed by cell elongation/expansion, which depends on the de novo biosynthesis and modification of cell wall (activated mainly by gene expression), the deposit orientation of nascent cellulose microfibrils (determined by cortical microtubules), and the internal turgor pressure (generated by water uptake). Both cellulose biosynthesis and reorganization of cortical microtubule were known to be important for BR-induced plant growth and it was thought that protein phosphorylation and microtubule-membrane interaction were involved in the process (Mayumi and Shibaoka, 1995). Recent studies using BR-deficient (Catterou et al., 2001) and BR-insensitive mutants (Yamamuro et al., 2000) confirmed the effect of BR on cortical microtubule in cell elongation and suggested the existence of a BR-signaling pathway for regulating microtubule reorganization independently of tubulin gene expression. Both ATPases and aquaporins could regulate turgor pressure to generate the driving force for cell elongation. It was reported that plasma membrane ATPases were involved in BR-induced plant growth. BR-treatment resulted in increased proton extrusion in treated root tissues (Mandava, 1988), and an inhibitor of the plasma membrane ATPase prevented BR-stimulated cell elongation (Katsumi, 1991). A recent study also suggested that BR could control aquaporin activities. Correlations were observed between the hypocotyl length and water permeability of protoplasts isolated from both non-treated or BR-treated dwarf mutants deficient in BR biosynthesis (Morillon et al., 2001).
We hypothesize that TTL might function as a negative regulator for one of the above-mentioned growth-promoting biochemical activities near the plasma membrane. In the absence of BR signaling, TTL is constitutively active in inhibiting such a growth-promoting activity by a yet unknown mechanism. In the presence of BR, BRI1 is activated, most likely through heterodimerization with BAK1 at the cell surface, which can then phosphorylate TTL and relieve its inhibitory effect on plant growth. It remains to be investigated whether the phosphorylation level of TTL is influenced by the plant steroid hormones and if the phosphorylation of TTL regulates its biological activity. More importantly, additional studies are needed to reveal its true biochemical function in modulating plant growth and provide useful information to study TRPs of other organisms.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana ecotype Columbia (Col-0) was used as the wild-type plants for generating transgenic plants and phenotype comparison. The ecotype Ws (Ws-0) was the wild type for phenotypic comparison with the ttl knockout mutants. The methods for seed sterilization and germination for plant growth were described in Li et al. (2001b).
Yeast Two-Hybrid Screening and Determination of the N-Terminal Binding Domain of TTL
Detailed methods for the yeast two-hybrid library screen were described previously (Nam and Li, 2002). Putative BRI1-interacting clones were screened by growth on the synthetic medium lacking His but containing 50 mM 3-amino-1,2,4-triazol and confirmed by blue color on medium containing the 5-bromo-4-chloro-3-indolyl-β-d-galactoside. pACT2-TTLΔN29 was constructed by deleting an 87-bp HpaI restriction fragment from the pACT2-TTL construct that contains the full-length TTL ORF. The yeast cells were transformed using the TRAFO method (http://www.umanitoba.ca/faculties/medicine/biochem/gietz/Trafo.html).
Measurement of β-Galactosidase Activity of Yeast Cells
Overnight cultures of yeast cells in selective medium were transferred to fresh YPD medium (containing 20 g/L peptone, 10 g/L yeast extract, and 2% [v/v] glucose) and grown at 30°C until the cells were in the mid-log growth phase (OD600 = 0.5 to 0.8). Yeast cells were broken by the freeze/thaw method and incubated with chlorophenol red-β-d-galactopyranoside in a reaction mixture containing 100 mM Hepes, pH 7.25, 150 mM NaCl, 22.5 mM l-aspartate, 1% (w/v) BSA, and 0.05% (v/v) Tween 20. Reactions were terminated by adding 3 mM ZnCl2, and the color change of the reaction was monitored at 578 nm using a Beckman DU520 UV/Vis spectrophotometer (Beckman Coulter, Fullerton, CA). One unit of β-galactosidase activity is defined as the amount of the enzyme that hydrolyzes 1 μmole of chlorophenol red-β-d-galactopyranoside to chlorophenol red and d-galactose per minute per cell.
Transphosphorylation Assay
The cytoplasmic kinase domain of BRI1 and the full-length TTL ORF were cloned into the pGEX-KG vector (Guan and Dixon, 1991) and the pMAL-c2 vector (New England Biolabs, Beverly, MA) for producing GST and MBP fusion proteins, respectively. Fusion protein induction and purification were performed according to the manufacturer's protocol (Amersham Biosciences, Piscataway, NJ for the GST-BRI1CK fusion proteins and New England Biolabs for the MBP-TTL fusion proteins). In vitro kinase assays were performed at 25°C for 60 min in a 20-μL reaction mixture containing 50 mM Hepes, pH 7.4, 10 mM MgCl2, 10 mM MnCl2, 1 mM DTT, 50 μM cold ATP, and 0.5 μL of [γ-32P]ATP (3000 Ci/mmole; ICN Biomedicals, Irvine, CA) with the approximately same amount of purified proteins determined by Coomassie Brilliant Blue staining. Kinase reactions were terminated by adding 5 μL of SDS sample buffer (0.35 M Tris-HCl, pH 6.8, 30% glycerol [v/v], 10% SDS [w/v], 0.6 M DTT, and 0.012% bromophenol blue [w/v]) and boiled for 3 min, and proteins were separated by a 8% SDS-PAGE. Gels were stained with Coomassie blue to reveal the amount of proteins and exposed to Kodak Biomax x-ray films (Eastman Kodak Company, Rochester, NY) to visualize phosphorylated bands.
Histochemical GUS Reporter Gene Expression Analysis
A 1.6-kb genomic fragment containing the TTL promoter sequence was amplified from the Arabidopsis genomic DNA and cloned into a modified pPZP222 binary vector (Hajdukiewicz et al., 1994) that contains the Escherichia coli GUS coding sequence. The resulting pPZP222-TTL-GUS construct was then transformed into wild-type Arabidopsis plants. Histochemical analysis of the GUS reporter gene was performed as described (Stomp, 1992) with T2 TTL-GUS transgenic plants of different developmental ages. After staining, samples were rinsed sequentially with 50 mM potassium phosphate, pH 7.0, acetic acid/methanol (1:3 [v/v]), and 70% (v/v) lactic acid to remove the chlorophyll and cytoplasm, allowing visualization of GUS staining of plant samples.
Confocal Microscopic Analysis of Subcellular Localization of BRI1 and TTL
The construct, pPZP212-BRI1-BRI1:GFP, used for generating BRI1:GFP transgenic plants was described previously (Friedrichsen et al., 2000). To generate TTL-TTL:GFP transgenic plants, we first fused the full-length TTL ORF with the GFP sequence in the pAVA393 vector (von Arnim et al., 1998) and cloned the resulting TTL:GFP fusion gene into the pPZP212 vector (Hajdukiewicz et al., 1994) that contains the 1.6-kb TTL promoter sequence to generate the pPZP212-TTL-TTL:GFP fusion construct. Both fusion constructs were transformed into Arabidopsis plants by the Agrobacterium tumefaciens–mediated method. Subcellular localization patterns of BRI1, TTL, and GFP alone were revealed by examining the green fluorescence patterns in the root elongation region right above the root apical meristem of 4-d-old vertically grown seedlings of transgenic lines containing a pPZP212-BRI1-BRI1:GFP, a pPZP212-TTL-TTL:GFP, or a 35S-GFP fusion construct using a Zeiss LSM510 confocal microscope (Jena, Germany) filtered with an FITC10 set (excitation 488 nm and emission 505 to 530 nm and 530 to 560 nm).
Cellular Fractionation Analysis for Localization of TTL
Cytosolic and microsomal fractions are separated according to the method previously reported (Kjellbom and Larsson, 1984) with modifications. One-week-old seedlings were homogenized in the hypertonic buffer (0.5 M sucrose, 50 mM Hepes, pH 7.5, 0.6% polyvinylpolypyrrolidone, and 5 mM ascorbic acid). After centrifugation at 10,000g for 20 min, crude extracts were centrifuged at 60,000g for 60 min. Supernatant was recovered as cytosolic fraction, and the pellet was resuspended in 5 mM potassium phosphate, pH 7.8, buffer containing 0.33 M sucrose and 3 mM KCl as microsomal fraction. All the procedures were performed at 4°C. Protein concentration was determined using the Bradford method, and the same amount of proteins for each sample were separated by SDS-PAGE, blotted onto a polyvinylidene difluoride membrane, and analyzed by immunoblotting with anti-GFP antibodies.
RNA Isolation and RNA Gel Blot Hybridization
Total RNAs were extracted from various tissues of 28-d-old soil-grown Arabidopsis plants or 10-d-old seedlings grown on synthetic medium as described previously (Li et al., 2001b). For RNA gel blot analysis, 5 μg of total RNAs were separated by a 1% formaldehyde-agarose gel, transferred to a Hybond-XR membrane (Amersham Pharmacia Biotech, Piscataway, NJ), and hybridized with a full-length TTL or 18s rDNA probe at 42°C for 22 h in a solution of 50% formamide (v/v), 250 mM sodium phosphate, pH 7.4, 1 mM EDTA, 1% casein, and 7% SDS (w/v).
Acknowledgments
We thank members of the Li lab for helpful discussions throughout this study, J. Chory for support with the initial yeast two-hybrid screen, A. von Arnim for the GFP construct, the ABRC (Ohio State University, Columbus, OH) for Arabidopsis seeds and DNA clones, and the Arabidopsis Gene Knockout Facility (University of Wisconsin, Madison, WI) for screening the ttl mutants. This work was supported by a research grant from the National Institutes of Health (GM60519) to J.L.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jianming Li (jian@umich.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.023903.
References
- Aranda, A., and Pascual, A. (2001). Nuclear hormone receptors and gene expression. Physiol. Rev. 81, 1269–1304. [DOI] [PubMed] [Google Scholar]
- Asami, T., Min, Y.K., Nagata, N., Yamagishi, K., Takatsuto, S., Fujioka, S., Murofushi, N., Yamaguchi, I., and Yoshida, S. (2000). Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol. 123, 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashultz, A.C., Nygaard, P., and Saxild, H.H. (2001). Functional analysis of 14 genes that constitute the purine catabolic pathway in Basillus subtilis and evidence for a novel regulon controlled by the PucR transcription activator. J. Bacteriol. 183, 3293–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson, M.D., and Uemichi, T. (1996). Transthyretin amyloidosis amyloid. Int. J. Exp. Clin. Invest. 3, 44–56. [Google Scholar]
- Caño-Delgado, A., Yin, Y., Yu, C., Vafeados, D., Mora-García, S., Cheng, J.-C., Nam, K.H., Li, J., and Chory, J. (2004). BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development, in press. [DOI] [PubMed]
- Cato, A.C.B., Nestl, A., and Mink, S. (2002). Rapid actions of steroid receptors in cellular signaling pathways. Sci. STKE 2002, RE9. [DOI] [PubMed] [Google Scholar]
- Catterou, M., Dubois, F., Schaller, H., Aubanelle, L., Vilcot, B., Sangwan-Norreel, B.S., and Sangwan, R.S. (2001). Brassinosteroids, microtubules and cell elongation in Arabidopsis thaliana. II. Effects of brassinosteroids on microtubules and cell elongation in the bul1 mutant. Planta 212, 673–683. [DOI] [PubMed] [Google Scholar]
- Choe, S., Fujioka, S., Noguchi, T., Takatsuto, S., Yoshida, S., and Feldmann, K.A. (2001). Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J. 26, 573–582. [DOI] [PubMed] [Google Scholar]
- Choe, S., Schmitz, R.J., Fujioka, S., Takatsuto, S., Lee, M.O., Yoshida, S., Feldmann, K.A., and Tax, F.E. (2002). Arabidopsis brassinosteroid-insensitive dwarf12 mutants are semidominant and defective in a glycogen synthase kinase 3 beta-like kinase. Plant Physiol. 130, 1506–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, S.E., Williams, R.W., and Meyerowitz, E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585. [DOI] [PubMed] [Google Scholar]
- Clouse, S.D. (2002. a). Brassinosteroid signal transduction: Clarifying the pathway from ligand perception to gene expression. Mol. Cell 10, 973–982. [DOI] [PubMed] [Google Scholar]
- Clouse, S.D. (2002. b). Brassinosteroid signaling: Novel downstream components emerge. Curr. Biol. 12, R485–R487. [DOI] [PubMed] [Google Scholar]
- Clouse, S.D., Langford, M., and McMorris, T.C. (1996). A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 111, 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eneqvist, T., Lundberg, E., Nilsson, L., Abagyan, R., and Sauer-Eriksson, A.E. (2003). The transthyretin-related protein family. Eur. J. Biochem. 270, 518–532. [DOI] [PubMed] [Google Scholar]
- Fankhauser, C., Yeh, K.C., Lagarias, J.C., Zhang, H., Elich, T.D., and Chory, J. (1999). PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284, 1539–1541. [DOI] [PubMed] [Google Scholar]
- Friedrichsen, D.M., Joazeiro, C.A., Li, J., Hunter, T., and Chory, J. (2000). Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 123, 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, K.L., and Dixon, J.E. (1991). Eukaryotic proteins expressed in Escherichia coli: An improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192, 262–267. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. [DOI] [PubMed] [Google Scholar]
- He, J.X., Gendron, J.M., Yang, Y., Li, J., and Wang, Z.Y. (2002). The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 99, 10185–10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Z., Wang, Z.Y., Li, J., Zhu, Q., Lamb, C., Ronald, P., and Chory, J. (2000). Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science 288, 2360–2363. [DOI] [PubMed] [Google Scholar]
- Katsumi, M. (1991). Physiological modes of brassinolide action in cucumber hypocotyls growth. In Brassinosteroids: Chemistry, Bioactivity, and Application, ACS Symposium Series 474, H.G. Cutler, T. Yokota, and G. Adam, eds (Washington, DC: American Chemical Society), pp. 246–254.
- Kauschmann, A., Jessop, A., Koncz, C., Szekeres, M., Willmitzer, L., and Altmann, T. (1996). Genetic evidence for an essential role for brassinosteroids in plant development. Plant J. 9, 701–713. [Google Scholar]
- Keller, T., Damude, H.G., Werner, D., Doerner, P., Dixon, R.A., and Lamb, C. (1998). A plant homolog of the neutrophil NADPH oxidase gp91phox subunit gene encodes a plasma membrane protein with Ca2+ binding motifs. Plant Cell 10, 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjellbom, P., and Larsson, C. (1984). Preparation and polypeptide composition of chlorophyll-free plasma membranes from leaves of light-grown spinach and barley. Physiol. Plant. 62, 501–509. [Google Scholar]
- Li, J., and Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938. [DOI] [PubMed] [Google Scholar]
- Li, J., Lease, K.A., Tax, F.E., and Walker, J.C. (2001. a). BRS1, a serine carboxypeptidase, regulates BRI1 signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 98, 5916–5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., and Nam, K.H. (2002). Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295, 1299–1301. [DOI] [PubMed] [Google Scholar]
- Li, J., Nam, K.H., Vafeados, D., and Chory, J. (2001. b). BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 127, 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Wen, J., Lease, K.A., Doke, J.T., Tax, F.E., and Walker, J.C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222. [DOI] [PubMed] [Google Scholar]
- Mandava, N.B. (1988). Plant growth-promoting brassinosteroids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39, 23–52. [Google Scholar]
- Mayumi, K., and Shibaoka, H. (1995). A possible double role for brassinolide in the reorientation of cortical microtubules in the epidermal cells of Azuki bean epicotyls. Plant Cell Physiol. 36, 173–181. [Google Scholar]
- Monaco, H.L. (2000). The transthyretin-retinol-binding protein complex. Biochim. Biophys. Acta 1482, 65–72. [DOI] [PubMed] [Google Scholar]
- Mora-Garcia, S., Vert, G., Yin, Y., Cano-Delgado, A., Cheong, H., and Chory, J. (2004). Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 18, 448–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillon, R., Catterou, M., Sangwan, R.S., Sangwan, B.S., and Lassalles, J.P. (2001). Brassinolide may control aquaporin activities in Arabidopsis thaliana. Planta 212, 199–204. [DOI] [PubMed] [Google Scholar]
- Nam, K.H., and Li, J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110, 203–212. [DOI] [PubMed] [Google Scholar]
- Noguchi, T., Fujioka, S., Choe, S., Takatsuto, S., Yoshida, S., Yuan, H., Feldmann, K.A., and Tax, F.E. (1999). Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 121, 743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palha, J.A. (2002). Transthyretin as a thyroid hormone carrier: Function revisited. Clin. Chem. Lab. Med. 40, 1292–1300. [DOI] [PubMed] [Google Scholar]
- Peng, P., and Li, J. (2003). Brassinosteroid signal transduction: A mix of conservation and novelty. J. Plant Growth Regul. 29, 298–312. [DOI] [PubMed] [Google Scholar]
- Perez-Perez, J.M., Ponce, M.R., and Micol, J.L. (2002). The UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev. Biol. 242, 161–173. [DOI] [PubMed] [Google Scholar]
- Stomp, A. (1992). Histochemical localization of β-glucuronidase. In GUS Protocols: Using the GUS Gene as a Reporter of Gene Expression, S.R. Gallagher, ed (London: Academic Press), pp. 103–113.
- Thummel, C.S., and Chory, J. (2002). Steroid signaling in plants and insects—Common themes, different pathways. Genes Dev. 16, 3113–3129. [DOI] [PubMed] [Google Scholar]
- von Arnim, A.G., Deng, X.W., and Stacey, M.G. (1998). Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene 221, 35–43. [DOI] [PubMed] [Google Scholar]
- Wang, Z.Y., Nakano, T., Gendron, J., He, J., Chen, M., Vafeados, D., Yang, Y., Fujioka, S., Yoshida, S., Asami, T., and Chory, J. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2, 505–513. [DOI] [PubMed] [Google Scholar]
- Wang, Z.Y., Seto, H., Fujioka, S., Yoshida, S., and Chory, J. (2001). BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410, 380–383. [DOI] [PubMed] [Google Scholar]
- Winkler, R.G., Frank, M.R., Galbraith, D.W., Feyereisen, R., and Feldmann, K.A. (1998). Systemic reverse genetics of transfer-DNA-tagged lines of Arabidopsis. Plant Physiol. 118, 743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X.W., Muzny, D.M., Lee, C.C., and Caskey, C.T. (1992). Two independent mutational events in the loss of urate oxidase during hominoid evolution. J. Mol. Evol. 34, 78–84. [DOI] [PubMed] [Google Scholar]
- Yamamoto, K., Kojima, Y., Kikuchi, T., Shigyo, T., Sugihara, K., Takashio, M., and Emi, S. (1996). Nucleotide sequence of the uricase gene from Bacillus sp. TB-90. J. Biochem. 119, 80–84. [DOI] [PubMed] [Google Scholar]
- Yamamuro, C., Ihara, Y., Wu, X., Noguchi, T., Fujioka, S., Takatsuto, S., Ashikari, M., Kitano, H., and Matsuoka, M. (2000). Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12, 1591–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Y., Wang, Z.Y., Mora-Garcia, S., Li, J., Yoshida, S., Asami, T., and Chory, J. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109, 181–191. [DOI] [PubMed] [Google Scholar]
- Zhao, J., Peng, P., Schmitz, R.J., Decker, A.D., Tax, F.E., and Li, J. (2002). Two putative BIN2 substrates are nuclear components of brassinosteroid signaling. Plant Physiol. 130, 1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]