Cortical microtubules (CMTs) are essential for normal plant morphogenesis because they affect the axes of cell elongation and predict the placement of cell division planes. The function of the CMTs is intimately linked to their organizational state, which is subject to spatial and temporal modifications by developmental and environmental cues. CMT assembly dynamics govern when, where, and how microtubules appear in a cell's cortex, and the regulation of these properties affects their organization. However, the principles that link microtubule assembly dynamics to cortical array organization and reconfiguration are not well understood. This essay focuses on recent advances in the understanding of CMT assembly dynamics and how this new information provides insight into the dynamic behavior and intermicrotubule interactions that affect the self-organizational state of CMTs.
BACKGROUND
Plant CMT arrays consist of a population of relatively short, overlapping microtubules that are predominantly plasma membrane bound (Hardham and Gunning, 1978). These microtubules are typically arranged in specific patterns that are important for their functions. For example, during interphase the CMTs in a rapidly elongating cell are typically arranged transverse to the elongation axis of the cell. Once the accelerative phase of cell elongation slows down, the transverse CMTs reorient into a predominantly oblique or longitudinal direction with respect to the cell's elongation axis (Sugimoto et al., 2000; Granger and Cyr, 2001).
Pioneering studies that directly quantified plant CMT assembly dynamics by following the recovery kinetics of photobleached CMTs labeled with fluorescent tubulin analogs revealed the highly dynamic nature of CMTs (Hush et al., 1994). These studies led the way to an appreciation of CMT arrays as highly dynamic structures. However, this understanding simultaneously raised the following question: How does a large population of dynamic CMTs become organized into specific arrays that can persist for considerable lengths of time during development?
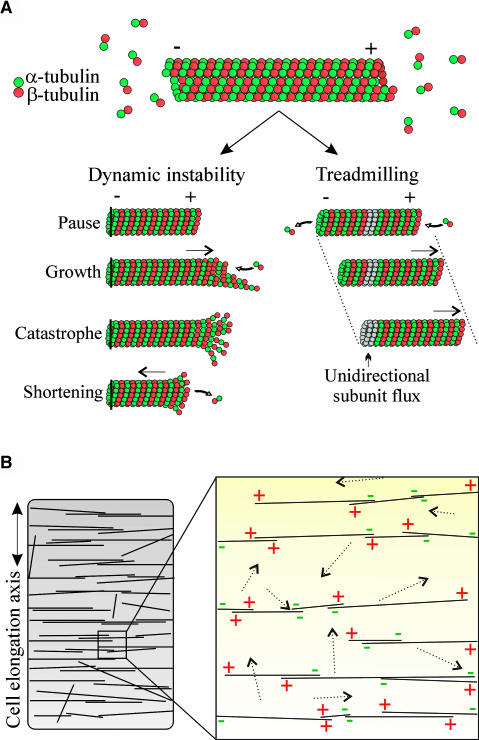
The polar nature of microtubules is an important consideration when trying to understand the relationship between microtubule assembly dynamics and organization. Microtubules possess an inherent polarity as a result of the head-to-tail assembly of the α/β-tubulin subunits during polymerization (Figure 1A). This structural polarity forms the basis for the kinetic polarity of microtubules, which possess a fast-growing end (designated as the plus-end) and a slow-growing end (designated as the minus-end). In animal cells, microtubules are typically nucleated from a centrally located centrosome that gives rise to a radial pattern of microtubules. Their minus-ends are typically anchored at the centrosome; therefore, the microtubules also display a uniform polarity with their plus-ends radiating toward the cell periphery. Therefore, in these cells, microtubule nucleation and organization are inextricably coupled.
Figure 1.
Microtubule Assembly Dynamics and Organization in Higher Plant Cells.
(A) Microtubules are composed of α/β-tubulin heterodimers that are assembled in a head-to-tail fashion to form linear protofilaments that associate laterally within the typical 13-protofilament, 25-nm microtubule. The β-tubulin end is fast growing and the more dynamic end (+end), whereas the α-tubulin end is slow growing and the less dynamic end (−end). Under conditions where the minus-ends of the microtubules are anchored, only the freely exposed plus-ends are dynamic and exhibit periods of growth and shortening with stochastic transitions between these two phases (dynamic instability). If both ends of the microtubules are freely accessible to the soluble tubulin subunits, then the assembly dynamics may be marked by net polymerization at the plus-ends and net depolymerization at the minus-ends, thereby leading to a directional subunit flux (shown by the gray subunits) and an apparent repositioning of the polymer over time (treadmilling).
(B) Plant CMTs form arrays in specific patterns. In a rapidly elongating cell, the CMTs are predominantly oriented transverse to the elongation axis. The individual CMTs are relatively short, and they bundle into a higher order structure that spans the circumference of the cell. CMTs in an elongating cell arise in random orientations throughout the cell's cortex (as depicted by the dotted arrows), but discordant microtubule orientations do not persist for long. Furthermore, the parallel alignment of CMTs is not based on a unidirectional orientation of their plus-ends. Therefore, CMT nucleation and organization are not tightly coupled in plant cells.
In contrast with animal cells, higher plant cells do not possess a centrosome as a microtubule organizing center. Instead, microtubule organizing center activity in these cells is localized to the nuclear surface and at dispersed sites in the cell's cortex (reviewed in Schmit, 2002). The dispersed nature of CMT organizing centers in higher plant cells accounts for the dispersed population of CMTs, but it is important to note that the ordered CMT array patterns are not associated with corresponding ordered patterns of CMT organizing centers. That is, CMT nucleation and organization are not tightly coupled events in higher plant cells: CMT organization occurs after microtubule nucleation (Wasteneys and Williamson, 1989; Hepler et al., 1993; Wasteneys et al., 1993; Yuan et al., 1994; Wymer et al., 1996). Additionally, recent studies have revealed that CMT arrays consist of microtubules with mixed plus-end orientations (Dhonukshe and Gadella, 2003; Shaw et al., 2003; Tian et al., 2004; Vos et al., 2004). Therefore, CMT array organization does not require a uniform orientation of the microtubule plus-ends (Figure 1B).
Given the absence of an ordered nucleation mechanism for CMT array organization, it seems reasonable to propose that CMTs self-organize into specific arrays. Self-organization is usually associated with systems consisting of a large number of nearly identical elements, existing in a nonequilibrium environment, undergoing numerous local interactions with each other. The CMT population satisfies these criteria, and information from recent studies of individual CMT assembly dynamics and their regulation through the activity of microtubule-associated proteins (MAPs) is providing the foundation for understanding CMT array organization in the context of CMT assembly dynamics.
CMT ASSEMBLY DYNAMICS
Microtubule assembly dynamics are characterized by periods of polymerization (microtubule growth) and depolymerization (microtubule shortening), with stochastic transitions between these two phases. This behavior is described quantitatively by the dynamic instability model in terms of at least four criteria: growth rate, shrinkage rate, rescue frequency (frequency of transitions from shrinkage to growth), and catastrophe frequency (frequency of transitions from growth to shrinkage). If the microtubule's minus-ends are anchored at their nucleating sites, only the freely exposed plus-ends of these microtubules exhibit dynamic instability (Figure 1A). Conversely, if the minus-ends of the microtubules are not anchored at their nucleating sites, then both microtubule ends are freely accessible to the soluble tubulin subunits and exhibit assembly dynamics. Regulation of the dynamic instability at the ends of unanchored microtubules (for example, as a result of the activity of MAPs) can lead to a specialized behavior termed microtubule treadmilling. Treadmilling microtubules typically display net polymerization at their plus-ends and net depolymerization at their minus-ends. This results in a subunit flux from the plus-end to the minus-end of treadmilling microtubules and a directional (with the plus-end leading) repositioning of the microtubules over time (Figure 1A).
Arabidopsis CMTs display a dual treadmilling/dynamic instability behavior: The minus-ends of the CMTs are freely exposed and display slow depolymerization interspersed with periods of inactivity, whereas their plus-ends exhibit polymerization-biased dynamic instability (Shaw et al., 2003). This type of CMT assembly dynamics is also seen in cultured tobacco cells (Vos et al., 2004), and it is therefore likely to be a common feature of higher plant cells.
This observation indicates that plant CMTs are not anchored at their minus-ends but rather are released from nucleation sites, presumably as a result of the action of the microtubule-severing protein katanin. Indeed, mutant Arabidopsis plants lacking an active catalytic katanin subunit show abnormally long microtubules radiating from the nuclei and microtubule converging centers in the cortex (Burk et al., 2001; Burk and Ye, 2002), consistent with the hypothesis that microtubule severing is required for the generation of unanchored CMTs.
In terms of microtubule organization, an important implication of the presence of free minus-ends is that these microtubules do not leave behind positional information after complete depolymerization. Therefore, the orientations of subsequent generations of CMTs are independent of the preexisting microtubules, and this property is likely to be important for the formation of ordered CMT arrays. The fact that Arabidopsis katanin mutants that possess abnormally long and presumably minus-end anchored CMTs show CMT array disorganization (Bichet et al., 2001; Burk et al., 2001; Burk and Ye, 2002; Bouquin et al., 2003) supports the hypothesis that the generation of unanchored minus-ends is important for facilitating ordered CMT arrays.
Microtubule translocation, as a result of motor activity, can result in the self-organization of microtubules into complex configurations such as astral arrays and bipolar spindles. Motor activity can cross-link and slide microtubules relative to each other, which can be envisioned to facilitate lateral associations of CMTs and result in parallel arrays. Plants possess a large number of uncharacterized motor proteins (Reddy, 2001), and it is possible that some of these have evolved to perform the specialized function of CMT organization. However, recent evidence indicates that motor-based translocation of plant CMTs does not occur. For example, experiments using photobleaching to mark Arabidopsis microtubules show that the motility of CMTs is not a result of motor-driven microtubule translocation but rather a result of microtubule treadmilling (Shaw et al., 2003). In addition, kymograph analysis of CMTs in cultured tobacco cells shows that they do not undergo lateral translocation (Vos et al., 2004). Although these reports do not conclusively rule out CMT translocation under certain circumstances, they suggest that motor activity may not be a significant component of CMT array organization.
Observations of CMT array organization, under both normal and experimental conditions, reveal that it occurs progressively, starting with a disorganized or regionally discordant CMT population, followed by new regional order, and resolving eventually into a new global order (Wasteneys and Williamson, 1989; Yuan et al., 1994; Granger and Cyr, 2001). This common mode of CMT organization suggests that there is one fundamental mechanistic route for CMT organization, and the progressive nature of this process is consistent with a self-organizing process.
In terms of self-organization models for the generation of ordered CMT arrays, one possibility is that intermicrotubule encounters between randomly nucleated CMTs deterministically modify the stochastic behavior of CMTs so as to foster a parallel arrangement. Here, the principal challenge will be to determine the rules that govern the outcome of the intermicrotubule encounters and decide whether they contribute to self-organization of CMT arrays. An alternative hypothesis envisions the self-organization of CMT arrays from branched (fractal) patterns of CMT assembly (Wasteneys, 2002). In this model microtubule nucleating complexes, generated through the periodic severing of CMT minus-ends, are transported along the lengths of preexisting CMTs and serve as templates for new CMT nucleation, thereby giving rise to clusters of branched CMTs. These clusters eventually dissociate into individual microtubules that become organized into parallel arrays. These two models are not mutually exclusive, and it is plausible that the organization of the individual CMTs, after cluster dissociation, occurs through intermicrotubule interactions.
REGULATION OF CMT ASSEMBLY DYNAMICS
The reconfiguration of the CMT arrays, in response to developmental and environmental cues, is probably as a result of regulated changes of their assembly dynamics. The direct regulation of microtubule assembly dynamics typically is achieved through the activity of MAPs; therefore, a key objective is to understand the mechanisms that regulate MAP activity and how these mechanisms are triggered in response to developmental and environmental stimuli. Several MAPs have been identified that are important for CMT organization, and these are providing the opportunity to dissect the relationship between CMT dynamics and organization.
An important MAP used to study CMT organization is Arabidopsis MICROTUBULE ORGANIZATION 1/GEMINI 1 (MOR1/GEM1), which is a member of the MAP215 family of microtubule stabilizing MAPs (Whittington et al., 2001). The tobacco homolog TOBACCO MICROTUBULE BUNDLING POLYPEPTIDE OF 200 kD (TMBP200) bundles microtubules (Yasuhara et al., 2002), and mutant Arabidopsis plants harboring temperature-sensitive mor1 alleles show short, disordered CMTs at the restrictive temperature (Whittington et al., 2001; Wasteneys, 2002). The mor1 mutant alleles result in single amino acid substitutions in an N-terminal HEAT repeat of the encoded MOR1 protein, which suggests that this domain is important for CMT organization (Hussey and Hawkins, 2001; Whittington et al., 2001). The correlation between shortened CMTs and array disorganization supports the notion that intermicrotubule encounters promote microtubule organization because a decrease in the average length of CMTs would be predicted to decrease the frequency of intermicrotubule encounters. Understanding the difference in microtubule assembly dynamics between mor1 mutant and wild-type plants may help to understand the mechanism of MOR1 action and how it contributes to microtubule organization. Interestingly, MOR1 bears a putative Cdk phosphorylation site (Hussey and Hawkins, 2001), which presents the possibility that it may be subject to cell cycle regulation.
From the perspective of the cell cycle, recent studies show that changes in CMT assembly dynamics are associated with the transition from interphase to G2/prophase (Dhonukshe and Gadella, 2003; Vos et al., 2004). In particular, these changes in CMT assembly dynamics are correlated with the transformation of the dispersed interphase cortical array to a highly bundled preprophase band of microtubules that typically encircles the nucleus and that predicts the site of cell division (Mineyuki, 1999). The molecular basis for the cell cycle, stage-specific changes in CMT dynamics is not known and may involve the regulation of MAP activity (such as MOR1) through the action of Cdk. In this regard, Cdk has been localized to the preprophase band (Colasanti et al., 1993; Weingartner et al., 2001), and the microinjection of an active Cdk complex into cells is associated with the stimulation of preprophase band breakdown (Hush et al., 1996).
MAPs that bundle or cross-link adjacent microtubules are of particular interest for CMT organization because microtubule bundling is an important feature of interphase cortical arrays and the preprophase band. CMT bundling is significant because it proposedly increases the stability of the bundled CMTs and allows relatively short CMTs to associate with one another to form a higher order structure that spans the circumference of the cell. Besides MOR1/TMBP200, some isoforms of the MAP65 family of MAPs also bundle microtubules (Chan et al., 1999; Smertenko et al., 2004; Wicker-Planquart et al., 2004). These MAP65 isoforms bind to CMTs and appear as regularly spaced, filamentous cross-bridges in electron micrographs of bundled microtubules (Chan et al., 1999; Smertenko et al., 2004). At least one of the MAP65 proteins forms homodimers that are essential for microtubule cross-linking (Smertenko et al., 2004). It will be interesting to determine whether the MAP65 isoforms that do not bundle microtubules fail to dimerize and whether different MAP65 isoforms can form heterodimers. Heterodimerization of different MAP65 isoforms represents a potential regulatory mechanism for coordinating the spatiotemporal pattern of activity of these proteins.
The plus-ends of the CMTs are highly dynamic and are involved in exploring the cortical space and in intermicrotubule encounters. Therefore, regulation of the plus-end dynamics can potentially control CMT stability and organization (Hashimoto, 2003). Several recent studies on microtubule dynamics have utilized proteins known to localize at microtubule plus-ends. Mammalian cytoplasmic linker protein-170 (Dhonukshe and Gadella, 2003) and two of the three Arabidopsis end binding1 (EB1) proteins (Chan et al., 2003; Mathur et al., 2003) localize to the plus-ends of growing CMTs and can facilitate the visualization of microtubule dynamics in dense arrays such as the preprophase band (Dhonukshe and Gadella, 2003). The EB1 proteins can potentially regulate microtubule plus-end dynamics and the targeted capture of microtubule ends through their interaction with EB1 binding partners. Such activities may selectively stabilize a subpopulation of CMTs and contribute to cortical array organization. Besides their plus-end localization, the EB1 proteins also appear to localize to the microtubule minus-ends (Chan et al., 2003) and an endomembrane compartment (Mathur et al., 2003). The localization of EB1 proteins to microtubule plus-ends depends on low-level expression; therefore, the significance of these novel localization patterns is not known because these non-plus-end localization patterns were obtained using the constitutive 35S promoter to drive transgene expression.
The importance of the microtubule plus-end in CMT organization is highlighted by the recent cloning of Arabidopsis SPIRAL1 (SPR1), which encodes a plant-specific, plus-end-enriched MAP (Nakajima et al., 2004; Sedbrook et al., 2004). The spr1 mutation results in aberrant right-hand twisting of cell files and greatly reduces anisotropic cell expansion in zones associated with rapid elongation (Furutani et al., 2000). These cellular aberrations are associated with irregular arrangements of the CMTs, which suggests that SPR1 encodes a MAP. Indeed, SPR1-GFP can bind to CMTs and is enriched at the growing ends of a subset of these microtubules (Nakajima et al., 2004; Sedbrook et al., 2004). The SPR1 protein has a bivalent microtubule binding domain, and overexpression of SPR1 results in increased resistance of the plants to long-term exposure to a microtubule-disrupting drug (Nakajima et al., 2004). These observations suggest that SPR1 regulates microtubule plus-end dynamics, perhaps by stabilizing the growing ends so as to foster CMT array organization and anisotropic cell expansion. A comparison between the CMT dynamics in spr1 mutants and wild-type plants would be valuable to determine if SPR1 regulates microtubule dynamics and/or microtubule interactions. This information may indicate the aspects of CMT dynamics that are important for their proper organization.
Most CMTs are bound to the plasma membrane, and recent work indicates that this attachment is important for CMT stability and organization. Phospholipase D (PLD) is one of the molecules involved in attaching the CMTs to the plasma membrane (Gardiner et al., 2001). Interestingly, activation of PLD by n-butanol results in the release of microtubules from the plasma membrane and a net decrease in average microtubule length, presumably as a result of a decrease in polymer stability (Dhonukshe et al., 2003; Gardiner et al., 2003). More importantly, these changes are associated with CMT disorganization, an inhibition of cell elongation, and abnormal plant morphology (Dhonukshe et al., 2003; Gardiner et al., 2003). Therefore, CMT attachment to the plasma membrane appears to be an important regulator of their organization. If true, PLD may mediate the reorganization of CMT arrays in response to various developmental and environmental cues. For example, it will be interesting to determine whether PLD activity is involved in the normally occurring transition from a transverse cortical array (e.g., in elongating root epidermal cells) to an oblique or longitudinal array (after cessation of cell elongation) during development. In addition, several plant hormones rapidly induce the organization or disorganization of CMT arrays (Nick, 1998), so it is possible that PLD acts as the transducing element to link hormone perception to a microtubule response via membrane cytoskeletal interactions.
Plasma membrane attachment of CMTs may be important for their organization because it determines the geometric space occupied by the CMTs: Plasma membrane-bound CMTs effectively occupy a two-dimensional space, whereas their detachment from the membrane results in their distribution into a three-dimensional space. The distribution of CMTs into a three-dimensional space would predictably reduce the frequency of intermicrotubule encounters, which may account for the disorganization of CMTs upon activation of PLD. Alternatively, it is possible that PLD activation also regulates the activity of certain MAPs that control CMT stability and organization.
CMTs AND CELL ELONGATION
During interphase, CMTs are typically coaligned with the cellulose microfibrils, and, therefore, it has long been thought that the organization of the interphase CMTs regulates the axis of cell elongation by guiding the oriented deposition of cellulose microfibrils in the apoplastic space (Green, 1980). The microfibrils, in turn, provide the biophysical constraint to restrict turgor-induced cell expansion to an axis perpendicular to the net orientation of the cellulose microfibrils.
However, recent studies have revealed that the relationship between CMT array organization, microfibril organization, and the axis of cell elongation is complex. In particular, the data show that mutation or drug-induced disruption of the transverse CMT array can disrupt anisotropic cell expansion without disruption of parallel cellulose microfibril organization (Baskin, 2001; Sugimoto et al., 2001, 2003; Himmelspach et al., 2003). These results suggest that CMTs do not directly regulate cellulose microfibril alignment and that CMT arrays may regulate the direction of cell expansion by regulation of other factors such as the deposition of other components of the cell wall matrix that contribute to wall strength (Sugimoto et al., 2001, 2003; Himmelspach et al., 2003). In the absence of CMTs, nascent cellulose microfibrils may become aligned based on information contained in the preexisting cell wall (Baskin, 2001).
In contrast with these reports, the disruption of the transverse CMT array in katanin mutants is associated with radial cell expansion and a concomitant disruption of cellulose microfibril orientation (Burk and Ye, 2002). More recently, the analysis of the effect of modest levels of a microtubule depolymerizing drug on Arabidopsis root cells indicates that the disruption of the transverse CMT array and cellular growth anisotropy is accompanied by a concomitant disruption of the global alignment of the cellulose microfibril array, although regions of patchy parallel microfibril arrangement can occur (Baskin et al., 2004). This observation led the authors to suggest that anisotropic cell growth is dependent on the degree of global, as opposed to local, microfibril alignment, and that the ordered interphase CMT arrays are important for this global microfibril alignment process.
Part of the complexity associated with the relationship between interphase CMT array organization and anisotropic cell growth may relate to the idea that CMTs form a continuum with the plasma membrane and the cell wall (Cyr, 1994; Fisher and Cyr, 1998). This scenario raises the possibility for a complex interplay between each of these components and the operation of redundant mechanisms.
CONCLUDING REMARKS
The recent discoveries highlighted in this essay represent significant steps toward understanding the complex problem of how hundreds to thousands of individual, dynamic CMTs are organized into specific arrays and how array transitions occur.
The fact that CMT arrays are not static, but rather continuously dynamic, suggests that CMTs may self-organize into specific arrays. A self-organizational mechanism can give rise to new array patterns via the tuning of system parameters instead of invoking new mechanisms for each new pattern. For example, the regulation of CMT stabilization/destabilization, CMT density, and CMT plasma membrane attachment by MAPs may result in new array patterns by regulating the frequency and/or the outcome of intermicrotubule interactions (e.g., microtubule bundling).
Microtubules that are selectively stabilized in specific orientations (e.g., the transverse orientation) could act as seeds to promote the establishment of a new, predominant orientation of CMTs. This hypothesis is supported by evidence that shows CMT stability is related to CMT orientation with respect to the cell's axis (Wiesler et al., 2002; Tian et al., 2004) and that stable, discordant microtubules precede the reorientation of the cortical array in that direction (Yuan et al.,1994; Granger and Cyr, 2001).
This self-organizational scheme may also account for the occurrence of helical cortical arrays that are associated with certain mutational or drug-induced perturbations of CMTs (Furutani et al., 2000; Sugimoto et al., 2003). The chirality of these helical arrays could arise from the inherent helical character of the microtubule, in which the tubulin subunits from adjacent protofilaments are not perfectly aligned but rather follow a helical path with a 10° pitch (Figure 1A). This pitch may be important to the binding of MAPs; therefore, changes in this pitch, resulting from experimental perturbations of microtubule structure, could alter the pattern of microtubule–MAP interactions and lead to a change in higher order organization of the cortical array (e.g., a shift from a transverse to helical presentation of the CMTs).
Acknowledgments
The work conducted in our lab, and that is cited herein, was supported by grants obtained from the U.S. Department of Agriculture and the Department of Energy. We thank Deb Fisher for critical reading of the manuscript.
References
- Baskin, T.I. (2001). On the alignment of cellulose microfibrils by cortical microtubules: A review and a model. Protoplasma 215, 150–171. [DOI] [PubMed] [Google Scholar]
- Baskin, T.I., Beemster, G.T.S., Judy-March, J.E., and Marga, F. (2004). Disorganization of cortical microtubules stimulates tangential expansion and reduces the uniformity of cellulose microfibril alignment among cells in the root of Arabidopsis. Plant Physiol. 135, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bichet, A., Desnos, T., Turner, S., Grandjean, O., and Höfte, H. (2001). BOTERO1 is required for normal orientation of cortical microtubules and anisotropic cell expansion in Arabidopsis. Plant J. 25, 137–148. [DOI] [PubMed] [Google Scholar]
- Bouquin, T., Mattsson, O., Naested, H., Foster, R., and Mundy, J. (2003). The Arabidopsis lue1 mutant defines a katanin p60 ortholog involved in hormonal control of microtubule orientation during cell growth. J. Cell Sci. 116, 791–801. [DOI] [PubMed] [Google Scholar]
- Burk, D.H., Liu, B., Zhong, R.Q., Morrison, W.H., and Ye, Z.H. (2001). A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13, 807–827. [PMC free article] [PubMed] [Google Scholar]
- Burk, D.H., and Ye, Z.H. (2002). Alteration of oriented deposition of cellulose microfibrils by mutation of a katanin-like microtubule-severing protein. Plant Cell 14, 2145–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J., Calder, G.M., Doonan, J.H., and Lloyd, C.W. (2003). EB1 reveals mobile microtubule nucleation sites in Arabidopsis. Nat. Cell Biol. 5, 967–971. [DOI] [PubMed] [Google Scholar]
- Chan, J., Jensen, C.G., Jensen, L.C.W., Bush, M., and Lloyd, C.W. (1999). The 65-kDa carrot microtubule-associated protein forms regularly arranged filamentous cross-bridges between microtubules. Proc. Natl. Acad. Sci. USA 96, 14931–14936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti, J., Cho, S.O., Wick, S., and Sundaresan, V. (1993). Localization of the functional p34cdc2 homolog of maize in root tip and stomatal complex cells: Association with predicted division sites. Plant Cell 5, 1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr, R.J. (1994). Microtubules in plant morphogenesis: Role of the cortical array. Annu. Rev. Cell Biol. 10, 153–180. [DOI] [PubMed] [Google Scholar]
- Dhonukshe, P., and Gadella, T.W., Jr. (2003). Alteration of microtubule dynamic instability during preprophase band formation revealed by yellow fluorescent protein-CLIP170 microtubule plus-end labeling. Plant Cell 15, 597–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe, P., Laxalt, A.M., Goedhart, J., Gadella, T.W.J., and Munnik, T. (2003). Phospholipase D activation correlates with microtubule reorganization in living plant cells. Plant Cell 15, 2666–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, D.D., and Cyr, R.J. (1998). Extending the microtubule/microfibril paradigm: Cellulose synthesis is required for normal cortical microtubule alignment in elongating cells. Plant Physiol. 116, 1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani, I., Watanabe, Y., Prieto, R., Masukawa, M., Suzuki, K., Naoi, K., Thitamadee, S., Shikanai, T., and Hashimoto, T. (2000). The SPIRAL genes are required for directional control of cell elongation in Arabidopsis thaliana. Development 127, 4443–4453. [DOI] [PubMed] [Google Scholar]
- Gardiner, J., Collings, D.A., Harper, J.D.I., and Marc, J. (2003). The effects of the phospholipase D-antagonist 1-butanol on seedling development and microtubule organisation in Arabidopsis. Plant Cell Physiol. 44, 687–696. [DOI] [PubMed] [Google Scholar]
- Gardiner, J.C., Harper, J.D.I., Weerakoon, N.D., Collings, D.A., Ritchie, S., Gilroy, S., Cyr, R.J., and Marc, J. (2001). A 90-kD phospholipase D from tobacco binds to microtubules and the plasma membrane. Plant Cell 13, 2143–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger, C.L., and Cyr, R.J. (2001). Spatiotemporal relationships between growth and microtubule orientation as revealed in living root cells of Arabidopsis thaliana transformed with green-fluorescent-protein gene construct GFP-MBD. Protoplasma 216, 201–214. [DOI] [PubMed] [Google Scholar]
- Green, P.B. (1980). Organogenesis: A biophysical view. Annu. Rev. Plant Physiol. 31, 51–82. [Google Scholar]
- Hardham, A.R., and Gunning, B.E.S. (1978). Structure of cortical microtubule arrays in plant cells. J. Cell Biol. 77, 14–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, T. (2003). Dynamics and regulation of plant interphase microtubules: A comparative view. Curr. Opin. Plant Biol. 6, 568–576. [DOI] [PubMed] [Google Scholar]
- Hepler, P.K., Cleary, A.L., Gunning, B.E.S., Wadsworth, P., Wasteneys, G.O., and Zhang, D.H. (1993). Cytoskeletal dynamics in living plant cells. Cell Biol. Int. 172, 127–142. [Google Scholar]
- Himmelspach, R., Williamson, R.E., and Wasteneys, G.O. (2003). Cellulose microfibril alignment recovers from DCB-induced disruption despite microtubule disorganization. Plant J. 36, 565–575. [DOI] [PubMed] [Google Scholar]
- Hush, J.M., Wadsworth, P., Callaham, D.A., and Hepler, P.K. (1994). Quantification of microtubule dynamics in living plant cells using fluorescence redistribution after photobleaching. J. Cell Sci. 107, 775–784. [DOI] [PubMed] [Google Scholar]
- Hush, J., Wu, L., John, P.C., Hepler, L.H., and Hepler, P.K. (1996). Plant mitosis promoting factor disassembles the microtubule preprophase band and accelerates prophase progression in Tradescantia. Cell Biol. Int. 20, 275–287. [DOI] [PubMed] [Google Scholar]
- Hussey, P.J., and Hawkins, T.J. (2001). Plant microtubule-associated proteins: The HEAT is off in temperature-sensitive mor1. Trends Plant Sci. 6, 389–392. [DOI] [PubMed] [Google Scholar]
- Mathur, J., Mathur, N., Kernebeck, B., Srinivas, B.P., and Hulskamp, M. (2003). A novel localization pattern for an EB1-like protein links microtubule dynamics to endomembrane organization. Curr. Biol. 13, 1991–1997. [DOI] [PubMed] [Google Scholar]
- Mineyuki, Y. (1999). The preprophase band of microtubules: Its function as a cytokinetic apparatus in higher plants. Int. Rev. Cytol. 187, 1–49. [Google Scholar]
- Nakajima, K., Furutani, I., Tachimoto, H., Matsubara, H., and Hashimoto, T. (2004). SPIRAL1 encodes a plant-specific microtubule-localized protein required for directional control of rapidly expanding Arabidopsis cells. Plant Cell 16, 1178–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick, P. (1998). Signaling to the microtubular cytoskeleton in plants. Int. Rev. Cytol. 184, 33–80. [Google Scholar]
- Reddy, A.S.N. (2001). Molecular motors and their functions in plants. Int. Rev. Cytol. 204, 97–178. [DOI] [PubMed] [Google Scholar]
- Schmit, A.C. (2002). A centrosomal microtubule nucleation in higher plants. Int. Rev. Cytol. 220, 257–289. [DOI] [PubMed] [Google Scholar]
- Sedbrook, J.C., Ehrhardt, D.W., Fisher, S.E., Scheible, W.R., and Somerville, C.R. (2004). The Arabidopsis SKU6/SPIRAL1 gene encodes a plus end-localized microtubule-interacting protein involved in directional cell expansion. Plant Cell 16, 1506–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, S.L., Kamyar, R., and Ehrhardt, D.W. (2003). Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science 300, 1715–1718. [DOI] [PubMed] [Google Scholar]
- Smertenko, A.P., Chang, H.-Y., Wagner, V., Kaloriti, D., Fenyk, S., Sonobe, S., Lloyd, C., Hauser, M.-T., and Hussey, P.J. (2004). The Arabidopsis microtubule-associated protein AtMAP65-1: Molecular analysis of its microtubule bundling activity. Plant Cell 16, 2035–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto, K., Himmelspach, R., Williamson, R.E., and Wasteneys, G.O. (2003). Mutation or drug-dependent microtubule disruption causes radial swelling without altering parallel cellulose microfibril deposition in Arabidopsis root cells. Plant Cell 15, 1414–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto, K., Williamson, R.E., and Wasteneys, G.O. (2000). New techniques enable comparative analysis of microtubule orientation, wall texture, and growth rate in intact roots of Arabidopsis. Plant Physiol. 124, 1493–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto, K., Williamson, R.E., and Wasteneys, G.O. (2001). Wall architecture in the cellulose-deficient rsw1 mutant of Arabidopsis thaliana: Microfibrils but not microtubules lose their transverse alignment before microfibrils become unrecognizable in the mitotic and elongation zones of roots. Protoplasma 215, 172–183. [DOI] [PubMed] [Google Scholar]
- Tian, G.-W., Smith, D., Glück, S., and Baskin, T.I. (2004). Higher plant cortical microtubule array analyzed in vitro in the presence of the cell wall. Cell Motil. Cytoskeleton 57, 26–36. [DOI] [PubMed] [Google Scholar]
- Vos, J.W., Dogterom, M., and Emons, A.M. (2004). Microtubules become more dynamic but not shorter during preprophase band formation: A possible “search-and-capture” mechanism for microtubule translocation. Cell Motil. Cytoskeleton 57, 246–258. [DOI] [PubMed] [Google Scholar]
- Wasteneys, G.O. (2002). Microtubule organization in the green kingdom: Chaos or self-order. J. Cell Sci. 115, 1345–1354. [DOI] [PubMed] [Google Scholar]
- Wasteneys, G.O., Gunning, B.E.S., and Hepler, P.K. (1993). Microinjection of fluorescent brain tubulin reveals dynamic properties of cortical microtubules in living plant cells. Cell Motil. Cytoskeleton 24, 205–213. [Google Scholar]
- Wasteneys, G.O., and Williamson, R.E. (1989). Reassembly of microtubules in Nitella tasmanica: Assembly of cortical microtubules in branching clusters and its relevance to steady-state microtubule assembly. J. Cell Sci. 93, 705–714. [Google Scholar]
- Weingartner, M., Binarova, P., Dryková, D., Schweighofer, A., David, J.-P., Heberle-Bors, E., Doonan, J., and Bögre, L. (2001). Dynamic recruitment of Cdc2 to specific microtubule structures during mitosis. Plant Cell 13, 1929–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittington, A.T., Vugrek, O., Wei, K.J., Hasenbein, N.G., Sugimoto, K., Rashbrooke, M.C., and Wasteneys, G.O. (2001). MOR1 is essential for organizing cortical microtubules in plants. Nature 411, 610–613. [DOI] [PubMed] [Google Scholar]
- Wicker-Planquart, C., Stoppin-Mellet, V., Blanchoin, L., and Vantard, M. (2004). Interactions of tobacco microtubule-associated protein MAP65-1b with microtubules. Plant J. 39, 126–134. [DOI] [PubMed] [Google Scholar]
- Wiesler, B., Wang, Q.-Y., and Nick, P. (2002). The stability of cortical microtubules depends on their orientation. Plant J. 32, 1023–1032. [DOI] [PubMed] [Google Scholar]
- Wymer, C.L., Fisher, D.D., Moore, R.C., and Cyr, R.J. (1996). Elucidating the mechanism of cortical microtubule reorientation in plant cells. Cell Motil. Cytoskeleton 35, 162–173. [DOI] [PubMed] [Google Scholar]
- Yasuhara, H., Muraoka, M., Shogaki, H., Mori, H., and Sonobe, S. (2002). TMBP200, a microtubule bundling polypeptide isolated from telophase tobacco BY-2 cells is a MOR1 homologue. Plant Cell Physiol. 43, 595–603. [DOI] [PubMed] [Google Scholar]
- Yuan, M., Shaw, P.J., Warn, R.M., and Lloyd, C.W. (1994). Dynamic reorientation of cortical microtubules, from transverse to longitudinal, in living plant cells. Proc. Natl. Acad. Sci. USA 91, 6050–6053. [DOI] [PMC free article] [PubMed] [Google Scholar]