Abstract
The modern patient is increasingly susceptible to bacterial infections including those due to multidrug-resistant organisms (MDROs). Noninvasive whole-body analysis with pathogen-specific imaging technologies can significantly improve patient outcomes by rapidly identifying a source of infection and monitoring the response to treatment, but no such technology exists clinically. Methods: We systematically screened 961 random radiolabeled molecules in silico as substrates for essential metabolic pathways in bacteria, followed by in vitro uptake in representative bacteria—Staphylococcus aureus, Escherichia coli, Pseudomonas aeruginosa, and mycobacteria. Fluorine-labeled analogs, that could be developed as PET-based imaging tracers, were evaluated in a murine myositis model. Results: We identified 3 novel, nontoxic molecules demonstrating selective bacterial uptake: para-aminobenzoic acid (PABA), with uptake in all representative bacteria including Mycobacterium tuberculosis; mannitol, with selective uptake in S. aureus and E. coli; and sorbitol, accumulating only in E. coli. None accumulated in mammalian cells or heat-killed bacteria, suggesting metabolism-derived specificity. In addition to an extended bacterial panel of laboratory strains, all 3 molecules rapidly accumulated in respective clinical isolates of interest including MDROs such as methicillin-resistant S. aureus, extended-spectrum β-lactamase–producing, and carbapenem-resistant Enterobacteriaceae. In a murine myositis model, fluorine-labeled analogs of all 3 molecules could rapidly detect and differentiate infection sites from sterile inflammation in mice (P = 0.03). Finally, 2-deoxy-2-[F-18]fluoro-d-sorbitol (18F-FDS) can be easily synthesized from 18F-FDG. PET, with 18F-FDS synthesized using current good manufacturing practice, could rapidly differentiate true infection from sterile inflammation to selectively localize E. coli infection in mice. Conclusion: We have developed a systematic approach that exploits unique biochemical pathways in bacteria to develop novel pathogen-specific imaging tracers. These tracers have significant potential for clinical translation to specifically detect and localize a broad range of bacteria, including MDROs.
Keywords: bacteria, PET, imaging, translational, drug-resistance
PET imaging for the diagnosis and therapeutic monitoring of infection is an emerging technology that offers advantages of speed, sensitivity, and whole-body analysis over traditional diagnostic methods. 18F-FDG is the most commonly used PET tracer; however, as an analog of glucose it is unable to differentiate between oncologic, inflammatory, or infectious processes. As a result, there is a need for novel, bacteria-specific PET imaging tracers. We present a systematic approach to identify, select, and develop novel PET probes for infectious diseases. Clinical microbiology has provided the means to differentiate microbes by selective metabolism (1). We hypothesized that a search strategy based on selective metabolism may be used to identify small molecules that are bacteria-specific. By using metabolic pathways selectively expressed by different classes of pathogens and not by mammalian cells, novel imaging tracers could be developed not only to detect the presence of bacterial infection, but also to identify the class of a clinical pathogen. This technology would allow for immediate and appropriate pathogen-directed antibacterial therapy, a critical component in sepsis management (2).
MATERIALS AND METHODS
All protocols were approved by the Johns Hopkins Biosafety, Radiation Safety, and Animal Care and Use Committees.
Radiolabeled Molecules
14C- and 3H-radiolabeled molecules were purchased from Moravek Biochemicals or American Radiolabeled Chemicals. Tritiated fluorine–labeled analogs of PABA and d-mannitol were developed by SRI International. 18F-FDS was synthesized from commercially available 18F-FDG (3,4) or using current good manufacturing practice at the Johns Hopkins PET Center.
In Silico Screen
Commercially available libraries of 14C, 3H, and 125I molecules were obtained from Moravek Biochemicals, American Radiolabeled Chemicals, Perkin Elmer, and ViTrax. Data were extracted from PubMed, PubChem, SciFinder, and Google Scholar. Escherichia coli folp (dihydropteroate synthase), mtlD (mannitol-1-phosphate dehydrogenase), or srlD (sorbitol-6-phosphate dehydrogenase) was queried against the UniProtKB database of bacterial species to calculate alignment and percentage identity using ClustalOmega (5).
In Vitro Uptake Assays
Bacterial strains (American Type Culture Collection); Mycobacterium tuberculosis H37Rv (SKJ Laboratory); and random, consecutive clinical isolates (Johns Hopkins Hospital) were used. All bacteria were aerobically grown to absorbance at 600 nm of 1.0 in Lysogeny Broth (Becton Dickinson) at 37°C. Murine macrophages J774.1 (American Type Culture Collection) were cultured in RPMI-GlutaMAX (Thermo Fisher Scientific) with 10% heat-inactivated fetal bovine serum at 37°C with 5% CO2. Macrophages were chosen because they are professional phagocytic cells, with promiscuous extracellular uptake mechanisms (6). Uptake assays were performed by incubating cultures with 14C and 3H radiotracer (1.85 kBq/mL) or 18F-FDS (10 kBq/mL) at 37°C with rapid agitation. Heat-killed (90°C for 30 min) bacteria were similarly incubated with each radiotracer. Bacteria were pelleted by centrifugation and washed 3 times with phosphate-buffered saline. Samples incubated with 14C and 3H tracers were mixed with 10 mL of liquid scintillation cocktail (MP Biomedicals). Total radioactivity was measured using a Beckman LS6500 Liquid Scintillation Counter (Beckman Coulter Inc.) or an automated γ-counter (1282 Compugamma CS Universal γ-counter; LKB Wallac). A minimum of 6 replicates was used for each assay and time point and presented as a percentage or normalized to total protein (Bradford Assay; Sigma-Aldrich).
Animal Models and Infection
Female, 6- to 8-wk-old CBA/J mice (Jackson Laboratory) were treated as previously described (7), injected with different strains of live or heat-inactivated bacteria in the right or left thigh muscles, respectively, and incubated for 10 h. 3H-labeled tracer (555 kBq) or 18F-FDS (7.4 MBq) was injected intravenously (via the tail), and the mice were sacrificed at 4 h for 3H-labeled tracer or 2 h for 18F-FDS after injection to obtain tissues for biodistribution (liquid scintillation or γ-counting) or bacterial colony-forming units enumerated by dilution and plating onto solidified medium. PET/CT was performed using the nanoScan PET/CT system (Mediso) 120 min after a 7.4-MBq tail vein injection of 18F-FDS (>99% purity) (4).
Statistical Analysis
Statistical comparisons were performed using a 2-tail, Mann–Whitney U test with Prism 6.07 (GraphPad Software Inc.).
RESULTS
In Silico Screen
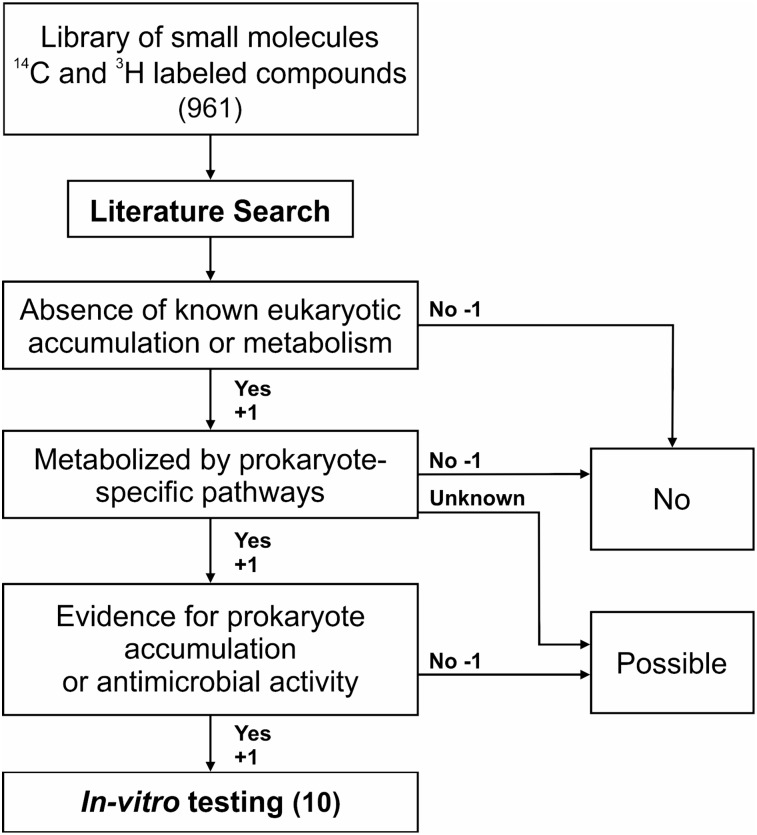
A total of 961 random radiolabeled small molecules (Supplemental Table 1; supplemental materials are available at http://jnm.snmjournals.org) were systematically screened and subjected to 3 selection criteria: absence of known mammalian cell (eukaryotic) metabolism, metabolism by prokaryotic-specific pathways, and evidence of prokaryote interaction such as accumulation or antibacterial activity (Fig. 1). For each selection criteria a score of +1 (criterion true for the molecule), −1 (criterion false), or 0 (unknown with no data available) was assigned to each molecule for a total score ranging from −3 to +3. The library screen spanned several classes of molecules including antivirals, antineoplastics, and antibiotics, though most could not be classified into any specific pharmacologic group (Supplemental Fig. 1). Ten molecules (1%), all identified as substrates for essential metabolic pathways in bacteria, earned a score of +3 (Table 1). They included 8 saccharides (l-arabinose, cellobiose, d-lyxose, d-mannitol, methyl-α-d-glucopyranoside, l-rhamnose, d-sorbitol, and d-xylose) used exclusively by bacteria, a cofactor precursor (PABA), and a component of the aromatic biosynthetic pathway leading to essential amino acids (shikimic acid).
FIGURE 1.
Selection criteria for developing bacteria-specific imaging tracers. Random radiolabeled small molecules were scored using the following 3 criteria: absence of known mammalian cell (eukaryotic) metabolism, metabolism by prokaryotic-specific pathways, and evidence of prokaryote interaction such as accumulation or antibacterial activity. A score of +1 if true, −1 if false, or 0 if unknown was assigned for each selection criteria to each molecule.
TABLE 1.
In Vitro Uptake of Promising Compounds in Model Bacteria Representing Major Classes of Pathogens
| Name | S. aureus (gram-positive) | E. coli (gram-negative) | P. aeruginosa | Mycobacteria | Macrophages (J774) |
| l-Arabinose [1-14C] | 0.41 ± 0.03 | 41.61 ± 9.91 | 0.21 ± 0.02 | 0.28 ± 0.01 (Mtb) | 0.18 ± 0.01 |
| Cellobiose [3H] | 1.81 ± 0.10 | 0.80 ± 0.05 | – | 0.13 ± 0.02 (Ms) | – |
| d-Lyxose [1-14C] | 0.03 ± 0.01 | 1.86 ± 0.14 | 0.12 ± 0.04 | 0.35 ± 0.08 (Mtb) | 0.04 ± 0.01 |
| d-mannitol [1-14C] | 68.40 ± 7.39 | 81.80 ± 1.96 | 0.69 ± 0.05 | 0.29 ± 0.13 (Mtb) | 0.12 ± 0.01 |
| Methyl-α-d-glucopyranoside [methyl-14C] | 11.01 ± 0.71 | 26.78 ± 0.59 | – | 0.11 ± 0.01 (Ms) | – |
| PABA [3,5-3H] | 16.82 ± 1.03 | 18.99 ± 5.80 | 4.02 ± 1.11 | 32.93 ± 4.73 (Mtb) | 0.11 ± 0.01 |
| l-Rhamnose [3H] | 4.96 ± 0.13 | 4.73 ± 0.07 | 0.24 ± 0.04 | 3.82 ± 0.84 (Mtb) | 0.60 ± 0.02 |
| Shikimic acid [3-3H] | 7.54 ± 0.01 | 1.52 ± 0.02 | 1.31 ± 0.02 | 0.17 ± 0.01 (Ms) | – |
| d-Sorbitol [14C] (18F-FDS)* | 0.47 ± 0.09 | 72.20 ± 9.09 | 0.52 ± 0.46 | – | 0.21 ± 0.01 |
| d-Xylose [1-14C] | 0.31 ± 0.01 | 73.94 ± 2.06 | 0.53 ± 0.08 | 0.18 ± 0.02 (Mtb) | 0.19 ± 0.01 |
18F-FDS used for uptake assays.
Mtb = Mycobacterium tuberculosis; Ms = Mycobacterium smegmatis.
Data are mean ± SD.
Rapid and Selective Accumulation in Bacteria
These 10 high-scoring radiolabeled small molecules were tested for in vitro accumulation in bacteria representing important pathogen classes: S. aureus (gram-positive), E. coli and P. aeruginosa (gram-negative), and mycobacteria (Mycobacterium smegmatis or M. tuberculosis) (Table 1). 3,5-3H-PABA (3H-PABA) was found to significantly and rapidly accumulate in all bacterial species tested. 1-14C-d-mannitol (14C-d-mannitol) accumulated rapidly in E. coli and S. aureus but not in P. aeruginosa or M. tuberculosis. Glucose-14C-(U)-methyl-α-d-glucopyranoside was also selectively accumulated by E. coli and S. aureus, but at levels lower than 14C-d-mannitol. 18F-FDS can be easily synthesized from 18F-FDG (3,4); therefore, radiofluorinated 18F-FDS was used instead of tritiated or 14C-labeled d-sorbitol. 1-14C-l-arabinose, 18F-FDS, and 1-14C-d-xylose selectively accumulated in E. coli but not in any other bacterial species tested. 3H-(G)-cellobiose, 1-14C-d-lyxose, and 3H-(G)-l-rhamnose were not accumulated significantly by any bacterial species tested. No uptake was noted by heat-killed bacteria for any molecule in any bacterial strain, suggesting metabolism-derived specificity. No significant uptake was noted in mammalian cells (J774 murine macrophages), indicating that the uptake was bacteria-specific (Table 1). We selected PABA, d-mannitol, and d-sorbitol for further evaluation because they provided a platform to detect and discriminate a range of clinically significant bacteria: PABA identifying all tested species, including P. aeruginosa and M. tuberculosis; d-mannitol detecting gram-positive (S. aureus) and gram-negative bacteria (E. coli); and d-sorbitol (18F-FDS) specifically targeting gram-negative bacteria (E. coli).
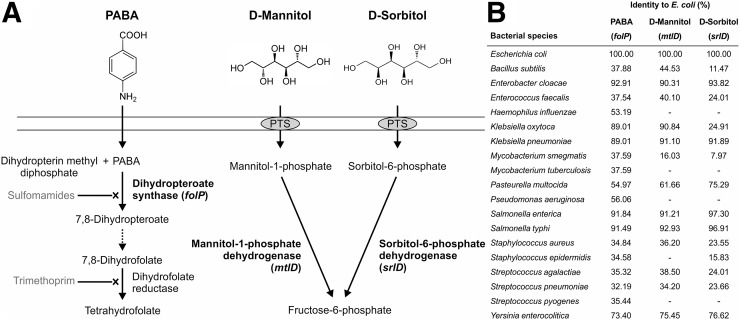
The mechanisms of bacterial uptake for PABA, d-mannitol, and d-sorbitol are shown (Fig. 2A). folP was present in all bacterial species, explaining PABA use by a wide range of bacterial species. However, mtlD and srlD were absent in several species, and only gram-negative bacteria from the Enterobacteriaceae family (e.g., E. coli, K. pneumoniae, S. enterica, E. cloacae, and Y. enterocolitica) had significant homology for srlD (Fig. 2B).
FIGURE 2.
Biologic mechanisms of tracer uptake. (A) PABA is used as substrate by dihydropteroate synthase (encoded by folP) in folate synthesis pathway and is subsequently trapped inside bacteria. d-mannitol and d-sorbitol are transported and phosphorylated inside the bacteria by a phosphoenolpyruvate-dependent carbohydrate phosphotransferase system (PTS). Mannitol-1-phosphate dehydrogenase (encoded by mtlD) and sorbitol-6-phosphate dehydrogenase (encoded by srlD) then convert their substrates into fructose-6-phosphate. (B) Comparison of homology of E. coli genes encoding key enzymes to other pathogenic bacteria in the UniProtKB database.
Uptake in Extended Bacterial Panel and Clinical Isolates Including MDROs
Time-dependent accumulation was determined by measuring in vitro uptake at various time points over 120 min for all tracers, using heat-killed bacterial cells as a negative control (Fig. 3A; Supplemental Fig. 2A).
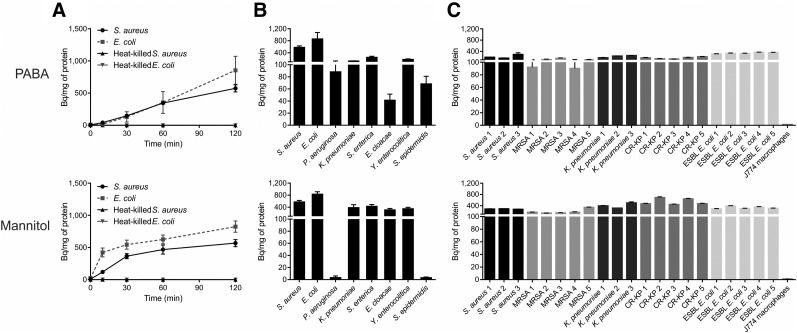
FIGURE 3.
Bacterial uptake of 3H-PABA and 14C-d-mannitol. (A) Uptake by S. aureus, E. coli, and corresponding heat-killed controls for 3H-PABA and 14C-d-mannitol, respectively. (B) Uptake of 3H-PABA and 14C-d-mannitol, respectively, at 120 min in extended panel of bacteria (reference laboratory strains) and by drug-susceptible and drug-resistant clinical isolates (C), compared with mammalian cells (J774 murine macrophages). Labels indicate methicillin-resistant S. aureus (MRSA), carbapenem-resistant K. pneumoniae (CR-KP), and extended-spectrum β-lactamase–producing E. coli (ESBL E. coli). Mean and SD are shown.
3H-PABA was rapidly accumulated in all laboratory and clinical bacterial isolates including MDROs: methicillin-resistant S. aureus, carbapenem-resistant K. pneumoniae, and extended-spectrum β-lactamase–producing E. coli (Figs. 3B and 3C) and was 100-fold higher or more than mammalian cells (murine J774 macrophages). 3H-PABA was also taken up specifically in M. tuberculosis (Supplemental Fig. 3). 14C-d-mannitol was accumulated in S. aureus and the Enterobacteriaceae family of bacteria that include E. coli (Fig. 3B). It was also rapidly taken up in all clinical isolates including MDROs (≥100-fold higher than mammalian cells) (Fig. 3C). 18F-FDS was only accumulated in Enterobacteriaceae family of bacteria, including MDRO clinical isolates (Supplemental Figs. 2B and 2C).
Fluorine-Labeled Analogs Uptake in Bacteria
18F is the most widely used PET isotope, with excellent imaging characteristics. Though PABA and d-mannitol do not inherently have any fluorine atoms, we custom-synthesized tritiated 2- and 3-fluoro analogs of PABA and d-mannitol and tested their uptake by E. coli and S. aureus to evaluate their potential as radiofluorinated PET tracers. For both PABA and d-mannitol, the analogs had higher uptake when fluorinated in the 2 position, as opposed to the 3 position, and were chosen for further development (Supplemental Fig. 4). Both 3H-2-F-PABA and 3H-2-deoxy-2-fluoro-d-mannitol (3H-2-F-mannitol) had a rapid, time-dependent uptake in S. aureus and E. coli (Figs. 4A and 4C). Furthermore, they were also rapidly accumulated by all clinical isolates including MDROs but not by mammalian cells (Figs. 4B and 4D). The uptake of 3H-2-F-mannitol by clinical isolates (S. aureus and methicillin-resistant S. aureus) was lower than 14C-d-mannitol, but still 12.5-fold and 8.3-fold higher than by mammalian cells, respectively (P < 0.01).
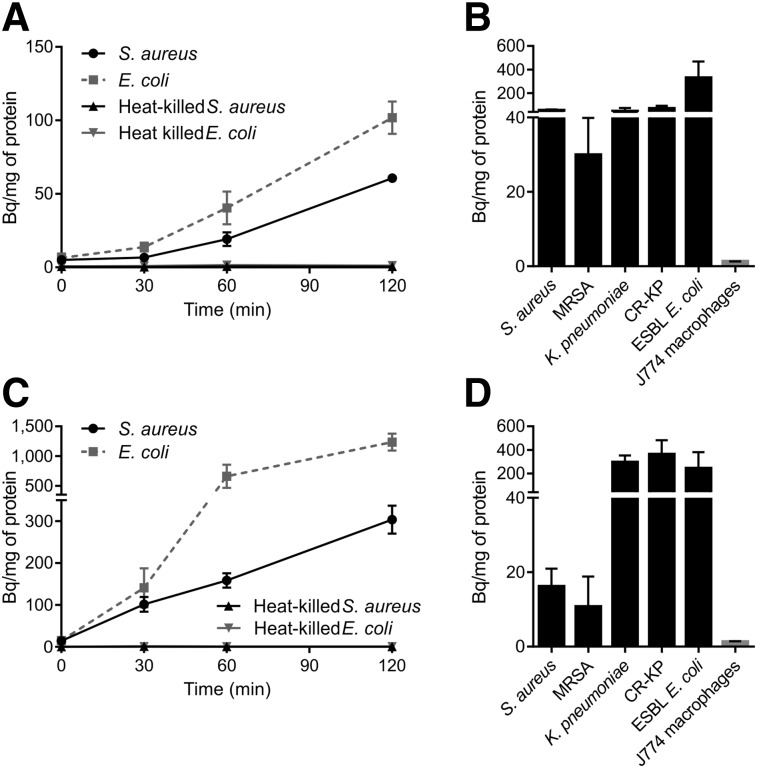
FIGURE 4.
Selective and specific accumulation of 3H-2-F-PABA and 3H-2-F-mannitol by bacteria. Uptake by S. aureus, E. coli, and corresponding heat-killed controls for 3H-2-F-PABA (A) and 3H-2-F-mannitol (C). Uptake of 3H-2-F-PABA (B) and 3H-2-F-mannitol (D) at 120 min by clinical bacterial isolates. Data represent uptake by at least 3 different clinical isolates for each species or strain type. Labels indicate methicillin-resistant S. aureus (MRSA), carbapenem-resistant K. pneumoniae (CR-KP), and extended-spectrum β-lactamase–producing E. coli (ESBL E. coli). Mean and SD are shown.
To test for specificity, we evaluated whether the addition of excess, unlabeled PABA and d-mannitol has the potential to block bacterial accumulation via competitive inhibition. Uptake of 3H-2-F-PABA in S. aureus was significantly reduced by coincubation with increasing concentrations of the unlabeled PABA, suggesting that uptake was saturable and specific (Supplemental Fig. 5A). The uptake of 3H-2-F-mannitol by S. aureus and E.coli increased with the addition of unlabeled mannitol at low concentrations but decreased at higher concentrations (Supplemental Figs. 5B and 5C). Finally, given that PABA is an essential nutrient required for bacterial growth and survival, we also evaluated its uptake by bacteria in different phases of growth. 3H-2-F-PABA was significantly accumulated by bacteria in all growth phases, with noteworthy uptake by stationary phase cultures (Supplemental Fig. 6).
Fluorine-Labeled Analogs Can Rapidly Detect Infection in Murine Myositis Model
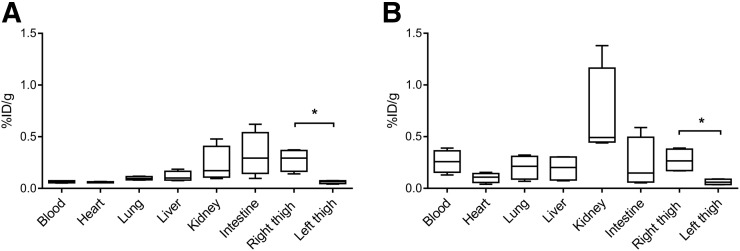
Mice were inoculated with live S. aureus (3H-2-F-PABA) or E. coli (3H-2-F-mannitol) into the right thigh and a 10-fold-higher burden of heat-killed bacteria into the left thigh (sterile inflammation). The tracer was administered intravenously. 3H-2-F-PABA was found to accumulate in the S. aureus–infected tissues significantly more than the inflammation control (4.5-fold; P = 0.03) (Fig. 5A). Bacterial burden in the infected thigh at the time of sacrifice was 1.1 × 106 colony-forming units of S. aureus. Similarly, 3H-2-F-mannitol was found to accumulate in the E. coli–infected tissues significantly more than the inflammation control (4.5-fold; P = 0.03) (Fig. 5B). Bacterial burden in the infected thigh at the time of sacrifice was 2.3 × 107 colony-forming units of E. coli. Selective uptake of 18F-FDS in E. coli–infected tissues (7.9-fold vs. the inflammation control; P < 0.01) was also noted (Supplemental Fig. 7 (4)). For all tracers, the uptake in (sterile) inflamed muscle was minimal. Low background, defined as uptake in healthy or (sterile) inflammatory tissues, was noted for all tracers, with the exception of kidneys due to renal excretion and the intestines due to uptake by normal gut flora.
FIGURE 5.
Tissue biodistribution of 3H-2-F-PABA and 3H-2-F-mannitol in murine myositis model. Mice were inoculated with live bacteria in right thigh and 10-fold-higher burden of heat-killed bacteria in left thigh (sterile inflammation). Tracer was injected intravenously and accumulation in tissues measured using scintillation counting from surgically resected tissues. Tissue biodistribution of 3H-2-F-PABA (A) (n = 4 animals) and 3H-2-F-mannitol (B) (n = 4 animals) is shown. *P = 0.03 (Mann–Whitney U test, 2-tail). Median, interquartile, and range are shown.
18F-FDS PET Imaging
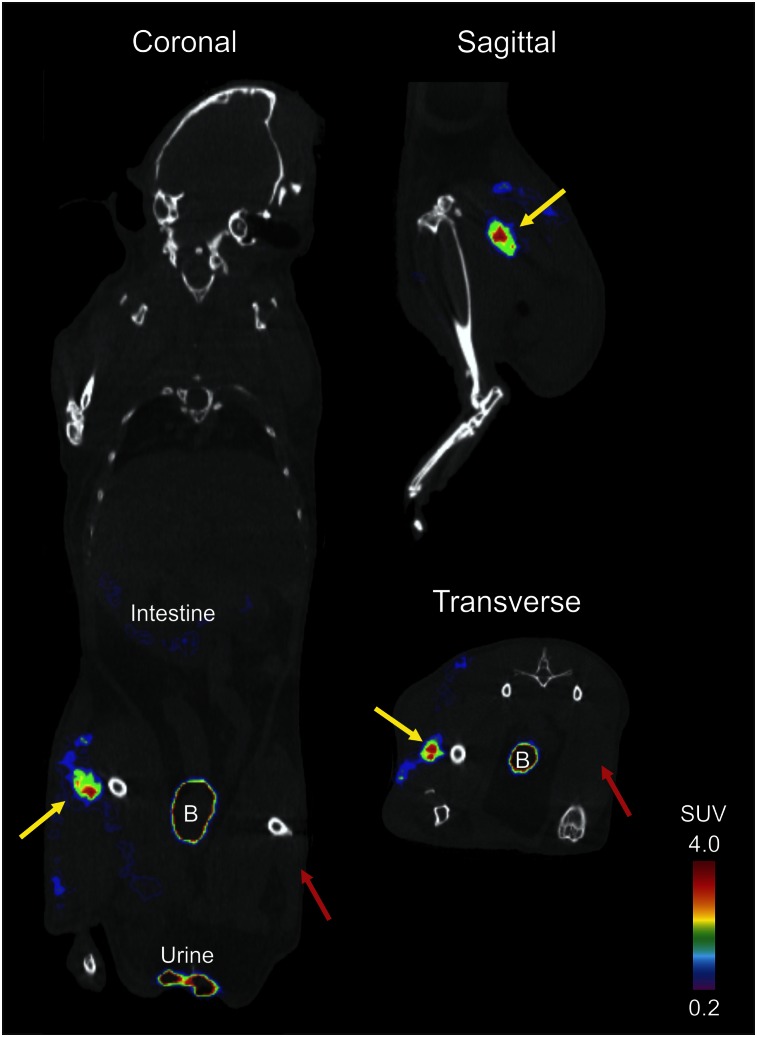
PET demonstrated selective uptake of clinical grade 18F-FDS in E. coli–infected tissues but not in the inflamed (control) sterile thigh (Fig. 6). Minimal PET signal was noted in healthy tissues with the exception of kidneys and the intestines.
FIGURE 6.
18F-FDS PET/CT imaging of E. coli murine myositis model. A 15-min acquisition window PET scan was performed 120 min after 7.4-MBq tail-vein injection of 18F-FDS. 18F-FDS signal is noted in infected (yellow arrow) but not in inflamed (control) sterile thigh (red arrow). Signal from bladder (B), extravasated urine, and intestine is also visible.
DISCUSSION
Rapid and accurate diagnosis of bacterial infections is essential for early therapeutic interventions. However, traditional diagnostic tools, namely microscopy, microbial culture, and molecular techniques, require sampling from suspected sites of infection. This approach is invasive, frequently dangerous in cases of deep-seated infections, labor intensive, time consuming, and subject to uncertainties from incorrect sampling and contamination. For example, blood cultures can be negative in the setting of deep-seated infections but can also result in false-positives (8). This has led to indiscriminate, empiric use of broad-spectrum antibacterials. Judicious antibiotic use could reduce the emergence of drug resistance, increase patient safety, and save billions of dollars (9).
Previous attempts at developing pathogen-specific imaging techniques have been based on radiolabeling existing antibiotics or antimicrobial peptides (10,11). Although antibiotics and antimicrobial peptides should bind specifically, they are designed to kill or disable bacteria at very minimal concentrations, thus potentially limiting their effectiveness as a radiotracer due to lack of signal amplification. For example, 99mTc-ciprofloxacin was evaluated in a clinical study for diagnosing infections (12), but follow-up studies demonstrated nonspecific binding and an inability to differentiate infection from sterile inflammation (13,14). Radiolabeled peptides have shown promise in preclinical models (15,16) but lack mechanistic binding or uptake, which could limit generalizability (17). In addition, 124I-FIAU, a nucleoside analog substrate for thymidine kinase, has been described as a γ-herpesvirus and bacteria-selective tracer (18). However, a challenging synthesis and lack of specificity (19), due to metabolism by host mitochondrial thymidine kinase, limits its use. Maltose and maltohexaose labeled with 18F are promising but have modest signal-to-noise ratios, likely due to host metabolism of the tracers (20,21). A 64Cu-NODAGA–labeled antibody was also evaluated as a PET tracer in a Yersinia murine model (22). Although this approach is promising, antibodies may require up to a week to clear from nontarget sites. Conversely, the rapid clearance and penetration of small molecules, such as radiofluorinated sugars, in diseased tissue combined with the short-half-life isotopes (e.g., 18F) make them ideal imaging candidates.
We analyzed almost 1,000 radiolabeled small molecules as substrates for essential metabolic pathways in bacteria and identified 3 novel, nontoxic analogs of U.S. Food and Drug Administration (FDA)–approved compounds. PABA is FDA approved for use in sunscreen, and is inert in humans (23). Bacteria use PABA as an intermediate to synthesize folate via the dihydropteroate synthase enzyme, which is acquired in the human diet. In fact, the sulfonamide class of antibiotics exploits this metabolic distinction to inhibit the growth of a wide range of bacterial pathogens (24). folP is present in most bacterial species analyzed, explaining the uptake of PABA by a wide range of bacterial species. d-mannitol and d-sorbitol are epimeric sugar alcohols, differing only in the orientation of the hydroxyl group on carbon 2. d-mannitol and d-sorbitol are FDA-approved osmotic diuretic and sugar-free sweetener/laxative, respectively, and not metabolized by mammalian cells. However, they are taken up by bacterial sugar transporters, phosphorylated by kinases, and oxidized by the respective dehydrogenases (25). The presence and percentage identity of the mtlD- or srlD-dehydrogenase cassettes was predictive of uptake by the different bacterial species. Finally, all 3 lead candidates were rapidly and significantly taken up by randomly selected clinical strains, including MDROs, reinforcing their clinical tractability. Although general bacteria-specific imaging tracers (e.g., PABA), that detect a wide range of bacterial species, would be valuable to determine bacterial infection, bacteria class–specific imaging tracers (d-mannitol, FDS) could inform the specific antibiotic treatments. For example, vancomycin is useful for treating infections due to gram-positive bacteria but ineffective against most gram-negative bacteria. Similarly, antibiotics used to treat mycobacteria are vastly different from those used to treat prominent bacteria pathogens such as S. aureus and E. coli.
Because PABA and d-mannitol do not inherently possess a fluorine atom, we evaluated 3H-labeled fluorinated analogs that could be developed as 18F PET–based imaging tracers. The analogs labeled in the 2 position, 3H-2-F-PABA and 3H-2-F-mannitol, retained the parent molecule characteristics of rapid, specific, and time-dependent bacterial accumulation. Interestingly, the uptake of 3H-2-F-mannitol in S. aureus and E. coli increased with coincubation of low concentrations of unlabeled mannitol, likely due to the induction of the sugar transporter and dehydrogenase (26) but decreased at higher concentrations. As mentioned above, the metabolic pathways for PABA, mannitol, and sorbitol have been well characterized. We hypothesized that the fluorine-labeled analogs are also metabolized by these (or similar) pathways. Competitive inhibition of bacterial uptake of the fluorine-labeled analog by the respective parent compound strengthens this assumption, though additional studies are required to prove this conclusively. Finally, uptake of 3H-2-F-PABA was high, regardless of growth phase, and accumulated by stationary phase (slowly dividing) bacterial cultures. Given that bacteria reside in different metabolic states in infectious lesions (27,28), 2-18F-PABA PET may have the potential to detect slowly dividing or dormant bacteria in vivo. Although the parent molecules (PABA, d-mannitol, and d-sorbitol) are FDA approved, preclinical toxicity studies will be needed to support an investigational new drug application for the fluorine-labeled analogs.
Using a murine model of myositis, we next tested whether the fluorine-labeled analogs could distinguish between bacterial infection and sterile inflammation. 3H-2-F-PABA, 3H-2-F-mannitol, and 18F-FDS significantly accumulated at the site of bacterial infection but not in tissues with sterile inflammation. Uptake in other normal, uninfected organs was minimal except for the kidneys due to renal excretion and the intestines due to uptake by normal gut flora. The synthesis of 18F versions of both 2-F-PABA and 2-F-mannitol are under development; however, 18F-FDS can be easily synthesized from 18F-FDG (3). We used clinical-grade 18F-FDS manufactured under current good manufacturing practice, and 18F-FDS PET rapidly differentiated true infection from sterile inflammation to selectively localize E. coli infection. Moreover, we have also demonstrated that 18F-FDS PET can rapidly and specifically localize infections in murine models of mixed gram-positive and gram-negative thigh coinfections, brain infection, Klebsiella pneumonia, and immunosuppressive chemotherapy (4). In addition, 18F-FDS PET also demonstrated a PET signal proportionate to the bacterial burden with the ability to monitor therapeutic failures associated with extended-spectrum β-lactamase–producing E. coli (clinical isolates) infections in real time (4). Recently, 18F-FDS was shown to have high renal excretion, low plasma protein binding, and high metabolic stability in rats (29). In a study of healthy volunteers, 18F-FDS was determined to be safe, well tolerated, and rapidly cleared, after a single, intravenous dose (30), suggesting promising potential for use in humans.
Although we have focused on developing PET tracers using 18F (due to easy translation to humans), other PET radioisotopes such as 11C as well as tracers used with SPECT could also be used.
CONCLUSION
We have developed a rational approach to discover novel candidate imaging tracers with the potential to specifically diagnose, localize, and monitor a broad range of bacterial infections. Our data show that noninvasive imaging can be useful not only to diagnose infections in situ, but also to rapidly identify the causative bacterial class ultimately aiding in the selection of appropriate antibiotic treatment. The high-scoring candidates from our approach, dependent on mechanism-based targeting of bacteria-specific pathways exhibited rapid penetration and clearance as well as high target-to-nontarget ratios, due to minimal metabolism by host-tissues. These tracers have significant potential for clinical translation for the rapid diagnosis of bacterial infections, including life-threatening infections due to MDROs, and may be especially critical in cancer and immunocompromised patients.
DISCLOSURE
This study was funded by the National Institutes of Health Director’s Transformative Research Award R01-EB-020539, Director’s New Innovator Award DP2-OD-006492, subcontract from Harvard University Center for AIDS Research 5P30AI060354-09, and R01-HL-116316 to Sanjay K. Jain. Funding from NIH-NIAID HHSN272201100022I is also acknowledged for the synthesis of 3H-fluorine–labeled analogs. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. No other potential conflict of interest relevant to this article was reported.
Supplementary Material
Acknowledgments
We thank Paula Mister and Dr. Karen Carroll (JHU) for providing the clinical isolates of bacteria.
REFERENCES
- 1.Mac Faddin JF. Biochemical Tests for Identification of Medical Bacteria. Baltimore, MD: The Williams & Wilkins Company; 1976. [Google Scholar]
- 2.Dellinger RP, Levy MM, Rhodes A, et al. Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock: 2012. Crit Care Med. 2013;41:580–637. [DOI] [PubMed] [Google Scholar]
- 3.Li ZB, Wu Z, Cao Q, et al. The synthesis of 18F-FDS and its potential application in molecular imaging. Mol Imaging Biol. 2008;10:92–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weinstein EA, Ordonez AA, DeMarco VP, et al. Imaging Enterobacteriaceae infection in vivo with 18F-fluorodeoxysorbitol positron emission tomography. Sci Transl Med. 2014;6:259ra146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sievers F, Wilm A, Dineen D, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. [DOI] [PubMed] [Google Scholar]
- 7.Zuluaga AF, Salazar BE, Rodriguez CA, Zapata AX, Agudelo M, Vesga O. Neutropenia induced in outbred mice by a simplified low-dose cyclophosphamide regimen: characterization and applicability to diverse experimental models of infectious diseases. BMC Infect Dis. 2006;6:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bates DW, Sands K, Miller E, et al. Predicting bacteremia in patients with sepsis syndrome. Academic Medical Center Consortium Sepsis Project Working Group. J Infect Dis. 1997;176:1538–1551. [DOI] [PubMed] [Google Scholar]
- 9.Scott RD. The direct medical costs of healthcare-associated infections in U.S. hospitals and the benefits of prevention. CDC website. http://www.cdc.gov/hai/pdfs/hai/scott_costpaper.pdf. Accessed October 6, 2016.
- 10.Gemmel F, Dumarey N, Welling M. Future diagnostic agents. Semin Nucl Med. 2009;39:11–26. [DOI] [PubMed] [Google Scholar]
- 11.van Oosten M, Hahn M, Crane LM, et al. Targeted imaging of bacterial infections: advances, hurdles and hopes. FEMS Microbiol Rev. 2015;39:892–916. [DOI] [PubMed] [Google Scholar]
- 12.Vinjamuri S, Hall AV, Solanki KK, et al. Comparison of 99mTc infection imaging with radiolabelled white-cell imaging in the evaluation of bacterial infection. Lancet. 1996;347:233–235. [DOI] [PubMed] [Google Scholar]
- 13.Sarda L, Cremieux AC, Lebellec Y, et al. Inability of 99mTc-ciprofloxacin scintigraphy to discriminate between septic and sterile osteoarticular diseases. J Nucl Med. 2003;44:920–926. [PubMed] [Google Scholar]
- 14.Palestro C, Love C, Caprioli R, et al. Phase II study of 99mTc-ciprofloxacin uptake in patients with high suspicion of osteomyelitis [abstract] J Nucl Med. 2006;47:152P. [Google Scholar]
- 15.Ebenhan T, Zeevaart JR, Venter JD, et al. Preclinical evaluation of 68Ga-labeled 1,4,7-triazacyclononane-1,4,7-triacetic acid-ubiquicidin as a radioligand for PET infection imaging. J Nucl Med. 2014;55:308–314. [DOI] [PubMed] [Google Scholar]
- 16.Vilche M, Reyes AL, Vasilskis E, Oliver P, Balter H, Engler H. 68Ga-NOTA-UBI-29-41 as a PET tracer for detection of bacterial infection. J Nucl Med. 2016;57:622–627. [DOI] [PubMed] [Google Scholar]
- 17.Welling M, Stokkel M, Balter J, Sarda-Mantel L, Meulemans A, Le Guludec D. The many roads to infection imaging. Eur J Nucl Med Mol Imaging. 2008;35:848–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bettegowda C, Foss CA, Cheong I, et al. Imaging bacterial infections with radiolabeled 1-(2′-deoxy-2′-fluoro-beta-D-arabinofuranosyl)-5-iodouracil. Proc Natl Acad Sci USA. 2005;102:1145–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang XM, Zhang HH, McLeroth P, et al. [124I]FIAU: human dosimetry and infection imaging in patients with suspected prosthetic joint infection. Nucl Med Biol. 2016;43:273–279. [DOI] [PubMed] [Google Scholar]
- 20.Gowrishankar G, Namavari M, Jouannot EB, et al. Investigation of 6-[18F]-fluoromaltose as a novel PET tracer for imaging bacterial infection. PLoS One. 2014;9:e107951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ning X, Seo W, Lee S, et al. PET imaging of bacterial infections with fluorine-18-labeled maltohexaose. Angew Chem Int Ed Engl. 2014;53:14096–14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiehr S, Warnke P, Rolle AM, et al. New pathogen-specific immunoPET/MR tracer for molecular imaging of a systemic bacterial infection. Oncotarget. 2016;7:10990–11001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansson G, Bingham S, Vahter M. A method to compensate for incomplete 24-hour urine collections in nutritional epidemiology studies. Public Health Nutr. 1999;2:587–591. [DOI] [PubMed] [Google Scholar]
- 24.Brown GM. The biosynthesis of folic acid: II—inhibition by sulfonamides. J Biol Chem. 1962;237:536–540. [PubMed] [Google Scholar]
- 25.Lengeler J. Nature and properties of hexitol transport systems in Escherichia coli. J Bacteriol. 1975;124:39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Figge RM, Ramseier TM, Saier MH., Jr The mannitol repressor (MtlR) of Escherichia coli. J Bacteriol. 1994;176:840–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ordonez AA, Tasneen R, Pokkali S, et al. Mouse model of pulmonary cavitary tuberculosis and expression of matrix metalloproteinase-9. Dis Model Mech. 2016;9:779–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murawski AM, Gurbani S, Harper JS, et al. Imaging the evolution of reactivation pulmonary tuberculosis in mice using 18F-FDG PET. J Nucl Med. 2014;55:1726–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wakabayashi H, Werner RA, Hayakawa N, et al. Initial preclinical evaluation of 18F-fluorodeoxysorbitol PET as a novel functional renal imaging agent. J Nucl Med. 2016;57:1625–1628. [DOI] [PubMed] [Google Scholar]
- 30.Zhu W, Yao S, Xing H, et al. Biodistribution and radiation dosimetry of the Enterobacteriaceae-specific imaging probe [18F]fluorodeoxysorbitol determined by PET/CT in healthy human volunteers. Mol Imaging Biol. 2016;5:782–787. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.