Abstract
Plant recognition of pathogen-derived molecules influences attack and counterattack strategies that affect the outcome of host–microbe interactions. To ascertain the global framework of host gene expression during biotrophic pathogen invasion, we analyzed in parallel the mRNA abundance of 22,792 host genes throughout 36 (genotype × pathogen × time) interactions between barley (Hordeum vulgare) and Blumeria graminis f. sp hordei (Bgh), the causal agent of powdery mildew disease. A split-split-plot design was used to investigate near-isogenic barley lines with introgressed Mla6, Mla13, and Mla1 coiled-coil, nucleotide binding site, Leu-rich repeat resistance alleles challenged with Bgh isolates 5874 (AvrMla6 and AvrMla1) and K1 (AvrMla13 and AvrMla1). A linear mixed model analysis was employed to identify genes with significant differential expression (P value < 0.0001) in incompatible and compatible barley-Bgh interactions across six time points after pathogen challenge. Twenty-two host genes, of which five were of unknown function, exhibited highly similar patterns of upregulation among all incompatible and compatible interactions up to 16 h after inoculation (hai), coinciding with germination of Bgh conidiospores and formation of appressoria. By contrast, significant divergent expression was observed from 16 to 32 hai, during membrane-to-membrane contact between fungal haustoria and host epidermal cells, with notable suppression of most transcripts identified as differentially expressed in compatible interactions. These findings provide a link between the recognition of general and specific pathogen-associated molecules in gene-for-gene specified resistance and support the hypothesis that host-specific resistance evolved from the recognition and prevention of the pathogen's suppression of plant basal defense.
INTRODUCTION
Active plant defense to microbial attack is highly dependent upon recognition events involving associated gene products in the host and the pathogen. Perception of both general and specific pathogen-associated molecules result in signal transduction cascades ultimately leading to disease resistance (Nürnberger and Brunner, 2002; Tyler, 2002; Jones and Takemoto, 2004; Zipfel et al., 2004). General elicitors, which include proteins, glycoproteins, peptides, carbohydrates, and lipids, signal the presence of the pathogen and are able to trigger defense responses in a non-cultivar–specific manner (Nürnberger, 1999; Heath, 2000; Nürnberger and Brunner, 2002; Tyler, 2002; Hahlbrock et al., 2003; Montesano et al., 2003). By contrast, specific effectors, encoded by pathogen avirulence genes, trigger cultivar-specific responses resulting in hyperactivation of basal defense, which is often accompanied by hypersensitive cell death (Dangl and Jones, 2001; Nimchuk et al., 2003). This specific recognition in plant–pathogen interactions conforms to the gene-for-gene hypothesis and is determined by direct (Tang et al., 1996; Jia et al., 2000) or indirect (Kim et al., 2002; Mackey et al., 2002, 2003; Axtell and Staskawicz, 2003) interaction of host resistance (R) proteins and cognate pathogen-derived avirulence (AVR) effectors (Dangl and Jones, 2001).
The most prevalent class of plant R proteins contain highly conserved motifs, including an N-terminal coiled-coil or Toll/Interleukin-1 receptor-like domain, a nucleotide binding site, and C-terminal, Leu-rich repeats (Dangl and Jones, 2001), whereas the structures of known pathogen AVR effectors lack significant similarity (Bonas and Lahaye, 2002; Collmer et al., 2002). Many pathogen AVR proteins not only elicit defense responses but also possess virulence functions that contribute to the induction of susceptibility (Abramovitch et al., 2003; Hauck et al., 2003; Jamir et al., 2004; Jones and Takemoto, 2004). With many different virulence effectors produced during pathogenesis, molecular recognition has been the key determinant of the possible outcome of plant–microbe interactions (Jones and Takemoto, 2004). Although many studies have been conducted on plant perception of pathogen-derived molecules, the link between the recognition of general and specific elicitors in the expression of compatibility and incompatibility responses remains poorly understood.
Powdery mildew of barley (Hordeum vulgare), caused by Blumeria graminis f. sp hordei (Bgh), is an ideal system to explore the interactions of obligate fungal biotrophs with their cereal hosts. Stages of Bgh infection in barley are well characterized (Ellingboe, 1972; Kunoh, 1982; Jørgensen, 1988; Clark et al., 1993; Hall et al., 1999), and each stage is a potential recognition point with the possible release of pathogen or plant-derived signaling molecules. Induction of localized responses in underlying barley cells has been shown as early as Bgh conidiospore germination, specifically during primary germ tube formation (Kunoh, 1982; Kruger et al., 2003). Although there is an indication of early fungal recognition by the host, initial conidiospore adhesion, germination, and development of appressoria are not fundamentally different in incompatible and compatible barley–Bgh interactions (Boyd et al., 1995). Significant variation occurs, however, at the later stages of infection with the termination of fungal growth in incompatible interactions as opposed to the successful penetration and formation of haustoria leading to conidiophore development in compatible interactions (Boyd et al., 1995; Panstruga and Schulze-Lefert, 2002, 2003).
Specific recognition in barley–Bgh interactions is triggered in a gene-for-gene manner by genes designated Ml (Jørgensen, 1994; Schulze-Lefert and Vogel, 2000; Wise, 2000). Approximately 30 distinct resistance specificities have been identified at the Mla locus on chromosome 5 (1H) (Jørgensen, 1994). Mla1, Mla6, and Mla13 normally confer rapid and absolute resistance, whereas others, such as Mla7, Mla10, and Mla12, confer an intermediate response (Wise and Ellingboe, 1983; Jørgensen, 1994; Wei et al., 1999; Shen et al., 2003; Halterman and Wise, 2004). Cloned Mla alleles belong to the coiled-coil, nucleotide binding site, Leu-rich repeat class of genes implicated in specific recognition between the host and pathogen (reviewed in Jones, 2001). A unique feature of this host–pathogen interaction is that 92 to 97% similar MLA proteins may or may not require the RAR1/SGT1/HSP90 Skp1-Cullin-F-box ubiquitin ligase complex to activate downstream components (Azevedo et al., 2002; Shen et al., 2003; Shirasu and Schulze-Lefert, 2003; Halterman and Wise, 2004).
The well-defined stages of powdery mildew disease development provide multiple possibilities to interrogate the regulation of host genes in response to Mla-specified incompatible and compatible barley–Bgh interactions (Ellingboe, 1972; Kunoh, 1982; Jørgensen, 1988; Clark et al., 1993). Information on transcript abundance can be used to describe a cellular state and predict functional involvement of genes in the interactions among plants and pathogens (Maleck et al., 2000; Schenk et al., 2000; Mysore et al., 2002; Wan et al., 2002; Puthoff et al., 2003; Tao et al., 2003; van Wees et al., 2003; Whitham et al., 2003; Eulgem et al., 2004). In this report, we analyzed the molecular mechanisms of gene-specific plant-biotrophic fungus interactions. The newly developed Barley1 GeneChip probe array (Close et al., 2004) was used in conjunction with a mixed linear model analysis to evaluate in parallel 22,792 barley genes over the course of powdery mildew infection. Twenty-two of the 22,792 host genes (P value < 0.0001) exhibited nearly identical expression patterns among all incompatible and compatible interactions up to 16 h after inoculation (hai), coinciding with Bgh conidiospore germination and appressorial germ tube growth. By contrast, divergent expression was observed from 16 to 32 hai during attempted penetration of host epidermal cells and Bgh haustorial formation, with notable suppression of most transcripts identified as differentially expressed in compatible interactions. Based on these results, we propose a model that links the recognition of general elicitors and specific avirulence proteins in the expression of plant defense responses, supporting the hypothesis that host-specific resistance evolved from the recognition and prevention of the pathogen's suppression of plant basal defense.
RESULTS
Analysis Strategy
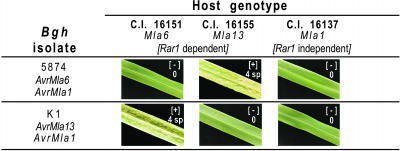
The abundance of host–pathogen interactions triggered by different alleles of Mla provide the means to address specific questions by choosing the appropriate host genotype combined with a suitable isolate of Bgh. Additionally, conditions have been established to achieve a high percentage of Bgh-infected epidermal cells with synchronous development of the pathogen (Ellingboe, 1972; Panstruga, 2004). We were therefore interested in making various expression profile comparisons over time to assess the robustness of our system and also to provide independent biological validation of related phenotypic outcomes. Our first goal was to focus on genes (other than R genes) whose expression might be used to distinguish incompatibility from compatibility. This was done in two different ways: (1) by comparing two near-isogenic host lines with contrasting Mla alleles challenged with one Bgh isolate, with or without the matching AvrMla gene, and (2) by comparing the effects of two alternate Bgh isolates, again with or without the corresponding AvrMla gene, inoculated onto one host harboring a single Mla allele. The second objective was to investigate genes that may distinguish Rar1-dependent versus Rar1-independent incompatible responses specified by different Mla alleles. Therefore, as shown in Figure 1, our experimental design contained three near-isogenic barley lines, with introgressed Mla6, Mla13, and Mla1 alleles, to be challenged with either Bgh isolate 5874 (AvrMla6 and AvrMla1) or K1 (AvrMla13 and AvrMla1). This design, similar to a classic quadratic check (Flor, 1971), resulted in two independent host-isolate combinations for each of the above questions we wished to address. The experiment was conducted in three independent biological replications using a standard split-split-plot design (Kuehl, 2000) with replications as blocks, Bgh isolate as the whole-plot factor, plant genotype as the split-plot factor, and time as the split-split-plot factor (see Methods). First leaves of inoculated barley seedlings were harvested at 0, 8, 16, 20, 24, and 32 hai. One Barley1 GeneChip (Close et al., 2004) was used for each of the 108 split-split-plot experimental units corresponding to 3 replications × 3 genotypes × 2 isolates × 6 time points.
Figure 1.
Infection Types of Six Barley Genotype-Isolate Combinations.
Phenotypes of the different barley powdery mildew interactions at 7 d after inoculation. An infection type of 0 is considered resistant ([−] designates incompatibility/no sporulation), whereas infection type of 4 sp is considered completely susceptible ([+] designates compatibility/abundant sporulation).
Because not all changes in gene expression are expected to be a direct consequence of pathogen infection, the statistical analysis used in this study was focused on the overall pattern of expression based on the kinetics of infection, instead of changes at a single time point. Our primary analysis strategy involved the identification of genes whose average pattern of expression in one host–pathogen interaction category (e.g., compatibility) differed significantly from its average pattern of expression in its contrasting category (e.g., incompatibility). To identify differences in patterns of expression over time, six time-specific differences between host–pathogen interaction categories were tested for equality. A lack of equality among these six differences indicated different patterns of expression (i.e., nonparallel time-course expression profiles) across categories. We identified many genes with differing patterns of expression between contrasting categories (compatibility versus incompatibility and Rar1-independent versus Rar1-dependent incompatibility) as detailed below. With the intent of focusing on the most biologically relevant of these genes, we further restricted our attention to genes whose within-category expression patterns were consistent among all individual Mla/AvrMla or Mla/avrMla combinations. Thus, although some other genes may be biologically relevant, we believe that genes with the most potential for differentiating host–pathogen interaction categories are those genes exhibiting significantly different expression patterns across categories and similar expression patterns across multiple host-isolate combinations within categories. The stringency of our criteria may eliminate some interesting genes, but this was counter balanced by the possibility of determining key pathways underlying the molecular basis of barley-obligate fungal pathogen interactions.
Significant Divergent Expression Occurs after Haustorial-Plasma Membrane Contact in Compatible and Incompatible Interactions
To narrow our focus to the genes having the highest potential for being involved in response pathways that distinguish compatibility from incompatibility, we conducted F-tests that compared the average time-course expression profile of each gene in incompatible interactions [Mla6/AvrMla6 (5874) and Mla13/AvrMla13 (K1)] to its average time-course expression profile in compatible interactions [Mla6/avrMla6 (K1) and Mla13/avrMla13 (5874)] as part of a mixed linear model analysis of the 22,792 barley probe sets. Note that this comparison focuses on the four leftmost cells in Figure 1. Thus, both the incompatible and compatible mRNA expression profiles are averages of Mla6 and Mla13 plant materials inoculated with Bgh isolates 5874 and K1. This quadratic check structure allows the comparison between compatible and incompatible interactions to potentially be free of both genotype and isolate main effects as an explanation for the pattern differences observed in this experiment.
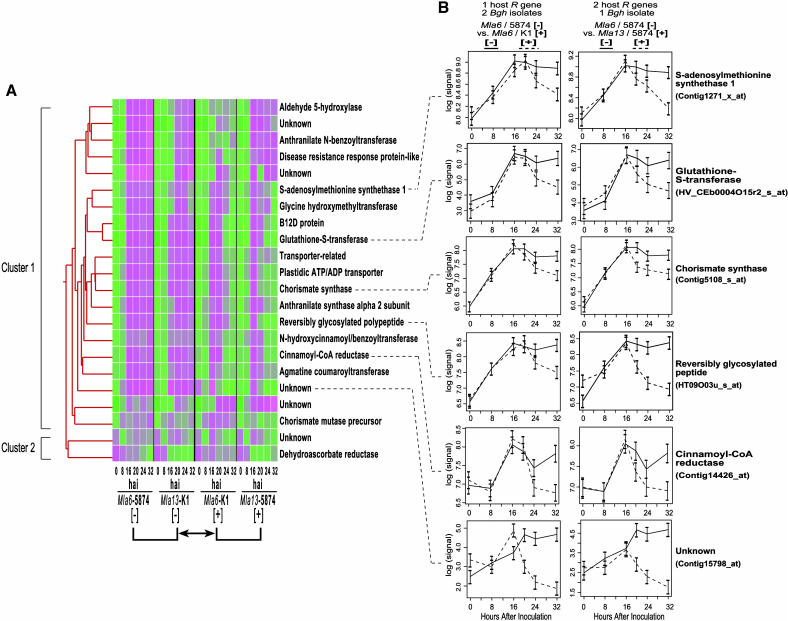
A total of 28 probe sets had P values < 0.0001 for the comparison of expression patterns across incompatible and compatible interactions. Using the method of Storey and Tibshirani (2003), this set of 28 genes was associated with a false discovery rate of under 7%. Transcript profiles of 22 of these 28 genes exhibited consistent patterns of expression within host–pathogen interaction categories and, thus, were selected for further analysis. The pattern of mRNA accumulation was highly similar in both incompatible and compatible interactions up to 16 hai and divergent thereafter (Table 1, Figure 2; see Supplemental Tables 1 and 2 online). Such a pattern is particularly interesting given that haustoria of avirulent Bgh make contact with the host cell plasma membrane at ∼16 h. These 22 genes exhibit the same basic divergence between incompatible and compatible expression patterns for all pairwise comparisons of contrasting barley–Bgh interactions in the four leftmost cells of Figure 1 (see Figure 2B for examples pertinent to Mla6-specified interactions). Thus, this further strengthens the argument that the expression of these genes is a feature that can be used to distinguish susceptible and resistant responses. Highly similar expression patterns were observed in genes of predicted function as well as the unknowns (Figure 2). Seven of the predicted proteins could be modeled onto the last step of shikimate pathway leading to the synthesis of phenylpropanoid phytoalexins and lignins (Figure 3). Other genes had predicted functions in ethylene biosynthesis, cellular metabolism, and oxidative stress. Three showed no significant sequence similarities in the public databases, whereas two shared significant similarities to genes with unknown function in the rice genome (Table 1).
Table 1.
Predicted Functions of 22 Genes Identified as Differentially Expressed and Their Corresponding P Values in the Comparison of the Mla-Specified Compatible and Incompatible Interactions with Bgh
| Affymetrix Probe Set IDa | Barley1 GeneChip Exemplar | Predicted Functionb | Predicted Functional Classification | Organism | E value | F-testc |
|---|---|---|---|---|---|---|
| Contig11969_at | 11969 | Aldehyde 5-hydroxylase | Cellular metabolism | Liquidambar styraciflua | 1e-110 | 1.1e-05 |
| Contig12219_at | 12219 | Unknown | Unknown | Oryza sativa | 2e-10 | 2.8e-06 |
| Contig15413_at | 15413 | Anthranilate N-benzoyltransferase | Phytoalexin synthesis | O. sativa | 2e-27 | 1.4e-05 |
| Contig15515_at | 15515 | Disease resistance response protein-like | Defense | O. sativa | 1e-25 | 2.7e-05 |
| Contig15861_at | 15861 | Unknown | Unknown | –d | – | 1.1e-06 |
| Contig1271_x_at | 01271 | S-adenosylmethionine synthetase 1 | Ethylene synthesis | H. vulgare | 1e-152 | 5.7e-05 |
| Contig2168_s_at | 02168 | Gly hydroxymethyltransferase | Cellular metabolism | A. thaliana | 0.0 | 6.1e-06 |
| Contig8605_s_at | 08605 | B12D protein | Unknown | O. sativa | 2e-35 | 1.3e-05 |
| HV_CEb0004O15r2_s_at | 39931 | Glutathione S-transferase | Oxidative stress | O. sativa | 1e-07 | 8.1e-06 |
| Contig24175_at | 24175 | Transporter-related | Cellular metabolism | A. thaliana | 3e-31 | 8.5e-05 |
| Contig4728_at | 04728 | Plastidic ATP/ADP-transporter | Cellular metabolism | O. sativa | 0.0 | 7.4e-06 |
| Contig5108_s_at | 05108 | Chorismate synthase | Shikimate pathway | A. thaliana | 2e-49 | 3.8e-05 |
| HY07P02u_at | 48443 | Anthranilate synthase α 2 subunit | Shikimate pathway | O. sativa | 2e-67 | 2.9e-05 |
| HT09O03u_s_at | 37781 | Reversibly glycosylated polypeptide | Cellular metabolism | Triticum aestivum | 0.0 | 2.6e-05 |
| Contig7815_s_at | 07815 | N-hydroxycinnamoyl/benzoyl transferase | Phytoalexin synthesis | O. sativa | 4e-69 | 3.3e-05 |
| Contig14426_at | 14426 | Cinnamoyl-CoA reductase | Lignin synthesis | Populus balsamifera | 8e-58 | 6.4e-06 |
| Contig20247_at | 20247 | Agmatine coumaroyltransferase | Phytoalexin synthesis | H. vulgare | 6e-36 | 5.4e-06 |
| Contig15798_at | 15798 | Unknown | Unknown | – | – | 3.0e-09 |
| Contig20747_at | 20747 | Unknown | Unknown | – | – | 2.5e-05 |
| Contig7705_at | 07705 | Chorismate mutase precursor | Shikimate pathway | O. sativa | 1e-125 | 2.4e-05 |
| Contig20954_at | 20954 | Unknown | Unknown | O. sativa | 1e-54 | 5.3e-05 |
| HI15L07r_s_at | 34930 | Dehydroascorbate reductase | Oxidative stress | T. aestivum | 1e-116 | 7.8e-05 |
Order of probe sets is identical to clustering in Figure 2.
BarleyBase (http://barleybase.org) annotations were based on the consensus of multiple searches. NCBI/TGIR/ATH1 searches were performed using HarvEST:Barley assembly 25 and best BLASTX nonredundant was performed using HarvEST:Barley assembly 31.
P values for the test of equality of the differences between compatible and incompatible interactions at six time points.
No organism designated for genes with nonsignificant E value.
Figure 2.
Expression Profiles of 22 Predicted Genes Differentially Expressed in Incompatible and Compatible Mla-Specified Interactions with Bgh.
(A) Scaled mean signal intensities in Mla6- and Mla13-specified incompatible and compatible interactions were used to determine the similarities of expression profiles through cluster analysis. A data matrix was constructed with genes in rows and time points of all genotype-isolate combinations in columns. A Pearson correlation was used to measure similarities of transcript accumulation in a pairwise manner. Hierarchical clustering was performed with GeneSpring 5.1 software. Cells lower than the reference (median) are designated in green, and cells higher than the reference (median) are shown in magenta.
(B) Reciprocal expression patterns of six differentially expressed genes in different incompatible and compatible combinations. The natural logarithm of signal intensities in C.I. 16151 (Mla6)/5874 [−] , C.I. 16151 (Mla6)/K1 [+], and C.I. 16155 (Mla13)/5874 [+] interactions were plotted in graphs. Standard errors were calculated based on three independent replications. Note scale differences in the graphs. Comparisons of one host R gene versus two Bgh isolates and two host R genes versus one Bgh isolate are shown.
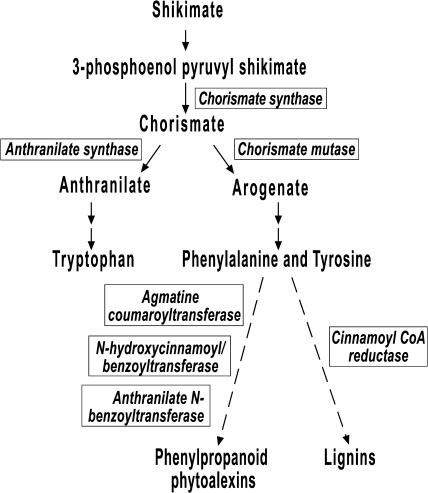
Figure 3.
Differentially Expressed Genes Modeled onto the Biosynthetic Pathway Leading to the Production of Phytoalexins and Lignins.
Genes identified differentially expressed in the comparison of compatible and incompatible interactions are designated in boxes. Seven highly significant genes are putatively involved in the last steps of shikimate pathway leading to the synthesis of defense-related compounds.
Expression Pattern Differences in Mla-Specified Rar1-Dependent and -Independent Incompatible Interactions
The Mla6, Mla13, and Mla1 alleles differ in their requirements for Rar1 to effect race-specific resistance (Halterman et al., 2001, 2003; Zhou et al., 2001; Shen et al., 2003; Halterman and Wise, 2004). We used the same basic strategy as described above to determine variations in transcript abundance after pathogen inoculation in host genotypes that differ in Rar1 dependency. The mean mRNA expression of each gene in the two Rar1-dependent incompatible interactions [Mla6/AvrMla6 (5874) + Mla13/AvrMla13 (K1)] was compared with the mean mRNA expression in the two Rar1-independent incompatible interactions [Mla1/AvrMla1 (5874) + Mla1/AvrMla1 (K1)] over the first 32 h of powdery mildew infection. Just as in the case of incompatible versus compatible interactions described above, this comparison is potentially free of Bgh isolate main effects. However, linkage drag and other possible genomic introgression in the unlinked regions could be partly responsible for the observed pattern differences because the Rar1-dependent versus Rar1-independent comparison is confounded with variations between the near-isogenic lines with introgressed Mla1, Mla6, and Mla13 alleles.
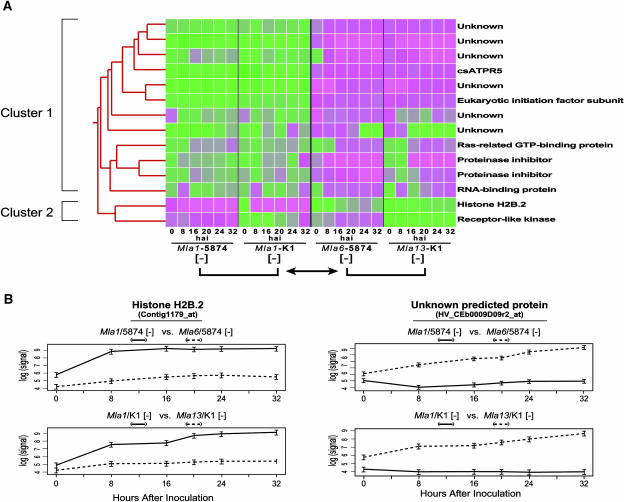
A total of 41 probe sets exhibited pattern differences significant at the 0.0001 level. Using the method of Storey and Tibshirani (2003), this set of 41 genes was associated with a false discovery rate of under 5%. Of these 41 genes, only 14 genes had consistent patterns of expression in two host-isolate combinations within Rar1-dependent and -independent categories (Figure 4; see Supplemental Tables 3 and 4 online). As shown in Table 2, most genes are predicted to function in signal transduction, regulation of gene expression, and plant defense. Six genes were of unknown function; two of these six shared significant similarities to annotated sequences in the rice genome. Genes encoding predicted histone H2B.2 and a receptor-like kinase were found to be highly upregulated in Mla-specified Rar1-independent incompatible interactions with Bgh. By contrast, transcripts of predicted genes that encode proteinase inhibitors, precursor of PR5 (csAtPR5), eukaryotic initiation factor subunit, RNA binding protein, Ras-related GTP binding protein, and six predicted proteins with unknown function were found to be highly abundant in Mla-specified Rar1-dependent interactions with Bgh.
Figure 4.
Expression Profiles of 14 Predicted Genes Differentially Expressed in the Comparison of Mla-Specified Rar1-Dependent (Mla6 and Mla13) and -Independent (Mla1) Incompatible Interactions.
(A) Average signal intensities for each time point in the Mla1-, Mla6-, and Mla13-specified interactions were used in the cluster analysis. A data matrix of signal intensities in genotype-isolate combinations involved in the comparisons was loaded in GeneSpring 5.1 software. Similarities of expression profiles were calculated using a Pearson correlation and presented in a dendrogram using the hierarchal clustering algorithm. Cells lower than the reference (median) are designated in green, and cells higher than the reference (median) are shown in magenta.
(B) Reciprocal expression profiles of selected genes in the different combinations of Rar1-dependent and -independent comparisons. Estimated mean natural log signal intensities are plotted for each isolate and time point along with their standard errors.
Table 2.
Predicted Functions of 14 Genes Identified as Differentially Expressed and Their Corresponding P Values in the Comparison of Mla-Specified Rar1-Dependent and -Independent Incompatible Interactions with Bgh
| Affymetrix Probe Set IDa | Barley1 GeneChip Exemplar | Predicted Functionb | Predicted Functional Classification | Organism | E value | F-testc |
|---|---|---|---|---|---|---|
| Contig24376_at | 24376 | Unknown | Unknown | O. sativa | 5e-53 | 7.7e-09 |
| HV_CEb0009D09r2_at | 39999 | Unknown | Unknown | –d | – | 0.0e+00 |
| HVSMEm0003C21r2_at | 45388 | Unknown | Unknown | – | – | 2.8e-07 |
| Contig16303_at | 16303 | csAtPR5 | Defense | Aegilops tauschii | 4e-24 | 5.9e-06 |
| Contig13292_at | 13292 | Unknown | Unknown | – | – | 1.7e-06 |
| Contig848_at | 00848 | Eukaryotic initiation factor subunit | Gene expression | O. sativa | 9e-48 | 2.0e-07 |
| Contig12044_at | 12044 | Unknown | Unknown | O. sativa | 9e-23 | 5.4e-05 |
| HVSMEa0004N12r2 | 40404 | Unknown | Unknown | – | – | 2.7e-05 |
| Contig3166_at | 03166 | Ras-related GTP binding protein | Signal transduction | O. sativa | 1e-110 | 1.9e-05 |
| Contig34_s_at | 00034 | Proteinase inhibitor | Defense | H. vulgare | 5e-33 | 0.0e+00 |
| EBro08_SQ005_A14_at | 31141 | Proteinase inhibitor | Defense | H. vulgare | 5e-33 | 4.7e-07 |
| Contig7649_at | 07649 | RNA binding protein | Gene expression | A. thaliana | 6e-60 | 6.0e-06 |
| Contig1179_at | 01179 | Histone H2B.2 | Gene expression | T. aestivum | 5e-42 | 2.9e-08 |
| Contig13692_x_at | 13692 | Receptor-like kinase ARK1AS | Signal transduction | H. vulgare | 0.0 | 9.9e-07 |
Order of probe sets is identical to clustering in Figure 4.
BarleyBase (http://barleybase.org) annotations were based on the consensus of multiple searches. NCBI/TGIR/ATH1 searches were performed using HarvEST:Barley assembly 25, and best BLASTX nonredundant was performed using HarvEST:Barley assembly 31.
P values for the test of equality of differences between Rar1-dependent and -independent interactions at six time points.
No organism designated for genes with nonsignificant E value.
Hierarchical Clustering of Temporal Responses in Mla-Specified Compatible versus Incompatible and Rar1-Dependent versus -Independent Interactions
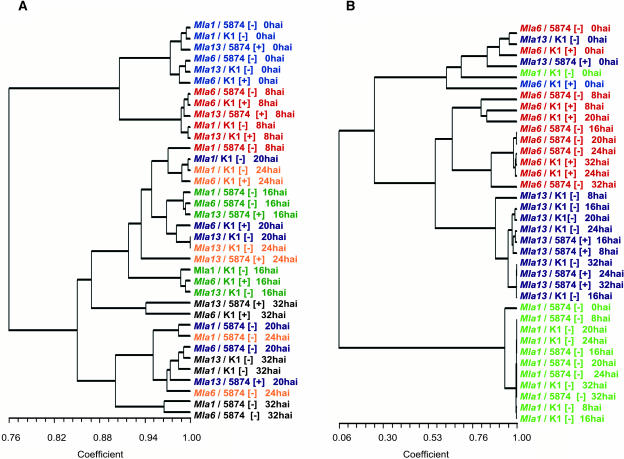
To determine the overall pattern of expression of the identified genes, dendrograms were constructed using the steady state transcript levels per time point of the 22 and 14 differentially expressed genes in Mla-specified compatible versus incompatible and Rar1-dependent versus -independent interactions, respectively (Figure 5). A Pearson correlation was used as the similarity measure for hierarchical clustering because it analyzes the pattern and not the magnitude of the expression. We also included the Mla1-specified mRNA profiles in clustering the 22 differentially expressed genes to determine the correlation of all resistant responses during powdery mildew infection.
Figure 5.
Temporal Expression Similarities of the Different Barley Powdery Mildew Combinations.
(A) Clustering of time point–specific responses using 22 differentially expressed genes in the comparison of compatible and incompatible interactions. The overall expression is color coded per time point: 0 hai, blue; 8 hai, red; 16 hai, green; 20 hai, purple; 24 hai, orange; 32 hai, black.
(B) Clustering of time point–specific responses based on 14 differentially expressed genes in the comparison of Rar1-dependent and -independent interactions. The overall expression is color coded by genotype: Mla6, red; Mla13, purple; Mla1, green. Data matrices of the expression profiles were constructed with genes in rows and time point-genotype-isolate combinations in columns. Similarities of the overall expression pattern per time point in the different genotype-isolate combinations were calculated in a pairwise manner using Pearson correlation. Cluster analysis was performed using the unweighted pair group mean arithmetic algorithm in NTSYSpc 2.1 software.
As shown in Figure 5A, mRNA levels of the 22 differentially expressed genes clustered together at each time point between 0 and 16 hai, regardless if the interaction was compatible or incompatible. This grouping suggests that the early expression changes in the selected genes are interaction independent (barley-Bgh nonspecific) and possibly modulated by the perception of general elicitors. At 16 hai, however, the clustering of compatible and incompatible expression was generally unique to Bgh isolates 5874 (AvrMla6 and AvrMla1) or K1 (AvrMla13 and AvrMla1), which may suggest that the pathogen perturbs the level of host early-induced mRNA expression in a race-specific manner. After 16 hai, the differences observed in the pattern of expression among compatible and incompatible interactions are likely attributable to the decreasing transcript levels of most identified genes in plants exhibiting susceptibility to powdery mildew infection shown in Figure 2. At 32 hai, compatible interactions clustered together regardless of genotype-isolate combinations and were separated from incompatible interactions. This separation of compatible from incompatible responses may reflect the dual role of avirulence effectors in barley–Bgh interactions: one in the suppression of defense in compatible interactions and the other in the enhancement of defense in incompatible interactions.
As illustrated in Figure 5B, time point–specific responses of the 14 genes after pathogen inoculation were grouped based on Rar1 dependency with Mla1/AvrMla1-specified mRNA expression patterns distinct from those specified for Mla6/AvrMla6 and Mla13/AvrMla13. The overall clustering of the selected genes separated expression at 0 hai from that of 8 to 32 hai, suggesting that the differential gene expression occurred at the early stages after pathogen inoculation. In addition, gene expression profiles after 0 hai within the Rar1-dependent group were found to be clustered based on genotypes, separating Mla6/AvrMla6 from Mla13/AvrMla13 interactions.
DISCUSSION
Modulation of Mla-Mediated Response Networks Is Dependent on the Kinetics of Biotrophic Infection
Obligate biotrophic fungi have evolved subtle mechanisms of invasion while producing minimal damage to host cells. Development of infective structures occurs extracellularly and is crucial to the establishment of pathogenesis (Mendgen and Hahn, 2002; Schulze-Lefert and Panstruga, 2003). For several plant pathogenic fungi, including Bgh, early stages of infection are not fundamentally different among incompatible and compatible interactions with the host (Boyd et al., 1995; Hardham, 2001; Tucker and Talbot, 2001; Mendgen and Hahn, 2002). However, differences become apparent during the more advanced stages of infection, resulting in termination of fungal growth in incompatible interactions as opposed to maturation of haustoria, establishment of secondary hyphae, and subsequent conidiophore development in compatible interactions (Wise and Ellingboe, 1983; Boyd et al., 1995; Panstruga and Schulze-Lefert, 2002, 2003).
In the analysis presented in this report, early recognition of Bgh resulted in nearly identical transcript accumulation of the identified genes up to 16 hai in all incompatible and compatible interactions (Figures 2, 5, and 6; see Supplemental Tables 1 and 2 online). This finding is consistent with the observations that fungal attachment and germination are accompanied by the release of proteins, carbohydrates, lipids, glycoproteins, and peptides from the spores (Tucker and Talbot, 2001), and many of these molecules can trigger general host defense responses (Kiba et al., 1999). In addition, primary germ tubes are also capable of breaching host epidermal walls leading to the initiation of cytoplasmic aggregates in underlying host cells (Kunoh, 1982). Not surprisingly, these general responses to the initial phases of fungal infection have been implicated in non-host resistance as well (Heath, 2000; Jones and Takemoto, 2004). Thus, it is likely that plants take advantage of this early detection of biotic elicitors to rapidly initiate responses that could lead to pathogen rejection. For example, perception of flagellin, a general elicitor in Arabidopsis thaliana, induces expression of numerous defense-related genes and contributes to bacterial disease resistance (Zipfel et al., 2004). Therefore, the initial induction of multicomponent defense responses most likely occurs at the early stages of infection and is possibly the result of synergistic effects of recognition of multiple pathogen-derived molecules.
Figure 6.
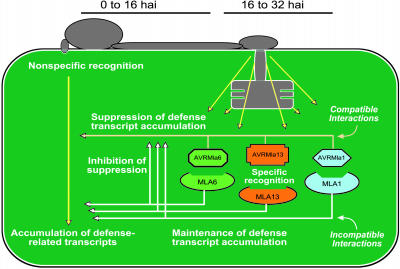
A Model for the Interplay of Recognition of General and Specific Elicitors in the Induction of Barley Gene-for-Gene Defense Responses.
In this model, detection of general elicitors by the host triggers nonspecific defense-related transcript accumulation at the early stages of fungal infection. During later stages of fungal development, the switch from the leaf surface to invasive growth leads to specific recognition at the fungal haustorial–host plasma membrane interface as a result of the release of pathogen-derived molecules. Lack of specific recognition of pathogen-derived molecules results in the suppression of defense-related transcript accumulation and subsequent disease development, whereas direct or indirect recognition of avirulence effectors by host R proteins sustains the level of nonspecific defense responses and triggers the accumulation of another layer of defense-related transcripts and subsequent disease resistance.
Coordinated reduction in mRNA abundance of most differentially expressed genes in compatible interactions implies active suppression of nonspecific defense responses (Figures 2 and 6). This suggests a mechanism where the lack of specific recognition of cognate pathogen AVR proteins allows the expression of pathogen-derived suppressor molecules. This may also explain the phenomenon of induced susceptibility in barley–Bgh interactions, wherein susceptibility is induced to an avirulent isolate by prior inoculation of a virulent isolate (Kunoh, 2002; Schulze-Lefert and Panstruga, 2003). Thus, the ability of the pathogen to effectively and coordinately inhibit host nonspecific defense is likely an essential component of successful pathogenesis. However, many suppressors not only block host defense responses but can also act, in contrary fashion, as an elicitor (Shiraishi et al., 1994). Supprescins produced by Mycosphaerella pinodes, the causal agent of Mycosphaerella blight on peas (Pisum sativum), inhibit host defense responses and alternatively function as avirulence factors when applied to plants other than pea (Yoshioka et al., 1990; Toyoda et al., 1993; Shiraishi et al., 1997; Toyoda et al., 2002). Indeed, elicitors of plant disease resistance are often virulence factors from the pathogen (Collmer, 1998; Kjemtrup et al., 2000; Tsiamis et al., 2000; White et al., 2000; Bonas and Lahaye, 2002; Nimchuk et al., 2003), many of which were recently shown to inhibit programmed cell death and cell wall–based extracellular defense (Abramovitch et al., 2003; Hauck et al., 2003; Jamir et al., 2004). Taken together, these examples illustrate the dual functions of many pathogen-derived molecules in plant–microbe interactions, suggesting dynamic evolution in the attack and counterattack strategies of the two organisms.
By contrast, expression of the identified genes generally increased or remained steady from 16 to 32 hai in incompatible interactions (Figure 2). This observation suggests that one of the outcomes of specific recognition of a cognate avirulence effector by the host is the maintenance of increased levels of defense-related transcripts. This is consistent with the phenomenon of induced resistance, wherein preinoculation of plants with a nonpathogen or an avirulent isolate often elicits resistance at the infection site to a virulent isolate (Kunoh, 2002). Because differences in incompatible and compatible responses occurred only after 16 hai, this would support the hypothesis that delivery and recognition of Bgh avirulence effectors most likely occurs during membrane-to-membrane contact after penetration and during early haustorial development (Halterman et al., 2003). Thus, to sustain defense transcript accumulation, the host appears to have evolved a mechanism to counteract the pathogen's suppression of basal defense or their cellular effects in a cultivar-specific manner (Figure 6; Ellis and Dodds, 2003; Parker, 2003).
The interaction between Bgh and attacked host cells is largely a cell-autonomous event (Shirasu et al., 1999; Panstruga, 2004); thus, the overall interpretation outlined in Figure 6 illustrates the immediate changes in gene expression that occur in the challenged epidermal cells. Our observed interaction-independent transcript accumulation is consistent with the fact that only infected cells are in direct contact with the spores and therefore are capable of perception of the pathogen-associated molecules. In addition, based on the evidence obtained from gene expression analysis using RNA isolated from epidermal cells, mesophyll cells, and whole leaf tissue (Gregersen et al., 1997), suppression of defense-related transcripts has been suggested to occur only in physical proximity to haustoria (Schulze-Lefert and Panstruga, 2003). Hence, although there are reports of the possible systemic spread of defense from infected cells in barley–Bgh interactions (Gregersen et al., 1997; Kunoh, 2002), the relationship between localized perception of signals from the pathogen and the subsequent systemic perception of signals from the attacked host cells remains unclear.
Interplay of Nonspecific and Specific Transcriptional Responses in Gene-for-Gene Resistance
Many race-specific, race-nonspecific, and non-host resistance responses share similar downstream components (Dangl and Jones, 2001; Nürnberger and Brunner, 2002; Tyler, 2002; Hahlbrock et al., 2003; Jones and Takemoto, 2004). The defense signaling proteins SGT1, NDR1, PAD4, and EDS1 are required in both host-specific and broad-spectrum disease resistance (Century et al., 1997; Xiao et al., 2001; Fellbrich et al., 2002; Peart et al., 2002a, 2002b; Peng et al., 2003; Yun et al., 2003). In addition, mitogen-activated protein kinase cascades are triggered not only by R–AVR interactions but also by general pathogen-derived molecules, such as flagellin, fungal cell wall fragments, elicitin (conserved 98–amino acid steorol binding protein from Phytophthora and Phythium species), Pep13 (a peptide fragment within cell wall glycoprotein from Phytophthora sojae), and necrosis-inducing Phytophthora protein 1 (reviewed in Zhang and Klessig, 2001; Brunner et al., 2002; Fellbrich et al., 2002; Jones and Takemoto, 2004; Menke et al., 2004).
In this study, early induction of nonspecific defense-related transcripts appears to be linked to Mla-specified gene-for-gene resistance. Indeed, seven genes that were coordinately upregulated are predicted to function in the last step of the shikimate pathway leading to the biosynthesis of phenylpropanoid phytoalexins and lignin, important chemical and structural defenses (Figure 3; Oelofse and Dubery, 1996; Tyler, 2002; Hahlbrock et al., 2003). These genes are downregulated 16 hai in compatible interactions, but accumulation is sustained in incompatible interactions. In the shikimate pathway, chorismate synthase catalyzes the conversion of 5-enol-pyruvylshikimate-3-phosphate to chorismate (Ahn et al., 2003). The resulting chorismate can then be catalyzed by chorismate mutase, leading to the production of Phe and Tyr (Guo et al., 2001). Alternatively, chorismate can also be converted to anthranilate by anthranilate synthase, leading to the production of Trp (Bohlmann et al., 1996). In the first step in the monolignol biosynthetic branch of the phenylpropanoid pathway, cinnamoyl-CoA reductase catalyzes the conversion of cinnamoyl-CoA into corresponding cinnamaldehydes for lignin biosynthesis (Lauvergeat et al., 2001). Regulation of the cinnamoyl CoA reductase encoding gene affects the levels of lignin in Arabidopsis (Lauvergeat et al., 2001; Goujun et al., 2003). In addition, N-hydroxycinnamoyl/benzoyl transferase and anthranilate N-benzoyl transferase convert N-benzoylanthranilate from anthranilate, a precursor of several sets of dianthramides in carnation (Dianthus caryophyllus) treated with fungal elicitor (Yang et al., 1997). Another antifungal protein, hordatine, is derived from barley hydroxycinnamoylagmatines, which are produced by the catalysis of agmatine and hydrocinnamoyl-CoA thiolesters with agmatine coumaroyl transferase (Burhenne et al., 2003). Preferential accumulation of p-coumaroyl-hydroxyagmatine after powdery mildew infection has been shown in barley plants undergoing both non-host and host-specific resistance (von Ropenack et al., 1998; Stein and Somerville, 2002).
Coordinated suppression of these nonspecific defense-related transcripts suggests that the potential targets of virulence functions are regulators of general or basal defense. It is therefore possible that host-specific resistance may have evolved from the recognition of the virulence effects on another host protein regulating early non-specific defense. This is in agreement with the guard hypothesis wherein the R protein is guarding another host protein that is the target of an avirulence effector (Van der Biezen and Jones, 1998; Dangl and Jones, 2001). This is also consistent with indirect pathogen recognition through disappearance of RIN4 (RPM1 interacting protein), a regulator of basal defense and a target of several unrelated avirulence effectors (Mackey et al., 2002, 2003; Axtell and Staskawicz, 2003). Perturbation of RIN4 by AvrRpm1/AvrB and AvrRpt2 modulates RPM1- and RPS2-mediated resistance, respectively. Taken together, the above examples support the idea that recognition of general and specific pathogen-derived molecules are linked in gene-for-gene disease resistance.
mRNA Expression in Mla-Specified Rar1-Dependent and -Independent Barley–Bgh Interactions
An additional layer of defense regulation was revealed by the comparison of Rar1-dependent and -independent incompatible interactions. Separation of mRNA expression patterns of the selected genes was dependent on whether or not the Mla allele required Rar1 to effect resistance (Figure 5). Some of the upregulated genes in Rar1-dependent interactions were found to have predicted functions in defense (Table 2, Figure 4; see Supplemental Tables 3 and 4 online). Proteinase inhibitors have been shown to have an antifungal activity and also to inhibit the growth of Botrytis cinerae, Fusarium solani f. sp pisi, and Alternaria brassicicola (Lorito et al., 1994; Joshi et al., 1999; Heath, 2000). Another gene associated with Rar1-dependent barley–Bgh interaction encodes a predicted Ras-related GTP binding protein, and overexpression of this class of gene in tobacco (Nicotiana tabacum) produced an abnormally high level of salicylic acid with associated increase in acidic pathogenesis-related proteins conferring resistance to tobacco mosaic virus infection (Sano et al., 1994). By contrast, genes associated with Rar1-independent interactions revealed sequence similarity to predicted receptor-like kinase and histone H2B-2. Ubiquitination of receptor-like kinases and histones (Cock et al., 2002; Jason et al., 2002) is very common in eukaryotic cells, but its association with the protein degradation process probably does not require RAR1 in Mla1–AvrMla1 interactions. Recently, phosphorylation of histone H2-B was correlated with cells undergoing programmed cell death in vertebrates (Cheung et al., 2003). Thus, functional analyses of these 14 differentially expressed genes are necessary to determine their possible involvement in Rar1-dependent and -independent plant–pathogen interactions.
In summary, we predict the possible interplay of plant recognition of general and specific pathogen-derived molecules in the expression and evolution of host-specific resistance. A rapid increase in mRNA accumulation of the identified genes was triggered early after pathogen challenge, with the significant suppression of these transcripts in plants undergoing susceptible reactions. Further study of the mechanism of suppression of the differentially expressed genes should provide new insights in the understanding of compatibility and incompatibility in host–pathogen interactions.
METHODS
Fungal Isolates
Blumeria graminis f. sp hordei (Bgh) isolates 5874 (Torp et al., 1978; Wei et al., 1999; AvrMla1 and AvrMla6) and K1 (Zhou et al., 2001; AvrMla1 and AvrMla13) were propagated on Hordeum vulgare cv Manchuria (C.I. 2330) in separate growth chambers at 18°C (16 h light/8 h darkness).
Plant Material
The Moseman Cereal Introduction (C.I.) lines used in these experiments each have unique Mla alleles introgressed into the universal susceptible cultivar Manchuria. Introgression was accomplished by backcrossing four times and selecting for the appropriate resistance specificity at each generation followed by selfing the heterozygous plants 12 to 15 generations, again selecting for the appropriate resistance specificity at each generation, resulting in ∼6% of the donor parent genome remaining in the backcrossed lines (Moseman, 1972).
Experimental Design
Incompatible and compatible barley powdery mildew interactions were generated by all pairwise combinations of the C.I. 16151, C.I. 16155, and C.I. 16137 near isogenic lines (containing Mla6, Mla13, and Mla1 resistance alleles, respectively) and the two Bgh isolates 5874 (AvrMla6 and AvrMla1) and K1 (AvrMla13 and AvrMla1). For each replication, individual genotypes were planted in separate 20 × 30-cm flats using sterilized potting soil. Each experimental flat consisted of six rows of 15 seedlings, with rows randomly assigned to one of six harvest times (0, 8, 16, 20, 24, and 32 hai). Seedlings were grown to the 2nd leaf stage with 1st leaf unfolded (GRO: 0007060), and inoculation was performed at 4 pm Central Standard Time by tipping the flats at 45° and dusting the plants with a high density of fresh conidiospores (84 ± 19 spores/mm2). This procedure was repeated from the opposite angle to ensure that a high proportion of the cells are in contact with the fungus. This conidial density per unit leaf area routinely results in >50% of epidermal cells that are successfully infected (Bushnell, 2002; Collinge et al., 2002). Groups of flats were placed at 18°C (8 h darkness, 16 h light, and 8 h darkness) in separate controlled growth chambers corresponding to the Bgh isolate. Rows of plants were harvested at their assigned harvest times and flash frozen in liquid nitrogen. The entire experiment was repeated three times in a standard split-split-plot design with 108 experimental units (Kuehl, 2000).
Barley1 GeneChip Probe Array
The Barley1 GeneChip probe array (part number 900515) is distributed by Affymetrix (Santa Clara, CA). The array includes 22,792 probe sets derived from worldwide contribution of 350,000 high-quality ESTs clustered from 84 cDNA libraries, in addition to 1145 barley gene sequences from the NCBI nonredundant database (Close et al., 2004). Three of the 84 libraries were derived from Mla6- and Mla13-specified race-specific incompatible interactions with Bgh, another library was derived from epidermal peels of mlo-5 broad-spectrum resistant plants challenged with Bgh, and two additional libraries were derived from susceptible interactions with Bgh and Fusarium graminearum (http://harvest.ucr.edu/Barley1.htm). The performance of the Barley1 GeneChip is consistent with other Affymetrix GeneChip probe arrays with respect to low false change rate for technical replicates and a broad linear detection range (Close et al., 2004). Array annotation information is hosted on the NetAffx data analysis center at affymetrix.com as well as the Harvest:Barley (http://harvest.ucr.edu/Barley1.htm) and BarleyBase (http://barleybase.org/) databases.
Target Synthesis and GeneChip Hybridization
Total RNA was isolated using a hot (60°C) phenol/guanidine thiocyanate method. Trizol-like reagent was made from 38% saturated phenol, pH 4.3 (Fisher Scientific, Pittsburg, PA), 1 M guanidine thiocyanate (Fisher Scientific), 1 M ammonium thiocyanate (Fisher Scientific), 0.1 M sodium acetate, pH 5.0, and 5% glycerol (Fisher Scientific). RNA purified further using the RNeasy Midi kit (Qiagen, Valencia, CA) yielded the most consistent cDNA synthesis and cRNA labeling among large numbers of samples. Probe synthesis, labeling, and hybridization protocols were followed as described in the Affymetrix manual (Affymetrix) and performed at the Iowa State University GeneChip Core facility (http://www.public.iastate.edu/∼qnzhou/Genechip.htm). Ten micrograms of purified RNA with a 260:280 ratio of 2.0 was used for cDNA synthesis using the Super-Script Choice system kit (Invitrogen, Carlsbad, CA) and GeneChip T7-oligo(dT) promoter primer kit (Affymetrix). Double-stranded cDNA was purified using the gene sample cleanup module, and 5 μL of purified cDNA was used to generate biotynilated cRNA target using the Enzo BioArray HighYield RNA transcript labeling kit (Affymetrix). Labeled cRNA was purified using the Affymetrix gene sample cleanup module, and the concentration of cRNA was determined using a Bio-Rad spectrophotometer (Hercules, CA) and adjusted based on the total RNA used as starting material. Twenty micrograms of cRNA at a final concentration of 0.5 μg/μL was fragmented in 5× fragmentation buffer at 94°C for 35 min. Quality of cDNA, cRNA, and fragmented cRNA was verified at each step on an Agilent 2100 bioanalyzer equipped with an RNA Nano LabChip (Agilent Technologies, Palo Alto, CA). Fifteen micrograms of fragmented cRNA was used to make each hybridization cocktail and 10 μg equivalent was hybridized to a GeneChip. Hybridization was performed at 60°C for 16 h in an Affymetrix hybridization oven model 640, GeneChips were washed and stained with streptavidin-phycoerythrin using the fluidics protocol EukGE-WS2 in the Affymetrix GeneChip fluidics station model 400, and stained chips were immediately scanned with an Agilent 2500A GeneArray scanner. All detailed protocols can be accessed online at http://barleypop.vrac.iastate.edu/BarleyBase/experiment_dataquery.php?class=protocolandname=any within the BarleyBase database (http://barleybase.org/).
Normalization
Before logging the data, MAS 5.0 signal measures on each GeneChip were scaled to a target intensity of 500. No additional normalization was employed. We chose to use this relatively simple normalization method to preserve independence among our measures of gene expression across GeneChips, which plays a key role in the mixed model analyses that we have implemented. More complex methods of normalization and expression calculation induce dependencies across GeneChips by making the transcript abundance measure for a gene on any one GeneChip a function of the perfect match and, in some cases, mismatch probe intensities observed for the gene on all GeneChips in the experiment. Examples of more complex normalization strategies that induce dependencies include the D-chip method (Li and Wong, 2001) and the robust multiarray average measure of expression (Irizarry et al., 2003). Although these methods have been shown to produce relatively good measures of expression, it is not clear how the dependencies created using these methods would impact the validity of subsequent statistical analyses. Furthermore, GeneChip versus GeneChip scatter plots of log signal measures (data not shown) provided no evidence that complex normalization was needed for our data.
Data Analysis
A mixed linear model analysis (Wolfinger et al., 2001) was conducted for each of the 22,792 probe sets on the Barley1 GeneChip using the SAS mixed procedure. The natural logarithm of the Affymetrix MAS 5.0 signal measure of gene expression was used as the response variable in the mixed linear model analyses to stabilize variance within genes and obtain approximate normality for random effects required for valid statistical inference. The mixed linear model included terms for the fixed effects of genotype, isolate, time point, and all interactions between these three factors as well as random effects for replications and random interactions corresponding to whole-plot, split-plot, and split-split-plot experimental units. We used contrast statements with the SAS mixed procedure to identify genes whose differences in average expression between compatible and incompatible interactions varied significantly across time. For each time point, we estimated the average expression of the gene in the compatible interactions [Mla6/virMla6 (K1) and Mla13/virMla13 (5874)] and compared that with the average expression of the gene in the incompatible interactions [Mla6/AvrMla6 (5874) and Mla13/AvrMla13 (K1)]. These time-specific differences between the average expressions (d0hai, d8hai, d16hai, d20hai, d24hai, and d32hai) were tested for equality using an F-statistic. Formally, the null hypothesis of this test may be written as H0: δ0hai = δ8hai = δ16hai = δ20hai = δ24hai = δ32hai, where δ is the true difference estimated by d. Genes whose time-specific differences varied significantly (P value < 0.0001) across time points were identified as differentially expressed. Note that genes for which H0 is true are those whose average expression patterns are the same in both compatible and incompatible interactions. Thus, rejection of H0 for a gene indicates a pattern of expression in compatible interactions that differs from its pattern in incompatible interactions. The P value < 0.0001 threshold for significance was chosen to obtain a list of which the proportion of false positive results would be low. We then estimated the proportion of false positive results to be <7% for the 0.0001 threshold using the method described by Storey and Tibshirani (2003).
The same basic strategy was used to identify genes involved in pathways that distinguish Mla-specified Rar1-dependent from Rar1-independent interactions. The mean mRNA expression levels in Mla6/AvrMla6 (5874) and Mla13/AvrMla13 (K1) were compared with the mean mRNA expression levels in Mla1/AvrMla1 (5874) and Mla1/AvrMla1 (K1) to identify genes whose transcriptional differences varied significantly (P value < 0.0001) across time.
Cluster Analysis
Average scaled signal intensities were calculated from three replications using Microsoft Excel 2002. Data matrices were constructed with genes in rows and time points of the different genotype-isolate combinations in columns. For the clustering of 22 (Figure 2) and 14 (Figure 4) gene sets, data matrices were loaded in the GeneSpring 5.1 (Silicon Genetics, Redwood City, CA) software. Signal intensities were standardized based on the median for each gene. A Pearson correlation and hierarchical clustering were both used in creating the gene tree. For the time point–specific clustering of 22 and 14 (Figure 5) gene sets, data matrices were loaded in the NTSYSpc statistical software version 2.1 (Exeter Software, Setauket, NY). Similarities were calculated using a Pearson correlation. Correlation matrix was used in the unweighted pair group mean arithmetic cluster algorithm.
Data Access
All detailed data and protocols from these experiments have been deposited in BarleyBase (http://barleybase.org/), a MIAME-compliant expression database for cereal GeneChips (Brazma et al., 2001; http://www.mged.org/Workgroups/MIAME/miame.html). Files are categorized under accession number BB4 and can be analyzed online using the current tools in BarleyBase, downloaded as batch files in MAGE-ML, CSV, CEL, DAT, or expression data formats at the Download Center or downloaded as individual CEL, CHP, DAT, or EXP files under “browse experiments.” Data files have also been deposited as accession number E-MEXP-142 in ArrayExpress (http://www.ebi.ac.uk/arrayexpress).
Supplementary Material
Acknowledgments
The authors thank J. Peng of the ISU GeneChip facility for significant assistance with the processing of the Barley1 probe arrays and S. Whitham, T. Baum, and D. Halterman for critical review of the manuscript. This research was supported by the USDA Initiative for Future Agriculture and Food Systems Grant 2001-52100-11346, USDA National Research Initiative Grant 02-35300-12619, and the USDA Cooperative State Research, Education, and Extension Service North American Barley Genome Project. This article is a joint contribution of the Corn Insects and Crop Genetics Research Unit, USDA-Agricultural Research Service, and the Iowa Agriculture and Home Economics Experiment Station.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Roger P. Wise (rpwise@iastate.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.023382.
References
- Abramovitch, R.B., Kim, Y.J., Chen, S., Dickman, M.B., and Martin, G.B. (2003). Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibition of host programmed cell death. EMBO J. 22, 60–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, H.J., Yang, J.K., Lee, B.I., Yoon, H.J., Kim, H.W., and Suh, S.W. (2003). Crystallization and preliminary X-ray crystallographic studies of chorismate synthase from Helicobacter pylori. Acta. Crystallogr. D. Biol. Crystallogr. 59, 569–571. [DOI] [PubMed] [Google Scholar]
- Axtell, M.J., and Staskawicz, B.J. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112, 369–377. [DOI] [PubMed] [Google Scholar]
- Azevedo, C., Sadanandom, A., Kitagawa, K., Freialdenhoven, A., Shirasu, K., and Schulze-Lefert, P. (2002). The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295, 2073–2076. [DOI] [PubMed] [Google Scholar]
- Bohlmann, J., Lins, T., Martin, W., and Eilert, U. (1996). Anthranilate synthase from Ruta graveolens. Duplicated AS alpha genes encode tryptophan-sensitive and tryptophan-insensitive isoenzymes specific to amino acid and alkaloid biosynthesis. Plant Physiol. 111, 507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonas, U., and Lahaye, T. (2002). Plant disease resistance triggered by pathogen-derived molecules: Refined models of specific recognition. Curr. Opin. Microbiol. 5, 44–50. [DOI] [PubMed] [Google Scholar]
- Boyd, L.A., Smith, P.H., Foster, E.M., and Brown, J.K.M. (1995). The effects of allelic variation at the Mla resistance locus in barley on the early development of Erysiphe graminis f. sp. hordei and host responses. Plant J. 7, 959–968. [Google Scholar]
- Brazma, A., et al. (2001). Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 29, 365–371. [DOI] [PubMed] [Google Scholar]
- Brunner, F., Rosahl, S., Lee, J., Rudd, J.J., Geiler, C., Kauppinen, S., Rasmussen, G., Sheel, D., and Nürnberger, T. (2002). Pep-13, a plant defense-inducing pathogen-associated pattern from Phytophthora transglutaminases. EMBO J. 21, 6681–6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burhenne, K., Kristensen, B.K., and Rasmussen, S.K. (2003). A new class of N-hydroxycinnamoyltransferases. Purification, cloning and expression of a barley agmatine coumaroyltransferase (EC 2.3.1.64). J. Biol. Chem. 278, 13919–13927. [DOI] [PubMed] [Google Scholar]
- Bushnell, W.R. (2002). The role of powdery mildew research in understanding host-parasite interaction: Past, present and future. In The Powdery Mildews: A Comprehensive Treatise, R.R. Bélanger, W.R. Bushnell, A.J. Dik, and T.L.W. Carver, eds (St. Paul, MN: APS Press), pp. 1–12.
- Century, K.S., Shapiro, A.D., Repetti, P.P., Dahlbeck, D., Holub, E., and Staskawicz, B.J. (1997). NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science 278, 1963–1965. [DOI] [PubMed] [Google Scholar]
- Cheung, W.L., Ajiro, K., Samejima, K., Kloc, M., Cheung, P., Mizzen, C.A., Beeser, A., Etkin, L.D., Chernoff, J., Earnshaw, W.C., and Allis, C.D. (2003). Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell 113, 507–517. [DOI] [PubMed] [Google Scholar]
- Clark, T.A., Zeyen, R.J., Smith, A.G., Bushnell, W.R., Szabo, L.J., and Vance, C.P. (1993). Host response gene transcript accumulation in relation to visible cytological events during Erysiphe graminis attack in isogenic barley lines differing at the Ml-a locus. Physiol. Mol. Plant Pathol. 43, 283–298. [Google Scholar]
- Close, T.J., Wanamaker, S., Caldo, R.A., Turner, S.M., Ashlock, D.A., Dickerson, J.A., Wing, R.A., Muehlbauer, G.J., Kleinhofs, A., and Wise, R.P. (2004). A new resource for cereal genomics: 22K Barley GeneChip comes of age. Plant Physiol. 134, 960–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock, J.M., Vanoosthuyse, V., and Gaude, T. (2002). Receptor kinase signaling in plants and animals: Distinct molecular systems with mechanistic similarities. Curr. Opin. Cell Biol. 14, 230–236. [DOI] [PubMed] [Google Scholar]
- Collinge, D.B., Gregersen, P.L., and Thordal-Christensen, H. (2002). The nature and role of defense response genes in cereals. In The Powdery Mildews: A Comprehensive Treatise, R.R. Bélanger, W.R. Bushnell, A.J. Dik, and T.L.W. Carver, eds (St. Paul, MN: APS Press), pp. 146–160.
- Collmer, A. (1998). Determinants of pathogenicity and avirulence in plant pathogenic bacteria. Curr. Opin. Plant Biol. 1, 329–335. [DOI] [PubMed] [Google Scholar]
- Collmer, A., Lindeberg, M., Petnicki-Ocwieja, T., Scheider, D.J., and Alfano, J.R. (2002). Genomic mining type III secretion system effectors in Pseudomonas syringae yields new picks for all TTSS prospectors. Trends Microbiol. 10, 462–469. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D.G. (2001). Plant pathogens and integrated defense responses to infection. Nature 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Ellingboe, A.H. (1972). Genetics and physiology of primary infection by Erysiphe graminis f.sp. hordei. Phytopathology 62, 401–406. [Google Scholar]
- Ellis, J., and Dodds, P. (2003). Plant pathology: Monitoring a pathogen-targeted host protein. Curr. Biol. 13, R400–R402. [DOI] [PubMed] [Google Scholar]
- Eulgem, T., Weigman, V.J., Chang, H.-S., McDowell, J.M., Holub, E.B., Glazebrook, J., Zhu, T., and Dangl, J.F. (2004). Gene expression signatures from three genetically separable resistance gene signaling pathways for downy mildew resistance. Plant Physiol. 135, 1129–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellbrich, G., Romanski, A., Varet, A., Blume, B., Brunner, F., Engelhardt, S., Felix, G., Kemmerling, B., Kryzmowska, M., and Nürnberger, T. (2002). NPP1, a Phytophthora-associated trigger of plant defense in parsley and Arabidopsis. Plant J. 32, 375–390. [DOI] [PubMed] [Google Scholar]
- Flor, H.H. (1971). Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9, 275–296. [Google Scholar]
- Goujun, T., Ferret, V., Mila, I., Pollet, B., Ruel, K., Burlat, V., Joseleau, J.P., Barrierre, Y., Lapierre, C., and Jouanin, L. (2003). Down-regulation of the AtCCR1 gene in Arabidopsis thaliana: Effects on phenotype, lignins and cell wall degradability. Planta 217, 218–228. [DOI] [PubMed] [Google Scholar]
- Gregersen, P.L., Thordal-Christensen, H., Forster, H., and Collinge, D.B. (1997). Differential gene transcript accumulation in barley leaf epidermis and mesophyll in response to attack by Blumeria graminis f.sp. hordei (syn. Erysiphe graminis f.sp. hordei). Physiol. Mol. Plant Pathol. 51, 85–97. [Google Scholar]
- Guo, H., Cui, Q., Lipscomb, W.N., and Karplus, M. (2001). Substrate conformational transitions in the active site of chorismate mutase: Their role in the catalytic mechanism. Proc. Natl. Acad. Sci. USA 98, 9032–9037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahlbrock, K., Bednarek, P., Ciolkowski, I., Hamberger, B., Heise, A., Liedgens, H., Logemann, E., Nurnberger, T., Schmelzer, E., Somssich, I.E., and Tan, J. (2003). Non-self recognition, transcriptional reprogramming, and secondary metabolite accumulation during plant/pathogen interactions. Proc. Natl. Acad. Sci. USA 100 (suppl. 2), 14569–14576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, A.A., Bindslev, L., Rouster, J., Rasmusses, S.W., Oliver, R.P., and Gurr, S.J. (1999). Involvement of cAMP and protein kinase A in conidial differentiation by Erysiphe graminis f. sp. hordei. Mol. Plant-Microbe Interact. 12, 960–968. [DOI] [PubMed] [Google Scholar]
- Halterman, D., Wei, F., and Wise, R.P. (2003). Powdery mildew induced Mla mRNAs are alternatively spliced and contain multiple upstream open reading frames. Plant Physiol. 131, 558–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halterman, D., and Wise, R.P. (2004). A single amino acid substitution in the sixth leucine-rich repeat of barley MLA6 and MLA13 alleviates dependence on RAR1 for disease resistance signaling. Plant J. 38, 215–226. [DOI] [PubMed] [Google Scholar]
- Halterman, D., Zhou, F.S., Wei, F., Wise, R.P., and Schulze-Lefert, P. (2001). The MLA6 coiled-coil, NBS-LRR protein confers AvrMla6-dependent resistance specificity to Blumeria graminis f. sp. hordei in barley and wheat. Plant J. 25, 335–348. [DOI] [PubMed] [Google Scholar]
- Hardham, A.J. (2001). Cell biology of fungal infection of plants. In The Mycota: Biology of the Fungal Cell, Vol. VIII, R.J. Howard and N.A.R. Gow, eds (New York: Springer-Verlag), pp. 91–124.
- Hauck, P., Thilmony, R., and He, S.Y. (2003). A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc. Natl. Acad. Sci. USA 100, 8577–8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath, M.C. (2000). Nonhost resistance and nonspecific plant defenses. Curr. Opin. Plant Biol. 3, 315–319. [DOI] [PubMed] [Google Scholar]
- Irizarry, R.A., Hobbs, B., Collin, F., Beazer-Barclay, Y.D., Antonellis, K.J., Scherf, U., and Speed, T.P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264. [DOI] [PubMed] [Google Scholar]
- Jamir, Y., Guo, M., Oh, H.S., Petnicki-Ocwieja, T., Chen, S., Tang, X., Dickman, M.B., Collmer, A., and Alfano, J.R. (2004). Identification of Pseudomonas syringae type III effectors that can suppress programmed cell death in plants and yeast. Plant J. 37, 554–565. [DOI] [PubMed] [Google Scholar]
- Jason, L.J.M., Moore, S.C., Lewis, J.D., Lindsey, G., and Ausio, J. (2002). Histone ubiquitination: A tagging tail unfolds? Bioessays 24, 166–174. [DOI] [PubMed] [Google Scholar]
- Jia, Y., McAdams, S.A., Bryan, G.T., Hershey, H.P., and Valent, B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19, 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D.A., and Takemoto, D. (2004). Plant innate immunity—Direct and indirect recognition of general and specific pathogen-associated molecules. Curr. Opin. Immunol. 16, 48–62. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G. (2001). Putting knowledge of plant disease resistance genes to work. Curr. Opin. Plant Biol. 4, 281–287. [DOI] [PubMed] [Google Scholar]
- Jørgensen, J.H. (1988). Genetic analysis of barley mutants with modifications of powdery mildew resistance gene Ml-a12. Genome 30, 129–132. [Google Scholar]
- Jørgensen, J.H. (1994). Genetics of powdery mildew resistance in barley. Crit. Rev. Plant Sci. 13, 97–119. [Google Scholar]
- Joshi, B.N., Sainani, M.N., Bastawade, B., Deshpande, V.V., Gupta, V.S., and Ranjekar, P.K. (1999). Pearl millet cysteine protease inhibitor. Evidence for the presence of two distinct sites responsible for anti-fungal and anti-feedent activities. Eur. J. Biochem. 265, 556–563. [DOI] [PubMed] [Google Scholar]
- Kiba, A., Takeda, T., Kanemitsu, T., Toyoda, K., Ichinose, Y., Yamada, T., and Shiraishi, T. (1999). Induction of defense responses by synthetic glycopeptides that have a partial structure of the elicitor in the spore germination fluid of Mycosphaerella pinodes. Plant Cell Physiol. 40, 978–985. [DOI] [PubMed] [Google Scholar]
- Kim, Y.J., Lin, N.C., and Martin, G.B. (2002). Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell 109, 589–598. [DOI] [PubMed] [Google Scholar]
- Kjemtrup, S., Nimchuk, Z., and Dangl, J.L. (2000). Effector proteins of phytopathogenic bacteria: Bifunctional signals in virulence and host recognition. Curr. Opin. Microbiol. 3, 73–78. [DOI] [PubMed] [Google Scholar]
- Kruger, W.M., Szabo, L.J., and Zeyen, R.J. (2003). Transcription of the defense response genes chitinase IIb, PAL and peroxidase is induced by the barley powdery mildew fungus and is only indirectly modulated by R genes. Physiol. Mol. Plant Pathol. 63, 167–178. [Google Scholar]
- Kuehl, R.O. (2000). Design of Experiments: Statistical Principles of Research Design and Analysis, 2nd ed. (Pacific Grove, CA: Duxbury Press).
- Kunoh, H. (1982). Primary germ tubes of Erysiphe graminis conidia. In Plant Infection: The Physiological and Biochemical Basis, Y. Asada, W.R. Bushnell, S. Ouchi, and C.P. Vance, eds (Tokyo: Japan Scientific Society Press), pp. 45–59.
- Kunoh, H. (2002). Localized induction of accessibility and inaccessibility by powdery mildew. In The Powdery Mildews: A Comprehensive Treatise, R.R. Bélanger, W.R. Bushnell, A.J. Dik, and T.L.W. Carver, eds (St. Paul MN: APS Press), pp. 126–133.
- Lauvergeat, V., Lacomme, C., Lacombe, E., Lasserre, E., Roby, D., and Grima-Pettenati, J. (2001). Two cinnamoyl-CoA reductase (CCR) genes from Arabidopsis thaliana are differentially expressed during development and in response to infection with pathogenic bacteria. Phytochemistry 57, 1187–1195. [DOI] [PubMed] [Google Scholar]
- Li, C., and Wong, W.H. (2001). Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 98, 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorito, M., Broadway, R.M., Hayes, C.K., Woo, S.L., Naviello, C., Williams, D.L., and Herman, G.E. (1994). Proteinase inhibitors in plants as a novel class of fungicides. Mol. Plant-Microbe Interact. 7, 525–527. [Google Scholar]
- Mackey, D., Belkhadir, Y., Alonso, J.M., Ecker, J.R., and Dangl, J.L. (2003). Arabidopsis RIN4 is a target of the type III virulence effectors AvrRpt2 and modulates RPS2-mediated resistance. Cell 112, 379–389. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Holt III, B.F., Wiig, A., and Dangl, J.L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108, 743–754. [DOI] [PubMed] [Google Scholar]
- Maleck, K., Levine, A., Eulgem, T., Morgan, A., Schmid, J., Lawton, K.A., Dangl, J.L., and Dietrich, R.A. (2000). The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26, 403–410. [DOI] [PubMed] [Google Scholar]
- Mendgen, K., and Hahn, M. (2002). Plant infection and the establishment of fungal biotrophy. Trends Plant Sci. 7, 352–356. [DOI] [PubMed] [Google Scholar]
- Menke, F.L.H., van Pelt, J.A., Pieterse, C.M.J., and Klessig, D.F. (2004). Silencing of the mitogen-activated protein kinase MPK6 compromises disease resistance in Arabidopsis. Plant Cell 16, 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesano, M., Brader, G., and Palva, E.T. (2003). Pathogen derived elicitors: Searching for receptors in plants. Mol. Plant Pathol. 4, 73–79. [DOI] [PubMed] [Google Scholar]
- Moseman, J.G. (1972). Isogenic barley lines for reaction to Erysiphe graminis f. sp. hordei. Crop Sci. 12, 681–682. [Google Scholar]
- Mysore, K.S., Crasta, O.R., Tuori, R.P., Folkerts, O., Swirsky, P.B., and Martin, G.B. (2002). Comprehensive transcript profiling of Pto- and Prf- mediated host defense responses to infection by Pseudomonas syringae pv. tomato. Plant J. 32, 299–315. [DOI] [PubMed] [Google Scholar]
- Nimchuk, Z., Eulgem, T., Holt III, B.F., and Dangl, J.L. (2003). Recognition and response in the plant immune system. Annu. Rev. Genet. 37, 579–609. [DOI] [PubMed] [Google Scholar]
- Nürnberger, T. (1999). Signal perception in plant pathogen defense. Cell. Mol. Life Sci. 55, 167–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nürnberger, T., and Brunner, F. (2002). Innate immunity in plants and animals: Emerging parallels between the recognition of general elicitors and pathogen-associated molecular patterns. Curr. Opin. Plant Biol. 5, 318–324. [DOI] [PubMed] [Google Scholar]
- Oelofse, D., and Dubery, I.A. (1996). Induction of defense responses in cultured tobacco cells by elicitors from Phytophthora nicotianae. Int. J. Biochem. Cell Biol. 28, 295–301. [DOI] [PubMed] [Google Scholar]
- Panstruga, R. (2004). A golden shot: How ballistic single cell transformation boosts the molecular analysis of cereal-mildew interactions. Mol. Plant Pathol. 5, 141–148. [DOI] [PubMed] [Google Scholar]
- Panstruga, R., and Schulze-Lefert, P. (2002). Live and let live: Insights into powdery mildew disease and resistance. Mol. Plant Pathol. 3, 495–502. [DOI] [PubMed] [Google Scholar]
- Panstruga, R., and Schulze-Lefert, P. (2003). Corruption of host seven-transmembrane protein by pathogenic microbes: A common theme in animals and plants? Microbes Infect. 5, 429–437. [DOI] [PubMed] [Google Scholar]
- Parker, J.E. (2003). Plant recognition of microbial patterns. Trends Plant Sci. 8, 245–247. [DOI] [PubMed] [Google Scholar]
- Peart, J.R., Cook, G., Feys, B.J., Parker, J.E., and Baulcombe, D.C. (2002. a). An EDS1 orthologue is required for N-mediated resistance against tobacco mosaic virus. Plant J. 29, 569–579. [DOI] [PubMed] [Google Scholar]
- Peart, J.R., et al. (2002. b). Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl. Acad. Sci. USA 99, 10865–10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J.L., Dong, H.S., Dong, H.P., Delaney, T.P., Bonasera, J.M., and Beer, S.V. (2003). Harpin-elicited hypersensitive cell death and pathogen resistance require the NDR1 and EDS1. Physiol. Mol. Plant Pathol. 62, 317–326. [Google Scholar]
- Puthoff, D.P., Nettleton, D., Rodermel, S.R., and Baum, T.J. (2003). Arabidopsis gene expression changes during cyst nematode parasitism revealed by statistical analyses of microarray expression profiles. Plant J. 33, 911–921. [DOI] [PubMed] [Google Scholar]
- Sano, H., Seo, S., Orudgev, E., Youssefian, S., Ishizuka, K., and Ohashi, Y. (1994). Expression of the gene for a small GTP binding protein in transgenic tobacco elevates endogenous cytokinin levels, abnormally induces salicylic acid in response to wounding, and increases resistance to tobacco mosaic virus infection. Proc. Natl. Acad. Sci. USA 91, 10556–10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk, P.M., Kazan, K., Wilson, I., Anderson, J.P., Richmond, T., Somerville, S.C., and Manners, J.M. (2000). Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 97, 11655–11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Lefert, P., and Panstruga, R. (2003). Establishment of biotrophy by parasitic fungi and reprogramming the host cells for disease resistance. Annu. Rev. Phytopathol. 41, 641–667. [DOI] [PubMed] [Google Scholar]
- Schulze-Lefert, P., and Vogel, J. (2000). Closing the ranks to attack by powdery mildew. Trends Plant Sci. 5, 343–348. [DOI] [PubMed] [Google Scholar]
- Shen, Q.-H., Zhou, F., Bieri, S., Haizel, T., Shirasu, K., and Shulze-Lefert, P. (2003). Recognition specificity and Rar1/Sgt1 dependence in barley Mla disease resistance genes to the powdery mildew fungus. Plant Cell 15, 732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi, T., Yamada, T., Ichinose, Y., Kiba, A., and Toyoda, K. (1997). The role of suppressor in determining host-parasite specificities in plant cells. Int. Rev. Cytol. 172, 55–93. [Google Scholar]
- Shiraishi, T., Yamada, T., Saitoh, K., Kato, T., Toyoda, K., Yoshioka, H., Kim, H.-M., Ichinose, Y., Tahara, M., and Oku, H. (1994). Suppressors: Determinants of specificity produced by plant pathogens. Plant Cell Physiol. 3, 1107–1119. [Google Scholar]
- Shirasu, K., Nielsen, K., Piffanelli, P., Oliver, R.P., and Schulze-Lefert, P. (1999). Cell-autonomous complementation of mlo resistance using a biolistic transient expression system. Plant J. 17, 293–299. [Google Scholar]
- Shirasu, K., and Schulze-Lefert, P. (2003). Complex formation, promiscuity and multi-functionality: Protein interactions in disease-resistance pathways. Trends Plant Sci. 8, 252–258. [DOI] [PubMed] [Google Scholar]
- Stein, M., and Somerville, S.C. (2002). MLO, a novel modulator of plant defense and cell death, binds calmodulin. Trends Plant Sci. 7, 379–380. [DOI] [PubMed] [Google Scholar]
- Storey, J.D., and Tibshirani, R. (2003). Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100, 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, X., Frederick, R.D., Zhou, J., Halterman, D.A., Jia, Y., and Martin, G.B. (1996). Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science 274, 2060–2063. [DOI] [PubMed] [Google Scholar]
- Tao, Y., Xie, Z., Chen, W., Glazebrook, J., Chang, H.-S., Han, B., Zhu, T., Zou, G., and Katagiri, F. (2003). Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torp, J., Jensen, H.P., Jørgensen, J.H., and Helms, J. (1978). Powdery midlew resistance genes in 106 Northwest European spring barley varieties. In The Royal Veterinary and Agricultural University Yearbook 1978. (Copenhagen: The Royal Veterinary and Agricultural University), pp. 27–44.
- Toyoda, K., Nicholas, C.C., Takahashi, A., and Shirasu, K. (2002). Resistance and susceptibility of plants to fungal pathogens. Transgenic Res. 11, 567–582. [DOI] [PubMed] [Google Scholar]
- Toyoda, K., Shiraishi, T., Yamada, T., Ichinose, Y., and Oku, H. (1993). Rapid changes in polyphosphoinositide metabolism in pea in response to fungal signals. Plant Cell Physiol. 34, 729–735. [Google Scholar]
- Tsiamis, G., Mansfield, J.W., Hockenhull, R., Jackson, R.W., Sesma, A., Athanassopoulos, E., Bennett, M.A., Stevens, C., Vivian, A., Taylor, J.D., and Murrilo, J. (2000). Cultivar-specific avirulence and virulence function assigned to avrPphF in Pseudomonas syringae pv. phaseolicola, the cause of bean halo-blight disease. EMBO J. 19, 3204–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, S.L., and Talbot, N.J. (2001). Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annu. Rev. Phytopathol. 39, 385–417. [DOI] [PubMed] [Google Scholar]
- Tyler, B.M. (2002). Molecular basis of recognition between Phytophthora pathogens and their hosts. Annu. Rev. Phytopathol. 40, 137–167. [DOI] [PubMed] [Google Scholar]
- Van der Biezen, E.A., and Jones, J.D.G. (1998). Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci. 23, 454–456. [DOI] [PubMed] [Google Scholar]
- van Wees, S.C., Chang, H.S., Zhu, T., and Glazebrook, J. (2003). Characterization of the early response of Arabidopsis to Alternaria brassicicola infection using expression profiling. Plant Physiol. 132, 606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Ropenack, E., Parr, A., and Schulze-Lefert, P. (1998). Structural analyses and dynamics of soluble and cell wall-bound phenolics in a broad spectrum resistance to the powdery mildew fungus in barley. J. Biol. Chem. 273, 9013–9022. [DOI] [PubMed] [Google Scholar]
- Wan, J., Dunning, F.M., and Bent, A.F. (2002). Probing plant-pathogen interactions and downstream defense signaling using DNA microarrays. Funct. Integr. Genomics 2, 259–273. [DOI] [PubMed] [Google Scholar]
- Wei, F., Gobelman-Werner, K., Morroll, S.M., Kurth, J., Mao, L., Wing, R., Leister, D., Schulze-Lefert, P., and Wise, R.P. (1999). The Mla (powdery mildew) resistance cluster is associated with three NBS-LRR gene families and suppressed recombination within a 240-kb DNA interval on chromosome 5S (1HS) of barley. Genetics 153, 1929–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, F.F., Yang, B., and Johnson, L.B. (2000). Prospects for understanding avirulence gene function. Curr. Opin. Plant Biol. 3, 291–298. [DOI] [PubMed] [Google Scholar]
- Whitham, S.A., Quan, S., Chang, H.S., Cooper, B., Estes, B., Zhu, T., Wang, X., and Hou, Y.M. (2003). Diverse RNA viruses elicit the expression of common sets of genes in susceptible Arabidopsis thaliana plants. Plant J. 33, 271–283. [DOI] [PubMed] [Google Scholar]
- Wise, R.P. (2000). Disease resistance: What's brewing in barley genomics. Plant Dis. 84, 1160–1170. [DOI] [PubMed] [Google Scholar]
- Wise, R.P., and Ellingboe, A.H. (1983). Infection kinetics of Erysiphe graminis f. sp. hordei on barley with different alleles at the Ml-a locus. Phytopathology 73, 1220–1222. [Google Scholar]
- Wolfinger, R.D., Gibson, G., Wolfinger, E.D., Bennett, L., Hamadeh, H., Bushel, P., Afshari, C., and Paules, R.S. (2001). Assessing gene significance from cDNA microarray expression data via mixed models. J. Comput. Biol. 8, 625–637. [DOI] [PubMed] [Google Scholar]
- Xiao, S., Ellwood, S., Calis, O., Patrick, E., Li, T., Coleman, M., and Turner, J.G. (2001). Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 291, 118–120. [DOI] [PubMed] [Google Scholar]
- Yang, Q., Reinhard, K., Schiltz, E., and Matern, U. (1997). Characterization and heterologous expression of hydroxycinnamoyl/benzoyl-CoA:anthranilate N-hydroxycinnamoyl/benzoyl-transferase from elicited cell cultures of carnation, Dianthus caryopyllus L. Plant Mol. Biol. 35, 777–789. [DOI] [PubMed] [Google Scholar]
- Yoshioka, H., Shiraishi, T., Yamada, T., Ichinose, Y., and Oku, H. (1990). Suppression of pisatin production and ATPase activity in pea plasma membranes by orthovanadate, verapamil and a suppressor from Mycosphaerella pinodes. Plant Cell Physiol. 31, 1139–1146. [Google Scholar]
- Yun, B.W., Atkinson, H.A., Gaborit, C., Greenland, A., Read, N., Pallas, J.A., and Loake, G.J. (2003). Loss of actin cytoskeletal function and EDS1 activity, in combination, severely compromises non-host resistance in Arabidopsis against wheat powdery mildew. Plant J. 34, 768–777. [DOI] [PubMed] [Google Scholar]
- Zhang, S., and Klessig, D.F. (2001). MAPK cascades in plant defense signaling. Trends Plant Sci. 6, 520–527. [DOI] [PubMed] [Google Scholar]
- Zhou, F.S., Kurth, J.C., Wei, F., Elliott, C., Vale, G., Yahiaoui, N., Keller, B., Somerville, S., Wise, R., and Schulze-Lefert, P. (2001). Cell-autonomous expression of barley Mla1 confers race-specific resistance to the powdery mildew fungus via a Rar1-independent signaling pathway. Plant Cell 13, 337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C., Robatzek, S., Navarro, L., Oakeley, E.J., Jones, J.D.G., Felix, G., and Boller, T. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428, 764–767. [DOI] [PubMed] [Google Scholar]
NOTE ADDED IN PROOF
- A similar association between Avr-dependent defense responses to Pseudomonas syringae and bacterial pathogenesis has been reported recently by Navarro et al. (2004).
- Navarro, L., Zipfel, C., Rowland, O., Keller, I., Robatzek, S., Boller, T., and Jones, J.D.G. (2004). The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 135, 1113–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.