Abstract
Dipyridamole (DIP) inhibits thrombus formation when given chronically, and causes vasodilation over a short time. To date, DIP can increase the anticancer drugs (5-fluorouracil, methotrexate, piperidine, vincristine) concentration in cancer cells and hence enhance the efficacy of treatment cancer. The inhibition of DIP may result in increased 5-fluorouracil efficacy and diminish the drug side effects. But the actual molecular targets remain unknown. In this study, reverse protein-ligands docking, and quantum mechanics were used to search for the potential molecular targets of DIP. The quantum mechanics calculation was performed by using Gaussian 03 program package. Reverse pharmacophore mapping was used to search for potential molecular target candidates for a given small molecule. The docking study was used for exploring the potential anti-cancer targets of dipyridamole. The two predicted binders with the statistically significant prediction are dihydropyrimidine dehydrogenase (DPD) (PDB Id: 1GTE) and human spindle checkpoint kinase Bub1 (PDB Id: 3E7E). Structure analysis suggests that electrostatic interaction and hydrogen bonding play an important role in their binding process. The strong functional linkage of DIP and 5FU supports our prediction. In conclusion, these results generate a tractable set of anticancer proteins. The exploration of polypharmacology will provide us new opportunities in treating systematic diseases, such as the cancers. The results would generate a tractable set of anticancer target proteins for future experimental validations.
Keywords: Dipyridamole, reverse protein-ligand docking, anticancer target proteins
Introduction
Dipyridamole (used as Persantine) is a medicine that inhibits thrombus formation when given chronically and causes vasodilation at a short time. It was well known that dipyridamole inhibits the phosphodiesterase enzymes that normally break down cAMP (increasing cellular cAMP levels and blocking the platelet response to ADP) and/or cGMP (resulting in added benefit when given together with nitric oxide [NO] or statins) [1,2]. Dipyridamole inhibits the cellular reuptake of adenosine into platelets, red blood cells and endothelial cells leading to increased extra-cellular concentrations of adenosine [3,4]. In addition, dipyridamole has been shown to lower pulmonary hypertension without significant drop of systemic blood pressure [3,4]. It inhibits proliferation of smooth muscle cells in vivo and has shown to prevent AV-shunt failure in dialysis patients [4].
Modified release dipyridamole is used in conjunction with aspirin in the secondary prevention of stroke and transient ischaemic attack. This practice has been confirmed by the ESPRIT trial [5]. However, it is notlicensed as mono-therapy for stroke prophylaxis, although a Cochrane Review has suggested that dipyridamole may reduce the risk of further vascular events in patients presenting after cerebral ischaemia [6].
Dipyridamole (DIP) is a weak basic drug with a pKa value of 6.4 and thereby exhibits a pH-dependent solubility with good solubility at a low pH value and poor solubility at a high pH value [7,8]. For its high permeability and low solubility, DIP is classified as class II drug according to the Biopharmaceutics Classification System (BSC) [9]. Recently, research data indicated that the individual difference in drug absorption and bioavailability of DIP is significant [10,11]. DIP can increase the anticancer drugs (5-fluorouracil, methotrexate, piperidine, vincristine) concentration in cancer cells and hence enhance the efficacy of treatment cancer [12-14]. Moreover, it is not clear whether proteins are involved in DIP enhancing the efficacy of treatment cancer.
In this study, proteome-wide ligand binding site analysis, reverse protein-ligand docking, and quantum mechanics are used to search for the potential molecular targets of metformin. The new structural insights obtained from this computational study are expected to stimulate further biochemical studies on the experimental validations of anti-neoplastic proteins.
Materials and methods
Cells, trial grouping and treatment
The human breast cancer cells, MCF7, T47D, MDA-MB-231 and MDA-MB-468, were purchased from the American Type Culture Collection (Manassas, VA, USA), and maintained in the DMEM (Gibco, Grand Island, NY, USA) supplemented with the 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% heat-inactivated fetal bovine serum (Gibco, Grand Island, NY, USA). The dimethyl sulfoxide (DMSO) was purchased from Sigma-Aldrich (CA, USA). The dipyridamole (Sigma-Aldrich) was freshly dissolved in the DMEM medium, neutralized, and filter sterilized immediately prior to the addition to the cell cultures. The breast cancer cells were randomly divided into two groups, including DIP group and control group. The DIP group was treated with the dipyridamole (at the final concentration of 5, 10, 15 and 20 μM, respectively), and control group was treated with DMSO (at the final concentration of 0.1%).
Cell cytotoxicity evaluation
The cell cytotoxicity was assessed according to the selective incorporation of propidium iodide (PI) into the MCF7 cells which lost the integrity of the plasma membrane by using the FACSCaliburflow cytometer. Then the data were analyzed by suing the CellQuest software package (BD Bioscience, CA, USA). The cell cytotoxicity experiment was independently performed at least for 6 repeats.
Quantum mechanics calculation
The quantum mechanics calculation was performed by using Gaussian 03 program package [31,32]. Density functional theory (DFT) was employed with the three-parameter hybrid exchanging function of Becke and Lee, Yang, and Parr correlation function (B3LYP) [33]. The 6-31G* basis set was applied for the DIP optimization. The HOMO and LOMO were generated by Gaussian view.
Reverse pharmacophore mapping
PharmMapper server is an open-source online platform, and it uses pharmacophore mapping strategy to search for potential molecular target candidates for a given small molecule [19,20]. It was reported that a large, in-house pharmacophore database derived from annotation of all the target information in TargetBank, BindingDB, and DrugBank was hosted by PharmMapper [19,20]. The mol2 files of crocetin, picrocrocin, safranal were submitted in PharmMapper and a list of possible binding receptors were received. The list was further annotated to screen out the putative target list pertaining to anti-tumor activity. In this study, Top 300 targets ranked by fit score in descending order was present. The identified 2 structures related to human cancer on the top 10 hits were selected.
Reverse docking procedure using idTarget
IdTarget is an reverse docking web platform for predicting possible binding targets of a small chemical molecule using a divide-and-conquer docking approach [20,23]. This web server requires an input ligand file for the target screening and it possesses two modes for searching binding poses namely: ① Scanning mode, where usual molecular docking procedures are carried out for each protein structure, ② Fast mode, where the binding sites of the homologous proteins are mapped to ligands by superposition of homologous protein structures with the pre-calculated structural alignment data. The next step is the optimization of binding poses by means of adaptive local searches to eliminate close contacts among protein atoms. The mol2 files were submitted in idTarget and a list of possible binding receptors were received. The list was further annotated to screen out the putative target list pertaining to anti-cancer activity of DIP. In this study, metformin are identified by searching against more than 5,000 non-redundant human protein structures using ligand binding site comparison software idTarget [20,23]. Total of 121 structures were found with the related to human disorder.
Docking studying for DIP
In its current iteration, AutoDock vina [34] was used in this study. AutoDock Vina is a new open-source program for drug discovery, molecular docking and virtual screening, offering multi-core capability, high performance and enhanced accuracy and ease of use [34]. These putative 123 off-targets are subject to further investigations using more computationally intensive protein-ligand docking software AutoDock vina [34]. Total of 109 structures are removed from the putative off-target list due to steric crashes.
Normalized docking score
50 structures similar to DIP were extracted from DUD-E database which is built to refine the decoy sets [35]. The protocol to generate decoys for DUD-E is made available online to generate decoys for any target given only a list of ligand structures, which enables extension of DUD-E to new targets of interest by individual investigators [35]. The decoy server extracts directly from the ZINC database [35].
These molecules are docked to 15 proteins (putative off-targets) from the lowest binding energy conformation using AutoDock vina [34]. The correlation of the docking score to the number of carbon atoms is derived from linear regression for each of the protein receptors. From the linear fitting curve, the average docking score for molecules with a certain number of carbon atoms can be estimated. Based on the fitted average docking score, a normalized docking score DS is calculated as a z-score:
DS = (Si-µi)/σ (1)
Where Si is the raw docking score for the molecule with i carbon atoms, µi is the fitted average docking score for the number of carbon atoms i, and σ is the standard deviation.
σ = √(∑N(Si-µi)2/N) (2)
Statistical analysis
The SPSS 19.0 software was used for the statistical analysis. Statistical analysis of data was performed using Students’ t test. Data is presented as a mean ± SEM with P<0.05 considered statistically significant.
Results and discussion
DIP is widely used to prevent strokes and vascular thrombosis, and it is here investigated for potential clinical use as a new treatment for cancer [15-18]. It was reported that DIP is a promising agent for breast-cancer treatment [15-18], therefore, which also implys its potential use in other cancers that show those highly activated pathways. Our results also found that the DIP significantly suppressed the proliferation of the breast cancer cell lines compared to the control group (Figure 1, P<0.05).
Figure 1.
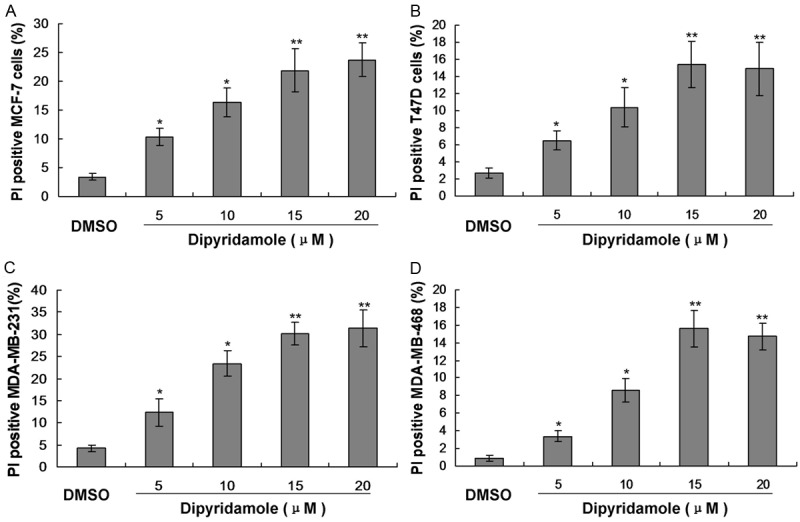
Observation for the cell cytotoxicity induced by the treatment of dipyridamole (5, 10, 15 and 20 μM) by using the FACS analysis in breast cancer cells. A. Cytotoxicity in MCF-7 cells. B. Cytotoxicity in T47D cells. C. Cytotoxicity in MDA-MB-231 cells. D. Cytotoxicity in MDA-MB-468 cells. *P<0.05 and **P<0.01 represent the percentage of PI positive cells in dipyridamole treated group compared to the DMSO control group.
In this study we have made an attempt to identify the therapeutic targets of the DIP. The chemical structure of DIP was displayed in Figure 1. Milliken charge is also showed in Figure 2.
Figure 2.
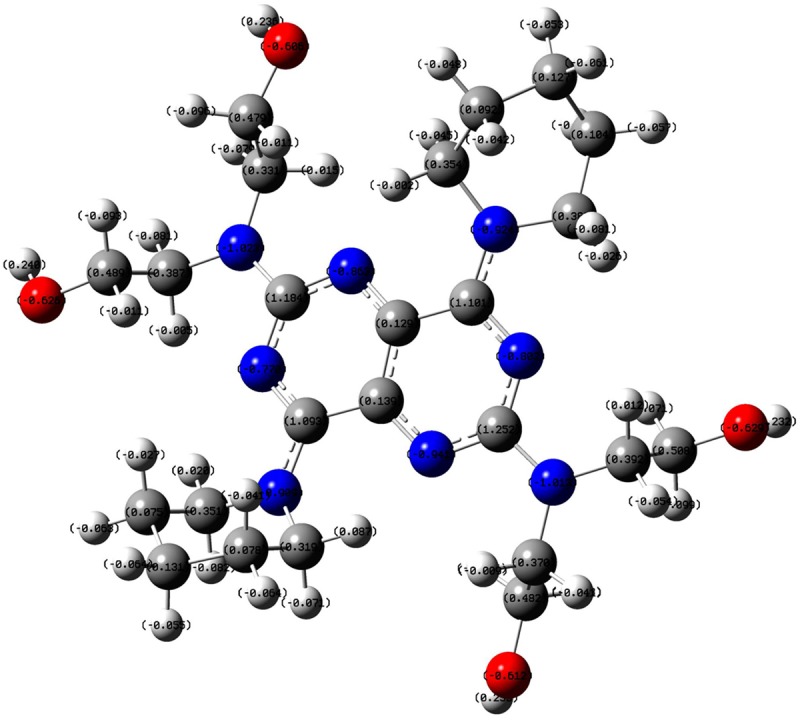
The 3D structure of DIP optimized with B3LYP 6-31G* set.
Seen from Figure 3, the HOMO orbit and he LUMO orbit of DIP indicated that the phenol ring is the active center of DIP. The energy between the HOMO and he LUMO (E gap) is 3.54 ev. The less energy indicated that DIP is easy to bind to the enzyme.
Figure 3.
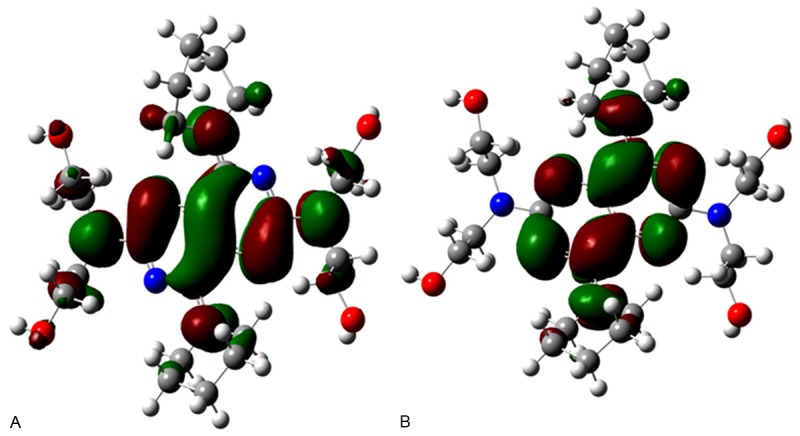
The HOMO and LUMO orbit of DIP. A. Illustration for the HOMO orbit of DIP. B. Illustration of the LUMO orbit of DIP.
Reverse docking through pharmMapper
Additional putative off-targets of DIP are obtained from PharmMapper server (http://59.78.96.61/pharmmapper/) [19-22]. PharmMapper finds the best mapping poses of the DIP against all the targets in PharmTarget DB [19] and top N potential drug targets as well as respective molecule’s aligned poses are outputted. Among 300 off-targets obtained, 2 proteins with high score related human cancers are selected (Table 1).
Table 1.
PharmMapper results
| Proteins | Z’-score | Fit score | PDB ID | Related disease |
|---|---|---|---|---|
| Dihydropyrimidine dehydrogenase | 2.21 | 5.19 | 1GTE | Cancer |
| GTPase HRas | 0.33 | 3.87 | 1P2S | Bladder cancer |
Reverse docking through idTarget
It was well known that identification of bimolecular targets of small chemical molecules is essential for unraveling their underlying causes of actions at the molecular level [20,23]. Many drugs are known to be a unpleasant adverse effects (off-target), but the molecular targets of such effects are largely unknown. The idTarget is a web server that can predict possible binding targets of a small chemical molecule via docking approach, in combination with our recently developed scoring functions and affinity profile of the protein target. The idTarget has been shown to be able to reproduce known off-targets of drugs or drug-like compounds [20,23].
Protein-ligand docking of putative off-targets
Fourteen putative off-targets predicted from idTarget and PharmMapper are subject to further study. Crystal structures of the 14 proteins come from PDB database. All of the potential off-targets are related to cancer, diabetes and other diseases, including 71.4% of the enzymes, 9.3% of the receptor, 9.3% of the regulatory factors.
Fourteen potential targets were identified for DIP after the dual reverse screening procedures by idTarget and PharmMapper as mentioned in Tables 1 and 2. The dihydropyrimidine dehydrogenase was related with the normal cancers, and GTPase HRas was related with the bladder cancer (Table 1). Many of these identified potential drug targets of DIP explained the mechanism of anti-cancer activity of DIP extracts in breast cancer induced animal models [24-27].
Table 2.
IdTarget results
| Proteins | Docking score | UniProt Id | PDB Id | Ki | Related disease |
|---|---|---|---|---|---|
| Human mitochondrial aldehyde dehydrogenase | -9.53 | P05091 | 1O04 | 103.2 nM | High incidence of acute alcohol intoxication |
| Human pyruvate kinase M2 | -8.25 | P14618 | 3GQY | 895.5 nM | Human cancer |
| Nuclear hormone receptor PPAR-gamma | -8.23 | P37231 | 2Q8S | 926.3 nM | Type 2 insulin-resistant diabetes, hyptertension and cancer |
| Phosphatidylinositol 4,5-bisphosphate 3-kinase | -8.13 | P48736 | 2CHX | 1.1 μM | Inflammatory diseases |
| Phosphotyrosyl phosphatase activator (PTPA) | -8.11 | Q15257 | 2HV7 | 1.1 μM | |
| Aminoadipate-Semialdehyde Dehydrogenase | -7.92 | P49419 | 2J6L | 1.6 μM | Pyridoxine-dependent epilepsy |
| human spindle checkpoint kinase Bub1 | -7.92 | O43683 | 3E7E | 1.6 μM | Human cancer |
| Disintegrin and metalloproteinase domain-containing protein 17 | -7.92 | P78536 | 3E8R | 1.6 μM | Neonatal inflammatory skin and bowel disease |
| Cyclin-dependent kinase 2 | -7.88 | P24941 | 2J9M | 1.7 μM | Human caner |
| Rho-related GTP-binding protein RhoB | -7.82 | P62745 | 2FV8 | 1.9 μM | Human ancer |
| crophage migration inhibitory factor | -7.74 | P14174 | 3L5P | 2.1 μM | Rheumatoid arthritis systemic juvenile |
| Tryptophan--tRNA ligase, cytoplasmic | -7.71 | P23381 | 1R6U | 2.2 μM | Possess angiostatic activity |
DIP and 50 decays generated from DUD-E are docked to 14 off-target protein with Autodock Vina. The statistical significance of the predicted binding affinity is estimated in this study. The normalized docking scores for proteins, including 1gte, 1o04, 3gqy, 1p2s, 3e7e, 3e8r, 2j9m, 2fv8, 3l5p and 1r6u, were listed in Table 3. One of predicted binder with the statistically significant prediction is dihydropyrimidine dehydrogenase (DPD) (PDB Id: 1gte) [21].
Table 3.
Normalized docking score
| Proteins | Docking score | The No. of the docked ligands | Mean | Standard deviations σ | Z-score |
|---|---|---|---|---|---|
| 1gte | -3.33 | 49 | -3.20 | 0.60 | -0.17 |
| 1o04 | -3.20 | 48 | -2.77 | 0.61 | 0.54 |
| 3gqy | -3.07 | 49 | -3.17 | 0.43 | 0.47 |
| 1p2s | - 3.03 | 47 | -3.11 | 0.74 | 0.33 |
| 3e7e | -2.98 | 50 | -2.70 | 0.61 | -0.11 |
| 3e8r | -2.82 | 49 | -2.15 | 0.64 | 0.17 |
| 2j9m | -1.61 | 48 | -1.70 | 0.60 | 0.19 |
| 2fv8 | -1.41 | 49 | -1.33 | 0.65 | 0.25 |
| 3l5p | -1.29 | 47 | -1.25 | 0.11 | 0.17 |
| 1r6u | -0.97 | 49 | -1.03 | 0.33 | 0.19 |
This enzyme has recently functioned as an adjunct target for anticancer drug design because it also inhibited by 5-fluorouracil (5FU), one of the most widely product for treatment of many common malignancies [24-27]. Severe life-threatening toxicities have been reported after treatment of cancer patients with 5FU [28-30]. To date, DIP is utilized as modulators of 5FU treatment [24-27]. Its inhibition may result in increased 5FU efficacy and diminished drug side effects.
Another statistically significant binder is human spindle checkpoint kinase Bub1 [28]. It is essential for spindle-assembly checkpoint signaling and for correct chromosome alignment. The normal mitotic checkpoints of cells displaying microsatellite instability become defective upon transfer of mutant human Bub1 alleles from either of the breast cancer [29,30]. And thence, we hypothesize DPD and hBub1 are two likely off-targets of DIP.
Binding pose analyze of DPD-DIP complex
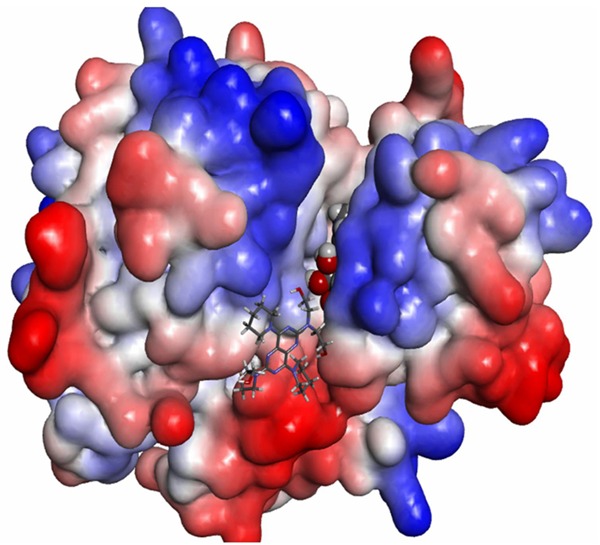
In this study, we identify that the proposed binding mode of DIP anti-cancer action in mammary and prostrate tissues is possibly because of binding of DIP to DPD. From Figure 4, it can be include that DIP was located in an allosteric site [20-22]. Our results suggest that substrate/inhibitor, 5FU, binding in the active-site pocket subsequently triggers closure of the active site loop (Figure 5).
Figure 4.
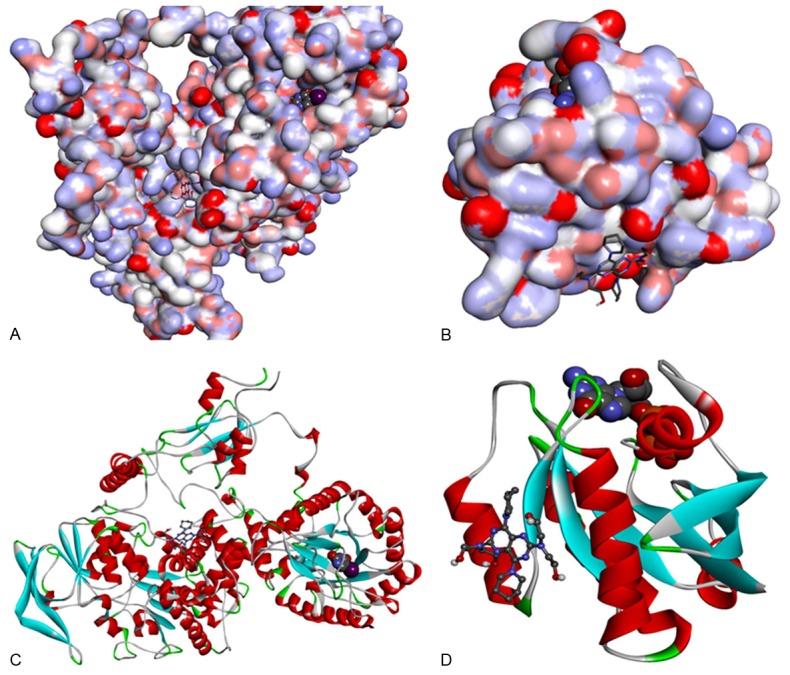
Target protein from PDB Id 1GET and 1P2S. A. Target protein from PDB Id 1GTE, Protein representation: surface. Original crystal complexed ligand representation: CPK. DIP representation: stick. B. Target protein from PDB Id 1P2S protein representation: surface. Original crystal complexed ligand representation: CPK. DIP representation: stick. C. Target protein from PDB Id 1GTE, protein representation: cartoon. Original crystal complexed ligand representation: CPK. DIP representation: ball and stick. D. Target protein from PDB Id 1P2S protein representation: cartoon. Original crystal complexed ligand representation: CPK. DIP representation: ball and stick.
Figure 5.
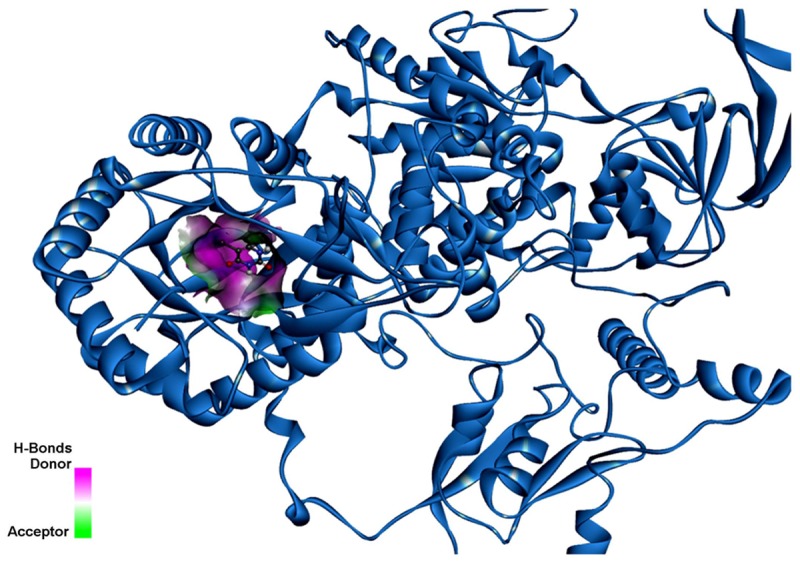
5FU binding in the DPD illustrating the H-Bonds donor and the acceptor.
Figure 6A indicated the important residues for 5FU binding. There are five residues (Asn609, Thr737, Asn736, Asn668, and Ser670) making hydrogen bonds with 5FU. Asn609, Thr737, Asn736, Asn668, Ser670, Glu611, Ile613, and Leu612 have electrostatic contact with 5FU, and Gly764 and Lys574 have van der Waals contacts with 5FU.
Figure 6.
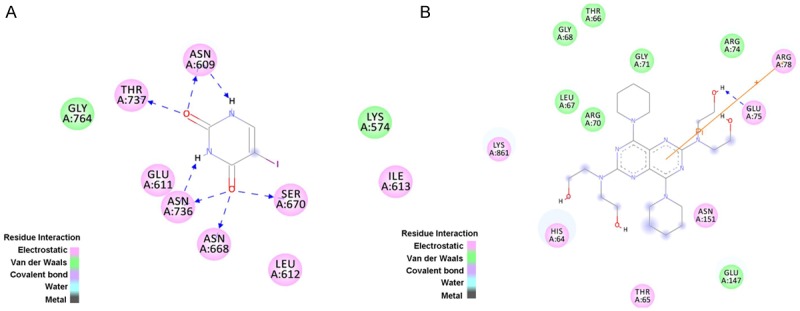
The active pocket residues around 5FU binding and Dip binding. A. The active pocket residues around 5FU binding. Color green represent for van der Waal contacts with 5FU, and color pink represent for electrostatic contacts with 5FU using with Discovery Studio 3.5 client. B. The active pocket residues of DPD around DIP binding. Color green represent for van der Waal contacts with 5FU, and color pink represent for electrostatic contacts with 5FU using with Discovery Studio 3.5 client.
From Figures 6B and 7, Glu75 forms a hydrogen bond (2.15 Å) with the OH group of the DIP. From Figure 5, it can seen that that the inhibitor, 5FU, binding pocket contains Asn609, Thr737, Asn736, Asn668, Ser670, Glu611, Ile613, Leu612, Gly764 and Lys574.
Figure 7.
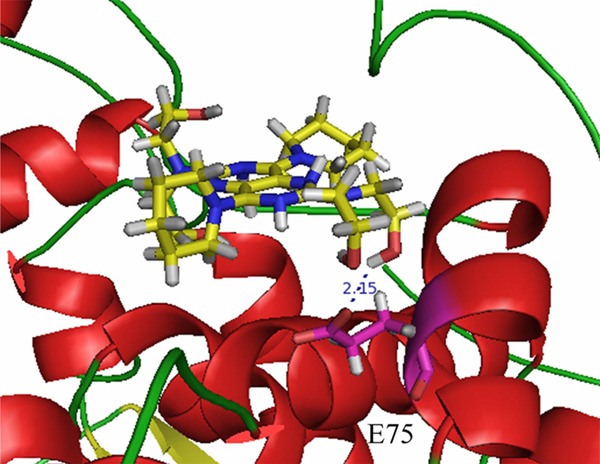
The hydrogen bond between DIP and E75.
As shown in Figure 6B, DIP binds to DPD in the binding pocket different from where 5FU is located. DIP is surrounded by amino acids such as His64, Thr65, Glu147, Asn151, Glu75, Arg78, Arg74, Gly71, Thr66, Gly68, Leu67, Arg70, and Lys861. In particular, Arg78 makes a π-cation contact with the phenol ring of DIP. Thus, Arg78 may be important for DIP binding. It was reported that DIP may increase the activity of 5-fluorouracil in a dose-dependent manner [24]. So it can be concluded that DIP functioned as allosterc activator for DPD. The predicted interaction between DIP and DPD is consistent with the existing biochemical and clinical evidences.
Binding pose analysis of DIP-Bub1 complex
From Figure 7, it can be seen that DIP and the substrate of Bub1, ATP, located in the different active pocket. Bub1 is ATP-dependent kinase. The docking result is shown in Figure 8. There is no hydrogen bond between DIP and Bub1.
Figure 8.
Bub1 protein representation: surface. Original crystal complexed ligand representation: CPK. DIP representation: stick.
As shown in Figure 9, DIP is in the binding pocket composed of Asn879, Leu875, Ala797, Glu795, Gly796, Tyr1003, Phe971, Lys919, Ser969, and Asp921.
Figure 9.
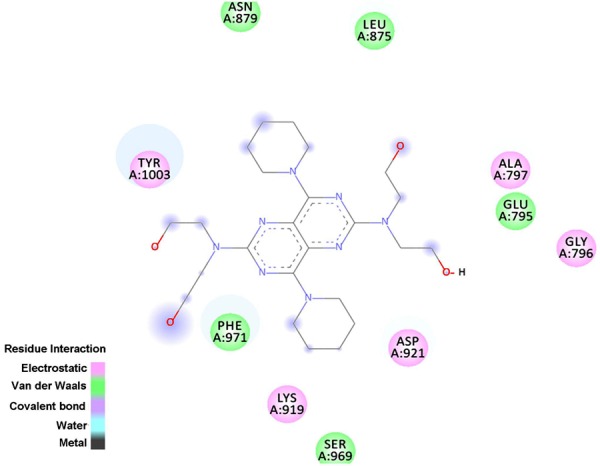
The active pocket residues of Bub1 around DIP binding. Color green represent for van der Waal contacts with DIP, and color pink represent for electrostatic contacts with DIP using with Discovery Studio 3.5 client.
In conclusion, in this study, the reverse dockings are applied to identify the potential off-targets for DIP. The results generate a tractable set of anticancer proteins. We further explore the binding mode between DIP and the two potential off-targets, DPD and Bub1. Structure analysis suggests that electrostatic interaction and hydrogen bonding play an important role in their binding process. The strong functional linkage of DIP and 5FU supports our prediction. The exploration of polypharmacology will provide us new opportunities in treating systematic diseases such as cancer.
Acknowledgements
This work was supported by the National Program on Key Basic Research Project (973 Program No. 2012CB721003), and the National Science Foundation of Jilin Province (201015109).
Disclosure of conflict of interest
None.
References
- 1.Kasprzak JD, Wejner-Mik P, Nouri A, Szymczyk E, Krzemińska-Pakuła M, Lipiec P. Transthoracic measurement of left coronary artery flow reserve improves the diagnostic value of routinedipyridamole-atropine stress echocardiogram. Arch Med Sci. 2013;9:802–807. doi: 10.5114/aoms.2013.38673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Del Ry S, Morales MA, Scali MC, Tafi A, Giustarini C, Posteraro F, Ambrosini V, Agrusta M, Picano E, Sicari R. Effect of concomitant oral chronic dipyridamole therapy on inflammatory cytokines in heart failure patients. Clin Lab. 2013;59:843–849. doi: 10.7754/clin.lab.2012.120816. [DOI] [PubMed] [Google Scholar]
- 3.Gomez G, Nardone V, Lotfi-Emran S, Zhao W, Schwartz LB. Intracellular adenosine inhibits IgE-dependent degranulation of human skin mast cells. J Clin Immunol. 2013;33:1349–1359. doi: 10.1007/s10875-013-9950-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vasu S, Bandettini WP, Hsu LY, Kellman P, Leung S, Mancini C, Shanbhag SM, Wilson J, Booker OJ, Arai AE. Regadenoson and adenosine are equivalent vasodilators and are superior than dipyridamole- a study of first pass quantitative perfusion cardiovascular magnetic resonance. J Cardiovasc Magn Reson. 2013;15:85–91. doi: 10.1186/1532-429X-15-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halkes PH, van-Gijn J, Kappelle LJ, Koudstaal PJ, Algra A. Aspirin plus dipyridamole versus aspirin alone after cerebral ischaemia of arterial origin (ESPRIT): randomised controlled trial. Lancet. 2006;367:1665–1673. doi: 10.1016/S0140-6736(06)68734-5. [DOI] [PubMed] [Google Scholar]
- 6.De Schryver EL, Algra A, van Gijn J. Dipyridamole for preventing stroke and other vascular events in patients with vascular disease. Cochrane Database Syst Rev. 2007;18:CD001820. doi: 10.1002/14651858.CD001820.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou R, Moench P, Heran C, Lu X, Mathias N, Faria TN, Wall DA, Hussain MA, Smith RL, Sun D. pH-dependent dissolution in vitro and absorption in vivo of weakly basic drugs: development of a canine model. Pharm Res. 2005;22:188–192. doi: 10.1007/s11095-004-1185-3. [DOI] [PubMed] [Google Scholar]
- 8.Zhang ZH, Nie SF, Wang QM, Peng B, Liu HF, Pan WS. Absorption kinetics of dipyridamole in rat gastrointestinal. Chin Pharm J. 2009;44:1229–1233. [Google Scholar]
- 9.Banker GS, Rhodes CT. Modern Pharmaceutics, Revised and Expanded. 4th edition. New York: Marcel Dekker; 2002. pp. 501–573. [Google Scholar]
- 10.Mahony C, Wolfram KM, Cocchetto DM, Bjornsson TD. Dipyridamole kinetics. Clin Pharmacol Ther. 1982;31:330–338. doi: 10.1038/clpt.1982.42. [DOI] [PubMed] [Google Scholar]
- 11.Terhaag B, Donath F, Le Petit G, Feller K. The absolute and relative bioavailability of dipyridamole from different preparations and the in vitro-in vivo comparison. Int J Clin Pharmacol Ther Toxicol. 1986;24:298–302. [PubMed] [Google Scholar]
- 12.Wang C, Schwab LP, Fan M, Seagroves TN, Buolamwini JK. Chemoprevention activity of dipyridamole in the MMTV-PyMT transgenic mouse model of breast cancer. Cancer Prev Res. 2013;6:437–447. doi: 10.1158/1940-6207.CAPR-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spano D, Marshall JC, Marino N, De Martino D, Romano A, Scoppettuolo MN, Bello AM, Di Dato V, Navas L, De Vita G, Medaglia C, Steeg PS, Zollo M. Dipyridamole prevents triple-negative breast-cancer progression. Clin Exp Metastasis. 2013;30:47–68. doi: 10.1007/s10585-012-9506-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goda AE, Yoshida T, Horinaka M, Yasuda T, Shiraishi T, Wakada M, Sakai T. Mechanisms of enhancement of TRAIL tumoricidal activity against human cancer cells of different origin bydipyridamole. Oncogene. 2008;27:3435–3445. doi: 10.1038/sj.onc.1211008. [DOI] [PubMed] [Google Scholar]
- 15.Spano D, Marshall JC, Marino N, De Martino D, Romano A, Scoppettuolo MN, Bello AM, Di Dato V, Navas L, De Vita G, Madaglia C, Steeg PS, Zollo M. Dipyridamole prevents triple-negative breast-cancer progression. Clin Exp Metastasis. 2013;30:47–68. doi: 10.1007/s10585-012-9506-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang C, Schwab LP, Fan M, Seagroves TN, Buolamwini JK. Chemoprevention activity of dipyridamole in the MMTV-PyMT transgenic mouse model of breast cancer. Cancer Prev Res. 2013;6:437–447. doi: 10.1158/1940-6207.CAPR-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodrigues M, Barbosa FJ, Perussi JR. Dipyridamole increases the cytotoxicity of cisplatin in human larynx cancer cells in vitro. Braz J Med Biol Res. 2004;37:591–599. doi: 10.1590/s0100-879x2004000400017. [DOI] [PubMed] [Google Scholar]
- 18.Tsavaris N, Kosmas C, Polyzos A, Genatas K, Vadiaka M, Paliaros P, Dimitrakopoulos A, Rokana S, Karatzas G, Vachiotis P, Fotiadis K. Leucovorin + 5-fluorouracil plus dipyridamole in leucovorin + 5-fluorouracil-pretreated patients with advanced colorectal cancer: a pilot study of three different dipyridamole regimens. Tumori. 2001;87:303–307. doi: 10.1177/030089160108700505. [DOI] [PubMed] [Google Scholar]
- 19.Liu X, Ouyang S, Yu B, Liu Y, Huang K, Gong J, Zheng S, Li Z, Li H, Jiang H. PharmMapper server: a web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Res. 2010;38:W609–614. doi: 10.1093/nar/gkq300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhattacharjee B, Vijayasarathy S, Karunakar P, Chatterjee J. Comparative reverse screening approach to identify potential anti-neoplastic targets of saffron functional components and binding mode. Asian Pac J Cancer Prev. 2012;13:5605–5611. doi: 10.7314/apjcp.2012.13.11.5605. [DOI] [PubMed] [Google Scholar]
- 21.Dobritzsch D, Ricagno S, Schneider G, Schnackerz KD, Lindqvist Y. Crystal structure ofthe productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5odouracil. Implications for mechanism of inhibitionand electron transfer. J Biol Chem. 2002;277:13155–13166. doi: 10.1074/jbc.M111877200. [DOI] [PubMed] [Google Scholar]
- 22.Buhrman G, de Serrano V, Mattos C. Organic solvents order the dynamic switch II in Ras crystals. Structure. 2003;11:747–751. doi: 10.1016/s0969-2126(03)00128-x. [DOI] [PubMed] [Google Scholar]
- 23.Wang JC, Chu PY, Chen CM, Lin JH. idTarget: a web server for identifying protein targets of small chemical molecules with robust scoring functions and a divide-and-conquer docking approach. Nucleic Acids Res. 2012;40:W393–399. doi: 10.1093/nar/gks496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsavaris N, Kosmas C, Polyzos A, Genatas K, Vadiaka M, Paliaros P, Dimitrakopoulos A, Rokana S, Karatzas G, Vachiotis P, Fotiadis K. Leucovorin + 5-fluorouracil plus dipyridamole in leucovorin + 5-fluorouracil-pretreated patients with advanced colorectal cancer: a pilot study of three different dipyridamole regimens. Tumori. 2001;87:303–307. doi: 10.1177/030089160108700505. [DOI] [PubMed] [Google Scholar]
- 25.Burch PA, Ghosh C, Schroeder G, Allmer C, Woodhouse CL, Goldberg RM, Addo F, Bernath AM, Tschetter LK, Windschitl HE, Cobau CD. Phase II evaluation of continuous-infusion 5-fluorouracil, leucovorin, mitomycin-C, and oral dipyridamole in advanced measurable pancreatic cancer: a North Central Cancer Treatment Group Trial. Am J Clin Oncol. 2000;23:534–537. doi: 10.1097/00000421-200010000-00021. [DOI] [PubMed] [Google Scholar]
- 26.Kohnoe S, Maehara Y, Takahashi I, Emi Y, Baba H, Sugimachi K. Treatment of advanced gastric cancer with 5-fluorouracil and cisplatin in combination with dipyridamole. Int J Oncol. 1998;13:1203–1206. doi: 10.3892/ijo.13.6.1203. [DOI] [PubMed] [Google Scholar]
- 27.Todd KE, Gloor B, Lane JS, Isacoff WH, Reber HA. Resection of locally advanced pancreatic cancer after downstaging with continuous-infusion 5-fluorouracil, mitomycin-C, leucovorin, and dipyridamole. J Gastrointest Surg. 1998;2:159–166. doi: 10.1016/s1091-255x(98)80008-5. [DOI] [PubMed] [Google Scholar]
- 28.Kang J, Yang M, Li B, Qi W, Zhang C, Shokat KM, Tomchick DR, Machius M, Yu H. Structure and substrate recruitment of the human spindle checkpoint kinase Bub1. Mol Cell. 2008;32:394–405. doi: 10.1016/j.molcel.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 30.Yuan B, Xu Y, Woo JH, Wang Y, Bae YK, Yoon DS, Wersto RP, Tully E, Wilsbach K, Gabrielson E. Increased expression of mitotic checkpoint genes in breast cancer cells with chromosomal instability. Clin. Cancer Res. 2006;12:405–410. doi: 10.1158/1078-0432.CCR-05-0903. [DOI] [PubMed] [Google Scholar]
- 31.Velichkova P, Himo F. Methyl Transfer in Glycine N-Methyltransferase, a Theoretical Study. J Phys Chem B. 2005;109:8216–8219. doi: 10.1021/jp0443254. [DOI] [PubMed] [Google Scholar]
- 32.Liao RZ, Yu JG, Himo F. Reaction mechanism of the trinuclear zinc enzyme phospholipase C: a density functional theory study. J Phys Chem B. 2010;114:2533–2540. doi: 10.1021/jp910992f. [DOI] [PubMed] [Google Scholar]
- 33.Wan H, Xu Y, Zhou G. Dual conductance, negative differential resistance, and rectifying behavior in a molecular device modulated by side groups. J Chem Phys. 2012;136:184704–184710. doi: 10.1063/1.4712615. [DOI] [PubMed] [Google Scholar]
- 34.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2011;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michael MM, Michael C, John JI, Brian KS. Directory of Useful Decoys, Enhanced (DUD-E): Better Ligands and Decoys for Better Benchmarking. J Med Chem. 2012;55:6582–6594. doi: 10.1021/jm300687e. [DOI] [PMC free article] [PubMed] [Google Scholar]


