Abstract
Alterations in intestinal microbiota composition could promote a proinflammatory state in adipose tissue that is associated with obesity and insulin resistance. Our aim was to identify the gut microbiota associated with insulin resistance in appendix samples from morbidly obese patients classified in 2 groups, high (IR-MO) and low insulin-resistant (NIR-MO), and to determine the possible association between these gut microbiota and variables associated with insulin resistance and the expression of genes related to inflammation and macrophage infiltration in adipose tissue. Appendix samples were obtained during gastric bypass surgery and the microbiome composition was determined by 16S rRNA pyrosequencing and bioinformatics analysis by QIIME. The Chao and Shannon indices for each study group suggested similar bacterial richness and diversity in the appendix samples between both study groups. 16S rRNA pyrosequencing showed that the IR-MO group had a significant increase in the abundance of Firmicutes, Fusobacteria, Pseudomonaceae, Prevotellaceae, Fusobacteriaceae, Pseudomonas, Catenibacterium, Prevotella, Veillonella and Fusobacterium compared to the NIR-MO group. Moreover, in the IR-MO group we found a significant positive correlation between the abundance of Prevotella, Succinovibrio, Firmicutes and Veillonella and the visceral adipose tissue expression level of IL6, TNF alpha, ILB1 and CD11b respectively, and significant negative correlations between the abundance of Butyricimonas and Bifidobacterium, and plasma glucose and insulin levels, respectively. In conclusion, an appendix dysbiosis occurs in IR-MO patients, with a loss of butyrate-producing bacteria, essential to maintenance of gut integrity, together with an increase in mucin-degrading bacteria and opportunistic pathogens. The microbiota present in the IR-MO group were related to low grade inflammation in adipose tissue and could be useful for developing strategies to control the development of insulin resistance.
Keywords: Microbiota, appendix, insulin resistance, gut integrity, inflammation, adipose tissue
Introduction
Obesity is characterized by chronic subclinical inflammation that affects insulin activity in metabolically sensitive tissues (liver, muscle and adipose tissues) [1]. Recent studies in the past ten years have shown that this metabolic inflammation is characterized by a moderate excess in cytokine production, including interleukin (IL) IL-6, IL-1 or tumor necrosis factor alpha (TNF alpha), that injures cellular insulin signals and contributes to insulin resistance and diabetes [2,3]. Recent research has highlighted links between the gut microbiota, obesity and insulin resistance [1,4-7]. Growing evidence suggests that the gut microbiota contribute to host metabolism through communication with adipose tissue, which influences the development of metabolic alterations associated with obesity [8].
The intestinal microbiota have been shown to influence intestinal permeability in obese mice, thereby promoting translocation of bacterial products and stimulating the low-grade inflammation characteristic of obesity and insulin resistance [9,10]. Verdam et al. showed that the human obesity-associated microbiota profile is associated with local and systemic inflammation, although they did not find an association between the obesity-related microbiota composition and intestinal permeability, suggesting that the obesity-related microbiota composition has a proinflammatory effect [11].
The physiologic function of the human appendix is largely unknown but several hypotheses involve interactions between the abundant lymphoid tissue in the appendix and the microbiota contained within the appendix [12,13]. In a recent study Guinane et al. concluded that the human appendix, although sharing a substantial amount of microbes with the intestinal tract, has its own defined microbiome. This microbial composition of the human appendix is subject to extreme variability and comprises a diversity of microbiota that may play an important role in human health [14]. The microbiota in appendix samples are a reflection of the microbiota present in the small intestine, which play an important role in host immunity and metabolism. In a preliminary study using polymerase chain reaction denaturing gradient gel electrophoresis (PCR-DGGE) we reported that intestinal bacterial DNA is a signature of insulin action in humans, but we did not identify the gut microbes associated with the insulin resistance phenotype [15].
The aim of the present study was to identify, using next-generation sequencing technologies, the precise gut microbiota associated with insulin resistance in appendix samples from morbidly obese patients, to provide a gut microbial signature for this phenotype, and also to determine the possible relationship between these gut microbiota and variables associated with insulin resistance and the expression of genes related to inflammation and macrophage infiltration in adipose tissue.
Material and methods
The homoeostasis model assessment of insulin resistance (HOMA-IR) was used to classify the morbidly obese female patients. Specifically, patients with a HOMA-IR score of <3 were considered to comprise the low-insulin-resistant (NIR-MO) group, according to the mean ± 2SD of a healthy control population. The morbidly obese patients with a HOMA-IR score of >7 were considered to be from the high insulin-resistant (IR-MO) group. Appendix samples from 5 IR-MO and 5 NIR-MO patients matched for body mass index (BMI), age, gender, race and dietary intake were obtained during bariatric surgery. The samples were washed, fragmented, and frozen in liquid nitrogen before being stored at -80°C. All subjects were of Caucasian origin with no systemic disease or infection during the month before the study. Liver disease and thyroid dysfunction were specifically excluded by biochemical work-up. Other patient exclusion criteria included type 2 diabetes mellitus treated with insulin or oral antidiabetics, cardiovascular disease in the 6 months prior to study inclusion, arthritis, evidence of acute or chronic inflammatory disease, infectious diseases, or receiving drugs that could alter the lipid profile or the metabolic parameters at the time of inclusion in the study, renal involvement, history of drug or alcohol abuse (defined as 80 g/day), or serum transaminase activity more than twice the upper limit of normal, and the patient’s decision not to participate in the study. The patients had not received any antibiotic, probiotic, or prebiotic agents in the 3-month period before the collection of appendix samples. The subjects were invited to participate by the Endocrinology Service of the Virgen de la Victoria Hospital (Malaga, Spain). Written informed consent was obtained in all cases and the protocol was approved by the Ethics Committee of Virgen de la Victoria Hospital.
Dietary assessment
A very-low-energy diet (Optifast® VLCD, Nestlé Health Science, Spain) was consumed by all the patients for a period of 8 weeks before gastric bypass surgery. Subjects ingested 3 shakes/day of Optifast®, which provided 456 kcal, 52 g protein, 7 g fat, and 45 g carbohydrates plus the recommended daily intake of vitamins, minerals, and trace elements. The patients were also permitted to eat other low-calorie foodstuffs (such as low-starch vegetables) to provide a total energy intake of 800 kcal/day. The dietary requirements were outlined by a dietitian before commencement of the diet, and all subjects attended for dietary counseling fortnightly thereafter. Any adverse effects of the diet were noted.
Analysis of biochemical variables
Blood samples were collected after an overnight fast. The serum was separated in aliquots and immediately frozen at -80°C. Serum levels of glucose, cholesterol, triglycerides and HDL cholesterol were analyzed using a Dimension autoanalyzer (Dade Behring Inc., Deerfield, IL) by enzymatic methods (Randox Laboratories Ltd., UK). LDL cholesterol was calculated from the Friedewald equation. Gamma-glutamyl transpeptidase (GGT), glutamate-oxaloacetate transaminase (GOT), and glutamic pyruvic transaminase (GPT) (Wako Bioproducts, Richmond, VA, USA) were all measured by standard enzymatic methods. Additionally, insulin was quantified by RIA provided by BioSource S.A. (Nivelles, Belgium). High-sensitivity C-reactive protein (CRP) levels were measured by ELISA kit from BLK Diagnostics (Badalona, Spain). Leptin and adiponectin were analyzed by enzyme immunoassay (ELISA) kits (DSL, Webster, TX, and DRG Diagnostics GmbH, Germany, respectively). Glucagon-like peptide-1 (GLP-1) was measured by a human GLP-1 enzyme immunoassay (EIA) kit from Phoenix Pharmaceuticals (Karlsruhe, Germany). Pancreatic peptide YY (PYY) was measured using a human PYY EIA kit from Phoenix Pharmaceuticals (Karlsruhe, Germany). The HOMA-IR was calculated from fasting insulin and glucose with the following equation: HOMA-IR = fasting insulin (mIU/mL)/fasting glucose (mol/L)/22.5.
Intravenous glucose tolerance test
An intravenous glucose tolerance test (IVGTT) was performed as previously described (Soriguer et al., 2009; Garcia-Serrano et al., 2015). The insulin sensitivity index (SI) was calculated after introduction of the results for glucose and insulin obtained during the IVGTT into the MINMOD program (version 3.0, 1994, Richard N. Bergman).
Anthropometric measurements
Body weight, height, waist and hip circumferences were measured according to standardized procedures [16]. BMI was calculated as weight (kilograms) divided by height (meters) squared.
Visceral adipose tissue mRNA
Visceral adipose tissue (VAT) was obtained during bariatric surgery in morbidly obese patients. The biopsy samples were washed in physiological saline buffer and immediately frozen in liquid nitrogen until analysis. Frozen adipose tissue was homogenized with an Ultra-Turrax 8 (Ika, Staufen, Germany). Total RNA was extracted by RNeasy lipid tissue midi kit (QIAGEN Science, Hilden, Germany), and treated with 55 U RNase-free deoxyribonuclease (QIAGEN Science, Hilden, Germany) following the manufacturer’s instructions. RNA purity was determined by 260/280 absorbance ratios on a Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific Inc. Waltham, MA). Total purified RNA integrity was checked by denaturing agarose gel electrophoresis and ethidium bromide staining. Total RNA was reverse transcribed to cDNA by a high-capacity cDNA reverse transcription kit with RNase inhibitor (Applied Biosystems, Foster City, CA). Quantitative real-time PCR with duplicates was done with the cDNA. The amplifications were performed using a MicroAmpH Optical 96-well reaction plate (Applied Biosystems, Foster City, CA) on an ABI 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA). Commercially available and pre-validated TaqMan® primer/probe sets were used as follows: cyclophilin A (4333763, RefSeq NM_002046.3), used as endogenous control for the target gene in each reaction, TNF alpha (Hs00174128_m1, RefSeq NM_000594.2), IL-6 (Hs00174131_m1, RefSeq NM_000600.2), IL-1β (Hs00174097_m1, RefSeq NM_000576.2), complement component 3 receptor 3 subunit (CD11b) (Hs01064804_m1, RefSeq NM_000632.3), insulin receptor substrate 1 (IRS-1) (Hs00178563_m1, RefSeq. NM_005544.2), insulin receptor substrate 2 (IRS-2) (Hs00275843_s1, RefSeq. NM_003749.2). A threshold cycle (Ct value) was obtained for each amplification curve and a ΔCt value was first calculated by subtracting the Ct value for human cyclophilin A cDNA from the Ct value for each sample and transcript. Fold changes compared with the endogenous control were then determined by calculating 2-ΔCt.
RNA extraction from cecal appendix
Total RNA was extracted from cecal appendix samples using a commercially available TriPure Isolation Reagent (Roche) and treated with DNase (RNase-free DNase Set; Qiagen). The RNA concentration was determined by absorbance at 260 nm (A260), and the purity was estimated by determining the A260/A280 ratio with a Nanodrop spectrophotometer (Nanodrop Technologies, Wilmington, DE). Denaturing agarose gel electrophoresis and ethidium bromide staining were used in order to check the integrity of total purified RNA. cDNA was synthesized using SuperScript II reverse transcriptase and random hexamer primers as described in the manufacturer’s protocol (Invitrogen Corp).
PCR amplification and analysis of 16S rRNA sequences
Variable regions 2-3 of the 16S rRNA gene were amplified using TaKaRa Ex Taq PCR mixture (TAKARA Bio USA, Madison, WI) and the PCR primers HDA1 (5’-GACTCCTACGGGAGGCAGCAGT-3’) and HDA2 (5’-GTATTACCGCGGCTGCTGGCAC-3’). Forward primers were designed with the adaptor A sequence (CGTATCGCCTCCCTCGCGCCA) plus a key sequence (TCAG) and reverse primers with the adaptor B sequence (CTATGCGCCTTGCCAGCCCG) plus a key sequence (TCGA). 454-adaptors were included in the forward primer followed by a 10 bp sample-specific Multiplex Identifier (MID). The PCR program was set as follows: 95°C 10 min and 30 cycles of 95°C 1 min, 50°C 1 min, 72°C 1.5 min followed by 72°C for 10 minutes. Agarose gel electrophoresis was performed and PCR products purified twice with Agencourt AMPure Kit (Beckman Coulter, Milan, Italy) and quantified using the Quant-iT™ PicoGreen® dsDNA Assay kit (Invitrogen, Burlington, ON). An equimolar pool was obtained prior to further processing. This equimolar pool was sequenced in a GS Junior 454 platform according to the manufacturer’s protocols using Titanium chemistry (Roche Applied Science, Indianapolis, IN).
Bioinformatics analysis
454 pyrosequencing data were analyzed using QIIME 1.8.0 software [17]. Raw reads were first filtered following the 454 amplicon processing pipeline. The pyrosequencing reads were demultiplexed and further filtered through the split_library.py script of QIIME. Reads with an average quality score lower than 25, ambiguous base calls, primer mismatches or shorter than 100 bp were excluded from the analysis in order to increase the level of accuracy. After the quality filter, the pipeline analysis used to analyze 16S gene reads was the following: sequences were denoised and singletons excluded. Operational taxonomic units (OTUs) were picked by clustering sequences at a similarity of >97% and the representative sequences, chosen as the most abundant in each cluster, were submitted to the UCLUST to obtain the taxonomy assignment and the relative abundance of each OTU using the Greengenes 16S rRNA gene database. Alpha and beta diversity were evaluated through QIIME, as described [18].
Statistical analysis
Group abundance was determined with a g-test using the group_significance.py script within QIIME in order to test whether the presence/abundance of any OTUs was significantly associated with a specific group. An Anosim statistical test through the compare_category.py script of QIIME was performed with a weighted UniFrac distance matrix in order to verify whether there were differences between the two types of patients. Comparisons between the results of the morbidly obese patients with high- and low-insulin-resistance were made with the Mann-Whitney test with a Bonferroni post hoc test. The Spearman correlation coefficients were calculated to estimate the correlations between variables. Statistical analyses were carried out with the statistical software package SPSS version 15.0 (SPSS Inc., Chicago, IL, USA). Values were considered to be statistically significant when P<0.05. The results are given as the mean ± SD.
Results
Biochemical and clinical characteristics together with gene expression in visceral adipose tissue in both study groups
The metabolic and anthropometric characteristics of the two study groups are shown in Table 1. The plasma levels of triglycerides, insulin, glucose, CRP, leptin and the HOMA-IR values were all significantly higher in the IR-MO group than in the NIR-MO group (P<0.05). Conversely, the SI, GLP-1, PYY and adiponectin levels were significantly higher in the NIR-MO group compared to the IR-MO group (P<0.05).
Table 1.
Biochemical and clinical characteristics of both study groups together with the expression of inflammatory cytokine and macrophage infiltration genes in visceral adipose tissue
| NIR-MO patients N=5 | IR-MO patients N=5 | * P | |
|---|---|---|---|
| Age (years) | 48.0±10.5 | 44.40±8.64 | 0.570 |
| BMI (kg/m2) | 59.18±4.71 | 58.20±4.12 | 0.735 |
| Waist circumference (cm) | 146.60±11.71 | 151.40±14.56 | 0.580 |
| Hip circumference (cm) | 162.67±14.36 | 162.20±17.03 | 0.964 |
| SBP (mmHg) | 132.00±23.28 | 141.0±27.87 | 0.595 |
| DBP (mmHg) | 82.40±10.90 | 86.60±10.13 | 0.546 |
| Total cholesterol (mg/dl) | 211.60±16.34 | 207.80±18.70 | 0.741 |
| HDL cholesterol (mg/dl) | 51.60±11.80 | 47.40±11.30 | 0.581 |
| LDL cholesterol (mg/dl) | 130.23±17.67 | 127.33±19.12 | 0.810 |
| Triglycerides (mg/dl) | 89.04±13.46 | 159.39±15.9 | 0.001 |
| Insulin (mg/dl) | 8.92±1.74 | 26.93±3.85 | 0.001 |
| Glucose (mg/dl) | 94.40±4.36 | 104.80±3.63 | 0.003 |
| HOMA-IR | 2.18±0.53 | 8.34±1.24 | 0.001 |
| SI (10-4 min-1/(μU/ml) | 2.96±2.33 | 0.84±0.80 | 0.020 |
| GOT (U/l) | 27.50±6.47 | 20.40±5.77 | 0.104 |
| GPT (U/l) | 39.00±15.05 | 42.80±16.76 | 0.716 |
| GGT (U/l) | 47.27±16.00 | 42.20±13.84 | 0.649 |
| CRP (mg/L) | 3.62±0.91 | 5.82±0.52 | 0.002 |
| Leptin (ng/ml) | 59.25±11.75 | 95.08±15.88 | 0.001 |
| Adiponectin (ug/ml) | 13.52±3.54 | 7.44±1.44 | 0.007 |
| PYY | 0.55±0.17 | 0.34±0.10 | 0.044 |
| GLP1 | 88.80±9.67 | 56.40±11.52 | 0.001 |
| IL6_V | 0.07±0.02 | 0.27±0.07 | 0.001 |
| IL1B_V | 0.08±0.02 | 0.28±0.07 | 0.001 |
| TNF_alpha_V | 0.002±0.0009 | 0.006±0.002 | 0.004 |
| IRS1_V | 0.014±0.001 | 0.005±0.001 | 0.001 |
| IRS2_V | 0.59±0.15 | 0.63±0.08 | 0.633 |
| CD11b_V | 0.12±0.04 | 0.35±0.10 | 0.001 |
Values are presented as means ± SD. N=5 subjects per group. DBP, Diastolic blood pressure; SBP, Systolic blood pressure; SI, Insulin sensitivity; GGT, Gamma-glutamyl transferase; GOT, Glutamic oxaloacetic transaminase; GPT, Glutamic pyruvic transaminase; CRP, C-reactive protein. Values are significantly different for
P<0.05.
We also found significantly higher mRNA expression levels in visceral adipose tissue of IL6, IL1B, TNF alpha, IRS2 and CD11b in the IR-MO group as compared with the NIR-MO group (P<0.05). Only the visceral adipose tissue expression levels of IRS1 were significantly higher in the NIR-MO group (P<0.05).
Analysis of the microbial community diversity between the study groups
A total 45,820 good quality 16S rRNA gene sequences with an average of 4,582±2,674.45 sequences per sample passed the filters applied through QIIME. To obtain a detailed structural overview of the microbiome of each study subject, an OTU analysis was performed. The microbiota of all appendix samples after QIIME were composed of 1128 OTUs with a relative abundance higher than 1% in at least two samples (97% similarity cut-off).
Prior to assessing alpha and beta diversity measures, samples were rarefied to 302 seq, which corresponded to the lowest number of quality reads obtained from any individual sample in the data set. The IR-MO group had a greater number of OTUs than the NIR-MO group (mean = 303 versus mean = 282, respectively; P>0.05).
The average diversity (Shannon index) and community richness (Chao index) were calculated to estimate the alpha diversity. The Chao and Shannon indices of each group suggested a similar bacterial richness and diversity in the appendix samples from the NIR-MO group compared to those from the IR-MO group, with no significant differences (P=0.89 and P=0.59, respectively) (Table 2). The rarefaction curve of observed OTUs calculated at 3% distance started to plateau at approximately 100 reads, suggesting that a higher number of reads per sample would not have provided a more comprehensive catalogue of bacterial taxa.
Table 2.
Chao1 richness estimator and Shannon diversity index among microbial communities obtained from appendix samples from IR-MO and NIR-MO patients
| IR-MO patients (n=5) | NIR-MO patients (n=5) | P | |
|---|---|---|---|
| Chao 1 | 99.19±29.03 | 102.0±36.51 | 0.89 |
| Shannon | 3.69±0.91 | 3.34±0.92 | 0.59 |
The Chao richness estimator and Shannon diversity index were calculated at 3% distance.
The unweighted UnifracPCoA Principal analysis based on the relative abundance of OTUs for each sample showed useful information about the phylogenetic relationships in the appendix bacterial microbiota in both study groups. Not all the samples from the IR-MO group were totally separate from the NIR-MO group in the combinations of coordinates, as demonstrated by the two principal component scores, which accounted for 23% and 16% of total variations. ANOSIM with permutations revealed no significant differences between groups (P=0.46), indicating no notable separation between the two groups (Figure 1).
Figure 1.
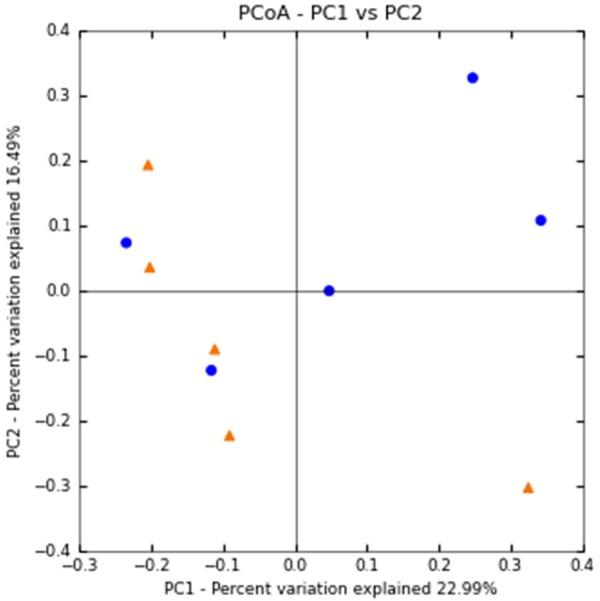
Principal component analysis of bacterial communities from appendix samples subjected to 454 sequencing. Positions of the bacterial communities for each species along the two first principal coordinate axes are illustrated, along with the percentage of variation explained by each axis. Results are based on unweighted Unifrac metrics. IR-MO samples (n=5) represented by red dots; NIR-MO samples (n=5) represented by blue dots.
16S rRNA gene pyrosequencing of samples from patients with high and low insulin resistance
In this study there were variations in the composition of appendix microbiota in the IR-MO and NIR-MO groups at different bacterial levels. At the phylum level, Firmicutes (47.98% IR vs. 21.45% NIR, P<0.001) and Fusobacteria (7.35% IR vs. 1.56% NIR, P<0.001) were significantly more abundant in the IR-MO patients, whereas Bacteroidetes (37.25% IR vs. 51.67% NIR, P=0.02) and Proteobacteria (7.23% IR vs. 25.90% NIR, P<0.001) were significantly predominant in the NIR-MO patients. The remainder of the bacterial population belonged to 9 other phyla that had a relative abundance lower than 1% (Figure 2). In addition, the Firmicutes/Bacteroidetes ratio was significantly different between study groups (1.28% IR vs. 0.41% NIR, P<0.001).
Figure 2.
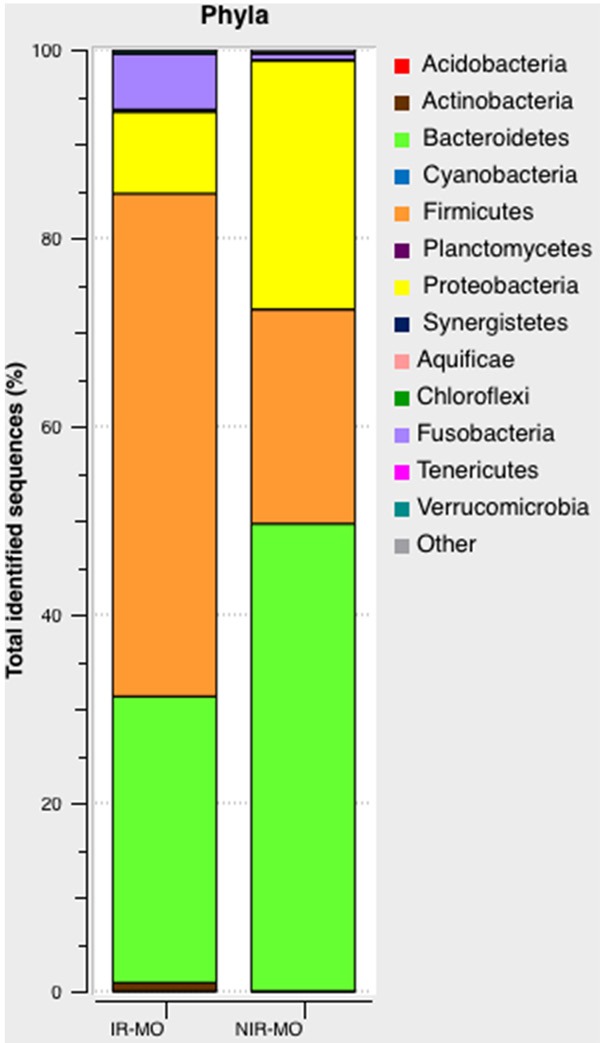
Pyrosequencing analysis of phyla in the IR-MO and NIR-MO groups. Data are shown as a percentage of the total identified sequences per group.
Within the 22 families detected among all the groups, 16 families were detected with a relative abundance greater than 1% in at least one group. In the IR-MO group we found a significant increase in Pseudomonaceae (27.83% IR vs. 20.72% NIR, P<0.001), Prevotellaceae (7.89% IR vs.0.46% NIR, P=0.01), and moreover in the unique family within the phyla Fusobacteria, Fusobacteriaceae (P<0.001); and a significant decrease in the abundance of Bacteroidaceae (49.92% IR vs. 70.41 % NIR, P=0.03), Lachnospiraceae (36.11% IR vs. 43.26% NIR, P<0.001), Ruminococcaceae (34.85% IR vs. 43.61% NIR, P<0.001) and Enterobacteriaceae (20.00% IR vs. 36.04% NIR, P<0.001). No significant differences between study groups were found in other abundant families, including Erysipelorichaceae (5.56% IR vs. 5.08% NIR, P=0.064), Odoribacteraceae (7.25% IR vs. 8.04% NIR, P=0.659), Porphyromonadaceae (0.48% IR vs. 2.65% NIR, P=0.108), Christensenellaceae (3.03% IR vs. 0.70% NIR, P=0.783), Rikenellaceae (34.30% IR vs. 17.53% NIR, P=0.297), Alcaligenaceae (20.87% IR vs. 4.68% NIR, P=589), Succinovibrionaceae (19.13% IR vs. 1.5% NIR, P=0.982), Veillonellaceae (14.39% IR vs. 5.08% NIR, P=0.984), and Peptococcaceae (1.26% IR vs. 0.53% NIR, P=0.668) (Figure 3).
Figure 3.
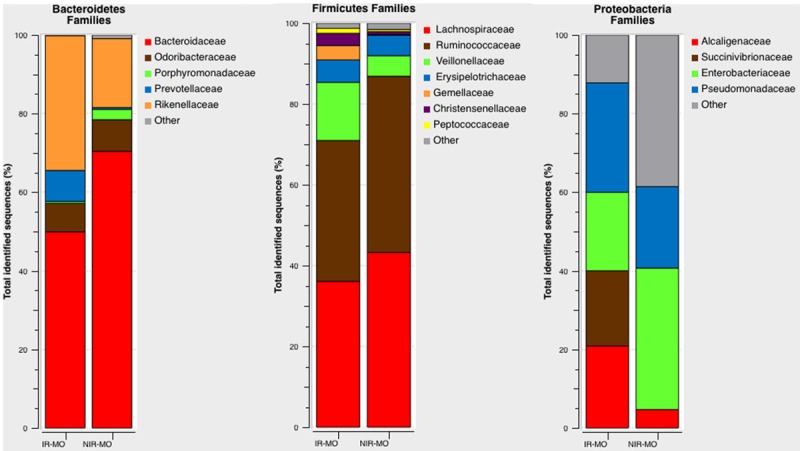
Family-level microbial classification of bacteria from the IR-MO and NIR-MO appendix samples. Data are shown as a percentage of the total identified sequences per group.
We also found significant differences in microbial composition at the genus level between the study groups. A total of 28 genera were identified among the appendix samples, with six significantly different genera between the IR-MO and the NIR-MO groups. Pseudomonas (27.83% IR vs. 20.72% NIR, P<0.001), Catenibacterium (3.54% IR vs. 0.88% NIR, P=0.027), Prevotella (7.89% IR vs. 0.46% NIR, P=0.01), Veillonella (2.27% IR vs. 0% NIR, P<0.001) and Fusobacterium (P<0.001) were significantly greater in the IR-MO group within the constitutive genera over 1% of the phylum bacteria in the appendix samples. On the other hand, Bacteroides (49.76% IR vs. 67.85% NIR, P=0.05) and Blautia (5.56% IR vs. 8.41% NIR, P=0.013) exhibited a significantly higher abundance in the NIR-MO patients compared to the IR-MO patients. In addition, the genera Odoribacter (6.12% IR vs. 5.48% NIR, P=0.702), Sutterella (20.87% IR vs. 4.68% NIR, P=0.589), Succinovibrio (19.13% IR vs. 0% NIR, P=1.000), Lachnospira (2.02% IR vs. 1.75% NIR, P=0.356), Oscillospira (4.04% IR vs. 1.05%, P=0.770), and Ruminococcus (3.03% IR vs. 0.35% NIR, P=0.637) dominated the IR-MO group, while Butyricimonas (1.13% IR vs. 2.56% NIR, P=0.395), Parabacteroides (0.48% IR vs. 2.65% NIR, P=0.107), Megamonas (0.25 IR vs. 1.75% NIR, P=0.650) and Phascolarctobacterium (1.26% IR vs. 3.33% NIR, P=0.956) dominated the NIR-MO group. Differences in abundance for all these minority genera between the IR-MO and NIR-MO groups were not significant (P>0.05 in all cases) (Figure 4).
Figure 4.
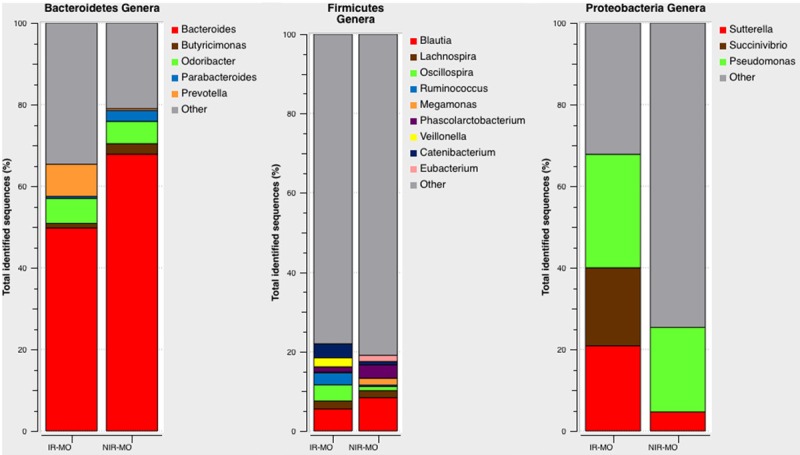
Relative abundance of bacterial genera in the microbiota of the IR-MO and NIR-MO patients. Data are shown as a percentage of the total identified sequences per group.
QIIME software generally provides confident phylogenetic assignments of DNA sequences down to the taxonomic level of the genus [19]. We were able to detect the following taxa: Prevotella stercorea, Mitsuokella multacida and Veilloneilla disparin in the IR-MO group.
Relationship between the gutmicrobiota composition, clinical parameters and inflammatory gene expression in the visceral adipose tissue from both study groups
The IR-MO patients had a significant univariate correlation between the abundance of specific bacterial groups and the plasma glucose levels (Butyricimonas r=-0.918, P=0.028), plasma insulin levels (Bifidobacterium r=-0.975, P=0.005), the expression levels of IL6 (Prevotella r=0.921, P=0.026), IL1B (Firmicutes r=0.963, P=0.009), TNF alpha (Succinovibrio r=0.975, P=0.005) and CD11b (Veillonella r=0.894, P=0.041). No significant correlation was found in the NIR-MO group (P<0.05).
Discussion
This study is the first to use appendix samples to establish that IR-MO and NIR-MO patients have a specific gut microbial profile using next-generation sequencing technologies. While limited data are available on the microbial composition of the appendix, it has been postulated that this organ could serve as a microbial reservoir for repopulating the gastrointestinal tract in times of necessity [12]. The associated lymphoid tissue of the appendix has been recognized to provide an ideal environment for bacterial growth in biofilms acting as an enteric reservoir [20,21]. The aforementioned arguments thus strongly suggest the importance of focusing on the appendix microbiota as a novel actor capable of modulating host metabolism via shaping the appendix immune function. In this study, analyzing therelationship between the microbiota composition and insulin resistance in both groups of morbidly obese patients, we considered confounding factors such as BMI, race, gender, age anddietary intake of the study subjects.
Our results indicate that the IR-MO patients have no increased bacterial diversity, as there was no separate cluster when compared to the NIR-MO group, clearly indicated by OTU based PCoA plot. However, the pyrosequencing analysis of the 16S rRNA gene sequences of the relative abundance of predominant phyla, family and genera taxa in the appendix samples revealed large significant differences between the IR-MO and the NIR-MO patients. At the family and genera level, we found that there was a significant increase in the abundance of Pseudomonaceae, Prevotellaceae, Fusobacteriaceae, Pseudomonas, Prevotella, Catenibacterium, Veillonella, and Fusobacterium and a decrease in Bacteroidaceae, Lachnospiraceae, Ruminococcaceae, Enterobacteriaceae, Bacteroides and Blautia in the appendix samples from the IR-MO group. These results suggest that the dominant microbiota are different in the IR-MO patients with respect to the NIR-MO patients. Alterations in the gut microbiota have also been suggested to occur in other human diseases such as Crohn’s disease, ulcerative colitis [22-24], celiac disease in children [25], and allergic inflammation in infants [26]. The morbidly obese patients with an IR microbiota phenotype also had a higher rate of systemic inflammation, with a significant increase in CRP and leptin levels and a decrease in serum adiponectin levels. Recent data have shown that variations in intestinal microbiota are associated with pro-inflammatory changes in adipose gene expression [27]. In this study we also found a significant increase in the expression level of inflammatory markers such as IL6, IL1B and TNF alpha in the VAT of the IR-MO patients. Moreover, there was a significant positive correlation between the abundance of Prevotella, Succinovibrio and Firmicutes and the VAT expression levels of IL6, TNF alpha and IL1B, respectively. Several years ago, Burcellin et al. suggested that a so-called ‘metabolic infection’ might exist, suggesting that the gut microbiota might be an important factor in low-grade systemic inflammation and the development of insulin resistance. In their study using animal models, the authors demonstrated that prior to the onset of diabetes, and soon after the mice are switched to a high-fat diet, intestinal bacteria translocate from the gut to the adipose tissue and blood, where they might induce low-grade inflammation [28]. In addition, TNF alpha is able to phosphorylate serine residue substrate from the IRS-1, leading to its inactivation [29].
Based on these data, the presence of a local appendix dysbiosis and the inflammation triggered by the intestinal bacteria in the IR-MO group might be pathological and could be linked to the development of insulin resistance in morbidly obese patients. In accordance with our results, Meadows described that Firmicutes are linked to weight gain and insulin resistance because they provide a source of extra calories by breaking down polysaccharides that are otherwise indigestible in mammals [30]. Verdam et al., with results similar to ours, showed that a decrease in Bacteriodetes and an increase in Firmicutes is associated with the presence of obesity and inflammation in humans [11]. Fusobacterium spp. has also been considered a pro-inflammatory passenger bacteria in the origin and proliferation of human colorectal cancer [31].
In this study, the significant increase in the abundance of Prevotella suggests an evolution of a mucin-degrading niche in the IR-MO group. Mucin degradation by bacteria is often regarded as an initial stage in pathogenesis [32], since it would disturb the protection of the host mucosal surfaces and potentially lead to a disruption in the epithelial barrier, increasing intestinal permeability, facilitating bacterial translocation and a paracellular flux of lipopolysaccharides (LPS) able to induce inflammation in the host [33,34], as we thought might possibly occur in the IR-MO patients. In addition, the significant increase in the VAT expression of CD11b in the IR-MO group may be due to this increase in permeability, which would trigger an immune response, inflammation, and immune cell (such as macrophages) infiltration in the adipose tissue. Moreover, we observed a significant positive correlation between the VAT expression of CD11b and the relative abundance of Veillonella in the IR-MO group. These bacteria are able to ferment glucose and lactate to propionate, acetate and succinate; however, these short fatty acids do not induce mucin synthesis [35], which could result in a reduction of the tight junction assembly, generating an increase in gut permeability. This situation is able to induce insulin resistance and also reduce the levels of gut-secreted hormones, such as GLP-1 and PYY, as we have described in the IR-MO patients in our study. One possible mechanism to explain the significant decrease in secretion of GLP1 and PYY in the IR-MO patients may be that its secretion is modulated by the short-chain fatty acids (SCFA) produced by the altered gut microbiota. The significant decrease in the abundance of Lachnospiraceae and Ruminococcaceae found in this study in the IR-MO group is very relevant because both families are able to degrade complex polysaccharides to SCFA, including acetate, butyrate, and propionate that can be used for energy by the host [36]. Moreover, these SCFA act as anti-inflammatory molecules, capable of inhibiting NF-κB activation in the host immune cells by binding to G-protein-coupled receptors (GPR43 and GPR41), thereby blocking inflammatory responses and suppressing TNF alpha and IL6 release [37]. In addition, previous studies have also shown that butyrate induces mucin synthesis [35], decreases bacterial transport across the epithelium [38] and decreases intestinal epithelial permeability by increasing the expression of tight junction proteins [39].
Butyrate producing intestinal bacteria also seem to play an important role in blood glucose regulation and lipid metabolism, as shown by fecal transplantation studies [40]. In the present study we found negative and significant correlations between the abundance of bacteria, such as Butyricimonas (butyrate producers with anti-inflammatory effects) and Bifidobacterium, and the plasma glucose and insulin levels, respectively, in the IR-MO patients. In previous studies, the levels of Bifidobacterium have also been related to improved glucose metabolism, insulin resistance and low-grade inflammation [11,41]. Furthermore, improved insulin sensitivity was found after infusion of butyrate-producing intestinal microbiota from lean donors to male subjects with the metabolic syndrome [40].
Our results also demonstrate that the abundance of bacteria that can act as opportunistic pathogens, such as Pseudomonas and Sutterella, was elevated in the appendix samples from the IR-MO patients compared with the NIR-MO patients. Williams et al. found high detection rates of Sutterella in gastrointestinal biopsies from autistic children with a gastrointestinal disturbance. These authors indicated that it is not yet evident what the consequences of this increase in fecal Sutterella population means, but it is possible that under specific conditions these bacteria could cause infection [42]. Previous reports have shown that Pseudomonadaceae were significantly increased in the stools of patients with end-stage renal disease [43] and Geng et al. considered the Pseudomonadaceae family as a new potential driver bacteria in human colorectal cancer progression [44].
In conclusion, our data support the hypothesis that appendix dysbiosis occurs in the IR-MO group. Moreover, the IR-MO patients showed a loss of butyrate-producing bacteria, essential to maintain gut integrity, together with an increase in mucin-degrading bacteria and opportunistic pathogens. Finally, the significant increase in the expression of inflammatory cytokines and macrophage infiltration genes in adipose tissue, possibly triggered by the microbiota present in the IR-MO group, may suggest that these specific bacteria could initiate the inflammation and the insulin resistance associated with obesity in these patients. These findings could be useful for developing strategies to control the development of insulin resistance by modifying the gut microbiota.
Acknowledgements
This work was supported by grant from the Fondo de Investigación Sanitaria of Instituto de Salud Carlos III and cofounded by Fondo Europeo de Desarrollo Regional FEDER (PI15/01114). The research group belongs to the “Centros de Investigación en Red” [CIBER, CB06/03/0018] of the “Instituto de Salud Carlos III”. Isabel Moreno Indias was supported by a “Sara Borrell” Postdoctoral contract (CD12/00530), Maria Isabel Queipo-Ortuño acknowledges support from the “Miguel Servet Type I” program (CP13/00065) and Fernando Cardona acknowledges support from the “Miguel Servet Type II” program (CP13/00023) from the Instituto de Salud Carlos III, Madrid Spain, co-founded by Fondo Europeo de Desarrollo Regional-FEDER.
Disclosure of conflict of interest
None.
Authors’ contribution
MI, SA, GF, CF, QO and FT contributed to the conception, design, analysis, and interpretation of data for the work. MI, SA, GF, CF, QO and FT drafted and revised the intellectual content of the work and approved the final version to be published. They are agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Abbreviations
- BMI
body mass index
- CRP
high-sensitivity C-reactive protein
- DBP
diastolic blood pressure
- GLP-1
glucagon-like peptide-1
- GPR43 and GPR41
G-protein-coupled receptor
- HOMA-IR
homoeostasis model assessment of insulin resistance
- IL
interleukin
- IR-MO
high-insulin-resistant group
- IRS-1
insulin receptor substrate
- IRS-2
insulin receptor substrate 2
- LPS
lipopolysaccharides
- NIR-MO
low-insulin-resistant group
- OTUs
operational taxonomic units
- PCR-DGGE
polymerase chain reaction denaturing gradient gel electrophoresis
- PYY
pancreatic peptide YY
- SBP
systolic blood pressure
- SCFA
short-chain fatty acids
- TNF alpha
tumor necrosis factor alpha
- VAT
Visceral adipose tissue
References
- 1.Carvalho BM, Saad MJ. Influence of gut microbiota on subclinical inflammation and insulin resistance. Mediators Inflamm. 2013;2013:986734. doi: 10.1155/2013/986734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Bandt JP, Waligora-Dupriet AJ, Butel MJ. Intestinal microbiota in inflammation and insulin resistance: relevance to humans. Curr Opin Clin Nutr Metab Care. 2011;14:334–340. doi: 10.1097/MCO.0b013e328347924a. [DOI] [PubMed] [Google Scholar]
- 3.Shoelson SE, Lee J, Goldfine AB. Inflammation and insulin resistance. J Clin Invest. 2006;116:1793–1801. doi: 10.1172/JCI29069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caricilli AM, Saad MJ. The role of gut microbiota on insulin resistance. Nutrients. 2013;5:829–851. doi: 10.3390/nu5030829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stachowicz N, Kiersztan A. [The role of gut microbiota in the pathogenesis of obesity and diabetes] . Postepy Hig Med Dosw (Online) 2013;67:288–303. doi: 10.5604/17322693.1044746. [DOI] [PubMed] [Google Scholar]
- 6.Tagliabue A, Elli M. The role of gut microbiota in human obesity: Recent findings and future perspectives. Nutr Metab Cardiovasc Dis. 2013;23:160–168. doi: 10.1016/j.numecd.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Tilg H, Moschen AR. Microbiota and diabetes: an evolving relationship. Gut. 2014;63:1513–21. doi: 10.1136/gutjnl-2014-306928. [DOI] [PubMed] [Google Scholar]
- 8.Cani PD, Osto M, Geurts L, Everard A. Involvement of gut microbiota in the development of low-grade inflammation and type 2 diabetes associated with obesity. Gut Microbes. 2012;3:279–288. doi: 10.4161/gmic.19625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amar J, Burcelin R, Ruidavets JB, Cani PD, Fauvel J, Alessi MC, Chamontin B, Ferrieres J. Energy intake is associated with endotoxemia in apparently healthy men. Am J Clin Nutr. 2008;87:1219–1223. doi: 10.1093/ajcn/87.5.1219. [DOI] [PubMed] [Google Scholar]
- 10.Cani PD, Possemiers S, Van de Wiele T, Guiot Y, Everard A, Rottier O, Geurts L, Naslain D, Neyrinck A, Lambert DM, Muccioli GG, Delzenne NM. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut. 2009;58:1091–1103. doi: 10.1136/gut.2008.165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verdam FJ, Fuentes S, de Jonge C, Zoetendal EG, Erbil R, Greve JW, Buurman WA, de Vos WM, Rensen SS. Human intestinal microbiota composition is associated with local and systemic inflammation in obesity. Obesity (Silver Spring) 2013;21:E607–615. doi: 10.1002/oby.20466. [DOI] [PubMed] [Google Scholar]
- 12.Randal Bollinger R, Barbas AS, Bush EL, Lin SS, Parker W. Biofilms in the large bowel suggest an apparent function of the human vermiform appendix. J Theor Biol. 2007;249:826–831. doi: 10.1016/j.jtbi.2007.08.032. [DOI] [PubMed] [Google Scholar]
- 13.Rhee KJ, Jasper PJ, Sethupathi P, Shanmugam M, Lanning D, Knight KL. Positive selection of the peripheral B cell repertoire in gut-associated lymphoid tissues. J Exp Med. 2005;201:55–62. doi: 10.1084/jem.20041849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guinane CM, Tadrous A, Fouhy F, Ryan CA, Dempsey EM, Murphy B, Andrews E, Cotter PD, Stanton C, Ross RP. Microbial composition of human appendices from patients following appendectomy. MBio. 2013;4 doi: 10.1128/mBio.00366-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Serino M, Fernandez-Real JM, Garcia-Fuentes E, Queipo-Ortuno M, Moreno-Navarrete JM, Sanchez A, Burcelin R, Tinahones F. The gut microbiota profile is associated with insulin action in humans. Acta Diabetol. 2013;50:753–761. doi: 10.1007/s00592-012-0410-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lohman TG, Roche AF, Martorell R. Human Kinetics Books. Champaign, IL: 1988. Anthropometric standardization reference manual. [Google Scholar]
- 17.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Filippis F, La Storia A, Villani F, Ercolini D. Exploring the sources of bacterial spoilers in beefsteaks by culture-independent high-throughput sequencing. PLoS One. 2013;8:e70222. doi: 10.1371/journal.pone.0070222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bollinger RR, Barbas AS, Bush EL, Lin SS, Lin SS, Parker W. Biofilms in the normal human large bowel: fact rather than fiction. Gut. 2007;56:1481–1482. [PMC free article] [PubMed] [Google Scholar]
- 21.Bollinger RR, Everett ML, Palestrant D, Love SD, Lin SS, Parker W. Human secretory immunoglobulin A may contribute to biofilm formation in the gut. Immunology. 2003;109:580–587. doi: 10.1046/j.1365-2567.2003.01700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willing B, Halfvarson J, Dicksved J, Rosenquist M, Jarnerot G, Engstrand L, Tysk C, Jansson JK. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn’s disease. Inflamm Bowel Dis. 2009;15:653–660. doi: 10.1002/ibd.20783. [DOI] [PubMed] [Google Scholar]
- 24.Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z, Jarnerot G, Tysk C, Jansson JK, Engstrand L. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–1854. e1841. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 25.Nadal I, Donat E, Ribes-Koninckx C, Calabuig M, Sanz Y. Imbalance in the composition of the duodenal microbiota of children with coeliac disease. J Med Microbiol. 2007;56:1669–1674. doi: 10.1099/jmm.0.47410-0. [DOI] [PubMed] [Google Scholar]
- 26.Kalliomaki M, Isolauri E. Role of intestinal flora in the development of allergy. Curr Opin Allergy Clin Immunol. 2003;3:15–20. doi: 10.1097/00130832-200302000-00003. [DOI] [PubMed] [Google Scholar]
- 27.Kong LC, Tap J, Aron-Wisnewsky J, Pelloux V, Basdevant A, Bouillot JL, Zucker JD, Doré J, Clément K. Gut microbiota after gastric bypass in human obesity: increased richness and associations of bacterial genera with adipose tissue genes. Am J Clin Nutr. 2013;98:16–24. doi: 10.3945/ajcn.113.058743. [DOI] [PubMed] [Google Scholar]
- 28.Amar J, Chabo C, Waget A, Klopp P, Vachoux C, Bermudez-Humaran LG, Smirnova N, Berge M, Sulpice T, Lahtinen S, Ouwehand A, Langella P, Rautonen N, Sansonetti PJ, Burcelin R. Intestinal mucosal adherence and translocation of commensal bacteria at the early onset of type 2 diabetes: molecular mechanisms and probiotic treatment. EMBO Mol Med. 2011;3:559–572. doi: 10.1002/emmm.201100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cani PD, Delzenne NM. The role of the gut microbiota in energy metabolism and metabolic disease. Curr Pharm Des. 2009;15:1546–1558. doi: 10.2174/138161209788168164. [DOI] [PubMed] [Google Scholar]
- 30.Meadows R. Gut Bacteria May Override Genetic Protections against Diabetes. PLoS Biol. 2011;9:e1001215. doi: 10.1371/journal.pbio.1001215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10:575–582. doi: 10.1038/nrmicro2819. [DOI] [PubMed] [Google Scholar]
- 32.Derrien M, van Passel MW, van de Bovenkamp JH, Schipper RG, de Vos WM, Dekker J. Mucin-bacterial interactions in the human oral cavity and digestive tract. Gut Microbes. 2010;1:254–268. doi: 10.4161/gmic.1.4.12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bischoff SC, Barbara G, Buurman W, Ockhuizen T, Schulzke JD, Serino M, Tilg H, Watson A, Wells JM. Intestinal permeability--a new target for disease prevention and therapy. BMC Gastroenterol. 2014;14:189. doi: 10.1186/s12876-014-0189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moreno-Indias I, Cardona F, Tinahones FJ, Queipo-Ortuno MI. Impact of the gut microbiota on the development of obesity and type 2 diabetes mellitus. Front Microbiol. 2014;5:190. doi: 10.3389/fmicb.2014.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burger-van Paassen N, Vincent A, Puiman PJ, van der Sluis M, Bouma J, Boehm G, van Goudoever JB, van Seuningen I, Renes IB. The regulation of intestinal mucin MUC2 expression by short-chain fatty acids: implications for epithelial protection. Biochem J. 2009;420:211–219. doi: 10.1042/BJ20082222. [DOI] [PubMed] [Google Scholar]
- 36.Biddle A, Stewart L, Blanchard J, Leschine S. Untangling the Genetic Basis of Fibrolytic Specialization by Lachnospiraceae and Ruminococcaceae in Diverse Gut Communities. Diversity. 2013;5:627–640. [Google Scholar]
- 37.Lewis K, Lutgendorff F, Phan V, Soderholm JD, Sherman PM, McKay DM. Enhanced translocation of bacteria across metabolically stressed epithelia is reduced by butyrate. Inflamm Bowel Dis. 2010;16:1138–1148. doi: 10.1002/ibd.21177. [DOI] [PubMed] [Google Scholar]
- 38.Peng L, Li ZR, Green RS, Holzman IR, Lin J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J Nutr. 2009;139:1619–1625. doi: 10.3945/jn.109.104638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Udayappan SD, Hartstra AV, Dallinga-Thie GM, Nieuwdorp M. Intestinal microbiota and faecal transplantation as treatment modality for insulin resistance and type 2 diabetes mellitus. Clin Exp Immunol. 2014;177:24–29. doi: 10.1111/cei.12293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vrieze A, Van Nood E, Holleman F, Salojarvi J, Kootte RS, Bartelsman JF, Dallinga-Thie GM, Ackermans MT, Serlie MJ, Oozeer R, Derrien M, Druesne A, Van Hylckama Vlieg JE, Bloks VW, Groen AK, Heilig HG, Zoetendal EG, Stroes ES, de Vos WM, Hoekstra JB, Nieuwdorp M. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143:913–916. e917. doi: 10.1053/j.gastro.2012.06.031. [DOI] [PubMed] [Google Scholar]
- 41.Philippe D, Favre L, Foata F, Adolfsson O, Perruisseau-Carrier G, Vidal K, Reuteler G, Dayer-Schneider J, Mueller C, Blum S. Bifidobacterium lactis attenuates onset of inflammation in a murine model of colitis. World J Gastroenterol. 2011;17:459–469. doi: 10.3748/wjg.v17.i4.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams BL, Hornig M, Parekh T, Lipkin WI. Application of novel PCR-based methods for detection, quantitation, and phylogenetic characterization of Sutterella species in intestinal biopsy samples from children with autism and gastrointestinal disturbances. MBio. 2012;3 doi: 10.1128/mBio.00261-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vaziri ND, Wong J, Pahl M, Piceno YM, Yuan J, DeSantis TZ, Ni Z, Nguyen TH, Andersen GL. Chronic kidney disease alters intestinal microbial flora. Kidney Int. 2013;83:308–315. doi: 10.1038/ki.2012.345. [DOI] [PubMed] [Google Scholar]
- 44.Geng J, Song Q, Tang X, Liang X, Fan H, Peng H, Guo Q, Zhang Z. Co-occurrence of driver and passenger bacteria in human colorectal cancer. Gut Pathogens. 2014;6:26–26. doi: 10.1186/1757-4749-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]


