Abstract
We have isolated a new mutant, hanaba taranu (han), which affects both flower and shoot apical meristem (SAM) development in Arabidopsis thaliana. Mutants have fused sepals and reduced organ numbers in all four whorls, especially in the 2nd (petal) and 3rd (stamen) whorls. han meristems can become flatter or smaller than in the wild type. HAN encodes a GATA-3–like transcription factor with a single zinc finger domain. HAN is transcribed at the boundaries between the meristem and its newly initiated organ primordia and at the boundaries between different floral whorls. It is also expressed in vascular tissues, developing ovules and stamens, and in the embryo. han interacts strongly with clavata (clv) mutations (clv1, clv2, and clv3), resulting in highly fasciated SAMs, and we find that WUS expression is altered in han mutants from early embryogenesis. In addition, HAN is ectopically expressed both in clv1 and clv3 mutants. We propose that HAN is normally required for establishing organ boundaries in shoots and flowers and for controlling the number and position of WUS-expressing cells. Ectopic HAN expression causes growth retardation, aberrant cell division patterns, and loss of meristem activity, suggesting that HAN is involved in controlling cell proliferation and differentiation.
INTRODUCTION
During the reproductive growth phase of Arabidopsis thaliana, flowers arise in a spiral phyllotaxis from the flanks of the shoot apical meristem (SAM), which is located at the tip of the stem and encompasses a stem cell population whose descendants essentially give rise to all of the aerial parts of the plant. The SAM can be divided into three zones: the central zone, which is composed of a small number of slowly dividing stem cells at the meristem apex, the rib meristem zone, which lies underneath the central zone and gives rise to the pith and vascular structure of the stem, and the peripheral zone, which surrounds the central zone and provides founder cells for the formation of new leaves and flowers (reviewed in Fletcher and Meyerowitz, 2000). Floral primordia originate as a buttress of undifferentiated cells (the floral meristem) growing at the peripheral zone of the SAM, which soon separates itself from the SAM (only connects to the stem through its peduncle) and sequentially gives rise to the sepals, the petals, and the stamens. Eventually the remaining inner floral meristem cells differentiate into two congenitally fused carpels (Smyth et al., 1990).
Two major pathways, the WUSCHEL (WUS)–CLAVATA (CLV) pathway and the SHOOT MERISTEMLESS (STM) pathway, have been characterized as pivotal for meristem establishment and maintenance. Both wus and stm mutants fail to initiate an embryonic SAM and have premature termination of adventitious shoot and floral meristems (Clark et al., 1996; Laux et al., 1996; Long et al., 1996). WUS encodes a homeodomain protein that is expressed near the boundary of the central zone and rib meristem in shoot and floral meristems and functions to promote meristem activity (Mayer et al., 1998). The clv mutations (clv1, clv2, and clv3) affect both SAM and floral meristem development, resulting in enlarged shoot apical and floral meristems as well as increased floral organ numbers (Clark et al., 1996, 1997; Kayes and Clark, 1998). CLV3 is normally expressed in the central zone, overlying the WUS domain (Fletcher et al., 1999), whereas CLV1 is expressed in the rib meristem zone, embracing the WUS domain basally and laterally (Clark et al., 1997). It has been suggested that CLV3, a secreted small protein (Fletcher et al., 1999; Rojo et al., 2002), interacts with the CLV1/CLV2 Leu-rich repeat receptor proteins as a ligand/receptor complex to activate a signal transduction cascade that limits WUS expression (Fletcher et al., 1999; Brand et al., 2000; Schoof et al., 2000). In the absence of CLV activity, WUS activity increases and causes accumulation of stem cells and thereby an enlarged, fasciated meristem. Conversely, it has been shown that ectopic WUS expression can induce ectopic CLV3 expression (Schoof et al., 2000). It has been proposed, therefore, that WUS regulates its activity so as to control SAM size by employing a negative feedback system involving the CLV proteins. STM encodes a member of the class 1 KNOX family of homeodomain proteins (Long et al., 1996). STM is expressed throughout the SAM where it appears to act through downregulating the ASYMMETRIC LEAVES genes (AS1 and AS2). Loss of AS1 activity restores meristem function to stm mutants by derepressing the activities of other KNOX class genes, such as KNAT1 (also named BREVIPEDICELLUS; Venglat et al., 2002) and KNAT2 (Byrne et al., 2000). More recently it has been shown that the STM and WUS pathways can act together to confer ectopic meristematic cell fate (Gallois et al., 2002; Lenhard et al., 2002).
Although much is known about genetic regulatory pathways within the SAM, little is known about the interactions between meristematic cells and their neighboring differentiating cells. Nevertheless, molecules coupling differentiating cells with meristem development have been uncovered in recent years. In Petunia hybrida, Hairy Meristem (HAM) encodes a GRAS family putative transcription factor that is expressed in provascular tissue, but ham mutations result in early termination of SAM activity. Hence, HAM may represent an extrinsic antidifferentiation factor that is required to maintain stem cells in the SAM (Stuurman et al., 2002). Additionally, lateral organ polarity genes, such as Antirrhinum majus PHANTASTICA and Arabidopsis PHABULOSA, also appear to have positive effects on the formation and maintenance of apical meristems (McConnell and Barton, 1998; Waites et al., 1998). Furthermore, members of the NAC-domain gene family expressed at the boundaries of meristems and primordia, including the Petunia NO APICAL MERISTEM (Souer et al., 1996), the Antirrhinum CUPULIFORMIS (Weir et al., 2004), and the Arabidopsis CUP-SHAPED COTYLEDON genes CUC1, CUC2, and CUC3 (Aida et al., 1999; Takada et al., 2001; Vroemen et al., 2003), regulate SAM development as well. In this report, we describe a new gene in Arabidopsis, HANABA TARANU (HAN; meaning fewer floral leaves in Japanese), that is required for normal flower and SAM development. This gene is expressed in the provascular tissues during embryogenesis and later is expressed at the boundary tissues between meristems and initiating organ primordia as well as in the vascular tissues. We demonstrate that han mutations interact strongly with clv mutations and that HAN is required for normal cell division and positioning of WUS-expressing cells in the SAM, suggesting that HAN also represents one of the extrinsic molecules linking the more mature boundary and vascular cells with SAM activities.
RESULTS
han Mutant Phenotypes and HAN Cloning
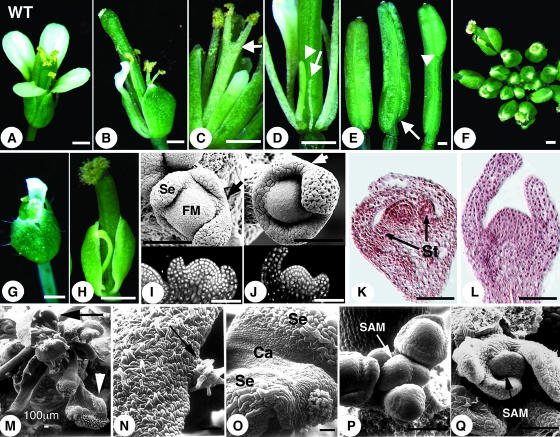
Four different han mutant alleles (han-1 to han-4) were isolated in three different mutagenesis screens in Arabidopsis. han-2 was isolated in an ethyl methanesulfonate mutagenesis screen, and the han-1, han-3, and han-4 alleles were generated in two separate Agrobacterium tumefaciens–mediated T-DNA screens (see Methods). All han mutants have similar flower defects, including fused sepals, reduced numbers of petals and stamens, and, sometimes, unfused carpels. Phenotypically han-2 is the weakest allele, and han-3 is the strongest, whereas han-4 and han-1 have similar, intermediate expressivity. Mutants homozygous for han-3, unlike lines homozygous for the other alleles, have fasciated SAMs. In addition, the han-3 allele shows slight semidominance, whereas the other han mutations are fully recessive. We will focus on the recessive mutations in this report. The majority of han-2 mutant flowers have two to four sepals (sepals fused together are counted as 1) in the 1st whorl, one or two organs in the 2nd whorl (including petals as well as filamentous structures), four or five stamens (including filaments lacking anthers) in the 3rd whorl, and two asymmetric carpels in the 4th whorl (Table 1, Figures 1B to 1E). Malformed organs observed include bifid stamens with two stalks fused partially or completely along their length but with anthers separated (Figure 1C, arrow), filamentous structures between whorls (Figure 1D), and stamens with abnormal anthers. In >50% (31/54) of the cases, the valves of the two fused carpels are not symmetrical, with one shorter than the other at the basal region (Figure 1E, arrow). At a much lower frequency (4/54), extra tissue is formed apically (Figure 1E, arrowhead). han-2 homozygous plants are partially fertile. han-1 flowers have stronger defects in all whorls when compared with han-2 flowers. In approximately half of han-1 flowers, all of the sepals are either partially or completely fused (Table 1, Figures 1F to 1H). Mature petals are rare and only appear in a few early flowers. The majority of han-1 flowers lack second whorl organs. In the third whorl, organ numbers are generally decreased to three or four, and stamens with normal morphology are rare. The frequency of filamentous structures occupying stamen positions in han-1 flowers is much higher than in han-2 flowers. han-1 homozygous plants have extremely low fertility. Consistent with their reduced floral organ numbers and sizes, han-1 mutants have smaller floral meristems than the wild type, as shown for a stage 3 flower in Figure 1J (arrow points to an area of congenital sepal fusion). In addition, inner whorl organ primordia either fail to develop or are delayed in appearance compared with the wild type in stage 6 flowers (Figures 1K and 1L).
Table 1.
han Flower Phenotypes Summary
| Organ Number | Sepal
|
Petal
|
Stamen
|
Carpel
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Flower Number | 0 | 1 | 2 | 3 | 4 | 5 | 0 | 1 | 2 | 3 | 4 | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 2 | Fused | Unfused |
| han-2 | 0 | 4 | 27 | 57 | 21 | 1 | 13 | 31 | 40 | 23 | 3 | 0 | 0 | 0 | 6 | 40 | 55 | 9 | 49b | 54 | 0 |
| 110a,b | (4) | (10) | (13) | (2) | (4) | (6) | (1) | ||||||||||||||
| han-1 | 0 | 53 | 37 | 13 | 4 | 0 | 77 | 24 | 4 | 2 | 0 | 0 | 8 | 12 | 37 | 32 | 16 | 1 | 107 | 91 | 16 |
| 107a | (6) | (3) | (3) | (11) | (77) | (90) | (62) | (4) | |||||||||||||
The numbers in parentheses represent the total number of filamentous structures in all flowers of each category.
Total number of flowers examined.
Only 54 flowers were examined for carpel number and fusion defects. Of the 54, 49 had two carpels, 1 had a single carpel, and 4 had extra carpel tissues positioned apically.
Figure 1.
han Mutant Flower and Floral Meristem Phenotypes.
(A) A wild-type flower.
(B) to (E) han-2 homozygotes.
(F) to (H) han-1 homozygotes.
(B) A han-2 flower.
(C) The arrow points to the fusion of two stamens.
(D) Filamentous structures in place of a petal or a stamen are indicated by the arrow and arrowhead, respectively. Sepals were removed.
(E) Carpel defects include asymmetric silique valves (arrow) and extra carpel tissue positioned at the apical region of the silique (arrowhead). Siliques shown are not age-matched.
(F) A han-1 inflorescence.
(G) An early-arising han-1 flower.
(H) A flower with sepals all fused into an open semicircle.
(I) A wild-type stage 3 flower. Se, sepal; FM, floral meristem.
(J) A han-1 stage 3 flower (because han floral meristem development is generally delayed, floral stage is determined based on sepal size).
(K) A wild-type stage 6 flower. St, stamens.
(L) A han-1 stage 6 flower.
(M) A han-1 inflorescence at late stage. The black arrow points to a carpelloid sepal, and the white arrowhead points to a sepalloid carpel.
(N) An enlarged view of the carpelloid sepal indicated in (M). Arrow points to stigmatic papillae at the edge of the sepal.
(O) An enlarged view of the sepal (Se) and carpel (Ca) fusion structure in (M).
(P) A wild-type SAM.
(Q) A han-1 mutant SAM.
Bars in (A) to (H) = 0.5 mm; bars in (I) to (Q) = 50 μm (unless indicated otherwise).
Vegetative growth and inflorescence structure of han mutants appears fairly normal. However, in some of the han-1 and han-4 plants, flowers that arise toward the end of the reproductive stage are morphologically more deformed than early flowers, composed either of carpels alone (sometimes subtended by leaf/sepal-like structures), carpelloid sepal/leaf-like structures (Figures 1M and 1N, arrows), or chimeric organs with carpelloid character (Figures 1M, arrowhead, and 1O). In addition, later-arising flowers fail to originate in a typical phyllotactic spiral. Perturbed floral phyllotaxy generally reflects a SAM defect. To determine if such a defect is visible in han-1 mutants, both early (postembryonic day 30) and late stage (after postembryonic day 40) inflorescence SAMs were examined using scanning electron microscopy. Early stage han SAMs are normal compared with the wild type, as also confirmed by confocal laser scanning microscopy (data not shown). However, for the late stage han inflorescences, five out of eight SAMs appeared flattened instead of dome-shaped as in the wild type (Figures 1P and 1Q) and in some cases appeared smaller and less distinct from the surrounding initiating organ primordia. In the flattened SAMs, early stage floral organ primordia were not the usual hemispherical masses of undifferentiated cells but were ridge-shaped. This indicates that there is a gradual transition toward an aberrant SAM morphology in han mutants during reproductive development.
The HAN gene was cloned by both positional mapping of the han-1 allele and by T-DNA tagging of the han-4 allele. HAN is located on chromosome 3 (At3g50870), and its full-length cDNA was isolated from a flower-specific λcDNA library. The protein sequence deduced from the open reading frame consists of 295 amino acids, encoded by two exons, and resembles a GATA-3–like protein with a single zinc finger motif (C-X2-C-X18-C-X2-C) (Figure 2). In addition, there is a stretch of 14 amino acids N-terminal to the zinc finger that appears highly conserved among some plant GATA transcription factors. The han-1 mutation results in a complete deletion of the gene, starting from 709 bp upstream of the translation initiation codon and ending at 1298 bp downstream of the stop codon. The mutation in han-2 results in a single amino acid change at position 179, Gly (GGC) to Ser (AGC). The mutation in han-4 is caused by the insertion of at least two copies of a T-DNA sequence into the intron at 112 bp, followed by a deletion of 20 bp. Both a 9- and 6-kb genomic fragment spanning the HAN gene fully rescues the han phenotypes (data not shown). Database searches identified 25 putative GATA genes in the Arabidopsis genome, among which are two close HAN relatives, located on chromosomes 2 and 4 (At2g18380 and At4g36620), and here named HANL1 and HANL2. The HAN gene shares 46 and 50% sequence similarity to its two close homologs HANL1 and HANL2, respectively.
Figure 2.
Protein and Gene Structure.
The HAN protein sequence deduced from the open reading frame of the full-length HAN cDNA. The position of the intron is indicated by the open triangle; a 14–amino acid domain that is highly conserved among some plant GATA factors is underlined twice; the zinc finger domain is underlined once. The mutation in han-1 results in deletion of the whole gene. The mutation in han-2 changes a Gly (GGC) to a Ser (AGC) at amino acid position 179 (asterisk). The mutation in han-4 results from a T-DNA insertion within the intron (arrow). The 5′ junction sequence is flanked by the Bar gene in the T-DNA, whereas the 3′ is flanked by the 4× 35S promoter in the T-DNA (near the right border).
HAN Expression
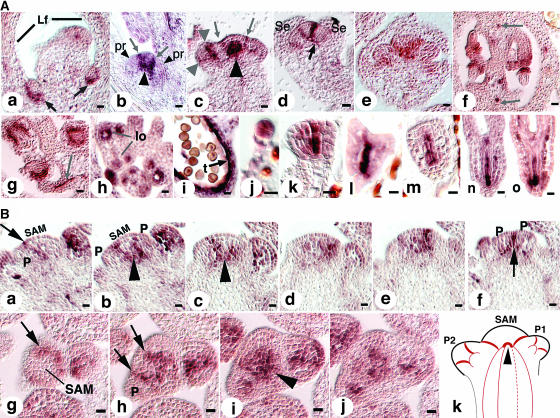
The HAN expression pattern was examined by in situ hybridization using a full-length cDNA probe and a HAN-specific 5′-cDNA probe, both of which are specific to HAN (see Methods) and which gave the same results. HAN is expressed in vegetative and inflorescence SAMs, axillary SAMs, floral meristems, developing ovules and stamens, vascular tissues, and in the embryo. In the developing axillary SAM, it is expressed at the boundary between nascent axillary meristems and the adaxial side of leaves (Figure 3A, a, arrows). Expression in all mature SAMs is similar, located at the boundaries between the central SAM and the initiating organ primordia, as well as between the neighboring initiating organ primordia (Figures 3A, b and c, and 3B).
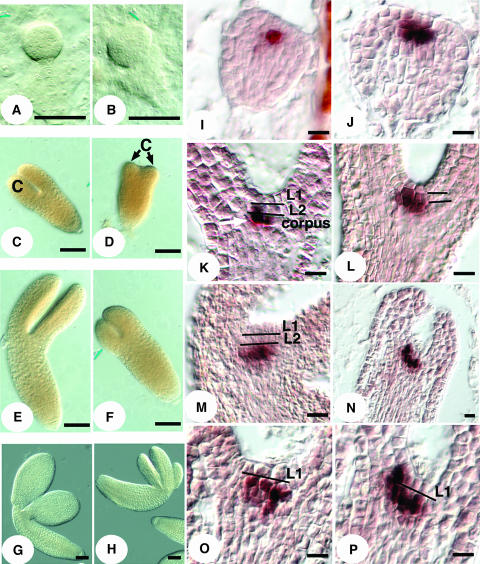
Figure 3.
HAN Expression Pattern.
(A) Budding axillary SAMs are shown in (a). Arrows point to the expression domains of HAN at the boundaries between the SAM and the adaxial sides of leaves. Lf, leaf. (b) Older axillary SAM. Arrows point to the expression domains of HAN at the boundaries between the SAM and the newly initiated organ primordia (pr, small arrowheads). Large arrowhead indicates the junction of the HAN expression domain in the SAM and its vascular expression in the stem. (c) Inflorescence SAM. Arrows point to two stripes of HAN expression at the boundaries between the SAM and newly initiated floral primordia. Small arrowheads indicate HAN expression in a stage 2 flower at the boundaries between the floral meristem and the soon-to-be-specified sepal primordia. Large arrowheads point to the junction of the HAN expression domain in the SAM and in the stem (note that the angle of this section is tilted toward the reader, and the domain indicated by the large arrow includes some of the expression of HAN between the SAM and a primordium pointing toward the reader). (d) Stage 5 flower. Arrow points to the connection arch of HAN expression in the floral meristem and its expression in the peduncle vascular tissue. Se, sepal. (e) Expression in the lateral and basal regions of carpel primordia in a stage 6 flower. (f) Early stage 12 ovary (cross section). HAN is expressed in initiating inner and outer integuments as well as in the vascular tissue (arrows). (g) Expression in integuments continues in the stage 13 ovary. Arrow points to expression in funiculus vascular tissue. (h) HAN expression in the stamens of a stage 8 flower. lo, locule. (i) HAN expression persists in the tapetum cell layer until it has degenerated and is absent in mature haploid pollen. t, tapetum. (j) Eight-cell stage embryo. HAN is expressed in all cells of the embryo proper. (k and l) Globular and transition stage embryos. HAN is expressed in the center files of cells. (m and n) Heart and torpedo stage embryos. HAN expression remains in the center cells destined to be provascular tissues. (o) Late torpedo stage. Bars = 10 μm.
(B) Expression in SAM in serial longitudinal (a to f) and cross sections (g to j). Arrows point to expression at the boundaries between the SAM and new organ primordia as well as between organ primordia. Expression of HAN in the SAM merges with its expression in the vascular strands in the stem (arrowheads) as illustrated in (k). In (k), red lines represent the expression domains of HAN in the SAM, stem, and developing organ primordia (P1 and P2). Arrowhead corresponds to the regions indicated by large arrowheads in Figure 3A (b and c). P, primordium. Bars = 10 μm.
Expression in the floral meristem reiterates the pattern seen in the SAM, with strong expression at the boundaries between the meristematic dome and the initiating floral organ primordia, and also at the boundaries between the primordia of different whorls (Figure 3A, d). Expression at the boundaries attenuates as the organ primordia grow apart. In stage 5 flowers, expression remains at the boundary between the central meristematic cells and differentiating stamen primordia (data not shown). In stage 6 flowers, expression is the strongest at the medial ridge region of the carpel (Figure 3A, e). In the developing ovule, HAN is expressed in the inner and outer integuments, with signal absent from the nucellus (Figure 3A, f and g). In all of the aerial tissues examined, including flowers, HAN is expressed strongly in cell types associated with the phloem tissues (arrows in Figure 3A, f and g). It is expressed strongly in the developing anthers in as early as stage 8 flowers when locules are initiated, in the tapetum cell layer, as well as the microsporocytes (Figure 3A, h). Expression persists until the tapetum layer degenerates and haploid pollen is mature (Figure 3A, i). Expression of HAN in the embryo is detected uniformly in the embryo proper of the eight-cell stage embryo (Figure 3A, j). It is then concentrated in the center cells of the embryo and absent from the epidermal cell layer at the globular stage (Figure 3A, k). This expression pattern persists in the center four files of cells; expression is not observed in the outermost two apical layers through the transition and late heart stages (Figure 3A, l to n). In the torpedo stage embryo, expression is detected in all provascular tissues (Figure 3A, o).
Genetic Interactions of han with Mutations Affecting Flower Development
Double mutants of han-1 with the floral organ identity mutations apetala 3, pistillata, and agamous are generally additive (data not shown). To determine if floral organ identity genes are affected in their expression in han mutants, AP1 and AP3 expression was examined in han-1 flowers. For both genes, expression was detected in the mutant in correct spatial and temporal patterns, but signal intensity was relatively low (data not shown). The lower expression intensity is potentially because of a smaller number of floral organ cells in han mutants. This indicates that the floral organ identity genes act independently of HAN in controlling flower development.
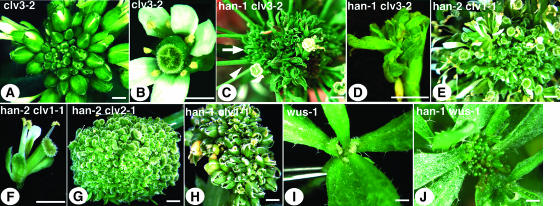
Double mutants of han and clv (including clv1, clv2, and clv3) or han and wus-1 were generated. Any combination of han/clv double mutants results in increased inflorescence fasciation and increased floral abnormalities as compared with either single mutant. Among all clv mutants, the clv3-2 mutants show the strongest phenotype with regards to SAM fasciation and increased floral organ numbers in all four whorls (Figures 4A and 4B). Interactions between han-2 and clv3-2 or han-1 and clv3-2 make for very short plants with fasciated stems thicker than those of clv3-2 single mutants. Early-arising flowers have elongated pedicels (Figure 4C, arrowhead) and are composed only of reduced sepals (or sepal-like tissue) and carpels, which are sometimes indeterminate and generate many sepal- or leaf-like structures from the base, or on top of an undifferentiated mass within the carpel valves (Figure 4D). Later-arising organs are either filamentous or are bracts tipped with stigmatic tissue, arising simultaneously in large numbers (Figure 4C, arrow). The han-2 clv1-1 double mutants have either a fairly normal stem or are shorter and more fasciated than clv1-1 and accumulate numerous flowers at the top of the inflorescence SAM (Figure 4E). These double mutant flowers generally have reduced organ numbers in the outer three whorls compared with han-2 mutant flowers (Figure 4F). The han-2 clv2-1 double mutants either appear similar to han-2 clv1-1 mutants or lack primary shoot dominance and have several unfasciated stems terminating in a ball of carpelloid structures (Figure 4G). han-1 clv1-1 and han-1 clv2-1 double mutants produce inflorescences more fasciated than clv single mutants, with flowers for the most part composed only of sepals and sometimes unfused carpels (Figure 4H). The phenotypes of han-4 clv double mutants are similar to those of han-1 clv double mutants.
Figure 4.
Genetic Interactions of han and clv.
(A) clv3-2 inflorescence, top view.
(B) clv3-2 flower, top view.
(C) han-1 clv3-2 double mutant inflorescence. Arrowhead points to an early-arising flower with long peduncle, and arrow points to late-arising bract-like or filamentous organs.
(D) han-1 clv3-2 double mutant flower.
(E) han-2 clv1-1 double mutant inflorescence.
(F) han-2 clv1-1 double mutant flower.
(G) han-2 clv2-1 double mutant inflorescence.
(H) han-1 clv1-1 double mutant inflorescence.
(I) wus-1 single mutant axillary shoot.
(J) han-1 wus-1 double mutant axillary shoot.
Bars = 1 mm.
han-1 wus-1 double mutants resemble wus-1 single mutants, except that they appear darker green in color and have many more radial-shaped leaf-like structures (with trichomes) arising from the leaf axils (Figure 4J). No flowers were observed in han-1 wus-1 double mutants (three plants). The han-1/han-1 wus-1/+ mutants are phenotypically similar to han-1 single mutants (data not shown). We further compared the number of floral organs present in han-1/+ wus-1/wus-1 flowers versus wus-1/wus-1 flowers. The former have an average of 3.2 (±1.1) sepals, 1.5 (±1.3) petals, and 0.5 (±0.9) stamens (including in 27% of cases filamentous structures) in 22 flowers counted, whereas the latter has an average of 3.9 (±1.0) sepals, 3.9 (±0.6) petals, and 1.1 (±0.3) stamens (no filamentous structures) of 10 flowers counted. The P value for each organ type difference based on an unpaired Student's t test is smaller than 0.05, suggesting that the difference is significant. This suggests that whereas wus is epistatic to han in overall development, flower development in wus mutants relies on HAN activity in a dose-dependent manner.
HAN Regulates WUS-Expressing Cells
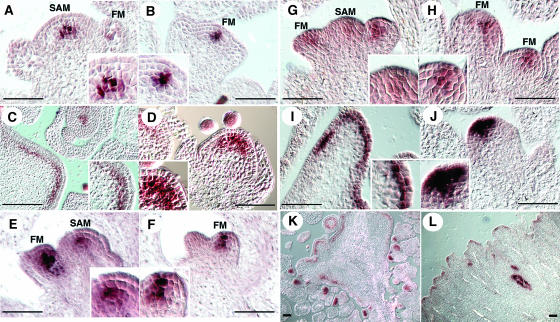
WUS serves as the earliest known marker for SAM development. It is detected in the inner two cells at the apical region of the 16-cell stage embryos, and its expression domain enlarges in embryonic and postembryonic SAMs of clv mutants (Mayer et al., 1998; Brand et al., 2000; Schoof et al., 2000). Because han mutations strongly enhance clv SAM phenotypes, we examined WUS expression in both han single mutants and han clv double mutants. In wild-type inflorescence meristems and floral meristems, WUS is expressed in the central region of the SAM, beneath the outermost two or three cell layers (Figures 5A and 5B). Expression in the floral meristem attenuates after stage 3 and is no longer detectable after stage 6. WUS expression in clv3-2 inflorescence and floral meristems expands outward along the fasciated meristems and upward into the L2 layer (Figures 5C and 5D). In most of han-1 and han-4 inflorescences, whether in early or late stages of development, the WUS expression domain appears diffuse compared with the wild type, with a few central cells showing the strongest expression and surrounding cells showing weaker expression (Figures 5E and 5G). In the floral meristem, WUS expression is clearly shifted to include the L2 and L1 layers (Figures 5F and 5H). As in the SAM, the signal appears diffuse, with the border between the WUS-expressing cells and the surrounding cells not as sharp as is observed in the wild type or in clv3-2 mutants. In some han meristems, however, WUS expression appears comparable to that in the wild type (see Supplemental Figure 1 online and Discussion).
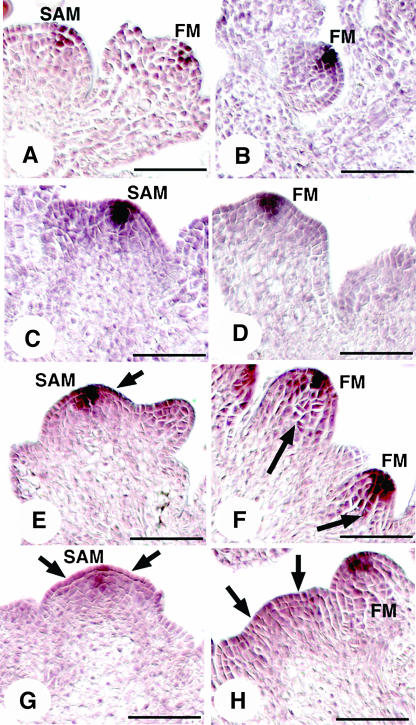
Figure 5.
HAN Regulates WUS-Expressing Cells.
(A) and (B) WUS expression in wild-type inflorescence SAM is below the outermost three (shown) or two (data not shown) layers.
(B) WUS expression in the wild-type floral meristem (FM) is below the outermost two layers.
(C) WUS expression in the clv3-2 inflorescence SAM concentrates in the cells below the L2 layer and at lower level in the L2 layer.
(D) WUS expression in the clv3-2 floral meristem expands into the L2 layer.
(E) and (G) WUS expression in most of the han-1 inflorescence SAMs is diffuse.
(F) and (H) WUS expression in the han-1 floral meristems is shifted to include the L2 and L1 layers.
(I) WUS expression in the han-1 clv3-2 double mutant inflorescence SAM is concentrated in the L2 layer.
(J) WUS expression in the han-1 clv3-2 double mutant floral meristem extends in some cases into the L1 layer.
(K) Overview of a clv3-2 inflorescence SAM.
(L) Partial view of a han-1 clv3-2 double mutant inflorescence SAM.
Bars = 50 μm.
In han-1 clv3-2 double mutants, the inflorescence SAM region is expanded compared with clv3-2 single mutants (Figures 5K and 5L). In contrast with its expression pattern in the clv3-2 single mutant, WUS expression in the SAMs of the double mutants is concentrated in the L2 layer cells (Figure 5I), and in the floral meristems, expression extends upward into the epidermal cell layer in some cases (Figure 5J). Ectopic expression of WUS in han-1 or han-4 flowers all along the inflorescences is also observed, appearing in undifferentiated cells in late stage flowers and in the sepals, which in some cases are clearly carpelloid as they produce WUS-expressing ovules (data not shown).
To find out how early the aberrant WUS expression occurs in han mutants, as well as how early its impact on morphological development is manifested, embryos from han-1/+ siliques were examined. Morphological defects can be observed as early as globular stage, when the shapes of what are likely the han-1 mutant embryos deviate from the wild type (Figures 6A and 6B). By the heart stage, some mutant embryos clearly fail to properly develop cotyledons, which appear as small stubs (Figure 6D) or are barely visible (data not shown). By the torpedo or walking stick stages, mutant embryos appear stunted to various degrees, with a majority having thickened hypocotyls and small cotyledons in numbers ranging from two to four (Figures 6F and 6H).
Figure 6.
han Mutant Embryo Defects and Perturbed WUS Expression in han Embryos.
(A) Globular stage wild-type embryo.
(B) Globular stage han-1 mutant embryo.
(C) Late heart stage wild-type embryo. C, cotyledon.
(D) Late heart stage han-1 embryo with stunted cotyledons.
(E) Walking stick stage wild-type embryo.
(F) Walking stick stage han-1 embryo.
(G) Mature wild-type embryo.
(H) Mature han-1 mutant embryo with three cotyledons.
(I) WUS expression in the transition stage wild-type embryo is concentrated in two cells in the subepidermal L2 layer.
(J) WUS expression in the transition stage han-1 embryo is located in more than two cells within and beneath the L2 layer.
(K) WUS expression in the heart stage wild-type embryo is shifted to two central cells in the corpus.
(L) WUS expression in the heart stage han-1 embryo in the L2 layer.
(M) WUS expression in the mature stage wild-type embryo is centered below the outermost two layers.
(N) WUS expression in the mature han-1 embryo.
(O) and (P) WUS expression in the L2 layer of mature han-1 embryos. (P) is an enlarged view of (N).
Bars = 50 μm in (A) to (H) and 10 μm in (I) to (P).
In wild-type embryos, WUS is expressed in the center two cells at the subepidermal layer of the apical region at the globular and transition stages (Figure 6I). At the heart stage, WUS expression shifts to the central two cells of the corpus (Figure 6K). In mature embryos, WUS is expressed even deeper in the meristem and is excluded from the L2 layer (Figure 6M). However, in apparent han mutant embryos, WUS expression is observed in both L2 and corpus cells and in more than two cells in the transition stage embryo, which corresponds to the enlarged size of the embryo (Figure 6J). At the heart stage, WUS expression is likewise concentrated in both L2 cells and deeper cells (Figure 6L). By the walking stick stage, expression in some apparent han-1 embryos stays strong in L2 cells (Figures 6N to 6P). Aberrant WUS expression in early stage han embryos suggests that HAN is normally required to control the number of WUS expressing cells and to correctly position these cells.
Because there is a feedback loop interaction between WUS and CLV3 in shoot and floral meristems (Brand et al., 2000; Schoof et al., 2000), we also examined CLV3 expression in han mutants in both embryonic and inflorescence stages. No obvious difference in CLV3 expression was observed between han mutant and wild-type plants during embryogenesis, but CLV3 expression becomes variable in han inflorescence meristems (Figure 7). In approximately half of the cases, CLV3 expression appears fairly normal as compared with the wild type (Figures 7C and 7D versus 7A and 7B). However, in the other half of the cases, CLV3 expression becomes slightly (Figure 7E, arrow) or more widely diffuse (Figures 7F to 7H, arrows). The effect of han on the CLV3 expression domain could be because of the alteration of the WUS expression domain that occurs in early embryogenesis before CLV3 expression is initiated.
Figure 7.
CLV3 Expression in han Mutants.
(A) and (B) CLV3 expression in the wild-type inflorescence SAM and floral meristem (FM) is concentrated in the central outermost three layers, at about three to four cells width.
(C) and (D) CLV3 expression appears fairly normal in some han inflorescence SAMs and floral meristems.
(E) CLV3 expression is slightly diffuse in some han mutants.
(F) CLV3 expression is expanded in some han floral meristems.
(G) and (H) CLV3 expression is markedly diffuse in some han mutants. Arrows point to the regions of diffuse expression.
Bars = 50 μm.
Interactions between HAN and the CLV Pathway
To uncover potential regulatory interactions between HAN and CLV, we also examined HAN expression in clv3-2 mutants. The HAN expression pattern in clv3-2 was unchanged in all tissues except for the meristems, where HAN was detected strongly in the L2 and corpus layers along the entire fasciated SAM as well as in the late stage floral meristems (Figures 8A, arrowheads, 8B, large arrow). This expression pattern mimicked the WUS expression pattern in clv3-2 mutants (Figure 8C, arrowhead, 8D, large arrow), suggesting that in the absence of CLV3 activity, HAN expression is ectopically induced in the apical region of the SAM. Because WUS is also ectopically expressed in clv1 mutants (Schoof et al., 2000), we also assessed whether HAN expression was altered in clv1 mutants. HAN shows a similar expression pattern in the SAM of clv1-4 mutants as in clv3-2 (data not shown). However, HAN is not ectopically expressed in the clv2-1 SAM (data not shown). To find out how early HAN expression is perturbed, we examined HAN expression in clv3-2 embryos. From globular stage to mature embryos, the HAN expression is similar in wild-type and clv3-2 mutants (Figures 8E to 8G). This suggests that HAN expression is altered in the clv3-2 SAM during postembryonic development.
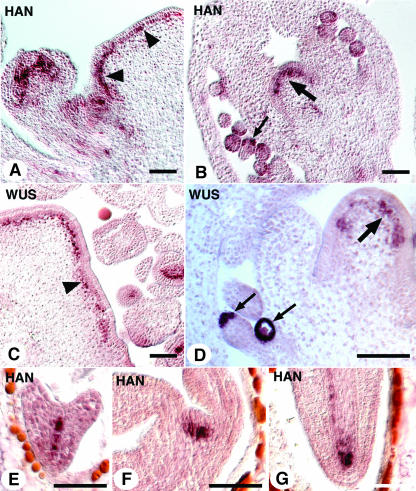
Figure 8.
HAN Is Ectopically Expressed in clv3-2 Mutants.
(A) HAN expression in a clv3-2 inflorescence SAM (arrowheads).
(B) HAN expression in the undifferentiated center dome cells in a late stage clv3-2 flower (large arrow). Small arrow points to HAN expression in the integuments of ovule.
(C) WUS expression in a clv3-2 inflorescence SAM (arrowhead).
(D) WUS expression in the undifferentiated cells in a late stage clv3-2 flower (large arrow). Small arrows point to WUS expression in the nucellus.
(E) HAN expression in a heart stage clv3-2 embryo.
(F) and (G) HAN expression in the SAM and the root of a clv3-2 embryo in late torpedo stage, respectively.
Bars = 50 μm.
Phenotypes Caused by Ectopic Expression of HAN
To examine the effects of HAN gain-of-function, we generated transgenic plants carrying 35S:HAN or 35S:HAN-GR (glucocorticoid receptor binding domain; Lloyd et al., 1994) transgenes. Among Landsberg erecta (Ler) plants transformed with a 35S:HAN construct, only four T1 transgenic plants were produced from >4 mL (∼1.5 × 104) of seeds screened, which is substantially lower than the normal transformation efficiency for the binary vector used (>0.05%). Of the four, one looked fairly normal, but the others were small in size, with unevenly shaped rosette leaves and cauline leaves that gradually turned purple, starting from the abaxial side (Figure 9A). The smallest one of the three died prematurely after making some miniature flowers that did not produce seeds. The remaining two plants had smaller inflorescences than the wild type (Figures 9B and 9C). Floral organs were shorter in length when compared with the wild type at similar stages. The gynoecium was short and also appeared bulky, with wavy surfaces and elongated peduncles compared with the wild type (Figures 9D and 9E). The scarcity of T1 transformants suggests that constitutive overexpression of HAN might be deleterious.
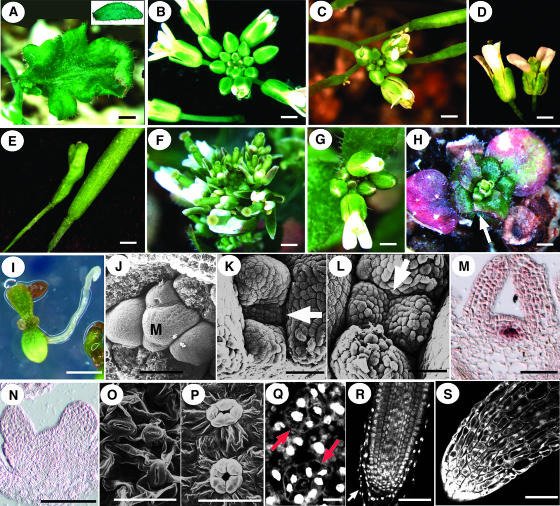
Figure 9.
Phenotypes of Ectopic HAN Expression.
(A) Lobed cauline leaf from a 35S:HAN plant. Inset shows a wild-type cauline leaf.
(B) Wild-type inflorescence.
(C) 35S:HAN inflorescence.
(D) Wild-type (left) and 35S:HAN flowers.
(E) Wild-type (right) and 35S:HAN siliques.
(F) to (H) 35S:HAN-GR transformants.
(F) DEX-treated inflorescence has early opening flowers and a short stem.
(G) Seedlings mock-treated from at 11 d of age, shown at 29 d old.
(H) Seedlings treated with DEX beginning at 11 d of age, shown at 29 d old. Arrow points to lobed leaf margins.
(I) Homozygous 35S:HAN-GR seedling germinated on a DEX plate at 9 d.
(J) SAM of 10-d-old wild-type seedling. M, SAM.
(K) and (L) SAMs of 10-d-old 35S:HAN-GR seedlings germinated on DEX plates. Arrows point to the SAMs.
(M) WUS expression in a 10-d-old 35S:HAN-GR seedling germinated on non-DEX plates.
(N) Absence of WUS expression in a 10-d-old 35S:HAN-GR seedling germinated on DEX plates.
(O) Non-DEX-induced control stomata with two guard cells.
(P) and (Q) Guard cells in 10-d-old and 6-d-old DEX-treated 35S:HAN-GR seedlings, respectively. Arrows point to stomata with five or seven guard cells.
(R) Mock-treated 35S:HAN-GR root tip. Arrow points to the root cap.
(S) DEX-induced 35S:HAN-GR root tip.
Bars in (A) to (I) = 1 mm; bars in (J) to (S) = 50 μm (except for [Q], in which the bar = 10 μm).
More than 50 T1 35S:HAN-GR transgenic plants were generated and all looked normal in the absence of dexamethasone (DEX). When treated three times at 1-d intervals with 10 μM DEX solution, the transgenic plants showed a distinctive morphology. Young flowers opened up precociously (Figure 9F), and stem elongation was retarded. In the 2-d interval between the 2nd and 3rd treatments, the mock-treated control plants increased in height an average of 2.1 cm, whereas the DEX-treated transgenic plants only increased an average of 0.8 cm. When seedlings from a homozygous T2 line were treated with 10 μM DEX four times at 1-d intervals, starting on postgermination day 11 (solution applied via soil), plants began to show accelerated leaf senescence after the second treatment. New leaves were still produced, but leaf expansion was inhibited, and leaf blades were serrated, similar to the leaves of the 35S:HAN transgenic lines (Figure 9H, arrow). Twelve days after the last treatment, the mock-treated control group plants had already bolted (Figure 9G), but approximately half of the DEX-treated plants were dead, and the surviving plants remained in miniature form (Figure 9H). The miniature plants were still able to produce flowers that grew extremely slowly and never reached normal size. These flowers had normal organ identities and numbers but were male sterile. When seeds from a T2 homozygous line were germinated on DEX plates, all seedlings ceased growth at the stage shown in Figure 9I and did not develop further. Transgenic seedlings on non-DEX MS plates grew normally, as did wild-type seedlings on DEX and non-DEX plates.
Stalled growth in 35S:HAN-GR DEX-treated transgenic seedlings suggests inhibited cell proliferation/growth and possible cessation of shoot and root meristem activity. To evaluate this, the SAM structures of homozygous 35S:HAN-GR seedlings that were germinated on either DEX or non-DEX plates were examined on postgermination day 10 using scanning electron microscopy. Whereas the non-DEX-treated control SAMs were dome-shaped like wild-type SAMs (Figure 9J), the DEX-treated SAMs either were flat (Figure 9K, arrow) or were no longer identifiable between the leaf primordia (Figure 9L, arrow). Because WUS serves as a marker of active meristematic cell activity in the SAM during normal development, we examined WUS expression in day 10 plants as well as in day 6 seedlings germinated on DEX plates. In both cases, no WUS expression was found in the DEX-treated seedlings (Figure 9N), suggesting that they had already lost active SAM activity. Similarly, STM expression is also lost in treated seedlings by day 6 (data not shown). HAN expression in the same plants was strong and ubiquitous (data not shown). DEX-treated 35S:HAN-GR seedlings had smaller cells in all cell layers of the cotyledons compared with untreated ones (data not shown), which explains at least in part why the treated plants have small cotyledon blades. In addition, stomatal pores in the cotyledons of induced seedlings were often composed of three or more guard cells instead of two cells as in the control (Figures 9O to 9Q), implying abnormal cell division of the guard cell mother cells or incomplete differentiation of the guard cells. In contrast with the cotyledon cells, the root cells of the induced seedlings were in general larger in size than those of the control group (Figures 9R and 9S). Additionally, in the induced roots, cell files were not as organized as in the control, and root caps were usually diminished or absent, further indicating aberrant cell division and positioning in HAN-overexpressing plants.
DISCUSSION
Functions of HAN in Flower Development
HAN mutations affect various aspects of flower development, including floral meristem size, floral organ separation, floral organ number and size, and floral organ identity. This indicates that HAN plays multiple developmental roles. Because the floral meristem size reduction precedes the other floral organ defects, it could be the primary cause of the other defects. Reduced floral meristem size suggests that HAN is important for controlling the proliferation of floral meristem cells, which are embraced by the cup-shaped HAN expression domain. WUS expression is more diffuse in han floral meristems than in the wild type, and it could be that a centered and high level of WUS expression, mediated by HAN, is required for promoting floral meristem cell proliferation. In addition, ectopic WUS expression in peripheral zone cells could hinder their progression toward cell differentiation and therefore repress organ primordium initiation. This might explain why floral organ initiation in han floral meristems is delayed. On the other hand, because the level of ectopic WUS expression in these cells is low, the peripheral zone cells may not be able to be transformed into central zone cells and thus do not contribute to a larger floral meristem. An alternative explanation for the reduced floral meristem size is that, as HAN is expressed in cells appearing to be the provascular tissue cells in as early as stage 1 flowers, the HAN-expressing cells may serve to transport nutrients or signals from other parts of the plant to the developing flower. In the case of insufficient supply of these factors, growth of the floral meristem is hindered.
However, unlike in SAM development where han mutations interact synergistically with clv mutations (see below), han appears epistatic to clv in flower development. han clv double mutants have similar or greater loss of floral organs than han single mutants, indicating that the mode of action of the HAN protein in controlling the proliferation of floral meristem cells may be different from that in the SAM, or HAN may have additional functions in flower development. The difference between HAN function in the development of SAM and flower is also reflected by the observation that wus is epistatic to han in SAM development but not in flower development in wus/wus han-1/+ mutants because these have reduced floral organ numbers compared with wus single mutants. This suggests that HAN is required for initiating and/or maintaining the proliferation of floral meristematic cells in wus mutants. By contrast, the increased leaf-like or shoot-like organs arising from the axils of leaves in wus han double mutants compared with wus single mutants suggests that HAN restricts meristematic cell activity in axillary meristem positions.
In addition to controlling floral meristem cell proliferation, HAN also appears to act in establishing boundaries between different whorls, as well as between different organ primordia in the same whorl. One possible mechanism for the boundary establishment is that HAN prevents cells expressing it from dividing, leading to the gap between organ primordia. In han mutants, gaps fail to form or be maintained, therefore resulting in one of its major floral defects: organ fusions. This theory is supported by the observation that overexpression of HAN causes growth retardation, small organs, and abolished meristem activity.
The other major floral defect in han mutants is decreased organ numbers in all four whorls, particularly in the 2nd and 3rd whorls. This could be a consequence of either reduced floral meristem size preceding organ initiation, diminished boundary formation, or both.
Interactions between HAN and the CLV-WUS Pathway in SAM Development
The HAN expression domain overlaps with the WUS expression domain during embryonic development, but HAN-expressing cells surround WUS-expressing cells in mature SAMs, with the possibility of some overlap. This suggests that, as in flower development, HAN's function in the SAM is to set up a boundary between the meristem and nascent organ primordia, which is required to confine WUS-expressing cells to a central domain. The WUS expression domain is spatially perturbed in han mutants beginning early in embryogenesis, before CLV3 and CLV1 expression initiates. This is consistent with the loss of WUS RNA in HAN overexpression.
Mutations in HAN greatly enhance clv phenotypes, implying that HAN may normally function to limit SAM size. Several mutations have been shown to enhance clv mutant phenotypes, including wiggum/enhanced response to abscisic acid1 and ultrapetala (Running et al., 1998; Fletcher, 2001). However, unlike the other enhancers, han loss of function does not lead to a SAM fasciation phenotype. Rather, some han mutants show flat or smaller SAMs toward the late stage of flowering. So how might perturbed WUS expression be tied to gradually flattened and reduced size of the SAM in han mutants? It is possible that diffuse WUS expression might result in a low level of WUS activity in individual cells, causing meristem cells gradually to lose their potency as stem cells, which may lead to flat and smaller SAMs. This possibility is supported by the observation that in some han inflorescence SAMs, the CLV3 domain also appears diffuse, which could have resulted from the low level of WUS expression. In contrast with han mutants, clv mutants have an expanded domain of WUS expression as a result of SAM enlargement. The different phenotypes of han and clv SAMs may be attributable to the fact that in the latter, WUS expression is not diffuse but is located in two cell layers below the epidermal cells of the SAM, and the expression level is fairly uniform across a single layer. Therefore, the mechanisms through which HAN and CLV control WUS-expressing cells are likely different. In either case, it is not clear whether the expanded WUS expression is caused by enhanced proliferation of WUS-expressing cells or by peripheral zone cells activating WUS expression.
Why then might the perturbed WUS expression in han mutants greatly enhance SAM fasciation in clv mutants? As mentioned above, HAN may play a partially CLV-independent role in restricting WUS expression to the L3 layer. A combination of han and clv mutations could lead to more WUS-expressing cells in the outer cell layers and peripheral zone, as well as increased levels of WUS expression, thus causing the formation of a more fasciated meristem. Although the above scenario may account for the increased SAM fasciation in han/clv double mutants, we cannot exclude the possibility that organ differentiation is further delayed in han/clv than in han mutants, which might also contribute to the accumulation of SAM cells.
The size variation of han mutant SAMs at different stages, as well as the variation of WUS and CLV3 expression in different han mutant meristems, suggests that the structure of the han SAM is dynamic and possibly self-correcting. Nevertheless, a more precise genetic control is required for a wild-type SAM, as han mutant SAMs become more irregularly shaped and produce more deformed organ primordia toward the later stages of development. The dynamic aspect of the SAM is shown by the observation that the WUS expression domain in wild-type inflorescence SAMs is variable either beneath two cell layers or (more often) three cell layers, suggesting that the WUS expression domain is fluctuating within an individual SAM during normal development. Another indication of regulatory activity in the SAM is the observation that in clv embryos, WUS expression is shifted to the L2 layer at the heart stage but moves to the layers underneath the L2 in mature embryos (Schoof et al., 2000). After postembryonic development, the WUS domain shifts back to include the L2 layer in clv inflorescence SAMs. We can speculate that in han mutants, the dynamics of WUS-expressing cells are imprecise, leading to a gradual disruption of the SAM structure.
In both clv1 and clv3 mutants, HAN expression is elevated and expanded along the fasciated SAM, overlapping with the WUS expression domain. Because HAN expression is not changed during clv3 mutant embryogenesis, the alteration of the HAN domain happens during postembryonic development. Is there any biological function of HAN expression in the outer layers of the SAMs in clv3 and clv1 mutants, or is it a mere effect of the altered SAM structure, causing the han domain to stretch laterally? It is possible that ectopic HAN expression in clv3 SAMs prevents WUS from further shifting toward the L2/L1 layer, making it still concentrated in the layer below the L2 layer. This explains why in the absence of both CLV and HAN functions, WUS becomes concentrated in the L2 layer and sometimes even shifted to the L1 layer. It is worth noting that HAN expression remains fairly normal in clv3 mutant flowers, further indicating that the mode of action of HAN in the floral meristem might be different from that in the SAM.
Effect of Differentiating Tissues on SAM Development
HAN is expressed in the provascular tissues below the WUS domain and begins to overlap with the WUS domain during the transition to the walking stick stage of embryogenesis. Later in the adult SAM, HAN expression marks the boundaries between the meristem and its initiating organs and between the WUS domain and the differentiated stem tissues below, with overlap between the HAN and WUS expression domains not being excluded. The effect of HAN on WUS-expressing cells could be direct during embryogenesis, but during postembryonic development it seems likely to be non-cell-autonomous. This adds another example of more mature cells exerting effects on meristematic cell activities (Stuurman et al., 2002). One can speculate that HAN-expressing cells could express certain secreted factor(s) or that the HAN protein could move from cell to cell via plasmodesmata to coordinate cell division/differentiation activity of WUS-expressing cells or to prevent non-WUS-expressing cells from activating WUS. In the absence of this coordination or prevention, cell division and differentiation become aberrant.
The han mutant phenotypes, HAN expression patterns, meristem marker analysis in han mutants, and overexpression of HAN all suggest that HAN is involved in regulating meristem cell activity by setting up a boundary between the meristem and differentiating organ primordia. In the event of HAN loss of function, the meristematic structure becomes disorganized, leading to meristem size reduction, reduced organ numbers and size, and organ fusion. Further molecular and biochemical studies will shed light on the mechanisms through which HAN regulates these important developmental processes.
METHODS
Plant Growth
Seeds were imbibed at 4°C cold room for 4 d before growth in constant light at 22°C. Plant age is calculated from the first day at 22°C. Seeds germinated on MS plates were sterilized as described at http://plantpath.wisc.edu/∼afb/vapster.html.
Isolation of han Alleles and Mapping of han Mutation
The han-1 allele (Clark et al., 1994) was isolated during T-DNA transformation mutagenesis of Wassilewskija ecotype seeds. The han-2 allele was isolated from an ethyl methanesulfonate mutagenesis screen in Landsberg erecta (Ler). The han-3 and han-4 alleles were both isolated from an activation-tagging screen in the clv1-1 mutant background (Weigel et al., 2000). Each single han mutant was backcrossed to Ler three times before phenotypic and genetic characterization.
The HAN gene was positionally mapped by crossing the han-1 mutant to the wild-type Columbia strain. DNA was prepared from 423 han-1 mutant plants in the T2 generation. Using cleaved amplified polymorphic sequence markers, recombination events between han-1 and surrounding regions were identified (Konieczny and Ausubel, 1993). Using this approach, the HAN region was narrowed down to BAC clone ATF18B3 on chromosome 3 (information about the polymorphic markers spanning HAN region is available in the supplemental data online). Sequencing of candidate genes revealed a 3262-bp DNA deletion at At3g50870 gene locus in han-1, when compared with wild-type sequences.
Morphological Analyses
Scanning electron microscope images were generated as described (Bowman et al., 1989). Confocal laser scanning microscopy images were generated as described (Running et al., 1995).
For 4′,6-diamidino-2-phenylindole staining, samples were fixed in ethanol/acetic acid (3:1) for 4 h on ice, washed twice in PBT (1× PBS with 0.1% Tween-20), each for 45 min, then stained with 1 μg/mL of 4′,6-diamidino-2-phenylindole in PBT for 15 min at room temperature. Samples were then rinsed four times with PBT and once with 25% glycerol. They were mounted on depression slides with glycerol and observed with a Zeiss LSM 510 META NLO confocal microscope (Jena, Germany) using a Chameleon titanium-sapphire laser at a wavelength of 760 nm.
In Situ Hybridization
HAN expression was examined by in situ hybridization using a full-length cDNA probe. The results were also confirmed using a 620-bp HAN DNA fragment starting from 170 bp upstream of the start codon and ending before the zinc finger domain (data not shown). Both HAN probes were tested on han-1 mutant samples and did not detect any specific signals. WUS, CLV3, and CLV1 probes were generated as described previously (Clark et al., 1997; Mayer et al., 1998; Brand et al., 2000). Apparent han-1 embryos were determined by the deviation of their morphology from the wild type in serial sections. For WUS and CLV3 probes, sections of the wild type and han mutant inflorescences were hybridized on the same slides or on different slides but were paired in sandwiches during probe hybridization, antibody incubation, and antibody detection. Conditions for all slides were identical in the remaining procedures. In situ hybridization was done by following the protocol at http://www.its.caltech.edu/∼plantlab/html/protocols.html with the following modifications. (1) After antibody reaction followed by four washes with antibody buffer, slides were equilibrated in detection buffer for 10 min (100 mM Tris-HCl, pH 9, 100 mM NaCl, and 50 mM MgCl2). Excess detection buffer was blotted off, and slides were then incubated with substrate solution (0.2 mM 5-bromo-4-chloro-3-indolyl phosphate and 0.2 mM nitro blue tetrazolium in detection buffer with 10% [w/v] polyvinyl alcohol [70 kD]) for 12 to 24 h at 30°C in darkness. (2) Slides were rinsed three times in distilled water to stop the detection reaction and mounted with 50% glycerol for observation under the microscope.
Construction of 35S:HAN and 35S:HAN-GR Lines
Both constructs were generated using the pGreen 0229 vector containing a 2× 35S promoter and a nopaline synthase terminator (http://www.pgreen.ac.uk/JIT/pGreen0000_fr.htm). Transformation was performed in Ler plants (see protocol at http://plantpath.wisc.edu/∼afb/protocol.html from Andrew Bent's lab in University of Illinois at Urbana-Champaign).
DEX Solution and Mock Solution
DEX stock solution (10 mM in ethanol) was stored at −20°C for up to 2 weeks. DEX solution (10 mM) was made by diluting the stock solution in pure water with or without 0.01% Triton X-100 (for inflorescences or soil, respectively). Mock solution consists of an equal concentration of ethanol and Triton X-100 in water per the final DEX solution.
Supplementary Material
Acknowledgments
We thank Ken Feldmann for allowing us to screen his T-DNA population, which resulted in the isolation of han-1, and Steven Clark for providing us with the han-2 allele. We also thank our laboratory colleagues and Robert Williams for their critical reading of the manuscript. This project is sponsored by National Science Foundation Grant IBN-0211670 to E.M.M.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Yuanxiang Zhao (zhaoyx@caltech.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.024869.
References
- Aida, M., Ishida, T., and Tasaka, M. (1999). Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: Interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126, 1563–1570. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Smyth, D.R., and Meyerowitz, E.M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1, 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand, U., Fletcher, J.C., Hobe, M., Meyerowitz, E.M., and Simon, R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Barley, R., Curtis, M., Arroyo, J.M., Dunham, M., Hudson, A., and Martienssen, R.A. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408, 967–971. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Jacobsen, S.E., Levin, J.Z., and Meyerowitz, E.M. (1996). The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 122, 1567–1575. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Sakai, H., and Meyerowitz, E.M. (1994). Inflorescence development in clavata mutants. In Arabidopsis: An Atlas of Morphology and Development, J.L. Bowman, ed (New York: Springer-Verlag), pp. 214–215.
- Clark, S.E., Williams, R.W., and Meyerowitz, E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585. [DOI] [PubMed] [Google Scholar]
- Fletcher, J.C. (2001). The ULTRAPETALA gene controls shoot and floral meristem size in Arabidopsis. Development 128, 1323–1333. [DOI] [PubMed] [Google Scholar]
- Fletcher, J.C., Brand, U., Running, M.P., Simon, R., and Meyerowitz, E.M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911–1914. [DOI] [PubMed] [Google Scholar]
- Fletcher, J.C., and Meyerowitz, E.M. (2000). Cell signaling within the shoot meristem. Curr. Opin. Plant Biol. 3, 23–30. [DOI] [PubMed] [Google Scholar]
- Gallois, J.L., Woodward, C., Reddy, G.V., and Sablowski, R. (2002). Combined SHOOT MERISTEMLESS and WUSCHEL trigger ectopic organogenesis in Arabidopsis. Development 129, 3207–3217. [DOI] [PubMed] [Google Scholar]
- Kayes, J.M., and Clark, S.E. (1998). CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 125, 3843–3851. [DOI] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Laux, T., Mayer, K.F., Berger, J., and Jurgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122, 87–96. [DOI] [PubMed] [Google Scholar]
- Lenhard, M., Jurgens, G., and Laux, T. (2002). The WUSCHEL and SHOOTMERISTEMLESS genes fulfil complementary roles in Arabidopsis shoot meristem regulation. Development 129, 3195–3206. [DOI] [PubMed] [Google Scholar]
- Lloyd, A.M., Schena, M., Walbot, V., and Davis, R.W. (1994). Epidermal cell fate determination in Arabidopsis: Patterns defined by a steroid-inducible regulator. Science 266, 436–439. [DOI] [PubMed] [Google Scholar]
- Long, J.A., Moan, E.I., Medford, J.I., and Barton, M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379, 66–69. [DOI] [PubMed] [Google Scholar]
- Mayer, K.F., Schoof, H., Haecker, A., Lenhard, M., Jurgens, G., and Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., and Barton, M.K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125, 2935–2942. [DOI] [PubMed] [Google Scholar]
- Rojo, E., Sharma, V.K., Kovaleva, V., Raikhel, N.V., and Fletcher, J.C. (2002). CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. Plant Cell 14, 969–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Running, M.P., Clark, S.E., and Meyerowitz, E.M. (1995). Confocal microscopy of the shoot apex. Methods Cell Biol. 49, 217–229. [DOI] [PubMed] [Google Scholar]
- Running, M.P., Fletcher, J.C., and Meyerowitz, E.M. (1998). The WIGGUM gene is required for proper regulation of floral meristem size in Arabidopsis. Development 125, 2545–2553. [DOI] [PubMed] [Google Scholar]
- Schoof, H., Lenhard, M., Haecker, A., Mayer, K.F., Jurgens, G., and Laux, T. (2000). The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644. [DOI] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souer, E., van Houwelingen, A., Kloos, D., Mol, J., and Koes, R. (1996). The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85, 159–170. [DOI] [PubMed] [Google Scholar]
- Stuurman, J., Jaggi, F., and Kuhlemeier, C. (2002). Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells. Genes Dev. 16, 2213–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada, S., Hibara, K., Ishida, T., and Tasaka, M. (2001). The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128, 1127–1135. [DOI] [PubMed] [Google Scholar]
- Venglat, S.P., Dumonceaux, T., Rozwadowski, K., Parnell, L., Babic, V., Keller, W., Martienssen, R., Selvaraj, G., and Datla, R. (2002). The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc. Natl. Acad. Sci. USA 99, 4730–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vroemen, C.W., Mordhorst, A.P., Albrecht, C., Kwaaitaal, M.A., and de Vries, S.C. (2003). The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15, 1563–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites, R., Selvadurai, H.R., Oliver, I.R., and Hudson, A. (1998). The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 93, 779–789. [DOI] [PubMed] [Google Scholar]
- Weigel, D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122, 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir, I., Lu, J., Cook, H., Causier, B., Schwarz-Sommer, Z., and Davies, B. (2004). CUPULIFORMIS establishes lateral organ boundaries in Antirrhinum. Development 131, 915–922. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.