Abstract
Plant photoreceptor phytochromes are phosphoproteins, but the question as to the functional role of phytochrome phosphorylation has remained to be elucidated. We investigated the functional role of phytochrome phosphorylation in plant light signaling using a Pfr-specific phosphorylation site mutant, Ser598Ala of oat (Avena sativa) phytochrome A (phyA). The transgenic Arabidopsis thaliana (phyA-201 background) plants with this mutant phyA showed hypersensitivity to light, suggesting that phytochrome phosphorylation at Serine-598 (Ser598) in the hinge region is involved in an inhibitory mechanism. The phosphorylation at Ser598 prevented its interaction with putative signal transducers, Nucleoside Diphosphate Kinase-2 and Phytochrome-Interacting Factor-3. These results suggest that phosphorylation in the hinge region of phytochromes serves as a signal-modulating site through the protein–protein interaction between phytochrome and its putative signal transducer proteins.
INTRODUCTION
Phytochromes are molecular light switches that regulate various aspects of plant growth and development (Kendrick and Kronenberg, 1994; Quail et al., 1995; Fankhauser, 2000; Smith, 2000). They are plant red/far-red photoreceptors that exist as dimeric photochromic proteins with covalently linked tetrapyrrole chromophore phytochromobilin. Of the two photointerconvertible species, Pr and Pfr forms, the latter is considered the active form of phytochrome because of the promotive effect of red light on most physiological responses. Although phytochromes are the first discovered photoreceptors and have been studied for several decades, their molecular mechanism and the downstream partner molecules through which phytochromes transmit a light signal were recently emerged. Yeast two-hybrid screens have revealed several phytochrome-interacting proteins (PIPs) as potential primary signaling partners, including Phytochrome-Interacting Factor-3 (PIF3; Ni et al., 1998) and Nucleoside Diphosphate Kinase-2 (NDPK2; Choi et al., 1999). PIF3 is a basic helix-loop helix transcription factor that exhibits phytochrome-mediated, light-dependent binding to promoter regions of various light-activated genes (Martínez-Garcia et al., 2000). NDPK2 is an enzyme that is activated by the Pfr form of phytochromes and appears to play a role in the cotyledon unfolding and greening response initiated by phytochromes (Choi et al., 1999). Although the discovery of these direct signaling partners of phytochromes is helpful for understanding of the phytochrome-mediated light signaling in plants, further studies are necessary to explain how these putative signal transducers and their interaction with phytochromes trigger the downstream signal transduction cascades in plant light signaling.
Posttranslational modification is important for the modulation of many signal transductions (e.g., rhodopsin is desensitized by phosphorylation by rhodopsin kinase) (Sokal et al., 2002). Phytochrome has been known as a phosphoprotein because it could be readily labeled with 32P isotope in vivo (Quail et al., 1978). Several observations and indirect lines of evidence for the possible role of protein phosphorylation downstream of the phytochrome-mediated light signal transduction pathway have been discussed (Singh and Song, 1990; Elich and Chory, 1997a; Fankhauser and Chory, 1999; Watson, 2000; Sharma, 2001). For example, mutation of N-terminal Ser to Ala results in an increased biological activity of phytochrome A (phyA), suggesting that phytochrome responses might be desensitized by this photoreceptor phosphorylation (Stockhaus et al., 1992; Jordan et al., 1997). However, the in vivo functional role of phytochrome phosphorylation is still unknown.
The sites of phytochrome phosphorylation in vivo and in vitro have been identified with oat (Avena sativa) phyA (McMichael and Lagarias, 1990; Lapko et al., 1996, 1997, 1999). There are two Ser sites, Serine-7 (Ser7) and Serine-598 (Ser598), that are phosphorylated in vivo. Phosphorylation at Ser7 in the N-terminal extension region (NTE) is similar in both Pr and Pfr forms, whereas Ser598 in the hinge region is phosphorylated in a Pfr preferential manner (Lapko et al., 1997, 1999). The in vitro phosphorylation sites of oat phyA have also been identified using protein kinase A (PKA). Two Ser, Serine-17 (Ser17) and Ser598, are detected as the in vitro phosphorylation sites by PKA. Ser17 is phosphorylated primarily in a Pr preferential manner, whereas Ser598 phosphorylation is preferred in the Pfr form (McMichael and Lagarias, 1990; Lapko et al., 1996). Because this Ser598 residue is phosphorylated only in the Pfr form that is considered the active form of phytochrome, it was suggested that the phosphorylation and dephosphorylation of Ser598 serves as a switch in phytochrome signaling (Park et al., 2000; Kim et al., 2002b).
Here, we demonstrate that phytochrome phosphorylation at Ser598 in the hinge region controls the interaction of phytochrome with its putative signal transducers, providing the functional role of a phytochrome phosphorylation site. The phosphorylation at Ser598 of oat phyA inhibits the interaction between phytochrome and its putative signal transducers, NDPK2 and PIF3. The transgenic plants with Ser598Ala mutant phyA are hypersensitive to light compared with the transgenic plants with wild-type oat phyA, suggesting that phytochrome phosphorylation at Ser598 plays an inhibitory role. Our findings suggest that the phosphorylation and dephosphorylation of phytochrome be a key regulatory mechanism in its light-signaling pathway.
RESULTS
Ser598Ala Mutant Transgenic Plants Show Hypersensitivity to Light
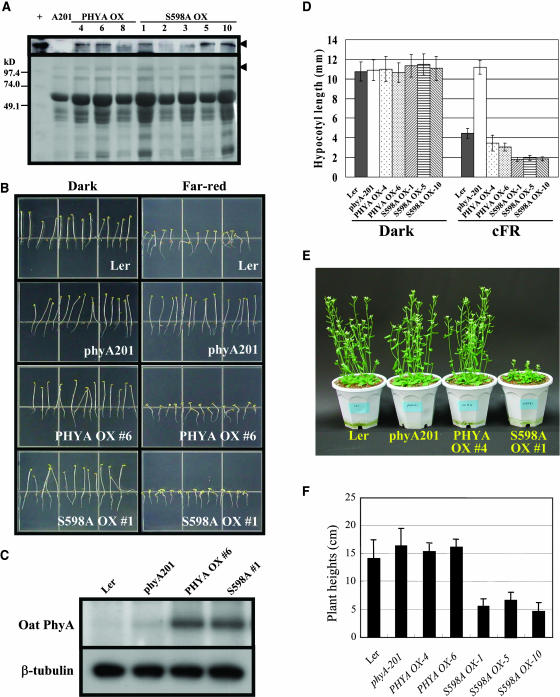
Ser598 of oat phyA is phosphorylated in a Pfr preferential manner both in vivo and in vitro (Lapko et al., 1996, 1999). Because oat phyA is physiologically active in transgenic plants, including Arabidopsis thaliana (Boylan and Quail, 1991; Boylan et al., 1994), in vivo functional assays of oat phyA mutants are possible by transforming the oat phyA mutant genes into phyA-deficient Arabidopsis (phyA-201, Landsberg erecta [Ler] ecotype). To elucidate the functional importance of phytochrome phosphorylation in plant light signaling, we mutagenized Ser598 to Ala to remove the Pfr-specific phosphorylation site and introduced oat wild-type phyA and Ser598Ala (S598A) mutant genes into phyA-201 for the functional analysis. We produced their T3 transgenic lines, and the expression levels of wild-type phyA and S598A genes and their proteins were confirmed in transgenic plants by RT-PCR (data not shown) and protein gel blot analysis, respectively. Protein gel blot analysis showed that the phyA expression levels in the oat phyA and S598A mutant transgenics were similar (Figures 1A and 1C). The transgenic lines were then chosen for further study, as illustrated with oat phyA-overexpressing lines (phyA OX) numbers 4 and 6 and S598A-overexpressing lines (S598A OX) numbers 1, 5, and 10 (Figure 1A).
Figure 1.
Analysis of Transgenic Arabidopsis (A-201) with Oat phyA or S598A Mutant.
(A) Protein expression confirmed by protein gel blot analysis. Leaves from light-grown plants were used for the protein extraction, and 50 μg of the extracts were loaded onto SDS-PAGE (see Methods). For the detection of oat phyA, a specific antibody oat22 was used for the protein gel blot analysis. Lane +, oat phyA (positive control); lane A201, protein sample from phyA-201 (negative control). The number represents independent transgenic seed lines of oat phyA OX and S598A OX lines. The transgenic lines that showed similar oat phyA protein levels were chosen for further study, as illustrated with oat phyA OX numbers 4 and 6 and S598A OX numbers 1, 5, and 10. The arrowheads show the protein band of phyA.
(B) Representative plant seedlings under light conditions. Ler, wild-type Arabidopsis; phyA201, phyA deficient Arabidopsis; S598A, number 1 S598A OX line; phyA, number 6 oat phyA OX line. The seeds were grown under far-red light (7 μmol/m2/s) or in darkness for 4 d (also see Figure 1D).
(C) Protein gel blot analyses of phytochromes prepared from seedlings in (B). Oat phyA specific antibody oat22 and β-tubulin specific antibody were used for this analysis. No oat phyA was detected in Ler and phyA-201 plants. Transgenic seedlings of wild-type oat phyA and S598A phyA showed similar level of oat phyA expression.
(D) The average hypocotyl lengths of seedlings. Each measurement was done with at least 30 seedlings
(E) Representative mature transgenic plants under long day condition for 5 weeks. Representative plants of Ler, phyA201, phyA OX (no. 4) and S598A OX (no. 1) were grown under white light condition (1.1 mW/cm2). The S598A transgenic plants show reduced inflorescence lengths (see also Figure 1F).
(F) The average heights of mature plants. Each measurement was done with at least 12 plants.
To examine the complementation of phyA function under phyA null background, the seedlings of T3 transgenic plants were grown under continuous far-red light (FRc) or in darkness, and the hypocotyl lengths were measured (Figures 1B and 1D). The wild-type Arabidopsis (Ler) seedlings showed typical FR high irradiance responses (shortening of hypocotyl elongation), but the phyA deficient Arabidopsis (phyA201) seedlings were insensitive to FRc light, exhibiting etiolated phenotypes under FR light (Whitelam et al., 1993). As previously reported (Boylan and Quail, 1991), wild-type oat phyA was functionally active in Arabidopsis, complementing the phyA deficiency and showing slightly shorter hypocotyls than Ler seedlings. The transgenic lines expressing the S598A mutant also complemented the FR high irradiance response of phyA (Figure 1B). Significantly, the S598A mutant transgenic seedlings produced shorter hypocotyl lengths than the oat phyA transgenic seedlings (Figure 1D), suggesting that the transgenic lines of this mutant are hypersensitive to light compared with those of wild-type oat phyA. The phenotypic contrast was even more obvious in the mature S598A transgenic plants (Figure 1E). The mature S598A transgenic plants showed reduced inflorescence lengths. The average heights of S598A phyA transgenic plants (S598A OX lines, nos. 1, 5, and 10) were found to be three to four times shorter than those of oat phyA transgenic plants (oat phyA OX lines, nos. 4 and 6) (Figure 1F). These results imply that phosphorylation at Ser598 plays a signal-attenuating role in the phytochrome-mediated light signal transduction.
Role of Phytochrome Phosphorylation in Light Signaling
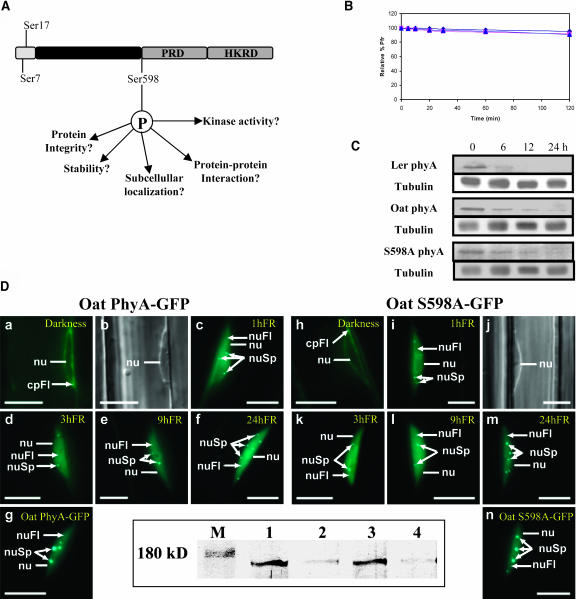
Because the protein expression levels of wild-type and S598A mutant phyA were similar in transgenic seedlings and mature plants, we tested several hypotheses on the phosphorylation of phytochromes and its functional role (Fankhauser, 2000) to understand the hypersensitivity of the S598A mutant transgenic plants to light (Figure 2A): (1) the photochemical and conformational changes by phosphorylation, (2) effects on phytochrome degradation, (3) dependence of nuclear localization of phyA on phosphorylation, (4) the autophosphorylation and kinase site of phyA, whether or not Ser598 is involved in phytochrome autophosphorylation and its kinase activity, and (5) effects of phytochrome phosphorylation on the protein–protein interaction between phyA and the phytochrome-interacting signal transducer proteins.
Figure 2.
Hypothesis and Examination for the Hypersensitivity to Light.
(A) Hypothesis for the role of Ser598 phosphorylation in phytochrome signaling.
(B) Dark reversion of phosphorylated phyA. Pr (square) and Pfr (triangle) forms of phyA were phosphorylated by PKA and compared with wild-type phyA (diamond). There was no difference in dark reversion among phosphorylated phyA and wild-type (unphosphorylated) phyA.
(C) In vivo phyA degradation. The 3.5-d-old seedlings were illuminated with red light (10 μmol/m2/s) and sampled at 0, 6, 12, and 24 h, and protein gel blot analyses were performed. β-Tubulin was used for a control. The degradation of oat wild-type oat phyA and S598A phyA is slower than Arabidopsis phyA, but there is no difference between wild-type oat phyA and S598A phyA. β-Tubulin was used for a control.
(D) Nuclear localization of oat phyA and S598A phyA. Representative nuclei of transgenic seedlings expressing oat phyA-GFP (left) and oat S598A-GFP (right) are shown. The seedlings were kept in darkness (a, b, and h) or transferred to FRc for 1 h (c, i, and j), 3 h (d and k), 9 h (e and l), or 24 h (f, g, m, and n). The subcellular localization of the GFP fusion proteins was investigated by fluorescence microscopy (a, c, d, e, f, g, h, i, k, l, m, and n). Differential interference contrast images (b and j) are included (a and b and i and j show identical nuclei). Nuclei (nu), nuclear fluorescence (nuFl), nuclear spots (nuSp), and cytoplasmic fluorescence (cpFl) are indicated. Bars = 10 μm. Protein gel blot analysis (below) for phyA-GFP and S598A-GFP protein expressions in the transgenic seedlings used for fluorescence microscopy was performed as follows. Crude protein extracts were prepared from transgenic seedlings expressing oat phyA-GFP (lanes 1 and 2) and S598A-GFP (lanes 3 and 4). Before protein extraction, dark-grown seedlings either were kept in darkness (lanes 1 and 3) or were irradiated for 8 h with continuous red light (lanes 2 and 4). Lanes 1 to 4 each contain 20 μg of protein. Reduced phyA signals in lanes 2 and 4 demonstrate the red light–induced degradation of both phyA GFP fusion proteins. M, molecular weight marker.
We previously reported that there were no significant spectroscopic and conformational changes in phyA by PKA-catalyzed phosphorylation in vitro (Lapko et al., 1996). A circular dichroism analysis showed no significant secondary structural changes upon phyA phosphorylation. Only proteolytic patterns with trypsin showed a subtle conformational change near the hinge region containing Ser598. The PKA-catalyzed phosphorylation of oat phyA inhibited protease accessibility at the Lys536-Asn537 bond (Lapko et al., 1996). Spectroscopic analyses of the purified S598A protein also displayed the absorbance and red/FR difference spectra identical with those of the wild-type phyA (see supplemental data online). These results suggest that the photochemical and conformational changes by phosphorylation and Ser598 mutation cannot account for the hypersensitivity of the mutant S598A phyA. The dark reversion of phytochromes is a possible mechanism to quench the phytochrome action (Elich and Chory, 1997b), so the rates of dark reversion of phosphorylated phytochromes were examined. The results showed no apparent difference between unphosphorylated and phosphorylated phyA (Figure 2B). As another hypothesis, phytochrome phosphorylation might trigger in vivo degradation of phyA upon light irradiation because in vivo phytochrome degradation could be related to the hypersensitivity (Clough et al., 1999). If this hypothesis were valid, degradation of S598A phyA would be suppressed under light treatments, resulting in hypersensitivity of the transgenic plants to light. Our results showed no significant difference in degradation between oat wild-type phyA and S598A mutant phyA in vivo (Figure 2C), although both wild-type and S598A mutant phyA degraded slower than did Arabidopsis phyA.
Phytochrome phosphorylation at Ser598 might inhibit the translocalization of phyA, and the phyA interaction with its positive signaling molecules in the nucleus could then be prevented (Smith, 2000). We tested the translocalization of oat phyA and S598A mutant phyA into the nucleus in a time-dependent manner (Figure 2D). To study the subcellular localization of oat phyA-green fluorescent protein (GFP) and S598A-GFP, transgenic seedlings expressing these phyA-GFP fusion proteins were kept in darkness or treated with FRc. In darkness, oat phyA-GFP and S598A-GFP showed no nuclear localization. In FRc, both phyA GFP fusion proteins showed nuclear localization and formed the intranuclear speckles. No qualitative or quantitative differences were detectable, in darkness or in FRc (Figure 2D), indicating that the S598A mutation did not affect the light-induced nuclear import and speckle formation of phyA.
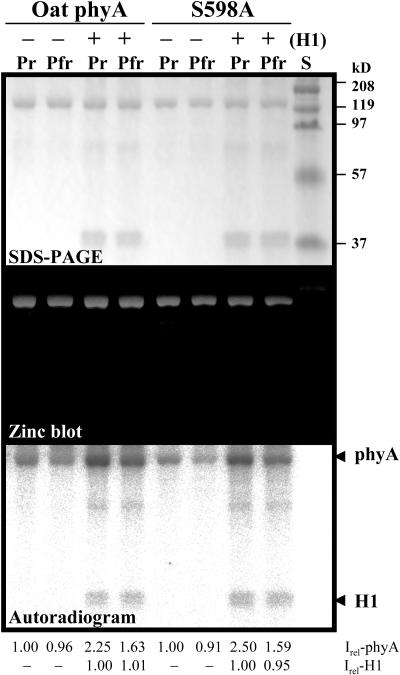
We next compared the phosphorylation of oat phyA versus S598A mutant because phytochromes are known as autophosphorylating Ser/Thr protein kinases (Yeh and Lagarias, 1998). For this purpose, recombinant phytochromes were expressed in the Pichia expression system and purified using streptavidin affinity chromatography (see Supplemental Figure 1 online). Because it is known that phytochrome phosphorylation is stimulated in the presence of histone H1 (Wong et al., 1986, 1989; Yeh and Lagarias, 1998), we also included it to check the stimulated phosphorylation of phyA by histone H1. The results showed that S598A mutant phyA still retained autophosphorylation and its phosphorylation was also stimulated by histone H1 (Figure 3), ruling out the possible involvement of Ser598 in the phytochrome autophosphorylation and phosphorylation activity on histone H1. These results also indicate that S598 is not the residue for phytochrome autophosphorylation and kinase activities.
Figure 3.
Phosphorylation of Oat phyA and S598A Mutant Proteins.
Histone H1 was included to check the stimulated phosphorylation activity of the phytochrome proteins. Amounts of phyA phosphorylation (Irel, relative intensity) are expressed relative to lane 1 or lane 5 (Pr forms of oat phyA and S598A, respectively).
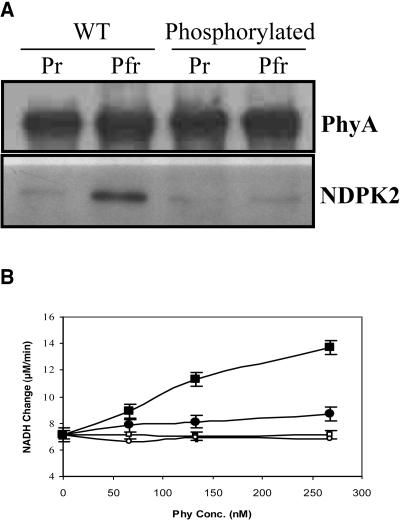
Finally, the interactions between phosphorylated phytochromes and PIPs were investigated. If phytochrome phosphorylation influences the protein–protein interaction between phytochrome and its signal transducers, the hypersensitivity of S598A mutant transgenic lines to light could be explained. NDPK2 was chosen to test this hypothesis at first because it is a positive regulator of phyA signaling and its enzymatic activity in vitro can be assayed by a spectroscopic method (Choi et al., 1999). The interaction between phyA and NDPK2 is Pfr specific, and only the Pfr form of phyA activates NDPK2 activity (i.e., GTP synthesis). To prepare the phosphorylated phyA for the study, PKA was used in the in vitro phosphorylation experiments. Results showed that the binding of the phosphorylated phyA to NDPK2 was significantly reduced compared with wild-type (unphosphorylated) phyA (Figure 4A). Moreover, the phosphorylated Pfr form of phyA showed a reduced NDPK2 activation compared with that of the Pfr form of wild-type phyA (Figure 4B), suggesting that the phosphorylation site in the Pfr form of phyA is responsible for the inhibition of the interaction. Because Ser598 is phosphorylated in a Pfr-specific manner, the Ser598 residue is likely the phosphorylation site. However, the reduction in NDPK2 activation by phosphorylation is not at the same level as NDPK2 by itself (basal level); this is attributable to a residual fraction of unphosphorylated protein by PKA. These results suggest that the phosphorylation plays a signal-modulating role through the interaction of phytochromes with their interacting protein(s), such as NDPK2.
Figure 4.
Effect of Phosphorylation on the Interaction between phyA and NDPK2.
(A) Immunoprecipitation of phyA and NDPK2. Native oat phyA was phosphorylated by PKA. Oat22 antibody was used in this immunoprecipitation reaction. Pfr-specific phyA interaction with NDPK2 was inhibited by its phosphorylation.
(B) NDPK2 activation assays by Pr/Pfr forms of wild-type (open square, Pr; closed square, Pfr) and phosphorylated phyA (open circle, Pr; closed circle, Pfr). The Pfr form of phosphorylated phyA showed reduced NDPK2 activation compared with that of wild-type phyA.
Ser598 Phosphorylation Blocks the Interaction between PhyA and NDPK2
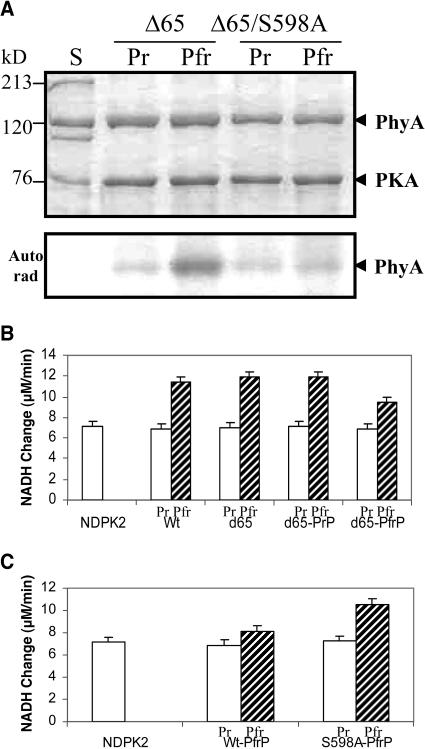
Because there are two Ser sites, Ser17 and Ser598, that can be phosphorylated by PKA (McMichael and Lagarias, 1990; Lapko et al., 1996), the inhibitory effect of phosphorylated phyA on the interaction with NDPK2 cannot be taken as unequivocal evidence for Ser598 to be the phosphorylation site. To establish the phosphorylation site, two mutants, N-terminal 65 amino acid–deleted phyA (Δ65, 66 to 1129 amino acids) and Δ65/S598A combination mutant phyA, were used for further study. These mutants interacted with NDPK2 normally unless they were phosphorylated (data not shown). Because Ser7 and Ser17 were removed in Δ65 mutants, only Ser598 could be phosphorylated by PKA. Results showed that PKA phosphorylated only the Pfr form of Δ65 mutant but not the Pr form (Figure 5A). Both Pr and Pfr forms of Δ65/S598A were not phosphorylated under the conditions employed. In NDPK2 activation assays, Δ65 phyA as well as full-length phyA (wild type) could activate the NDPK2 activity. However, only the phosphorylated Pfr of Δ65 mutant (Δ65 PfrP) showed a reduction in the level of NDPK2 activation (Figure 5B), indicating that Ser598 phosphorylation suppressed NDPK2 activation as expected. The inhibition was not complete because the percentage of phosphorylation on Ser598 was not stoichiometrically complete. These results indicate that Ser598 phosphorylation in fact inhibits the interaction of phytochrome with its signal transducer(s), such as NDPK2.
Figure 5.
Ser598 as the Site for Controlling the phyA Interaction with NDPK2.
(A) Ser598 as the phosphorylation site by PKA. SDS-PAGE (top) and autoradiogram (bottom; Autorad) of Δ65 and Δ65/S598A. Only the Pfr form of Δ65 was significantly phosphorylated by PKA, and the site was confirmed as Ser598 using the Δ65/S598A mutant.
(B) NDPK2 activation assays by wild-type phyA (Wt) and Δ65 mutant phyA (d65) proteins. The NDPK2 activation assays were performed with Pr (open bars) and Pfr (hatched bars) forms of each protein. PKA was used for the phosphorylation of Pr and Pfr forms of Δ65. Only the phosphorylated Pfr of Δ65 (d65-PfrP) showed reduced NDPK2 activation, while the Δ65 and the phosphorylated Pr of Δ65 (d65-PfrP) showed similar activation to the wild type, which means that Ser598 phosphorylation inhibits NDPK2 activation. In these assays, 67-nM phytochromes and 27-nM NDPK2 were used. NDPK2, NDPK2 only (control); Wt, wild-type oat phyA; d65, Δ65 phyA; d65-PrP, phosphorylated Pr form of Δ65; d65-PfrP, phosphorylated Pfr form of Δ65.
(C) NDPK2 activation by S598A phyA. The Pfr forms of wild-type phyA and S598A mutant phyA were phosphorylated at the same time using PKA and were used for the NDPK2 activation assays. The assays were performed with Pr (open bars) and Pfr (lined bars) forms of each protein. The phosphorylated Pfr forms of S598A phyA (S598A-PfrP) could increase NDPK2 activation, whereas those of wild-type phyA (Wt-PfrP) could not. This result suggested that the S598 phosphorylation plays a role as a modulation site for the interaction of phytochromes with NDPK2 and its activation. NDPK2, NDPK2 only (control); Wt-PfrP, phosphorylated Pfr form of wild-type oat phyA; S598A-Pfr, phosphorylated Pfr form of S598A.
The purified S598A mutant proteins were also used for NDPK2 activation assays. The wild-type oat phyA and S598A mutant proteins were phosphorylated at the same time using PKA and used for these assays. Results showed that the phosphorylated Pfr form of S598A mutant protein (S598A PfrP) activated NDPK2, whereas the phosphorylated Pfr form of wild-type phyA (Wt PfrP) showed little NDPK2 activation (Figure 5C). Thus, Ser598 phosphorylation is a signal-modulating site for the interaction between phytochrome and its positive phytochrome signal transducer(s), such as NDPK2.
Phytochrome Phosphorylation Also Influences Its Interaction with PIF3
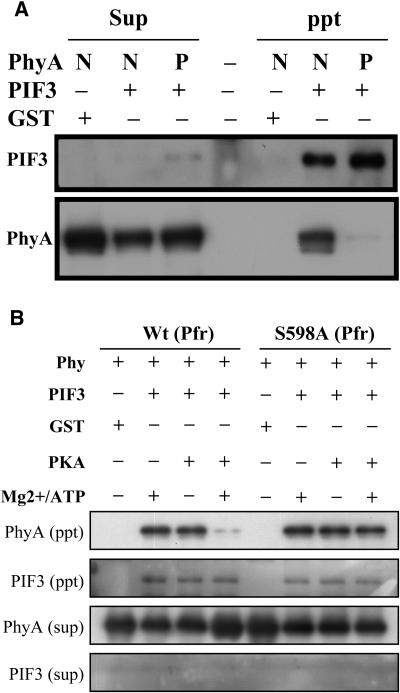
Although Ser598 phosphorylation inhibits the interaction between phyA and NDPK2, the phenotypes of S598A transgenic plants could not be explained in terms of NDPK2 knockout or overexpression. There could be other mechanism(s) involving Ser598 phosphorylation, (i.e., multiple regulation of signaling by Ser598 phosphorylation). Thus, we also tested the phytochrome interaction with another phytochrome-interacting protein, PIF3, that is also known as a putative signal transducer of phytochrome and interacts with the Pfr form of phyA and phyB (Ni et al., 1998). Using unphosphorylated (wild-type) and PKA-phosphorylated Pfr-phyA, we assessed the interaction with PIF3 by pull-down assay (Figure 6). The PKA-phosphorylated Pfr-phyA was purified using a Bio-gel P-6 column before this pull-down assay. Each Pfr form of native and phosphorylated phyA was incubated with glutathione S-transferase (GST)-PIF3, and protein gel blot analysis was performed with supernatant and precipitate fractions of the reactions. The results showed that phytochrome phosphorylation also prevented its interaction with PIF3 (Figure 6A). The unphosphorylated Pfr-phyA interacted with GST-PIF3 as reported (Ni et al., 1998), whereas phosphorylated Pfr-phyA did not interact with PIF3. We also demonstrated the interaction of PIF3 with PKA-phosphorylated S598A mutant phyA (Figure 6B). To rule out the possible nonspecific effects of PKA or ATP/Mg2+, the phyA:PIF3 interactions were also examined in the presence of ATP/Mg2+ or PKA as controls. These results show that neither PKA nor ATP/Mg2+ affected the interaction between phyA and PIF3. The PKA phosphorylation of Pfr-S598A did not show any difference in the phyA:PIF3 interaction, but that of the wild-type Pfr form prevented its interaction with PIF3. This is consistent with the Ser598 to be the signal-modulating site. Thus, we suggest that phytochrome phosphorylation in the hinge region controls interactions of the photoactivated phyA with its putative signal transducers.
Figure 6.
Effect of Phytochrome Phosphorylation on the Interaction with PIF3.
(A) The interaction between PKA-phosphorylated phyA and PIF3. The Pfr forms of phytochromes and GST-PIF3 were used for this experiment. After incubation, supernatants and precipitates were separated and analyzed by protein gel blot analysis. GST was used for a negative control. Sup, supernatant fraction of each reaction; ppt, precipitated fraction of each reaction; N, native phyA; P, PKA-phosphorylated phyA.
(B) The interaction between PKA-phosphorylated S598A phyA and PIF3. The effects of PKA or ATP/Mg2+ were also tested. Each reaction was performed by incubating phyA proteins with components indicated, PKA or ATP/Mg2+ or both, at 30°C for 30 min, then GST-PIF3 was added to the reactions, and pull-down analyses were performed. There were no effects of PKA or ATP/Mg2+ in the phyA:PIF3 interaction. The phosphorylation of S598A Pfr-phyA showed normal interaction with PIF3, whereas that of wild-type Pfr-phyA showed little interaction with PIF3 at the same condition.
DISCUSSION
Despite considerable progress in defining the photosensory roles of phytochromes based on the structure-function relation, the primary biochemical mechanism by which the phytochrome molecule transduces the perceived light signals into cellular responses remains to be elucidated (Elich and Chory, 1997a; Park et al., 2000; Smith, 2000; Kim et al., 2002b). This study provides evidence for the regulatory function of phytochrome phosphorylation in the phytochrome-mediated light signaling in plants. We demonstrated that phytochrome phosphorylation modulates the interaction of phytochrome with its putative signal transducers. It is thus possible that reversible phosphorylation/dephosphorylation of the photoreceptor protein is a key biochemical mechanism for the phytochrome-mediated light signaling.
Ser598 phosphorylation occurs only in the Pfr form of phyA in red light–treated oat seedlings (McMichael and Lagarias, 1990; Lapko et al., 1999), whereas phosphorylation of other sites (Ser7 and Ser17) was found either Pr/Pfr indifferent or Pr specific (Lapko et al., 1996, 1997). In fact, the Ser598 phosphorylation was thought to play an active role in the phyA-mediated light-signal transduction (Lapko et al., 1999; Park et al., 2000; Kim et al., 2002b). Here, we showed that the transgenic plants of the S598A mutant were hypersensitive to light. In the case of oat wild-type phyA transgenics, their seedlings under FRc displayed shorter hypocotyls than wild-type Arabidopsis (Ler), suggesting that they are hypersensitive to light. However, when they were grown under white light, their phenotype is similar to Ler plants. These results are consistent with the previous report (Boylan and Quail, 1991). In the case of S598A transgenics, they exhibited reduced inflorescence lengths under white light. The heights of S598A transgenics under white light were also much shorter (three to four times) than those of Ler and oat phyA transgenic plants. Their hypocotyls under FRc were also much shorter than wild-type Arabidopsis and oat phyA transgenics. Comparing these S598A transgenic phenotypes with wild-type Arabidopsis and oat phyA transgenic plants, we conclude that the Ser598 mutation confers hypersensitivity to light in the transgenic plants, suggesting that S598A phyA is hyperactive in plants.
We tested the possible hypotheses to explain the hypersensitive phenotype of S598A in terms of protein integrity, stability (degradation), nuclear localization, phosphorylation (kinase site), and protein–protein interaction. Our results are consistent with the last hypothesis (i.e., the interaction between the phytochrome and its positive signal transducers is favored by the phosphorylation-deficient mutant phyA). Our data clearly showed that the phosphorylation of phyA influences the protein–protein interaction between phyA and its putative signal transducers, NDPK2 and PIF3, whose binding sites are near to the hinge region and to PER-ARNT-SIM–related domains implicated in mediating protein–protein interaction in other systems (Lindebro et al., 1995; Choi et al., 1999; Zhu et al., 2000). Another phytochrome-interacting protein, PKS1, which is known to bind the His kinase-related domain at the C-terminal end region (Fankhauser et al., 1999), normally bound with phosphorylated phyA (data not shown). Thus, the Ser598 phosphorylation might influence the phyA interaction with its signal transducers whose binding regions are near to the hinge region. However, how this inhibition of protein–protein interaction can be related to the hypersensitivity to light remains to be answered. NDPK2 is known as a positive signaling component of phytochrome signaling, so the removal of the inhibitory site of phytochrome interaction with this positive component can make phytochrome hyperactive. On the contrary, PIF3 was recently reported to play dual roles depending on the type of light response and the light conditions: negative roles in phyB- but not phyA-mediated inhibition of hypocotyl elongation and in both phyA- and phyB-induced cotyledon expansion and a positive role in phyA- and phyB-induced anthocyanin accumulation (Kim et al., 2003). Thus, phytochrome phosphorylation at the hinge region may positively or negatively influence on the PIF3-mediated signaling, which remained to be elucidated.
Recently, we found that an Arabidopsis Ser/Thr-specific protein phosphatase 2A (FyPP) interacts with phyA (Kim et al., 2002a). The FyPP-overexpressing transgenic plants stimulated phytochrome activity in flowering and hypocotyl shortening, whereas the antisense repression of FyPP transgenic plants displayed reduced phytochrome activity. These results are consistent with the negative regulation of phytochrome signaling in plants through protein phosphorylation. Specifically, phytochrome phosphorylation at Ser598 attenuates the signaling, whereas dephosphorylation by phytochrome phosphatase, such as FyPP, can amplify the signaling. It is reminiscent of rhodopsin signaling, also modulated by phosphorylation and dephosphorylation. On stimulation, rhodopsin is phosphorylated at several sites on its C terminus as the first step in deactivation (Lee et al., 2002). The phytochrome signal transduction is thus modulated by protein phosphorylation and dephosphorylation; the phosphorylation blocks the interaction with its signal transducers, whereas the dephosphorylation enforces the interaction. S598A mutation might assure maintaining a higher level of the active pool of the unphosphorylated phytochrome molecules in displaying the hypersensitive phenotype to light. Ser598 can also be considered a switching site for modulating the interaction of phytochrome with its signal transducers, NDPK2 and PIF3.
Because Ser598 is not autophosphorylated to a significant extent, a protein kinase(s) is likely to play a regulatory role by catalyzing the phosphorylation of S598 residue. A phytochrome-associated kinase in oat seedlings that specifically and substantially phosphorylated Ser598 was detected during the preparation of native oat phyA (V.N. Lapko and P.-S. Song, unpublished data). Therefore, there may be several protein kinases and phosphatases for the phytochrome phosphorylation and dephosphorylation, but we still do not know what specific kinases and phosphatases are involved in phosphorylation/dephosphorylation at specific sites and their importance in the signaling.
We wondered whether the corresponding site to Ser598 of oat phyA exists in other phyA. So far, the phosphorylation sites of oat phyA have only been determined, so there is no available information for the phosphorylation site(s) of other phytochromes. Actually, the amino acid sequences are poorly conserved in the hinge region of phyA (Leu585 to Gly604 region in oat phyA). When we analyzed the amino acid sequences for possible phosphorylation sites using the NetPhos 2.0 server (http://www.cbs.dtu.dk/services/NetPhos), only Ser598 showed a high score (>0.9) for a phosphorylation site in the 20–amino acid hinge region of oat phyA. Although the amino acid sequences in the hinge region are not conserved among the phytochromes, we found that phyA usually has a possibly reactive phosphorylation site in the hinge region, for example, Ser600 in rice (Oryza sativa) phyA and Thr592 in Arabidopsis phyA that shows scores of >0.9 in the NetPhos search results. There are two Ser residues at 589 and 601 in the hinge region of Arabidopsis phyA, however. Further studies will be necessary to confirm whether the same mechanism based on phosphorylation/dephosphorylation at the hinge is applicable to other phytochromes, including phyB.
For the phytochrome Pr→Pfr phototransformation, we proposed that the primary step of phytochrome signaling involves phototransformation-dependent conformational changes (Park et al., 2000; Kim et al., 2002b). On Pr→Pfr phototransformation, the NTE region undergoes a conformational change from random coil to amphiphilic α-helix, which then interacts with the chromophore in the Pfr form (Parker et al., 1992; Deforce et al., 1994). Also, two Trp residues near the core regulatory region of oat phyA become preferentially exposed in the Pfr form (Wells et al., 1994). Based on these results, it was proposed that part of the NTE chain interacts with the regulatory C-terminal core motif in the Pr form (switch off conformation) (Park et al., 2000; Kim et al., 2002b). After the Pr→Pfr phototransformation (switch on conformation), the NTE chain is withdrawn by its interaction with the chromophore, exposing and thus activating the regulatory core motif (or Quail box) for the interaction with PIPs to initiate the signal transduction cascade. In this model, the phosphorylation in the hinge region can block the access of phytochrome signal transducers to the regulatory domain. We propose that the phosphorylated Pfr phytochrome represents a switch off conformation, and the phosphorylation/dephosphorylation of phytochrome provides a way to modulate the phytochrome-mediated light signaling (signal attenuation and amplification, respectively); light absorption by the photoreceptor generates an active Pfr signal, and the Ser598 phosphorylation by a kinase provides a signal-attenuating mode in the phytochrome signaling.
METHODS
Plant Materials and Phenotypic Measurements
The full-size cDNA of oat (Avena sativa) phyA was cloned into pGEM-11zf(+) (Promega, Madison, WI) from pFY122 (Boylan and Quail, 1989) by digesting with BamHI and EcoRI. The site-directed mutagenesis was performed to create S598A Avena phyA mutant using the GeneEditor in vitro site-directed mutagenesis system (Promega) according to the manufacturer's instructions. The oligonucleotide sequence of mutagenic primer was phosphorylated 5′-GCGGGAAGCTGCTCTAGATAACCAGATTGG-3′, and the mutagenized plasmids were confirmed by direct DNA sequencing with a Sequenase version 2.0 DNA sequencing kit (Amersham Pharmacia Biotech, Uppsala). In the numbering of amino acids, Ser598 was numbered by counting the first N-terminal Ser because the first amino acid as the first Met is posttranslationally removed in the cells. The oat wild-type phyA and S598A mutant genes were subcloned into the 35S promoter of Cauliflower mosaic virus containing pBI121 binary plasmid (Clontech, Palo Alto, CA) for transformation into phyA-deficient Arabidopsis thaliana (phyA-201). For the cloning into pBI121, the inserts were prepared by EcoRI digestion, T4 polymerase treatment, and BamHI digestion, and the vector was prepared by BamHI/EcoICRI digestion. The constructed pBI121 binary vector containing oat phyA or S598A mutant genes was introduced into phyA-201 using the Agrobacterium tumefaciens (strain GV3101)–mediated transformation method as described previously (Clough and Bent, 1998). The T3 transgenic plants were obtained from T2 transformants showing a 3:1 segregation ratio.
To assess the expression of oat phyA and S598A mutant proteins in adult plants, protein gel blot analysis was performed as described (Jordan et al., 1995): four leaves were removed before bolting, the leaves were incubated between soaked Whatman papers for at least 12 h in the dark condition, leaf tissues were ground with sea sand, and 50 μg of protein samples were loaded onto 10% SDS-PAGE gels for electrophoresis (Laemmli, 1970). The protein bands on the SDS-PAGE gel were transferred to a polyvinylidene difluoride membrane (Hybond-P; Amersham Pharmacia Biotech), and the membrane was incubated with oat phyA-specific monoclonal antibody, oat22 or oat25 (Cordonnier et al., 1983), for 2 h and developed using an ECL protein gel blotting analysis system (Amersham Pharmacia Biotech). To assess the expression of oat phyA and S598A mutant proteins in 4-d-old dark-grown seedlings, crude extracts were purified by the EZ method as described (Martínez-García et al., 1999). Five micrograms of crude extracts were loaded onto 10% SDS-PAGE gels. The membrane was incubated with oat phyA-specific oat22 or β-tubulin–specific monoclonal antibody (Sigma, St. Louis, MO) and developed using an ECL advanced protein gel blotting analysis system (Amersham Pharmacia Biotech).
Hypocotyl lengths were measured as described (Boylan and Quail, 1991). The seeds were sown onto MS media (Sigma) and cold treated for 2 d. The seeds were exposed to white light for 12 h to promote germination and then grown under far-red light (7 μmol/m2/s) or in darkness for 4 d. The hypocotyl lengths were photographed with a digital camera (Nikon, Tokyo) and then analyzed with the image analysis software (NIH Image; Bethesda, MD). Five-week-old mature plants were used for the height measurements.
Phytochrome in Vivo Degradation Assay
The transgenic plant seeds were obtained, germinated, and grown for 3.5 d in the dark. Seedlings were then illuminated with red light (10 μmol/m2/s) and harvested at time intervals (0, 6, 12, and 24 h) for 24 h. The harvested seedlings were stored in liquid nitrogen, and the protein samples were prepared as described (Jordan et al., 1995). Five micrograms of crude extracts were loaded onto SDS-PAGE gels, and protein gel blot analysis was performed to detect phytochromes. For the detection of Arabidopsis phyA, regulatory domain–specific P25 antibody (Cordonnier, 1989) was used.
Nuclear Localization
The subcellular localization of oat wild-type phyA-GFP and S598A phyA-GFP were examined as described (Kim et al., 2000). Dark-grown seedlings were either kept in the dark or exposed to 1, 3, 9, or 24 h of FRc (3.5 W/m2). All subsequent manipulations were performed under dim green light. The seedlings were transferred to glass slides and analyzed with an Axioskop microscope (Zeiss, Oberkochem, Germany). Excitation and detection of GFP was performed with a standard GFP filter set (AHF Analysentechnik, Tübingen, Germany). Representative nuclei were photographed with an Axiocam camera and Axiovision software (Zeiss). Only epidermal cells of the hypocotyls were analyzed. To minimize nuclear import of phyA GFP fusion proteins induced by microscopic light, photographs of the GFP fluorescence were taken during the first minute of microscopic analysis.
Phytochrome Protein Preparations
Native 124-kD phytochrome was purified from 3.5-d-old etiolated oat seedlings (A. sativa) in the Pfr form as described (Lapko and Song, 1995). The specific absorbance ratio of native phytochrome preparations was >1.00.
To express recombinant phytochrome proteins, phytochrome constructs were subcloned into a Pichia expression vector, pPIC3.5K (Invitrogen, Carlsbad, CA). Ten–amino acid streptavidin affinity-tag from pASK75 vector (Biometra, Goettingen, Germany) was attached to the 3′ end of the oat phyA gene. The primers 5′-CGGGATCCACCATGGCTTCCTCAAGGCCTGCTTCC-3′ (forward, BamHI) and 5′-TCGCGTCGACTTGTCCCATTGCTGTTGGAGC-3′ (reverse, SalI), were used for the subcloning of phyA genes into pPIC3.5K. For the first 65 amino acids, deleted phyA (Δ65, 1 to 65–amino acid deletion) and a combination mutant of S598A and Δ65 (S598A/Δ65), the forward primer was 5′-CGGGATCCACCATGGTCATAGCCTACTTACAGCAC-3′.
The pPIC3.5K constructs with phytochrome genes were transformed into Pichia cells using a Micropulser electroporation apparatus (Bio-Rad, Hercules, CA). Recombinant phytochrome proteins were expressed in the Pichia expression system, according to the manufacturer's recommendations (Invitrogen), and purified using streptavidin affinity chromatography (Sigma-Genosys, Haverhill, UK). Phytochromobilin and phycoerythrobilin were extracted from the red alga Porphyridium cruentum by methanolysis and subsequently purified by chromatography as previously described (Beale and Cornejo, 1991). Phycocyanobilin was purified using spirulina extracts (Sigma) by methanolysis. Holophytochromes were prepared by adding chromophores in DMSO to apoproteins at a final concentration of 20 μM, and the mixture was incubated on ice for 1 h. From the harvested Pichia cells, crude extract were prepared by breaking cells in liquid nitrogen using a homogenizer (Nihonseiki Kaisha, Tokyo, Japan). The phytochrome samples were precipitated by adding 0.23 g/L of ammonium sulfate, resuspended in a buffer (100 mM Tris, pH 7.8, and 1 mM EDTA), and then chromophores were added to the samples for in vitro reconstitution. The direct addition of chromophores to ammonium sulfate fraction makes better reconstitution and removal of free chromophores, compared with the addition to purified apoproteins. After dialysis to remove free chromophores, the samples were loaded to streptavidin affinity chromatography and purified holophytochromes without free chromophores.
Preparations for PIPs
Two known putative phytochrome signal transducers (PIF3 and NDPK2) were subcloned into pGEX 4T (Amersham Pharmacia Biotech) for the protein expression in Escherichia coli. Because the first 57 amino acids of NDPK2 are postulated to be a signal peptide for protein localization, they were removed for recombinant protein expression. The primers used were 5′-CTCGGATCCATGGAGGACGTTGAGGAGACTTAC-3′ (BamHI) and 5′-CGGAATTCTCACTCCCTTAGCCATGTAGC-3′ (EcoRI) for NDPK2 (80 to 231 amino acids) and 5′-CTCGGATCCATGCCTCTGTTTGAGCTTTTCAG-3′ (BamHI) and 5′-CGGAATTCTCACGACGATCCACAAAACTG-3′ (EcoRI) for full-length PIF3 (1 to 450 amino acids). The constructs were transformed into E. coli strain BL21 (Invitrogen) and used for the protein induction. Protein purification was performed using glutathione affinity chromatography (Amersham Pharmacia Biotech).
Phytochrome Autophosphorylation and Kinase Assay
Phytochrome autophosphorylation experiments were performed as described (Yeh et al., 1997; Yeh and Lagarias, 1998) with minor modifications. The reaction mixtures (total volume of 30 μL) contained kinase buffer (25 mM Tris-HCl, pH 7.5, 0.2 mM EDTA, 4 mM DTT, and 5 mM MgCl2) and 1 μg of purified recombinant phytochromes in either Pr or Pfr. Phytochrome samples were irradiated with red or far-red light for 2 min before the start of reaction. A fiber optic illuminator system (Cole-Parmer, Vernon Hills, IL) equipped with 656- and 730-nm interference filters (Oriel, Franklin, MA) was used as a light source. The light intensity was 8 W/m2 for red light and 6 W/m2 for far-red light. Histone H1 is known to stimulate phytochrome kinase activity and is also a phosphate acceptor from phytochromes. Depending on the experiments, histone H1 was included in the phosphorylation reaction (1 μg histone H1 per 1 μg phytochrome) to see the stimulation of phytochrome phosphorylation activity.
The PKA-phosphorylated phytochromes were obtained by incubating PKA (Pierce, Rockford, IL; 80 units/μL) and phytochromes, as either Pr or Pfr, at 30°C for 30 min in a kinase buffer (25 mM Tris-HCl, pH 7.5, 0.2 mM EDTA, 4 mM DTT, and 5 mM MgCl2) with 100 μM ATP. The reaction was initiated by adding 1 μL of PKA (10 times diluted in 0.1% BSA) to 1 μg of phytochrome samples and then quenched by adding 25 mM EDTA in 20 mM Tris buffer, pH 7.5. The phosphorylated phytochromes were purified using a Bio-gel P-6 column (Bio-Rad) for native phyA or streptavidin affinity column for recombinant phytochromes and dialyzed against 20 mM Tris, pH 7.8. After exposure on x-ray films, the bands were quantified using ImageMaster VDS (Pharmacia Biotech, Piscataway, NJ).
Immunoprecipitation and Pull-Down Analysis
Ten micrograms of purified phytochrome and 20 μg of NDPK2 were incubated in TBS (50 mM Tris-HCl, pH 7.5, and 150 mM NaCl) containing 1 mM dCDP and protease inhibitors at 4°C for 30 min. Oat phyA specific antibody (oat22) was then added to the reaction mixtures. The antibody/phytochrome complexes were recovered by incubating with one-tenth volumes of Protein A/G beads (Oncogene, San Diego, CA) for an additional 30 min with occasional mixing and then collected by centrifugation. The beads were washed five times in TBS containing 0.1% (v/v) Nonidet P-40. The attached proteins were solubilized by boiling for 3 min in 1× SDS sample buffer. The proteins of the pellet were resolved on 10% (w/v) SDS-polyacrylamide gels, and protein gel blot analysis was performed. For the interaction assay between phyA and PIF3, a pull-down analysis was performed with phyA and GST-PIF3 using glutathione beads. Two micrograms of phyA and 0.5 μg of PIF3 were incubated for 30 min at 4°C and then the glutathione resin was added and incubated for 20 min (50 μL reaction mixture in PBS and 1% [v/v] Triton X-100). The supernatant and precipitate were separated by centrifugation, and the same amount (50 μL) of 1× SDS sample buffer was added to precipitates. Both supernatants and precipitates were analyzed with phyA-specific antibody (oat22) or GST-specific monoclonal antibody (Oncogene).
NDPK2 Activation Assay by PhyA
The NDPK2 enzymatic activity can be activated by the Pfr form of phytochrome (Choi et al., 1999). The γ-phosphate-exchanging activity of NDPK was measured as described with minor modifications (Choi et al., 1999). The reaction buffer contained 100 mM Tris-HCl, pH 7.5, 100 mM KCl, 25 mM MgCl2, 3 mM phosphoenolpyruvate, 2 mM ATP, 0.3 mM NADH, 2.5 units of pyruvate kinase, 2.5 units of lactate dehydrogenase, and 1 mM dCDP. The reaction was started by adding purified NDPK2 to a final concentration of 3 nM. The NDPK activity was measured by monitoring the absorbance decrease at 340 nm for 10 min in the lactate dehydrogenase-pyruvate kinase-coupled reaction using a UV-VIS spectrophotometer. The γ-phosphate-exchange activity of NDPK2 was analyzed in the presence of dephosphorylated or phosphorylated wild-type phyA with a concentration of ∼268 nM. For the NDPK2 activation using mutant phytochromes, either phosphorylated or unphosphorylated, 67-nM phytochromes and 27-nM NDPK2 were used.
Supplementary Material
Acknowledgments
This work was supported by Korea Kumho Petrochemical (publication number 68) and in part by grants from the National Research Laboratory/Korea Institute of Science and Technology Evaluation and Planning, BioGreen 21 program of the Rural Development Administration (to P.-S.S.), the Crop Functional Genomics Center of the 21st Century Frontier Research Program by the Ministry of Science and Technology and the Rural Development Administration (code M101KG010001-03K0701-02910), and the Korea Science and Engineering Foundation/Ministry of Science and Technology to the Environmental Biotechnology National Core Research Center (to J.-I.K. and P.-S.S; Grant R15-2003-012-01003-0). We thank Veniamin N. Lapko for unpublished data and discussion. We also thank Andras Viczian for the cloning of the phytochrome-GFP constructs and the production of the phytochrome-GFP transgenic plants.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Pill-Soon Song (pssong@kkpc.com).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.023879.
References
- Beale, S.I., and Cornejo, J. (1991). Biosynthesis of phycobilins. 3(Z)-phycoerythrobilin and 2(Z)-phycocyanobilin are intermediates in the formation of 3(E)-phycocyanobilin from biliverdin IXα. J. Biol. Chem. 266, 22333–22340. [PubMed] [Google Scholar]
- Boylan, M.T., Douglas, N., and Quail, P.H. (1994). Dominant negative suppression of Arabidopsis photoresponses by mutant phytochrome A sequences identifies spatially discrete regulatory domains in the photoreceptor. Plant Cell 6, 449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan, M.T., and Quail, P.H. (1989). Oat phytochrome is biologically active in transgenic tomatoes. Plant Cell 1, 765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan, M.T., and Quail, P.H. (1991). Phytochrome A overexpression inhibits hypocotyl elongation in transgenic Arabidopsis. Proc. Natl. Acad. Sci. USA 88, 11806–11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, G., Yi, H., Lee, J., Kwon, Y.-K., Soh, M.-S., Shin, B., Luka, Z., Hahn, T.-R., and Song, P.-S. (1999). Phytochrome signaling is mediated through nucleoside diphosphate kinase 2. Nature 401, 610–613. [DOI] [PubMed] [Google Scholar]
- Clough, R.C., Jordan-Beebe, E.T., Lohman, K.N., Marita, J.M., Walker, J.M., Gatz, C., and Vierstra, R.D. (1999). Sequences within both the N- and C-terminal domains of phytochrome A are required for PFR ubiquitination and degradation. Plant J. 17, 155–167. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cordonnier, M.-M. (1989). Monoclonal antibodies: Molecular probes for the study of phytochrome. Photochem. Photobiol. 49, 821–831. [Google Scholar]
- Cordonnier, M.-M., Smith, C., Greppin, H., and Pratt, L.H. (1983). Production and purification of monoclonal antibodies to Pisum and Avena phytochrome. Planta 158, 369–376. [DOI] [PubMed] [Google Scholar]
- Deforce, L., Tokutomi, S., and Song, P.-S. (1994). Phototransformation of pea phytochrome A induces an increase in α-helical folding of the apoprotein: Comparison with a monocot phytochrome A and CD analysis by different methods. Biochemistry 33, 4918–4922. [DOI] [PubMed] [Google Scholar]
- Elich, T.D., and Chory, J. (1997. a). Phytochrome: If it looks and smells like a histidine kinase, is it a histidine kinase? Cell 91, 713–716. [DOI] [PubMed] [Google Scholar]
- Elich, T.D., and Chory, J. (1997. b). Biochemical characterization of Arabidopsis wild-type and mutant phytochrome B holoproteins. Plant Cell 9, 2271–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser, C. (2000). Phytochromes as light-modulated protein kinases. Semin. Cell Dev. Biol. 11, 467–473. [DOI] [PubMed] [Google Scholar]
- Fankhauser, C., and Chory, J. (1999). Photomorphogenesis: Light receptor kinases in plants! Curr. Biol. 9, R123–R126. [DOI] [PubMed] [Google Scholar]
- Fankhauser, C., Yeh, K.-C., Lagarias, J.C., Zhang, H., Elich, T.D., and Chory, J. (1999). PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284, 1539–1541. [DOI] [PubMed] [Google Scholar]
- Jordan, E.T., Hatfield, P.M., Hondred, D., Talon, M., Zeevaart, J.A., and Vierstra, R.D. (1995). Phytochrome A overexpression in transgenic tobacco. Correlation of dwarf phenotype with high concentrations of phytochrome in vascular tissue and attenuated gibberellin levels. Plant Physiol. 107, 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, E.T., Marita, J.M., Clough, R.C., and Viestra, R.D. (1997). Characterization of regions within the N-terminal 6-kilodalton domain of phytochrome A that modulate its biological activity. Plant Physiol. 115, 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick, R.E., and Kronenberg, G.H.M. (1994). Photomorphogenesis in Plants, 2nd ed. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- Kim, D.-H., Kang, J.-G., Yang, S.-S., Chung, K.-S., Song, P.-S., and Park, C.-M. (2002. a). A phytochrome associated protein phosphatase 2A modulates light signals in flowering time control in Arabidopsis. Plant Cell 14, 3043–3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Yi, H., Choi, G., Shin, B., Song, P.-S., and Choi, G. (2003). Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15, 2399–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.-I., Kozhukh, G.V., and Song, P.-S. (2002. b). Phytochrome-mediated signal transduction pathways in plants. Biochem. Biophys. Res. Commun. 298, 457–463. [DOI] [PubMed] [Google Scholar]
- Kim, L., Kircher, S., Tóth, R., Ádám, É., Schäfer, E., and Nagy, F. (2000). Light-induced nuclear import of phytochrome-A:GFP fusion proteins is differentially regulated in transgenic tobacco and Arabidopsis. Plant J. 22, 125–134. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lapko, V.N., Jiang, X.-Y., Smith, D.L., and Song, P.-S. (1996). Protein kinase A-catalyzed phosphorylation and conformational changes in phytochrome A. Biochemistry 35, 6585–6594. [DOI] [PubMed] [Google Scholar]
- Lapko, V.N., Jiang, X.-Y., Smith, D.L., and Song, P.-S. (1997). Post-translational modification of oat phytochrome A: Phosphorylation of a specific serine in a multiple serine cluster. Biochemistry 36, 10595–10599. [DOI] [PubMed] [Google Scholar]
- Lapko, V.N., Jiang, X.-Y., Smith, D.L., and Song, P.-S. (1999). Mass spectrometric characterization of oat phytochrome A: Isoforms and posttranslational modifications. Protein Sci. 8, 1032–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapko, V.N., and Song, P.-S. (1995). A simple and improved method of isolation and purification for native oat phytochrome. Photochem. Photobiol. 62, 194–199. [DOI] [PubMed] [Google Scholar]
- Lee, K.A., Craven, K.B., Niemi, G.A., and Hurley, J.B. (2002). Mass spectrometric analysis of the kinetics of in vivo rhodopsin phosphorylation. Protein Sci. 11, 862–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindebro, M.C., Poellinger, L., and Whitelaw, M.L. (1995). Protein-protein interaction via PAS domains: Role of the PAS domain in positive and negative regulation of the bHLH/PAS dioxin receptor-Arnt transcription factor complex. EMBO J. 14, 3528–3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Garcia, J.F., Huq, E., and Quail, P.H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288, 859–863. [DOI] [PubMed] [Google Scholar]
- Martínez-García, J.F., Monte, E., and Quail, P.H. (1999). A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J. 20, 251–257. [DOI] [PubMed] [Google Scholar]
- McMichael, R.W., Jr., and Lagarias, J.C. (1990). Phosphopeptide mapping of Avena phytochrome phosphorylated by protein kinase in vitro. Biochemistry 29, 3872–3878. [DOI] [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95, 657–667. [DOI] [PubMed] [Google Scholar]
- Park, C.-M., Bhoo, S.-H., and Song, P.-S. (2000). Inter-domain crosstalk in the phytochrome molecules. Semin. Cell Dev. Biol. 11, 449–456. [DOI] [PubMed] [Google Scholar]
- Parker, W., Partis, M., and Song, P.-S. (1992). N-terminal domain of Avena phytochrome: Interactions with sodium dodecyl sulfate micelles and N-terminal chain truncated phytochrome. Biochemistry 31, 9413–9420. [DOI] [PubMed] [Google Scholar]
- Quail, P.H., Boylan, M.T., Parks, B.M., Short, T.W., Xu, Y., and Wagner, D. (1995). Phytochromes: Photosensory perception and signal transduction. Science 268, 675–680. [DOI] [PubMed] [Google Scholar]
- Quail, P.H., Briggs, W.R., and Pratt, L.H. (1978). In vivo phosphorylation of phytochrome. Carnegie Institution Yearbook 77, 342–344. [Google Scholar]
- Sharma, R. (2001). Phytochrome: A serine kinase illuminates the nucleus. Curr. Biol. 80, 178–188. [Google Scholar]
- Singh, B.R., and Song, P.-S. (1990). Phytochrome and protein phosphorylation. Photochem. Photobiol. 52, 249–254. [DOI] [PubMed] [Google Scholar]
- Smith, H. (2000). Phytochromes and light signal perception by plants-An emerging synthesis. Nature 407, 585–591. [DOI] [PubMed] [Google Scholar]
- Sokal, I., Pulvermuller, A., Buczylko, J., Hofmann, K.P., and Palczawski, K. (2002). Rhodopsin and its kinase. Methods Enzymol. 343, 578–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockhaus, J., Nagatani, A., Halfter, U., Kay, S., Furuya, M., and Chua, N.-H. (1992). Serine-to-alanine substitutions at the amino-terminal region of phytochrome A result in an increase in biological activity. Genes Dev. 6, 2364–2372. [DOI] [PubMed] [Google Scholar]
- Watson, J.C. (2000). Light and protein kinases. Adv. Bot. Res. 32, 149–184. [Google Scholar]
- Wells, T.A., Nakazawa, M., Manabe, K., and Song, P.-S. (1994). A conformational change associated with the phototransformation of Pisum phytochrome A as probed by fluorescence quenching. Biochemistry 33, 708–712. [DOI] [PubMed] [Google Scholar]
- Whitelam, G.C., Johnson, E., Peng, J., Carol, P., Anderson, M.L., Cowl, J.S., and Harberd, N.P. (1993). Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5, 757–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, Y.-S., Cheng, H.-C., Walsh, D.A., and Lagarias, J.C. (1986). Phosphorylation of Avena phytochrome in vitro as a probe of light-induced conformational changes. J. Biol. Chem. 261, 12089–12097. [PubMed] [Google Scholar]
- Wong, Y.-S., McMichael, R.W., Jr., and Lagarias, J.C. (1989). Properties of a polycation-stimulated protein kinase associated with purified Avena phytochrome. Plant Physiol. 91, 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, K.-C., and Lagarias, J.C. (1998). Eukaryotic phytochromes: Light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc. Natl. Acad. Sci. USA 95, 13976–13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, K.-C., Wu, S.-H., Murphy, J.T., and Lagarias, J.C. (1997). A cyanobacterial phytochrome two-component light sensory system. Science 277, 1505–1508. [DOI] [PubMed] [Google Scholar]
- Zhu, Y., Tepperman, J.M., Fairchild, C.D., and Quail, P.H. (2000). Phytochrome B binds with greater apparent affinity than phytochrome A to the basic helix-loop-helix factor PIF3 in a reaction requiring the PAS domain of PIF3. Proc. Natl. Acad. Sci. USA 97, 13419–13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.