Abstract
Classical arabinogalactan proteins (AGPs) are an abundant class of cell surface proteoglycans widely distributed in flowering plants. We have used a combination of enhancer detection tagging and RNA interference (RNAi)–induced posttrancriptional silencing to demonstrate that AGP18, a gene encoding a classical arabinogalactan protein, is essential for female gametogenesis in Arabidopsis thaliana. AGP18 is expressed in cells that spatially and temporally define the sporophytic to gametophytic transition and during early stages of seed development. More than 75% of the T1 transformants resulted in T2 lines showing reduced seed set during at least three consecutive generations but no additional developmental defects. AGP18-silenced T2 lines showed reduced AGP18 transcript levels in female reproductive organs, the presence of 21-bp RNA fragments specific to the AGP18 gene, and the absence of in situ AGP18 mRNA localization in developing ovules. Reciprocal crosses to wild-type plants indicate that the defect is female specific. The genetic and molecular analysis of AGP18-silenced plants containing a single T-DNA RNAi insertion suggests that posttranscriptional silencing of AGP18 is acting both at the sporophytic and gametophytic levels. A cytological analysis of all defective AGP18-RNAi lines, combined with the analysis of molecular markers acting at key stages of female gametogenesis, showed that the functional megaspore fails to enlarge and mitotically divide, indicating that AGP18 is essential to initiate female gametogenesis in Arabidopsis. Our results assign a specific function in plant development to a gene encoding a classical AGP.
INTRODUCTION
The life cycle of flowering plants consists of a diploid sporophytic phase and two morphologically different haploid gametophytic phases taking place in specialized reproductive organs. Distinct types of meiotically derived cells give rise to the male and female gametophytic phases. In the anther, many microsporocytes develop into pollen grains, which harbor the sperm cells and represent the male gametophyte. In the ovule, usually a single sporophytic cell (the megaspore mother cell [MMC]) undergoes meiosis and gives rise to four haploid products (the megaspores) during a process referred to as megasporogenesis. While three of the megaspores undergo programmed cell death, a single functional megaspore enlarges and gives rise to the female gametophyte (or megagametophyte). In Arabidopsis thaliana, female gametogenesis initiates when the single functional megaspore divides mitotically to form an eight-nucleate syncitium. Subsequent cellularization partitions the eight nuclei into seven cells: an egg cell and two synergids at the distal (or micropylar) pole of the female gametophyte, three antipodals at the proximal (or chalazal) pole, and a binucleated central cell whose nuclei fuse before fertilization. This type of development and organization of the female gametophyte defines the Polygonum type that prevails in >70% of the species examined (Maheshwari, 1950; Willemse and van Went, 1984; Reiser and Fischer, 1993; Drews and Yadegari, 2002). Whereas the fusion of a sperm with the egg cell forms a zygote that subsequently develops into an embryo, fertilization of the binucleated central cell eventually gives rise to the endosperm, a triploid tissue essential for seed viability.
Little is known about the genetic basis and molecular mechanisms that regulate the initiation of female gametogenesis in the ovule. A large collection of both sporophytic and gametophytic mutants defective in female gametophyte development has been identified in Arabidopsis (Schneitz et al., 1997; Christensen et al., 1998; Howden et al., 1998; Grini et al., 1999; Drews and Yadegari, 2002). Whereas sporophytic mutations act at the diploid level and are inherited in a Mendelian 3:1 ratio, gametophytic mutants are poorly transmitted through either one or both types of gametes and exhibit distorted segregation patterns. Many mutants that disrupt meiosis have been isolated (Klimyuk and Jones, 1997; Siddiqi et al., 2000; Yang and Sundaresan, 2000; Reddy et al., 2003), but little is known about other developmental aspects of megasporogenesis. To date, SPOROCYTELESS (SPO)/NOZZLE is the only gene shown to be required for the initiation of microsporogenesis and megasporogenesis (Schiefthaler et al., 1999; Yang et al., 1999). SPO encodes a nuclear protein related to MADS box transcription factors that is expressed during early anther and ovule development. In plants homozygous for hadad (hdd) and prolifera (prl), female gametophyte development is arrested at either the two-nucleate or the four-nucleate stage, respectively. Whereas the gene responsible for the mutation in hdd has yet to be identified (Moore et al., 1997), PRL encodes a Mcm7-like licensing factor essential for DNA replication (Springer et al., 1995). Two insertional alleles in CYTOKININ-INDEPENDENT 1, a gene encoding a putative Arabidopsis His kinase, were shown to cause nuclear degeneration in the female gametophyte as early as the four-nucleate stage (Christensen et al., 1998; Pischke et al., 2002). Recently, the NOMEGA gene was shown to be required for cell cycle progression beyond the two-nucleate stage; NOMEGA encodes a putative APC6/CDC16 component of the anaphase promoting complex in Arabidopsis (Kwee and Sundaresan, 2003). Several additional gametophytic mutants that fail to progress beyond the one-nucleate haploid stage have been identified (Christensen et al., 1998), but the corresponding genes have yet to be isolated and characterized.
Numerous studies showing that modifications of plant growth conditions can alter the sporophyte to gametophyte transition (Bell, 1989) or even the whole plant reproductive outcome (Knox, 1967) indicate that the presence of molecular signals that determine the fate of competent cells is fundamental for switching from a sporophytic into a gametophytic developmental pathway. Several regulatory proteins have been shown to have important functions in cell signaling and recognition during plant development. Arabinogalactan proteins (AGPs) are an abundant and heterogeneous class of highly glycosylated Hyp-rich glycoproteins widely distributed in the plant kingdom (Fincher et al., 1974; Clarke et al., 1979; Kreuger and van Holst, 1996; Sommer-Knudsen et al., 1998; Gaspar et al., 2001; Showalter, 2001). The recent characterization of genes encoding different AGP backbones in several species gave rise to the current distinction between classical and nonclassical AGPs (Chen et al., 1994; Du et al., 1994; Mau et al., 1995; Knox, 1999). Classical AGPs contain a domain responsible for attaching the protein backbone to a glycosylphosphotidylinositol (GPI) membrane anchor (Schultz et al., 1998; Youl et al., 1998; Sherrier et al., 1999; Schindelman et al., 2001; Borner et al., 2002; Sun et al., 2004). By contrast, nonclassical AGPs lack the GPI anchor signal and are soluble components of the extracellular matrix often containing Asn- or Hyp-rich domains (Majewska-Sawka and Nothnagel, 2000; Schultz et al., 2000; Gaspar et al., 2001). Molecular and biochemical evidence indicates that AGPs have specific functions during root formation (Willats and Knox, 1996; Casero et al., 1998; van Hengel and Roberts, 2002), the promotion of somatic embryogenesis (Serpe and Nothnagel, 1994; Kreuger and van Holst, 1993, 1996; van Hengel et al., 2001; van Hengel and Roberts, 2002), or the attraction of pollen tubes in the style (Du et al., 1994; Cheung et al., 1995; Wu et al., 1995; Jauh and Lord, 1996; Roy et al., 1998). The use of monoclonal antibodies directed against carbohydrate epitopes provided evidence suggesting that AGPs play an important role during the alternation between sporophytic and gametophytic transitions in the ovule (Pennell and Roberts, 1990; Pennell et al., 1991; McCabe et al., 1997). Although these studies elegantly showed that the establishment of a female reproductive lineage is associated with changes in the distribution of AGP epitopes (Pennell et al., 1992), they did not identify a specific AGP protein or the corresponding gene acting during ovule development or early embryo formation. A few mutations altering the activity of specific genes encoding AGPs in Arabidopsis have been described. Homozygous plants for resistant to agrobacterium transformation 1 are resistant to root-dependent transformation via Agrobacterium tumefaciens (Nam et al., 1999); however, mutant plants are phenotypically indistinguishable from the wild type, and no developmental defects associated with the mutation have been described. The mutation is caused by a T-DNA insertion within the promoter region of the Arabidopsis AGP17 gene (Gaspar et al., 2001). A second insertional mutant in a gene encoding a nonclassical AGP (AGP30) has recently been shown to be involved in root regeneration and seed germination (van Hengel and Roberts, 2003). Recently, hybrid-type proteoglycans having properties of both AGPs and lipid-transfer proteins have been shown to be essential for the differentiation of tracheary elements in Zinnia elegans and Arabidopsis (Motose et al., 2004).
In this study, we report the enhancer detection-based identification of AGP18, a classical AGP gene that specifically acts during female gametophyte development in Arabidopsis. To determine the function of AGP18, we introduced double-stranded RNA in wild-type plants and specifically degraded the endogenous AGP18 transcript by RNA interference (RNAi). More than 75% of the primary transformants resulted in lines showing reduced seed set but no additional developmental abnormalities. Reciprocal crosses to wild-type plants suggested that the defect is female specific. The genetic and molecular analysis of a line containing a single T-DNA RNAi insertion suggests that posttranscriptional silencing of AGP18 is acting both at the sporophytic and gametophytic levels. The cytological analysis of all defective AGP18-RNAi lines indicates that AGP18 is essential to initiate female gametogenesis in Arabidopsis. Our results assign a specific function in plant development to a gene encoding a classical AGP.
RESULTS
Enhancer Detection Tagging of AGP18
Using the system established by Sundaresan et al. (1995), we have generated a Ds enhancer detector and a gene trap population to identify patterns of expression associated with genes acting during female gametophyte development in Arabidopsis. The enhancer detection vector relies on a maize (Zea mays) Ds transposon carrying a β-glucoronidase reporter gene (uidA or GUS) under the control of a minimal promoter. Such a reporter construct is not trapping genes but rather integrating into genomic sequences to serve as a detector of any given regulatory sequence that is acting as an enhancer of promoter activity at the specific location of the insertion (Bellen, 1999; Springer, 2000). The Ds enhancer detector element (DsE) also contains the neomycin phosphotransferase II (NPTII) gene (conferring resistance to kanamycin); NPTII acts as a selectable marker and facilitates the genetic analysis of segregating enhancer detector or gene trap lines.
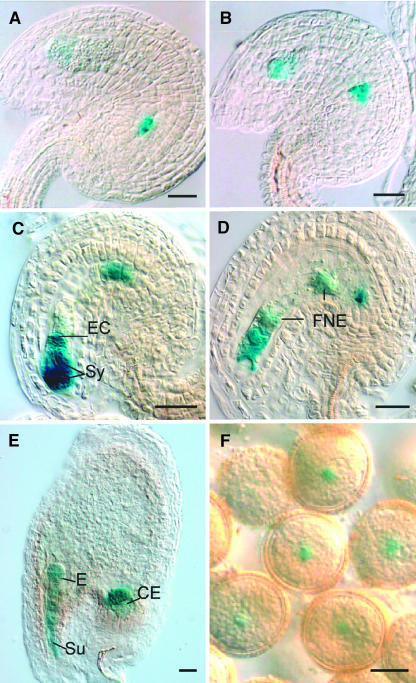
Whole-mount staining and clearing procedures allow screening for reporter gene expression (GUS) at different developmental stages encompassing megasporogenesis and female gametogenesis, from the time when the ovule primordium has just started its elongation (before MMC differentiation) to stages where the female gametophyte is fully differentiated (J.-P. Vielle-Calzada and U. Grossniklaus, unpublished results; Vielle-Calzada et al., 2000). Figure 1 shows the pattern of GUS expression identified in MET333. MET333 shows initial GUS expression in the chalazal region of the four-nucleate female gametophyte (Figure 1A). At the eight-nucleate stage, expression is restricted to the young antipodal cells and the cellularizing egg apparatus (Figure 1B). At maturity, the female gametophyte shows GUS expression in the synergids, the egg cell, and the antipodals but not in the central cell (Figure 1C). This pattern of expression persists after fertilization (Figure 1D); however, GUS is also expressed in the free nuclear endosperm after fertilization of the central cell (Figure 1D). During early seed development, GUS is expressed in the embryo proper, the suspensor, and the chalazal endosperm (Figure 1E). Interestingly, MET333 also shows GUS expression in cytoplasmic domains closely associated with the vegetative nucleus of mature pollen grains and in pollen tubes (Figure 1F) but not in microsporocytes at earlier stages of development. A detailed analysis of MET333 plants homozygous for kanamycin resistance did not reveal a mutant phenotype at any stage of plant reproductive development.
Figure 1.
Pattern of GUS Expression in the Enhancer Detector Line MET333.
(A) Female gametophyte at four-nucleate stage.
(B) Cellularized female gametophyte.
(C) Mature female gametophyte before fertilization.
(D) Female gametophyte after fertilization.
(E) Embryo at four-cell stage.
(F) Mature pollen with GUS expression associated with the vegetative nucleus.
Sy, synergids; EC, egg cell; E, embryo; FNE, free nuclear endosperm; Su, suspensor; CE, chalazal endosperm. Bars in (A) to (E) = 20 μm; bar in (F) = 10 μm.
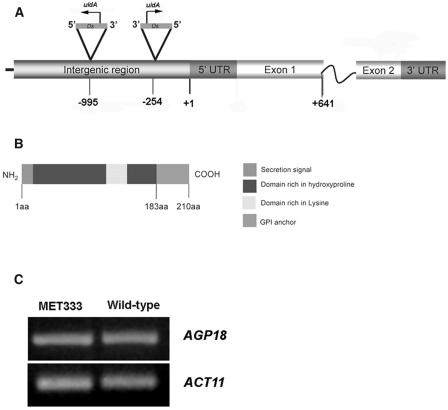
DNA gel blot analysis using a DsE-specific probe that includes a portion of NPTII showed that two copies of the DsE element were present in MET333 heterozygotes but were absent from wild-type siblings in which GUS expression was not detected (data not shown). Genomic sequences flanking both DsE elements were rescued using thermal asymmetric interlaced PCR (Liu et al., 1995). Sequence analysis of the PCR products showed that the DsE elements were inserted 254 and 995 bp upstream of the transcription initiation site of AGP18 (At4g37450). Figure 2A illustrates the molecular structure of AGP18 and the localization of both DsE insertion sites. AGP18 encodes a classical AGP containing a C-terminal domain responsible for anchoring the protein to GPI. In animals, GPI anchors have been shown to provide an alternative to transmembrane proteins for anchoring proteins to components of the cell surface (Takos et al., 1997, 2000; Schultz et al., 1998; Svetek et al., 1999; Borner et al., 2002; Sharma et al., 2004). Additionally, AGP18 contains an N-terminal secretory signal predicted to direct the secretion of the protein via the endoplasmic reticulum, two Pro-rich domains that are possible targets of glycosylation (Tan et al., 2003), and a Lys-rich domain predicted to interact with negatively charged molecules (Figure 2B; Gilson et al., 2001; Schultz et al., 2002). The Lys-rich domain is present in only 3 of 15 classical AGPs found to be encoded in the genome of Arabidopsis (Schultz et al., 2002). RT-PCR analysis revealed that the levels of AGP18 transcription in MET333 are similar to the wild type, confirming that AGP18 expression is not diminished by the presence of the DsE insertions (Figure 2C).
Figure 2.
Genomic Structure and Protein Organization of AGP18.
The enhancer detector line MET333 has two DsE elements inserted in the 5′ regulatory region of AGP18.
(A) Genomic structure of AGP18. The arrows show the direction of transcription of uidA (GUS).
(B) Predicted protein structure of AGP18. aa, amino acids.
(C) RT-PCR analysis shows that the levels of transcription of AGP18 are identical in MET333 and wild-type plants.
AGP18 Is Expressed in Adjacent Sporophytic and Gametophytic Cells
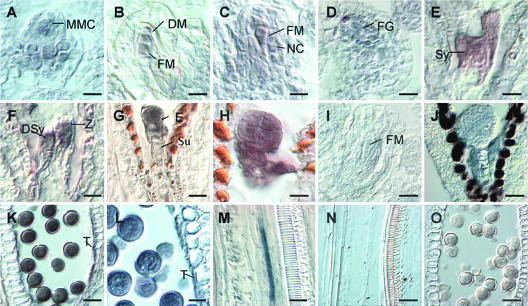
To determine if the pattern of GUS expression identified in MET333 reflected the pattern of expression of AGP18, we determined the localization of AGP18 mRNA by in situ hybridization. To avoid the detection of mRNA corresponding to other AGP transcripts structurally resembling AGP18, sense and antisense digoxygenin-labeled probes were generated using a specific portion of the first exon that shows no homology with other AGP genes. The results are summarized in Figure 3. Detailed analysis of all aerial parts of Arabidopsis demonstrated that AGP18 mRNA could be localized only in developing anthers and ovules and transiently in clusters of companion cells closely associated with vascular elements of the stem. AGP18 is initially expressed in the MMC and the neighboring nucellar cells of the young ovule primordium (Figure 3A). AGP18 expression persists in all four products of female meiosis (Figure 3B). At the end of megasporogenesis, when the three nonfunctional megaspores have already degenerated, AGP18 mRNA is abundant in the functional megaspore but also in the adjacent nucellar cells (Figure 3C). During female gametogenesis, AGP18 is expressed in the developing female gametophyte (Figure 3D). At maturity, abundant AGP18 mRNA can be detected in the synergids (Figure 3E), the egg cell, and the antipodals but not in the central cell. After fertilization, AGP18 mRNA is present in the developing embryo as well as in the free nuclear endosperm (Figures 3F to 3H). Abundant levels of AGP18 mRNA persist in the embryo until the late globular stage and subsequently start to decrease. No AGP18 mRNA can be detected in seeds containing torpedo or cotyledonary embryos. In the anther, AGP18 is expressed in the tapetum and the mature pollen grain (Figures 3K and 3L). These results indicate that GUS expression in MET333 overlaps with the localization of AGP18 mRNA, confirming that DsE elements partially detect the expression of AGP18; however, the absence of GUS expression in MET333 sporophytic cells (the developing nucellus and the mature tapetum) suggests that additional regulatory elements driving the expression of AGP18 are not detected by either DsE element.
Figure 3.
Localization of AGP18 mRNA by in Situ Hybridization.
(A) MMC stage. Bar = 9 μm.
(B) Female meiosis stage. Bar = 8 μm.
(C) Functional megaspore stage with young nucellus. Bar = 8.5 μm.
(D) Two-nucleate stage female gametophyte. Bar = 14 μm.
(E) Mature female gametophyte. Bar = 8 μm.
(F) Zygote stage. Bar = 9μm.
(G) Embryo four-cell stage. Bar = 15 μm.
(H) Embryo at early globular stage. Bar = 11 μm.
(I) Functional megaspore and young nucellus, sense probe. Bar = 15 μm.
(J) Embryo at early globular stage, sense probe. Bar = 20 μm.
(K) Mature pollen. Bar = 24 μm.
(L) Anther showing the tapetum. Bar = 20 μm.
(M) Longitudinal section of a stem. Bar = 20 μm.
(N) Longitudinal section of a stem, sense probe. Bar = 20 μm.
(O) Mature pollen, sense probe. Bar = 20 μm.
(A) to (H) and (K) to (M) hybridizations with antisense probe; (I), (J), (N), and (O) hybridizations with sense probe. NC, nucellar cells; FM, functional megaspore; DM, degenerating megaspores; Sy, synergids; DSy, degenerating synergid; FG, female gametophyte; Z, zygote; E, embryo; Su, suspensor; T, tapetum.
Generation of AGP18-RNAi Plants and Analysis of RNA Levels
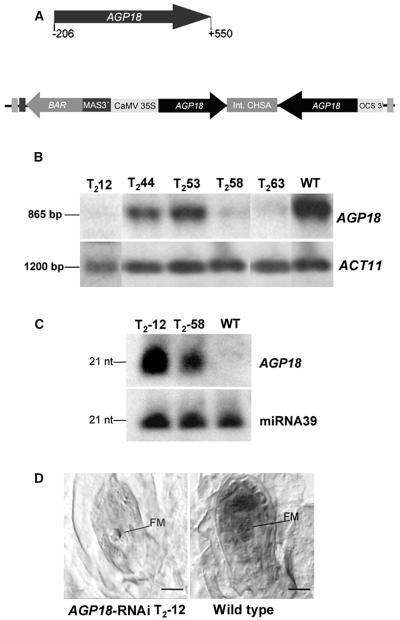
To determine the role of AGP18 in Arabidopsis, a 740-bp fragment of the AGP18 cDNA was cloned into a pFGC5941 RNAi vector (Kerschen et al., 2004) in both sense and antisense orientations and used to transform wild-type Columbia plants. Figure 4A illustrates the RNAi construct that was used to conduct these experiments. pFGC5941 contains a 35S promoter of Cauliflower mosaic virus (CaMV35S) that drives the transcription of a partial AGP18 sequence cloned in both sense and antisense orientations and separated by an intron of the chalcone synthase gene. After formation of hairpin RNA structures, the resulting double-stranded RNA transcripts can cause posttranscriptional silencing of endogenous gene activity (Waterhouse et al., 1998; Chuang and Meyerowitz, 2000; Smith et al., 2000). Although a detailed pattern of CaMV35S promoter activity during male and female gametogenesis has yet to be determined in Arabidopsis, we reasoned that AGP18 transcripts localized in sporophytic cells can be the target of RNAi-dependent silencing driven by CaMV35S. After floral-dipping transformation, 75 primary transformants were generated, none of which showed visible defects during vegetative growth, root development, or floral organogenesis; however, 58 out of 75 adult T1 transformants showed semisterility defects. All 58 transformants maintained a reduced fertility phenotype in the T2 generation. To determine a possible relationship between a decrease in AGP18 transcript levels and the defective phenotype, RNA was extracted from developing gynoecia of BASTA-resistant AGP18-RNAi T2 lines and used for RNA gel blot analysis. The results are shown in Figure 4B, with actin as a constitutive control to show that equal amounts of RNA were used. Compared with wild-type plants, all 10 T2 lines tested showed a substantial decrease in the transcripts levels of AGP18. Among those lines, T2-12, T2-44, and T2-58 showed significantly >50% reduction in seed set, whereas T2-63 and T2-53 had a 27.4 and 49.8% reduction, respectively. No correlation was found between the level of AGP18 expression determined by RNA gel blot analysis and the degree of sterility; this absence of correlation has been documented in previous studies showing that abnormal phenotypes induced by RNAi are not always associated with a detectable decrease in transcript levels (Kerschen et al., 2004; C. Napoly and R. Jorgensen, personal communication). The induction of posttrancriptional gene silencing has been shown to result in the production of 21- to 23-bp RNA fragments with a sequence identical to a portion of the silenced gene (Elbashir et al., 2000). To determine if the production of 21- to 23-bp RNA fragments could be associated with the degradation of the AGP18 transcript, polyacrylamide gels were used to detect the presence of small RNA fragments corresponding to AGP18. As shown in Figure 4C, small RNAs corresponding to AGP18 were detected in lines in which sufficient quantities of total RNA were available, including lines having strong fertility defects as T2-12 and T2-58 but not in wild-type plants. Finally, to determine if a decrease in transcript levels was associated with a decrease in AGP18 expression within the ovule or the anther, we performed in situ hybridization in selected lines showing the lowest levels of AGP18 transcript. As shown in Figure 4D, no AGP18 mRNA could be detected in the young ovule at the functional megaspore stage of AGP18-RNAi transformants, indicating that the normal expression of AGP18 during female gametogenesis is severely impaired in AGP18-RNAi lines.
Figure 4.
Accumulation of AGP18 Transcript, Presence of 21-bp Small RNAs, and Absence of AGP18 Expression in the Gynoecium of AGP18-RNAi T2 Lines.
(A) Schematic diagram of the vector used to posttrancriptionally silence AGP18. The arrow indicates the sequence cloned in the RNAi silencing vector. Numbers indicate nucleotide positions with respect to initiation of the AGP18 mRNA.
(B) Expression analysis of four AGP18-RNAi T2 lines and a wild-type control. RNA was isolated from mature gynoecia in both silenced and wild-type plants. A portion of the AGP18 cDNA was used as a probe. RNA gel blots were subsequently rehybridized with a specific actin probe (ACT11) as a loading control.
(C) A polyacrylamide gel of 100 μg of low molecular weight RNA extracted from gynoecia of AGP18-RNAi T2 lines and wild-type plants was blotted and hybridized with a portion of the AGP18 cDNA. The blot was rehybridized with a probe specific to the constitutively expressed microRNA 39 (miR39) as a control. nt, nucleotides.
(D) Localization of AGP18 mRNA in developing ovules at the functional megaspore stage. In situ hybridization with specific AGP18 digoxygenin-labeled antisense probes was performed on gynoecia of both silenced (AGP18-RNAi T2-12; bar = 8.5 μm) and wild-type plants (bar = 10 μm). FM, functional megaspore.
Posttranscriptional Gene Silencing Is Specific to AGP18
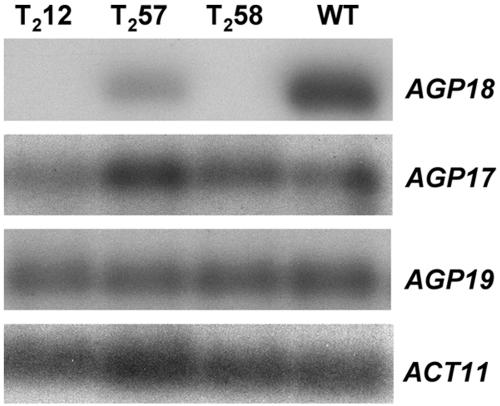
Plant transformation with RNAi vectors targeting a conserved gene family has been shown to often result in simultaneous posttranscriptional gene silencing of several family members. In Arabidopsis, 16 genes are predicted to encode the protein backbones of classical AGPs (Schultz et al., 2000). AGP17 and AGP19 encode classical AGPs with 56 and 42% amino acid similarity to AGP18, respectively. All three proteins are the only Arabidopsis AGP members containing a Lys-rich domain. At the DNA level, AGP17 and AGP19 share 55 and 52% homology with AGP18 in the 740-bp cDNA fragment that was used to generate the RNAi construct. To determine if the transcript levels of any of these two genes were also decreased in AGP18-RNAi lines, total RNA extracted from developing gynoecia of three T2 AGP18-RNAi lines showing decreased levels of AGP18 mRNA accumulation were used to perform RT-PCR analysis. As shown in Figure 5, both AGP17 and AGP19 are expressed in the gynoecia of wild-type plants; however, none of the T2 AGP18-RNAi lines analyzed showed a significant decrease in either AGP17 or AGP19 expression. In lines T2-12 and T2-58, no amplification signal could be detected after blotting RT-PCR gels and hybridizing with the corresponding AGP18 probe, confirming that in these lines AGP18 expression is almost completely silenced. These results demonstrate that in AGP18-RNAi lines posttranscriptional gene silencing is specific to AGP18.
Figure 5.
Posttranscriptional Gene Silencing Is Specific to AGP18.
RNA extracted from developing gynoecia of selected AGP18-RNAi T2 lines (T2-12, T2-57, and T2-58) was used for cDNA synthesis. PCR amplification was performed with primers specific to AGP17, AGP18, or AGP19 (Schultz et al., 2002) using as a template samples corresponding to the same cDNA synthesis. Agarose gels were blotted on nitrocellulose membranes and probed with a corresponding AGP probe. Wild-type cDNA and amplification of ACT11 were used as positive controls.
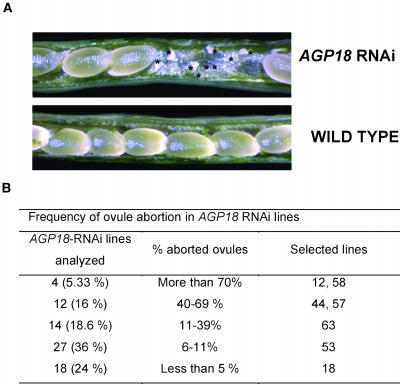
Ovule Abortion in AGP18-RNAi Lines Is Controlled at the Sporophytic and Gametophytic Levels
As shown in Figure 6A, the siliques of AGP18-RNAi T2 lines contain a variable number of aborted ovules that do not show signs of early seed formation. This defect can also be observed in the form of empty spaces within siliques of self-fertilized flowers from AGP18-RNAi plants. Gametophytic defects affecting the female gametophyte but not the male are expected to show a decrease in seed set of ∼50%. As shown in Figure 6B, T2 lines showed a wide range of frequencies of ovule abortion, with >20% showing a frequency significantly >50% and four lines showing >70% of aborted ovules. To determine the nature of the reproductive defect found in RNAi lines, we conducted reciprocal crosses between lines showing >50% ovule abortion and wild-type plants. When T2 lines were used as female parents, the same percentage of ovule abortion was obtained for all lines tested (data not shown); however, in crosses where T2 lines were used as male parents, full fertility was recovered, suggesting that the sterility defect is female specific.
Figure 6.
Siliques of AGP18-RNAi Lines Show Aborted Ovules.
(A) Micrographs of AGP18-RNAi and wild-type siliques. The asterisks indicate the aborted ovules observed in AGP18-RNAi lines.
(B) An average of 250 ovules was scored for each AGP18-RNAi line. A χ2 statistical analysis showed that lines having >5% ovule abortion were significantly different from the wild type (2.5% ovule abortion).
DNA gel blot analysis was used to determine that the number of RNAi T-DNA insertions present in the genome of T2 lines varied between 1 and 6 (data not shown). Because the T-DNA RNAi construct used is marked with a BASTA herbicide resistance marker, its segregation pattern can easily be followed in seedlings. We characterized the segregation of BASTA resistance in a group of AGP18-RNAi lines showing high levels of ovule abortion but different numbers of T-DNA insertions. A summary of these results are presented in Table 1. Interestingly, a single T-DNA RNAi insertion was present in the genome of T2-12, a line showing 74.9% of aborted ovules and the highest levels of AGP18 silencing in female reproductive organs. As shown in Table 1, T2-12 segregated BASTA-resistant (BASTAr) and BASTA-sensitive (BASTAs) seedlings in a distorted ratio of 1:1 BASTAr:BASTAs (χ2 = 2.41 < χ20.05[1] = 3.84) as compared with 3:1 expected for normally transmitted insertions. In addition, no homozygous individuals have been identified in the T4 progeny resulting from self-pollination of 20 heterozygous T3-12 plants, suggesting that the transgene is poorly or not transmitted through the gametophytic phase of the Arabidopsis life cycle. The frequency of aborted ovules and the distorted segregation ratio in T2-12 suggests that posttranscriptional silencing of AGP18 is affecting female reproductive development both at the sporophytic and gametophytic levels. In lines containing more than one T-DNA RNAi insertion, the number of BASTAr seedlings is significantly increased, suggesting that not all insertions are abnormally transmitted through the gametophytic phase and that the degree of semisterility found in AGP18-RNAi lines is not directly proportional to the number of introduced T-DNA insertions. These results are in agreement with recent estimations of the efficiency of RNAi in transgenic plants (Kerschen et al., 2004).
Table 1.
Number of T-DNA Insertions, Segregation Analysis, and Percentage of Ovule Abortion in Selected T2 AGP18RNAi Lines
| Line | Number of Insertions | BASTAr | BASTAs | Ratio | Viable Ovules | Aborted Ovules | Percentage of Ovule Abortion |
|---|---|---|---|---|---|---|---|
| T2-12 | 1 | 127 | 153 | 1.0:1.2 | 62 | 185 | 74.90 |
| T2-57 | 2 | 143 | 53 | 2.6:1.0 | 100 | 125 | 55.55 |
| T2-58 | 2 | 186 | 15 | 12.4:1.0 | 67 | 167 | 71.36 |
| T2-44 | 5 | 182 | 46 | 3.9:1.0 | 125 | 99 | 49.01 |
To determine if fertility defects were consistently inherited, we quantified the sterility phenotype during the first three consecutive generations of four selected lines. As shown in Table 2, the percentage of aborted ovules is maintained in the T3 generation; however, a small decrease in the frequency of ovule abortion was observed in all lines evaluated. Statistical analysis of variance showed that this decrease is not statistically significant during the generations tested.
Table 2.
Inheritance of Ovule Abortion in AGP18-RNAi Lines
| Line
|
||||
|---|---|---|---|---|
| Generation | 12 | 44 | 57 | 58 |
| T1 | 74.9% (247) | 44.1% (224) | 55.50% (225) | 71.36% (234) |
| T2 | 73.2% (220) | 45.2% (271) | 54.90% (232) | 74.00% (255) |
| T3 | 67.5% (234) | 39.0% (230) | 46.15% (244) | 70.80% (222) |
Female Gametophyte Development Is Defective in AGP18-RNAi Lines
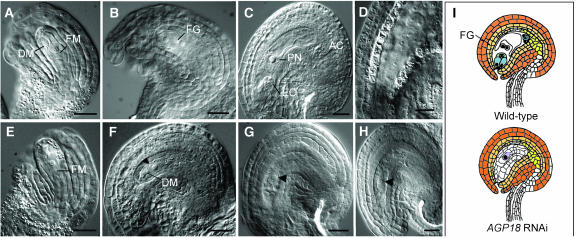
To determine the cellular nature of the defect, whole-mounted cleared anthers and ovules were analyzed during male and female reproductive development. We initially determined the terminal phenotypes of all AGP18-RNAi lines by examining mature organs at developmental stages in which male and female gametogenesis have normally resumed and fully differentiated cellularized gametophytes are already formed. All 58 lines examined had a variable proportion of ovules showing a similar abnormal phenotype but no defects in pollen formation. Our observations are summarized in Figure 7. In the large majority of defective ovules, a single conspicuous cell with a centrally located nucleus was present in the nucellus. In all lines, the conspicuous cell did not show signs of degeneration, and the adjacent nucellar tissue was not reabsorbed; in rare cases, its nucleus was not observed. To determine the developmental stage at which female gametophyte development first departs from the wild type, we analyzed female gametogenesis at earlier developmental stages in ovules of line T2-12 (showing 74.9% of ovule abortion). All ovules of T2-12 undergo normal and synchronized megasporogenesis. Shortly after the differentiation of the functional megaspore (Figures 7A and 7E), a minority of ovules divide mitotically and give rise to two nuclei located at opposite ends of an enlarged cell showing a central vacuole. This type of ovule does not show differences with wild-type development (Figure 7B). By contrast, within the same gynoecium, the nucellus of the majority of ovules contains a single cell that closely resembles the differentiated functional megaspore but that shows no signs of subsequent enlargement or vacuolization (Figure 7F). At later stages of development, whereas some ovules undergo two additional divisions and form a normal female gametophyte identical to the wild type (Figure 7C), the majority contains a single cell that does not divide mitotically (Figures 7G and 7H). To determine if the frequency of aborted ovules scored in dissected siliques is similar to the frequency of ovules showing abnormal female gametophyte development, we quantified the number of ovules showing a single conspicuous cell in the nucellus of fully differentiated ovules. The results are shown in Table 3. For all lines examined, there is a close correlation between the two values, suggesting that defects in female gametophyte development are sufficient to explain the abnormal phenotype observed in AGP18-RNAi lines.
Figure 7.
Female Gametophyte Development Is Defective in Ovules of AGP18-RNAi Lines.
Wild-type and AGP18-RNAi T2 gynoecia were fixed, cleared, whole mounted, and viewed under Nomarsky optics.
(A) to (D) Development of wild-type ovules.
(A) Functional megaspore in a developing ovule.
(B) Female gametophyte at the two-nucleate stage.
(C) Mature female gametophyte.
(D) Young embryo at the two-cellular stage.
(E) to (H) Development of AGP18-RNAi T2-12 defective ovules.
(E) Functional megaspore in the developing T2-12 ovule.
(F) Arrested cell in defective T2-12 mature ovule (arrowhead); normal ovules in the same gynoecium are at the two-nucleate stage.
(G) Arrested cell in defective T2-12 mature ovule (arrowhead); normal ovules in the same gynoecium contain a mature female gametophyte.
(H) Arrested cell in T2-12 mature ovule; normal ovules in the same gynoecium contain seeds undergoing early stages of embryogenesis.
(I) Schematic representation compares a mature wild-type ovule to a mature AGP18-RNAi T2-12 defective ovule.
FM, functional megaspore; DM, degenerating megaspore cells; EC, egg cell; AC, antipodal cells; FG, female gametophyte; PN, polar nuclei. Arrowheads indicate the presence of an arrested cell at the one-nucleate stage. Bars = 20 μm.
Table 3.
Frequency of Ovules Showing an Arrested Cell Phenotype
| AGP18-RNAi Line | Viable Ovules | Aborted Ovules | Ovules with Arrested Cells |
|---|---|---|---|
| T2-12 | 138 (27.7%) | 185 (74.9%) | 359 (72.23%) |
| T2-57 | 196 (43.3%) | 125 (55.5%) | 257 (56.73%) |
| Wild type | 443 (99.4%) | 6 (2.4%) | 3 (0.60%) |
The Functional Megaspore Does Not Initiate Female Gametogenesis in AGP18-RNAi Ovules
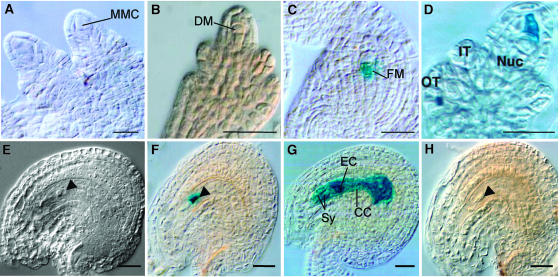
To determine the identity of the cell that persists in the nucellus of defective ovules, we took advantage of additional enhancer detection and gene trap lines that show GUS expression in specific cells of the developing female gametophyte. Some of them represent ideal molecular markers to conduct crosses with AGP18-RNAi lines showing a high proportion of defective ovules. Figure 8 illustrates the pattern of GUS expression obtained in ovules of F1 plants resulting from crosses of line AGP18-RNAi T2-12 with individuals homozygous either for the ET499 or for the ET2209 enhancer detection element. In ET499, GUS is only expressed in the functional megaspore and not in the three dying megaspores or at earlier stages of megasporogenesis (Figures 8A to 8C; J.-P. Vielle-Calzada and U. Grossniklaus, unpublished results). This observation was confirmed in 100 whole-mounted and cleared ovules of homozygous ET499 plants. Other enhancer detection lines show GUS expression in the dying megaspores but not in the functional megaspore (Figure 8D), indicating that ET499 is an appropriate marker to characterize megaspores that have acquired a functional identity at the end of megasporogenesis. By constrast, ET2209 shows GUS expression at the onset of the second haploid mitotic division of the uncellularized female gametophyte and subsequently in all haploid differentiated cells: the synergids, the egg cell, the central cell, and the antipodals (Figure 8G; Vielle-Calzada et al., 2000). In defective AGP18-RNAi/+ ET499/+ F1 ovules, GUS expression is restricted to the conspicuous cell that persists in the nucellus after meiosis (Figures 8E and 8F), suggesting that in AGP18-RNAi lines female gametophyte development is arrested after the differentiation of the functional megaspore. By contrast, defective AGP18-RNAi/+ ET2209/+ F1 do not show GUS expression (Figure 8H), indicating that the arrested functional megaspore does not acquire the identity of a multinucleated female gametophyte or of any of the haploid gametophytic cells. These results indicate that defective AGP18-RNAi ovules fail to undergo haploid mitosis after differentiation of the functional megaspore.
Figure 8.
The Functional Megaspore Does Not Initiate Female Gametogenesis in AGP18-RNAi Ovules.
Ovules of F1 plants resulting from crosses of line AGP18-RNAi T2-12 with individuals homozygous either for the ET499 or for ET2209 were either fixed and whole-mount cleared or processed for histochemical localization of GUS activity. Shown are patterns of GUS expression in ET499 ([A] to [C]), ET4127 (D), F1 plants resulting from the cross of AGP18-RNAi T2-12 and homozygous ET499 plants ([E] and [F]), ET2209 (G), and F1 plants resulting from the cross of AGP18-RNAi T2-12 and homozygous ET2209 plants (H).
(A) Ovules of ET499 at MMC not showing GUS expression.
(B) Ovules of ET499 showing absence of expression in all three dying megaspores.
(C) Functional megaspore showing GUS expression in ET499.
(D) Ovule of ET4127 showing GUS expression in a dying megaspore.
(E) Whole-mounted cleared ovule showing the phenotype observed in defective ET499;AGP18-RNAi T2-12 F1 ovules.
(F) GUS expression in the arrested functional megaspore of defective ET499;AGP18-RNAi T2-12 F1 ovules. The arrowhead shows the position of the arrested functional megaspore.
(G) Pattern of GUS expression in the mature female gametophyte of ET2209.
(H) Absence of GUS expression in defective ET2209;AGP18-RNAi T2-12 F1 ovules.
FM, functional megaspore; IT, inner integument; OT, outer integument; Nuc, nucellus; DM, dying megaspore; EC, egg cell; Sy, synergid cells; CC, central cell. The arrowheads indicate the arrested cells. Bars = 20 μm.
DISCUSSION
Here, we report the successful use of RNAi-induced posttranscriptional silencing to inactivate the AGP18 gene and show that it plays an essential role during the initiation of female gametogenesis in Arabidopsis. AGP18 encodes a classical AGP shown to be expressed in cells that spatially and temporally defines the sporophytic to gametophytic transition, but also during early stages of embryogenesis. More than 77% of independent transgenic Arabidopsis lines expressing the AGP18-RNAi construct showed moderate to severe fertility defects reminiscent of semisterile gametophytic mutants in Arabidopsis. Although in other experiments RNAi-dependent silencing is not always associated with mRNA turnover (Kerschen et al., 2004), all lines tested showed a decrease in AGP18 transcript accumulation during female reproductive development in the T2 generation. In at least three lines, AGP18 expression was almost completely suppressed. T2 lines with fertility defects showed ovules impaired in female gametogenesis but normal male gametophytic development and pollen formation. The use of molecular markers expressed at key stages of female gametogenesis determined that in defective ovules meiosis gives rise to a differentiated functional megaspore that is unable to give rise to a two-nucleate female gametophyte.
In plants developing a female gametophyte of the Polygonum type, megasporogenesis ends with the initiation of the haploid phase of the life cycle during the mitotic division of the functional megaspore nucleus (Huang and Russell, 1992). Although little is known about cellular communication during early ovule development, the interaction between sporophytic and gametophytic tissues has been suggested to be essential for female gametogenesis. For example, the isolation of the meiotic precursors and young tetrads by the accumulation of callosic walls has been interpreted as an interfacial reaction leading to the necessary separation of the two generations and the consequent protection of the haploid phase in ferns, mosses, and flowering plants (Dickinson, 1994; Bell, 1995). In Arabidopsis, the deposition of callose in dying meiotic products separates these cells from the functional megaspore (Webb and Gunning, 1990); however, the frequent formation of plasmodesmata connecting the functional megaspore to its adjacent nucellar cells indicates that cell-to-cell communication at the sporophytic–gametophytic transition is important during female gametophyte development (Bajon et al., 1999). In wild-type plants of Arabidopsis, several changes occur during the cytoplasmic maturation of the functional megaspore, including the polarized enlargement of the cell after the micropylar chalazal axis, the formation of a central vacuole, and the concomitant division of the nucleus (Webb and Gunning, 1990; Grossniklaus and Schneitz, 1998; Schneitz, 1999). No signs of cell enlargement or initial vacuolization were detected in arrested functional megaspores of AGP18-RNAi plants. In combination with results from expression analysis of molecular markers acting at specific stages of development, these observations indicate that the function of AGP18 is required after differentiation of the functional megaspore for the initiation of female gametogenesis.
Several interpretations of the functional activity of AGP18 during ovule development can be proposed based on the analysis of heterozygous AGP18-RNAi lines containing a single T-DNA insertion. Although T2-12 shows a 1:1 segregation ratio of BASTAr to BASTAs seedlings expected for defective traits causing gametophytic lethality, it also shows >70% of arrested female gametophytes, indicating that the gametophytic activity of RNAi-mediated silencing of AGP18 is not sufficient to explain the defective phenotype. As shown by in situ hybridization, AGP18 is expressed in the MMC and the adjacent nucellar cells; therefore, it is possible that, to ensure the initiation of female gametogenesis, the activity of AGP18 is required in both sporophytic as well as gametophytic cells. Differences on the degree of penetrance of the RNAi effect at the diploid and haploid levels could explain the variable but incomplete sterility shown by all AGP18-RNAi lines. Recent studies suggest that the maximal reduction of target transcript levels is obtained in RNAi lines containing a single T-DNA insertion (Kerschen et al., 2004). Although each target sequence is characterized by an inherent degree of susceptibility to RNAi-dependent silencing, a systematic study to assess the efficiency of posttranscriptional gene silencing in the gametophytic phase has not been conducted. Therefore, it is currently not possible to assess the effectiveness of AGP18 silencing in the female gametophyte. A second possibility is that key RNA factors generated by RNAi-mediated AGP18 silencing are produced at the diploid level and meiotically transmitted to a variable number of functional megaspores not carrying a T-DNA RNAi insertion; alternatively, these factors could be transported from adjacent nucellar cells to the functional megaspore via plasmodesmata. Although the transport of mRNA or proteins has not been reported in female meiotic products, detailed ultrastructural studies of megasporogenesis in Arabidopsis have shown that multiple plasmodesmata form between the functional megaspore and its adjacent nucellar cells (Bajon et al., 1999). Under this hypothesis, the function of AGP18 could be strictly gametophytic; however, the non-fully penetrant effect of RNAi-mediated silencing factors generated at the diploid level would be responsible for the abortion of female gametophytes at a frequency significantly higher than 50%. A third alternative includes the possibility that gametophytic lethality in T2-12 results from differences in the degree of RNAi silencing mediated by the CaMV35S promoter. A potential lack of CaMV35S activity during microsporogenesis could partially explain the absence of pollen abnormalities associated with the expression of AGP18 during anther development. To date, a detailed pattern of the CaMV35S promoter activity during male and female gametogenesis has yet to be reported in Arabidopsis. Although it is generally believed that this promoter is not active during the gametophytic phase (Bechtold et al., 2000), it is not clear at which developmental stage of the sporophytic to gametophytic transition its activity is no longer detected in either male or female gametes. Although this lack of quantitative information on the pattern of CaMV35S complicates the elucidation of the role played by AGP18 during the alternation of diploid and haploid phases, experiments showing that RNAi factors transmitted during meiosis can trigger posttranscriptional silencing in the female gametophyte support the hypoythesis that the CaMV35S promoter can be used to successfully target gametophytically expressed genes (our unpublished results). The use of specific promoters to drive RNAi-mediated silencing exclusively in the gametophyte or in the nucellus should lead to the elucidation of the specific role of gametophytic and sporophytic AGP18 activity during the initiation of female gametogenesis.
Classical AGPs play a role in mechanisms as distinct as cell division, cell expansion, or cell determination (Nothnagel, 1997; Schultz et al., 1998; van Hengel and Roberts, 2003). Monoclonal antibodies (MAbs) raised against AGP epitopes have been extensively used to investigate the cellular localization of AGPs (VandenBosch et al., 1989; Knox, 1992); however, these probes do not easily allow the elucidation of single AGP distribution patterns because they can recognize many different AGPs containing a conserved sugar epitope but different protein backbones. Elegant immunolocalization studies have shown that the establishment of a reproductive lineage in certain species was associated with changes in the distribution of AGP epitopes (Knox et al., 1989; Pennell et al., 1992). In Pisum sativum, the determination of reproductive cells in male and female gametes is associated with the loss of a cell surface arabinose-containing epitope recognized by the Mab MAC207 (Pennell and Roberts, 1990). Interestingly, a second AGP epitope recognized by MAb JIM8 became detectable only during differentiation of anthers and ovules in Brassica napus (Pennell et al., 1991). During ovule development, this epitope was initially detected in the wall of the two- to four-nucleate female gametophyte, in nucellar cells adjacent to the developing female gametophyte, and later in the plasma membrane of all cells forming the egg apparatus, but not the central cell. The presence of this epitope was also detected in the tapetum, in developing pollen grains, and in the stem vasculature (Pennell et al., 1991). Our results show that AGP18 is expressed in reproductive tissues, particularly in cell types that are involved in establishing the sporophytic to gametophytic transitions. During female gametophyte development, AGP18 is initially expressed in the MMC, in all four meiotically derived megaspores, the functional megaspore, and the adjacent nucellar cells. AGP18 expression persists in all female gametophyte cells except the central cell. During male gametophytic development, AGP18 is expressed in pollen grains and the cells of the tapetum. Developing seeds also express AGP18 during the first stages of embryogenesis; interestingly, abundant AGP18 mRNA is detected at early stages of endosperm development, indicating that transcription in the central cell occurs only after double fertilization. We only detected additional AGP18 expression in restricted clusters of companion cells present in the vasculature of the stem. The pattern of AGP18 mRNA localization is almost identical to the pattern of localization of the epitope recognized by JIM8, strongly suggesting that an AGP encoded by a gene homologous to AGP18 is detected by JIM8 in B. napus. The generation of AGP18-RNAi lines opens new possibilities for immunolocalization analysis with JIM8 and new antibodies raised against AGP18 to further elucidate the function and distribution of AGPs during reproductive development.
Our results indicate that AGP18 plays a crucial role during interactions between sporophytic and gametophytic cells in the young ovule and that these interactions are essential for the establishment of the female gametophytic phase in Arabidopsis. Although the nature of this communication has yet to be characterized at the genetic and molecular levels, the elucidation of the developmental function of a gene encoding a classical AGP creates new perspectives for the understanding of cell surface signaling and the molecular mechanisms that regulate sexual reproduction in flowering plants.
METHODS
Plant Material and Growth Conditions
The transposant line MET333 (Arabidopsis thaliana Heynh. var Landsberg erecta) was identified in a collection of enhancer detector lines generated in our laboratory, using the system implemented by Sundaresan et al. (1995), looking for expression patterns during female gametophyte development. To select plants carrying the Ds transposon, 50 mg/L of kanamycin and 0.66 μg/mL of 1-naphtalenacetamide were added to MS solid medium (Sigma, St. Louis, MO). Resistant seedlings were transferred to soil and grown in a greenhouse under long-day conditions.
Generation of RNAi Lines
A 740-bp fragment containing the first exon and a region of 5′ untranslated region of AGP18 was amplified by RT-PCR using RNA extracted from wild-type developing gynoecia using the following primers containing restriction sites as indicated in boldface: 5′-AATCTAGAGGCGCGCCACGGCTACATCTGTCTGT-3′ (XbaI and AscI; sense primer) and 5′-AAGGATCCATTTAAATAATGTACCTGATCGTCGG-3′ (BamHI and SwaI; antisense primer). The PCR fragment generated with the sense/antisense primer combination was cloned in pCRII TOPO (Invitrogen, Carlsbad, CA) and subsequently digested with SwaI and AscI restriction enzymes. The resulting DNA fragment was cloned using appropriate restriction sites in the silencing vector pFGC5941 (kindly donated by Carolyn Napoli and Rich Jorgensen, www.chromdb.org). The PCR fragment cloned in pCRII TOPO was digested with XbaI and BamHI (to obtain the antisense fragment) and cloned in appropriate restriction sites of pFGC5941. The resulting pFGC5941 vector contained AGP18 in both sense and antisense orientations separated by a chalcone synthase intron and under the control of the CaMV35S promoter. Four-week-old Arabidopsis plants (Columbia-0) were transformed by floral dipping as previously described (Clough and Bent, 1998). Seeds from Agrobacterium tumefaciens–treated plants were selected and directly grown under greenhouse long-day conditions (16 h light). Resistant seedlings were selected by spraying the herbicide BASTA (50 mg/L; Finale; AgrRvo, Montvale, NJ) three times each week for 2 weeks. Presence of AGP18-RNAi insertions was confirmed by PCR amplification on DNA extracted from seedlings. Seeds from mature plants were collected and plated onto MS medium supplemented with glufosinate ammonium (10 μg/mL; Crescent Chemical, Augsburg, Germany).
RNA Analysis
For RNA gel blots, total RNA was extracted from developing gynoecia from selected AGP18-RNAi transformants and wild-type plants using Trizol (Invitrogen) and following the manufacturer's instructions. Fifty micrograms of RNA were separated in a 1.3% agarose gel containing 17% formaldehyde and blotted onto hybond N+ membranes. Blots were hybridized with random-primed (Amersham Biosciences, Buckinghamshire, UK) 32P-labeled 810-bp probe corresponding to the complete AGP18 cDNA (At4g37450) and an ACT11 (At3g12110) probe generated by PCR using the following primers: ACT11-S (5′-TTCAACACTCCTGCCATG-3′) and ACT11-AS (5′-TGCAAGGTCCAAACGCAG-3′). The temperature of hybridization was 65°C in Church's buffer.
For small RNA analysis, total RNA from developing gynoecia was enriched for low molecular weight RNAs using Hamilton's homogenization solution (Hamilton and Baulcombe, 1999) as described in Mette et al. (2000). Low molecular weight RNA was normalized by spectrophotometry to 100 μg/lane, separated by electrophoresis through 15% polyacrylamide, 7 M urea, 0.5× Tris-borate EDTA gel, and transferred to Zeta-Probe GT membranes (Bio-Rad, Hercules, CA). After transfer, membranes were cross-linked with 200 mJ of UV and baked at 80°C for 1 h. To detect small RNAs in AGP18-RNAi lines, an AGP18 cDNA probe as described above was randomly labeled and hybridized at 62°C in Church's buffer. An oligonucleotide probe corresponding to the sequence of miR39 (5′- GATATTGGCGCGGCTCAAGCA-3′; Llave et al., 2002) was 5′-end labeled with [γ 32P]ATP and hybridized as a loading control. For RT-PCR analysis, total RNA was isolated from developing gynoecia of wild-type and AGP18-RNAi lines using Trizol following the manufacturer's instructions. First-strand cDNA was synthesized using 2 μg of total RNA and Superscript II reverse transcriptase (Invitrogen). For semiquantitative RT-PCR, 3 μL of the first-strand cDNA reaction served as a template in a PCR reaction that used specific primers for AGP18 (At4g37450), AGP17 (At2g23130), AGP19 (At1g68725), and ACT11. After estimating the amount of amplified DNA produced at different rounds of PCR cycles, we determined that 20 cycles ensured that the amplified product was proportional to the initial concentration of template present in the reaction. After electrophoresis on a 1% agarose gel and blotting into hybond N+ membranes, hybridization was performed with 32P cDNA probes specific to each AGP gene tested and labeled with the random primer method (Amersham Biosciences). Hybridization was performed at 62°C as described (Maniatis et al., 1989). AGP17 primers were as follows: sense (5′-GCTTTTAAGCCCGCCTGCTCC-3′) and antisense (5′-CTGAATACAAAATGTGAGCTG-3′). AGP19 primers were as follows: sense (5′-AAGTTGCACCAGTAATCAGCC-3′) and antisense (5′-TCCTTTAAGCTGATTTAAGGC-3′). AGP18 primers were as follows: sense (5′-CACGCTTGGTTAAACTCC-3′) and antisense (5′-TTTTTTCATCACTGACAG-3′).
Whole-Mount Preparations and Histological Analysis
Wild-type and AGP18-RNAi siliques were dissected longitudinally with hypodermic needles (1-mL insulin syringes) and fixed with FAA buffer (50% ethanol, 5% acetic acid, and 10% formaldehyde), dehydrated in increasing ethanol concentration, cleared in Herr's solution (phenol:chloral hydrate:85% lactic acid:xylene:oil of clove [1:1:1:0.5:1]), and observed on a Leica microscope (Wetzlar, Germany) under Nomarski optics. GUS staining assays for stages before fertilization were conducted as described by Vielle-Calzada et al. (2000). For developmental stages after fertilization, we used the protocol described by Köhler et al. (2003) with slight modifications. Longitudinally dissected siliques were fixed for 2 h at −20°C in 90% acetone and subsequently immersed in GUS staining buffer (10 mM EDTA, 0.1% Triton, 0.5 mM Fe2+CN, 0.5 mM Fe3+CN, 100 μg m−1 chloranphenicol, and 1 mg mL−1 5-bromo-4-chloro-3-indolyl-β-d-galactoside in 50 mM sodium phosphate buffer, pH 7.0) for 1 to 3 d at 37°C. The tissue was cleared in Hoyer's solution and observed under Nomarski optics.
In Situ Hybridization
Developing flower buds, developing flowers, isolated gynoecia, and siliques of wild-type and AGP18-RNAi T2-12 plants were fixed in 4% paraformaldehyde and embedded in Paraplast (Fisher Scientific, Fair Lawn, NJ). Sections of 12-μm thickness were cut using a Leica microtome and mounted on ProbeOnPlus slides (Fischer Biotech, Pittsburgh, PA). A fragment of 180 bp that included a portion of the first exon of AGP18 was amplified using sense (5′-CGACGATCAGGTACATTAG-3′) and the antisense (5′-CATCACTGACAGATATGAA-3′) primers and subsequently cloned in the pCRII TOPO vector (Invitrogen). The resulting construct was digested with NotI and BamHI to synthesize sense and antisense digoxygenin-labeled probes, respectively, and hybridization was conducted as described by Vielle-Calzada et al. (1999).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number NM119909 (At4g37450).
Acknowledgments
We thank Carolyn Napoli, Richard Jorgensen, and Ueli Grossniklaus for sharing materials before publication. We also thank Plinio Guzmán, Luis Herrera-Estrella, and Daphné Autran for critically reading the manuscript. We are grateful to past and current members of our laboratory for their help generating the Mexican MET/MGT transposon collection. Marcelina García-Aguilar and Andrés Estrada-Luna provided assistance with histological analysis; Santos García-Aguilar helped with plant handling. This work was supported by the Consejo Nacional de Ciencia y Tecnología (B-34324 and Z-029) and the Howard Hughes Medical Institute. G.A.G. is the recipient of a graduate scholarship from the Consejo Nacional de Ciencia y Tecnología. J.-P.V.C. is an International Scholar of the Howard Hughes Medical Institute.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jean-Philippe Vielle-Calzada (vielle@ira.cinvestav.mx).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.024588.
References
- Bajon, C., Horlow, C., Motamayor, J.C., Sauvanet, A., and Robert, D. (1999). Megasporogenesis in Arabidopsis thaliana L.: An ultrastructural study. Sex. Plant Reprod. 12, 99–109. [Google Scholar]
- Bechtold, N., Jaudeau, B., Jolivet, S., Maba, B., Vezon, D., Voisin, R., and Pelletier, G. (2000). The maternal chromosome set is the target of the T-DNA in the in planta transformation of Arabidopsis thaliana. Genetics 155, 1875–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, P.R. (1989). The alternation of generations. Adv. Bot. Res. 16, 55–93. [Google Scholar]
- Bell, P.R. (1995). Incompatibility in flowering plants: Adaptation of an ancient response. Plant Cell 7, 5–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen, H.J. (1999). Ten years of enhancer detection: Lessons from the fly. Plant Cell 11, 2271–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner, G.H.H., Sherrier, D.J., Stevens, T.J., Arkin, I.T., and Dupree, P. (2002). Prediction of glycosylphophatidylinositol-anchored proteins in Arabidopsis. A genomic analysis. Plant Physiol. 129, 486–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casero, P.J., Casimiro, I., and Knox, J.P. (1998). Occurrence of cell surface arabinogalactan-protein and extensin epitopes in relation to pericycle and vascular tissue development in the root apex of four species. Planta 204, 252–259. [Google Scholar]
- Chen, C.G., Pu, Z.Y., Moritz, R.L., Simpson, R.J., Bacic, A., Clarke, A.E., and Mau, S.L. (1994). Molecular cloning of a gene encoding for an arabinogalactan-protein from pear (Pyrus communis) cell suspension culture. Proc. Natl. Acad. Sci. USA 91, 305–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, A.Y., Wang, H., and Wu, H.M. (1995). A floral transmitting tissue-specific glycoprotein attracts pollen tubes and stimulates their growth. Cell 82, 383–393. [DOI] [PubMed] [Google Scholar]
- Christensen, C.A., Subramanian, S., and Drews, G.N. (1998). Identification of gametophytic mutations affecting female gametophyte development in Arabidopsis. Dev. Biol. 202, 136–151. [DOI] [PubMed] [Google Scholar]
- Chuang, C.F., and Meyerowitz, E. (2000). Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97, 4985–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, A.E., Anderson, R.L., and Stone, B.A. (1979). Form and function of arabinogalactans and arabinogalactan proteins. Phytochemistry 18, 521–540. [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Dickinson, H.G. (1994). The regulation of alternation of generation in flowering plants. Biol. Rev. 69, 419–422. [Google Scholar]
- Drews, G.N., and Yadegari, R. (2002). Development and function of the angiosperm female gametophyte. Annu. Rev. Genet. 36, 99–124. [DOI] [PubMed] [Google Scholar]
- Du, H., Simpson, R.J., Moritz, R.L., Clarke, A.E., and Bacic, A. (1994). Isolation of the protein backbone of an arabinogalactan-protein from the styles of Nicotiana alata and characterization of a corresponding cDNA. Plant Cell 6, 1643–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Lendeckel, W., and Tuschl, T. (2000). RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15, 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher, G.B., Sawyer, W.H., and Stone, B.A. (1974). Chemical and physical properties of an arabinogalactan-peptide from wheat endosperm. Biochem. J. 139, 535–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar, Y., Johnson, K., McKenna, J.A., Bacic, A., and Schultz, C. (2001). The complex structures of arabinogalactan-proteins and the journey towards understanding function. Plant Mol. Biol. 47, 161–176. [PubMed] [Google Scholar]
- Gilson, P., Gaspar, Y., Oxley, D., Youl, J.J., and Bacic, A. (2001). NaAGP4 is an arabinogalactan protein whose expression is suppressed by wounding and fungal infection in Nicotiana alata. Protoplasma 215, 128–139. [DOI] [PubMed] [Google Scholar]
- Grini, P.E., Schnittger, A., Schwarz, H., Zimmermann, I., Schwab, B., Jurgens, G., and Hulskamp, M. (1999). Isolation of ethyl methanesulfonate-induced gametophytic mutants in Arabidopsis thaliana by a segregation distortion assay using the multimarker chromosome 1. Genetics 151, 849–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus, U., and Schneitz, K. (1998). The molecular and genetic basis of ovule and megagametophyte development. Semin. Cell Dev. Biol. 9, 227–238. [DOI] [PubMed] [Google Scholar]
- Hamilton, A.J., and Baulcombe, D.C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286, 950–952. [DOI] [PubMed] [Google Scholar]
- Howden, R., Park, S.K., Moore, J.M., Orme, J., Grossniklaus, U., and Twell, D. (1998). Selection of T-DNA-tagged male and female gametophytic mutants by segregation distortion in Arabidopsis. Genetics 149, 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, B.Q., and Russell, S.D. (1992). Female germ unit: Organization, isolation and function. Int. Rev. Cytol. 140, 233–293. [Google Scholar]
- Jauh, G.Y., and Lord, E.M. (1996). Localization of pectins and arabinogalactan-proteins in lily (Lilium longiflorum L.) pollen tube and style, and their possible roles in pollination. Planta 199, 251–261. [Google Scholar]
- Kerschen, A., Napoli, C.A., Jorgensen, R.A., and Müller, A.E. (2004). Effectiveness of RNA interference in transgenic plants. FEBS Lett. 566, 223–228. [DOI] [PubMed] [Google Scholar]
- Klimyuk, V.I., and Jones, J.D.G. (1997). AtDMC1, the Arabidopsis homologue of the yeast DMC1 gene: Characterization, transposon-induced allelic variation and meiosis-associated expression. Plant J. 11, 1–14. [DOI] [PubMed] [Google Scholar]
- Knox, J.P. (1992). Cell adhesion, cell separation and plant morphogenesis. Plant J. 2, 137–141. [Google Scholar]
- Knox, J.P. (1999). Intriguing, complex and everywhere: Getting to grips with arabinogalactan-proteins. Trends Plant Sci. 4, 123–125. [Google Scholar]
- Knox, J.P., Day, S., and Roberts, K. (1989). A set of cell surface glycoproteins forms an early marker of cell position, but not cell type, in the root apical meristem of Daucus carota L. Development 106, 47–56. [Google Scholar]
- Knox, R.B. (1967). Apomixis. Seasonal and population differences in a grass. Science 157, 325–326. [DOI] [PubMed] [Google Scholar]
- Köhler, C., Hennig, L., Spillane, C., Pien, S., Gruissem, W., and Grossniklaus, U. (2003). The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev. 17, 1540–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuger, M., and van Holst, G.J. (1993). Arabinogalactan proteins are essential in somatic embryogenesis of Daucus carota L. Planta 189, 243–248. [Google Scholar]
- Kreuger, M., and van Holst, G.J. (1996). Arabinogalactan proteins and plant differentiation. Plant Mol. Biol. 30, 1077–1086. [DOI] [PubMed] [Google Scholar]
- Kwee, H.S., and Sundaresan, V. (2003). The NOMEGA gene required for female gametophyte development encodes the putative APC6/CDC16 component of the Anaphase Promoting Complex in Arabidopsis. Plant J. 36, 853–866. [DOI] [PubMed] [Google Scholar]
- Llave, C., Xie, Z., Kasschau, D.K., and Carrington, J.C. (2002). Cleavage of Scarecrow-like mRNA target directed by a class of Arabidopsis miRNA. Science 297, 2053–2056. [DOI] [PubMed] [Google Scholar]
- Liu, Y.G., Mitsukawa, N., Oosumi, T., and Whittier, R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8, 457–463. [DOI] [PubMed] [Google Scholar]
- Maheshwari, P. (1950). An Introduction to the Embryology of Angiosperms. (New York: McGraw-Hill).
- Majewska-Sawka, A., and Nothnagel, E.A. (2000). The multiple roles of Arabinogalactan proteins in plant development. Plant Physiol. 122, 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis, T., Fritsch, T., and Sambrook, J. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 7.37–7.39.
- Mau, S.L., Chen, C.G., Pu, Z.Y., Moritz, R.L., Simpson, R.J., Bacic, A., and Clarke, A.E. (1995). Molecular cloning of cDNAs encoding the protein backbones of arabinogalactan proteins from the filtrate of suspension-cultured cells of Pyrus communis and Nicotiana alata. Plant J. 8, 269–281. [DOI] [PubMed] [Google Scholar]
- McCabe, P.F., Valentine, T.A., Forsberg, L.S., and Pennell, R.I. (1997). Soluble signals from cells identified at the cell wall establish a developmental pathway in carrot. Plant Cell 9, 2225–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mette, M.F., Aufsatz, W., van der Winden, J., Matzke, M.A., and Matzke, A.J.M. (2000). Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 19, 5194–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, J.M., Vielle-Calzada, J.P., Gagliano, W., and Grossniklaus, U. (1997). Genetic characterization of hadad, a mutant disrupting female gametogenesis in Arabidopsis thaliana. Cold Spring Harb. Symp. Quant. Biol. 62, 35–47. [PubMed] [Google Scholar]
- Motose, H., Suriyama, M., and Fukuda, H. (2004). A proteoglycan mediates inductive interaction during plant vascular development. Nature 429, 873–878. [DOI] [PubMed] [Google Scholar]
- Nam, J., Mysore, K.S., Zheng, C., Knue, M.K., Matthysse, A.G., and Gelvin, S.B. (1999). Identification of T-DNA tagged Arabidopsis mutants that are resistant to transformation by Agrobacterium. Mol. Gen. Genet. 261, 429–438. [DOI] [PubMed] [Google Scholar]
- Nothnagel, E.A. (1997). Proteoglycans and related components in the plant cells. Int. Rev. Cytol. 174, 195–291. [DOI] [PubMed] [Google Scholar]
- Pennell, R.I., Janniche, L., Kjellbom, P., Scofield, G.N., Peart, J.M., and Roberts, K. (1991). Developmental regulation of a plasma membrane arabinogalactan protein epitope in oilseed rape flowers. Plant Cell 3, 1317–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell, R.I., Janniche, L., Scofield, G.N., Booij, H., de Vries, S.C., and Roberts, K. (1992). Identification of a transitional cell state in the developmental pathway to carrot somatic embryogenesis. J. Cell Biol. 119, 1371–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell, R.I., and Roberts, K. (1990). Sexual development in the pea is presaged by altered expression of arabinogalactan protein. Nature 344, 547–549. [Google Scholar]
- Pischke, M.S., Jones, L.G., Otsuga, D., Fernandez, D.E., Drews, G.N., and Sussman, M.R. (2002). An Arabidopsis histidine kinase is essential for megagametogenesis. Proc. Natl. Acad. Sci. USA 26, 15800–15805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, T.V., Kaur, J., Agashe, B., Sundaresan, V., and Siddiqi, I. (2003). The DUET gene is necessary for chromosome organization and progression during male meiosis in Arabidopsis and encodes a PHD finger protein. Development 130, 5975–5987. [DOI] [PubMed] [Google Scholar]
- Reiser, L., and Fischer, R.L. (1993). The ovule and the embryo sac. Plant Cell 5, 1291–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, S., Jauh, G.Y., Hepler, P.K., and Lord, E.M. (1998). Effects of Yariv phenylglycoside on cell wall assembly in the lily pollen tube. Planta 204, 450–458. [DOI] [PubMed] [Google Scholar]
- Schiefthaler, U., Balasubramanian, S., Sieber, P., Chevalier, D., Wisman, E., and Schneitz, K. (1999). Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96, 11664–11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelman, G., Morikami, A., Jung, J., Baskin, T.I., Carpita, N.C., Derbyshire, P., McCann, M.C., and Benfey, P.N. (2001). COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev. 15, 1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneitz, K. (1999). The molecular and genetic control of ovule development. Curr. Opin. Plant Biol. 2, 13–17. [DOI] [PubMed] [Google Scholar]
- Schneitz, K., Hülskamp, M., Kopczak, S.D., and Pruitt, R.E. (1997). Dissection of sexual organ ontogenesis: A genetic analysis of ovule development in Arabidopsis thaliana. Development 124, 1367–1376. [DOI] [PubMed] [Google Scholar]
- Schultz, C., Gilson, P., Oxley, D., Youl, J., and Bacic, A. (1998). GPI-anchors on arabinogalactan-proteins: Implications for signaling in plants. Trends Plant Sci. 3, 426–431. [Google Scholar]
- Schultz, C., Johnson, K.L., Currie, G., and Bacic, A. (2000). The classical arabinogalactan protein gene family of Arabidopsis. Plant Cell 12, 1751–1767.11006345 [Google Scholar]
- Schultz, C.J., Rumsewicz, M.P., Johnson, K.L., Jones, B.J., Gaspar, Y.M., and Bacic, A. (2002). Using genomic resources to guide research directions. The arabinogalactan protein gene family as a test case. Plant Physiol. 129, 1448–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serpe, M.D., and Nothnagel, E.A. (1994). Effects of Yariv phenylglycosides on Rosa cell suspensions: Evidence for the involvement of arabinogalactan-proteins in cell proliferation. Planta 193, 542–550. [Google Scholar]
- Sharma, P., Varma, R., Sarasij, R.C., Gousset, K., Khrihnamoorthy, G., Rao, M., and Mayor, S. (2004). Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell 116, 577–589. [DOI] [PubMed] [Google Scholar]
- Sherrier, D.J., Prime, T.A., and Dupree, P. (1999). Glycosylphosphatidylinositol-anchored cell-surface proteins from Arabidopsis. Electrophoresis 20, 2027–2035. [DOI] [PubMed] [Google Scholar]
- Showalter, A.M. (2001). Arabinogalactan-proteins: Structure, expression, and function. Cell. Mol. Life Sci. 58, 1399–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi, I., Ganesh, G., Grossniklaus, U., and Subbiah, V. (2000). The dyad gene is required for progression through female meiosis in Arabidopsis. Development 127, 197–207. [DOI] [PubMed] [Google Scholar]
- Smith, N.A., Singh, S.P., Wang, M.B., Stoutjesdijk, P.A., Green, A.G., and Waterhouse, P.M. (2000). Total silencing by intron-spliced hairpin RNAs. Nature 407, 319–320. [DOI] [PubMed] [Google Scholar]
- Sommer-Knudsen, J., Bacic, A., and Clarke, A.E. (1998). Hidroxyproline-rich plant glycoproteins. Phytochemistry 47, 483–497. [Google Scholar]
- Springer, P.S. (2000). Gene traps: Tools for plant development and genomics. Plant Cell 12, 1007–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer, P.S., McCombie, W.R., Sundaresan, V., and Martienssen, R.A. (1995). Gene trap tagging of PROLIFERA, an essential MCM2-3-5-like gene in Arabidopsis. Science 268, 877–880. [DOI] [PubMed] [Google Scholar]
- Sun, W., Zhao, Z.D., Hare, M.C., Kieliszewski, M.J., and Showalter, A.M. (2004). Tomato LeAGP-1 is a plasma membrane-bound, glycosylphosphatidylinositol-anchored arabinogalactan-protein. Physiol. Plant. 120, 319–327. [DOI] [PubMed] [Google Scholar]
- Sundaresan, V., Springer, P., Volpe, T., Haward, S., Jones, J.D., Dean, C., Ma, H., and Martienssen, R. (1995). Patterns of the gene action in plant development is revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9, 1797–1810. [DOI] [PubMed] [Google Scholar]
- Svetek, J., Yadav, M.P., and Nothnagel, E.A. (1999). Presence of a glycosylphosphatidylinositol lipid anchor on rose arabinogalactan proteins. J. Biol. Chem. 274, 14724–14733. [DOI] [PubMed] [Google Scholar]
- Takos, A.D., Dry, I.B., and Soole, K.L. (1997). Detection of glycosyl-phosphatidylinositol-anchored proteins on the surface of Nicotiana tabacum protoplasts. FEBS Lett. 405, 1–4. [DOI] [PubMed] [Google Scholar]
- Takos, A.M., Dry, I.B., and Soole, K.L. (2000). Glycosyl-phosphatidylinositol-anchor addition signals are processed in Nicotiana tabacum. Plant J. 21, 43–52. [DOI] [PubMed] [Google Scholar]
- Tan, L., Leykam, J.F., and Kieliszewski, M.J. (2003). Glycosylation motifs that direct arabinogalactan addition to arabinogalactan-proteins. Plant Physiol. 3, 1362–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandenBosch, K.A., Bradley, D.J., Perotto, S., Butcher, G.W., and Brewin, J.J. (1989). Common components of the infection thread matrix and the intercellular space identified by immunocytochemical analysis of pea nodules and uninfected roots. EMBO J. 8, 335–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hengel, A.J., and Roberts, K. (2002). Fucosylated arabinogalactan-proteins are required for full root cell elongation in Arabidopsis. Plant J. 32, 105–113. [DOI] [PubMed] [Google Scholar]
- van Hengel, A.J., and Roberts, K. (2003). AtAGP30, an arabinogalactan-protein in the cell walls of the primary root, plays a role in the root generation and seed germination. Plant J. 36, 256–270. [DOI] [PubMed] [Google Scholar]
- van Hengel, A.J., Tadesse, Z., Immerzeel, P., Schols, H., van Kammen, A., and de Vries, S.C. (2001). N-acetylglusamine and glucosamine-containing arabinogalactan proteins control somatic embryogenesis. Plant Physiol. 125, 1880–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielle-Calzada, J.P., Baskar, R., and Grossniklaus, U. (2000). Delayed activation of the paternal genome during seed development. Nature 404, 91–94. [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada, J.P., Thomas, J., Spillane, C., Coluccio, A., Hoeppner, M.A., and Grossniklaus, U. (1999). Maintenance of genomic imprinting at the Arabidopsis medea locus requires zygotic DDM1 activity. Genes Dev. 13, 2971–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse, P.M., Graham, M.W., and Wang, M.B. (1998). Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc. Natl. Acad. Sci. USA 95, 13959–13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb, M.C., and Gunning, E.S. (1990). Embryo sac development in Arabidopsis thaliana. I. Megasporogenesis, including the microtubular cytoskeleton. Sex. Plant Reprod. 3, 244–256. [Google Scholar]
- Willats, W.G.T., and Knox, J.P. (1996). A role for arabinogalactan-proteins in plant cell expansion: Evidence from studies on the interaction of β-glucosyl Yariv reagent with seedlings of Arabidopsis thaliana. Plant J. 9, 919–925. [DOI] [PubMed] [Google Scholar]
- Willemse, M.T.M., and van Went, J.L. (1984). The female gametophyte. In Embryology of Angiosperms, B.H. Johri, ed (New York: Springer-Verlag), pp. 159–196.
- Wu, H., Wang, H., and Cheung, A.Y. (1995). A pollen tube growth simulator glycoprotein is deglycosylated by pollen tubes and displays a glycosylation gradient in the flower. Cell 82, 395–403. [DOI] [PubMed] [Google Scholar]
- Yang, W.C., and Sundaresan, V. (2000). Genetics of gametophyte biogenesis in Arabidopsis. Curr. Opin. Plant Biol. 3, 53–57. [DOI] [PubMed] [Google Scholar]
- Yang, W.C., Ye, D., Xu, J., and Sundaresan, V. (1999). The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 15, 2108–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youl, J.J., Bacic, A., and Oxley, O. (1998). Arabinogalactan-proteins from Nicotiana alata and Pyrus communis contain glycosylphophatidylinositol membrane anchors. Proc. Natl. Acad. Sci. USA 95, 7921–7926. [DOI] [PMC free article] [PubMed] [Google Scholar]