Abstract
Pronounced variability of transgene expression and transgene silencing are commonly observed among independent plant lines transformed with the same construct. Single-copy T-DNA lines harboring reporter genes of various kind and number under the control of a strong promoter were established in Arabidopsis thaliana for a comprehensive analysis of transgene expression. Characterization of 132 independent transgenic lines revealed no case of silencing as a result of site of T-DNA integration. Below a certain number of identical transgenes in the genome, gene copy number and expression were positively correlated. Expression was high, stable over all generations analyzed, and of a comparable level among independent lines harboring the same copy number of a particular transgene. Conversely, RNA silencing was triggered if the transcript level of a transgene surpassed a gene-specific threshold. Transcript level–mediated silencing effectively accounts for the pronounced transgene expression variability seen among transformants. It is proposed that the RNA sensing mechanism described is a genome surveillance system that eliminates RNA corresponding to excessively transcribed genes, including transgenes, and so plays an important role in genome defense.
INTRODUCTION
Independent transgenic lines generated with the same construct often vary >100-fold with respect to transgene expression levels (Jones et al., 1985; Peach and Velten, 1991; Holtorf et al., 1995), and gene silencing is frequently observed. Recently, great progress has been made in elucidating the molecular mechanisms of gene silencing. Silencing is caused by epigenetic effects rather than alterations of the transgene sequence: it either results from abolished transcription of the introduced gene (transcriptional gene silencing [TGS]) or the degradation of transgene RNA (posttranscriptional gene silencing [PTGS]). PTGS is established during plant development and may spread systemically, and after meiosis, the process is reset. Conversely, TGS is mitotically and meiotically heritable, but reversion has been observed. Methylation of transgene sequences has often been found in silenced lines. Methylation of the transcribed region of the transgene is associated with PTGS, whereas methylation of promoter sequences points to TGS (summarized in Vaucheret et al., 1998). Small RNAs in sense and antisense orientations (small interfering RNAs [siRNAs]) specific for the transcribed transgene sequence are a molecular hallmark of PTGS (Hamilton and Baulcombe, 1999). Similarly, a correlation between siRNAs specific for the promoter region and TGS has been described (Mette et al., 2000; Sijen et al., 2001).
Epigenetic regulation has been studied extensively in transgenic systems, but gene silencing is not restricted to transgenes. PTGS and the related process RNA interference require a set of related proteins in protozoa, fungi, plants, and animals. This suggests an ancient origin for these RNA silencing pathways that probably evolved as defense mechanisms against viruses and transposons (summarized in Zamore, 2002). Furthermore, it is becoming increasingly apparent that the siRNA-related microRNAs (miRNAs), produced by and feeding into similar pathways as siRNAs, play an important role in endogenous gene regulation (summarized in Bartel and Bartel, 2003).
With the plant transformation methods currently used, it is neither possible to introduce a defined number of transgenes into the genome of a higher plant nor feasible to target efficiently the foreign DNA to specific positions in the genome. As one result, repeat arrangements and truncated and/or rearranged transgene copies are often observed in transgenic lines. Hence, independent transgenic lines differ in respect to number, arrangement, and position of transgene copies they harbor in the genome.
The study of a large set of petunia (Petunia hybrida) transformants carrying Chs sense transgenes showed that neither position effects by flanking plant DNA sequences nor read-through from such sequences were the primary determinant of silencing (Jorgensen et al., 1996). Silencing was frequently associated with repetitive T-DNA structures (Jorgensen et al., 1996; summarized in Muskens et al., 2000), but neither inverted repeat T-DNA configurations nor arrangements of tandemly repeated transgenes as such were sufficient to trigger transgene silencing (Lechtenberg et al., 2003). Two distinct modes of silencing may account for these findings. On the one hand, a causal relationship between transcribed inverted repeat structures and silencing has been documented (Waterhouse et al., 1998; Chuang and Meyerowitz, 2000; Mette et al., 2000; Sijen et al., 2001). RNA capable of duplex formation is processed to siRNA, and homologous mRNA is targeted for degradation. RNA-directed de novo methylation of cytosine residues is often observed within regions of RNA-DNA sequence identity. Thus, compelling evidence exists that RNA/RNA and RNA/DNA interactions mediate PTGS and TGS in plants (summarized in Matzke et al., 2001). On the other hand, several studies lent support to the hypothesis that silencing is triggered by threshold concentrations of transgene transcript or another product of transgene expression (Lindbo et al., 1993), for example, the frequency of silencing positively correlated with the promoter strengths of introduced genes (Que et al., 1997), and silencing was more pronounced in homozygous than in corresponding hemizygous plants (summarized in Vaucheret et al., 1998).
It remains to be shown whether the described silencing phenomena fully account for the very pronounced variability of transgene expression level in populations of transgenic plants, especially because apparently conflicting results were reported when the analysis of transformants was devoted to the study of transgene expression level. On the one hand, it has been proposed that variability of transgene expression level among transformants may be attributable to different positions of transgene integration (Jones et al., 1985; Peach and Velten, 1991; Day et al., 2000). On the other hand, the analysis of a small number of single-copy T-DNA transformants did not reveal evidence that transgene transcript levels were affected by position effects (Hobbs et al., 1990). Direct proportionality between transgene copy number and expression level has been reported (Stockhaus et al., 1987; Hobbs et al., 1993; Ku et al., 1999), whereas several other studies described a correlation between silencing and high transgene doses and/or repeat arrangements of transgenes (Hobbs et al., 1990; Jorgensen et al., 1996; Que et al., 1997).
A systematic study of transgene expression in Arabidopsis thaliana enabled us to determine to what extent number and arrangement of transgene copies, the position of T-DNA integration in the genome, and transgene nature contribute to the variability of transgene expression and/or cause silencing. The results obtained identify transcript level–mediated PTGS as the main cause for the large variability in transgene expression seen among transformants and predict an important role for the described RNA sensing mechanism in genome defense.
RESULTS
No Case of Silencing Because of Site of T-DNA Integration
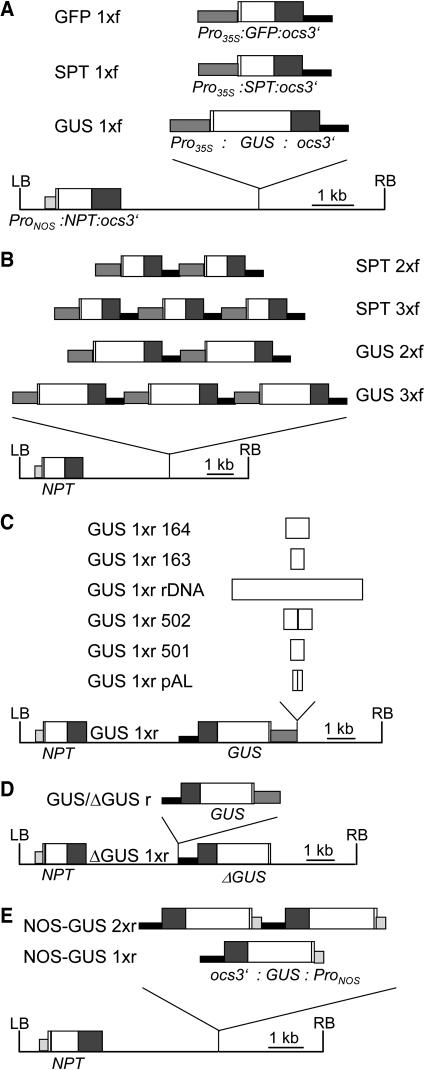
For a comprehensive study of transgene expression, derivatives of pMDL-T-DNA (Forsbach et al., 2003) were introduced into Arabidopsis by Agrobacterium tumefaciens–mediated transformation. The T-DNAs harbored a chimeric neomycin phosphotransferase (NPT) gene conferring kanamycin resistance and reporter gene cassettes of various kind and number. The reporter gene cassettes consisted of the 35S promoter of Cauliflower mosaic virus (CaMV) (Pro35S), a modified Ω-leader of Tobacco mosaic virus, the coding sequence of a reporter gene, and the 3′-untranslated region of the octopine synthase gene (ocs3′) (Lechtenberg et al., 2003). The genes for β-glucuronidase (GUS) (Jefferson et al., 1986), streptomycin phosphotransferase (SPT) (Maliga et al., 1988), and green fluorescent protein (GFP) (Siemering et al., 1996; Haseloff et al., 1997) were used as reporters (Figure 1A). The different T-DNAs were designated according to kind, number, and orientation of the reporter genes they harbored (Figures 1A and 1B). The direction of transcription of the reporter gene was either pointing toward the left (r-constructs) or right border (f-constructs). T-DNAs containing different repetitive sequences from Arabidopsis (Martinez-Zapater et al., 1986; Simoens et al., 1988; Thompson et al., 1996; Wanzenböck et al., 1997) immediately upstream of the GUS gene (Figure 1C), a promoterless copy of the GUS gene (ΔGUS, Figure 1D), or the GUS reporter gene under the control of the nopaline synthase promoter (ProNOS) were also introduced into Arabidopsis (NOS-GUS, Figure 1E).
Figure 1.
T-DNA Maps.
(A) T-DNAs carrying one copy of a particular chimeric reporter gene cassette. The different T-DNAs were derivatives of vector pMDL-T-DNA that harbored a chimeric NPT gene under the control of ProNOS near the left border (LB). HindIII fragments corresponding to GFP, SPT, and GUS reporter gene cassettes were inserted in both orientations into the HindIII site of vector pMDL-T-DNA to generate constructs GFP 1x, SPT 1x, and GUS 1x. The maps of the f-constructs in which the transcription of the reporter gene was pointing toward the right border are shown. The reporter gene cassettes consisted of the CaMV 35S promoter (Pro35S), a modified Tobacco mosaic virus Ω-leader (shown as a white horizontal bar), the coding sequences of a reporter gene, ocs3′, and Tn5 sequences (depicted as a black vertical bar).
(B) T-DNAs carrying reporter genes in a tandem arrangement. The maps of the f-constructs in which the direction of transcription of the reporter genes was the same as that of the NPT gene are given.
(C) T-DNAs carrying repetitive elements from Arabidopsis upstream of the GUS reporter gene. Construct GUS 1xr pAL contained two copies of the 180-bp HindIII tandem repeat (Martinez-Zapater et al., 1986). GUS 1xr 501 and GUS 1xr 502 carried one and two copies, respectively, of the 500-bp HindIII tandem repeat (Simoens et al., 1988). A fragment corresponding to the intergenic spacer of rDNA sequences (Wanzenböck et al., 1997) was present in T-DNA vector GUS 1xr rDNA. In constructs GUS 1xr 163 and GUS 1xr 164, the sequences cloned upstream of the CaMV 35S promoter corresponded to different dispersed repetitive sequences (Thompson et al., 1996).
(D) T-DNAs carrying a promoterless copy of the GUS gene (ΔGUS). In T-DNA GUS/ΔGUS r, a promoterless copy of the GUS gene adjoined a GUS gene under the control of the CaMV 35S promoter.
(E) T-DNAs carrying the GUS gene under the control of ProNOS.
Molecular and genetic analyses established 157 single-copy transgenic lines harboring MDL-T-DNA derivatives. Plants homozygous for the T-DNA were identified for 149 lines, the remaining eight lines carried gametophytic or embryo lethal defects. The T-DNA insertions of 107 lines were mapped on the sequence maps of the Arabidopsis chromosomes and were found distributed on all five chromosomes. Two of the insertions were located in the pericentromeric heterochromatin of chromosome I (Forsbach et al., 2003).
Seeds of homozygous single-copy T-DNA lines were germinated on kanamycin-containing medium, and resistance of the resulting seedlings to kanamycin was evaluated to assess NPT gene expression under the control of the weak nopaline synthase promoter in subsequent generations. For each of 132 independent transgenic lines, progeny of several homozygous plants (3 to 29 per line) derived from consecutive generations (2 to 7) were assayed. On average, 651 seedlings per line were analyzed, and only fully kanamycin-resistant seedlings were observed. Thus, among the transgenic lines tested, not a single case of silencing of the NPT gene because of site of T-DNA integration in the genome was identified. However, it should be noted that our selection of primary transformants on kanamycin-containing medium would preclude recovery of any insertions in which the NPT gene was subjected immediately to TGS.
Comparable Transgene Expression Levels in Independent Single-Copy T-DNA Transformants
Fluorometric assays were used to quantify reporter gene expression of 12 independent single-copy T-DNA lines that harbored GUS 1x T-DNAs (Figure 1A). Repeated GUS activity measurements of 2-week-old and 8-week-old plants homozygous for the GUS 1x T-DNAs revealed high and stable expression throughout plant development and different generations for all lines. Given that the plants were grown and analyzed under identical conditions, similar GUS activity values were established for independent transgenic lines: twofold differences in expression level were the highest observed in the vast majority of cases tested (data not shown).
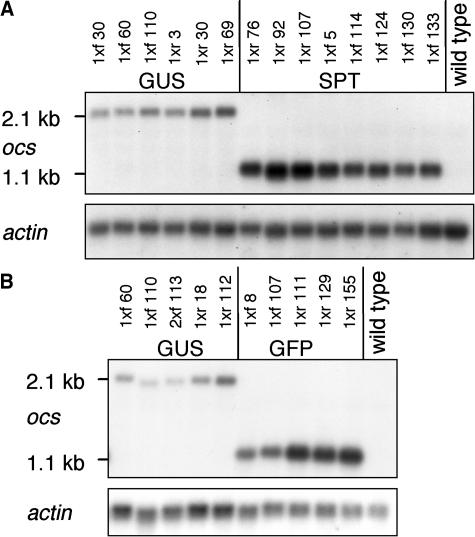
A comparison of GUS transcript levels revealed a similarly low variability between independent transgenic lines (Figure 2). Likewise, only small differences of steady state transcript levels were found when independent transgenic lines carrying two copies of the SPT or GFP reporter genes were compared. The results shown in Figure 2 are representative for 19 lines containing SPT transgenes and 21 lines harboring chimeric GFP genes. GFP fluorescence measurements provided further evidence that twofold differences in transgene expression level were the highest observed for independent single-copy T-DNA lines. The results for eight lines, each homozygous for a GFP 1x T-DNA (Figure 1A), are exemplified in Figure 3A. Repeated GFP fluorescence measurements documented stable expression throughout plant development and different generations for independent lines (data not shown), consistent with the results obtained for lines harboring two copies of the GUS gene.
Figure 2.
Comparably High Transcript Levels in Independent Transgenic Lines Harboring Two Copies of a Particular Reporter Gene under the Control of the CaMV 35S Promoter.
RNA gel blot analyses of plants carrying GUS ([A] and [B]), SPT (A), or GFP (B) genes. Hemizygous plants were assayed for line GUS 2xf 113; in all other cases, 8-week-old homozygous plants were examined. The blots were hybridized with a probe corresponding to the ocs3′ end and with a probe derived from Arabidopsis actin genes 2 and 8 (An et al., 1996).
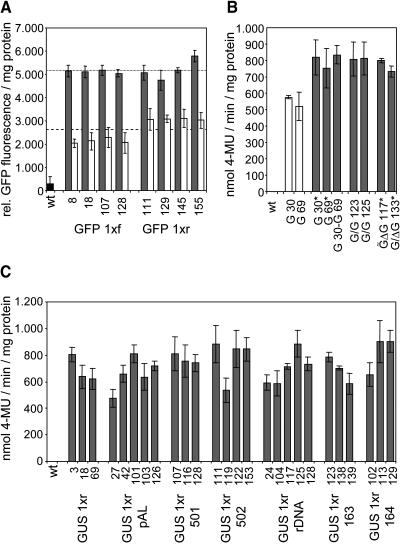
Figure 3.
Neither the Arrangement of Transgenes Relative to Each Other nor the Presence of Repetitive Sequences Immediately Adjacent to the Reporter Genes Affected Transgene Expression Levels.
(A) Results of GFP fluorescence measurements of 8-week-old transgenic plants containing one and two copies of a GFP 1x T-DNA (Figure 1A) are shown as white and gray bars, respectively. Average values of GFP fluorescence/mg protein obtained for homozygous and hemizygous plants of eight independent lines are indicated by a dotted and a dashed line, respectively.
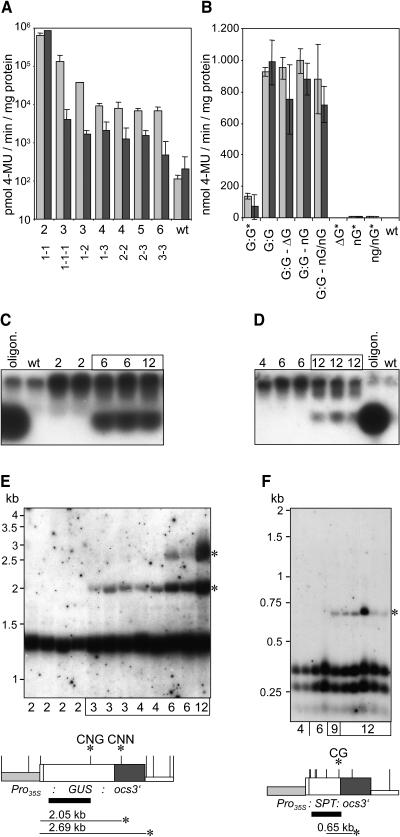
(B) Results of GUS activity measurements of 8-week-old transgenic plants harboring one and two copies of the GUS reporter gene under the control of the CaMV 35S promoter are shown as white and gray bars, respectively. Either transgenic lines homozygous for a particular T-DNA were analyzed, or F1 progeny derived from crosses of homozygous transgenic lines among each other or with wild-type plants. T-DNAs GUS 1xr, GUS 2xr, and GUS/ΔGUS r (Figure 1) are abbreviated G, G/G, and G/ΔG, respectively. Different loci are divided by dashes, and asterisks mark plants homozygous for a particular T-DNA.
(C) Arabidopsis repetitive sequences adjacent to a GUS gene did not influence reporter gene expression. Transgenic lines homozygous for a single copy of T-DNA GUS 1xr (Figure 1A) and derivatives thereof that contained different repetitive elements in the immediate vicinity of the GUS reporter gene (Figure 1C) were analyzed for GUS activity at the age of 8 weeks.
Using the ocs3′ sequences common to all three reporter genes (Figure 1A) as probe for RNA gel blot and slot blot analyses, it was possible to compare steady state transgene transcript levels of lines harboring two copies of the GUS, GFP, or SPT genes. Without exception, GUS-containing lines had lower steady state transgene transcript levels than lines carrying SPT or GFP genes (Figure 2), even though the three reporter genes were driven by the same regulatory elements.
Neither Transgene Repeat Arrangements nor Vicinity to Repetitive Elements Affect Reporter Gene Expression Levels
All transgenic plants harboring two copies of a GUS gene under the control of the CaMV 35S promoter had a comparable expression level, regardless of whether the copies were present at the same locus of the homologous chromosomes (39 lines; Figure 3B, GUS 1xr 30* and GUS 1xr 69*), at two different loci (F1 progeny derived from 17 crosses of different homozygous lines; Figure 3B, GUS 1xr 30 and GUS 1xr 69), or at one locus as a tandem repeat (20 lines; Figure 3B, GUS 2xr 123 and GUS 2xr 125). Furthermore, plants homozygous for the GUS/ΔGUS r T-DNA (Figure 1D) that harbored a promoterless copy of the GUS gene next to a copy under the control of the CaMV 35S promoter showed as high GUS activity (Figure 3B, GUS/ΔGUS r 117 and GUS/ΔGUS r 133) as plants containing only two highly expressed copies of the GUS gene. Thus, a direct repeat arrangement of transgene sequences did not affect reporter gene expression. This finding was corroborated by the expression analysis of 15 transgenic lines homozygous for T-DNAs with two or three copies of the SPT gene in a tandem arrangement. High SPT transcript levels were found in all cases (Figure 4A; data not shown).
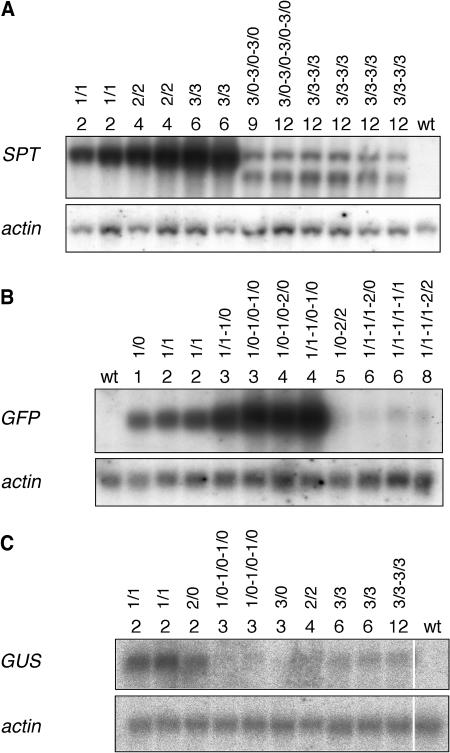
Figure 4.
Silencing by Exceeding a Certain Copy Number of a Particular Transgene under the Control of the Strong CaMV 35S Promoter in a Transformant.
RNA gel blot analyses of 8-week-old plants harboring different copy numbers of the SPT (A), GFP (B), or GUS genes (C). Transgenic lines homozygous for a particular T-DNA locus or progeny plants derived from crosses among homozygous transgenic lines or with wild-type plants were analyzed. For each line, numbers of transgene copies present in the genome and at the different T-DNA loci are given. Forward slashes separate copies of a particular locus that are present on the homologous chromosomes, and dashes divide copies of different loci. Single plants were analyzed for GFP reporter gene expression, whereas leaves of several plants were pooled for the analysis of SPT and GUS transcript levels. The resulting blots were hybridized with probes corresponding to the different reporter genes and with a probe derived from Arabidopsis actin genes 2 and 8 (An et al., 1996).
It was analyzed whether the presence of Arabidopsis repetitive sequences immediately upstream of the GUS gene would affect reporter gene expression. All 27 transgenic lines harboring different repetitive elements in the immediate vicinity of the promoter sequences of the GUS gene (Figure 1C) showed high and stable GUS expression (Figure 3C; data not shown). Moreover, expression levels of homozygous lines harboring repetitive sequences adjacent to the GUS gene were as high as the values of GUS 1xr lines (Figure 3C). Notably, this was also the case for line GUS 1xr rDNA 125, for which the site of T-DNA integration had been mapped to the pericentromeric heterochromatin of chromosome I (Forsbach et al., 2003).
The study of 115 independent transgenic lines and progeny of 17 crosses among different transformants documented a low variability of transgene expression between plants carrying the same copy number of a particular reporter gene, irrespective of the arrangement of the transgenes and the position of integration in the genome.
Below a Certain Number of Transgene Copies under the Control of the CaMV 35S Promoter in the Genome, a Positive Correlation between Gene Copy Number and Expression Was Observed
Comparative expression analysis of hemizygous and homozygous plants harboring one or two copies of the GUS gene, respectively, was performed for 35 transgenic lines. On average, the expression level of plants containing two copies of the GUS gene was approximately twofold higher than in plants with one copy. The results for two lines are shown in Figure 3B (GUS 1xr 30 and GUS 1xr 69). Similar results were obtained for 16 independent transgenic lines when hemizygous and homozygous plants harboring one or two copies of the GFP gene, respectively, were assayed for GFP fluorescence (Figure 3A; data not shown).
Intercrosses of independent transgenic lines containing one (Figure 1A) or two GFP genes at one locus (Lechtenberg et al., 2003) were performed to generate plants harboring different copy numbers of the GFP gene. Analysis of plants containing one to four copies of the GFP gene revealed a positive correlation between the number of transgenes in the genome and the expression level (Figure 4B). This was also noted when SPT transcript levels of transgenic lines harboring two (9 lines), four (9 lines), or six copies (6 lines) of the SPT gene were compared (Figure 4A; data not shown).
Above a Certain Number of Identical Transgene Copies under the Control of the CaMV 35S Promoter in the Arabidopsis Genome Gene Silencing Was Triggered
High GFP transcript levels were observed for plants harboring up to four copies of the GFP gene, whereas GFP transcripts were barely detectable in 8-week-old plants carrying five or more reporter genes (Figure 4B). GFP fluorescence was only observed in plants with high GFP transcript levels (data not shown). Thus, the combination of five or more transgene loci under the control of the CaMV 35S promoter resulted in silencing of the GFP gene.
A comparison of SPT transcript levels of plants that harbored 2, 4, 6, 9, or 12 copies of the SPT gene revealed that the amount of SPT transcripts in 8-week-old plants harboring 9 or 12 copies of the SPT gene was severely reduced when compared with those of lines carrying two, four, or six transgene copies. SPT gene silencing was associated with the appearance of truncated transcripts (Figure 4A).
Plants harboring three or more copies of the GUS gene under the control of the CaMV 35S promoter revealed a pronounced reduction of GUS transcript level and activity when compared with lines with two copies (Figures 4C and 5A). Silencing was triggered regardless of whether the reporter gene copies under the control of the CaMV 35S promoter were present at one locus as a tandem repeat (12 lines), at the same locus of the homologous chromosomes (25 lines), or at different loci (progeny derived from 23 crosses of different homozygous lines) (Figures 4C, 5A, and 5B; data not shown). By contrast, plants harboring one or two promoterless copies of the GUS gene or one or two GUS genes under the control of the weak nopaline synthase promoter in addition to two transgenes under the control of the CaMV 35S promoter showed GUS activity as high as plants containing only two highly expressed copies of this gene (Figures 3B and 5B). Hence, the presence of at least three GUS genes under the control of the CaMV 35S promoter was a prerequisite for the onset of GUS gene silencing.
Figure 5.
Silencing by a Posttranscriptional Mechanism.
(A) GUS activity assays revealed silencing of the GUS gene in transgenic lines harboring three or more copies of the GUS reporter gene under the control of the CaMV 35S promoter. Extracts were prepared from F1 progeny plants derived from crosses among homozygous transgenic lines harboring T-DNAs GUS 1xr, GUS 2xr, and GUS 3xr. The results for 2-week-old and 8-week-old plants are shown as light and dark gray bars, respectively. Copy numbers of GUS genes in the genome and at the different T-DNA loci are indicated for the different plants. Dashes divide copies of different loci.
(B) Lowly expressed GUS genes do not trigger transgene silencing. Either transgenic lines homozygous for a particular T-DNA were analyzed, or F1 progeny plants derived from crosses of homozygous transgenic lines among each other or with wild-type plants. Light and dark gray bars indicate the results for 2-week-old and 8-week-old plants. T-DNA loci GUS 2*xr 114, ΔGUS 1xr 102, NOS-GUS 1xr 111, and NOS-GUS 2xr 108 are abbreviated G:G, ΔG, nG, and nG/nG, respectively. Construct GUS 2*xr is a derivative of construct GUS 2xr that contains the SPT reporter gene cassette as a spacer between two GUS reporter gene cassettes under the control of the CaMV 35S promoter in a tandem arrangement. Different loci are divided by dashes, and asterisks mark plants homozygous for a particular T-DNA.
(C) and (D) Detection of siRNAs in 8-week-old plants of lines displaying silencing of the GUS (C) or SPT gene (D). A GUS-oligonucleotide (20 nucleotides, 75 ng) and an ocs-oligonucleotide (21 nucleotides, 75 ng) mixed with wild-type RNA served as size standard and hybridization control for the detection of siRNAs with probes corresponding to the GUS reporter gene and ocs3′ sequences, respectively. Numbers of transgene copies in the genome are given for each line.
(E) and (F) Methylation analysis of plants carrying different copy numbers of the GUS (E) or SPT genes (F). DNA of 8-week-old plants harboring GUS and SPT genes was cut with AluI and Bsp143II, respectively. The resulting DNA gel blots were hybridized with the probes that are indicated as solid black bars. AluI and Bsp143II restriction sites are shown as vertical bars on the maps of the GUS and the SPT reporter gene cassettes, respectively. Asterisks indicate hybridizing fragments indicative of methylation as well as the methylated sites. It is shown whether CG, CNG, or CNN methylation took place.
siRNAs specific for the transcribed region of the reporter genes were only detected in plants displaying transgene silencing (Figures 5C and 5D). These results are consistent with a posttranscriptional mechanism of gene silencing (Hamilton and Baulcombe, 1999). This finding was corroborated by methylation studies. Methylation of AluI sites located in the transcribed region of the GUS gene (Figure 5E) was only found in plants that displayed GUS gene silencing. The use of different probes confirmed that methylation in silenced lines was restricted to the transcribed region of the transgene (data not shown). Likewise, evidence for methylation of Bsp143II sites located in the transcribed region of the SPT gene was only observed in plants showing SPT gene silencing (Figure 5F).
A high transcript level conferred by multiple copies of a particular reporter gene under the control of the CaMV 35S promoter resulted in RNA silencing; however, the copy number at which silencing was triggered was different for the GFP, GUS, and SPT genes (Figure 4).
Variability of Transgene Expression between Different Leaves and among Isogenic Plants
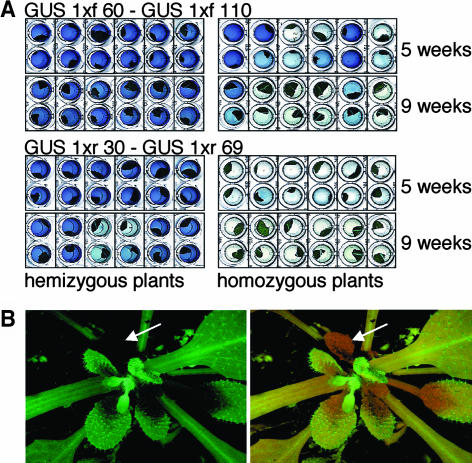
Plants containing three or four copies of the GUS gene showed silencing, but the mean GUS activity was in many cases not as low as in plants harboring 6 or 12 GUS genes, especially when 2-week-old plants were analyzed. Colorimetric and quantitative GUS assays of single explants and/or plants revealed that these differences reflected a deviating onset of silencing among isogenic plants and between leaves of a plant. This is exemplified for 5-week-old plants harboring loci GUS 1xf 60 and GUS 1xf 110 homozygously (Figure 6A). Analysis of the same set of plants at the age of 5 and 9 weeks revealed loss of GUS activity throughout development (Figure 6A).
Figure 6.
Variability of Transgene Expression between Different Leaves and among Isogenic Plants.
(A) Difference in the onset of silencing among plants harboring four copies of the GUS gene under the control of the CaMV 35S promoter. Colorimetric GUS activity assays were performed for transgenic plants that were either hemizygous or homozygous for two different GUS 1x T-DNA loci. Two leaf discs per plant were analyzed, and the results for a particular plant at the age of 5 and 9 weeks are shown underneath each other. The intensity of the blue staining reflected the GUS expression level.
(B) Systemic spread of GFP gene silencing in a 6-week-old plant homozygous for three different GFP 1x T-DNA loci. Fluorescence was detected using different filter sets. Red corresponds to chlorophyll fluorescence, whereas the green signal is specific for GFP fluorescence. The leaf marked by an arrow did not show any GFP fluorescence.
To compare the onset of silencing in plants that carried the same number of GUS genes, but which differed in transgene expression levels, we exploited the finding that plants harboring loci GUS 1xr 30 and GUS 1xr 69 had higher GUS transcript and activity levels than those carrying loci GUS 1xf 60 and GUS 1xf 110 (Figure 2A; data not shown). In plants harboring two copies of the GUS gene, evidence for reduced GUS expression was noted in single 9-week-old plants carrying loci GUS 1xr 30 and GUS 1xr 69, whereas all plants containing loci GUS 1xf 60 and GUS 1xf 110 showed high expression (Figure 6A). Almost no GUS activity was found in 5-week-old plants carrying four of the more highly expressed GUS gene copies, whereas high GUS activity was observed for several explants of plants harboring four of the GUS gene copies conferring a lower expression level (Figure 6A). Thus, we noted that the onset of silencing was generally earlier in plants in which more highly expressed loci had been combined.
Evidence for the commencement of silencing during development was also found for lines displaying silencing of the SPT or GFP genes. For example, high expression was found in 2-week-old plants harboring 12 copies of the SPT gene, whereas 8-week-old plants displayed silencing (Figure 4A; data not shown). Moreover, analysis of plants harboring six copies of the GFP gene for GFP fluorescence revealed patterns of GFP fluorescence consistent with the systemic spread of silencing (Figure 6B; data not shown).
DISCUSSION
Populations of independent lines transformed with the same T-DNA construct have been indispensable to the analysis of the extent of transgene expression variability and/or gene silencing, but the use of different experimental systems often yielded apparently conflicting results, and different hypotheses have been put forward to explain this phenomenon. Additionally, case studies of transformants have provided a detailed description of specific transgene insertions in silenced petunia and tobacco (Nicotiana tabacum) lines (Pröls and Meyer, 1992; Iglesias et al., 1997). However, case studies are not suitable for drawing general conclusions because it is impossible to judge the frequency at which insertion events, such as the ones described, occur.
Therefore, we have chosen to conduct a systematic and comprehensive study of transgene expression in Arabidopsis. This analysis enabled us to determine to what extent number and arrangement of transgene copies, the position of T-DNA integration in the genome, and transgene nature contribute to the variability of transgene expression and/or cause silencing. However, the approach we have taken was not suitable to assess the effect of truncated and/or rearranged T-DNA copies on transgene expression.
Neither Position Effects, Repetitive Elements, nor Repeat Arrangements of Transgenes per se Account for the Large Variability of Transgene Expression Seen among Transformants
To assess the influence of the position of T-DNA integration on transgene expression, expression analysis was focused on 79 independent, single-copy T-DNA lines mapping to different positions in the Arabidopsis genome. Without exception, differences in expression levels less than twofold were found between plants harboring two copies of a particular transgene under the control of the strong CaMV 35S promoter (Figures 2 and 3) in agreement with data from a small number of single-copy transgenic lines in tobacco (Hobbs et al., 1990). Thus, the documented several 100-fold differences in expression level (e.g., independent transgenic tobacco and Arabidopsis lines harboring GUS genes under the control of the CaMV 35S promoter [Figure 5]; Hobbs et al., 1990; Holtorf et al., 1995) cannot be attributed to position effects. These results are especially noteworthy because the Arabidopsis and tobacco genomes markedly differ in size (Arumuganathan and Earle, 1991). However, genomic sequences flanking transgene insertion sites can significantly influence expression of lowly transcribed transgenes as is evident from studies of transgenic lines carrying enhancer trap T-DNA constructs (Campisi et al., 1999).
In Drosophila melanogaster, a clonally inherited variegated expression pattern is observed when genes are translocated from a euchromatic environment into the vicinity of heterochromatin (Wakimoto, 1998). In our work, analysis of reporter gene expression revealed stable expression of the NPT gene under the control of the weak nopaline synthase promoter in all 132 transgenic lines tested, regardless of the positions of T-DNA integration in the Arabidopsis genome. Remarkably, position effects by flanking plant DNA sequences were also ruled out as the primary determinant of silencing in P. hybrida (Jorgensen et al., 1996), a species that has a genome ∼10-fold larger than that of Arabidopsis (Arumuganathan and Earle, 1991). Plants containing two GUS genes in the pericentromeric region of chromosome I (GUS 1xr rDNA 125 and GUS 2xr 126) showed similarly high expression levels when compared with other transgenic lines harboring two copies of the GUS reporter gene in other parts of the genome. Moreover, repetitive sequences from Arabidopsis, some of which contribute to heterochromatic regions (summarized in Schmidt, 1998), did not affect transgene expression when placed immediately upstream of the GUS gene, regardless of whether high-copy tandem repeat sequences (Martinez-Zapater et al., 1986) or less common dispersed repetitive elements (Thompson et al., 1996) were analyzed (Figure 3C).
We cannot rule out the possibility that a subset of T-DNA insertions might have been immediately subjected to TGS. Such events would not have been recovered in our study because all primary transformants were initially selected for expression of the NPT gene. The results of our study are nevertheless of broad significance because expression of a selectable and/or screenable marker is the basis for identification of transgenic plants in nearly all experiments in which transgenic plants are generated for use in biotechnology and academic research.
Given the high frequency of genes in repeat arrangements in the Arabidopsis genome (Arabidopsis Genome Initiative, 2000), it is particularly important to assess whether repeat structures per se influence gene expression. Transcribed inverted repeat structures have been found to trigger gene silencing efficiently (Waterhouse et al., 1998; Chuang and Meyerowitz, 2000; Mette et al., 2000; Sijen et al., 2001). Thus, in transgene populations in which inverted repeat T-DNA structures result in read-through transcripts capable of duplex formation, such T-DNA arrangements may contribute significantly to gene silencing, whereas inverted repeat T-DNA structures unlikely to produce double-stranded RNA (dsRNA) did not mediate reporter gene silencing (Lechtenberg et al., 2003). In none of 37 transgenic Arabidopsis lines studied in the context of this work did a direct repeat arrangement of transgene sequences alter reporter gene expression. Gene silencing of transgene copies arranged as concatemeric array was observed in Drosophila (Dorer and Henikoff, 1994) but not for all genes studied (Clark et al., 1998), clearly documenting that transgene repetitiveness as such is not sufficient for gene silencing in Drosophila, similar to our findings in Arabidopsis.
These results rule out that the position of transgene integration, vicinity to repetitive element sequences, and repetitive transgene arrangements per se are major determinants of the high variability of transgene expression in independent transformants.
Silencing by Surpassing a Threshold
It is important to determine the influence of gene copy number on expression because only 35% of the predicted protein-coding genes are single copy in the Arabidopsis genome (Arabidopsis Genome Initiative, 2000), and gene copy number changes are particularly frequent events in plant genome evolution (summarized in Schmidt, 2002).
We have documented a direct proportionality between transgene copy number and expression for the three reporter genes tested (Figures 3 and 4) unless the copy number of the introduced genes exceeded a gene-specific threshold (Figures 4 and 5); in these cases, silencing was observed. These findings reconcile the seemingly contradictory data that were obtained in previous studies of different transgene populations in plants. On the one hand, a positive correlation between transgene copy number and expression was reported (Stockhaus et al., 1987; Hobbs et al., 1993; Ku et al., 1999); on the other hand, many transformants containing multiple copies of an introduced gene showed low levels of transgene expression and/or silencing (Hobbs et al., 1990; Jorgensen et al., 1996).
Methylation and siRNAs specific for the transcribed regions (Figure 5), onset and systemic spread of silencing during development (Figure 6), and meiotic reversibility of the process (data not shown) have been documented for the silenced transgenes studied in this work. All these features support a posttranscriptional mechanism of gene silencing (summarized in Vaucheret et al., 1998; Hamilton and Baulcombe, 1999). Plants harboring two GUS transgenes under the control of the CaMV 35S promoter showed silencing in the enhancer of gene silencing2 mutant background (D. Schubert and R. Schmidt, unpublished results) consistent with the result that PTGS is more efficient in this mutant than in wild-type plants (Dehio and Schell, 1994).
Because stable expression and silencing of reporter genes did not correlate with the orientation of the reporter genes relative to the T-DNA vector in the transgenic lines studied in this work (data not shown), the possibility that unintended T-DNA vector antisense transcripts act as triggers for silencing is excluded. The GUS, GFP, and SPT reporter genes were controlled by the same set of regulatory elements (Figure 1A), yet silencing was observed at a different copy number threshold for the three transgenes (Figure 4). These results, taken together with the observation that silencing was neither triggered by promoterless copies nor copies of the GUS gene under the control of the weak nopaline synthase promoter (Figures 3B and 5B), made clear that it is equally unlikely that unintended antisense transcripts derived from the reporter gene cassettes contribute to gene silencing in the lines studied. A central role has been proposed for dsRNA in RNA silencing (summarized in Vaucheret et al., 2001); however, because dsRNA does not appear to be a direct product of the highly transcribed transgenes studied in this work, we infer that dsRNA is produced by another means, for example, by the RNA-dependent RNA polymerase encoded by the SGS2/SDE1 gene (Dalmay et al., 2000; Mourrain et al., 2000). Vaucheret et al. (2001) proposed that SGS2/SDE1, as well as SDE3, SGS3, and AGO1, are essential for the production of dsRNA in transgenic lines displaying PTGS of highly transcribed transgenes, whereas transgene loci producing dsRNA do not have this requirement. Our results are consistent with this model.
Several lines of evidence suggest that sense RNA–mediated silencing is triggered by threshold concentrations of transgene transcript or another product of transgene expression. First, silencing susceptibility depended on transgene dose rather than on the interaction of alleles at the same locus (Figures 4 and 5). Second, promoterless copies or copies of the GUS gene under the control of the weak nopaline synthase promoter failed to trigger silencing (Figures 3B and 5B). Third, comparisons of plants harboring the same number of GUS genes but differing in expression levels revealed an overall earlier onset of silencing in plants in which more highly expressed loci were present (Figure 6A). The findings that the frequency of silencing positively correlates with the promoter strengths of introduced genes (Que et al., 1997) and that silencing is more pronounced in homozygous than in corresponding hemizygous plants (summarized in Vaucheret et al., 1998) also support the model that sense RNA–mediated silencing is triggered by threshold concentrations of transgene transcript or another product of transgene expression. Furthermore, the threshold model is consistent with results of gene expression studies of full-length Adh transgenes in Drosophila (Pal-Bhadra et al., 2002).
The Nature of the Coding Regions Determines Gene-Specific Thresholds
In the transgenic lines studied in this work, the GUS, GFP, and SPT reporter genes were controlled by the same set of regulatory elements (Figure 1A). Nonetheless, lines containing two copies of the GUS transgene had lower steady state transgene transcript levels than lines carrying two copies of the SPT or GFP genes (Figure 2). Thus, silencing was not only triggered at a different copy number threshold for the three transgenes (Figure 4) but also at different steady state transcript levels. This reveals that the nature of the coding region is an important determinant of the gene-specific threshold for the onset of silencing and that differences between genes are recognized by an RNA sensing mechanism.
The GFP, GUS, and SPT coding sequences have different contents of guanine and cytosine (GC) residues (44.7, 52.2, and 68.4% GC content, respectively). However, no correlation between the GC content of the coding sequences and the copy number threshold at which the different transgenes were silenced (Figure 4) was observed. The GFP and SPT transgenes have an almost identical transcript length, yet silencing was triggered at different copy numbers for these transgenes (Figure 4). Thus, transcript length and GC content of coding sequences are unlikely to be primary determinants of the gene-specific threshold.
Arabidopsis genes cluster into two groups that differ in codon usage. In highly expressed genes, a C is preferentially found at the codon silent sites, whereas in genes with lower expression levels a T is predominantly used (Mathé et al., 1999). The SPT, GFP, and GUS open reading frames carry C at ∼44, 34, and 31% of codon silent sites, respectively. Adaptation of GFP gene codon usage toward a higher proportion of codons with a C or a G in the third position resulted in a more frequent occurrence of highly expressing plants in a population of transgenic lines (Rouwendal et al., 1997). It is tempting to speculate that codon usage plays a role in determining the gene-specific silencing threshold.
Less than 10-fold reduction in steady state RNA level was found for silenced GUS transgenes, whereas GUS activity was reduced >100-fold when compared with plants with stable GUS reporter gene expression (Figures 4 and 5). A reduced protein/transcript ratio has also been documented for posttranscriptionally silenced NPT transgenes (Van Houdt et al., 1997). Possibly, these findings suggest that PTGS operates through translational inhibition as well as by siRNAs directing cleavage of complementary RNAs. In view of the recent findings summarized below, it will be important to assess whether the reduced protein/transcript ratio in lines harboring posttranscriptionally silenced GUS and NPT genes might be brought about by a dual role of siRNAs. The Caenorhabditis elegans miRNA let-7 functions in translational repression by partially base pairing to the 3′-untranslated region of target mRNAs but is also capable of directing cleavage of a perfectly complementary target mRNA (Hutvágner and Zamore, 2002). Conversely, siRNAs can function in a similar way to miRNAs via translational repression if they do not contain perfect complementary target sequences (Doench et al., 2003). In Arabidopsis, several miRNAs match targets with a high degree of sequence complementarity. Like siRNAs, they direct the cleavage of their target RNAs (summarized in Bartel and Bartel, 2003). By contrast, miRNA172, which is highly complementary to a region in the mRNA of APETALA2, regulates expression of this Arabidopsis floral homeotic gene primarily through translational inhibition rather than by RNA cleavage (Aukerman and Sakai, 2003; Chen, 2004).
Based on our comprehensive study of highly transcribed transgenes, we propose the following model that is immediately relevant to all overexpression studies in Arabidopsis and to studies that exploit gene fusions to GFP and GUS (e.g., for localization). The expression of highly similar genes is additive unless transgene transcript levels exceed a gene-specific threshold and consequently trigger PTGS. Positive gene dosage effects and, in particular, the stochastic onset of silencing effectively account for the large variability of transgene expression seen among transformants. Reduced protein/mRNA ratios in silenced lines may also contribute to the observed large differences in transgene expression. Thus, the use of weaker promoters and/or appropriate modification of codon usage should result in a reduced variability of transgene expression in populations of transformants. It may be equally useful to transform transgenes of interest into mutants with impaired PTGS (Butaye et al., 2004) or into lines expressing proteins that suppress PTGS (Vaucheret et al., 2001). Importantly, our data indicate that a sensitive monitoring system suitable to identify cells exhibiting transgene silencing would allow for an efficient screening of plants showing high and stable transgene expression. Such a system would be exceptionally valuable for the analysis of transgenic lines in research and biotechnology.
The transgene system described mimics the effects of gene copy number changes that have been particularly frequent events in plant genome evolution (summarized in Schmidt, 2002). Endogenous genes should have become adapted such that transcript threshold levels are not exceeded, but it is tempting to speculate that the RNA sensing mechanism would counterbalance an excessive amplification of highly transcribed genes. Studies of the natural Chs mutants Red Star and Velvet Picotee that both contain a tandem arrangement of two closely linked almost identical Chs genes support the hypothesis that changes in copy number of endogenous genes may also be subject to the RNA sensing mechanism. Posttranscriptional silencing of the Chs genes in these mutants resembles Chs transgene silencing in several respects (Stam et al., 1998). The RNA sensing mechanism may play a particularly important role as a genome surveillance system capable of defending a genome against molecular parasites, such as transposons and viruses.
METHODS
T-DNA Constructs
The SPT, GFP, and GUS reporter gene cassettes were retrieved as HindIII fragments from plasmids SLJ1491 (Jones et al., 1992), pMDL-mgfp5-ER, and pMDL-GUS (Lechtenberg et al., 2003), respectively. The GFP reporter gene cassette was cloned into the HindIII site of pMDL-T-DNA (Forsbach et al., 2003) to generate plasmids GFP 1xf and GFP 1xr (Figure 1A). Constructs SPT 1xf, SPT 1xr, SPT 2xf, SPT 2xr, and SPT 3xf (Figures 1A and 1B) were established by cloning one to three copies of the SPT reporter gene cassette into pMDL-T-DNA. One to three copies of the GUS reporter gene cassette were cloned into pMDL-T-DNA to generate constructs GUS 1xf, GUS 1xr, GUS 2xf, GUS 2*xf, GUS 2xr, GUS 3xf, and GUS 3xr (Figures 1A and 1B). In construct GUS 2*xr, the SPT reporter gene cassette has been inserted between two copies of the GUS reporter gene. Several constructs were made that harbored repetitive Arabidopsis thaliana sequences in the HindIII site immediately upstream of the CaMV 35S promoter sequences of construct GUS 1xr (Figure 1C). The 360-bp HindIII fragment containing two copies of the 180-bp HindIII tandem repeat sequence was retrieved from vector pAL (Martinez-Zapater et al., 1986). One copy and two copies of the 500-bp HindIII tandem repeat sequence (Simoens et al., 1988) were present in constructs GUS 1xr 501 and GUS 1xr 502, respectively. Two different dispersed repetitive element sequences were retrieved as HindIII fragments from clones 163A and 164A (Thompson et al., 1996). To generate construct GUS 1xr rDNA, a 4.8-kb XhoI/XbaI fragment corresponding to the intergenic spacer of the rDNA sequences (Wanzenböck et al., 1997) was cloned immediately upstream of the CaMV 35S promoter sequences. Construct GUS 1xr was digested with XhoI and subsequently religated to yield a T-DNA vector that carried a promoterless GUS reporter gene (ΔGUS 1xr, Figure 1D). The Pro35S:GUS:ocs3′ reporter gene cassette was cloned into the HindIII site of vector ΔGUS 1xr to establish construct GUS/ΔGUS r (Figure 1D). The SalI/XhoI fragment from vector pMDL-GUS (Lechtenberg et al., 2003), which contained the CaMV 35S promoter sequences, was replaced with an SmaI/SalI fragment corresponding to ProNOS sequences to yield a plasmid carrying a ProNOS:GUS:ocs3′ reporter gene cassette. The ProNOS:GUS:ocs3′ reporter gene cassette was cloned as a HindIII fragment into pMDL-T-DNA (Forsbach et al., 2003) to generate constructs NOS-GUS 1xr and NOS-GUS 2xr (Figure 1E).
Molecular-Genetic Analysis of Transgenic Plants
The different T-DNA vectors were used for Agrobacterium tumefaciens–mediated transformation of Arabidopsis Columbia-0 and single-copy T-DNA lines were identified as described (Forsbach et al., 2003; Lechtenberg et al., 2003). Integrity of the reporter gene cassettes was evaluated for plants of different generations using DNA gel blot and PCR analyses. This study demonstrated the stable inheritance of the different T-DNA insertions. The plant growth conditions have been described previously (Lechtenberg et al., 2003). Germination of seeds in the presence of kanamycin revealed plants homozygous for a single-copy T-DNA locus. A plant homozygous for a particular T-DNA was crossed with a wild-type plant to yield progeny hemizygous for this locus. Intercrosses of different homozygous transgenic lines were performed to combine different T-DNA loci. PCR analyses were used to establish the presence of the different T-DNA loci in the progeny plants.
Expression Analysis of Transgenic Lines
RNA preparations and RNA gel blot analyses were performed as previously described (Lechtenberg et al., 2003). Hybridization results were quantified with a phosphor imager. For each transgenic line, the ratio of the transgene and actin gene specific signals was calculated. Glucuronidase activity was analyzed using either colorimetric (Lechtenberg et al., 2003) or fluorometric assays (Jefferson et al., 1986). For fluorometric measurements, two different protein extracts were prepared from 5 to 10 small rosette leaves of up to 10 plants for each transgenic line, and two aliquots were assayed for each of the extracts to determine protein concentration and GUS activity. Protein concentrations of plant extracts were determined spectrophotometrically using the Bio-Rad protein assay (Bio-Rad, Richmond, CA) based on the Bradford method. Fluorescence of 4-methylumbelliferone (4-MU) was measured with excitation at 360 nm and emission at 455 nm with the Versa-Fluor fluorimeter (Bio-Rad). Calibration of the instrument was performed using freshly prepared standards of 4-MU in 0.2 M Na2CO3. For each transgenic line, the mean of four GUS activity measurements and the according standard deviation were calculated. GUS activity is given as nmol or pmol 4-MU/min/mg protein. A fluorometric assay was performed to assess GFP fluorescence quantitatively (Lechtenberg et al., 2003). For each transgenic line, two different protein extracts were prepared from 10 to 15 small rosette leaves of up to 10 plants, and the mean of four fluorescence measurements and the standard deviation were determined. GFP fluorescence is given as relative fluorescence units/mg protein. A fluorescence stereomicroscope (Leica MZ FL III, GFP2 filter set [excitation at 480 nm and emission at 510 nm], GFP3 filter set [excitation at 470 nm and emission at 525 nm]; Leica, Mannheim, Germany) was used to detect GFP fluorescence in transgenic plants.
Acknowledgments
The research was initiated at the Max Delbrück Laboratory with funding granted by the German Federal Ministry of Education and Research (BMBF, Grant 0311107). D.S., B.L., and S.B. were supported by fellowships from the Max Planck Society. We thank the greenhouse staff at the Max Delbrück Laboratory and the Max-Planck-Institute of Molecular Plant Physiology for taking care of the plants. Skillful technical assistance by S. Stegemann is gratefully acknowledged. We thank M. McKenzie for carefully editing the manuscript and C. Dean, I. Witt, J. Goodrich, N. Johnsson, J. Lunn, R. Simon, and C. Wood for comments on the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Renate Schmidt (rschmidt@mpimp-golm.mpg.de).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.024547.
References
- An, Y.Q., McDowell, J.M., Huang, S., McKinney, E.C., Chambliss, S., and Meagher, R.B. (1996). Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 10, 107–121. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Sequence and analysis of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Arumuganathan, K., and Earle, E.D. (1991). Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9, 208–218. [Google Scholar]
- Aukerman, M.J., and Sakai, H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15, 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, B., and Bartel, D.P. (2003). MicroRNAs: At the root of plant development? Plant Physiol. 132, 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butaye, K.M.J., Goderis, I.J.W.M., Wouters, P.F.J., Pues, J.M.-T.G., Delauré, S.L., Broekaert, W.F., Depicker, A., Cammue, B.P.A., and De Bolle, M.F.C. (2004). Stable high-level transgene expression in Arabidopsis thaliana using gene silencing mutants and matrix attachment regions. Plant J. 39, 440–449. [DOI] [PubMed] [Google Scholar]
- Campisi, L., Yang, Y., Yi, Y., Heilig, E., Herman, B., Cassista, A.J., Allen, D.W., Xiang, H., and Jack, T. (1999). Generation of enhancer trap lines in Arabidopsis and characterization of expression patterns in the inflorescence. Plant J. 17, 699–707. [DOI] [PubMed] [Google Scholar]
- Chen, X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang, C.F., and Meyerowitz, E.M. (2000). Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97, 4985–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, D.V., Sabl, J.F., and Henikoff, S. (1998). Repetitive arrays containing a housekeeping gene have altered polytene chromosome morphology in Drosophila. Chromosoma 107, 96–104. [DOI] [PubMed] [Google Scholar]
- Dalmay, T., Hamilton, A., Rudd, S., Angell, S., and Baulcombe, D.C. (2000). An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101, 543–553. [DOI] [PubMed] [Google Scholar]
- Day, C.D., Lee, E., Kobayashi, J., Holappa, L.D., Albert, H., and Ow, D.W. (2000). Transgene integration into the same chromosome location can produce alleles that express at a predictable level, or alleles that are differentially silenced. Genes Dev. 14, 2869–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehio, C., and Schell, J. (1994). Identification of plant genetic loci involved in a posttranscriptional mechanism for meiotically reversible transgene silencing. Proc. Natl. Acad. Sci. USA 91, 5538–5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench, J.G., Petersen, C.P., and Sharp, P.A. (2003). siRNAs can function as miRNAs. Genes Dev. 17, 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorer, D.R., and Henikoff, S. (1994). Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell 77, 993–1002. [DOI] [PubMed] [Google Scholar]
- Forsbach, A., Schubert, D., Lechtenberg, B., Gils, M., and Schmidt, R. (2003). A comprehensive characterization of single-copy T-DNA insertions in the Arabidopsis thaliana genome. Plant Mol. Biol. 52, 161–176. [DOI] [PubMed] [Google Scholar]
- Hamilton, A.J., and Baulcombe, D.C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286, 950–952. [DOI] [PubMed] [Google Scholar]
- Haseloff, J., Siemering, K.R., Prasher, D.C., and Hodge, S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94, 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs, S.L., Kpodar, P., and DeLong, C.M. (1990). The effect of T-DNA copy number, position and methylation on reporter gene expression in tobacco transformants. Plant Mol. Biol. 15, 851–864. [DOI] [PubMed] [Google Scholar]
- Hobbs, S.L., Warkentin, T.D., and DeLong, C.M. (1993). Transgene copy number can be positively or negatively associated with transgene expression. Plant Mol. Biol. 21, 17–26. [DOI] [PubMed] [Google Scholar]
- Holtorf, S., Apel, K., and Bohlmann, H. (1995). Comparison of different constitutive and inducible promoters for the overexpression of transgenes in Arabidopsis thaliana. Plant Mol. Biol. 29, 637–646. [DOI] [PubMed] [Google Scholar]
- Hutvágner, G., and Zamore, P.D. (2002). A microRNA in a multiple-turnover RNAi enzyme complex. Science 297, 2056–2060. [DOI] [PubMed] [Google Scholar]
- Iglesias, V.A., Moscone, E.A., Papp, I., Neuhuber, F., Michalowski, S., Phelan, T., Spiker, S., Matzke, M., and Matzke, A.J. (1997). Molecular and cytogenetic analyses of stably and unstably expressed transgene loci in tobacco. Plant Cell 9, 1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R.A., Burgess, S.M., and Hirsh, D. (1986). β-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc. Natl. Acad. Sci. USA 83, 8447–8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G., Dunsmuir, P., and Bedbrook, J. (1985). High-level expression of introduced chimaeric genes in regenerated transformed plants. EMBO J. 4, 2411–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G., Shlumukov, L., Carland, F., English, J., Scofield, S.R., Bishop, G.J., and Harrison, K. (1992). Effective vectors for transformation, expression of heterologous genes, and assaying transposon excision in transgenic plants. Transgenic Res. 1, 285–297. [DOI] [PubMed] [Google Scholar]
- Jorgensen, R.A., Cluster, P.D., English, J., Que, Q., and Napoli, C.A. (1996). Chalcone synthase cosuppression phenotypes in petunia flowers: Comparison of sense vs. antisense constructs and single-copy vs. complex T-DNA sequences. Plant Mol. Biol. 31, 957–973. [DOI] [PubMed] [Google Scholar]
- Ku, M.S., Agarie, S., Nomura, M., Fukayama, H., Tsuchida, H., Ono, K., Hirose, S., Toki, S., Miyao, M., and Matsuoka, M. (1999). High-level expression of maize phosphoenolpyruvate carboxylase in transgenic rice plants. Nat. Biotechnol. 17, 76–80. [DOI] [PubMed] [Google Scholar]
- Lechtenberg, B., Schubert, D., Forsbach, A., Gils, M., and Schmidt, R. (2003). Neither inverted repeat T-DNA configurations nor arrangements of tandemly repeated transgenes are sufficient to trigger transgene silencing. Plant J. 34, 507–517. [DOI] [PubMed] [Google Scholar]
- Lindbo, J.A., Silva-Rosales, L., Proebsting, W.M., and Dougherty, W.G. (1993). Induction of a highly specific antiviral state in transgenic plants: Implications for regulation of gene expression and virus resistance. Plant Cell 5, 1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliga, P., Svab, Z., Harper, E.C., and Jones, J.D. (1988). Improved expression of streptomycin resistance in plants due to a deletion in the streptomycin phosphotransferase coding sequence. Mol. Gen. Genet. 214, 456–459. [DOI] [PubMed] [Google Scholar]
- Martinez-Zapater, J.M., Estelle, M.A., and Somerville, C.R. (1986). A highly repeated DNA sequence in Arabidopsis thaliana. Mol. Gen. Genet. 204, 417–423. [Google Scholar]
- Mathé, C., Peresetsky, A., Déhais, P., Van Montagu, M., and Rouzé, P. (1999). Classification of Arabidopsis thaliana gene sequences: Clustering of coding sequences into two groups according to codon usage improves gene prediction. J. Mol. Biol. 285, 1977–1991. [DOI] [PubMed] [Google Scholar]
- Matzke, M.A., Matzke, A.J., Pruss, G.J., and Vance, V.B. (2001). RNA-based silencing strategies in plants. Curr. Opin. Genet. Dev. 11, 221–227. [DOI] [PubMed] [Google Scholar]
- Mette, M.F., Aufsatz, W., van der Winden, J., Matzke, M.A., and Matzke, A.J. (2000). Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 19, 5194–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain, P., et al. (2000). Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101, 533–542. [DOI] [PubMed] [Google Scholar]
- Muskens, M.W., Vissers, A.P., Mol, J.N., and Kooter, J.M. (2000). Role of inverted DNA repeats in transcriptional and post-transcriptional gene silencing. Plant Mol. Biol. 43, 243–260. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra, M., Bhadra, U., and Birchler, J.A. (2002). RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol. Cell 9, 315–327. [DOI] [PubMed] [Google Scholar]
- Peach, C., and Velten, J. (1991). Transgene expression variability (position effect) of CAT and GUS reporter genes driven by linked divergent T-DNA promoters. Plant Mol. Biol. 17, 49–60. [DOI] [PubMed] [Google Scholar]
- Pröls, F., and Meyer, P. (1992). The methylation patterns of chromosomal integration regions influence gene activity of transferred DNA in Petunia hybrida. Plant J. 2, 465–475. [DOI] [PubMed] [Google Scholar]
- Que, Q., Wang, H.-Y., English, J.J., and Jorgensen, R.A. (1997). The frequency and degree of cosuppression by sense chalcone synthase transgenes are dependent on transgene promoter strength and are reduced by premature nonsense codons in the transgene coding sequence. Plant Cell 9, 1357–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouwendal, G.J., Mendes, O., Wolbert, E.J., and de Boer, A.D. (1997). Enhanced expression in tobacco of the gene encoding green fluorescent protein by modification of its codon usage. Plant Mol. Biol. 33, 989–999. [DOI] [PubMed] [Google Scholar]
- Schmidt, R. (1998). The Arabidopsis thaliana genome: Towards a complete physical map. In Arabidopsis, Annual Plant Reviews, Vol. 1, M. Anderson and J.A. Roberts, eds (Sheffield: Sheffield Academic Press), pp. 1–30.
- Schmidt, R. (2002). Plant genome evolution: Lessons from comparative genomics at the DNA level. Plant Mol. Biol. 48, 21–37. [PubMed] [Google Scholar]
- Siemering, K.R., Golbik, R., Sever, R., and Haseloff, J. (1996). Mutations that suppress the thermosensitivity of green fluorescent protein. Curr. Biol. 6, 1653–1663. [DOI] [PubMed] [Google Scholar]
- Sijen, T., Vijn, I., Rebocho, A., van Blokland, R., Roelofs, D., Mol, J.N., and Kooter, J.M. (2001). Transcriptional and posttranscriptional gene silencing are mechanistically related. Curr. Biol. 11, 436–440. [DOI] [PubMed] [Google Scholar]
- Simoens, C.R., Gielen, J., Van Montagu, M., and Inzé, D. (1988). Characterization of highly repetitive sequences of Arabidopsis thaliana. Nucleic Acids Res. 16, 6753–6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam, M., Viterbo, A., Mol, J.N., and Kooter, J.M. (1998). Position-dependent methylation and transcriptional silencing of transgenes in inverted T-DNA repeats: Implications for posttranscriptional silencing of homologous host genes in plants. Mol. Cell. Biol. 18, 6165–6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockhaus, J., Eckes, P., Blau, A., Schell, J., and Willmitzer, L. (1987). Organ-specific and dosage-dependent expression of a leaf/stem specific gene from potato after tagging and transfer into potato and tobacco plants. Nucleic Acids Res. 15, 3479–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, H.L., Schmidt, R., and Dean, C. (1996). Identification and distribution of seven classes of middle-repetitive DNA in the Arabidopsis thaliana genome. Nucleic Acids Res. 24, 3017–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houdt, H., Ingelbrecht, I., Van Montagu, M., and Depicker, A. (1997). Post-transcriptional silencing of a neomycin phosphotransferase II transgene correlates with the accumulation of unproductive RNAs and with increased cytosine methylation of 3′ flanking regions. Plant J. 12, 379–392. [Google Scholar]
- Vaucheret, H., Béclin, C., Elmayan, T., Feuerbach, F., Godon, C., Morel, J.B., Mourrain, P., Palauqui, J.C., and Vernhettes, S. (1998). Transgene-induced gene silencing in plants. Plant J. 16, 651–659. [DOI] [PubMed] [Google Scholar]
- Vaucheret, H., Béclin, C., and Fagard, M. (2001). Post-transcriptional gene silencing in plants. J. Cell Sci. 114, 3083–3091. [DOI] [PubMed] [Google Scholar]
- Wakimoto, B.T. (1998). Beyond the nucleosome: Epigenetic aspects of position-effect variegation in Drosophila. Cell 93, 321–324. [DOI] [PubMed] [Google Scholar]
- Wanzenböck, E.-M., Schöfer, C., Schweizer, D., and Bachmair, A. (1997). Ribosomal transcription units integrated via T-DNA transformation associate with the nucleolus and do not require upstream repeat sequences for activity in Arabidopsis thaliana. Plant J. 11, 1007–1016. [DOI] [PubMed] [Google Scholar]
- Waterhouse, P.M., Graham, M.W., and Wang, M.B. (1998). Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc. Natl. Acad. Sci. USA 95, 13959–13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore, P.D. (2002). Ancient pathways programmed by small RNAs. Science 296, 1265–1269. [DOI] [PubMed] [Google Scholar]