Abstract
WRKY transcription factors form a large family that plays a role in plant responses to biotic stress and during senescence. Defining in vivo relevant WRKY/promoter relationships has been hampered by the factors' indiscriminate binding to known W box DNA elements and their possible genetic redundance. Employing chromatin immunoprecipitations (ChIP) of cultured cells, we show that parsley (Petroselinum crispum) WRKY1 protein binds to the W boxes of its native promoter as well as to that of PcWRKY3 and the defense-related PR10-class marker gene Pathogenesis-Related1-1 (PcPR1-1). Although present at low concentrations in resting cells, WRKY1 does not appear to play a role in the immediate early gene response upon elicitation because it does not bind to the promoter at this time. Paradoxically, in vivo binding at the PcWRKY1 promoter correlates more with downregulation of gene expression, whereas previous overexpression studies suggested an activating function of WRKY1 on PcWRKY1 expression. By contrast, PcPR1-1 expression remains strong when its promoter is occupied in vivo by WRKY1. Unexpectedly, ChIP revealed that W boxes at promoter sites are constitutively occupied by other WRKY transcription factors, indicating that site recruitment does not seem to play a major role in their regulation. Rather, WRKY proteins very likely act in a network of mutually competing participants with temporal displacement occurring at defined preoccupied sites by other family members in a stimulus-dependent manner.
INTRODUCTION
WRKY proteins are a major family of plant transcription factors implicated in the regulation of various processes, including pathogen defense (Eulgem et al., 1999; Chen and Chen, 2000, 2002; Du and Chen, 2000; Kim et al., 2000; Maleck et al., 2000; Robatzek and Somssich, 2001, 2002; Yu et al., 2001; Chen et al., 2002; Cormack et al., 2002; Deslandes et al., 2002; Yoda et al., 2002), wound response (Dellagi et al., 2000; Hara et al., 2000; Yoda et al., 2002), trichome development (Johnson et al., 2002a), and senescence (Hinderhofer and Zentgraf, 2001; Robatzek and Somssich, 2001, 2002; Johnson et al., 2002a). In Arabidopsis thaliana, WRKY proteins consist of 74 members, each defined by the presence of at least one DNA binding WRKY domain, a 60–amino acid stretch that contains the amino acid sequence WRKYGQK and a zinc-finger–like motif (Eulgem et al., 2000). Based on sequence comparisons of WRKY domains, a further grouping into three major subfamilies (groups I to III) was proposed (Eulgem et al., 2000).
High conservation of the WRKY domain is mirrored by a remarkable conservation of the W box as the binding site for WRKY proteins. Gel-shift experiments (Chen and Chen, 2000), DNA-ligand binding screens (Rushton et al., 1996), random binding site selection (Du and Chen, 2000), yeast one-hybrid studies (Cormack et al., 2002), and cotransfection assays (Rushton et al., 1996) performed with different WRKY proteins have demonstrated that this cis-acting element with its minimal consensus TGAC-C/T is the preferred cognate binding site. However, little is known about the specific interaction of a given WRKY protein with a defined promoter. The importance of WRKY proteins in pathogen responses is highlighted by the fact that the TGAC-C/T motif is significantly overrepresented in promoters of Arabidopsis genes that are coregulated with PR1, a robust marker gene associated with systemic acquired resistance (Maleck et al., 2000). Although some of these promoters display up to eight W boxes, a single W box can be sufficient to render transcription WRKY factor dependent (Eulgem et al., 1999).
Previous work has shown that the elicitor-mediated transcriptional activation of the parsley (Petroselinum crispum) PcWRKY1 gene is of the immediate-early response type (Cormack et al., 2002). Despite containing several TGAC-C/T consensus W box core motifs (Eulgem et al., 1999), a composite element within the PcWRKY1 promoter composed of two adjacent W boxes in inverted orientation, designated WBC, is necessary and sufficient for the elicitor-dependent induction of reporter gene activity in transiently transfected parsley protoplasts (Eulgem et al., 1999). A homologous W box arrangement is found in the parsley PcWRKY3 promoter that has equally been shown to confer elicitor inducibility in reporter gene assays (Eulgem et al., 1999). Cotransfection with an expression plasmid encoding WRKY1 induces the WBC-dependent reporter gene activity independent of elicitor treatment (Eulgem et al., 1999). However, several other WRKY proteins not highly related to WRKY1 are capable of binding equally well to the WBC element (Cormack et al., 2002).
From such studies a model was derived that proposed WRKY-dependent transcriptional activation to follow a two step procedure. First, upon perception of the signal, a pool of preformed WRKY proteins undergo posttranslational modifications, thereby activating genes of the immediate-early response type. Second, because several WRKY genes belong to this class of induced genes, a massive subsequent increase of WRKY proteins occurs that would partly be responsible for a slower but prolonged activation of downstream target genes. It remains unresolved whether WRKY proteins, such as WRKY1, can play roles in both steps and if so, how the positive feedback they confer on their own expression can be controlled.
Nearly all current data regarding WRKY protein–W box interactions involve in vitro assays, such as electrophoretic mobility shift assays and DNA-ligand binding or in vivo experiments under nonphysiological conditions employing strong overexpression of the respective proteins. Although such studies have yielded important information, they may not necessarily reflect how WRKY factors modulate gene expression in vivo nor can they detect specific WRKY protein interactions with endogenous plant promoters. For this, chromatin immunoprecipitation (ChIP) after formaldehyde cross-linking provides an experimental tool that allows one to monitor protein–DNA and protein–protein interactions in vivo under physiological conditions in a dynamic manner (Orlando, 2000). Originally employed in studies in metazoae and yeast, the method has recently been successfully applied to plants particularly to study the genomic distribution of histones and their modified forms (Chua et al., 2001; Johnson et al., 2002b) but also to study transcription factor/promoter interactions (Johnson et al., 2001; Lopez-Molina et al., 2002).
Using ChIP, we demonstrate that the parsley PcWRKY3 and Pathogenesis-Related1-1 (PcPR1-1) genes are in vivo WRKY1 targets as is PcWRKY1 itself. However, contrary to previous studies that showed WRKY1 to be an activator of its own gene expression, in vivo WRKY1 binding correlated more with downregulation of PcWRKY1 transcription. By contrast, WRKY1 binding to the PR10 class marker gene, PcPR1-1 (also designated Ypr10*Pc.1; van Loon et al., 1994), coincided with high expression levels of PcPR1-1. WRKY1 binds preferentially to W boxes at the PcPR1-1 promoter, as compared with such elements found at the PcWRKY1 promoter if present in submaximal concentrations within the cells. ChIP resolution scanning of the PcWRKY1 locus with an antiserum that recognizes most of the WRKY protein family revealed that W boxes at promoter sites are constitutively occupied by WRKY proteins. Based on our results, we propose a site occupation/site displacement model where WRKY proteins act in a network of mutually competing participants.
RESULTS
Preformed and de Novo Synthesized WRKY Proteins in Stimulated Parsley Cells
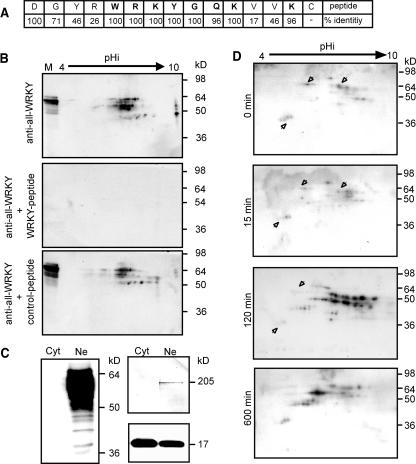
To demonstrate the presence of a pool of preformed WRKY proteins in parsley cells, we generated antipeptide antibodies directed against a stretch of amino acids highly conserved in all WRKY proteins. The peptide was designed on the basis of all 74 Arabidopsis WRKY protein sequences known so far. Because the nearly invariant peptide, WRKYGQK, appeared to be too short to provide sufficient immunogenicity, the sequence was extended four positions toward the N terminus and three positions toward the C terminus (Figure 1A). The derived conserved sequence has highest similarity to group I WRKY proteins (Eulgem et al., 2000). The specificity of the antiserum toward the peptide was assessed by peptide competition assays. Protein gel blots of nuclear extracts from Arabidopsis suspension cultured cells separated by two-dimensional PAGE show ∼50 spots of molecular sizes between 40 and 90 kD (Figure 1B, top panel). Incubation in the presence of immunogenic peptide substantially decreases the signals from all spots to background levels (Figure 1B, middle panel), whereas parallel incubation with an equal amount of unrelated control peptide has no effect (Figure 1B, bottom panel). Similar competition protein gel blots performed with one-dimensionally separated nuclear extracts revealed that the serum specifically recognizes proteins with similar size distribution from both parsley and Arabidopsis (see Supplemental Figure 1A online). Further validation that the observed signals represented WRKY proteins were obtained from the analysis of different cellular fractions from parsley cells by protein gel blotting. In accordance with experiments performed with WRKY-green fluorescent protein fusion proteins, which showed that PcWRKY1 and AtWRKY6 proteins are localized in the nucleus (Eulgem et al., 1999; Robatzek and Somssich, 2001), signals detected by the anti-all-WRKY serum were exclusively found in the nuclear fraction, whereas they were absent in cytosolic fractions (Figure 1C). Furthermore, bacterially expressed representative members from all WRKY subgroups were recognized by the serum that did not cross-react with bacterial proteins (see Supplemental Figure 1B online).
Figure 1.
Characterization of WRKY Protein Pools.
(A) Immunogenic peptide for the generation of anti-all-WRKY serum. Rabbits were immunized with the depicted 15–amino acid long peptide that spans the most conserved part of the WRKY domain. The overall identities of sequential amino acids to all 74 known Arabidopsis WRKY proteins are indicated.
(B) Specificity of the generated serum. Arabidopsis nuclear extracts were separated by two-dimensional PAGE and probed with the anti-all-WRKY serum alone (anti-all-WRKY, top panel), in the presence of immunogenic peptide (+ WRKY-peptide, middle panel), or in the presence of the same amount of unrelated peptide (+ control-peptide, bottom panel). At the left border of the gel, a mixture of prestained protein gel markers and Arabidopsis nuclear extract was loaded for one-dimensional separation (M).
(C) Cytoslic/nuclear distribution of WRKY proteins. The anti-all-WRKY serum was used to probe parsley cytosolic (Cyt) and nuclear (Ne) extracts from 1 h pep25 stimulated parsley cells for the presence of WRKY proteins (left panel). The quality of cell fractionation was assessed by probing parallel blots with monoclonal antibodies against the Ser-5 phosphorylated heptapeptide repeat YSPTSPS of RNA polymerase II (right, top panel) or a serum raised against PcPR1-1 (right, bottom panel).
(D) Preformed and induced WRKY protein pools in parsley. Nuclear extracts from resting parsley cells (0 min, first panel) or cells stimulated for 15 min (15 min, second panel), 120 min (120 min, third panel), or 600 min (600 min, fourth panel) with pep25 elicitor were separated by two-dimensional PAGE and analyzed by protein gel blots using the anti-all-WRKY serum. Arrowheads denote proteins showing reduced levels in elicitor treated cells. Molecular mass range is indicated at the right. The entire time course was analyzed twice, and time points from 0 to 120 min were analyzed five times in independent experiments.
Analysis of nuclear extracts from parsley suspension cultured cells demonstrated the existence of a preformed WRKY protein pool (Figure 1D, first panel). The pattern was complex with at least 20 distinguishable signals and a molecular size distribution from 40 to 90 kD. Several WRKY proteins migrate as adjacent pearl strings, suggesting that the same WRKY protein exists in different posttranslationally modified forms. Notably, within the separated range, WRKY protein levels are low in resting parsley cells. Immediately after stimulation with the peptide elicitor pep25, only minor changes in the WRKY migration pattern can be observed (Figure 1D, second panel). Furthermore, these minor changes can be interpreted as a redistribution of signals to adjacent spots within a pearl string (Figure 1D, first and second panels, arrowheads). Expression of numerous WRKY family members have been shown to be rapidly induced under pathogen-mimicking conditions (Cormack et al., 2002; Dong et al., 2003; Kalde et al., 2003). Therefore, as expected, the pool of detectable WRKY proteins was greatly enhanced in complexity as well as in quantity 2 h poststimulation of cells with pep25 (Figure 1D, third panel). In addition to the appearance of several novel WRKY proteins, most of the preexisting spots also increase in signal strength. However, in at least two cases, a WRKY protein signal does decrease after stimulation (Figure 1D, third panel, arrowheads). At a later phase of the elicitor response, the signals for the majority of novel proteins decline, whereas a signal trail situated at the center of the two-dimensional gel is further enhanced and becomes prominent (Figure 1D, fourth panel).
This data demonstrate the presence of a complex pool of low-abundant preformed WRKY proteins, some of which may undergo rapid posttranslational modifications. Elicitor treatment of the cells dramatically alters the profile of this pool, most likely because of de novo and increased WRKY protein synthesis at early stages, followed by a transition back toward the resting state at later stages.
WRKY1 Protein Levels Strongly Increase after Elicitor Stimulation
WRKY1 mRNA cannot be detected in resting control cells by RNA gel blot analysis, but it is rapidly and transiently induced by pep25 (Rushton et al., 1996). Furthermore, PcWRKY1 transcriptional activation does not require de novo protein synthesis (Cormack et al., 2002) and is mediated through W box elements within its promoter (Eulgem et al., 1999). Moreover, it was shown that WRKY1 can bind to the WBC box within the PcWRKY1 promoter and can activate transcription (Eulgem et al., 1999). To monitor WRKY1 protein levels, we generated specific peptide antibodies against WRKY1. Database searches revealed that a suitable peptide derived from the C terminus of WRKY1 does not display significant homology to any other known plant protein except to its orthologs from Arabidopsis (AtWRKY33) and Pimpenella brachycarpa (ZPF1; AF080595). This is consistent with the fact that in Arabidopsis, homology is low at the C terminus of WRKY proteins even among closely related family members. The specificity of the signal detected by the antiserum in protein gel blots with WRKY1 was validated by peptide competition assays (data not shown).
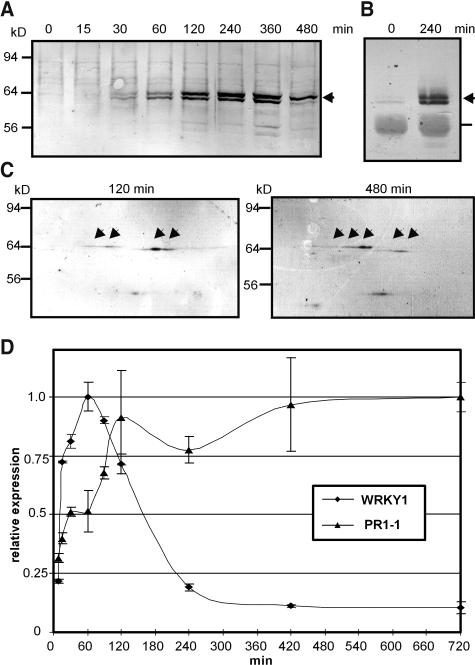
Protein gel blot analysis of WRKY1 protein levels over a time course of elicitor stimulation in parsley cells revealed that the protein is not detectable in control cells and in cells that have been stimulated for 15 min with pep25 elicitor but increases markedly 30 min after elicitation (Figure 2A). However, immunoprecipitation enrichment performed with covalently cyanogen bromide-agarose coupled anti-WRKY1 resulted in detection of WRKY1 in resting cells (Figure 2B). We observed that the protein migrates in at least two distinguishable coexisting forms, which could be indicative of posttranslational modifications. Furthermore, two-dimensional PAGE of WRKY1 reveals that both doublets are composed of several separable spots that show the typical pearl string migration pattern (Figure 2C, arrows). Whereas the most acidic of the faster migrating WRKY1 forms is dominant early in the elicitor response, at later times it is the most basic of the slower migrating WRKY1 forms that is most prominent (Figure 2C, compare left and right panels). All WRKY1-specific signals detected on two-dimensional protein gel blots can be overlapped with protein spots from parallel blots probed with the anti-all-WRKY serum, indicating that this serum recognizes various modified WRKY1 forms (see Supplemental Figure 1C online). Interestingly, a temporal comparison of WRKY1 mRNA and protein levels revealed that the protein is by far less transient in nature than its mRNA transcript, with maximum protein level reached 4 h after elicitor treatment and dropping markedly only after 8 h. Reverse-transcribed real-time PCR analysis confirmed previous RNA gel blot data (Rushton et al., 1996) that a marked increase in mRNA can be detected 15 min after elicitation with a transient peak maximum at 60 min (Figure 2D). By contrast, elicitor induced expression of PcPR1-1, whose promoter contains two widely spaced W boxes that can be equally bound by WRKY1 protein in vitro (Rushton et al., 1996), shows a closer correlation to altered WRKY1 protein levels (Figure 2D).
Figure 2.
Alterations of WRKY1 in Stimulated Parsley Cells.
(A) Direct protein gel blot analysis of parsley nuclear extracts during an 8 h time course of stimulation with pep25 elicitor using a WRKY1 peptide specific antiserum. The position of the WRKY1 protein doublet is marked by an arrow. Other visible signals on the blot correspond with background detected by the serum because they are not competed out by the immunogenic peptide (see Methods). Molecular mass range is indicated at the left. The experiment was performed five times with similar results.
(B) Protein gel blot analysis of immuno-enriched nuclear extracts. After immunoprecipition of WRKY1 from nuclear extracts of resting or 2 h elicitor treated cells with the help of cyanogen bromide-agarose coupled anti-WRKY1–specific serum, precipitates were separated on one-dimensional gels and reprobed with the same antibodies. WRKY1 protein doublet bands are marked by an arrow and IgG background by a line. The experiment was performed twice with similar results.
(C) Two-dimensional protein gel blot of WRKY1. Nuclear extracts of parsley cells that were pep25 elicitor treated for the indicated time were separated on two-dimensional gels. Pictures show portions of the original gels covering the pH range of 7 to 9. The position of WRKY1 signals are indicated by arrows. This experiment was performed twice with similar results.
(D) Analysis of WRKY1 and PR1-1 expression by RT-PCR. Changes in mRNA levels were assayed over a time course of elicitor stimulation of parsley cells with pep25 as indicated. Expression levels were analyzed by SYBR Green-based real-time PCR and expression values normalized to those of PcUbiquitin4. Values on the graph are normalized to the point of highest expression for each gene. Expression analyses of the full time course were performed twice with similar results.
WRKY1 Protein Binds to the PcWRKY1 Promoter in Vivo
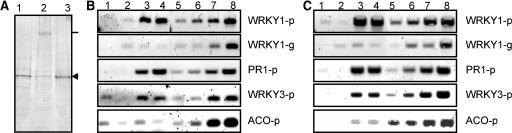
Because WRKY1 protein is present in the preformed WRKY factor pool, although at very low levels, it may directly mediate immediate-early induction of its own gene expression. On the other hand, WRKY1 mRNA levels are already strongly reduced later during elicitation at a time when WRKY1 protein is still present in the cells. This may argue against a direct protein/promoter interaction. We established a formaldehyde-mediated cross-linking approach in our cell culture system to evaluate whether WRKY1 protein/PcWRKY1 promoter interactions occur in the genomic context in vivo. Cells cultivated in suspension were cross-linked after 2 h elicitor stimulation by direct addition of formaldehyde to the culture medium. Nuclei were purified through a glycerol cushion before sonication in high detergent buffer. Proteins of parallel samples from noncross-linked control cells and cells that had been cross-linked for as little as 7 min were separated by SDS-PAGE and analyzed for WRKY1 by protein gel blots (Figure 3A). In contrast with the control sample (Figure 3A, lane 1), ∼50% of the protein in the cross-linked sample remains trapped in the pocket of the SDS-PAGE gel (Figure 3A, lane 2). We interpret this to be because of cross-linking of this proportion of WRKY1 protein to the DNA and possibly other proteins, resulting in large complexes that are excluded from the gel. Harsh heat treatment, which breaks the covalent bonds within such complexes, allows recovery of this proportion as de-cross-linked WRKY1 in the gel (Figure 3A, lane 3). Longer incubations with formaldehyde will trap all WRKY1 into high molecular weight complexes (data not shown). Because the resulting chromatin is so highly linked that it cannot be sheared into suitably sized fragments, such material is not suited for the subsequent immunoprecipitation of WRKY1 with WRKY1-specific antiserum. After immunoprecipitation of DNA/protein complexes, the DNA was recovered and analyzed by PCR. Primer combinations were chosen that amplify fragments of ∼300 bp either within the promoter of PcWRKY1, also encompassing the W box WBC (Figure 3B, WRKY1-p), or within the transcribed region, 1.3 kb downstream of the promoter (Figure 3B, WRKY1-g). Only background levels of PCR signal were detected if preimmune serum (Figure 3B, lane 1) or affinity-purified anti-WRKY1 antibodies in the presence of an excess of immunogenic WRKY1 peptide (Figure 3B, lane 2) were used in the incubations. The same levels were detected if the WRKY1-g primer combination was used on samples that were incubated with anti-WRKY1 antibodies alone (Figure 3B, lane 3) or with anti-WRKY1 antibodies in the presence of an excess of unrelated peptide (Figure 3B, lane 4). By contrast, at least a 10-fold enrichment, determined by comparing the signals to twofold dilutions of reference samples, of the PCR signals was seen in these incubations if the WRKY1-p primer combination was used (Figure 3B, lanes 3 and 4, WRKY1-p). Similar enrichments could be observed with primer combinations that covered the two W boxes contained in the parsley PcPR1-1 promoter (Figure 3B, PR1-p). Slightly lower enrichments were obtained when amplifying the PcWRKY3 promoter, which is coregulated with the PcWRKY1 promoter and includes structurally related W boxes (Figure 3B, WRKY3-p) (Eulgem et al., 1999). The W box free control promoter of the parsley acyl-CoA oxidase gene (ACO; Logemann et al., 2000) was not significantly enriched over background levels in immunoprecipitations (Figure 3B, ACO-p). Our data demonstrate that WRKY1 binds to the W box containing promoters of PcWRKY1, PcPR1-1, and PcWRKY3 in vivo under conditions where it is present in the cells and transcription of all three genes occurs.
Figure 3.
Detection of WRKY1 in Vivo Target Sites.
(A) Analysis of WRKY1 protein. Chromatin was prepared from cells treated with pep25 elicitor for 2 h (lane 1) or from cells subsequently fixed with formaldehyde (0.5%) for 7 min (lane 2) and analyzed by protein gel blots for the detection of WRKY1 protein. The cross-links from a parallel sample as in lane 2 were reversed by heat treatment (lane 3) and analyzed. The position of free WRKY1 is indicated by an arrowhead and the position of cross-linked complexes at the height of the gel pocket by a straight line. This experiment was performed three times with similar results.
(B) Analysis of DNA after anti-WRKY1 immunoprecipitation. Chromatin preparations from elicitor-treated and formaldehyde fixed cells as in (A) were immunoprecipitated with WRKY1-specific antibody. The DNA was recovered after reversal of the cross-links and analyzed for the enrichment of WRKY1 promoter (WRKY1-p), WRKY1 intragenic region (WRKY1-g), PR1-1 promoter (PR1-p), WRKY3 promoter (WRKY3-p), and parsley ACO promoter (ACO-p) by PCR. Immunoprecipitations were performed with anti-WRKY1 preimmune serum (lane 1), with affinity-purified anti-WRKY1 antibodies in the presence of immunogenic peptide (lane 2), with anti-WRKY1 antibodies in the presence of the same amount of unrelated peptide (lane 3), or with anti-WRKY1 antibodies alone (lane 4). As a control for quantities amplified during the PCR, sequential twofold dilutions of input material from chromatin preparations were processed in parallel. Lane 5, 1.25 × 10−4; lane 6, 2.5 × 10−4; lane 7, 0.5 × 10−3; lane 8, 1.0 × 10−3.
(C) Analysis of DNA after anti-all-WRKY immunoprecipitation. Parallel incubations were performed as in (B) except that anti-all-WRKY serum and immunogenic peptide were used in the incubations. The input dilutions were as follows: lane 5, 0.5 × 10−3; lane 6, 1.0 × 10−3; lane 7, 2.0 × 10−3; lane 8, 4.0 × 10−3. The experiment was performed twice with similar results.
Parallel ChIP experiments were also performed using the anti-all-WRKY serum. As for the WRKY1 specific antibodies, preimmune serum (Figure 3C, lane 1), immune serum in the presence of an excess of immunogenic peptide (Figure 3C, lane 2), immune serum alone (Figure 3C, lane 3), or immune serum in the presence of unrelated competitor peptide (Figure 3C, lane 4) was used for immunoprecipitation. The pattern of amplification is identical to the anti-WRKY1 pattern, indicating that the anti-all-WRKY serum can be used to immunoprecipitate cross-linked material.
Resolution of ChIP Analyses
Apart from the well-characterized WBC composite element, the PcWRKY1 promoter contains several additional W box consensus motifs upstream in its promoter. The presence of these upstream elements is not required for elicitor responsiveness of the promoter, but their presence may play a role in modulating the overall strength of the response (T. Eulgem and I.E. Somssich, unpublished data). In addition, PcWRKY1 possesses three perfect W box consensus motifs at the very end of the coding region, two of which are closely spaced and form an inverted repeat that is comparable in structure to WBC (F. Turck, unpublished data).
We assessed the resolution of the ChIP method in our experimental system via a PCR amplification scan throughout the genomic PcWRKY1 locus after elicitation of the cells (Figure 4A, boxes a to j). WRKY1 protein precipitates fragments throughout the 1.7-kb promoter region (Figures 4A and 4B). However, there seems to be substantially less enrichment if primer combination b is used that is positioned 1.3 kb away from the transcriptional start site and covers the second W box consensus. Significantly stronger enrichment is detected from the primer combination a, encompassing the first W box consensus motif. As expected, enrichments are detected from primer combinations d and e, which include the third and fourth W box consensus motif and the WBC composite element, respectively (Figures 4A and 4B). By contrast, the first primer combination within the transcribed region (f), which is situated 500 bp away from the WBC composite element, and all subsequent primer combinations (g to j), show no significant enrichment of PCR products above background. We conclude that the resolution of our ChIP experiments is roughly 500 bp and that the first, third, and fourth W boxes as well as the WBC element cluster are bound by WRKY1 1 h after elicitation, whereas the second W box may not be an in vivo target for WRKY1. Furthermore, although they possess perfect consensus motifs, the W boxes at the 3′-end of PcWRKY1 are not significantly bound by WRKY1.
Figure 4.
Resolution of ChIP.
(A) Schematic representation of the parsley WRKY1 locus. PcWRKY1 promoter and terminator regions are indicated by horizontal lines and transcribed sequences by closed boxes (untranslated regions), open boxes (exons), and bent lines (introns). The positions of W box consensus motifs are indicated by gray transversal lines, and the position of the well-characterized functional WBC cluster is labeled. The positions and relative sizes of the different PCR fragments covering the locus are indicated by labeled boxes (a to j).
(B) WRKY1 ChIP resolution scan. Formaldehyde cross-linked chromatin preparations (derived from 1 h elicitor treated cells) were immunoprecipitated with anti-WRKY1 preimmune serum (top panels, lane 1), anti-WRKY1–specific affinity-purified antibodies in the presence of immunogenic peptide (top panels, lane 2), anti-WRKY1 antibodies in the presence of unrelated peptide (top panels, lane 3), or with anti-WRKY1 antibodies alone (top panels, lane 4). DNA was recovered and analyzed on agarose gels after performing PCR with the different primer combinations yielding the appropriate fragments as indicated above the panels. As a control for quantities amplified during PCR, sequential twofold dilutions of input material from chromatin preparations were processed in parallel and are shown in the bottom panels. Lane 1, 1.25 × 10−4; lane 2, 2.5 × 10−4; lane 3, 0.5 × 10−3; lane 3, 1.0 × 10−3.
(C) All-WRKY ChIP scan. ChIP and analysis were performed as in (B) except that the anti-all-WRKY serum and specific peptide were used in the incubations. Parallel PCR of input dilutions are shown in the bottom panels. Lane 1, 0.5 × 10−3; lane 2, 1.0 × 10−3; lane 3, 2.0 × 10−3; lane 4, 4.0 × 10−3. The experiment was performed three times with similar results.
Interestingly, differences can be detected using the anti-all WRKY serum in parallel analyses (Figure 4C). Here the results indicate that all promoter regions containing W boxes, including the second W-box, are equally well bound by WRKY proteins. Again, no significant binding above background can be observed within the transcribed genomic region, but there appears to be a minor but clearly detectable enrichment of PCR product j, suggesting, that WRKY proteins interact with the W boxes at the end of the coding region (Figure 4C).
WRKY1 Binding to Its Own Promoter Is Not an Immediate-Early Event
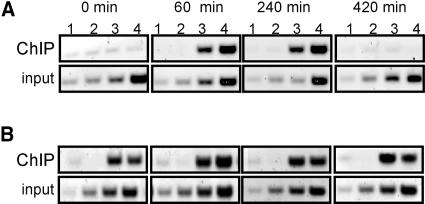
We analyzed the temporal behavior of WRKY1 binding to its own promoter during the response of parsley cells to elicitor. Despite the presence of WRKY1 protein in the cells before elicitor treatment (Figure 2B), no binding can be detected at the PcWRKY1 promoter before elicitor treatment or immediately afterwards (Figure 5A, top panel set, 0 to 15 min). Steady state WRKY1 mRNA levels are maximal 60 min after elicitor treatment, at a time when the level of WRKY1 protein has already increased but has not yet reached its peak (Figure 2). By this time, weak but clearly detectable binding of WRKY1 to its own promoter is first observed (Figure 5A, 60 min). The amount of immunoprecipitated PcWRKY1 promoter DNA markedly increases 120 to 240 min after elicitor stimulation, which correlates with the highest amount of detectable protein within the cells but also to the rapid drop of steady state WRKY1 mRNA levels (Figures 2D and 5A, 120 to 240 min). At later stages, the binding of WRKY1 to its own promoter diminishes to undetectable levels, which does not overlap with the disappearance of the protein from the cells but with the reduction of the faster migrating form of WRKY1 (Figure 2A). Identical temporal binding patterns were observed for all promoter fragments previously identified as WRKY1 target sites (data not shown for PCR fragments a, c, and d). No WRKY1 binding was observed to the W box elements at the end of the coding region (Figure 5A, box j). In conclusion, WRKY1 does not play a role in the immediate-early induction of its own gene expression, and strongest detected binding to its own promoter correlates with the highest levels of WRKY1 protein.
Figure 5.
Elicitor-Dependent Time-Course Analysis of WRKY Factor Binding to the WRKY1 Promoter.
(A) In vivo WRKY1 binding to parsley WRKY1. Pep25-treated parsley cells were formaldehyde fixed over a time course of 12 h and analyzed by ChIP using anti-WRKY1 preimmune serum (lane 1), anti-WRKY1–specific antibodies in the presence of immunogenic peptide (lane 2), anti-WRKY1–specific antibodies in the presence of unrelated peptide (lane 3), or with anti-WRKY1–specific antibodies alone (lane 4). PCR was performed using primer combinations e (top panel set) and j (bottom panel set). Diluted samples from ChIP inputs were analyzed by PCR in parallel (bottom row of each set, lane 1, 1.25 × 10−4; lane 2, 2.5 × 10−4; lane 3, 0.5 × 10−3; lane 3, 1.0 × 10−3).
(B) Overall WRKY factor binding to parsley WRKY1. ChIP was performed from the same chromatin samples as in (A) with anti-all-WRKY preimmune serum (lane 1), anti-all-WRKY immune serum in the presence of immunogenic peptide (lane 2), anti-all-WRKY immune serum in the presence of unrelated peptide (lane 3), or with anti-all-WRKY immune serum alone (lane 4). PCR was performed using primer combinations e (top panel set), j (middle panel set), and h (bottom panel set). Diluted samples from ChIP inputs were analyzed by PCR in parallel (bottom row of each panel set, lane 1, 0.5 × 10−3; lane 2, 1.0 × 10−3; lane 3, 2.0 × 10−3; lane 4, 4.0 × 10−3). The experiment covering the entire time course was performed twice with similar results, whereas the time points from 0 to 240 min were analyzed in five independent experiments with similar results.
WRKY Proteins Constitutively Occupy the W Boxes within the PcWRKY1 Promoter
We next assessed the binding of other WRKY proteins to the PcWRKY1 promoter. Immunoprecipitations of extracts from in vivo cross-linked cells with the anti-all-WRKY serum revealed unchanging coverage of the PcWRKY1 promoter by WRKY proteins, independent of the status of elicitation of the cells and, consequently, transcription rate of the gene (WRKY1-e, Figure 5B, top panel set). As illustrated in Figure 4C, a weaker but significant enrichment of binding at the W box consensus site at the end of the coding region is observed (WRKY1-j, Figure 5B, middle panel set), whereas no binding is detected in the middle of the coding region (WRKY1-h, Figure 5B, bottom panel set). Thus, it appears that accessible W box elements, such as found in promoter regions, are constitutively occupied by WRKY proteins but that the WRKY composition at a given promoter is dynamic.
WRKY1 Binding to the PcPR1-1 Promoter
The binding of WRKY1 specifically and WRKY proteins in general to the PcPR1-1 promoter was also assessed by ChIP. As for the PcWRKY1 promoter, WRKY1 protein is not bound to promoter sites before elicitor treatment; whereas other WRKY proteins are clearly present (Figure 6, cf. top to bottom panel sets, 0 min). Binding of WRKY1 at the PcPR1-1 promoter is observed from 60 min up to 240 min after elicitor stimulation (Figure 6A, top panel set), which is comparable to the binding observed at the PcWRKY1 promoter (Figure 5A, top panel set). By contrast, binding to PcPR1-1 W boxes is equally strong at 60 min and 240 min with a fold enrichment at 60 min that is twofold to fourfold higher for PcPR1-1 than for PcWRKY1. Because the amount of WRKY protein has not yet reached maximal levels 60 min after elicitation, WRKY1 appears to preferentially target PcPR1-1 binding sites over the PcWRKY1 binding sites if present in limiting amounts. However, in both cases, W box promoter elements are constitutively occupied by WRKY proteins, irrespective of the expression status of the gene. Thus, the presence of a particular WRKY protein at a given W box correlates with high concentration of this WRKY protein within the cell but to a certain degree also on preference of the specific element.
Figure 6.
Elicitor-Dependent Time-Course Analysis of WRKY Factor Binding in Vivo to the Parsley PR1-1 Promoter.
(A) WRKY1-specific binding. Pep25 treated parsley cells were formaldehyde treated over a time course of 7 h and analyzed by ChIP using anti-WRKY1 preimmune serum (lane 1), anti-WRKY1–specific antibodies in the presence of immunogenic peptide (lane 2), anti-WRKY1–specific antibodies in the presence of unrelated peptide (lane 3), or with anti-WRKY1–specific antibodies alone (lane 4). PCR was performed using a primer combination that covered the W box–containing region of PR1-1. Diluted input samples from ChIP were analyzed by PCR in parallel (bottom row of panel set, lane 1, 1.25 × 10−4; lane 2, 2.5 × 10−4; lane 3, 0.5 × 10−3; lane 3, 1.0 × 10−3).
(B) Overall WRKY factor binding. Samples were processed and analyzed as in (A) except that anti-all-WRKY serum and immunogenic peptide were used in the incubations. Diluted samples from ChIP inputs were analyzed by PCR in parallel (bottom row of each panel set, lane 1, 0.5 × 10−3; lane 2, 1.0 × 10−3; lane 3, 2.0 × 10−3; lane 4, 4.0 × 10−3). The entire time-course experiment was performed twice with similar results, whereas time points from 0 to 240 min were analyzed in five independent experiments with similar results.
DISCUSSION
Several reports have highlighted the importance of W boxes in rapid, immediate-early transcriptional responses to various stimuli (Eulgem et al., 1999; Asai et al., 2002). This led to the postulation that a pool of preformed WRKY proteins exists in the cells, which would be responsive to signaling events, such as posttranslational modifications. In apparent contrast with this is the finding that, so far, all reported WRKY proteins have the potential to be stimulus-independent transcription factors (de Pater et al., 1996; Eulgem et al., 1999; Hara et al., 2000; Asai et al., 2002; Robatzek and Somssich, 2002; Yoda et al., 2002). Because such studies were all performed with transiently overexpressed proteins, it was proposed that WRKY proteins may be controlled in their activities by the close association with an inhibitory partner that is titrated out by an excess of WRKY protein (Asai et al., 2002), a mechanism termed squelching (Cahill et al., 1994). As shown in several plant species, a significant number of WRKY genes are themselves part of the W box–mediated early gene activation response, implying that a stimulus may greatly increase the pool of these proteins within the cells. Indeed, in Arabidopsis, 49 out of 72 tested WRKY genes responded to pathogen-mimicking stimuli (Dong et al., 2003). Because the increase in transcription of most of these early response WRKY genes is transient, the current model fails to take into account how the positive feedback on WRKY gene expression is circumvented at high concentrations of WRKY proteins.
Our study is the first to visualize the different WRKY pools and their changes at the protein level. We show that the transition from the preformed pool of WRKY proteins to the induced pool is indeed dramatic (Figure 1) and most likely also involves protein posttranslational modifications. The nature of these modified forms and their functional importance for WRKY protein activity will be the focus of future work. Closer analysis of WRKY1 protein in particular revealed that it is present at very low levels in resting cells, but its abundance increases significantly upon elicitation (Figure 2). Based on its migration pattern on two-dimensional PAGE, modifications of WRKY1 may be functionally important for the regulation of its activity as a transcription factor. We cannot completely rule out the possibility that a WRKY1-like protein is synthesized upon elicitation that may cross-react with the WRKY1 antiserum. However, 3′ rapid amplification of cDNA ends performed on RNA derived from parsley cells 1 h after elicitation, in combination with an oligo(dT) primer and five different degenerate primers corresponding to various parts of the WRKY1 peptide sequence, resulted exclusively in amplification products that were sequence identical to PcWRKY1 (data not shown).
Because the physiological relevance of data obtained in experiments employing protein overexpression remain highly questionable (Zhang, 2003), we monitored the in vivo binding behavior of endogenous WRKY proteins to W boxes. Our experiments confirmed that WRKY1 indeed binds to its own promoter; however, clear binding only occurred at later stages when the protein was present at relatively high levels in the cell. This finding has several implications on the postulated working model as outlined below.
Elicitor-Dependent Regulation of WRKY1 Target Sites in Vivo
WRKY1 protein is present in the preformed WRKY pool, and overexpressed WRKY1 can stimulate transcription of its own promoter (Eulgem et al., 1999). Despite these facts, we propose that it does not play a direct role in the immediate-early gene response in vivo. We postulate that before elicitor stimulation, other WRKY factors occupy the W boxes of the PcWRKY1 promoter and are thus more likely candidates to function as activators/derepressors via a posttranslational mechanism. Constitutive occupancy of W boxes in the promoter may fulfill several functions. First, the presence of an inactive transcription factor at a potential positive promoter element ensures effective protection against basal expression of the regulated gene. Similar molecular switches are exemplified in studies in metazoae and yeast (Barolo and Posakony, 2002). Second, preoccupancy of the promoter site may be of importance for the delay of induction conferred by the promoter because posttranslational modifications on prebound factors should be extremely rapid. It has recently been demonstrated in parsley cells that elicitor-dependent activation of the immediate-early genes PcWRKY1 and PcPR2 occurs via a defense pathway that is dependent on posttranslational phosphorylation and activation of a distinct mitogen-activated protein kinase cascade (Kroj et al., 2003). The mitogen-activated protein kinase components of this pathway, PcMPK3a,/b and PcMPK6 are translocated to the nucleus in an elicitor-dependent manner and function as response activators (Lee et al., 2004). One intriguing possibility is that such kinases directly modify selective prebound WRKY factors at the PcWRKY1 promoter site in analogy to the mode of action of the Hog1 kinase in yeast (De Nadal et al., 2004). These prebound WRKY proteins may have escaped attention because their synthesis is not significantly enhanced upon elicitation or because they do not act as transcriptional activators in transient overexpression studies.
In such a scenario, overexpression of defined WRKY factors, such as WRKY1, may lead to displacement of the in vivo regulating prebound WRKY proteins from the promoter, thereby allowing nonregulated activation of the target gene. This may partly explain why WRKY1, which has transactivation capability, was previously assumed to be part of the transcription complex required for immediate-early activation of PcWRKY1 (Eulgem et al., 1999). In light of our in vivo findings, it may be necessary to reevaluate several previous reports on direct WRKY transcription factor/target gene or transcription factor/biological function relationships that have been postulated solely on the basis of ectopic overexpression studies (Asai et al., 2002; Chen and Chen, 2002; Robatzek and Somssich, 2002).
Elicitor-mediated activation of PcWRKY1 results in increased WRKY1 mRNA and subsequent protein levels. Because the arrangement of W boxes and their surrounding DNA sequences in the PcWRKY1 and PcPR1-1 promoters are significantly different (Eulgem et al., 1999), the composition of the associated protein complexes to which WRKY1 is recruited at these sites may also differ. In the case of the PcPR1-1 promoter, WRKY1 may indeed positively affect gene expression. This is supported by the fact that the time point of WRKY1 appearance at this promoter closely coincides with the elicitor-dependent protein/DNA alterations observed at the W box element by in vivo genomic footprinting and with the concomitant onset of PcPR1-1 transcription (Meier et al., 1991). Several transcription factors can act either as activators or as repressors of gene expression in a tissue-specific and promoter-dependent context (Latchman, 2001; Barolo and Posakony, 2002). In Arabidopsis, AtWRKY6 was shown to negatively regulate its own promoter both in plants and in protoplasts but to positively effect SIRK gene expression (Robatzek and Somssich, 2002; S. Robatzek and I.E. Somssich, unpublished data). Whether the elicitor-dependent recruitment of WRKY1 to the PcWRKY1 promoter influences expression in a positive manner or acts as a repressor, as is the case for rice (Oryza sativa) OsWRKY71 (Zhang et al., 2004), requires further in-depth analysis.
Clearly, as revealed by ChIP, we expect other WRKY factors along with as yet unidentified regulatory factors to also be involved in coregulating both PcWRKY1 and PcPR1-1. It is important to note that the PR10 class parsley gene PcPR1-1 share neither sequence relationship nor subcellular localization with the PR1 class of genes and their encoded products (van Loon et al., 1994). Currently, regulatory factors involved in PR10 gene regulation are ill defined. Although TGA factors have been shown to be important in pathogen-mediated activation of certain PR genes (Després et al., 2003), the promoter regions of PcWRKY1 and PcPR1-1 critical for elicitor-induced expression both lack cognate TGA/OBF factor binding sites (AS1 elements). Recently, a novel single-stranded DNA binding factor, designated Whirly (StWhy1), was found to be involved in the regulation of the PR-10a gene in potato (Solanum tuberosum), and its Arabidopsis ortholog, AtWhy1, was demonstrated to be required for salicylic acid–dependent disease resistance (Desveaux et al., 2004). StWhy1 binds to a PR-10a promoter sequence termed PB element (GTCAAAAA). PB elements potentially overlap with both TGA/OBF and WRKY binding sites, suggesting a possible interplay between Whirly factors and these two types of transcription factors at distinct defense-response promoters.
WRKY1 Is Displaced from in Vivo Target Sites at Later Stages during the Elicitor Response
Loss of in vivo binding to both the PcWRKY1 and PcPR1-1 promoters later than 420 min after elicitation is unlikely attributable to WRKY1 degradation because the protein can still be detected on protein gel blots. Rather, additional WRKY1 modifications may be involved. Indicative for this is the observed alteration of the migration profile of WRKY1 on one-dimensional and two-dimensional PAGE (Figures 2A and 2C). A similar shift in electrophoretic mobility was recently observed for the Arabidopsis transcription factor TGA1, caused by an intramolecular conformational change associated with alterations in the cellular redox state occurring upon pathogen attack or elicitor treatment (Després et al., 2003). In this case, binding of TGA1 protein to its target sites was dependent on being in the reduced state. In contrast with TGA1, treatment with reducing agents does not convert the WRKY1 doublet to a single band during SDS-PAGE (data not shown). Future experiments will be directed to the isolation of the modified WRKY1 forms and their biochemical characterization.
Accessibility and Discrimination of W Boxes
The resolution of ChIP was ∼500 bp, which does not allow discrimination between adjacent W boxes (Figure 4). Nevertheless, it does allow us to conclude that W boxes at a given promoter are widely associated with WRKY proteins and that different WRKY factors may be found at a given promoter simultaneously. We interpret the higher fold enrichment of the ChIP signal when using the anti-all-WRKY serum in immunoprecipitations as compared with the WRKY1-specific serum (Figure 4) to indicate that WRKY proteins, other than WRKY1, bind to and coregulate the promoter. Alternatively, this could depend entirely on the higher affinity of the anti-all-WRKY serum to cross-linked protein/DNA complexes. Although our results confirm the promiscuous binding of WRKY proteins to several W boxes (Figure 4), they also demonstrate that not all W box elements are accessible for WRKY factors. The three W box sequences present in the transcribed region of PcWRKY1 fit perfectly to the consensus and are indeed strongly bound by several bacterial expressed WRKY proteins in in vitro gel-shift experiments (data not shown). Yet, only a very minor signal is enriched from these non-promoter-derived W boxes in ChIP experiments with the anti-all-WRKY serum (Figure 5B, middle panel set). The higher fold enrichment of the PcPR1-1 promoter compared with that of PcWRKY1 at 60 min after elicitation indicates that WRKY factors may have specific target preferences in vivo, a finding not observed in other experimental set ups so far.
In summary, we could demonstrate elicitor-mediated recruitment of the sequence-specific transcription factor WRKY1 to in vivo cognate binding sites at two distinct parsley genomic loci. This work, apart from being a starting point for identifying additional target genes of WRKY1, highlights the universal power of ChIP to provide new in vivo insights concerning the spatial and temporal occupancy of promoters by DNA binding proteins also in nonmodel plant species. For WRKY factors, this approach unexpectedly revealed that W boxes at promoter sites are constantly occupied by regulatory WRKY proteins that compete for binding at selective W box elements with other de novo synthesized family members, thereby illustrating the need of a more global approach to understand WRKY protein functions. Given the complexity of the protein family's response to pathogen-related stimuli, it is obvious that a more extended analysis of WRKY proteins is required to understand their mode of actions in the defense-triggered WRKY-dependent transcriptional regulatory network. Fortunately, a protein family encompassing approach is becoming feasible in Arabidopsis where the ChIP technique can be applied to an entire set of wrky knockout mutants expressing functional epitope-tagged WRKY proteins driven by their native promoters or to probe for specific WRKY promoter interactions on a genome-wide level.
METHODS
A detailed protocol of the ChIP method employed in this work is available in the supplemental data online.
Antibody Generation and Purification
Antibodies with specificity to parsley WRKY1 or family specificity to all WRKY proteins were generated by immunizing rabbits with the keyhole limpet hemocyanin–coupled synthetic peptide CKDEPDVDSFFDSFLA or DGYRWRKYGQKVVKC, respectively. The Cys residues were added to the peptides to ensure efficient coupling to keyhole limpet hemocyanin carrier protein. Antibodies to WRKY1 were further affinity purified using bacterial expressed, His tagged WRKY1 protein as previously published (Gu et al., 1994). WRKY1 antiserum was used in protein gel blots (1/2000 dilution), whereas affinity-purified antibodies were used in immunoprecipitations. Anti-all-WRKY family serum was used without additional purifications in protein gel blots (1/4000 dilution) and immunoprecipitations. Successful cell fractionation was verified by protein gel blot with serum raised against bacterially expressed PcPr1-1 (1/3000 dilution) or with commercially available monoclonal antibodies (COVANCE, Berkeley, CA) that recognize the S-5 phosphorylated form of the YSPTSPS repeat of eukaryotic RNA-polII (1/1000 dilution).
Cell Cultures, Elicitor Treatment, and Formaldehyde Fixation
Cell suspension cultures of parsley (Petroselinum crispum) were maintained in modified B5 medium as described (Kombrink and Hahlbrock, 1986). Cell cultures were elicitor treated 5 d after passage by the addition of pep25 elicitor (300 ng/mL) for the indicated time. For ChIP, cells were fixed 7 min in the presence of 0.5% formaldehyde under constant agitation (120 rpm). Fixation was stopped by the addition of 100 mM Gly and incubation for 5 min under constant shaking. Cells were then harvested by filtration, flash-frozen in liquid N2, and stored at −80°C until further processing.
RNA Isolation and Reverse Transcription
Total RNA was extracted from 200 mg of frozen ground cells with the RNAwizz solution (Ambion, Austin, TX) according to the manufacturer's protocol. Reverse-transcribed PCR was performed in a two step method using 450 ng of total RNA as template.
Two-Dimensional PAGE
Acetone-precipitated nuclear protein extracts of parsley cells (100 μg) were resuspended in rehydration buffer (8 M urea, 1% CHAPS, 100 mM DTT, 0.2% pharmalytes 2-10, and 0.001% orange G) and separated on 7-cm immobilized pH gradient strips with a pH range of 2 to 10 (Bio-Rad, Hercules, CA) on a Protein II IEF cell according to the manufacturer's instructions using an active rehydration protocol. The strips were equilibrated in equilibration buffers I (6 M urea, 2% SDS, 0.375 M Tris/HCl, pH 8.8, 20% glycerol, and 130 mM DTT) and II (6 M urea, 2% SDS, 0.375 M Tris/HCl, pH 8.8, 20% glycerol, and 135 mM iodoacetamide) for 10 min each before separation by SDS-PAGE. Proteins were transferred onto polyvinylidene fluoride membranes and analyzed by protein gel blots according to standard protocols.
Nuclear Extractions and Chromatin Preparations
Cells (5 g from 35 mL of liquid culture) were ground to fine powder in liquid N2, suspended in 40 mL of 70% glycerol buffer (20 mM Hepes/NaOH, pH 7.4, 5 mM MgCl2, 5 mM KCl, 50 mM saccharose, 70% glycerol, 1 mM DTT, 1 μg/mL of leupeptine, 1 μg/mL of pepstatin, and 200 μM phenylmethylsulfonyl fluoride), and incubated for 20 min at 4°C under mild shaking. After filtration of the slurry through a double layer of 70 μM and 20 μM nylon mesh, nuclei were collected by centrifugation (3000g, 40 min, 4°C). Aliquots of the supernatant were collected as the cytosolic fraction. Nuclei were washed once in 10% glycerol buffer by gentle resuspension followed by centrifugation. To obtain nuclear extracts from uncross-linked cells, proteins were extracted for 30 min in 500 μL high salt buffer (20 mM Hepes/NaOH, pH 7.4, 5 mM MgCl2, 0.5 M NaCl, 1 mM DTT, 1 μg/mL of leupeptin, 1 mg/mL of pepstatin, and 200 μM phenylmethylsulfonyl fluoride) at 4°C on a rotary shaker, followed by 20 min centrifugation (13000g, 4°C). Formaldehyde cross-linked nuclei were resuspended in 500 μL of sonication buffer (10 mM Hepes/NaOH, pH 7.4, 1 mM EDTA, and 0.5% SDS), incubated at 4°C for 30 min on a rotary shaker, and diluted with 500 μL sonication buffer without SDS. Chromatin was sheared to an average size of 1.5 kb by repetitive sonication with a Branson 450 sonicator (Branson Ultrasonics, Danbury, CT) equipped with a microtip in an ethanol/ice bath (output position 3.5, five times 30 s followed by cooling for 2 min). The sonicated chromatin was cleared by centrifugation (13,000g, 10 min, 4°C), followed by ultracentrifugation in a TLA100.2 rotor (Beckman Coulter, Fullerton, CA; 70,000 rpm, 4°C, 20 min). The sonicated chromatin was flash-frozen in liquid N2 and could be stored for several weeks. Protein analysis of sheared, cross-linked chromatin was performed after concentration of samples by acetone precipitation (80% acetone, −20°C, overnight). Reversal of cross-links was obtained by incubation of samples in 1× Laemmli buffer with β-mercaptoethanol (1 h, 96°C).
Immunoprecipitations
Before ChIP, DNA content of the preparations was measured as OD260 after reversal of cross-links by incubation of 50 μL of aliquots in the presence of 10 μg of Proteinase K for 12 h at 37°C, followed by incubation at 65°C for 8 h. Free DNA was purified from the solution by phenol/Tris-EDTA buffer extraction followed by phenol/chloroform extraction. RNase A was added to the aqueous phase and incubated for 10 min at ambient temperature before the DNA was precipitated by the addition of one-tenth volume of 3 M Na acetate and 2.5 volumes ethanol. The size distribution of DNA fragments was monitored for each sample by agarose-gel electrophoresis. For ChIP, equal amounts of chromatin preparations as measured by DNA content (8 to 16 μg) were adjusted to equal concentrations with dilution buffer (10 mM Hepes/NaOH, pH 7.4, 1 mM EDTA, and 0.25% SDS), mixed with 1.5 volumes of RIPA buffer (50 mM Hepes/NaOH, pH 7.4, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, and 0.1% deoxycholate), and incubated in the presence of preimmune serum (5 μL) and Protein A agarose beads (30 μL) for 2 h at 4°C on a rotary shaker. Beads were pelleted, and supernatants were split in four equal volumes and incubated with preimmune serum (3 μL), immune serum (3 μL), or affinity-purified antibodies (<1 μg) in the presence of free immunogenic or control peptide (50 μg) or without immunogenic peptide in the presence of specific antiserum for 4 h on ice. Samples were cleared from unspecific precipitations by centrifugation (13000g, 10 min, 4°C), the supernatant transferred to fresh Eppendorf tubes, supplemented with 30 μL Protein A agarose beads, and incubated for 1 h on a rotary shaker at 4°C. Immunoprecipitates were washed four times in 1 mL of RIPA buffer supplemented with SDS (0.1%), followed by an additional wash and transfer of the liquid suspensions to fresh tubes. Immunocomplexes were then eluted from the beads by two sequential incubations in 200 μL of Gly elution buffer (0.1 M Gly, 0.5 M NaCl, and 0.05% Tween 20, pH 2.8) followed by centrifugation. After the addition of 100 μL 1 M Tris/HCl, pH 9.7, to the pooled eluates, free DNA was obtained from 400 μL of eluate (80% volume) as described above, except that 20 μg of glycogen was added for precipitation at −20°C for at least 12 h. Precipitated DNA was resuspended in 100 μL of water for PCR. As input control, 8% volume of the supernatant from all preimmune incubations was processed in parallel and resuspended in 1000 μL of water for subsequent PCR.
PCR Reactions and Oligonucleotides
PCR amplifications were performed in the presence of 0.4 μM of each primer, 200 μM deoxynucleotide triphosphate and 1 M betaine with Taq polymerase (Boehringer Mannheim, Basel) in the supplemented buffer. For chromatin immunoprecipitations, 5 μL of immunoprecipitated sample DNAs were amplified for 33 cycles of 94°C, 55°C, and 72°C steps of 30 s. In each experiment, an appropriate series of four sequential twofold dilutions of the corresponding input was run in parallel to ensure that PCR reactions were in a semiquantitative range.
The following oligonucleotide sequences were used: WRKY1p, 5′-TGCAAGTCACACATGATGGGGGG-3′ and 5′-TCATCATTGTTTGGTTGAGCAAGAAGG-3′; WRKY1a, 5′-CAGGCTGAACTTGACCCAAAGTGCTGG-3′ and 5′-AGCTCAGATGCTTGAGCCCTTTCGAAG-3′; WRKY1b, 5′-GTTGTCTCGAGTTGTTGGCAAAGC-3′ and 5′-GGCTTACTATCTTGTGCGACGACC-3′; WRKY1c, 5′-GGTCGAGCTCTTGTCCCCTTGCGG-3′ and 5′-GCAACATAATCACATTCCGGGATCA-3′; WRKY1d, 5′-TCTACATCACATGATCCCGGAATG-3′ and 5′-TGGGGTGTAAGATCAACGG-3′; WRKY1e, 5′-GGTTCTTGATTAATCAATCTCCACCGTTGA-3′ and 5′-CTTTTGGTTTTCAAATCCCCAG-3′; WRKY1f, 5′-TGATGAGTTGAGTAAACCGAAAGC-3′ and 5′-ATTAGACTGCAGAGCAGCATTGCTC-3′; WRKY1g, 5′-GAGATCAAAGTAAATTGGACGATGG-3′ and 5′-GCGAAGAATTTTCAGGGGTTG-3′; WRKY1h, 5′-TTCTTCGCTTTCTTTTGGCGAGG-3′ and 5′-GCCTTACCTAGGATTTGGATTGCC-3′; WRKY1i, 5′-GGCAAGCAGTGCTTGAACCCTCG-3′ and 5′-CGTTGGCCTTATAGCCGTAGGAG-3′; WRKY1j, 5′-AAATGCTGCAACGTCCACGAAGC-3′ and 5′-AGCGCTTCATATTTCAAACAAGGTACAC-3′; PR1-1p, 5′-AGTCATAGATTATTGTCTCCGTGG-3′ and 5′-GAACAATTTCTCTGCTGAGACGG-3′; WRKY3p, 5′-AGATATTCTCAAAATCTTTCCACG-3′ and 5′-GGCGGCGGCCGCTTCTTG-3′; ACOp, 5′-GGTTCACCGGCGGCGAATGAGGTGAG-3′ and 5′-CTTGACTTGAACCTCAACACCCAC-3′.
All RT-PCR amplifications were performed for 40 cycles in a Real Time Thermocycler (Applied Biosystems, Foster City, CA) and analyzed with the GeneAmp software supplied. PCR reactions were performed in triplicates with lab-made SYBR green buffer using 1% of the reverse transcribed cDNA reaction per sample. Reaction buffer conditions were 2 mM MgCl2, 50 mM KCl, 10 mM Tris, pH 8.3, 0.01% gelatine, 0.05% Tween 20, 25 μg/mL of BSA, 2.5 μM ROX, 1/2000 SYBR Green I (Molecular Probes, Eugene, OR), 1 μM of each oligonucleotide, 250 μM deoxynucleotide triphosphate and 0.625 units of Taq polymerase in reaction volume of 20 μL. Primers that span portions of the coding sequence of the following genes were used for RT-PCR analysis: for WRKY1, WRKY1h, and WRKY1j; for PR1-1, 5′-CGAAACTACTTCTTCCGTCTCAGC-3′ and 5′-TCATCGTCTTGAATGGGCTCG-3′. All values were normalized to PcUbiquitin 4 expression that was amplified using the following oligonucleotide sequences: 5′-CTTCGTCTCCGTGGTGGTAt-3′ and 5′-GCTAGGGTCCTTCCATCCTC-3′.
Supplementary Material
Acknowledgments
We thank Elke Logemann for excellent technical assistance and Sabine Zachgo and Paul Schulze-Lefert for critical reading of the manuscript and for valuable comments. This work was supported by research fellowship TU-126 from the Deutsche Forschungsgemeinschaft to F.T.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Imre E. Somssich (somssich@mpiz-koeln.mpg.de).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.024810.
References
- Asai, T., Tena, G., Plotnikova, J., Willmann, M.R., Chiu, W.L., Gomez-Gomez, L., Boller, T., Ausubel, F.M., and Sheen, J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983. [DOI] [PubMed] [Google Scholar]
- Barolo, S., and Posakony, J.W. (2002). Three habits of highly effective signaling pathways: Principles of transcriptional control by developmental cell signaling. Genes Dev. 16, 1167–1181. [DOI] [PubMed] [Google Scholar]
- Cahill, M.A., Ernst, W.H., Janknecht, R., and Nordheim, A. (1994). Regulatory squelching. FEBS Lett. 344, 105–108. [DOI] [PubMed] [Google Scholar]
- Chen, C., and Chen, Z. (2000). Isolation and characterization of two pathogen- and salicylic acid-induced genes encoding WRKY DNA-binding proteins from tobacco. Plant Mol. Biol. 42, 387–396. [DOI] [PubMed] [Google Scholar]
- Chen, C., and Chen, Z. (2002). Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol. 129, 706–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W., et al. (2002). Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14, 559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua, Y.L., Brown, A.P., and Gray, J.C. (2001). Targeted histone acetylation and altered nuclease accessibility over short regions of the pea plastocyanin gene. Plant Cell 13, 599–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack, R.S., Eulgem, T., Rushton, P.J., Kochner, P., Hahlbrock, K., and Somssich, I.E. (2002). Leucine zipper-containing WRKY proteins widen the spectrum of immediate early elicitor-induced WRKY transcription factors in parsley. Biochim. Biophys. Acta 1576, 92–100. [DOI] [PubMed] [Google Scholar]
- De Nadal, E., Zapater, M., Alepuz, P.M., Sumoy, L., Mas, G., and Posas, F. (2004). The MAPK Hog1 recruits Rpd3 histone deacetylase to activate osmoresponsive genes. Nature 427, 370–374. [DOI] [PubMed] [Google Scholar]
- de Pater, S., Greco, V., Pham, K., Memelink, J., and Kijne, J. (1996). Characterization of a zinc-dependent transcriptional activator from Arabidopsis. Nucleic Acids Res. 24, 4624–4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellagi, A., Helibronn, J., Avrova, A.O., Montesano, M., Palva, E.T., Stewart, H.E., Toth, I.K., Cooke, D.E., Lyon, G.D., and Birch, P.R. (2000). A potato gene encoding a WRKY-like transcription factor is induced in interactions with Erwinia carotovora subsp. atroseptica and Phytophthora infestans and is coregulated with class I endochitinase expression. Mol. Plant-Microbe Interact. 13, 1092–1101. [DOI] [PubMed] [Google Scholar]
- Deslandes, L., Olivier, J., Theulieres, F., Hirsch, J., Feng, D.X., Bittner-Eddy, P., Beynon, J., and Marco, Y. (2002). Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. USA 99, 2404–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després, C., Chubak, C., Rochon, A., Clark, R., Bethune, T., Desveaux, D., and Fobert, P.R. (2003). The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15, 2181–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desveaux, D., Subramaniam, R., Despres, C., Mess, J.N., Levesque, C., Fobert, P.R., Dangl, J.L., and Brisson, N. (2004). A “Whirly” transcription factor is required for salicylic acid-dependent disease resistance in Arabidopsis. Dev. Cell 6, 229–240. [DOI] [PubMed] [Google Scholar]
- Dong, J., Chen, C., and Chen, Z. (2003). Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 51, 21–37. [DOI] [PubMed] [Google Scholar]
- Du, L., and Chen, Z. (2000). Identification of genes encoding receptor-like protein kinases as possible targets of pathogen- and salicylic acid-induced WRKY DNA-binding proteins in Arabidopsis. Plant J. 24, 837–847. [DOI] [PubMed] [Google Scholar]
- Eulgem, T., Rushton, P.J., Robatzek, S., and Somssich, I.E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. [DOI] [PubMed] [Google Scholar]
- Eulgem, T., Rushton, P.J., Schmelzer, E., Hahlbrock, K., and Somssich, I.E. (1999). Early nuclear events in plant defence signalling: Rapid gene activation by WRKY transcription factors. EMBO J. 18, 4689–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, J., Stephenson, C.G., and Iadarola, M.J. (1994). Recombinant proteins attached to a nickel-NTA column: Use in affinity purification of antibodies. Biotechniques 17, 257–262. [PubMed] [Google Scholar]
- Hara, K., Yagi, M., Kusano, T., and Sano, H. (2000). Rapid systemic accumulation of transcripts encoding a tobacco WRKY transcription factor upon wounding. Mol. Gen. Genet. 263, 30–37. [DOI] [PubMed] [Google Scholar]
- Hinderhofer, K., and Zentgraf, U. (2001). Identification of a transcription factor specifically expressed at the onset of leaf senescence. Planta 213, 469–473. [DOI] [PubMed] [Google Scholar]
- Johnson, C., Boden, E., Desai, M., Pascuzzi, P., and Arias, J. (2001). In vivo target promoter-binding activities of a xenobiotic stress-activated TGA factor. Plant J. 28, 237–243. [DOI] [PubMed] [Google Scholar]
- Johnson, C.S., Kolevski, B., and Smyth, D.R. (2002. a). TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14, 1359–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, L., Cao, X., and Jacobsen, S. (2002. b). Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 12, 1360–1367. [DOI] [PubMed] [Google Scholar]
- Kalde, M., Barth, M., Somssich, I.E., and Lippok, B. (2003). Members of the Arabidopsis WRKY group III transcription factors are part of different plant defense signaling pathways. Mol. Plant-Microbe Interact. 16, 295–305. [DOI] [PubMed] [Google Scholar]
- Kim, C.Y., Lee, S.H., Park, H.C., Bae, C.G., Cheong, Y.H., Choi, Y.J., Han, C., Lee, S.Y., Lim, C.O., and Cho, M.J. (2000). Identification of rice blast fungal elicitor-responsive genes by differential display analysis. Mol. Plant-Microbe Interact. 13, 470–474. [DOI] [PubMed] [Google Scholar]
- Kombrink, E., and Hahlbrock, K. (1986). Responses of cultured parsley cells to elicitors from phytopathogenic fungi. Timing and dose dependency of elicitor-induced reactions. Plant Physiol. 81, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroj, T., Rudd, J.J., Nurnberger, T., Gabler, Y., Lee, J., and Scheel, D. (2003). Mitogen-activated protein kinases play an essential role in oxidative burst-independent expression of pathogenesis-related genes in parsley. J. Biol. Chem. 278, 2256–2264. [DOI] [PubMed] [Google Scholar]
- Latchman, D.S. (2001). Transcription factors: Bound to activate or repress. Trends Biochem. Sci. 26, 211–213. [DOI] [PubMed] [Google Scholar]
- Lee, J., Rudd, J.J., Macioszek, V.K., and Scheel, D. (2004). Dynamic changes in the localization of MAP kinase cascade components controlling pathogenesis-related (PR) gene expression during innate immunity in parsley. J. Biol. Chem. 279, 22440–22448. [DOI] [PubMed] [Google Scholar]
- Logemann, E., Tavernaro, A., Schulz, W., Somssich, I.E., and Hahlbrock, K. (2000). UV light selectively coinduces supply pathways from primary metabolism and flavonoid secondary product formation in parsley. Proc. Natl. Acad. Sci. USA 97, 1903–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina, L., Mongrand, S., McLachlin, D.T., Chait, B.T., and Chua, N.H. (2002). ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32, 317–328. [DOI] [PubMed] [Google Scholar]
- Maleck, K., Levine, A., Eulgem, T., Morgan, A., Schmid, J., Lawton, K.A., Dangl, J.L., and Dietrich, R.A. (2000). The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet 26, 403–410. [DOI] [PubMed] [Google Scholar]
- Meier, I., Hahlbrock, K., and Somssich, I.E. (1991). Elicitor-inducible and constitutive in vivo DNA footprints indicate novel cis-acting elements in the promoter of a parsley gene encoding pathogenesis-related protein 1. Plant Cell 3, 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando, V. (2000). Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. Sci. 25, 99–104. [DOI] [PubMed] [Google Scholar]
- Robatzek, S., and Somssich, I.E. (2001). A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J. 28, 123–133. [DOI] [PubMed] [Google Scholar]
- Robatzek, S., and Somssich, I.E. (2002). Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 16, 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton, P.J., Torres, J.T., Parniske, M., Wernert, P., Hahlbrock, K., and Somssich, I.E. (1996). Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 15, 5690–5700. [PMC free article] [PubMed] [Google Scholar]
- van Loon, L.C., Pierpont, W.S., Boller, T., and Conejero, V. (1994). Recommendations for naming plant pathogenesis-related proteins. Plant Mol. Biol. Rep. 12, 245–264. [Google Scholar]
- Yoda, H., Ogawa, M., Yamaguchi, Y., Koizumi, N., Kusano, T., and Sano, H. (2002). Identification of early-responsive genes associated with the hypersensitive response to tobacco mosaic virus and characterization of a WRKY-type transcription factor in tobacco plants. Mol. Genet. Genomics 267, 154–161. [DOI] [PubMed] [Google Scholar]
- Yu, D., Chen, C., and Chen, Z. (2001). Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell 13, 1527–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J.Z. (2003). Overexpression analysis of plant transcription factors. Curr. Opin. Plant Biol. 6, 430–440. [DOI] [PubMed] [Google Scholar]
- Zhang, Z.L., Xie, Z., Zou, X., Casaretto, J., Ho, T.H., and Shen, Q.J. (2004). A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol. 134, 1500–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.