Abstract
DNA gyrase, which catalyzes topological transformation of DNA, plays an essential role in replication and transcription in prokaryotes. Virus-induced gene silencing of NbGyrA or NbGyrB, which putatively encode DNA gyrase subunits A and B, respectively, resulted in leaf yellowing phenotypes in Nicotiana benthamiana. NbGyrA and NbGyrB complemented the gyrA and gyrB temperature-sensitive mutations of Escherichia coli, respectively, which indicates that the plant and bacterial subunits are functionally similar. NbGyrA and NbGyrB were targeted to both chloroplasts and mitochondria, and depletion of these subunits affected both organelles by reducing chloroplast numbers and inducing morphological and physiological abnormalities in both organelles. Flow cytometry analysis revealed that the average DNA content in the affected chloroplasts and mitochondria was significantly higher than in the control organelles. Furthermore, 4′,6-diamidino-2-phenylindole staining revealed that the abnormal chloroplasts contained one or a few large nucleoids instead of multiple small nucleoids dispersed throughout the stroma. Pulse-field gel electrophoresis analyses of chloroplasts demonstrated that the sizes and/or structure of the DNA molecules in the abnormal chloroplast nucleoids are highly aberrant. Based on these results, we propose that DNA gyrase plays a critical role in chloroplast nucleoid partitioning by regulating DNA topology.
INTRODUCTION
DNA gyrase belongs to the type II class of DNA topoisomerases that catalyze DNA supercoiling/relaxation and catenation/decatenation (Wang, 1996; Champoux, 2001). It is exclusively found in prokaryotes and is essential for their survival. Escherichia coli DNA gyrase is composed of A and B subunits and is functional when it is a heterotetramer (A2B2). DNA gyrase is unique among the type II DNA topoisomerases in being able to negatively supercoil DNA. This activity is largely responsible for maintaining bacterial DNA within a physiological range of negative superhelical density in vivo (Zechiedrich et al., 2000). DNA gyrase is also involved in removing positive supercoils generated by DNA replication and transcription and in separating the decatenated daughter chromosomes during cell division (Wang, 1996; Champoux, 2001). As a result of this multiplicity of functions, bacteria with mutations in either gyrase subunit show typically pleiotropic defects in DNA supercoiling, DNA replication, and partitioning (Levine et al., 1998).
Increasing evidence indicates that there is DNA gyrase activity in the chloroplasts and mitochondria of plants. First, the inhibitors of bacterial DNA gyrase novobiocin and nalidixic acid inhibit chloroplast DNA synthesis in higher plants (Heinhorst et al., 1985; Mills et al., 1989). The same drugs also affect the accumulation of specific chloroplast transcripts in higher plants (Lam and Chua, 1987). Moreover, Arabidopsis thaliana genome analyses have predicted that the Arabidopsis DNA gyrase A and B subunits are targeted to organelles (Elo et al., 2003). Indeed, the DNA gyrase A and B subunits were found in the mitochondrial proteome of Arabidopsis (Heazlewood et al., 2004). Very recently the function of the Arabidopsis DNA gyrase subunit genes was analyzed (Wall et al., 2004). T-DNA knockout mutants of gyrA (subunit A) exhibited an embryo-lethal phenotype, whereas knockouts of the gyrB (subunit B) genes showed either a seedling-lethal phenotype or severely stunted growth and development (Wall et al., 2004). Moreover, it was found that the GyrA protein appears to be targeted to both chloroplasts and mitochondria, whereas two of the GyrB homologs are instead targeted separately to chloroplasts and mitochondria (Wall et al., 2004). These results demonstrate that among eukaryotic organisms, plants appear to be unique in possessing genes encoding the prokaryotic type DNA gyrase A and B subunits. In addition, DNA gyrase activity appears to be essential for plant growth and development. However, it is still not clear how the loss-of-function mutations in DNA gyrase affect the development and physiology of plant organelles or what role plant DNA gyrase plays in regulating organellar DNA metabolism.
To address these questions, we used virus-induced gene silencing (VIGS) to study the functions of DNA gyrase in the plant organelles of Nicotiana benthamiana. The basis of VIGS is that gene expression is suppressed in a sequence-specific manner by infection with viral vectors carrying a host gene. As a result, the symptoms in the infected plant phenocopy loss-of-function or reduced-expression mutants in the host gene (Waterhouse et al., 2001). VIGS has proved to be a particularly powerful tool in the analysis of embryo- or seedling-lethal genes because it permits the investigation of mutant phenotypes in other tissues and developmental stages beyond the embryo and seedling stages. In this article, we show that depletion of DNA gyrase leads to defective chloroplast nucleoid partitioning in chloroplasts and increased DNA content in chloroplasts and mitochondria. We also show that the DNA gyrase A and B subunits of N. benthamiana are both targeted to chloroplasts and mitochondria using an ambiguous signal that is recognized by the transport machinery of both organelles.
RESULTS
N. benthamiana Homologs of the DNA Gyrase A and B Subunits
Previously, we have used tobacco rattle virus (TRV)-based VIGS in N. benthamiana to assess the functions of various signaling genes and other genes that may cause embryo or seedling lethality when their expression is suppressed (Kim et al., 2003; Lee et al., 2003). Our screening procedure revealed that gene silencing of plant homologs of bacterial DNA gyrase A and B subunits causes a severe leaf yellowing phenotype. Based on genomic DNA gel blot analysis, the N. benthamiana genome contains one copy of the gene for DNA gyrase subunit A and two copies of the gene for subunit B (data not shown). The full-length N. benthamiana cDNA encoding DNA gyrase subunit A was obtained by 5′-rapid amplification of cDNA ends (RACE) PCR and was designated as NbGyrA. A cDNA of only one of the two subunit B genes was isolated. Its full-length form was obtained by 5′-RACE PCR and was designated as NbGyrB. Sequence information for the other gene encoding DNA gyrase subunit B is not available. NbGyrA and NbGyrB encode polypeptides of 949 and 731 amino acids, respectively, corresponding to theoretical molecular masses of 105,199 and 81,599 D, respectively. NbGyrA, NbGyrB, and other plant DNA gyrase subunits contain an N-terminal extension that features chloroplast and mitochondria targeting signals (Emanuelsson and von Heijne, 2001). Of the prokaryotic GyrA and GyrB sequences, those of cyanobacterial species are most closely related to NbGyrA and NbGyrB (50 to 61% sequence identity), which suggests that the plant GyrA and GyrB genes have an endosymbiotic origin (Osteryoung and Nunnari, 2003).
Complementation Analysis of E. coli Temperature-Sensitive gyrA and gyrB Mutations
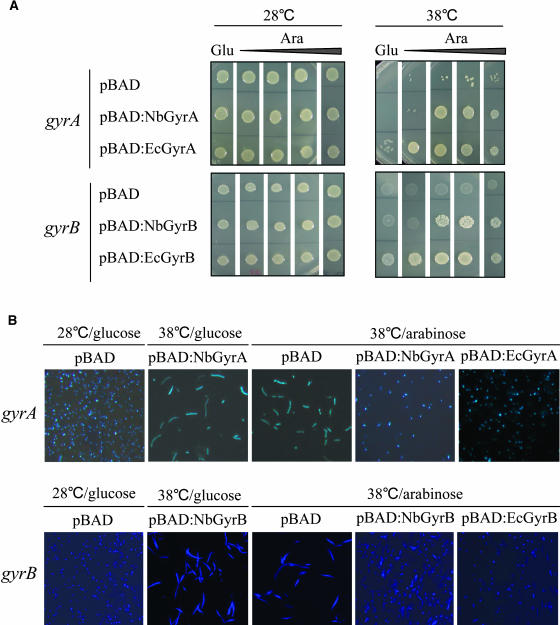
To examine the functional similarities between plant DNA gyrase subunits and their bacterial counterparts, complementation analysis was performed with E. coli strains KNK452 (Soussy et al., 1993) and N4177 (Gross et al., 2003), which carry a temperature-sensitive gyrA and gyrB mutation, respectively (Figure 1). The cDNA fragments encoding the NbGyrA and NbGyrB proteins without the N-terminal extensions were cloned into pBAD18 (Blanc-Potard and Bossi, 1994) such that expression of NbGyrA and NbGyrB was under the control of the arabinose-dependent promoter. As controls, the wild-type gyrA and gyrB genes of E. coli were also cloned into pBAD18. The recombinant plasmids and the pBAD vector control were transformed into KNK452 and N4177 cells, and the transformants were grown in media containing glucose or arabinose, which repress and induce, respectively, the expression of genes from the PBAD promoter. The growth and cell morphology of the transformants were then determined at 28 and 38°C in the presence of glucose or increasing concentrations of arabinose.
Figure 1.
Functional Complementation of E. coli gyrA and gyrB Mutants.
(A) Suppression of gyrA and gyrB thermosensitive growth phenotypes. Top, the E. coli strain harboring a temperature-sensitive gyrA mutation (KNK452) was transformed with the pBAD vector control, pBAD:NbGyrA, or pBAD:EcGyrA. The transformants were plated on NZY agar containing glucose (0.2%) or arabinose (0.0002, 0.002, 0.02, and 0.2%) to repress and induce, respectively, the expression of the genes from the PBAD promoter and incubated for 2 d at the indicated temperatures. Bottom, complementation of the E. coli gyrB mutant (N4177) was performed in the same way.
(B) Bacterial cell shapes. Overnight cultures of E. coli gyrA (KNK452) and gyrB (N4177) mutants and their various transformants grown in NZY broth (+ 0.2% glucose) at 28°C were diluted 1:100 in NZY (+ 0.2% glucose or + 0.2% arabinose) and incubated at 28 or 38°C for 10 h. The cells were then stained with DAPI and observed under a fluorescent microscope (40×).
The KNK452 and N4177 strains could grow at 28°C but not at 38°C and exhibited elongated cell shapes at the nonpermissive temperature (Figures 1A and 1B). However, the wild-type gyrA and gyrB genes of E. coli on the plasmid allowed the temperature-sensitive strains to grow at 38°C in the presence of arabinose regardless of its concentration (Figure 1A). Because of their leaky expression, E. coli gyrA and gyrB on the plasmid also partially restored the growth of the mutant cells cultured with glucose. The arabinose-dependent expression of NbGyrA in the KNK452 strain completely restored the growth of the mutant at 38°C, although it required higher concentrations of arabinose than the expression of E. coli gyrA. This indicates that the lethal effect of the gyrA mutation is suppressed by the expression of the plant GyrA subunit. In addition, the expression of NbGyrB in the N4177 strain restored growth at 38°C, albeit not completely (Figure 1A). Thus, the structural similarities between NbGyrB and the bacterial B subunit seem to be more restricted. The vector control could not rescue the temperature-sensitive lethality of the mutant strains under any conditions.
Cell morphology was examined by 4′,6-diamidino-2-phenylindole (DAPI) staining (Figure 1B). Whereas KNK452 and N4177 cells were morphologically normal at 28°C, they produced long filaments with nucleoid segregation defects when the temperature was shifted to 38°C. Expression of E. coli gyrA and gyrB in KNK452 and N4177 strains, respectively, completely restored normal cell morphology, unlike the vector control. Expression of NbGyrA and NbGyrB in the KNK452 and N4177 strains, respectively, also corrected the abnormal morphology of the mutants, although some slightly elongated cells were still observed in the NbGyrB-expressing N4177 strain (Figure 1B). These results demonstrate that DNA gyrase subunits of prokaryotes and plants are functionally conserved.
Suppression of the NbGyrA and NbGyrB Transcripts by VIGS Causes a Leaf Yellowing Phenotype
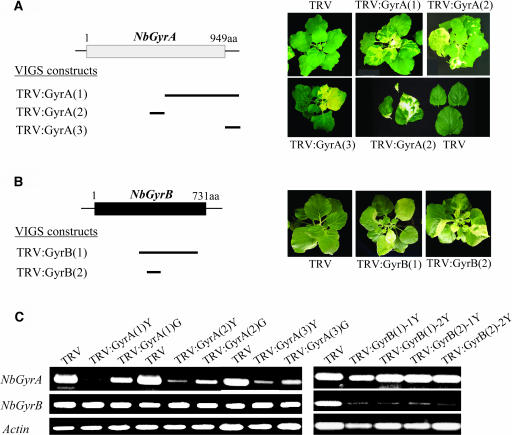
To induce gene silencing of NbGyrA and NbGyrB, we cloned different fragments of the NbGyrA and NbGyrB cDNAs into the TRV-based VIGS vector pTV00 (Ratcliff et al., 2001) and infiltrated N. benthamiana plants with Agrobacterium tumefaciens containing each plasmid (Figure 2). TRV:GyrA(1) and TRV:GyrA(2) contain the 1.5-kb C-terminal and 0.3-kb central regions of the NbGyrA cDNA, respectively, whereas TRV:GyrA(3) contains the 0.3-kb 3′-untranslated region of the cDNA. Similarly, TRV:GyrB(1) and TRV:GyrB(2) contain the 1.2- and 0.3-kb fragments of the NbGyrB cDNA, respectively. VIGS with each of these constructs resulted in the same phenotypes in newly emerged leaves, namely, severe yellowing and morphological abnormalities (Figures 2A and 2B).
Figure 2.
VIGS Phenotypes and Suppression of the Endogenous Transcripts.
(A) Schematic representation of the structure of NbGyrA, the cDNA regions used in the VIGS constructs, and the VIGS phenotypes of the three TRV:GyrA VIGS lines. The box indicates the protein-coding region of NbGyrA. The three VIGS constructs containing different regions of the NbGyrA cDNA are marked by bars. N. benthamiana plants were infected with Agrobacterium containing the TRV control or one of the TRV:GyrA constructs. The plants were photographed 20 d postinoculation. The whole plants and individual leaves from the TRV control and TRV:GyrA lines are shown. aa, amino acids.
(B) Schematic representation of the structure of NbGyrB, the cDNA regions used in the VIGS constructs, and the VIGS phenotypes of the two TRV:GyrB VIGS lines. The box indicates the protein-coding region of NbGyrB. The two VIGS constructs containing different regions of the NbGyrB cDNA are marked by bars. The plants were photographed 20 d postinoculation and exhibited the same phenotype as the TRV:GyrA VIGS lines.
(C) Semiquantitative RT-PCR analysis to examine the transcript levels of NbGyrA and NbGyrB. The primers were designed to exclude the cDNA regions used in the VIGS constructs. RNA was extracted from the yellow (Y) or green (G) sectors of the leaves from the VIGS plants. Two independent plants were each analyzed for TRV:GyrB(1) and TRV:GyrB(2). As a control, the actin mRNA levels were examined.
The amounts of endogenous NbGyrA and NbGyrB mRNAs in the VIGS lines were measured by semiquantitative RT-PCR to determine the effect of gene silencing (Figure 2C). The RT-PCR primers used were designed to exclude the cDNA regions used in the VIGS constructs. RT-PCR using the GyrA primers showed that the TRV:GyrA(1), TRV:GyrA(2), and TRV:GyrA(3) VIGS lines had significantly lower amounts of PCR products in the green and yellow sectors of the leaves compared with the TRV control, indicating that the endogenous level of NbGyrA transcripts is greatly reduced in these plants (Figure 2C). Furthermore, the different degrees of gene silencing observed in the green and yellow sectors showed there was a correlation between the transcript levels and the severity of the phenotype. The transcript levels of NbGyrB and actin, which served as controls, remained constant.
The TRV:GyrB VIGS line showed the same phenotype as the TRV:GyrA line (Figure 2B). Genomic DNA gel blot analysis indicated that the two gyrase B genes of N. benthamiana are highly similar (data not shown), and in Arabidopsis, two organelle-targeted DNA gyrase B subunits show >90% amino acid sequence identity to each other (Wall et al., 2004). Although we lack the sequence of the second copy of the subunit B gene in N. benthamiana, these observations indicate a possibility that VIGS using the NbGyrB cDNA suppresses the expression of both subunit B genes in N. benthamiana. RT-PCR using the GyrB primers showed that the yellow sectors of the leaves from individual plants of the TRV:GyrB(1) and TRV:GyrB(2) lines had significantly reduced amounts of PCR products, which indicates that the endogenous level of the NbGyrB transcript is significantly reduced in these plants (Figure 2C). By contrast, the transcript levels of NbGyrA and actin remained constant.
Numbers of Chloroplasts and Mitochondria
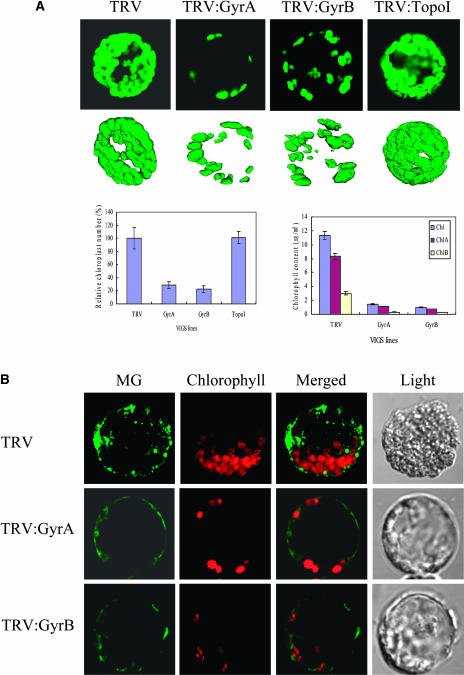
To analyze the effect of TRV:GyrA and TRV:GyrB VIGS on chloroplast number and size, protoplasts were generated from newly emerged leaves of TRV:GyrA and TRV:GyrB lines with the yellowing phenotype. These were then examined by confocal laser scanning microscopy (Figure 3A). Protoplasts from the TRV:TopoI line, in which topoisomerase I expression is suppressed by VIGS, and protoplasts from the TRV line served as controls. Topoisomerase I has been predicted to be targeted to the nucleus in Arabidopsis (Takahashi et al., 2002). In both TRV:GyrA and TRV:GyrB, almost all of the protoplasts, most of which originated from leaf mesophyll cells, exhibited drastically reduced chloroplast numbers (∼28 and 22% of those in the TRV control, respectively). Consistent with this, the chlorophyll contents in the leaves were much lower in the gyrase VIGS lines than in the TRV control (Figure 3A). Furthermore, confocal microscopy revealed that a large portion of the chloroplasts from the TRV:GyrA and TRV:GyrB lines are highly variable in size and shape and contain dumbbell-shaped chloroplasts (Figure 3A).
Figure 3.
Numbers of Chloroplasts and Mitochondria.
(A) Confocal laser scanning microscopy of chloroplasts in protoplasts isolated from the TRV, TRV:GyrA, TRV:GyrB, and TRV:TopoI VIGS lines. The upper panels show one- and three-dimensional images of typical chloroplasts. Confocal microscopy was used to determine the number of chloroplasts per protoplast (left histogram). The TRV control had on average 53 chloroplasts per protoplast. The data points represent means ± sd of 40 to 50 individual protoplasts. Chlorophyll contents in leaves from the VIGS lines (right histogram) were measured as described (Porra et al., 1989).
(B) To visualize mitochondria, protoplasts isolated from leaves of the VIGS lines were stained with 200 nM MG for 1 to 2 min.
Mitochondria in leaf protoplasts isolated from the TRV control, TRV:GyrA, and TRV:GyrB lines were examined by MitoTracker Green FM (MG) (Figure 3B). MG accumulates in mitochondria regardless of the mitochondrial membrane potential and becomes fluorescent in the lipid environment of mitochondria (Oubrahim et al., 2001). MG is widely used for determining mitochondrial mass. The MG fluorescence of protoplasts from TRV:GyrA and TRV:GyrB leaves was significantly lower than that of protoplasts from the TRV control (Figure 3B). This indicates that mitochondrial numbers and/or mass are reduced in the gyrase VIGS lines.
Ultrastructural Analysis of Chloroplasts and Mitochondria
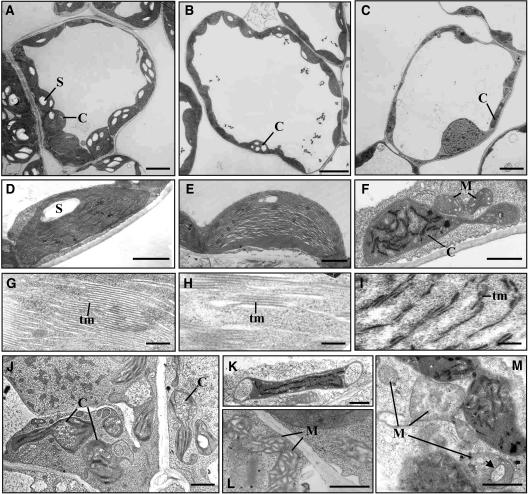
Transmission electron microscopy of transverse leaf sections of the TRV:GyrA and TRV:GyrB VIGS lines showed that their chloroplast numbers and morphology were markedly different from those of the TRV control (Figure 4). In the TRV control leaves, the mesophyll cell chloroplasts displayed a well-developed thylakoid membrane system that was distributed regularly throughout the stroma (Figures 4A, 4D, and 4G). Large starch granules displaced the thylakoid membranes and distorted the chloroplast outline (Figures 4A and 4D). The green sector of the leaves from the TRV:GyrB line exhibited somewhat reduced numbers of chloroplasts with atypical thylakoid membrane stacking (Figures 4B, 4E, and 4H), whereas the yellow sector showed a severe reduction of chloroplast number and morphological abnormalities (Figures 4C, 4F, and 4I). The chloroplasts in the yellow sector were elongated and irregularly shaped and contained few starch grains (Figures 4C and 4F). The thylakoid membranes were either poorly developed or in the process of disintegrating as shown by their radical alteration in arrangement and distribution (Figures 4F and 4I). Most of the chloroplasts in the TRV:GyrA line were also irregularly shaped and appeared to be degenerating (Figures 4J and 4K). Interestingly, some of the chloroplasts in the TRV:GyrA and TRV:GyrB lines exhibited a dumbbell-shaped morphology with central constriction. This phenotype mimics that of chloroplasts undergoing division, except that the daughter chloroplasts of the TRV:GyrA and TRV:GyrB lines looked abnormal and were asymmetrically sized (Figures 4F and 4J). The general shape and size of most of the mitochondria in the TRV:GyrA and TRV:GyrB lines appeared to be largely unaffected (data not shown). However, some of the mitochondria did exhibit morphological abnormalities, including less pronounced inner cristae, an enlarged size, and an inner region that appeared to be disintegrating (marked by the arrowhead in Figure 4M, cf. control Figure 4L).
Figure 4.
Ultrastructural Analysis of Mesophyll Cell Chloroplasts and Mitochondria.
(A) to (I) Transmission electron micrographs of a leaf mesophyll cell ([A] to [C]), chloroplasts ([D] to [F]), and chloroplast thylakoid membranes ([G] to [I]). TRV control ([A], [D], and [G]) and green ([B], [E], and [H]) and yellow sectors ([C], [F], and [I]) of leaves from the TRV:GyrB line are shown. Note that the TRV:GyrB chloroplast exhibits dumbbell-shaped morphology with central constriction (F).
(J) and (K) Transmission electron micrographs of the dumbbell-shaped (J) and degrading chloroplasts (K) from the TRV:GyrA line.
(L) and (M) Transmission electron micrograph of mesophyll mitochondria from the TRV control (L) and the TRV:GyrB line (M). The arrowhead indicates the region that appears to be disintegrating.
C, chloroplast; S, starch; M, mitochondrion; tm, thylakoid membrane. Bars = 5 μm in (A) to (C), 1 μm in (D) to (F) and (J) to (M), and 0.1 μm in (G) to (I).
Expression of NbGyrA and NbGyrB Transcripts
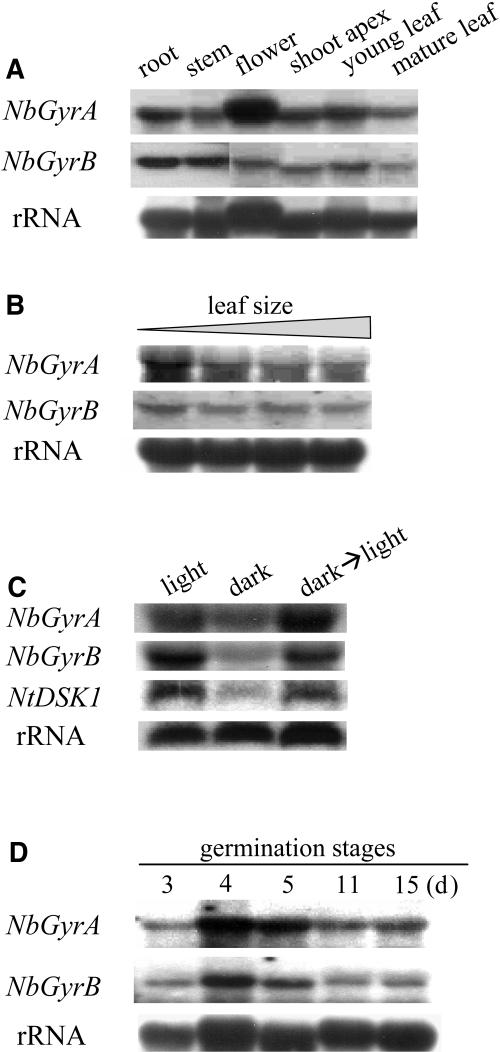
RNA gel blot analyses were performed using the 1.1-kb NbGyrA and the 1.2-kb NbGyrB cDNA fragments as hybridization probes, both of which correspond to the C-terminal ends of the coding regions of these genes. Transcripts of ∼3.4 kb (NbGyrA) and 2.7 kb (NbGyrB) were detected in all of the N. benthamiana tissues examined, including roots, stems, flowers, shoot apex regions, and young and mature leaves (Figure 5A). During leaf development, the NbGyrA and NbGyrB mRNA levels were slightly higher at the early stage than at later stages (Figure 5B). NbGyrA and NbGyrB mRNA levels were also examined in 7-d-old N. benthamiana seedlings grown under normal light conditions (16 h light/8 h dark), under dark conditions, or under dark conditions followed by a transfer to light for 1 h (Figure 5C). The levels of both mRNAs were lower in dark-grown seedlings than in light-grown seedlings, but exposure to light for 1 h increased the levels of both mRNAs to the levels of the light-grown seedlings. As a positive control, NtDSK1 expression in response to light was also monitored. NtDSK1 encodes a chloroplast-targeted dual-specificity kinase that phosphorylates both Tyr and Ser/Thr residues and whose expression is regulated by light (Cho et al., 2001). With regard to NbGyrA and NbGyrB expression during seedling development, the levels of both mRNAs increased during seed germination, particularly 4 d after sowing (Figure 5D). Taken together, these results demonstrate that expression of these subunits is stimulated during seed germination and leaf expansion and in response to light.
Figure 5.
Expression of the NbGyrA and NbGyrB Genes.
RNA gel blot analysis was performed with total RNA from N. benthamiana plants. Each lane represents 30 μg of total RNA.
(A) Expression in various tissues.
(B) Expression in leaves of various sizes. From left, 0.5, 2, 5, and 9 cm.
(C) Light-stimulated expression. Seven-day-old seedlings grown with 16 h light and 8 h dark (light), in the dark (dark), or grown in the dark and then transferred to light for 1 h (dark→light) were used. As a control for the light regulation, the expression of NtDSK1, which encodes a chloroplast-targeted dual-specificity protein kinase (Cho et al., 2001), was examined.
(D) Expression in seedlings at various stages of growth. The transcript levels were measured in germinated seedlings 3 to 15 d after sowing.
Targeting of DNA Gyrase Subunits A and B to Both Chloroplasts and Mitochondria
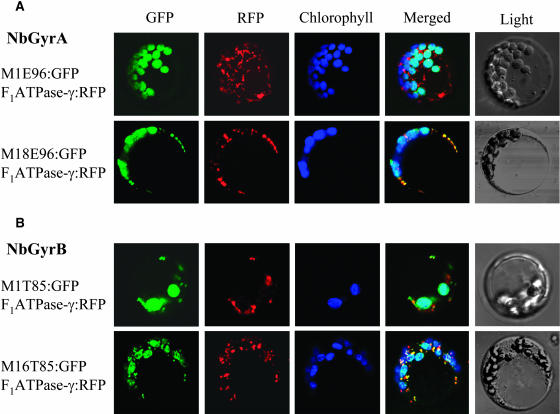
Interestingly, the N-terminal extensions of the two proteins contain two in-frame translation initiation codons. We therefore tested whether NbGyrA and NbGyrB are targeted to both organelles by producing two protein products from the alternative translation start sites. Thus, we generated fusion proteins in which different N-terminal regions of the NbGyrA and NbGyrB proteins are fused to green fluorescent protein (GFP). For NbGyrA, the N-terminal 96–amino acid (Met-1 to Glu-96 [M1E96]) and 79–amino acid (Met-18 to Glu-96 [M18E96]) regions were used. For NbGyrB, the N-terminal 85–amino acid (Met-1 to Thr-85 [M1T85]) and 70–amino acid (Met-16 to Thr-85 [M16T85]) regions were used. The DNA constructs encoding these different forms of NbGyrA-GFP and NbGyrB-GFP under the control of the 35S promoter of Cauliflower mosaic virus were introduced into protoplasts isolated from Arabidopsis seedlings (Figure 6). To track mitochondria, a DNA construct encoding F1ATPase-γ:RFP, a fusion protein of the mitochondrial F1ATPase γ subunit (Lee et al., 2001) and red fluorescent protein (RFP), was cotransformed into the protoplasts. After incubation at 25°C for 24 h, expression of the introduced genes was examined under a confocal laser scanning microscope using different filters to capture the fluorescence of GFP and RFP and chlorophyll autofluorescence.
Figure 6.
Targeting of NbGyrA and NbGyrB to Both Chloroplasts and Mitochondria.
Arabidopsis protoplasts were transformed with the GFP fusion constructs as well as with the F1ATPase-γ:RFP construct to mark the mitochondria, and the localization of the fluorescent signals was examined 24 h after transformation under a confocal laser scanning microscope. Chloroplasts and mitochondria were visualized by chlorophyll autofluorescence and the red fluorescence of F1ATPase-γ:RFP, respectively. The false color (blue) was used for chlorophyll autofluorescence to distinguish it from the red fluorescence of RFP. The merged images of GFP, RFP, and chlorophyll autofluorescence as well as bright-field images are shown. The M18E96:GFP fusion protein of NbGyrA and the M16T85:GFP fusion protein of NbGyrB used the alternative in-frame translation sites.
(A) GFP was fused to the N-terminal peptides of NbGyrA, resulting in M1E96:GFP and M18E96:GFP.
(B) GFP was fused to the N-terminal peptides of NbGyrB, resulting in M1T85:GFP and M16T85:GFP.
For GyrA(M1E96)-GFP, most of the green fluorescent signals overlapped with the chloroplast autofluorescence, demonstrating that the fusion protein was targeted to the chloroplasts, although a minor signal was also detected in the mitochondria (Figure 6A; data not shown). For GyrA(M18E96)-GFP, the green fluorescent signal completely overlapped with both chloroplast autofluorescence and the red fluorescent signal of F1ATPase-γ:RFP, indicating that the protein is targeted to both chloroplasts and mitochondria. These results demonstrate that the N-terminal extension of NbGyrA contains a signal that targets the protein to both chloroplasts and mitochondria. For both GyrB(M1T85)-GFP and GyrB(M16T85)-GFP, the green fluorescent signal overlapped with both chloroplast autofluorescence and the RFP signal of mitochondria (Figure 6B). Thus, both of the N-terminal peptides contain sufficient information for the protein to be targeted to both chloroplasts and mitochondria. Taken together, these data demonstrate that DNA gyrase A and B subunits of N. benthamiana are targeted to both chloroplasts and mitochondria using ambiguous sequences in their N termini that are recognized by the transport machinery of both organelles, instead of by making two protein products from two alternative translation start sites.
Increased DNA Content in Abnormal Chloroplasts and Mitochondria
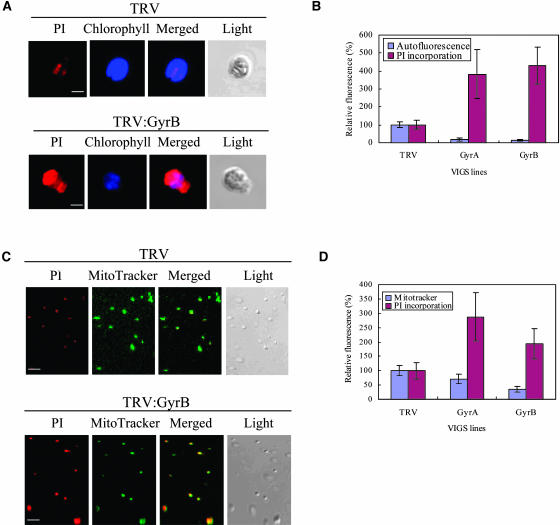
To examine the effects of gene silencing of the DNA gyrase subunits on mitochondrial and chloroplast DNA metabolism, we measured genomic DNA contents in the chloroplasts and mitochondria of TRV, TRV:GyrA, and TRV:GyrB cell lines at the same developmental stage by propidium iodide (PI) staining (Figures 7 to 9). Chloroplasts and mitochondria were isolated from leaf protoplasts and fresh leaves, respectively, PI stained, and subjected to confocal laser scanning microscopy. In seed plants, chloroplast DNA is arranged into DNA/protein complexes called chloroplast nucleoids. The nucleoids of the leaf chloroplasts are dispersed in the stroma as small particles with a diameter of ∼0.2 μm, which are sometimes fused to form a larger particle or a filament (Sato, 2001). PI staining of the TRV control choloroplasts exhibited the typical pattern of nucleoids in mature chloroplasts (Figure 7A, top). However, many chloroplasts from the GyrA- or GyrB-suppressed lines exhibited significantly enlarged nucleoids (Figure 7A, bottom; data not shown) with a much higher intensity (3.7- to 4.3-fold) of PI fluorescence (Figure 7B).
Figure 7.
Increased DNA Content in Abnormal Chloroplasts and Mitochondria.
Chloroplasts isolated from the fourth to sixth leaves above the infiltrated leaf of each VIGS line were stained with PI, and the mitochondria were stained with PI and MG, as described in Methods. The false color (blue) was used for chlorophyll autofluorescence to distinguish it from the red PI fluorescence.
(A) PI staining and chlorophyll autofluorescence images of representative chloroplasts from the TRV (top) and TRV:GyrB (bottom) lines. Bars = 5 μm.
(B) The average PI fluorescence and chlorophyll autofluorescence of individual chloroplasts. PI incorporation in chloroplasts from the TRV:GyrA and TRV:GyrB lines is ∼3.8- to 4.3-fold higher than in the TRV control chloroplasts. The data points represent means ± sd of 30 to 32 selected single chloroplasts.
(C) PI- and MG-staining images of mitochondria from the TRV (top) and TRV:GyrB (bottom) lines. Bars = 10 μm.
(D) The average PI and MG fluorescence of individual mitochondria. PI incorporation in mitochondria from the TRV:GyrA and TRV:GyrB lines was 2.8-fold and twofold higher than in the TRV control mitochondria, respectively. MG incorporation in mitochondria from the TRV:GyrA and TRV:GyrB lines was 69 and 37% of that in the TRV control mitochondria, respectively. The data points represent means ± sd of 60 to 80 selected single mitochondria.
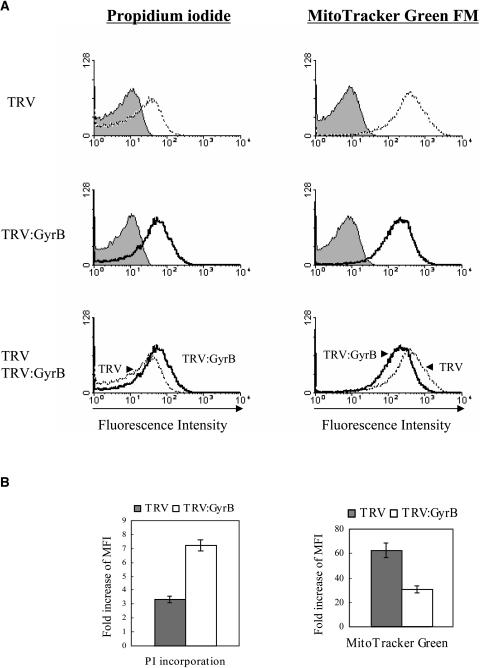
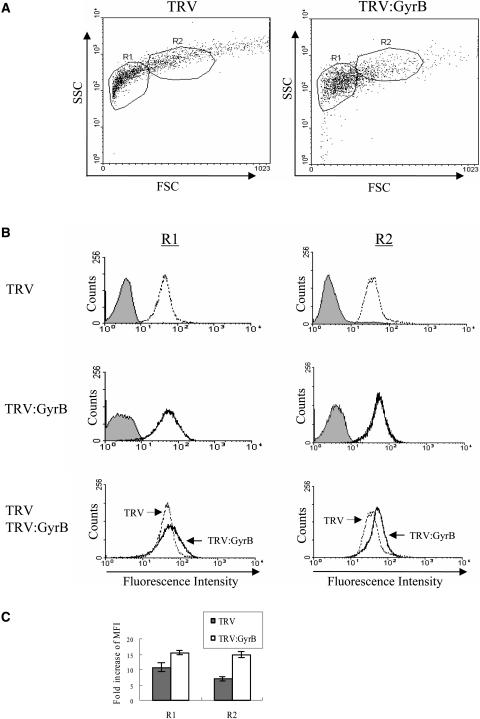
Figure 9.
Flow Cytometry Profiles of Mitochondria.
Twenty thousand mitochondria isolated from the fourth to sixth leaves above the infiltrated leaf of each VIGS line were fixed, stained with PI and MG, and analyzed by flow cytometry.
(A) DNA histogram of mitochondria of the TRV and TRV:GyrB lines. The fluorescence profiles of PI (indicating DNA contents) and MG staining (indicating mitochondrial mass) were analyzed. The filled histogram represents the fluorescence profile before the staining of mitochondria. The mitochondria from the TRV:GyrB line had higher DNA contents but lower masses than those from the TRV control.
(B) Fold increase of MFI compared with the MFI before PI and MG staining. The fold increase of MFI of PI staining in TRV:GyrB is 2.1-fold higher than that of the TRV control, which indicates the TRV:GyrB mitochondria have on average 2.1-fold higher DNA contents than TRV mitochondria. The fold increase of MFI of MG staining in TRV:GyrB is twofold lower than that in the TRV control, which indicates the TRV:GyrB mitochondria have on average a twofold lower mitochondrial mass. The data points represent the means ± sd of three independent experiments.
The mitochondria from each VIGS line were stained with PI and MG and visualized by confocal laser scanning microscopy. Figure 7C shows colocalization of PI and MG fluorescence in representative mitochondria from the TRV (top) and TRV:GyrB (bottom) lines. PI incorporation into mitochondria of the TRV:GyrA and TRV:GyrB lines was ∼2.8-fold and twofold higher compared with that of the TRV control, respectively (Figure 7D). By contrast, MG incorporation into mitochondria from the TRV:GyrA and TRV:GyrB lines was reduced to ∼69 and 37% of that of the TRV control, respectively. These results demonstrate that the affected mitochondria have higher DNA contents but a lower mitochondrial mass than control mitochondria.
Flow Cytometry of Chloroplasts and Mitochondria
Because the degree of gene silencing may vary between cells, defects in DNA metabolism in individual chloroplasts and mitochondria may be heterogeneous. To test this, we used flow cytometry to compare the DNA contents of a large population of chloroplasts (50,000) and mitochondria (20,000) from TRV:GyrB and TRV lines (Figures 8 and 9). Chloroplasts and mitochondria from VIGS lines of the same developmental stage were isolated from leaf protoplasts and fresh leaves, respectively. Light microscopy revealed that the isolated chloroplasts consisted of single chloroplasts and aggregates of two to four chloroplasts. The average PI staining intensity of the chloroplast from the TRV:GyrB line was significantly higher than that of the TRV control, namely, ∼1.5-fold and 2.1-fold higher than that of the TRV control in the R1 (mostly containing single chloroplasts) and R2 (mostly containing aggregates of two to four chloroplasts) regions, respectively (Figure 8C). Furthermore, R1 chloroplasts from the TRV:GyrB line showed a broader flow cytometry profile compared with the control, which indicates that the DNA content of this population of chloroplasts is more heterogenous than that of control chloroplasts.
Figure 8.
Flow Cytometry Profiles of Chloroplasts.
Fifty thousand chloroplasts isolated from the fourth to sixth leaves above the infiltrated leaf of each VIGS line were fixed, stained with PI, and analyzed by flow cytometry.
(A) Light scatter cytogram. FSC and SSC represent forward light scatter and side scatter, respectively. The single chloroplasts and aggregates of two to four chloroplasts are mostly enclosed in the R1 and R2 regions, respectively.
(B) DNA histogram of the R1 and R2 regions of TRV and TRV:GyrB. The filled histogram represents the fluorescence profile before PI staining of the chloroplasts. Chloroplasts from the TRV:GyrB line contain higher DNA contents than the TRV control chloroplasts in both R1 and R2.
(C) Fold increase of mean fluorescence intensity (MFI) in R1 and R2 compared with the MFI before PI staining. The fold increase of MFI in TRV:GyrB is 1.5- and 2.1-fold higher than that in the TRV control in R1 and R2, respectively, which indicates that the DNA contents in the chloroplasts in the R1 and R2 regions are on average 1.5- and 2.1-fold higher, respectively, compared with the DNA contents of TRV control chloroplasts. The data points represent means ± sd of three independent experiments.
Flow cytometry analysis of mitochondria was performed using PI and MG staining (Figure 9). As with chloroplasts, DNA contents in TRV:GyrB mitochondria were higher than in TRV mitochondria (Figure 9B). The fold increase in mean fluorescence intensity (MFI) of PI-stained TRV:GyrB mitochondria was 2.1-fold higher than that of TRV control mitochondria, which indicates that the TRV:GyrB mitochondria have on average 2.1-fold higher DNA contents than mitochondria of the TRV control. The fold increase of MFI of the MG-stained TRV:GyrB mitochondria was twofold lower than that of TRV control mitochondria, which indicates that TRV:GyrB mitochondria have on average twofold lower mitochondrial masses than TRV mitochondria (Figure 9B). These results indicate that depletion of DNA gyrase leads to increased DNA content in chloroplasts and mitochondria.
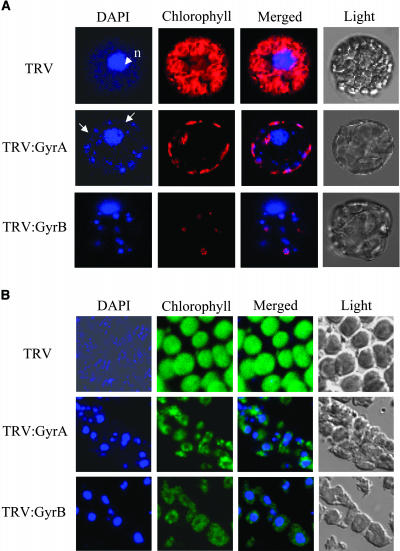
DAPI Staining of Chloroplast Nucleoids
Leaf protoplasts were stained with DAPI and examined under a confocal laser scanning microscope (Figure 10A). In the TRV control, the nucleus was prominent with faint staining of the cytosol. However, TRV:GyrA and TRV:GyrB lines exhibited a large number of fluorescent spots of variable sizes in the cytosol in addition to the nucleus. The spots probably represent individual or aggregated populations of chloroplasts and mitochondria. These results suggest that the genomic DNA contents in organelles were increased. Individual chloroplasts and chloroplast nucleoids were visualized by squashing the DAPI-stained protoplasts (Figure 10B). The TRV control chloroplasts exhibited moderate numbers of chloroplast nucleiods that were randomly distributed as small particles in the stroma (Figure 10B). However, many chloroplasts of TRV:GyrA and TRV:GyrB lines contained one or only a few enlarged nucleoids that had significantly increased fluorescence. By contrast, some chloroplasts of TRV:GyrA and TRV:GyrB lines showed weak or no DAPI staining, which indicates that they contained little or no chloroplast DNA. It has been previously reported that chloroplast nucleoids in seed plants grow in size in parallel with chloroplast growth and that they divide just before chloroplastkinesis, thereby ensuring the equal segregation of daughter chloroplast nucleoids into daughter chloroplasts (Kuroiwa et al., 1981; Kuroiwa, 1991). Thus, the size, number, and distribution of chloroplast nucleoids in gyrase VIGS lines drastically deviate from the normal pattern. These enlarged chloroplast nucleoids are probably caused by defects in chloroplast nucleoid division that result from impaired chloroplast DNA replication and partitioning under conditions of reduced DNA gyrase activity.
Figure 10.
DAPI Staining of Organelle Genomes.
(A) DAPI staining of protoplasts isolated from the fourth leaf above the infiltrated leaf of TRV, TRV:GyrA, and TRV:GyrB VIGS lines. Fluorescent signals of DAPI and chlorophyll were examined under a confocal laser scanning microscope. Arrows indicate the cytosolic fluorescent spots that probably represent individual and aggregated organelles. n, nucleus.
(B) DAPI staining of individual chloroplasts from TRV, TRV:GyrA, and TRV:GyrB VIGS lines.
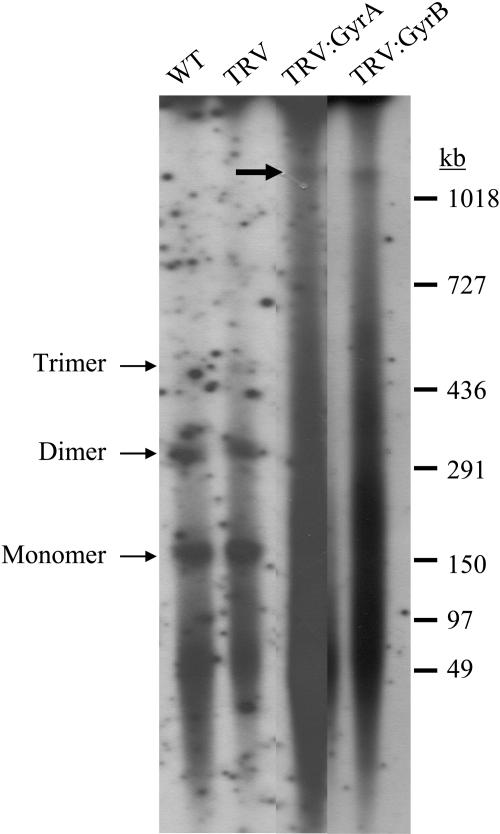
Pulse-Field Gel Electrophoresis Analysis of Chloroplast DNA
We used pulse-field gel electrophoresis (PFGE) to estimate the sizes of chloroplast DNA molecules in leaf chloroplasts isolated from uninfected N. benthamiana plants (wild type), TRV, TRV:GyrA, and TRV:GyrB VIGS lines (Figure 11). Hybridization of the rbcL gene probe to DNA gel blots from PFGE revealed two prominent bands that correspond to monomeric and dimeric chloroplast DNA in the wild-type and TRV controls, which is consistent with previous observations of tobacco chloroplasts (Lilly et al., 2001). Significant hybridization signals below the monomeric band may represent both sheared chloroplast DNA and possibly molecules of subgenomic size (Lilly et al., 2001). By contrast, PFGE analysis of chloroplasts from TRV:GyrA and TRV:GyrB lines using the rbcL probe generated smears. No distinct monomeric or dimeric chloroplast DNA bands were observed, which indicates that the sizes and/or structures of chloroplast DNA molecules are highly variable. In addition, a distinct band corresponding to chloroplast DNA of >1000 kb was observed in gyrase VIGS lines (large arrow in Figure 11). These multimeric and/or atypical molecules in TRV:GyrA and TRV:GyrB lines may represent randomly cleaved or fused DNA molecules that result from the entanglement of unresolved replication intermediates in abnormal chloroplast nucleoids. Similar results were obtained from PFGE analyses with the rpoA gene probe (data not shown).
Figure 11.
PFGE Analysis of Chloroplast DNA.
PFGE of chloroplast DNA was performed as described (Lilly et al., 2001). The membrane was hybridized with the radiolabeled rbcL gene probe. Small arrows indicate the bands representing monomeric, dimeric, and trimeric molecules of chloroplast DNA. The big arrow indicates a large chloroplast DNA band (>1000 kb). DNA size markers are indicated in kilobases.
DISCUSSION
We show in this article that N. benthamiana DNA gyrase subunits are functionally active as DNA gyrase subunits in E. coli because their expression rescues the temperature-sensitive lethal phenotypes of E. coli gyrA and gyrB mutants. We also found that the two DNA gyrase subunits of N. benthamiana are targeted to both chloroplasts and mitochondria and that the disruption of either DNA gyrase subunit by VIGS severely affected the morphology of both organelles. The abnormal chloroplasts and mitochondria had higher DNA contents compared with the TRV control organelles. Furthermore, the abnormal chloroplasts contained one or only a few large chloroplast nucleoids instead of multiple small nucleoids randomly dispersed throughout the chloroplast, indicating defective nucleoid division. These loss-of-function phenotypes in planta and the capability of the plant gyrase subunits to substitute for their E. coli counterparts strongly suggest that DNA gyrase plays an essential role in plant cells by organizing DNA topology during DNA replication and partitioning in chloroplasts and possibly also in mitochondria.
Phylogenic and developmental differences in the morphology of chloroplast nucleoids have been reported (Kuroiwa, 1991). Proplastids in meristemic cells contain a single ovoid chloroplast nucleoid in their center (Sato, 2001). In developing chloroplasts, the chloroplast nucleoid is extensively replicated while attached to the envelope membrane before dividing into multiple small chloroplast nucleoids. In the mature chloroplast, chloroplast nucleoids are dispersed within the stroma as small particles mostly associated with thylakoids. In most higher plants, including Nicotiana species, the estimated number of chloroplast nucleoids in mature chloroplasts ranges from 8 to 40 (Kuroiwa et al., 1981), and each nucleoid is estimated to contain ∼10 copies of chloroplast DNA (Kuroiwa, 1991). However, the detailed mechanisms of chloroplast genome replication through the chloroplast nucleoid structure and the mechanism by which chloroplast nucleoids divide and become distributed within a chloroplast are still unknown.
This study reveals that reduced DNA gyrase activity in chloroplasts results in distinctive nucleoid-partitioning defects. In E. coli, a large number of conditional lethal mutations that cause defects in the segregation of bacterial nucleoids have been identified. These are designated as partition (par) mutants, and most map to genes coding for DNA gyrases or topoisomerases. Loss-of-function mutations of parD (gyrase A), parA (gyrase B), parC (topoisomerase IVA), and parE (topoisomerase IVB) all result in large nucleoid masses in the middle of filamentous cells (Kato et al., 1990; Sun and Margolin, 2001). Conditional lethality caused by mutations in these Par genes is also associated with incomplete replication. Thus, the par− phenotype appears to result from combined defects in replication elongation, replication termination, and daughter chromosome segregation. Consistent with this, the structure of chloroplast DNA in the large chloroplast nucleoids of the gyrase VIGS lines appears to be highly aberrant, including very large molecules (Figure 11). These multimeric and aberrantly sized molecules may result from random cleavage and fusion of unresolved replication intermediates that accumulate in chloroplasts under conditions of reduced DNA gyrase activity.
Interestingly, some chloroplasts in TRV:GyrA and TRV:GyrB lines exhibited tadpole-like doublet shapes with a central constriction (Figure 4). Similar phenotypes are observed in the gyrase mutants of E. coli, where the inhibition of gyrase leads to a 3.5-fold increase in the percentage of nucleoids possessing a doublet morphology (Steck and Drlica, 1984). Furthermore, elongated cells of an E. coli gyrase mutant bud off DNA-less minicells, indicating that unsegregated chromosomes may block normal division and force the cells to use secondary division sites (Orr et al., 1979). More recently, it has been revealed that the nucleoid indeed negatively regulates cell division by preventing the formation of the FtsZ ring (Margolin, 2001). The nucleoid inhibits FtsZ ring formation at the regions it occupies (nucleoid occlusion), which normally includes the entire length of the cell except for the cell poles and the region between the newly segregated nucleoids. Thus, mutations such as gyrA (parD), gyrB (parA), minB, parC, and parE, which delay the proper separation of the nucleoids, delay and/or misplace cell division (Nordström and Dasgupta, 2001). It has been further revealed that the chromosome-partitioning defects of E. coli gyrase mutants are mainly caused by defects in superhelicity and condensation of the nucleoid rather than disrupted decatenation of daughter chromosomes. This indicates that DNA compaction by supercoiling is critical for chromosome partitioning in prokaryotes (Holmes and Cozzarelli, 2000). It is tempting to speculate that the chloroplasts in the gyrase VIGS lines may undergo asymmetric division because the nucleoids fail to segregate at the cell center and cell division is forced to occur at the cell periphery. As a result of this, some progeny may have incomplete or missing chromosomes. Consistent with this, some of the chloroplasts in the gyrase VIGS lines showed little or no DAPI staining (Figure 10).
Animal mitochondria possess type IB DNA topoisomerase (Tua et al., 1997; Zhang et al., 2001). Several studies have also indicated the presence of a type IIA DNA topoisomerase in the mitochondria of several organisms, the strongest evidence coming from the parasite Trypanosoma. Inhibition of trypanosomal DNA topoisomerase II by RNA interference led to shrinkage and loss of the kinetoplast DNA network in the mitochondria and cessation of trypanosome growth (Wang et al., 2002). Our study demonstrates that plants are unique among eukaryotic organisms in using bacterial DNA gyrase for DNA metabolism in mitochondria. Intriguingly, topoisomerase VI, a unique type II topoisomerase found in archaebacteria but not in most eukaryotic phyla, was recently found to play an important role in nuclear DNA replication and gene expression in Arabidopsis (Corbett and Berger, 2003). These unique features of DNA topoisomerases in plants may have been gained through horizontal gene transfer from prokaryotes after the evolutionary diversion of plants, or alternatively, they might have been lost in the common ancestor of yeast and animals.
With respect to the morphological changes associated with defective gyrase activity, the data we have presented here are mostly consistent with the results of a recently published article that analyzed Arabidopsis DNA gyrase (Wall et al., 2004). However, the mechanism by which the gyrase subunits in N. benthamiana are targeted to the organelles appears to differ from that in Arabidopsis. In N. benthamiana, NbGyrA and NbGyrB are both targeted to chloroplasts and mitochondria by a single transit peptide. Thus, the transit peptides of NbGyrA and NbGyrB apparently belong to a group of transit peptides that are denoted as ambiguous targeting signals. The sequences of these targeting signals are intermediate in character between mitochondrial and chloroplast targeting sequences and are recognized by the transport machinery of both organelles. By contrast, Arabidopsis GyrA protein seems to be targeted to both chloroplasts and mitochondria using alternative translation initiation sites in a protein that contains in tandem a mitochondrial and a chloroplast targeting sequence at its N terminus (Wall et al., 2004). With regard to Arabidopsis GyrB, two of the GyrB homologs possess unambiguous targeting sequences that determine their chloroplast or mitochondrial targeting (Wall et al., 2004). This discrepancy may suggest that the organelle targeting mechanism for DNA gyrase subunits may be species specific. Interestingly, GyrA in rice (Oryza sativa) (GenBank accession number AAP78360) does not have two in-frame translation start sites in its N extension, which indicates that, like NbGyrA and NbGyrB, its targeting to both organelles probably involves ambiguous signals.
We demonstrated in this study that DNA gyrase activity is critically required for nucleoid partitioning in chloroplasts. Increased DNA contents in both chloroplasts and mitochondria in the gyrase VIGS lines are probably caused by the defects in organelle genome partitioning. Defective chloroplast nucleoid partitioning appeared to affect chloroplast division in the gyrase VIGS lines, as shown by the dumbbell-shaped chloroplasts and chloroplasts with little DNA, and this was probably responsible for the reduction in chloroplast numbers. These results also suggest that bacterial and plant organelles share conserved mechanisms of DNA replication and partitioning. Plant DNA gyrase may be involved in maintaining negative supercoiling of organelle DNA, in relaxing positive supercoils ahead of the replication fork, and in DNA condensation necessary for proper genome partitioning during organelle division.
METHODS
VIGS
The various cDNA fragments of NbGyrA and NbGyrB were PCR amplified and cloned into the pTV00 vector containing part of the TRV genome (Ratcliff et al., 2001) using BamHI and ApaI sites. To generate the TRV:TopoI VIGS line, a 0.6-kb Nicotiana benthamiana cDNA fragment encoding a homolog of Arabidopsis thaliana topoisomerase I (Takahashi et al., 2002) was cloned into the pTV00 vector using BamHI and ApaI sites. The recombinant pTV00 plasmids and the pBINTRA6 vector containing RNA1, which is required for virus replication, were separately transformed into Agrobacterium tumefaciens GV3101. The Agrobacterium culture was prepared for infiltration as described by Ratcliff et al. (2001). The third leaf of N. benthamiana (3 weeks old) was then pressure infiltrated with the Agrobacterium suspension. For RT-PCR, the fourth leaf above the infiltrated leaf was used.
5′-RACE PCR of NbGyrA and NbGyrB
The partial NbGyrA and NbGyrB cDNAs were 1.5 and 1.2 kb in length, respectively. To obtain the full-length NbGyrA and NbGyrB cDNAs, 5′-RACE PCR reactions were performed using the SMART RACE cDNA amplification kit (BD Biosciences, Franklin Lakes, NJ) according to the manufacturer's instructions. In total, 1.9- and 1.5-kb 5′ cDNA ends of NbGyrA and NbGyrB, respectively, were synthesized from mRNAs isolated from N. benthamiana seedlings. The gene-specific primers for NbGyrA and NbGyrB are 5′-TTCAGGGATCTT GTAAGCCGGAGAAG-3′ and 5′-CTCTCTTACATGCTCGCCACTTAAGC-3′, respectively.
RT-PCR and RNA Gel Blot Analysis
Total RNA was prepared using TRIzol reagent (Gibco BRL, Cleveland) according to the manufacturer's instructions. RT-PCR was performed with 5 μg of total RNA as described (Lee et al., 2003). The primers for RT-PCR were as follows. To detect the endogenous NbGyrA transcripts, the primers 5′-GAAGGCTTACTGCACTTGA-3′ and 5′-TGATGGCGAGAGAATAACT-3′ for the TRV:GyrA(1) and TRV:GyrA(3) lines and 5′-AAGTCGCTAGCATGGACA-3′ and 5′-GGTATGCGAGAGGAGTAA-3′ for the TRV:GyrA(2) line were used. To detect the endogenous NbGyrB transcripts, the primers 5′-CAGCGAGCATGATGAA-3′ and 5′-GACAGAGCGCACAAAG-3′ were used. For RNA gel blot analysis, ∼30 μg of total RNA was electrophoresed on an agarose gel containing 5.1% (v/v) formaldehyde and blotted onto a Zeta-probe GT membrane (Bio-Rad, Hercules, CA). Prehybridization and hybridization were performed according to the manufacturer's instructions. The 1.1-kb NbGyrA and 1.2-kb NbGyrB cDNA fragments were used as probes.
Transmission Electron Microscopy
For transmission electron microscopy, the fourth leaf above the infiltrated leaf was collected from the TRV, TRV:GyrA, and TRV:GyrB lines. It was fixed first with 2.5% (v/v) glutaraldehyde and then 1% osmium tetraoxide, followed by dehydration with an ethanol series and embedding in Spurr's resin (EM Sciences, Gibbstown, NJ). Thin sections were made with the LKB III ultramicrotome (Leica, Deerfield, IL) and stained sequentially with 5% uranyl acetate and 3% lead citrate and then observed under a JEOL 1200 EXII transmission electron microscope (Tokyo, Japan).
Confocal Laser Scanning Microscopy
To stain mitochondria with MG, protoplasts isolated from the fourth leaf above the infiltrated leaf of the VIGS lines were resuspended in an appropriate volume of TEX buffer (B5 salts, 500 mg/L of Mes, 750 mg/L of CaCl2·2H2O, 250 mg/L of NH4NO3, and 0.4 M sucrose, pH 5.7). The protoplasts were then fixed with 0.5% glutaraldehyde in TEX buffer for 30 min, washed twice with TEX buffer by centrifugation at 60g for 10 min, and stained with 200 nM MG (Molecular Probes, Eugene, OR) to visualize the mitochondria. The samples were observed under a confocal microscope (Carl Zeiss LSM 510; Jena, Germany) with optical filters BP 505-530 (excitation 488 nm, emission 505 to 530 nm) and LP650 (excitation 488 nm, emission 650 nm) (Carl Zeiss) to detect the green MG fluorescence and the autofluorescence of chlorophyll, respectively.
For DAPI staining, protoplasts isolated from the fourth leaf above the infiltrated leaf were fixed with 0.5% glutaraldehyde in TEX buffer for 30 min, washed twice with TEX buffer by centrifugation at 60g for 10 min, and stained with 1 μg/mL of DAPI (Molecular Probes) in TEX buffer for 10 min. After staining, the protoplasts were washed twice with TEX buffer to remove the unbound dye. The samples were observed under a confocal microscope (Carl Zeiss LSM 510) with the optical filter LP420 (excitation 359 nm, emission 441 nm) to visualize the nuclear and organelle genomes. To examine chloroplast DNA, the DAPI-stained protoplasts were gently squashed under a cover slide.
Subcellular Localization of NbGyrA and NbGyrB
The NbGyrA cDNA fragments corresponding to the N-terminal 96–amino acid (residues 1 to 96) and 79–amino acid (residues 18 to 96) regions were cloned into the 326-GFP plasmid (Lee et al., 2001) using BamHI sites to generate the NbGyrA-GFP fusion proteins. The NbGyrB cDNA fragments corresponding to the N-terminal 85–amino acid (residues 1 to 85) and 70–amino acid (residues 16 to 85) regions of NbGyrB were cloned into the same vector to produce the NbGyrB-GFP fusion proteins. The various GFP fusion constructs and the F1ATPase-γ:RFP fusion construct were introduced into Arabidopsis protoplasts prepared from whole seedlings by polyethylene glycol–mediated transformation (Lee et al., 2001). Expression of the fusion constructs was monitored 24 h after transformation by confocal laser scanning microscopy (Carl Zeiss LSM 510). The filter sets used were BP505-530 (excitation 488 nm, emission 505 to 530 nm), BP560-615 (excitation 543 nm, emission 560 to 615 nm), and LP650 (excitation 488 nm, emission 650 nm) (Carl Zeiss) to detect GFP, RFP, and the chlorophyll autofluorescence, respectively.
Flow Cytometry
Chloroplasts were prepared from protoplasts isolated from the fourth to sixth leaves above the infiltrated leaf of the VIGS lines as described (Slaymaker et al., 2002), and mitochondria were prepared from the same leaves as described (Douce et al., 1987). For flow cytometry, chloroplasts and mitochondria were fixed in 1% formaldehyde for 30 min on ice and washed briefly with MB buffer (300 mM sucrose, 1 mM EDTA, 0.1% BSA [defatted], and 10 mM potassium phosphate, pH 7.2). The organelles (at a concentration of 4 to 5 × 106/mL) were then treated with RNase (0.1 mg/mL) for 30 min at room temperature and stained with PI (25 μg/mL for chloroplasts and 50 μg/mL for mitochondria) for 1 h at room temperature, followed by one wash for 10 min with MB buffer. During PI staining, the mitochondria were simultaneously stained with 200 nM MG (Molecular Probes). PI incorporation into DNA was quantified in the FL-2 channel, and MG incorporation into mitochondria was quantified in the FL-1 channel of FACSCalibur (BD Biosciences) employing the Win MDI version 2.8 program (Scripps Research Institute, La Jolla, CA).
Complementation of Escherichia coli gyrA and gyrB Mutants
Complementation analysis was performed with the E. coli strain KNK452 [F− gyrA42(ts) polA0 thyA45 uvrA103 deo-225] (E. coli Genetic Stock Center no. 6971) (Soussy et al., 1993), which carries a temperature-sensitive gyrA mutation, and with strain N4177 [F− gyrB203(ts) strA galK gyrB221(couR)] (E. coli Genetic Stock Center no. 6462) (Gross et al., 2003), which carries a temperature-sensitive gyrB mutation. The NbGyrA and NbGyrB cDNA fragments encoding the full-length NbGyrA and NbGyrB proteins but lacking the N-terminal extensions were cloned into the pBAD18 vector to generate pBAD-NbGyrA and pBAD-NbGyrB. As controls, the gyrA and gyrB cDNAs from E. coli were also cloned into pBAD18 to produce pBAD-EcGyrA and pBAD-EcGyrB, respectively. KNK452 and N4177 cells were transformed with the pBAD18 vector control or the recombinant plasmids carrying the E. coli and plant subunit genes and selected on NZY medium (supplemented with 10μg/L of thymine) containing ampicillin (50 mg/L), streptomycin (30 mg/L, for GyrB 4771 mutant only), and 0.2% glucose at 28°C. The transformants were then grown on NZY media (supplemented with 10 μg/L of thymine) containing glucose (0.2%) or arabinose (0.0002, 0.002, 0.02, and 0.2%) and ampicillin (50 mg/L).
To test cell growth, single colonies of the various transformants were inoculated into 3 mL of NZY liquid medium (+ 0.2% glucose) and grown overnight at 28°C with shaking (250 rpm) to an OD590 exceeding 1.0. The cells were then precipitated and resuspended in NZY broth (without glucose) to an OD590 of 0.5. Five microliters of this cell suspension was then plated on NZY agar media (+ 0.2% glucose, or + 0.0002, 0.002, 0.02, or 0.2% arabinose) and grown for 2 d at 28 and 38°C. To examine cell morphology, overnight cultures of the E. coli gyrA and gyrB mutants and their various transformants grown in NZY broth (+ 0.2% glucose) at 28°C were diluted 1:100 in NZY broth (+ arabinose) and incubated at 38°C for 10 h. To visualize the cell shape, bacterial preparations were stained with DAPI and examined under a fluorescence microscope (40×).
PFGE Analysis
Chloroplasts were prepared from protoplasts isolated from the fourth to sixth leaves above the infiltrated leaf of the VIGS lines as described (Slaymaker et al., 2002). Chloroplast embedding and PFGE were performed as described (Lilly et al., 2001). PFGE was performed using Chef Mapper III (Bio-Rad), 1% gels, 0.5× TBE buffer cooled to 12°C, and conditions of 5- to 120-s pulses at 4.5 V/cm with a total run time of 46 h. After PFGE, the DNA was transferred to a Zetaprobe GT membrane (Bio-Rad) according to the manufacturer's instructions. The labeling of probe DNA, membrane hybridization, and autoradiography was performed according to Lilly et al. (2001). The rbcL and rpoA cDNAs were used as probes.
Sequence data for the full-length sequences of NbGyrA and NbGyrB have been deposited with the EMBL/GenBank data libraries under accession numbers AY351386 and AY351387, respectively.
Acknowledgments
The authors wish to thank David C. Baulcombe (John Innes Center, Norwich, UK) for providing VIGS vectors, Zee Won Lee (Korea Basic Science Institute, Taejon, Korea) for technical help with confocal scanning microscopy, and Un-Ho Jin (Dong-A University, Busan, Korea) for performing the VIGS screening. This research was supported by grants from the Plant Diversity Research Center of the 21st Century Frontier Research Program and the Molecular and Cellular BioDiscovery Research Program, which are funded by the Ministry of Science and Technology of the Korean government.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Hyun-Sook Pai (hspai@mju.ac.kr).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.024281.
References
- Blanc-Potard, A.-B., and Bossi, L. (1994). Phenotypic suppression of DNA gyrase deficiencies by a deletion lowering the gene dosage of a major tRNA in Salmonella typhimurium. J. Bacteriol. 176, 2216–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux, J.J. (2001). DNA topoisomerases: Structure, function, and mechanism. Annu. Rev. Biochem. 70, 369–413. [DOI] [PubMed] [Google Scholar]
- Cho, H.S., Yoon, G.M., Lee, S.S., Kim, Y.A., Hwang, I., Choi, D., and Pai, H.S. (2001). A novel dual-specificity protein kinase targeted to the chloroplast in tobacco. FEBS Lett. 497, 124–130. [DOI] [PubMed] [Google Scholar]
- Corbett, K.D., and Berger, J.D. (2003). Emerging roles for plant topoisomerase VI. Chem. Biol. 10, 107–111. [DOI] [PubMed] [Google Scholar]
- Douce, R., Bourguignon, J., Brouquisse, R., and Neuburger, M. (1987). Isolation of plant mitochondria. Methods Enzymol. 148, 403–415. [Google Scholar]
- Elo, A., Lyznik, A., Gonzalez, D.O., Kachman, S.D., and Mackenzie, S.A. (2003). Nuclear genes that encode mitochondrial proteins for DNA and RNA metabolism are clustered in the Arabidopsis genome. Plant Cell 15, 1619–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O., and von Heijne, G. (2001). Prediction of organellar targeting signals. Biochim. Biophys. Acta 1541, 114–119. [DOI] [PubMed] [Google Scholar]
- Gross, C.H., Parsons, J.D., Grossman, T.H., Charifson, P.S., Bellon, S., Jernee, J., Dwyer, M., Chambers, S.P., Markland, W., Botfield, M., and Raybuck, S.A. (2003). Active-site residues of Echerichia coli DNA gyrase required in coupling ATP hydrolysis to DNA supercoiling and amino acid substitutions leading to novobiocin resistance. Antimicrob. Agents Chemother. 47, 1037–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood, J.L., Tonti-Filippini, J.S., Gout, A.M., Day, D.A., Whelan, J., and Millar, A.H. (2004). Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16, 241–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinhorst, S., Cannon, G., and Weissbach, A. (1985). Chloroplast DNA synthesis during the cell cycle in cultured cells of Nicotiana tabacum: Inhibition of nalidixic acid and hydroxylurea. Arch. Biochem. Biophys. 239, 475–479. [DOI] [PubMed] [Google Scholar]
- Holmes, V.F., and Cozzarelli, N.R. (2000). Closing the ring: Links between SMC proteins and chromosome partitioning, condensation, and supercoiling. Proc. Natl. Acad. Sci. USA 97, 1322–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, J., Nishimura, Y., Imamura, R., Niki, H., Hiraga, S., and Suzuki, H. (1990). New topoisomerase essential for chromosome segregation in E. coli. Cell 63, 393–404. [DOI] [PubMed] [Google Scholar]
- Kim, M., Ahn, J.-W., Jin, O., Paek, K.-H., and Pai, H.-S. (2003). Activation of the programmed cell death pathway by inihibition of proteasome function in plants. J. Biol. Chem. 278, 19406–19415. [DOI] [PubMed] [Google Scholar]
- Kuroiwa, T. (1991). The replication, differentiation, and inheritance of plastids with emphasis on the concept of organelle nuclei. Int. Rev. Cytol. 128, 1–62. [Google Scholar]
- Kuroiwa, T., Suzuki, T., Ogawa, T., and Kawano, S. (1981). The chloroplast nucleus: Distribution, number, size and shape, and a model for the multiplication of the chloroplast genome during chloroplast development. Plant Cell Physiol. 22, 381–396. [Google Scholar]
- Lam, E., and Chua, N.-H. (1987). Chloroplast DNA gyrase and in vitro regulation of transcription by template topology and novobiocin. Plant Mol. Biol. 8, 415–424. [DOI] [PubMed] [Google Scholar]
- Lee, S.S., Cho, H.S., Yoon, G.M., Ahn, J.-W., Kim, H.H., and Pai, H.-S. (2003). Interaction of NtCDPK1 calcium-dependent protein kinase with NtRpn3 regulatory subunit of the 26S proteasome in Nicotiana tabacum. Plant J. 33, 825–840. [DOI] [PubMed] [Google Scholar]
- Lee, Y.J., Kim, D.H., Kim, Y.-W., and Hwang, I. (2001). Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell 13, 2175–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, C., Hiasa, H., and Marians, K.J. (1998). DNA gyrase and topoisomerase IV: Biochemical activities and physiological roles during chromosome replication, and drug sensitivities. Biochim. Biophys. Acta 1400, 29–43. [DOI] [PubMed] [Google Scholar]
- Lilly, J.W., Havey, M.J., Jackson, S.A., and Jiang, J. (2001). Cytogenomic analyses reveal the structural plasticity of the chloroplast genome in higher plants. Plant Cell 13, 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin, W. (2001). Spatial regulation of cytokinesis in bacteria. Curr. Opin. Microbiol. 4, 647–652. [DOI] [PubMed] [Google Scholar]
- Mills, W.R., Reeves, M., Fowler, D.L., and Capo, S.F. (1989). DNA synthesis in chloroplasts. J. Exp. Bot. 40, 425–429. [Google Scholar]
- Nordström, K., and Dasgupta, S. (2001). Partitioning of the Escherichia coli chromosome: Superhelicity and condensation. Biochimie 83, 41–48. [DOI] [PubMed] [Google Scholar]
- Orr, E., Fairweather, N.F., Holland, I.B., and Pritchard, R.H. (1979). Isolation and characterization of a strain carrying a conditional lethal mutation in the cou gene of Escherichia coli K-12. Mol. Gen. Genet. 177, 103–112. [DOI] [PubMed] [Google Scholar]
- Osteryoung, K.W., and Nunnari, J. (2003). The division of endosymbiotic organelles. Science 302, 1698–1704. [DOI] [PubMed] [Google Scholar]
- Oubrahim, H., Stadtman, E.R., and Chock, P.B. (2001). Mitochondria play no roles in Mn(II)-induced apoptosis in HeLa cells. Proc. Natl. Acad. Sci. USA 98, 9505–9510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra, R.J., Thompson, W.A., and Kriedemann, P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975, 384–394. [Google Scholar]
- Ratcliff, F., Martin-Hernansez, A.M., and Baulcombe, D.C. (2001). Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25, 237–245. [DOI] [PubMed] [Google Scholar]
- Sato, N. (2001). Was the evolution of plastid genetic machinery discontinuous? Trends Plant Sci. 6, 151–155. [DOI] [PubMed] [Google Scholar]
- Slaymaker, D.H., Navarre, D.A., Clark, D., del Pozo, O., Martin, G.B., and Klessig, D.F. (2002). The tobacco salicylic acid-binding protein 3 (SABP3) is the chloroplast carbonic anhydrase, which exhibits antioxidant activity and plays a role in the hypersensitive defense response. Proc. Natl. Acad. Sci. USA 99, 11640–11645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soussy, C.J., Wolfson, J.S., Ng, E.Y., and Hooper, D.C. (1993). Limitations of plasmid complementation test for determination of quinolone resistance due to changes in the gyrase A protein and identification of conditional quinolone resistance locus. Antimicrob. Agents Chemother. 37, 2588–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steck, T.R., and Drlica, K. (1984). Bacterial chromosome segregation: Evidence for DNA gyrase involvement in decatenation. Cell 36, 1081–1088. [DOI] [PubMed] [Google Scholar]
- Sun, Q., and Margolin, W. (2001). Influence of the nucleoid on placement of FtsZ and MinE rings in Escherichia coli. J. Bacteriol. 183, 1413–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, T., Matsuhara, S., Mitsumoto, A., and Komeda, Y. (2002). Disruption of a DNA topoisomerase I gene affects morphogenesis in Arabidopsis. Plant Cell 14, 2085–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tua, A., Wang, J., Kulpa, V., and Wernette, C.M. (1997). Mitochondrial DNA topoisomerase I of Saccharomyces cerevisiae. Biochimie 79, 341–350. [DOI] [PubMed] [Google Scholar]
- Wall, M.K., Mitchenall, L.A., and Maxwell, A. (2004). Arabidopsis thaliana DNA gyrase is targeted to chloroplasts and mitochondria. Proc. Natl. Acad. Sci. USA 101, 7821–7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Drew, M.E., Morris, J.C., and Englund, P.T. (2002). Asymmetric division of the kinetoplast DNA network of the trypanosome. EMBO J. 21, 4998–5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J.C. (1996). DNA topoisomerases. Annu. Rev. Biochem. 65, 635–692. [DOI] [PubMed] [Google Scholar]
- Waterhouse, P.M., Wang, M.-B., and Lough, T. (2001). Gene silencing as an adaptive defence against viruses. Nature 411, 834–842. [DOI] [PubMed] [Google Scholar]
- Zechiedrich, E.L., Khodursky, A.B., Bachellier, S., Schneider, R., and Chen, D. (2000). Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J. Biol. Chem. 275, 8103–8113. [DOI] [PubMed] [Google Scholar]
- Zhang, H., Barcelo, J.M., Lee, B., Kohlhagen, G., Zimonjic, D.B., Popescu, N.C., and Pommier, Y. (2001). Human mitochondrial topoisomerase I. Proc. Natl. Acad. Sci. USA 98, 10608–10613. [DOI] [PMC free article] [PubMed] [Google Scholar]