Abstract
Recent studies have demonstrated that the crucial regulatory roles of long noncoding RNAs (lncRNAs) in tumorigenesis. Expression levels of several lncRNAs are abnormally up-regulated or down-regulated and play a primary role in colorectal cancer (CRC) cell tumorigenesis. However, the potential role and regulatory mechanisms of the novel human lncRNA, LINC00152, in CRC cells are poorly understood. Here, we found that LINC00152 expression was significantly decreased in CRC tissues and CRC cell lines, and this change was more frequent in patients with advanced stage (tumor-node-metastasisi (TNM) III and IV). Overexpression of LINC00152 (LINC000152over) resulted in decreased cell viability and increased apoptosis in CSC cell lines (HT-29 and SW480). Furthermore, decreased Ki-67 and B-cell lymphoma-2 (Bcl-2), and increased Fas, were observed in CSC cells. However, a change in Bax expression was undetected. Interestingly, microRNA (miR)-376c-3p down-regulated the expression of LINC00152 in CSC cells. Overexpression of miR-376c-3p (miR-376c-3pover) enhanced viability and limited apoptosis of CSC cells. In addition, miR-376c-3pover suppressed the effect of LINC00152over on the viability and apoptosis of CSC cells. Taken together, these data indicate that LINC00152 in CSC cells negatively regulated by miR-376c-3p, restricts cell viability and stimulates cell apoptosis, possibly by modulating the expression of Ki-67, Bcl-2, and Fas. MiR-376c-3p/LINC00152 plays an important role in the pathogenesis of CRC and may serve as a potential target for its diagnosis and treatment.
Keywords: LINC00152, miR-376c-3p, colorectal cancer cells, viability, apoptosis
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumors worldwide, and is highly lethal with an increasing incidence annually [1,2]. Although current detection methods and therapeutic targets for CRC are under development, the overall patient survival rate of CRC patients remains poor [3]. Consequently, there exists a great challenge in exploring novel biomarkers and/or therapeutic targets for early detection and treatment of CRC.
Recently, a variety of molecular markers for the characterization and prognosis of CRC have been identified [4-6]. Among these, long noncoding RNAs (lncRNAs) act as regulators in the origin and progression of human CRC and may represent novel therapeutic targets [6-9]. LncRNA is recently discovered RNA that is larger than 200 nucleotides with no protein-coding capacity [10]. LncRNAs play important regulatory roles in diverse cellular processes, including cell growth, apoptosis, embryonic development, and tumorigenesis, by modulating gene expression at the chromatin organization, epigenetic control, and transcriptional and post-transcriptional levels [11,12].
Based on the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/gene/112597), lncRNA-LINC00152 is an intergenic lncRNA located on 2p11.2. Previous studies have demonstrated that LINC00152 functions as an oncogene in gastric cancer, hepatocellular, and clear cell renal cell carcinomas [13-17]. It can stimulate the proliferation and growth in these cancer cells by the epidermal growth factor receptor (EGFR) or mammalian target of rapamycin (mTOR) signaling pathway [15,16]. In a recent study using several datasets, CRC patients with higher expression of LINC00152 showed significantly better overall survival than those with lower expression, suggesting LINC00152 may be a negative prognostic factor for CRC [4]. However, the information on the expression and role of LINC00152 in CRC is limited.
The present study aimed to investigate whether LINC00152 is aberrantly expressed in CRC tissue and cells, to further analyze the regulation mechanism for LINC00152 expression, and to identify the role of LINC00152 on the viability and apoptosis of CRC cell lines in vitro.
Materials and methods
Tissue collection
All tissue samples were collected with written informed consent in accordance with the requirements of the Research Ethics Committee in Shanghai Eighth People’s Hospital Affiliated Jiangsu University. In this study, CRC tissues and their paired adjacent normal tissues (located >5 cm away from the tumor border) were collected from 49 CRC patients. All tissues were confirmed by histopathological evaluation, and none of the included patients had received any therapy before surgery.
Cell lines
All human CRC cell lines (CACO2, HCT-116, HT-29, SW480, and SW620) were purchased from the American Type Culture Collection (ATCC, USA). The human embryonic kidney 293 cell line (HER293) was obtained from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China), and the human normal intestinal mucous cell line (CCC-HIE-2) was purchased from Type Culture Collection of the Chinese Academy of Medical Sciences (Beijing, China).
All culture media (Gibco, USA) were supplemented with 10% fetal bovine serum (FBS, Hyclone, Logan, UT, USA). Cells were incubated at 37°C in a humidified at mosphere containing 5% CO2.
Quantitative real-time polymerase chain reaction (qRT-PCR)
qRT-PCR was performed to determine the expression levels of LINC00152. Total RNA was isolated from tissues or cells using RNAiso Plus (TaKaRa, Otsu, Japan), and reverse transcribed into cDNA using a PrimeScriptTM II 1st Strand Synthesis Kit (TaKaRa) according to the manufacturer’s instructions. The qRT-PCR reactions were performed using an ABI7500 System (Applied Biosystems, Foster City, CA, USA) and SYBR Green PCR Master Mix (TaKaRa). MiR-376c-3p miRNA was harvested using the PureLink™ miRNA Isolation Kit (Invitrogen, CA, USA) and miR-376c-3p expression was detected by the TaqMan MicroRNA Assay Kit (Applied Biosystems). The relative expression levels of LINC00152 and miR-376c-3p were analyzed using the 2-ΔΔCT relative quantification method with human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and small nuclear 6 (U6) snRNA as internal controls, respectively. Primer sequences: 1) LINC00152: forward, 5’-CTCCAGCACCTCTACCTGTTG-3’ and reverse, 5’-GGACAAGGGATTAAGACACACA-3’; 2) GAPDH: forward, 5’-GTCAACGGATTTGGTCTGTATT-3’ and reverse, 5’-AGTCTTCTGGGTGGCAGTGAT-3’; 3) miR-376c-3p: forward, 5’-AACATAGAGGAAATTCCACG-3’; 4) U6 snRNA: forward, 5’-CGCAAGGATGACACGCAAATTC-3’. All assays were performed in triplicate.
Overexpression of LINC00152 and miR-376c-3p in HT-29 and SW480 cells
Lentiviruses expressing the LINC00152 sequence (LINC00152-overexpression: pWPXL-LINC00152) and the negative control lentivirus (Vector: pWPXL); the miR-376c-3p mimic lentivirus (miR-376c-3p-mimics) and its corresponding control miRNA lentivirus (C-miRNA: Negative ctrl) were constructed by GenePharma (Shanghai, China). LINC00152-overexpression, miR-376c-3p-overexpression and the corresponding control stable cell lines were then established. Additionally, the LINC00152 and miR-376c-3p co-overexpressed cell lines were also constructed. The efficiency of overexpression was verified by qRT-PCR.
The cell-counting kit-8 (CCK-8) assay and apoptosis assay
HT-29 and SW480 cells (pWPXL, pWPXL-LINC00152, Negative ctrl and miR-376c-3p-mimics) were seeded into 96-well plates (5×103 cells/well). After culture for 24 h, 48 h or 72 h, cell viability was analyzed by the CCK-8 assay (Dojindo, Japan). CCK-8 reagent was added to each well and cells were incubated at 37°C for 1-4 h according to the manufacturer’s protocol. The absorbance at 450 nm was measured and used to represent cell viability. Each experiment was performed in six parallel wells and repeated in triplicate.
To determine apoptosis, HT-29 and SW480 cells were seeded in 24-well plates (2×105 cells/well) for 48 h. The level of apoptosis was then analyzed by the Annexin V-FITC apoptosis assay (Invitrogen).
Flow cytometry (FCM)
HT-29 and SW480 cells were digested with trypsin, gently aspirated and washed with phosphate-buffered saline. After blocking with 10% FBS, the recovered cells were mixed with anti-human Ki-67 (Biolegend, San Diego, USA), Bcl-2 (BD Biosciences, USA), Bax (Santa Cruz Biotechnology, USA), or Fas (Biolegend) antibodies in darkness for 30 min at room temperature. An isotope control was used as a negative control. After incubation, the cells were washed and analyzed immediately by FCM analysis using a Beckman-Coulter CyAN ADP Analyzer (Beckman Coulter, Inc. Kraemer Boulevard Brea, CA, USA). Data were analyzed with FlowJo Version 6.1 software (TreeStar, Asland, OR, USA). Statistical analysis was conducted using isotype-matched controls as references.
Statistics
All values are shown as the means ± standard error of the mean (SEM). Data were analyzed with GraphPad Prism version 5 by t-test, one-way analysis of variance (ANOVA) or two-way ANOVA. Differences were considered statistically significant at P<0.05.
Results
LINC00152 expression in CRC tissues and cell lines was aberrantly decreased
We measured the expression level of LINC00152 in 49 CRC patients. The results in Figure 1 show that LINC00152 was significantly down-regulated in CRC tissues compared with the adjacent normal tissues (P<0.001) (Figure 1A). Based on the tumor-node-metastasis (TNM) stage, low-level LINC00152 expression was more frequently observed in CRC patient in advanced stage (TNM: III-IV) (P<0.001) (Figure 1B). These data indicate that LINC00152 level is negatively correlated with TNM stage. However, there was no obvious difference in LINC00152 expression in patients with or without distant metastasis (P>0.05) (Figure 1C).
Figure 1.
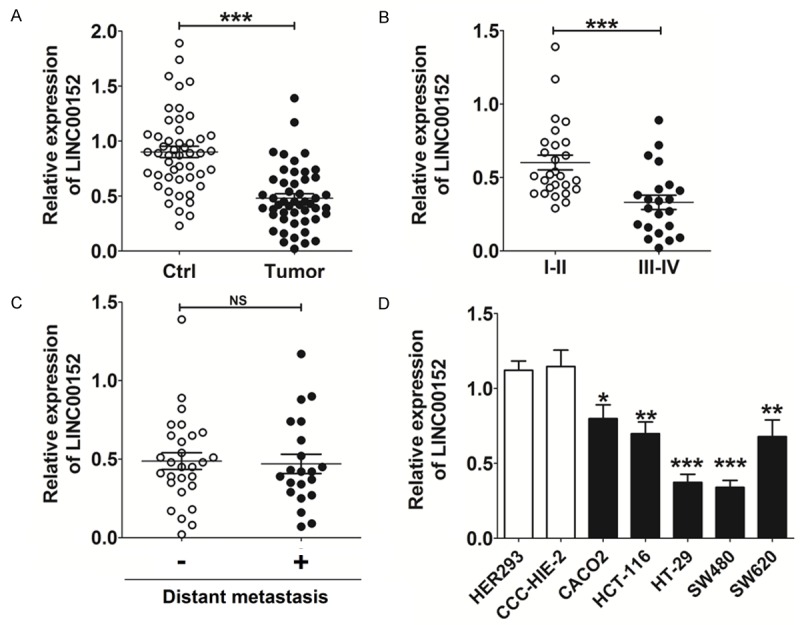
The expression of LINC00152 on CRC tissue and cell lines is aberrantly decreased. A-C. qRT-PCR analysis for LINC00152 expression in CRC tissues (n=49) and paired adjacent normal tissues. Ctrl: adjacent normal tissues; tumor: CRC tissues; I-II: patient with the early stage (TNM: I-II); III-IV: patient with advanced stage (TNM: III-IV). D. qRT-PCR analysis for LINC00152 expression in CRC cell lines (CACO2, HCT-116, HT-29, SW480 and SW620) and non-tumorigenic cell lines (CCC-HIE-2 and HER293). Data are expressed as means ± SEM. *P<0.05, **P<0.01 and ***P<0.001 (t-test or one-way ANOVA). NS: no statistically significant difference.
Similarly, there was a marked decrease in LINC00152 levels in all five CRC cells (CACO2, HCT-116, HT-29, SW480 and SW620) compared with the non-tumorigenic cell lines (CCC-HIE-2 and HER293) (P<0.05, P<0.01 or P<0.001) (Figure 1D), especially HT-29 and SW480 cells (P<0.001) (Figure 1D). Because of the relatively low level of LINC00152 in HT-29 and SW480 cells, these cell lines were chosen to further study the potential function of LINC00152 in CRC cells in vitro.
LINC00152 suppresses viability and promotes apoptosis of CRC cells
To investigate the potential effect of LINC00152 in CRC cells, we constructed LINC00152-overexpression HT-29 and SW480 cells by transfection (P<0.01 or P<0.001) (Figure 2A). As shown, overexpression of LINC00152 (pWPXL-LINC00152) resulted in decreased viability of HT-29 and SW480 cells (P<0.05 or P<0.01) (Figure 2B), especially at 72 h (P<0.01) (Figure 2B). Further analysis showed that LINC00152 overexpression significantly stimulated apoptosis in HT-29 (P<0.01) (Figure 2C and 2D) and SW480 (P<0.001) cells (Figure 2C and 2D). Collectively, these data suggest that LINC00152 is a suppressor regulator for CRC cell growth.
Figure 2.
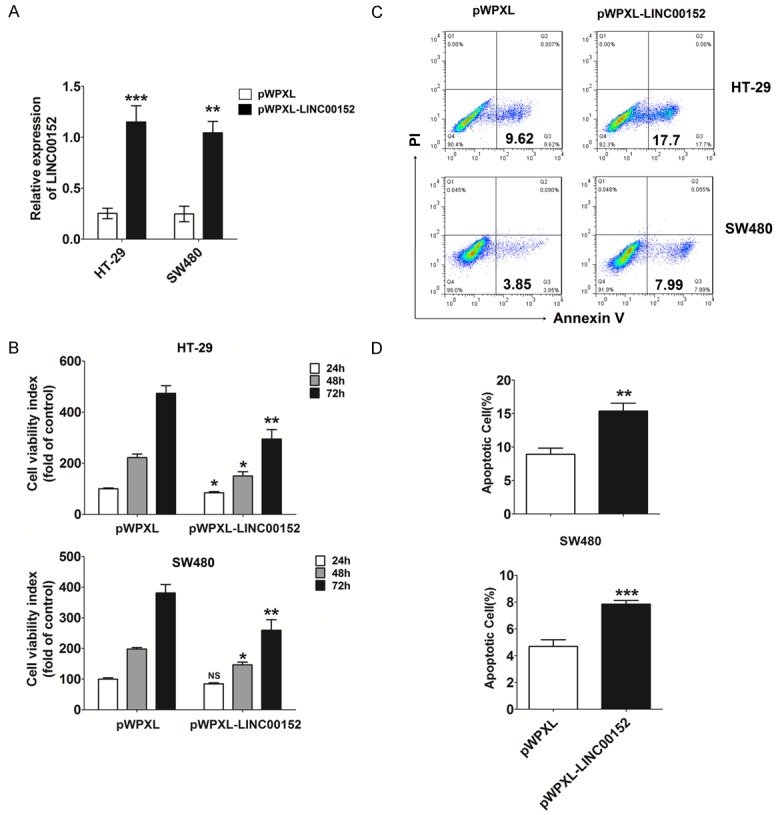
LINC00152 suppresses viability and promotes apoptosis of CRC cells. (A) The construction of LINC00152 overexpression in HT-29 and SW480 cells by transfection. CCK-8 (B) and apoptosis (C, D) assays were performed to analyze viability and apoptosis of HT-29 and SW480 cells with or without LINC00152 overexpression. pWPXL: HT-29 or SW480 cells transfected with the negative control lentivirus (Vector: pWPXL); pWPXL-LINC00152: HT-29 or SW480 cells transfected with lentiviruses expressing the LINC00152 sequence. Data are expressed as means ± SEM. *P<0.05, **P<0.01 and ***P<0.001 (t-test or two-way ANOVA).
LINC00152 down-regulates Ki-67, and Bcl-2, and up-regulates Fas expression in CRC cells
To explore the potential mechanism of LINC00152 on the growth of CRC cells, levels of the proliferation-related molecule Ki-67, and apoptosis-related molecules Bcl-2, Bax and Fas, were analyzed in HT-29 and SW480 cells. As shown in Figure 3, expression levels of Ki-67 and Bcl-2 in HT-29 (P<0.01) (Figure 3A and 3B) and SW480 (P<0.001) cells (Figure 3C and 3D) with overexpression of LINC00152 were significantly higher than in the control HT-29 and SW480 cells. On the contrary, the level of Fas in HT-29 (P<0.001) (Figure 3A and 3B) and SW480 (P<0.001) cells (Figure 3C and 3D) was significantly increased after overexpression of LINC00152. However, there was no significant change of Bax expression in HT-29 and SW480 cells between groups (P<0.05) (Figure 3A-D). These results indicate that the inhibitory effect of LINC00152 on the growth of CRC cells may be dependent on the regulation of Ki-67, Bcl-2, and Fas.
Figure 3.
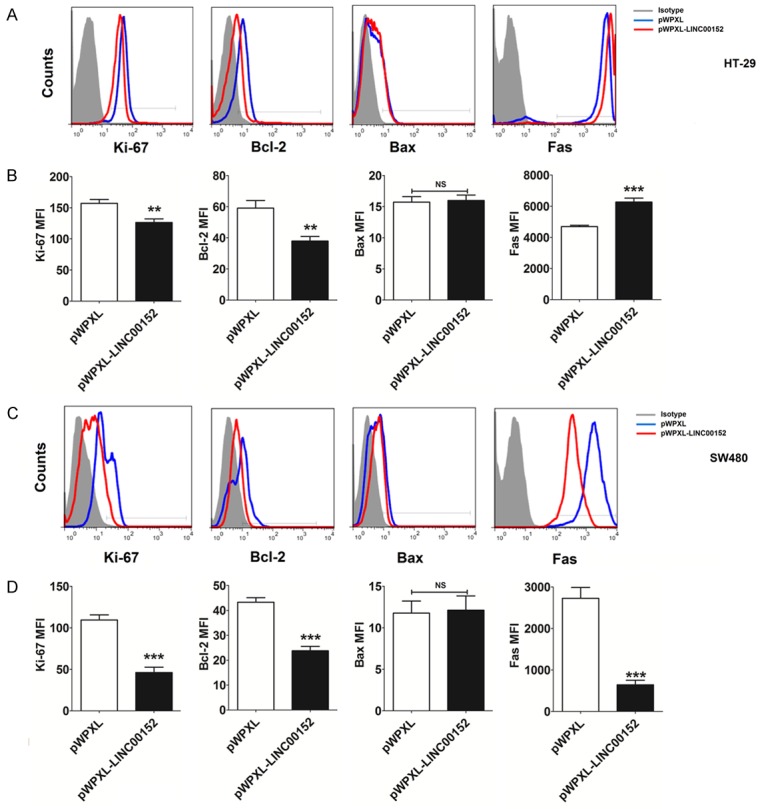
LINC00152 down-regulates Ki-67 and Bcl-2, and up-regulates Fas expression in CRC cells. The expression of Ki-67, Bcl-2, Bax and Fas in HT-29 (A, B) and SW480 (C, D) cells after transfection with pWPXL or pWPXL-LINC00152 was detected by FCM. MFI: Median Fluorescence Intensity. Data are expressed as means ± SEM. **P<0.01 and ***P<0.001 (t-test).
MiR-367c-3p is negatively correlated with LINC00152 in CRC tissues and cells
To further investigate the possible mechanisms underlying the abnormal level of LINC00152 in CRC cells, we performed a search for potential binding sites of miRNA-LncRNA using the online software programs miRcode 11 (ht-tp://mircode.org) and starBase v2.0 (http://starbase.sysu.edu.cn/mirLncRNA.php), and found that LINC00152 contains complementary sites related miRNAs such as miR-376c-3p [13,18]. We then detected the expression of miR-376c-3p in CRC tissues and found that the level of miR-376c-3p was significantly increased in CRC tissues relative to adjacent normal control tissues (P<0.001) (Figure 4A). Further analysis of the relationship between miR-376c-3p and LINC00152 showed there was a significant negative correlation in the expression of miR-376c-3p and LINC00152 (R2=0.511, P<0.001) (Figure 4B). Subsequently, we overexpressed the level of miR-376c-3p in HT-29 and SW480 cells (Figure 4C).
Figure 4.
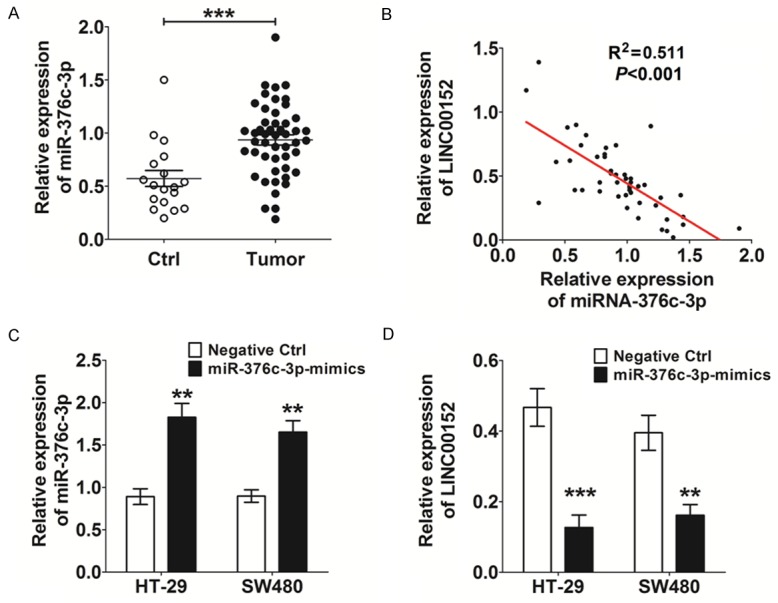
miR-367c-3p negatively correlates with LINC00152 in CRC tissue and cells. A. qRT-PCR analysis for miR-367c-3p expression in CRC tissues (n=49) and adjacent normal tissues. B. Relationship analysis between miR-376c-3p and LINC00152 in CRC tissues (R2=0.511). C. Construction of miR-367c-3p overexpression in HT-29 and SW480 cells by transfection. Negative Ctrl: HT-29 or SW480 cells transfected with the control miRNA lentivirus (C-miRNA); miR-376c-3p-mimics: HT-29 or SW480 cells transfected with the miR-376c-3p mimic lentivirus. D. qRT-PCR analysis for miR-367c-3p expression in HT-29 and SW480 cells with or without miR-367c-3p overexpression. Data are expressed as means ± SEM. **P<0.01 and ***P<0.001 (t-test).
The qRT-PCR results demonstrated the negative regulatory effect of miR-376c-3p on LINC00152 in CRC cells (P<0.01 or P<0.001) (Figure 4D).
MiR-376c-3p enhances the growth of CRC cells by down-regulating LINC00152
We next analyzed the potential role of miR-376c-3p in the biological behavior of CRC cells. Compared to the control group (Negative ctrl), HT-29 and SW480 cell viability was obviously increased after overexpression of miR-376c-3p (miR-376c-3p-mimics) for 48 h and 72 h (P<0.05 or P<0.01) (Figure 5A and 5B). Moreover, overexpression of miR-376c-3p led to a low level of apoptosis in HT-29 and SW480 cells (P<0.01) (Figure 5C and 5D). Together, these results suggest that miR-376c-3p represents an oncogene in CRC cells.
Figure 5.
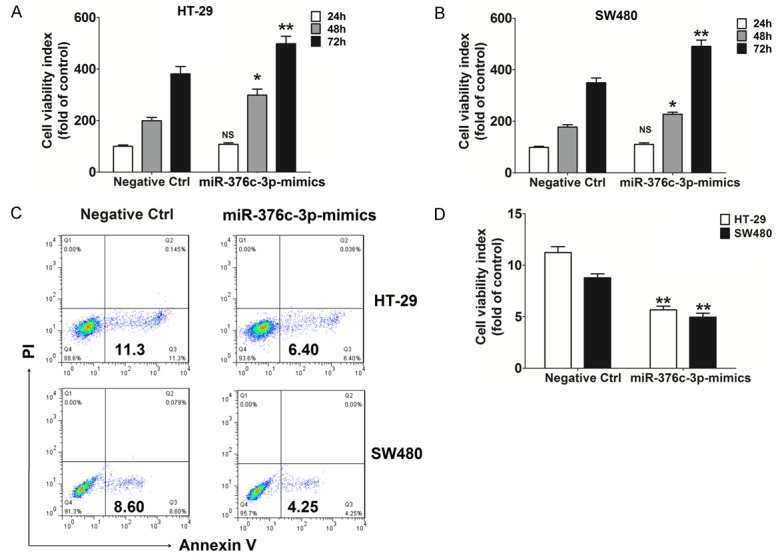
miR-367c-3p enhances viability and restricts apoptosis of CRC cells. CCK-8 (A, B) and apoptosis (C, D) assays were performed to analyze the viability and apoptosis of HT-29 and SW480 cells with or without miR-367c-3p overexpression. Data are expressed as means ± SEM. *P<0.05 and **P<0.01 (t-test or two-way ANOVA).
To confirm whether the effect of miR-376c-3p on CRC cells is dependent on LINC00152, we analyzed whether overexpression of miR-376c-3p and LINC00152 could promote and inhibit HT-29 and SW480 cell viability, respectively (P<0.05, P<0.01 or P<0.001) (Figure 6A and 6B). However, co-overexpression of miR-376c-3p restricted the inhibitory effect of LINC00152 overexpression on the viability of HT-29 and SW480 cells (P>0.05) (Figure 6A and 6B). We then observed a similar role in cell apoptosis (P>0.05) (Figure 6C and 6D), suggesting that miR-376c-3p promotes the growth in CRC cells by down-regulating LINC00152.
Figure 6.
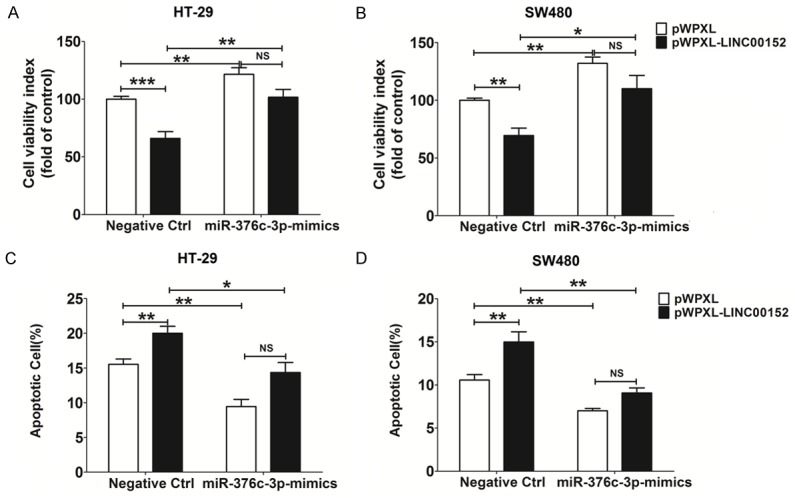
miR-367c-3p enhances the growth of CRC cells by down-regulating LINC00152. CCK-8 (A, B) and apoptosis (C, D) assays were performed to analyze the viability and apoptosis of HT-29 and SW480 cells with or without co-overexpression of LINC00152 and/or miR-367c-3p. Data are expressed as means ± SEM. *P<0.05, **P<0.01 and ***P<0.001 (two-way ANOVA).
Discussion
LINC00152 is confirmed to be aberrantly and highly express in several cancer cells, such as gastric and hepatocellular carcinomas [13-17]. LINC00152 can also be detected in plasma, and one possible mechanism for its stable expression in peripheral blood is dependent on protection by exosomes [19]. Many studies have indicated a positive relationship between LINC00152 and the pathogenesis of gastric cancers [14,16,17]. LINC00152 is reported to be involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition (EMT), cell migration and invasion in gastric cancer [16,20]. However, the expression level and role of LINC00152 in CRC are not well-understood. In the current study, LINC00152 was detected at an abnormally low expression level in CRC tissues and cells, and correlated negatively with TNM stage. These results are comparable with a previous CRC study [4].
Here, the in vitro results showed that LINC00152 significantly suppressed viability and promoted apoptosis of CRC cell lines (HT-29 and SW-480). Ki-67 is a known nuclear protein that is associated with cellular proliferation and ribosomal RNA transcription [21,22]. Usually, the fraction of Ki-67-positive tumor cells is often correlated with the clinical course of cancer. As an anti-apoptosis molecule, Bcl-2 plays an important role in inhibiting the actions of pro-apoptotic proteins and promoting cellular survival [23]. In the BCL-2 family, the pro-apoptotic proteins Bax and Bak, which often promote permeabilization and release of cytochrome C and reactive oxygene species (ROS) are important signals in the apoptosis cascade. In turn, these pro-apoptotic proteins are inhibited by the function of Bcl-2 and its relative, Bcl-Xl [23]. Fas (also known as CD95 or APO-1) is considered to be the prototypic and major member of the death-receptor family. Interaction of Fas with its cognate ligand (Fas ligand, FasL) results in the process of cell apoptosis [24]. Thus, we next analyzed the effect of LINC00152 on proliferation- and apoptosis-related molecules (Ki-67, Bcl-2, Bax and Fas), and found that overexpression of LINC00152 obviously down-regulated the expression of Ki-67 and Bcl-2, and up-regulated the expression of Fas in CRC cells. In gastric and hepatocellular carcinomas, LINC00152 promotes cell cycle progression and proliferation possibly via binding to specific proteins, similar to ZFAS1 [25], MINCR [26], and GAS5 [27] in an EGFR or mTOR-dependent manner [15,16]. However, LINC00152 inhibited CRC cell growth in vitro, and this effect may be associated with the regulation of Ki-67, Bcl-2, and Fas. The detailed mechanisms for the different roles of LINC00152 in CRC and gastric cancer warrant further research.
It is reported that LINC00152 responds highly to chemical stresses [28]. Furthermore, LINC00152 may interact with thrombospondin (THBS1) mediated by miR-18a-5p [29]. However, the regulatory mechanism for LINC00152 in cancer cells is largely unknown. To fully understand CRC pathogenesis, we next focused on the mechanism of low-level LINC00152 in CRC cells, based on bioinformatics analysis, and found that miR-376c-3p was highly expressed in CRC tissue and cells. Further analysis showed an inverse correlation between miR-376c-3p and LINC00152.
MiR-368, known as miR-376c, belongs to an evolutionary conserved miRNA family that also includes miR-376a, miR-376a* and miR-376b, and these genes are characterized in a syntenic cluster on human chromosome 14 [30]. MiR-376c suppresses the proliferation and invasion of osteosarcoma and non-small-cell lung cancer cells by targeting transforming growth factor (TGF)-alpha or liver receptor homolog-1 (LRH-1) [31,32], respectively. However, as a potential biomarker for preeclampsia, the abnormal decrease of miR-376c in the placental preeclampsia tissue may be involved in the impairment of trophoblast cell invasion [33,34]. In addition, miR-376c is reported to be up-regulated in various acute myeloid leukemia specimens [35,36]. Similarly, miR-376c exerts pro-survival functions in ovarian cancer cells by suppressing the expression of activin receptor-like kinase 7 (ALK7) [18]. Here, we also found miR-376c-3p down-regulated the expression of LINC00152, and promoted the viability and restricted the apoptosis of CRC cells in vitro. Furthermore, the overexpression of miR-376c-3p significantly reversed the role of LINC00152 on the viability and apoptosis of CRC cells, suggesting a stimulatory effect of miR-376c-3p on the growth of CRC cells possibly by down-regulating the expression of LINC00152.
It has been reported that the miR-376 cluster transcripts can undergo adenosin-to-inosine editing in a tissue-specific manner, leading to silencing of a different set of genes [37]. Therefore, the detailed mechanisms for the diverse effect of miR-376c in different cancer cells may be due to tissue specificity. Hypoxia condition can up-regulate the expression of miR-376c in glioblastoma (GBM) tumors [38]. Tumor cells usually respond to hypoxia through activation of several different pathways, leading to many biological consequences [39], including angiogenesis, proliferation, glycolytic tumor metabolism, metastasis, autophagy, and apoptosis regulation [40-42]. The above results suggest that the low level of miR-376c-3p in CRC cells may be induced by hypoxia. The up-regulation of miR-376c-3p triggered by hypoxia may lead to the high level of LINC00152 and the rapid growth of CRC cells. This complex role of hypoxia/miR-376c-3p and LINC00152 requires further research.
Taken together and as shown in Figure 7, it can be concluded that LINC00152 can increase expression of the proliferation-related molecule Ki-67 and anti-apoptosis molecule Bcl-2, and decrease expression of the pro-apoptosis molecule Bax in CRC cells to suppress viability and promote apoptosis of CRC cells. The abnormally low level of LINC00152, possibly induced by hypoxia/miR-376c-3p, likely lead to increased CRC cell growth. With this rapid growth, the local hypoxia condition would aggravate and further induced the up-regulation of miR-376c-3p and the down-regulation of LINC00152 in CRC cells. These processes form a vicious circle finally leading to tumorigensis and progression of CRC. Therefore, targeting hypoxia/miR-376c-3p/LINC00152 may be a potential treatment strategy for inhibiting the growth of CRC cells.
Figure 7.
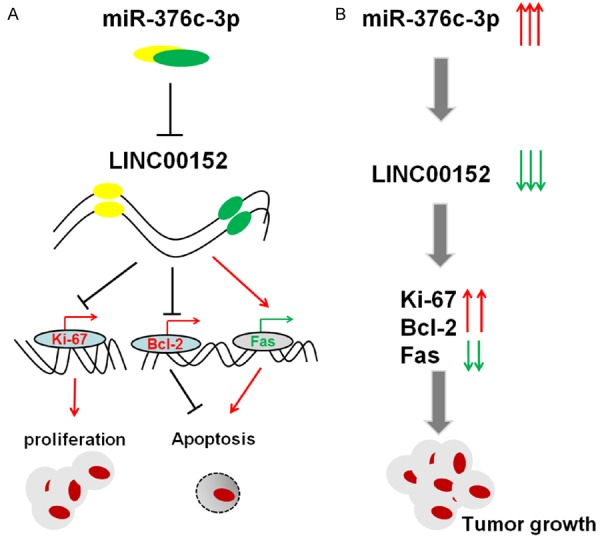
The role of LINC00152 and miR-367c-3p in CRC. A. LINC00152 down-regulated by miR-367c-3p decreases the expression of proliferation-related molecule Ki-67 and anti-apoptosis molecule Bcl-2, and increases the expression of pro-apoptosis molecule Bax in CRC cells, further suppressing viability and promoting apoptosis in CRC cells. B. The abnormally low level of LINC00152, possibly induced by the aberrant up-regulation of miR-367c-3p, will lead to an increase of CRC cell growth, and may be involved in the tumorigenesis and progression of CRC.
Acknowledgements
This study was supported by the Training Program for Young Talents of Xuhui District Health System.
Disclosure of conflict of interest
None.
References
- 1.Karsa LV, Lignini TA, Patnick J, Lambert R, Sauvaget C. The dimensions of the CRC problem. Best Pract Res Clin Gastroenterol. 2010;24:381–396. doi: 10.1016/j.bpg.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Edwards BK, Noone AM, Mariotto AB, Simard EP, Boscoe FP, Henley SJ, Jemal A, Cho H, Anderson RN, Kohler BA, Eheman CR, Ward EM. Annual report to the nation on the status of cancer, 1975-2010, featuring prevalence of comorbidity and impact on survival among persons with lung, colorectal, breast, or prostate cancer. Cancer. 2014;120:1290–1314. doi: 10.1002/cncr.28509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qiu JJ, Yan JB. Long non-coding RNA LINC01296 is a potential prognostic biomarker in patients with colorectal cancer. Tumour Biol. 2015;36:7175–7183. doi: 10.1007/s13277-015-3448-5. [DOI] [PubMed] [Google Scholar]
- 5.Witalison EE, Cui X, Causey CP, Thompson PR, Hofseth LJ. Molecular targeting of protein arginine deiminases to suppress colitis and prevent colon cancer. Oncotarget. 2015;6:36053–36062. doi: 10.18632/oncotarget.5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang G, Wu X, Li S, Xu X, Zhu H, Chen X. The long noncoding RNA CASC2 functions as a competing endogenous RNA by sponging miR-18a in colorectal cancer. Sci Rep. 2016;6:26524. doi: 10.1038/srep26524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin D, He X, Zhang E, Kong R, De W, Zhang Z. Long noncoding RNA GAS5 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Med Oncol. 2014;31:253. doi: 10.1007/s12032-014-0253-8. [DOI] [PubMed] [Google Scholar]
- 8.Han Y, Yang YN, Yuan HH, Zhang TT, Sui H, Wei XL, Liu L, Huang P, Zhang WJ, Bai YX. UCA1, a long non-coding RNA up-regulated in colorectal cancer influences cell proliferation, apoptosis and cell cycle distribution. Pathology. 2014;46:396–401. doi: 10.1097/PAT.0000000000000125. [DOI] [PubMed] [Google Scholar]
- 9.Ding J, Lu B, Wang J, Wang J, Shi Y, Lian Y, Zhu Y, Wang J, Fan Y, Wang Z, De W, Wang K. Long non-coding RNA Loc554202 induces apoptosis in colorectal cancer cells via the caspase cleavage cascades. J Exp Clin Cancer Res. 2015;34:100. doi: 10.1186/s13046-015-0217-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sana J, Faltejskova P, Svoboda M, Slaby O. Novel classes of non-coding RNAs and cancer. J Transl Med. 2012;10:103. doi: 10.1186/1479-5876-10-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butler AA, Webb WM, Lubin FD. Regulatory RNAs and control of epigenetic mechanisms: expectations for cognition and cognitive dysfunction. Epigenomics. 2016;8:135–151. doi: 10.2217/epi.15.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Y, Tan C, Weng WW, Deng Y, Zhang QY, Yang XQ, Gan HL, Wang T, Zhang PP, Xu MD, Wang YQ, Wang CF. Long non-coding RNA Linc00152 is a positive prognostic factor for and demonstrates malignant biological behavior in clear cell renal cell carcinoma. Am J Cancer Res. 2016;6:285–299. [PMC free article] [PubMed] [Google Scholar]
- 14.Chen WM, Huang MD, Sun DP, Kong R, Xu TP, Xia R, Zhang EB, Shu YQ. Long intergenic non-coding RNA 00152 promotes tumor cell cycle progression by binding to EZH2 and repressing p15 and p21 in gastric cancer. Oncotarget. 2016;7:9773–9787. doi: 10.18632/oncotarget.6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji J, Tang J, Deng L, Xie Y, Jiang R, Li G, Sun B. LINC00152 promotes proliferation in hepatocellular carcinoma by targeting EpCAM via the mTOR signaling pathway. Oncotarget. 2015;6:42813–42824. doi: 10.18632/oncotarget.5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou J, Zhi X, Wang L, Wang W, Li Z, Tang J, Wang J, Zhang Q, Xu Z. Linc00152 promotes proliferation in gastric cancer through the EGFR-dependent pathway. J Exp Clin Cancer Res. 2015;34:135. doi: 10.1186/s13046-015-0250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pang Q, Ge J, Shao Y, Sun W, Song H, Xia T, Xiao B, Guo J. Increased expression of long intergenic non-coding RNA LINC00152 in gastric cancer and its clinical significance. Tumour Biol. 2014;35:5441–5447. doi: 10.1007/s13277-014-1709-3. [DOI] [PubMed] [Google Scholar]
- 18.Ye G, Fu G, Cui S, Zhao S, Bernaudo S, Bai Y, Ding Y, Zhang Y, Yang BB, Peng C. MicroRNA 376c enhances ovarian cancer cell survival by targeting activin receptor-like kinase 7: implications for chemoresistance. J Cell Sci. 2011;124:359–368. doi: 10.1242/jcs.072223. [DOI] [PubMed] [Google Scholar]
- 19.Li Q, Shao Y, Zhang X, Zheng T, Miao M, Qin L, Wang B, Ye G, Xiao B, Guo J. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biol. 2015;36:2007–2012. doi: 10.1007/s13277-014-2807-y. [DOI] [PubMed] [Google Scholar]
- 20.Zhao J, Liu Y, Zhang W, Zhou Z, Wu J, Cui P, Zhang Y, Huang G. Long non-coding RNA Linc00152 is involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition, cell migration and invasion in gastric cancer. Cell Cycle. 2015;14:3112–3123. doi: 10.1080/15384101.2015.1078034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol. 2000;182:311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 22.Bullwinkel J, Baron-Lühr B, Lüdemann A, Wohlenberg C, Gerdes J, Scholzen T. Ki-67 protein is associated with ribosomal RNA transcription in quiescent and proliferating cells. J Cell Physiol. 2006;206:624–635. doi: 10.1002/jcp.20494. [DOI] [PubMed] [Google Scholar]
- 23.Hardwick JM, Soane L. Multiple functions of BCL-2 family proteins. Cold Spring Harb Perspect Biol. 2013;5:a008722. doi: 10.1101/cshperspect.a008722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gajate C, Mollinedo F. Lipid raft-mediated Fas/CD95 apoptotic signaling in leukemic cells and normal leukocytes and therapeutic implications. J Leukoc Biol. 2015;98:739–759. doi: 10.1189/jlb.2MR0215-055R. [DOI] [PubMed] [Google Scholar]
- 25.Thorenoor N, Faltejskova-Vychytilova P, Hombach S, Mlcochova J, Kretz M, Svoboda M, Slaby O. Long non-coding RNA ZFAS1 interacts with CDK1 and is involved in p53-dependent cell cycle control and apoptosis in colorectal cancer. Oncotarget. 2016;7:622–637. doi: 10.18632/oncotarget.5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doose G, Haake A, Bernhart SH, López C, Duggimpudi S, Wojciech F, Bergmann AK, Borkhardt A, Burkhardt B, Claviez A, Dimitrova L, Haas S, Hoell JI, Hummel M, Karsch D, Klapper W, Kleo K, Kretzmer H, Kreuz M, Küppers R, Lawerenz C, Lenze D, Loeffler M, Mantovani-Löffler L, Möller P, Ott G, Richter J, Rohde M, Rosenstiel P, Rosenwald A, Schilhabel M, Schneider M, Scholz I, Stilgenbauer S, Stunnenberg HG, Szczepanowski M, Trümper L, Weniger MA ICGC MMML-Seq Consortium. Hoffmann S, Siebert R, Iaccarino I. MINCR is a MYC-induced lncRNA able to modulate MYC’s transcriptional network in Burkitt lymphoma cells. Proc Natl Acad Sci U S A. 2015;112:E5261–5270. doi: 10.1073/pnas.1505753112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, Zhao J, Zhang W, Gan J, Hu C, Huang G, Zhang Y. lncRNA GAS5 enhances G1 cell cycle arrest via binding to YBX1 to regulate p21 expression in stomach cancer. Sci Rep. 2015;5:10159. doi: 10.1038/srep10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tani H, Onuma Y, Ito Y, Torimura M. Long non-coding RNAs as surrogate indicators for chemical stress responses in human-induced pluripotent stem cells. PLoS One. 2014;9:e106282. doi: 10.1371/journal.pone.0106282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi Y, Guo J. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep. 2014;4:6088. doi: 10.1038/srep06088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seitz H, Royo H, Bortolin ML, Lin SP, Ferguson-Smith AC, Cavaille J. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res. 2004;14:1741–1748. doi: 10.1101/gr.2743304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang W, Tian Y, Jiang S, Liu S, Zhao X, Tian D. MicroRNA-376c suppresses non-small-cell lung cancer cell growth and invasion by targeting LRH-1-mediated Wnt signaling pathway. Biochem Biophys Res Commun. 2016;473:980–986. doi: 10.1016/j.bbrc.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 32.Jin Y, Peng D, Shen Y, Xu M, Liang Y, Xiao B, Lu J. MicroRNA-376c inhibits cell proliferation and invasion in osteosarcoma by targeting to transforming growth factor-alpha. DNA Cell Biol. 2013;32:302–309. doi: 10.1089/dna.2013.1977. [DOI] [PubMed] [Google Scholar]
- 33.Baker AH, Delles C. Is microRNA-376c a biomarker or mediator of preeclampsia? Hypertension. 2013;61:767–769. doi: 10.1161/HYPERTENSIONAHA.111.00087. [DOI] [PubMed] [Google Scholar]
- 34.Fu G, Ye G, Nadeem L, Ji L, Manchanda T, Wang Y, Zhao Y, Qiao J, Wang YL, Lye S, Yang BB, Peng C. MicroRNA-376c impairs transforming growth factor-β and nodal signaling to promote trophoblast cell proliferation and invasion. Hypertension. 2013;61:864–872. doi: 10.1161/HYPERTENSIONAHA.111.203489. [DOI] [PubMed] [Google Scholar]
- 35.Dixon-McIver A, East P, Mein CA, Cazier JB, Molloy G, Chaplin T, Andrew Lister T, Young BD, Debernardi S. Distinctive patterns of microRNA expression associated with karyotype in acute myeloid leukaemia. PLoS One. 2008;3:e2141. doi: 10.1371/journal.pone.0002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z, Lu J, Sun M, Mi S, Zhang H, Luo RT, Chen P, Wang Y, Yan M, Qian Z, Neilly MB, Jin J, Zhang Y, Bohlander SK, Zhang DE, Larson RA, Le Beau MM, Thirman MJ, Golub TR, Rowley JD, Chen J. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci U S A. 2008;105:15535–15540. doi: 10.1073/pnas.0808266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agrawal R, Pandey P, Jha P, Dwivedi V, Sarkar C, Kulshreshtha R. Hypoxic signature of microRNAs in glioblastoma: insights from small RNA deep sequencing. BMC Genomics. 2014;15:686. doi: 10.1186/1471-2164-15-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hockel M, Vaupel P. Tumor hypoxia: definitions and current clinical, biologic, and molecular aspects. J Natl Cancer Inst. 2001;93:266–276. doi: 10.1093/jnci/93.4.266. [DOI] [PubMed] [Google Scholar]
- 40.Semenza GL. Regulation of cancer cell metabolism by hypoxia-inducible factor 1. Semin Cancer Biol. 2009;19:12–16. doi: 10.1016/j.semcancer.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 41.Meijer TW, Kaanders JH, Span PN, Bussink J. Targeting hypoxia, HIF-1, and tumor glucose metabolism to improve radiotherapy efficacy. Clin Cancer Res. 2012;18:5585–5594. doi: 10.1158/1078-0432.CCR-12-0858. [DOI] [PubMed] [Google Scholar]
- 42.Ulivi P, Marisi G, Passardi A. Relationship between hypoxia and response to antiangiogenic therapy in metastatic colorectal cancer. Oncotarget. 2016;7:46678–46691. doi: 10.18632/oncotarget.8712. [DOI] [PMC free article] [PubMed] [Google Scholar]


