Abstract
The MYC-like sequence CATGTG plays an important role in the dehydration-inducible expression of the Arabidopsis thaliana EARLY RESPONSIVE TO DEHYDRATION STRESS 1 (ERD1) gene, which encodes a ClpA (ATP binding subunit of the caseinolytic ATP-dependent protease) homologous protein. Using the yeast one-hybrid system, we isolated three cDNA clones encoding proteins that bind to the 63-bp promoter region of erd1, which contains the CATGTG motif. These three cDNA clones encode proteins named ANAC019, ANAC055, and ANAC072, which belong to the NAC transcription factor family. The NAC proteins bound specifically to the CATGTG motif both in vitro and in vivo and activated the transcription of a β-glucuronidase (GUS) reporter gene driven by the 63-bp region containing the CATGTG motif in Arabidopsis T87 protoplasts. The expression of ANAC019, ANAC055, and ANAC072 was induced by drought, high salinity, and abscisic acid. A histochemical assay using PNAC-GUS fusion constructs showed that expression of the GUS reporter gene was localized mainly to the leaves of transgenic Arabidopsis plants. Using the yeast one-hybrid system, we determined the complete NAC recognition sequence, containing CATGT and harboring CACG as the core DNA binding site. Microarray analysis of transgenic plants overexpressing either ANAC019, ANAC055, or ANAC072 revealed that several stress-inducible genes were upregulated in the transgenic plants, and the plants showed significantly increased drought tolerance. However, erd1 was not upregulated in the transgenic plants. Other interacting factors may be necessary for the induction of erd1 in Arabidopsis under stress conditions.
INTRODUCTION
Plants grow in a dynamic environment that frequently imposes constraints on growth and development. Among the adverse environmental factors commonly encountered by land plants are extremes in temperature and osmotic stress, which can result in either water deficit or salinity stress. Molecular and cellular responses to these stresses have been analyzed extensively at the biochemical level (for reviews, see Ingram and Bartels, 1996; Thomashow, 1999; Bray et al., 2000; Shinozaki and Yamaguchi-Shinozaki, 2000; Zhu, 2002). The early events of plant adaptation to environmental stresses are perception and subsequent stress-signal transduction, leading to the activation of various physiological and metabolic responses, including stress-responsive gene expression. Many genes are induced by osmotic stress or low temperature (Thomashow, 1999; Bray et al., 2000; Zhu, 2002; Seki et al., 2003; Shinozaki et al., 2003). Gaining an understanding of the mechanisms that regulate the expression of these genes is a fundamental issue in plant biology and will be necessary for the genetic improvement of plants cultivated in extreme environments.
Plant breeding for improved stress tolerance has consistently demonstrated that plant vigor under a range of environmental conditions is governed by multiple loci. Thus, stress tolerance is an inherently multigenic trait in nature. Although the vast majority of these genes remain to be identified, some transcription factors and regulatory sequences in plant promoters have been described. In Arabidopsis thaliana, cis-elements and corresponding binding proteins, each containing a distinct type of DNA binding domain, such as AP2/ERF, basic leucine zipper, HD-ZIP, MYB, MYC, and several classes of zinc finger domains, have been implicated in plant stress responses because their expression is induced or repressed under different stress conditions (Shinozaki and Yamaguchi-Shinozaki, 2000; Pastori and Foyer, 2002). Altering the expression of certain transcription factors can greatly influence plant stress tolerance (Jaglo-Ottosen et al., 1998; Liu et al., 1998; Kasuga et al., 1999).
There are at least four independent regulatory systems for gene expression in response to drought stress in Arabidopsis. Two are abscisic acid (ABA) dependent and two are ABA independent (Shinozaki and Yamaguchi-Shinozaki, 2000). Dehydration-responsive element/C-repeat (DRE/CRT) has been identified as a cis-acting element involved in one of the ABA-independent regulatory systems. DRE/CRT also functions in cold- and high-salt-responsive gene expression. When the DRE/CRT binding protein DREB1/CBF was overexpressed in the transgenic Arabidopsis plants, altered expression of more than 40 stress-inducible genes was observed, leading to increase freezing, salt, and drought tolerance (Seki et al., 2001; Fowler and Thomashow, 2002; Maruyama et al., 2004). Other transcriptional regulators, such as the MYC and MYB proteins, are activators in one of the ABA-dependent regulatory systems (Abe et al., 2003). ABA-responsive element functions as a cis-acting element in the other ABA-dependent regulatory system. ABA-responsive element binding basic leucine zipper–type proteins known as AREBs/ABFs have been identified as transcriptional activators in this ABA-dependent regulatory system (Choi et al., 2000; Uno et al., 2000).
The least understood regulatory system is the ABA-independent one, which functions in the activation of drought-inducible genes that do not respond to either cold or ABA treatment, such as erd1, which encodes a protein with homology to the ATP binding subunit of the Clp ATP-dependent protease from Escherichia coli (Kiyosue et al., 1993; Nakashima et al., 1997). To further dissect this ABA-independent regulatory system, much research, including promoter analysis, has been conducted on erd1, which is upregulated in response to drought, high salinity, and dark-induced senescence (Kiyosue et al., 1993; Nakashima et al., 1997). Recently, Simpson et al. (2003) demonstrated that erd1 expression during dehydration depends on the integrity of both the 14-bp rps1 site 1–like sequence and the putative MYC-like (CATGTG) sequence in the promoter region.
To elucidate the trans-acting factors that interact with the putative cis-acting motifs found in the erd1 promoter region, and thus further dissect the signal transduction machinery involved in the early response to dehydration in Arabidopsis, we cloned three cDNAs encoding proteins that can bind to the 63-bp fragment of the erd1 promoter from −434 to −497 containing the CATGTG motif. Sequence analyses revealed that the three newly identified proteins belong to a large multigene family of plant-specific NAC transcription factors with more than 100 members (Riechmann et al., 2000). We analyzed the function of the three NAC proteins as trans-acting factors by transient expression in Arabidopsis T87 protoplasts. By analysis of the NAC proteins using the yeast one-hybrid protocol, we identified the complete DNA binding sequence recognized by the NAC proteins, which we named NAC recognition sequence (NACRS), and the core binding site. We studied the expression of the associated NAC genes by RNA gel blot analysis and promoter–β-glucuronidase (GUS) assay. We also studied the gene expression profile of Arabidopsis transgenic plants ectopically expressing ANAC019, ANAC055, and ANAC072 using a 7000-cDNA microarray and identified several target genes of the ANAC019, ANAC055, and ANAC072 transcriptional activators. These transgenic plants showed improved drought tolerance.
RESULTS
Isolation of cDNAs Encoding DNA Binding Proteins That Recognize MYC-Like CATGTG Motif in the 63-bp DNA Fragment of the erd1 Promoter
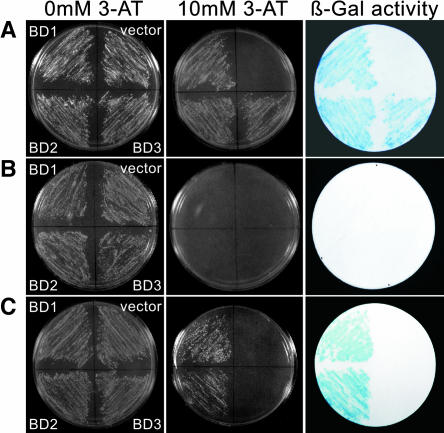
To isolate cDNAs encoding DNA binding proteins that interact with the 63-bp fragment containing the CATGTG motif, we used the yeast one-hybrid screening system. First, we constructed the YSMHL2 yeast strain carrying, as dual reporter genes, integrated copies of HIS3 and lacZ fused to a four-times tandemly repeated 63-bp DNA fragment of the erd1 promoter containing the CATGTG motif. The yeast strain was tested for HIS3 and lacZ background expression: (1) YSMHL2 transcribes the HIS3 gene at basal levels and grows on medium lacking His, but not in the presence of 10 mM 3-aminotriazole (3-AT); (2) it forms white colonies on filter papers that had been prewetted with 5-bromo-4-chloro-3-indoyl-β-d-galactopyranoside. To screen for cDNAs encoding DNA binding proteins of interest, we separately transformed the target reporter YSMHL2 strain with two pAD-GAL4 cDNA libraries, which were constructed from cDNA fragments of mRNAs prepared from unstressed and 3-h-dehydrated Arabidopsis rosette plants. Forty-nine 10 mM 3-AT–resistant clones were isolated from the library prepared from dehydrated Arabidopsis plants. All of these clones induced lacZ activity and formed blue colonies on filter paper containing 5-bromo-4-chloro-3-indoyl-β-d-galactopyranoside (Figure 1A). The cDNA fragments on the isolated plasmids were analyzed by restriction enzyme digestion and DNA sequencing, which led to the classification of these 49 clones into three close cDNA groups designated BD1 (11 clones), BD2 (35 clones), and BD3 (three clones; Figure 1A).
Figure 1.
Isolation of cDNA Encoding CATGTG Motif Binding Proteins.
(A) Forty-nine positive clones were isolated using the yeast one-hybrid system and classified into three groups by sequence analysis: BD1, BD2, and BD3.
(B) Three pAD-GAL4 plasmid derivatives carrying cDNA clones encoding BD1, BD2, and BD3 were transformed into a yeast strain carrying the reporter genes under the control of four tandemly repeated 63-bp promoter fragments containing AAAAAA in place of CATGTG. The transformants were examined for growth in the presence of 3-AT and β-galactosidase (β-Gal) activity, demonstrating binding specificity of BD1, BD2, and BD3.
(C) The cDNA fragments encoding BD1, BD2, and BD3 were cloned into the constitutively expressed YepGAP vector, and the resulting plasmids were introduced into a yeast strain carrying the reporter genes under the control of four tandemly repeated 63-bp promoter fragments containing the CATGTG motif. BD1 and BD2, but not BD3, were able to transactivate the expression of the HIS3 and lacZ reporter genes.
When the BD1, BD2, and BD3 clones were transformed into the YSMHLMA yeast strain carrying the two reporter genes fused to the 63-bp DNA fragment with base substitution in the CATGTG motif, the recombinant yeast strains neither grew on medium lacking His in the presence of 3-AT nor induced lacZ activity (Figure 1B). To confirm whether the three clones were transcriptional activators, we cloned the insert cDNA fragments encoding the intact BD1, BD2, and BD3 proteins into the yeast expression vector YepGAP (Liu et al., 1998). The resulting plasmids were transformed into the YSMHL2 reporter strain. Yeast cells carrying the plasmid containing cDNA of either BD1 or BD2 grew on the medium lacking His in the presence of 3-AT and induced lacZ activity, but those carrying the plasmid containing the cDNA of BD3 did not (Figure 1C). The result indicated that BD1 and BD2, but not BD3, could function as transcriptional activators, at least in yeast.
Structural Analysis of the BD cDNAs
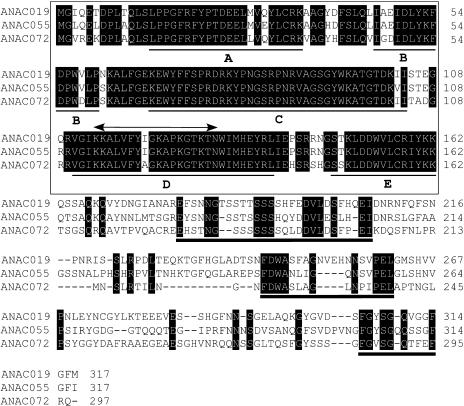
To study the structure of the BD1, BD2, and BD3 clones, we sequenced the inserted cDNA fragments, measuring 1.1, 1.2, and 0.9 kb, respectively. The amino acid sequence of each encoded protein was subjected to a database search. The N-terminal half of the three proteins was found to share substantial sequence similarity with all members of the NAC family of proteins (Ooka et al., 2003). BD1, BD2, and BD3 were therefore renamed ANAC055, ANAC019, and ANAC072 according to the nomenclature established for the family of Arabidopsis NAC proteins (Ooka et al., 2003). The MYC-like CATGTG motif was thus designated NACRS. At the nucleotide sequence level, ANAC019 has 77% sequence similarity to ANAC055 and 56% to ANAC072, and ANAC055 has 51% sequence similarity to ANAC072. The isolated cDNAs each contains a single open reading frame that encodes a novel protein of 317, 317, and 297 amino acids, respectively (Figure 2). At the amino acid level, ANAC019 has 72% sequence similarity to ANAC055 and 65% to ANAC072, and ANAC055 has 62% sequence similarity to ANAC072. The observed regions of homology among ANAC019, ANAC055, and ANAC072 were found not only in the NAC domain but also in three short regions, one a Ser-rich region and one near the C terminus (Figure 2).
Figure 2.
Comparison of the Amino Acid Sequences of ANAC019, ANAC055, and ANAC072.
Identical amino acids are indicated by white letters on a black background. The highly conserved region found in NAC family members is boxed. The putative nuclear localization signal is shown by a double-headed arrow above the sequence. The consensus subdomains in the NAC binding domain are shown by thin underlines. The consensus regions in the C-terminal part are shown by thick underlines.
To date the biological function of these NAC transcription factors remains unknown. The amino acid sequence of the ANAC055 protein is identical to that of AtNAC3 (Takada et al., 2001), which is a putative jasmonic acid regulatory protein with a molecular mass of 35.4 kD. ANAC019 is identical to a recently identified ABA-responsive protein, ANAC (Greve et al., 2003), with a molecular mass of 35.8 kD. ANAC072, or the complete version of RD26, is a drought-inducible protein (Yamaguchi-Shinozaki et al., 1992) with an apparent molecular mass of 32.7 kD.
Purified ANAC019, ANAC055, and ANAC072 Bind Specifically to the NACRS Motif
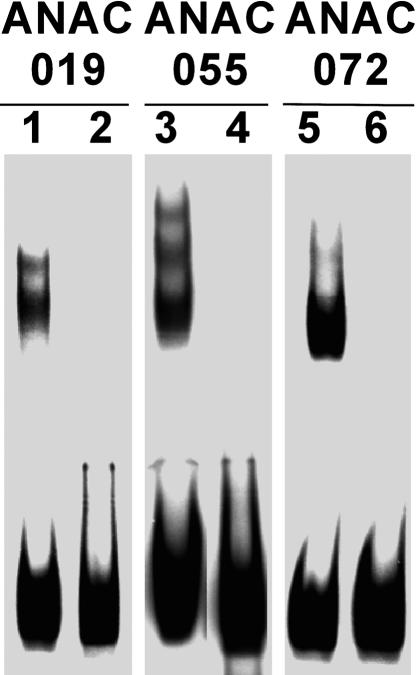
To validate the genetic data in vitro, we attempted to see whether the purified ANAC019, ANAC055, and ANAC072 proteins specifically interacted with the NACRS. To accomplish this, we used E. coli to express recombinant NAC–glutathione S-transferase (NAC-GST) fusion proteins, which we then purified. These proteins were incubated with both the wild-type 63-bp fragment containing the intact NACRS motif and the base-substituted 63-bp fragment in which the CATGTG motif was replaced with the sequence AAAAAA. The incubation mixtures were then analyzed by gel retardation assay. Whereas the wild-type fragment caused a distinct shift in the molecular weight of the NAC fusion proteins, the base-substituted fragment produced no such shift (Figure 3). The GST protein did not bind to either the wild-type or base-substituted 63-bp fragments (data not shown). This result demonstrates that the ANAC019, ANAC055, and ANAC072 proteins specifically bind to the 63-bp DNA fragment carrying the NACRS motif.
Figure 3.
Purified NAC Proteins Bind Specifically to the CATGTG Motif.
In vitro DNA binding reactions were performed with the wild-type 63-bp fragment containing the CATGTG motif (lanes 1, 3, and 5) and the base-substituted 63-bp fragment in which the CATGTG motif was replaced by AAAAAA (lanes 2, 4, and 6).
ANAC019, ANAC055, and ANAC072 Transactivate the erd1 Promoter–GUS Fusion in Arabidopsis T87 Protoplasts
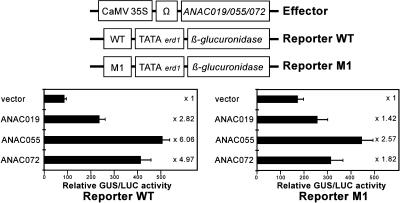
To determine whether the ANAC019, ANAC055, and ANAC072 proteins are capable of transactivating NACRS-dependent transcription in plant cells, we performed transactivation assays using Arabidopsis T87 protoplasts. Protoplasts were cotransfected with a GUS reporter gene fused to a copy of the 63-bp fragment containing the NACRS motif (reporter WT) and an effector plasmid (Figure 4). The effector plasmid consisted of the 35S promoter of Cauliflower mosaic virus (CaMV) fused to either ANAC019, ANAC055, or ANAC072 cDNAs. Expression of each of these constructs in T87 protoplasts significantly transactivated the expression of the GUS reporter gene (Figure 4). By contrast, when protoplasts were cotransfected with a GUS reporter gene fused to a copy of the base substituted 63-bp fragment described above (reporter M1) and either a ANAC019, ANAC055, or ANAC072 effector plasmid, the degree of induction of the GUS reporter gene was significantly lower (Figure 4). These results demonstrate that ANAC019, ANAC055, and ANAC072 indeed function as transcriptional activators involved in the NACRS-dependent expression of erd1 in Arabidopsis and confirm the pivotal role of the CATGTG motif on the binding activity of the NAC genes. Interestingly, in Arabidopsis the ANAC072 construct significantly induced the NACRS-dependent expression of erd1, but in yeast it was not able to transactivate either HIS or lacZ reporter genes fused with up to four tandemly repeated copies of the 63-bp fragment (Figure 1).
Figure 4.
Transactivation Assay of ANAC019, ANAC055, and ANAC072 in T87 Protoplasts.
cDNA fragments encoding ANAC019, ANAC055, and ANAC072 were cloned into plant expression vector pBI35SΩ and cotransformed into T87 protoplasts with the reporter gene plasmids (reporter WT or reporter M1). To normalize for transformation efficiency, the CaMV 35S-luciferase plasmid was also cotransferred in all cases. Bars indicate the standard errors of three replicates. Multiplication values refer to the ratio of expression relative to the value obtained with the pBI35SΩ vector.
Determination of Complete NACRS and Core DNA Binding Sequence Using Yeast One-Hybrid System
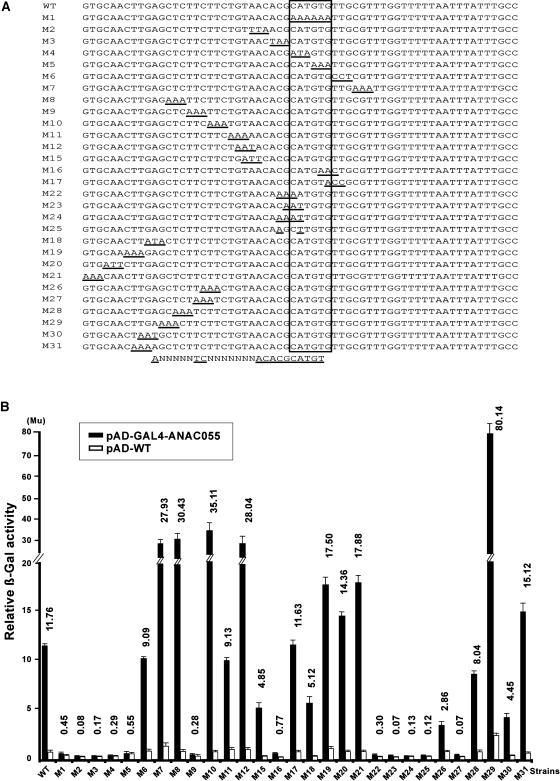
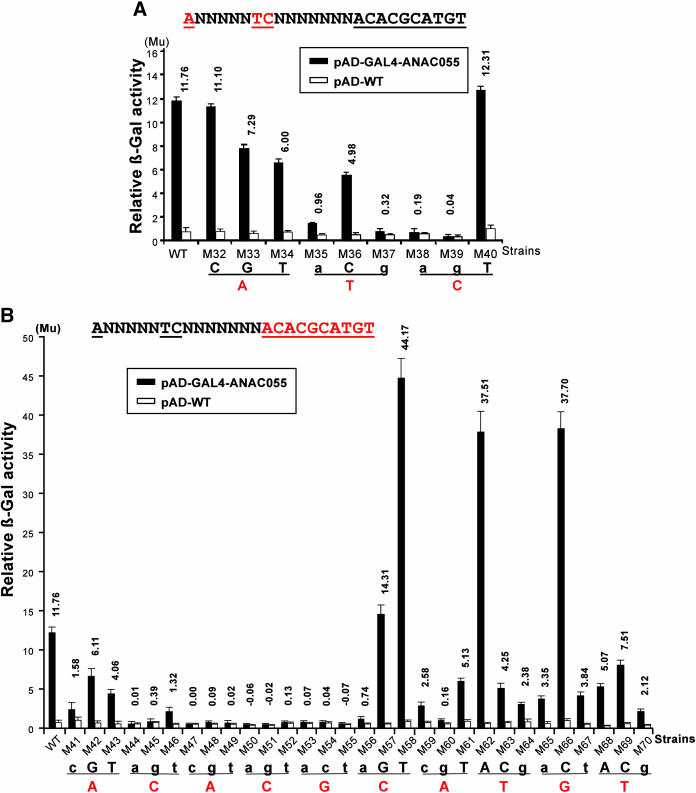
To localize the complete NACRS sequence and the core DNA sequence within the 63-bp fragment required for the DNA binding of the NAC proteins, we used the GAL4AD-ANAC055 fusion for yeast one-hybrid system. First, we analyzed the influence of discrete, 3- to 4-bp sections on β-galactosidase activity using a series of nonoverlapping and overlapping base substitutions (Figure 5A). Base substitutions were not performed further on the 3′-side of the CATGTG motif because those nucleotides do not influence the drought-responsive expression of erd1 (Simpson et al., 2003). Analysis of the transcriptional activation measured in relative β-galactosidase activity using the criterion of six units' difference, which is the difference in the β-galactosidase activity observed when comparing the pAD-GAL4-ANAC055 and pAD-WT plasmids, was performed to define the significance of the observed binding affinity. This analysis revealed that ANNNNNTCNNNNNNNACACGCATGT contains the complete NACRS (Figures 5A and 5B). To specify the NACRS more precisely and to localize the core DNA binding motif, we generalized a second series of single base substitutions of the nucleotides in the above sequence (Figures 6A and 6B). β-Galactosidase activity was assessed in recombinant yeast strains, using the criterion of four units' difference to indicate significant binding affinity. Results indicated that TCNNNNNNNACACGCATGT and CACG are the minimal NACRS and the core DNA binding motif of the ANAC055 protein, respectively (Figures 6A and 6B).
Figure 5.
Localization of Complete NACRS Using GAL4AD-ANAC055 Fusion.
(A) Base substitution analysis of the 63-bp promoter fragment. Twenty-nine constructs were made for localization of the complete NACRS in the 63-bp fragment. Each base substitution construct was initially fused to the lacZ gene and integrated into the YM4271 chromosome. Yeast recombinant strains were then transformed with pAD-GAL4-ANAC055 or pAD-WT plasmids. The position of the MYC-like sequence is boxed. Mutated nucleotides are underlined.
(B) Results of liquid culture assays. The criterion of six units' difference, which is approximately half of the units' difference in the wild type, was used for analysis of the differences of β-galactosidase (β-Gal) activities given by pAD-GAL4-ANAC055 and pAD-WT in each mutated construct. Mu, Miller units.
Figure 6.
Determination of the Core DNA Binding Motif Using GAL4AD-ANAC055 Fusion.
Thirty-nine constructs were made to define the core motif of NACRS. Each base substitution construct was initially fused to the lacZ gene and integrated into the YM4271 chromosome. Yeast recombinant strains were then transformed with pAD-GAL4-ANAC055 or pAD-WT plasmids. β-Gal, β-galactosidase; Mu, Miller units.
(A) Results of liquid culture assays of nine constructs designed to facilitate an investigation of the first three nucleotides (red letters).
(B) Results of liquid culture assays of 30 constructs designed to facilitate an investigation of the next 10 nucleotides (red letters). The criterion of four units' difference, which is approximately one-third of the units' difference in the wild type, was used for analysis of the differences of β-galactosidase activities given by pAD-GAL4-ANAC055 and pAD-WT in each mutated construct. Nucleotide changes in the NACRS that had an adverse effect on the binding activity of ANAC055 protein are indicated by small letters.
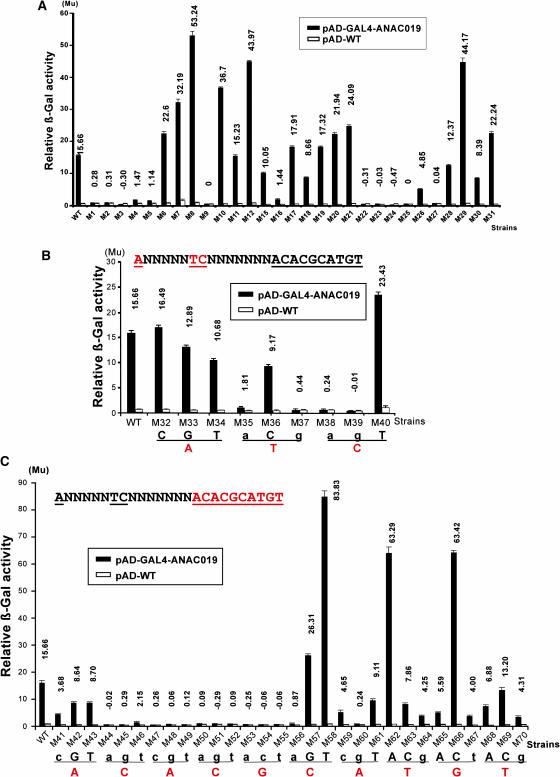
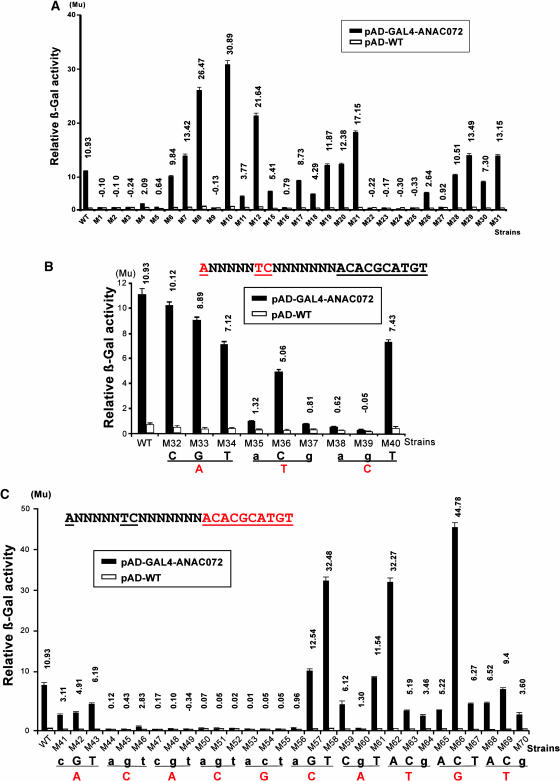
To determine that whether TCNNNNNNNACACGCATGT and CACG are also the NACRS and the core DNA binding motif of ANAC019 and ANAC072, we introduced the GAL4AD-ANAC019 and GAL4AD-ANAC072 fusions to all the 69 yeast reporter strains. The results of yeast one-hybrid analyses are shown in Figures 7 and 8. The profiles of activation of GAL4AD-ANAC055, GAL4AD-ANAC019, and GAL4AD-ANAC072 measured in relative β-galactosidase activity are very similar (Figures 5, 6, 7, and 8), suggesting that the NAC proteins share a highly conserved binding site. With the wild-type 63-bp fragment, GAL4AD-ANAC019 showed a greater activation level than either GAL4AD-ANAC055 or GAL4AD-ANAC072, which gave similar levels of activation. This might be because of difference in binding affinity of the NAC proteins because of the different nature of the NAC domain of each NAC protein or because of the additional contribution of the NAC activation domain to the relative β-galactosidase activity. These results indicated that the three NAC proteins have the same core DNA binding sequence, CACG, and led us to a hypothesis that CACG may also be the core DNA binding sequence for other NAC proteins.
Figure 7.
Localization of Complete NACRS and Core DNA Binding Motif Using GAL4AD-ANAC019 Fusion.
(A) Localization of complete NACRS. Results of liquid culture assays of 29 constructs (Figure 5A) designed for localization of the complete NACRS in the 63-bp promoter fragment. The criterion of eight units' difference, which is approximately half of the units' difference in the wild type, was used for analysis of the differences of β-galactosidase (β-Gal) activities given by pAD-GAL4-ANAC019 and pAD-WT in each mutated construct. Mu, Miller units.
(B) and (C) Localization of core DNA binding motif. Results of liquid culture assays of nine (B) and of 30 (C) constructs designed to facilitate an investigation of the first three and the next 10 nucleotides (red letters), respectively, to define the core motif of NACRS. The criterion of 5.5 units' difference, which is approximately one-third of the units' difference in the wild type, was used for analysis of the differences of β-galactosidase activities given by pAD-GAL4-ANAC019 and pAD-WT in each mutated construct. Nucleotide changes in the NACRS that had an adverse effect on the binding activity of ANAC019 protein are indicated by small letters.
Figure 8.
Localization of Complete NACRS and Core DNA Binding Motif Using GAL4AD-ANAC072 Fusion.
(A) Localization of complete NACRS. Results of liquid culture assays of 29 constructs (Figure 5A) designed for localization of the complete NACRS in the 63-bp promoter fragment. The criterion of 5.5 units' difference, which is approximately half of the units' difference in the wild type, was used for analysis of the differences of β-galactosidase (β-Gal) activities given by pAD-GAL4-ANAC072 and pAD-WT in each mutated construct. Mu, Miller units.
(B) and (C) Localization of core DNA binding motif. Results of liquid culture assays of nine (B) and of 30 (C) constructs designed to facilitate an investigation of the first three and the next 10 nucleotides (red letters), respectively, to define the core motif of NACRS. The criterion of 3.7 units' difference, which is approximately one-third of the units' difference in the wild type, was used for analysis of the differences of β-galactosidase activities given by pAD-GAL4-ANAC072 and pAD-WT in each mutated construct. Nucleotide changes in the NACRS that had an adverse effect on the binding activity of ANAC072 protein are indicated by small letters.
Expression of the ANAC019, ANAC055, and ANAC072 Genes
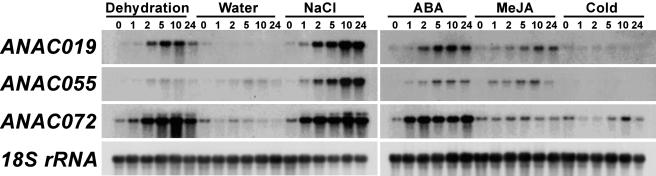
The expression patterns of ANAC019, ANAC055, and ANAC072 were analyzed under various stresses and hormone treatments, including jasmonic acid (JA), because the ANAC055 product is a putative JA regulatory protein (Figure 9). ANAC055 expression was slightly induced after 2 h of dehydration, ABA, and JA treatments and strongly induced by salt treatment. ANAC019 transcription was more strongly induced by dehydration, ABA, and salt within 2 h than that of ANAC055 and slightly induced by JA after 5 h. The strongest and fastest accumulation of mRNA, within 1 h, was that of ANAC072, when plants were subjected to dehydration stress, high salinity stress, or ABA treatment. Expression of ANAC072 was mostly unchanged by JA treatment. There was a slight accumulation of ANAC072 mRNA after 10 h of cold treatment, whereas ANAC019 and ANAC055 mRNAs were not accumulated. These results demonstrate that expression of ANAC019, ANAC055, and ANAC072 was induced mainly by drought and high salinity stresses. Previously, it was reported that increased levels of erd1 mRNA were observed 1 to 2 h after dehydration or high salinity treatment but not after low temperature treatment (Nakashima et al., 1997). Thus, the ANAC019, ANAC055, and ANAC072 proteins most likely function directly upstream of erd1.
Figure 9.
Expression of ANAC019, ANAC055, and ANAC072.
Each lane was loaded with 5 μg of total RNA from 3-week-old Arabidopsis plants that had been dehydrated (Dehydration), transferred for hydroponic growth in distilled water (Water), transferred for hydroponic growth in 250 mM NaCl (NaCl), transferred for hydroponic growth in 100 μM ABA (ABA), transferred for hydroponic growth in 20 μM methyl jasmonate (MeJA), or transferred for hydroponic growth at 4°C (Cold). Numbers above each lane indicate the number of hours after the initiation of treatment before isolation of RNA. mRNA transcription was analyzed by RNA gel hybridization with specific probes from the 3′ flanking sequence of the NAC genes.
Promoter Regions of the NAC Genes and Histochemical Assay
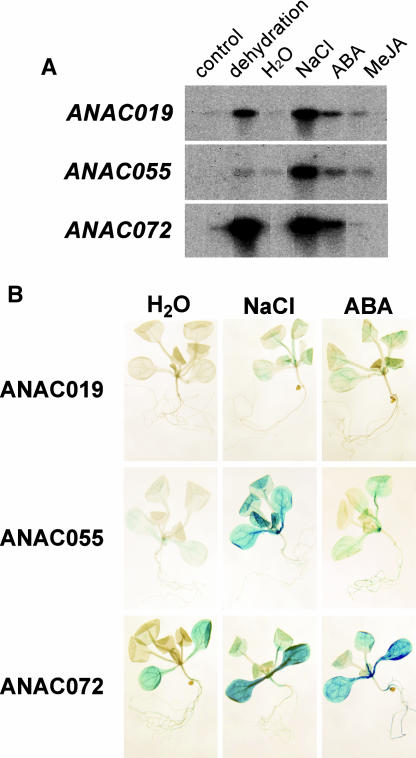
To gain a better understanding of the upstream signaling function of the NAC transcription factors in the activation of erd1 (Shinozaki and Yamaguchi-Shinozaki, 2000), we examined whether the cis-acting elements involved in the dehydration-responsive expression of the NAC genes are located in the isolated promoter regions of these genes. Chimeric constructs, consisting of the NAC promoter fragments and GUS reporter gene (PNAC-GUS), were introduced into Arabidopsis, and stable transformant lines were analyzed. Promoter activity was examined in various treatments using RNA gel blot analysis (Figure 10A). The GUS expression profile was similar to that of the NAC gene expression (Figures 9 and 10A), suggesting that the isolated 5′ upstream region of the NAC genes contains cis-acting elements involved in the dehydration-, high-salt-, and ABA-responsive transcription of these genes. A MEME search (http://meme.sdsc.edu) revealed that except for the G-box homologous sequences (ACACGTGTCA at position −126 for the ANAC019 promoter, CCACGTGTCA at position −107 for the ANAC055 promoter, and ACACGTGTCG at position −472 for the ANAC072 promoter), no other significant known motif was found, suggesting that there may be unknown cis-element(s) in the NAC promoter region involved in drought- and high-salt-inducible transcription.
Figure 10.
Expression of the GUS Reporter Gene under the Control of NAC Promoters in Transgenic Plants.
(A) RNA gel blot analysis. Each lane was loaded with 10 μg of total RNA from 3-week-old transgenic Arabidopsis plants containing PANAC019-GUS (ANAC019), PANAC055-GUS (ANAC055), or PANAC072-GUS (ANAC072) fusions, which were either untreated controls (control), or dehydrated (dehydration), transferred for hydroponic growth in distilled water (H2O), transferred for hydroponic growth in 250 mM NaCl (NaCl), transferred for hydroponic growth in 100 μM ABA (ABA), or transferred for hydroponic growth in 20 μM methyl jasmonate (MeJA) for 10 h.
(B) Histochemical assay. Two-week-old transgenic Arabidopsis plants containing either PANAC019-GUS (ANAC019), PANAC055-GUS (ANAC055), or PANAC072-GUS (ANAC072) fusions were treated with distilled water (H2O), 250 mM NaCl (NaCl), or 100 μM ABA (ABA) for 10 h before being subjected to histochemical analysis.
In addition, we also studied the localization of GUS reporter gene expression under the control of the NAC promoter fragments in transgenic Arabidopsis plants. Figure 10B shows the histochemical analysis of GUS activity of the transgenic plants. We found that during high-salinity stress the GUS enzyme accumulated exclusively in the leaves of transgenic plants containing the PNAC-GUS fusions. In the case of ABA treatment, the GUS enzyme not only accumulated in leaves, but also at lower levels in roots of transgenic plants containing the PANAC055-GUS or PANAC072-GUS fusions.
Arabidopsis Plants Overexpressing ANAC019, ANAC055, and ANAC072: Morphological Effects and Microarray Analysis
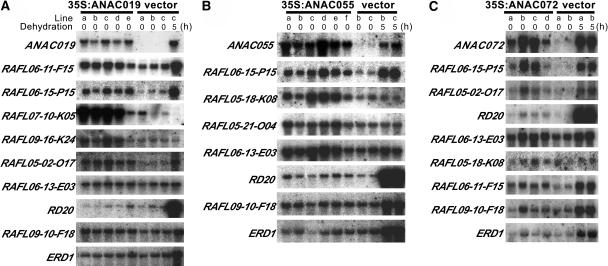
To analyze the function of the NAC proteins in plants, we generated transgenic plants in which the NAC protein was overexpressed (35S:ANAC019, 35S:ANAC055, and 35S:ANAC072). The ANAC019, ANAC055, and ANAC072 cDNAs were overexpressed under the control of the CaMV 35S promoter. To check the expression level of ANAC019, ANAC055, and ANAC072 in transgenic plants, total RNA prepared from 15 lines of the 35S:ANAC019 plants, 15 lines of the 35S:ANAC055 plants, and 15 lines of the 35S:ANAC072 plants was subjected to RNA gel blot analyses using ANAC019-, ANAC055-, and ANAC072-specific probes. As a control, total RNA prepared from two or three transgenic lines containing the pBI35SHyg vector was used. The results of RNA gel blot hybridization of several representative 35S:ANAC019, 35S:ANAC055, and 35S:ANAC072 transgenic lines with ANAC019, ANAC055, or ANAC072 probes are shown in Figure 11. ANAC019, ANAC055, and ANAC072 were significantly overexpressed in these transgenic lines.
Figure 11.
Confirmation of Microarray Data by RNA Gel Blot Analysis.
Each lane was loaded with 10 μg of total RNA from 3-week-old transgenic Arabidopsis plants containing pBI35S:ANAC019Hyg (35S:ANAC019) (A), pBI35S:ANAC055Hyg (35S:ANAC055) (B), pBI35S:ANAC072Hyg (35S:ANAC072) (C), or pBI35SHyg (vector) as a control. Transgenic Arabidopsis plants were untreated or dehydrated as indicated before analysis.
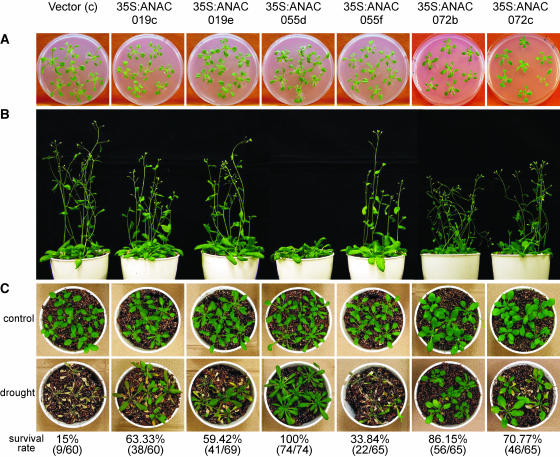
The effects of overexpression of ANAC019, ANAC055, and ANAC072 on the morphology of 35S:ANAC019, 35S:ANAC055, and 35S:ANAC072 plants, respectively, were carefully watched in time-course experiments. Transgenic plants expressing ANAC055 germinated and grew at the same rate as vector control plants until they reached the rosette stage (Figure 12A). At this point, whereas the 35S:ANAC055f line, representing a group in which ANAC055 was expressed at a medium level, proceeded to bolt with little delay compared with the vector control, the 35S:ANAC055d line, in which ANAC055 was overexpressed at a high level, remained at the rosette stage for an additional 10 to 15 d before the first bolting appeared (Figure 12B). During this period, leaf size, leaf number, and root mass continued to increase. This phenotype was also observed when BnNAC14 of Brassica napus was overexpressed in Arabidopsis (Hegedus et al., 2003). Two 35S:ANAC019 transgenic plants, 35S:ANAC019c and 35S:ANAC019e, overexpressing ANAC019 at a high level exhibited a phenotype and a time course of growth similar to those of vector control. The same were observed with 35S:ANAC072b and 35S:ANAC072c transgenic plants confirmed to express ANAC072 strongly by RNA gel blot analysis (Figures 11, 12A, and 12B).
Figure 12.
Phenotype of the 35S:ANAC019, 35S:ANAC055, and 35S:ANAC072 Plants.
The effect of the overexpression of NAC genes on plant morphology was studied as follows:
(A) 35S:ANAC019, 35S:ANAC055, 35S:ANAC072, and vector control plants were germinated on selective germination medium and grown for 3 weeks. The control and transgenic plants grew at the same rate.
(B) Three-week-old plants were transferred to pots and grown for two additional weeks. Bolting of the 35S:ANAC055d plant was delayed by strong ectopic expression of ANAC055.
(C) Drought tolerance phenotype of the 35S:ANAC019, 35S:ANAC055, and 35S:ANAC072 transgenic plants was investigated as described in Methods. Control, 4-week-old plants growing under normal conditions; drought, water withheld from plants for 9 d, then rewatering for 3 d. Percentage of surviving plants and numbers of surviving plants per total number of tested plants are indicated under the photographs. Vector line c (Figure 11) was used as a control plant.
To identify genes upregulated in the 35S:ANAC019, 35S:ANAC055, and 35S:ANAC072 plants, we used a cDNA microarray containing ∼7000 Arabidopsis full-length cDNAs (Seki et al., 2002a), among which ANAC019 and ANAC055 cDNAs are not found. mRNAs prepared from the 35S:ANAC019, 35S:ANAC055, 35S:ANAC072, and control plants were used for the generation of Cy5-labeled and Cy3-labeled cDNA probes. These cDNA probes were mixed and hybridized with the cDNA microarray. Microarray data analysis revealed that several genes were upregulated in the 35S:ANAC019, 35S:ANAC055, and 35S:ANAC072 plants. A summary of the most significantly upregulated genes is given in Tables 1, 2, and 3. Many of these upregulated genes are stress inducible based on the data published by Seki et al. (2002a, 2002b) (Tables 1, 2, and 3). Overexpression of ANAC072 induced the highest number of stress-related genes. The complete microarray data sets can be seen in Supplemental Table s1 online. ERD1 was not among the upregulated genes (with an expression level of 1.2-, 1.1-, and 1.9-fold in the 35S:ANAC019, 35S:ANAC055, and 35S:ANAC072 plants, respectively, when compared with the control plants). The reliability of the microarray data was confirmed by RNA gel blot analyses of the upregulated genes. The RNA gel blot analyses were performed using five 35S:ANAC019 lines and three control lines in the case of ANAC019, six 35S:ANAC055 lines and two control lines in the case of ANAC055, and four 35S:ANAC072 lines and two control lines in the case of ANAC072 (Figure 11). Because the number of upregulated genes listed in Tables 1, 2, and 3 is relatively high, five, three, and seven genes were randomly chosen from Tables 1, 2, and 3, respectively, for RNA gel blot analysis. The results shown in Figure 11 strongly supported the reliability of the microarray data. We searched the Arabidopsis genome database to obtain the promoter regions of the upregulated genes listed in Tables 1, 2, and 3. Most of them have the NAC core motif, CACG, in their promoter regions.
Table 1.
ANAC019-Dependent Genes Detected through Microarray Analysis
| Gene Namea | MIPS IDb | Ratioc | Descriptiond | Stress Responsee | CACGf |
|---|---|---|---|---|---|
| RAFL06-11-F15 | At5g01600 | 4.2 | Ferritin 1 precursor | D | Yes |
| RAFL06-15-P15 | At1g80160 | 3.3 | Glyoxalase I family protein | A, D, S | Yes |
| RAFL09-10-B07 | At3g52260 | 2.9 | Pseudouridine synthase family protein | –g | No |
| RAFL05-07-D07 | At5g65930 | 2.8 | Kinesin-like calmodulin binding protein | D, S | Yes |
| RAFL05-07-D22 | At2g01030 | 2.7 | 26S ribosomal RNA | D, S | Yes |
| RAFL05-07-C09 | At2g01020 | 2.6 | 5.8S ribosomal RNA | – | Yes |
| RAFL11-01-A10 | At3g62650 | 2.6 | Putative mitochondrial carrier protein | D | Yes |
| RAFL09-12-B03 | At3g60140 | 2.5 | β-Glucosidase | A, D, S | Yes |
| RAFL11-01-J02 | At3g10860 | 2.4 | Putative ubiquinol-cytochrome C reductase complex | – | Yes |
| RAFL09-16-K24 | At1g10070 | 2.4 | Putative branched-chain amino acid aminotransferase | A, D, S | Yes |
| RAFL09-18-N08 | At5g23590 | 2.4 | DNAJ heat shock N-terminal domain-containing protein | – | Yes |
| RAFL07-10-K05 | At4g14130 | 2.3 | Xyloglucan endotrans-glycosylase-related protein | A | No |
| RAFL06-09-H20 | At2g40300 | 2.3 | Putative ferritin | – | Yes |
| RAFL06-16-O09 | At5g20150 | 2.2 | ids4-like protein | – | Yes |
| RAFL02-01-K01 | At4g25100 | 2.2 | Superoxide dismutase | – | Yes |
| RAFL05-02-O17 | At4g33150 | 2.2 | Lys-ketoglutarate reductase/saccharopine dehydrogenase | A, D, S | Yes |
| RAFL09-13-J20 | At1g69410 | 2.2 | Putative eukaryotic initiation factor 5A | – | Yes |
According to Seki et al. (2002c).
Munich Information Center for Protein Sequences (MIPS) entry codes for the cDNAs used in this study.
Ratio is given in median of fold changes of three repeats (see supplemental data online). Fold change was defined as flourescence intensity [FI] of each cDNA of 35S:ANAC019/FI of each cDNA of vector control line. Genes expressed in 35S:ANAC019 with upregulation ratio higher than 2.2 are shown.
Description as given by the MIPS database.
According to Seki et al. (2002a, 2002b). A, ABA treatment; D, dehydration; S, high salinity.
Existence of NACRS core sequence in 1000 bp of sequence.
None.
Table 2.
ANAC055-Dependent Genes Detected through Microarray Analysis
| Gene Namea | MIPS IDb | Ratioc | Descriptiond | Stress Responsee | CACGf |
|---|---|---|---|---|---|
| RAFL06-09-D14 | At4g27150 | 4.3 | NWMU2-2S albumin 2 precursor | –g | Yes |
| RAFL06-13-C15 | At5g44120 | 2.9 | Legumin-like protein | – | Yes |
| RAFL05-17-A02 | At3g20810 | 2.8 | Transcription factor jumonji (jmjC) domain-containing protein | – | No |
| RAFL04-09-K06 | At5g20630 | 2.7 | Germin-like protein (GLP3b) | – | Yes |
| RAFL09-11-P10 | At1g05560 | 2.7 | UDP-glucose transferase (UGT75B2) | A, S | Yes |
| RAFL06-07-B18 | At4g27160 | 2.6 | NWMU3-2S albumin 3 precursor | – | Yes |
| RAFL06-15-P15 | At1g80160 | 2.6 | Glyoxalase I family protein | A, D, S | Yes |
| RAFL04-10-D06 | At5g17170 | 2.5 | Rubredoxin family protein | – | Yes |
| RAFL05-14-B09 | At4g27140 | 2.5 | NWMU1-2S albumin 1 precursor | – | Yes |
| RAFL05-18-K08 | At2g43510 | 2.4 | Putative trypsin inhibitor | A | No |
| RAFL06-16-O09 | At5g20150 | 2.3 | ids4-like protein | – | Yes |
| RAFL08-13-D16 | At3g29646 | 2.2 | Putative protein | – | No |
| RAFL05-21-O04 | At3g08860 | 2.2 | Putative aminotransferase | A | Yes |
| RAFL06-09-G04 | At1g03880 | 2.2 | Putative cruciferin 12S seed storage protein | – | Yes |
According to Seki et al. (2002c).
MIPS entry codes for the cDNAs used in this study.
Ratio is given in median of fold changes of three repeats (see supplemental data online). Fold change was defined as FI of each cDNA of 35S:ANAC055/FI of each cDNA of vector control line. Genes expressed in 35S:ANAC055 with upregulation ratio higher than 2.2 are shown.
Description as given by the MIPS database.
According to Seki et al. (2002a, 2002b). A, ABA treatment; D, dehydration; S, high salinity.
Existence of NACRS core sequence in 1000 bp of sequence.
None.
Table 3.
ANAC072-Dependent Genes Detected through Microarray Analysis
| Gene Namea | MIPS IDb | Ratioc | Descriptiond | Stress Responsee | CACGf |
|---|---|---|---|---|---|
| RAFL06-15-P15 | At1g80160 | 9.8 | Glyoxalase I family protein | A, D, S | Yes |
| ANAC072/RD26 | At4g27410 | 9.0 | Stress-inducible NAC protein | A, D, S, C | Yes |
| RAFL06-11-B11 | At1g73260 | 5.7 | Putative trypsin inhibitor | A, D | Yes |
| RAFL05-02-O17 | At4g33150 | 4.0 | Lys-ketoglutarate reductase/saccharopine dehydrogenase | A, D, S | Yes |
| RAFL02-05-F08 | At4g23680 | 3.6 | Major latex protein-related | A | Yes |
| RAFL06-13-E03 | At3g44300 | 3.4 | Nitrilase 2 | A, D, S | Yes |
| RAFL04-09-O24 | At4g30270 | 3.2 | Xyloglucan endo-1,4-β-d-glucanase precursor | A | Yes |
| RD20 | At2g33380 | 3.0 | Putative calcium-binding EF-hand protein | A, D, S | Yes |
| RAFL09-15-K15 | At3g18080 | 3.0 | Glycosyl hydrolase family 1 | –g | Yes |
| RAFL05-18-K08 | At2g43510 | 3.0 | Putative trypsin inhibitor | A | No |
| RAFL04-09-D07 | At1g54100 | 2.9 | Aldehyde dehydrogenase homolog, putative | A, D, S | Yes |
| RAFL05-12-I22 | At3g44310 | 2.7 | Nitrilase 1 | A | Yes |
| RAFL05-19-C11 | At2g25450 | 2.7 | Putative dioxygenase | – | Yes |
| RAFL06-11-F15 | At5g01600 | 2.6 | Ferritin 1 precursor | D | Yes |
| RAFL05-02-G08 | At3g59930 | 2.6 | Putative protein | A, D, S | No |
| RAFL05-18-E12 | At3g19770 | 2.6 | Vacuolar sorting protein 9 domain-containing protein | – | No |
| RAFL05-08-B17 | At1g16410 | 2.5 | Putative cytochrome P450 | – | No |
| RAFL05-18-O20 | At1g29395 | 2.5 | Similar to the cold acclimation protein WCOR413 | D, C | Yes |
| RAFL05-14-O18 | At4g22240 | 2.5 | Fibrillin precursor-like protein | – | Yes |
| RAFL07-13-A19 | At1g04400 | 2.5 | Putative cryptochrome 2 apoprotein | – | Yes |
| RAFL05-14-M10 | At5g14180 | 2.5 | Lipase family protein | A, D, S | Yes |
| RAFL08-12-I18 | At2g37760 | 2.5 | Aldo/keto reductase family | – | Yes |
| RAFL05-15-K08 | At5g57655 | 2.4 | Xylose isomerase | A, D, S | Yes |
| RAFL11-01-D24 | At3g01420 | 2.4 | Feebly-like protein | A, D | Yes |
| RAFL05-21-O04 | At3g08860 | 2.3 | Putative aminotransferase | A | Yes |
| RAFL08-17-B19 | At1g77840 | 2.3 | Putative eukaryotic translation initiation factor 5 | – | Yes |
| RAFL07-15-F13 | At2g40840 | 2.3 | Glycosyl hydrolase family 77 (4-α-glucanotransferase) | – | Yes |
| RAFL09-10-F18 | At1g58360 | 2.3 | Amino acid permease I | A, D | Yes |
| RAFL05-16-G04 | At2g30140 | 2.3 | Putative glucosyl transferase | A, D, S | No |
According to Seki et al. (2002c).
MIPS entry codes for the cDNAs used in this study.
Ratio is given in median of fold changes of three repeats (see supplemental data online). Fold change was defined as FI of each cDNA of 35S:ANAC072/FI of each cDNA of vector control line. Genes expressed in 35S:ANAC072 with upregulation ratio higher than 2.3 are shown.
Description as given by the MIPS database.
According to Seki et al. (2002a, 2002b). A, ABA treatment; D, dehydration; S, high salinity; C, cold.
Existence of NACRS core sequence in 1000 bp of sequence.
None.
Effect of ANAC019, ANAC055, and ANAC072 Overexpression on Drought Stress Tolerance
The effect of ANAC019, ANAC055, and ANAC072 overexpression on stress tolerance, particularly drought tolerance, was evaluated by considering whole-plant survival. Four-week-old transgenic and control plants of similar size (Figure 12C) were subjected to 9 d of drought treatment. Three days after rewatering, 15% of control plants survived, but 63.33% of 35S:ANAC019c, 59.42% of 35S:ANAC019e, 100% of 35S:ANAC055d, 33.84% of 35S:ANAC055f, 86.15% of 35S:ANAC072b, and 70.77% of 35S:ANAC072c plants survived (Figure 12C). These results demonstrate that overexpression of either ANAC019, ANAC055, or ANAC072 significantly improved drought tolerance, which appeared to be associated with the expression level of the NAC genes under unstressed condition. The survival rate of 35S:ANAC055d (100%) was significantly higher than that of 35S:ANAC055f (33.84%), which has a lower ANAC055 expression. Similarly, the survival rate of 35S:ANAC072b (86.15%) was higher than that of 35S:ANAC072c (70.77%), which exhibits lower ANAC072 expression (Figures 11 and 12C).
DISCUSSION
Using the yeast one-hybrid screening method, we identified three cDNAs encoding ANAC019, ANAC055, and ANAC072 transcription factors that specifically interact with the NACRS sequence in the erd1 promoter region through the conserved NAC binding domain. ANAC019, ANAC055, and ANAC072 can function as activators in plants, suggesting that they have a role in the activation of downstream target genes in tissues in which NAC proteins are present and/or induced. In yeast, ANAC019 and ANAC055, but not ANAC072, were shown to function as transcriptional activators for NACRS-dependent transcription. However, in Arabidopsis T87 protoplasts all three NAC proteins could transactivate NACRS-dependent expression of a GUS reporter gene under the control of the erd1 promoter, indicating that to function as a transcriptional activator, ANAC072 may require the presence of factors not available in yeast.
ANAC019, ANAC055, and ANAC072 have significant sequence similarity in the NAC binding domain, which can be divided into five subdomains (A to E) according to Ooka et al. (2003) (Figure 2). Moreover, the three proteins have conserved sequences in the C-terminal half (Figure 2), and there are acidic and Gln-rich regions in the C-terminal portion of the predicted sequences that may be involved in transcriptional activation (Künzler et al., 1994). Furthermore, ANAC019, ANAC055, and ANAC072 might be assigned to the class of Ser-rich activation domains (Triezenberg, 1995). However, we cannot exclude the possibility that the newly identified NAC proteins may also feature a novel activation domain. In addition, the amino acid sequences of ANAC019 and ANAC055 show greater sequence similarity to each other than either does to ANAC072. This may further explain why ANAC072 was active as a transcriptional activator only in Arabidopsis, whereas ANAC019 and ANAC055 were functionally active in both Arabidopsis and yeast.
The NAC proteins bound specifically to the NACRS, which contains the CATGTG motif, both in vivo and in vitro (Figures 1B and 3). In addition, the gel mobility shift experiment showed that the NAC proteins could bind to NACRS as multimers (Figure 3). Structural studies of the NAC domain of ANAC019 also suggest that the NAC domain of ANAC019 mediated dimerization of the NAC proteins through conserved interactions (Ernst et al., 2004). It is worthy to note that the NAC domains of ANAC019, ANAC055, and ANAC072 are highly conserved; thus, they might also be able to heterodimerize. This possible heterodimerization might potentiate transcriptional activity of the NAC proteins, which would have a significant effect on expression of target genes, consequently on stress response. Using the yeast one-hybrid system, we identified the complete NACRS binding sequence and the core DNA binding motif (CACG) for the newly identified NAC proteins within the 63-bp fragment of the erd1 promoter (Figures 5, 6, 7, and 8). The Arabidopsis NAC1 protein has been shown to bind to a 21-bp segment (CTGACGTAAGGGATGACGCAC) within the 35S −90 promoter fragment (Xie et al., 2000). Independently, Duval et al. (2002) demonstrated that purified AtNAM recombinant protein, another NAC protein, protected the region of the CaMV 35S promoter between −70 and −76, which is located and underlined in the 21-bp segment. The newly identified CACG core motif is not found within the 21-bp segment, but we cannot exclude the possibility that ANAC019, ANAC055, and ANAC072 can also bind to that segment. Indeed, at least for ANAC019, Ernst et al. (2004) have reported that the NAC domain of ANAC019 could bind to the −90 to +9 fragment of the CaMV promoter as both monomer and dimer. In addition, among the upregulated genes listed in Tables 1, 2, and 3, nine genes do not contain the CACG core motif in their promoter region. However, the promoter regions of these genes have similar motifs with high homology to the AGGGATG sequence (data not shown).
Expression of ANAC019 and especially of ANAC072 was strongly induced by dehydration and high-salinity treatment within 1 to 2 h, whereas that of ANAC055 was only induced markedly by high-salt stress. Expression of erd1 was also observed 1 to 2 h after dehydration and high-salt stress (Nakashima et al., 1997). Thus, the NAC proteins likely function upstream of the erd1 gene and transactivate gene expression in response to these stresses. Because the ANAC019 and ANAC055 transcripts accumulated to a higher level in plants under high-salt stress than in those under dehydration stress, these two transcription factors probably play a major role in high-salt stress rather than in drought conditions. The ANAC072 mRNA, meanwhile, accumulated to higher levels in plants experiencing dehydration rather than under high-salt stress, suggesting that ANAC072's role is associated mainly with the plant response to dehydration. To more precisely elucidate the function of the NAC genes in the signal transduction pathway leading to the activation of erd1, we are currently analyzing anac019, anac055, and anac072 mutants. Nakashima et al. (1997) showed that the erd1 promoter was not strongly affected by ABA treatment because induction of erd1 mRNA accumulation occurred earlier than did the increase of endogenous ABA, whose accumulation began to increase 2 h after Arabidopsis plants had been subjected to dehydration stress (Kiyosue et al., 1994). Expression of the NAC genes was observed also within 1 to 2 h after ABA treatment, suggesting that dehydration-regulated expression of the NAC genes and of erd1 does not require endogenous ABA. The NAC genes, especially ANAC072, do respond to exogenous ABA. These NAC proteins may also regulate ABA-responsive genes as target genes. In addition, histochemical assays showed that in response to high salinity and ABA treatment, the NAC promoters direct protein accumulation mainly in the leaves of affected Arabidopsis plants (Figure 10B). Similarly, Nakashima et al. (1997) reported that in transgenic plants containing an erd1 promoter-GUS fusion construct, GUS gene expression was localized to the leaves in plants experiencing abiotic stresses.
Microarray analysis of the 35S:ANAC019, 35S:ANAC055, and 35S:ANAC072 plants revealed that several genes were upregulated by overexpression of ANAC019, ANAC055, and ANAC072, respectively (Tables 1, 2, and 3). One gene, RAFL06-15-P15, was overexpressed by either ANAC019, ANAC055, or ANAC072 overproduction. RAFL06-16-O09 was upregulated by both ANAC019 and ANAC055, RAFL05-18-K08 by both ANAC055 and ANAC072, RAFL06-11-F15 and RAFL05-02-O17 by both ANAC019 and ANAC072 overexpression, and the others were either ANAC019, ANAC055, or ANAC072 dependent (Tables 1, 2, and 3). This result indicated that there may be nonoverlapping and overlapping targets for the NAC proteins. We used three nonoverlapping target genes—RAFL06-13-E03, RD20, and RAFL09-10-F18, which are upregulated by ANAC072 overproduction (Figure 11)—as probes for RNA gel blot analysis to assess the ability of these three NAC proteins to recognize target genes. With the exception of RAFL06-13-E03, whose expression was clearly induced by overexpression of ANAC055 but not of ANAC019, the transcription of the other two genes remained mostly unchanged after the overexpression of either ANAC019 or ANAC055 (Figure 11).
Transcript levels of erd1 were largely unaffected in transgenic plants overexpressing the NAC genes (Figure 11). Moreover, the expression levels of erd1 in the transgenic plants that were exposed to drought stress also remained relatively unchanged when compared with the vector control plants (data not shown). Simpson et al. (2003) showed that during water stress, erd1 transcription is controlled by coordinated activity between two cis-acting elements, the CATGTG motif and the 14-bp rps1 site 1–like sequence. On the other hand, the several-fold induction of the GUS reporter gene driven by the erd1 promoter region containing NACRS in the transient transactivation assay using Arabidopsis T87 protoplasts (Figure 4) may be because of osmotic stress imposed on the T87 protoplasts themselves. Because the overexpression of NAC does not induce the transcription of erd1, we hypothesize that ANAC019 and/or ANAC055 and/or ANAC072 bind to the NACRS sequence, and transcription factors that bind the 14-bp rps1 site 1–like sequence cooperatively form a structure called an enhanceosome to control the dehydration-inducible transcription of erd1. To gain a better understanding of the signaling pathway that leads to the activation of erd1, we recently isolated zinc-finger homeodomain transcription factors containing a homeodomain that can bind to the rps1 site 1–like sequence using the yeast one-hybrid system. Overexpression of both NAC and zinc-finger homeodomain proteins activated the expression of erd1 under unstressed normal growth conditions in the transgenic Arabidopsis plants (our unpublished data).
In Arabidopsis transgenic plants, ANAC019, ANAC055, and ANAC072 overexpression increased drought stress tolerance (Figure 12C). This may be because of the overexpression of stress-inducible genes that are controlled by ANAC019, ANAC055, and ANAC072 under unstressed conditions because several stress-inducible genes were upregulated by overexpression of ANAC019, ANAC055, and ANAC072 among the 7000 cDNAs screened (Tables 1, 2, and 3). Of these identified target genes, the expression of RAFL06-15-P15, which encodes glyoxalase I family protein, was clearly correlated with the relative expression level of ANAC019, ANAC055, and ANAC072 (Figure 11). Glyoxalase enzymes are important for the glutathione-based detoxification of methylglyoxal, which is formed primarily as a byproduct of carbohydrate and lipid metabolism (Thornalley, 2003). Singla-Pareek et al. (2003) have reported that overexpression of Brassica juncea glyoxalase I and Oryza sativa glyoxalase II, either alone or together, confers improved salinity tolerance in tobacco (Nicotiana tabacum). Moreover, reduction of the cytotoxic methylglyoxal by overexpression of a drought-inducible alfalfa (Medicago sativa) NADPH-dependent aldose/aldehyde reductase protects transgenic tobacco against drought stress (Oberschall et al., 2000). Therefore, we postulate that the improved resistance of the ANAC019, ANAC055, and ANAC072 transgenic lines against drought stress is primarily derived from the reduction and, thus, detoxification of toxic aldehydes through the glyoxalase pathway. Alternatively, ANAC019-, ANAC055-, and ANAC072-dependent expression levels of other unknown target genes may have an impact on drought stress tolerance as well because we could expect to find more target genes of ANAC019, ANAC055, and ANAC072 if a higher number of genes were screened.
METHODS
Plant Materials and Stress Treatments
Plants (Arabidopsis thaliana ecotype Columbia) were grown on germination medium agar plates for 3 weeks as described (Nakashima et al., 1997). Various treatments were applied according to Nakashima et al. (1997). T87 cultured cells, derived from Arabidopsis, were maintained, grown, and treated as described previously (Axelos et al., 1992).
Yeast One-Hybrid Screening of Arabidopsis cDNA Libraries
Reporter plasmids were constructed and the yeast reporter strain was selected as described (Liu et al., 1998). Approximately 1.8 × 106 and 4 × 106 yeast transformants were screened according to the manufacturer's protocol (Clontech Matchmaker one-hybrid system; Palo Alto, CA) using 40 μg of AD-cDNA libraries prepared from unstressed and 3-h-dehydrated Arabidopsis plants as previously described (Liu et al., 1998). The unstressed and 3-h-dehydrated primary libraries have 5.8 × 106 pfu and 6.2 × 106 pfu, respectively. Forty-nine positive colonies were obtained from selective plates containing 10 mM 3-AT. When a colony-lift filter assay was performed as described in the Yeast Protocols Handbook (Clontech) to verify the DNA–protein interaction, all 49 colonies conferred β-Gal activity. The cDNA isolation, subcloning, and sequencing of these 49 clones were performed as described (Liu et al., 1998). To analyze the binding specificity of isolated cDNA clones, we used four tandemly repeated copies of a mutated 63-bp fragment in which the CATGTG motif was replaced with AAAAAA for the construction of the reporter plasmids.
RNA Gel Blot Analyses
RNA extraction and gel blot hybridization were performed as described (Nakashima and Yamaguchi-Shinozaki, 2002).
Construction of Transgenic Plants Containing the PNAC-GUS Fusion and Histochemical Assay
The 1030-, 983-, and 868-bp fragments upstream of the starting codon ATG containing the promoter regions of the ANAC019, ANAC055, and ANAC072 genes were cloned by PCR and inserted into GUS expression vector pBI101.1. The sequence of the promoter fragments was then verified by sequencing. Plasmids containing the NAC promoter–GUS fusions were introduced into Arabidopsis to make transgenic plants. Histochemical assay was done essentially as described previously (Nakashima and Yamaguchi-Shinozaki, 2002).
Transactivation Experiment with T87 Protoplasts
cDNA fragments encoding ANAC019, ANAC055, and ANAC072 were cloned into plant expression vector pBI35SΩ (Abe et al., 1997) and cotransformed into T87 protoplasts with the reporter gene plasmid pBIerd1NACRSWT (reporter WT) or pBIerd1NACRSM1 (reporter M1). To construct these reporter plasmids, we ligated one copy of the 63-bp wild-type or M1 fragments containing the CATGTG or AAAAAA motifs in place of CATGTG, respectively, to the HindIII site of pSKTATA (Simpson et al., 2003) to make WT-Perd1 and M1-Perd1 fused fragments, which were then used to replace the CaMV 35S promoter in pBI221. T87 Arabidopsis protoplasts were transactivated according to Satoh et al. (2004).
Determination of the Complete NACRS and Core Sequence by Yeast One-Hybrid System
All the fragments, wild-type as well as single- or triple-base-substituted 63-bp fragments, were bluntly generated by PCR and cloned directly into the SmaI site of reporter plasmid pLacZi. The sequence of the inserts was confirmed, and then the derived plasmids were linearized and integrated into the genome of yeast strain YM4271 to construct the respective reporter strains. pAD-GAL4 derivatives, the pAD-GAL4-ANAC019, the pAD-GAL4-ANAC055, and the pAD-GAL4-ANAC072 were used as binding and activating plasmids in one-hybrid experiments. For control, plasmid pAD-WT containing a fragment encoding amino acids 132 to 236 of wild-type λ cI repressor was used (HybriZAP-2.1 XR library construction kit and HybriZAP-2.1 XR cDNA synthesis kit; Stratagene, La Jolla, CA).
Liquid Culture Assay for β-Galactosidase Activity
β-Galactosidase activity, expressed in Miller units, was measured as described in the Yeast Protocols Handbook (Clontech) using o-nitrophenyl-β-d-galactopyranoside as a substrate.
Preparation of GST Fusion Proteins and Gel Retardation Assay
Fragments encoding ANAC019, ANAC055, and ANAC072 were PCR amplified and fused to the pGEX-4T-2 vector (Amersham Biosciences, Buckinghamshire, UK). The recombinant pGEX-4T-2 plasmids were introduced into Escherichia coli strain BL21. GST fusion proteins were produced and purified as described (Urao et al., 1993). Gel shift assays were conducted according to Sakuma et al. (2002).
Arabidopsis Full-Length cDNA Microarray Analysis
cDNAs encoding ANAC019, ANAC055, and ANAC072 were inserted into plasmid pBI35SΩHyg (Abe et al., 2003), and the resulting plasmids pBI35SΩ:ANAC019Hyg (35S:ANAC019), pBI35SΩ:ANAC055Hyg (35S:ANAC055), and pBI35SΩ:ANAC072Hyg (35S:ANAC072) were introduced into Arabidopsis to generate transgenic plants overexpressing ANAC019, ANAC055, or ANAC072. As the control, transgenic plants containing pBI35SΩHyg without the insert were used. Total RNA was isolated from plants using TRIZOL reagent (Invitrogen, Carlsbad, CA). One milligram of total RNA was used for isolation of mRNA by PolyATtract mRNA isolation systems (Promega, Madison, WI). One microgram of mRNA was employed on microarray analysis. Microarray analysis was performed as described previously (Seki et al., 2002a, 2002b). mRNA samples from plants overexpressing NAC were fluorescently labeled with Cy5-dUTP and samples from vector control plants labeled with Cy3-dUTP. λ-DNA (Takara, Tokyo, Japan) was used as an internal control because its fluorescence level is almost the same in the two conditions. To assess the reproducibility of microarray analysis, each experiment was repeated three times.
Drought Stress Tolerance of Transgenic Plants
Plants were grown aseptically in Petri dishes containing selective agar germination medium for 3 weeks, then transferred to 8-cm pots filled with a 1:1 mixture of perlite and vermiculite, and grown for one more week before exposure to drought stress. Drought stress was imposed by withholding water for 9 d in a growth chamber (22°C, 50 to 60% relative humidity, continuous 55 μmol m−2 s−1 photon flux density) until the lethal effect of dehydration was observed on most of the control plants. After rewatering for 3 d, the numbers of plants that survived and continued to grow were counted.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY117224, AB049070, and AY091428 for ANAC019 (At1g52890), ANAC055 (At3g15500), and ANAC072 (At4g27410), respectively.
Supplementary Material
Acknowledgments
We are grateful for the excellent technical support provided by Ekuko Ohgawara, Kyoko Yoshiwara, Kaori Amano, and Hiroko Sado of the Japan International Research Center for Agricultural Sciences (JIRCAS). This work was supported in part by the Program for Promotion of Basic Research Activities for Innovative Biosciences. It was also supported in part by a project grant from the Ministry of Agriculture, Forestry, and Fisheries, Japan. L.-S.P.T. was supported by a JIRCAS fellowship under the JIRCAS Visiting Research Fellowship Program at Tsukuba.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Kazuko Yamaguchi-Shinozaki (kazukoys@jircas.affrc.go.jp).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.022699.
References
- Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15, 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe, H., Yamaguchi-Shinozaki, K., Urao, T., Iwasaki, T., Hosokawa, D., and Shinozaki, K. (1997). Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9, 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelos, M., Curie, C., Mazzolini, L., Bardet, C., and Lescure, B. (1992). A protocol for transient gene expression in Arabidopsis thaliana protoplasts isolated from cell suspension cultures. Plant Physiol. Biochem. 30, 123–128. [Google Scholar]
- Bray, E., Bailey-Serresand, J., and Weretilnyk, E. (2000). Responses to abiotic stresses. In Biochemistry and Molecular Biology of Plants, B. Buchanan, W. Gruissem, and J.R. Rockville, eds (Rockville, MD: American Society of Plant Biologists), pp. 1158–1203.
- Choi, H., Hong, J., Ha, J., Kang, J., and Kim, S.Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275, 1723–1730. [DOI] [PubMed] [Google Scholar]
- Duval, M., Hsieh, T.F., Kim, S.Y., and Thomas, T.L. (2002). Molecular characterization of AtNAM: A member of the Arabidopsis NAC domain superfamily. Plant Mol. Biol. 50, 237–248. [DOI] [PubMed] [Google Scholar]
- Ernst, H.A., Olsen, A.N., Larsen, S., and Lo Leggio, L. (2004). Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 5, 297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler, S., and Thomashow, M.F. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14, 1675–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve, K., La Cour, T., Jensen, M.K., Poulsen, F.M., and Skriver, K. (2003). Interactions between plant RING-H2 and plant-specific NAC (NAM/ATAF1/2/CUC2) proteins: RING-H2 molecular specificity and cellular localization. Biochem. J. 371, 97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedus, D., Yu, M., Baldwin, D., Gruber, M., Sharpe, A., Parkin, I., Whitwill, S., and Lydiate, D. (2003). Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Mol. Biol. 53, 383–397. [DOI] [PubMed] [Google Scholar]
- Ingram, J., and Bartels, D. (1996). The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 377–403. [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen, K.R., Gilmour, S.J., Zarka, D.G., Schabenberger, O., and Thomashow, M.F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280, 104–106. [DOI] [PubMed] [Google Scholar]
- Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17, 287–291. [DOI] [PubMed] [Google Scholar]
- Kiyosue, T., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1993). Characterization of a cDNA for a dehydration-inducible gene that encodes a Clp A, B-like protein in Arabidopsis thaliana L. Biochem. Biophys. Res. Commun. 196, 1214–1220. [DOI] [PubMed] [Google Scholar]
- Kiyosue, T., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). Cloning of cDNAs for genes that are early-responsive to dehydration stress (ERDs) in Arabidopsis thaliana L.: Identification of three ERDs as HSP cognate genes. Plant Mol. Biol. 25, 791–798. [DOI] [PubMed] [Google Scholar]
- Künzler, M., Braus, G.H., Georgiev, O., Seipel, K., and Schaffner, W. (1994). Functional differences between mammalian transcription activation domains at the yeast GAL1 promoter. EMBO J. 13, 641–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama, K., Sakuma, Y., Kasuga, M., Ito, Y., Seki, M., Goda, H., Shimada, Y., Yoshida, S., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2004). Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 38, 982–993. [DOI] [PubMed] [Google Scholar]
- Nakashima, K., Kiyosue, T., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1997). A nuclear gene encoding a chloroplast-targeted Clp protease regulatory subunit homolog is not only induced by water stress but also developmentally up-regulated during senescence in Arabidopsis thaliana. Plant J. 12, 851–861. [DOI] [PubMed] [Google Scholar]
- Nakashima, K., and Yamaguchi-Shinozaki, K. (2002). Use of β-glucuronidase to show dehydration and high-salt gene expression. In Molecular Methods of Plant Analysis: Testing for Genetic Manipulation in Plants, Vol. 22, J.F. Jackson, H.F. Linsken, and R.B. Inman, eds (Berlin: Springer-Verlag), pp. 37–61.
- Oberschall, A., Deak, M., Torok, K., Sass, L., Vass, I., Kovacs, I., Feher, A., Dudits, D., and Horvath, G.V. (2000). A novel aldose/aldehyde reductase protects transgenic plants against lipid peroxidation under chemical and drought stresses. Plant J. 24, 437–446. [DOI] [PubMed] [Google Scholar]
- Ooka, H., et al. (2003). Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 20, 239–247. [DOI] [PubMed] [Google Scholar]
- Pastori, G.M., and Foyer, C.H. (2002). Common components, networks, and pathways of cross-tolerance to stress. The central role of “redox” and abscisic acid-mediated controls. Plant Physiol. 129, 460–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann, J.L., et al. (2000). Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. [DOI] [PubMed] [Google Scholar]
- Sakuma, Y., Liu, Q., Dubouzet, J.G., Abe, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009. [DOI] [PubMed] [Google Scholar]
- Satoh, R., Fujita, Y., Nakashima, K., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2004). A novel subgroup of bZIP proteins functions as transcriptional activators in hypoosmolarity-responsive expression of the ProDH gene in Arabidopsis. Plant Cell Physiol. 45, 309–317. [DOI] [PubMed] [Google Scholar]
- Seki, M., et al. (2002. a). Monitoring the expression pattern of around 7000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct. Integr. Genomics 2, 282–291. [DOI] [PubMed] [Google Scholar]
- Seki, M., Kamei, A., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2003). Molecular responses to drought, salinity and frost: Common and different paths for plant protection. Curr. Opin. Biotechnol. 14, 194–199. [DOI] [PubMed] [Google Scholar]
- Seki, M., Narusaka, M., Abe, H., Kasuga, M., Yamaguchi-Shinozaki, K., Carninci, P., Hayashizaki, Y., and Shinozaki, K. (2001). Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13, 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, M., et al. (2002. b). Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 31, 279–292. [DOI] [PubMed] [Google Scholar]
- Seki, M., et al. (2002. c). Functional annotation of a full-length Arabidopsis cDNA collection. Science 296, 141–145. [DOI] [PubMed] [Google Scholar]
- Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3, 217–223. [PubMed] [Google Scholar]
- Shinozaki, K., Yamaguchi-Shinozaki, K., and Seki, M. (2003). Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6, 410–417. [DOI] [PubMed] [Google Scholar]
- Simpson, S.D., Nakashima, K., Narusaka, Y., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2003). Two different novel cis-acting elements of erd1, a clpA homologous Arabidopsis gene, function in induction by dehydration stress and dark-induced senescence. Plant J. 33, 259–270. [DOI] [PubMed] [Google Scholar]
- Singla-Pareek, S.L., Reddy, M.K., and Sopory, S.K. (2003). Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proc. Natl. Acad. Sci. USA 100, 14672–14677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada, S., Hibara, K., Ishida, T., and Tasaka, M. (2001). The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128, 1127–1135. [DOI] [PubMed] [Google Scholar]
- Thomashow, M.F. (1999). Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571–599. [DOI] [PubMed] [Google Scholar]
- Thornalley, P.J. (2003). Glyoxalase I—Structure, function and a critical role in the enzymatic defence against glycation. Biochem. Soc. Trans. 31, 1343–1348. [DOI] [PubMed] [Google Scholar]
- Triezenberg, S.J. (1995). Structure and function of transcriptional activation domains. Curr. Opin. Genet. Dev. 5, 190–196. [DOI] [PubMed] [Google Scholar]
- Uno, Y., Furihata, T., Abe, H., Yoshida, R., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 97, 11632–11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao, T., Yamaguchi-Shinozaki, K., Urao, S., and Shinozaki, K. (1993). An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5, 1529–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Q., Frugis, G., Colgan, D., and Chua, N.H. (2000). Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 14, 3024–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki, K., Koizumi, M., Urao, S., and Shinozaki, K. (1992). Molecular cloning and characterization of 9 cDNAs for genes that are responsive to desiccation in Arabidopsis thaliana: Sequence analysis of one cDNA clone that encodes a putative transmembrane channel protein. Plant Cell Physiol. 33, 217–224. [Google Scholar]
- Zhu, J.K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 53, 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.