Abstract
Transgenic Arabidopsis thaliana plants overproducing the E2Fa-DPa transcription factor have two distinct cell-specific phenotypes: some cells divide ectopically and others are stimulated to endocycle. The decision of cells to undergo extra mitotic divisions has been postulated to depend on the presence of a mitosis-inducing factor (MIF). Plants possess a unique class of cyclin-dependent kinases (CDKs; B-type) for which no ortholog is found in other kingdoms. The peak of CDKB1;1 activity around the G2-M boundary suggested that it might be part of the MIF. Plants that overexpressed a dominant negative allele of CDKB1;1 underwent enhanced endoreduplication, demonstrating that CDKB1;1 activity was required to inhibit the endocycle. Moreover, when the mutant CDKB1;1 allele was overexpressed in an E2Fa-DPa–overproducing background, it enhanced the endoreduplication phenotype, whereas the extra mitotic cell divisions normally induced by E2Fa-DPa were repressed. Surprisingly, CDKB1;1 transcription was controlled by the E2F pathway, as shown by its upregulation in E2Fa-DPa–overproducing plants and mutational analysis of the E2F binding site in the CDKB1;1 promoter. These findings illustrate a cross talking mechanism between the G1-S and G2-M transition points.
INTRODUCTION
Mitosis and endoreduplication are two different modes of the cell cycle. The mitotic cell cycle comprises the duplication and subsequent distribution of chromosomes between two daughter cells, whereas endoreduplication involves repetitive chromosomal DNA replication without intervening mitosis or cytokinesis, leading to an increase in the ploidy level (D'Amato, 1964; Brodsky and Uryvaeva, 1977). In plants, endoreduplication is common and assumed to be the most prevalent process for increasing the nuclear ploidy. The level of ploidy varies between species and tissues. In Arabidopsis thaliana, up to 32C nuclei have been detected (Galbraith et al., 1991).
Until now, the physiological significance of endoreduplication is poorly understood. A positive correlation commonly exists between the ploidy level of cells and their size, and it was postulated that endoreduplication is required for maintaining an optimal ratio between genome and cell size (Melaragno et al., 1993; Folkers et al., 1997; Traas et al., 1998; Sugimoto-Shirasu and Roberts, 2003). Alternative hypotheses link endoreduplication with increased metabolic activity, maintenance of the optimal ratio between the nuclear and organellar DNA, cellular differentiation, and resistance against irradiation (Traas et al., 1998; Joubès and Chevalier, 2000; Kondorosi et al., 2000; Larkins et al., 2001).
Not only the functional role of endoreduplication is unclear, but also the molecular mechanism triggering the process remains elusive. The continuity of the meristematic zone with the region of endoreduplication suggested that the endocyle is achieved by a modification of the mitotic cell cycle (Jacqmard et al., 1999). In the maize (Zea mays) endosperm, the onset of endoreduplication is correlated with the inhibition of M-phase–associated cyclin-dependent kinase (CDK) activity by a yet unidentified component (Grafi and Larkins, 1995). Similarly, in developing tomato (Lycopersicon esculentum) fruits, M-phase–specific CDK activity is highly reduced during the endoreduplication process (Joubès et al., 1999). These data indicate that in analogy to what has been found previously in yeast and Drosophila melanogaster, an inhibition of the G2-M transition might be sufficient to initiate the endoreduplication program. Several strategies have been proposed by which M-phase CDK activity could be reduced. Currently, the best-documented mechanism involves the CCS52 protein, which activates the anaphase-promoting complex, thereby triggering the destruction of M-phase–specific cyclins. In alfalfa (Medicago sativa), CCS52 expression correlates with the onset of endoreduplication. Moreover, CCS52 overexpression in yeast transforms the mitotic cycles into endocycles, whereas plants with downregulated levels of CCS52 display a decrease in ploidy level (Cebolla et al., 1999). Another likely candidate to inhibit M-phase–specific CDK activity in endoreduplicating cells is the WEE1 kinase, for which transcript levels have been demonstrated to accumulate during the endoreduplication process in maize endosperm (Sun et al., 1999). Nevertheless, the identity of the CDK/cyclin complex to be inhibited to trigger endoreduplication remains to be elucidated.
Recently, we have identified the E2Fa-DPa complex as a positive regulator of the endocycle (De Veylder et al., 2002). E2F-DP transcription factors regulate the onset of S-phase by controlling the expression of genes needed for DNA replication (Trimarchi and Lees, 2002). In E2Fa-DPa–overexpressing plants, DNA replication is strongly activated, resulting in a mixed phenotype: some cells are triggered to undergo supernumerous mitotic divisions; in others, the endoreduplication cycle is stimulated, resulting in a dramatic increase in ploidy level and formation of giant nuclei (De Veylder et al., 2002; Kosugi and Ohashi, 2003). The decision of E2Fa-DPa–overproducing cells to undergo extra cell division or endoreduplication depends on the cell type and tissue, as clearly seen in the epidermal cell files of the hypocotyls: stomata forming cell files are stimulated to undergo ectopic divisions, whereas cell files without stomata mainly consist of bulged cells containing huge nuclei (De Veylder et al., 2002). Because the formation of stomata is linked with competence to divide, we have postulated that the decision of E2Fa-DPa–overproducing cells to undergo extra cell divisions depends on the presence of a yet to be identified mitosis-inducing factor (MIF; De Veylder et al., 2002).
As in all eukaryotic organisms, cell division in plants is regulated by CDKs (De Veylder et al., 2003). In the model plant Arabidopsis, 12 CDKs have been annotated to date, grouped into six classes (Vandepoele et al., 2002). Curiously, plants possess a unique class of CDKs (B-type) for which no counterpart is found in other organisms, suggesting that they regulate plant-specific aspects of the cell cycle. A role of CDKB1;1 in skotomorphogenesis has been proposed, based on the observation that dark-grown CDKB1;1 antisense lines display a short hypocotyl and open cotyledons (Yoshizumi et al., 1999). Recently, CDKB1;1 activity has been demonstrated to be essential for correct stomatal development (Boudolf et al., 2004). Here, we highlight an additional role for CDKB1;1 in plant development: plants with reduced CDKB1;1 activity prematurely exit the mitotic cell cycle and have elevated ploidy levels. Moreover, overexpression of a dominant negative CDKB1;1 allele in an E2Fa-DPa–overproducing background enhanced the endoreduplication phenotype triggered by E2Fa-DPa, whereas ectopic cell divisions were suppressed, demonstrating that CDKB1;1 determines the fate of E2Fa-DPa–expressing cells to divide mitotically or to endoreduplicate. We demonstrate that CDKB1;1 transcription is regulated through an E2F cis-acting element in its promoter, providing a mechanism that links G1-S and G2-M transition points.
RESULTS
CDKB1;1.N161 Overexpression Stimulates Exit from Mitosis
To address the role of CDKB1;1 in plant growth and development, transgenic lines were generated that overproduce the wild-type or a dominant negative allele of CDKB1;1 (CDKB1;1.N161). The CDKB1;1.N161 allele encodes a CDK in which the amino acid Asp at position 161 is changed into Asn, resulting into a kinase-dead protein (van den Heuvel and Harlow, 1993; Labib et al., 1995). Previously, we have demonstrated that overexpression of this mutant kinase gene results in a decrease in CDK activity, probably because the mutant and endogenous wild-type proteins compete for the association with rate-limiting interacting proteins such as cyclins (Porceddu et al., 2001; Boudolf et al., 2004).
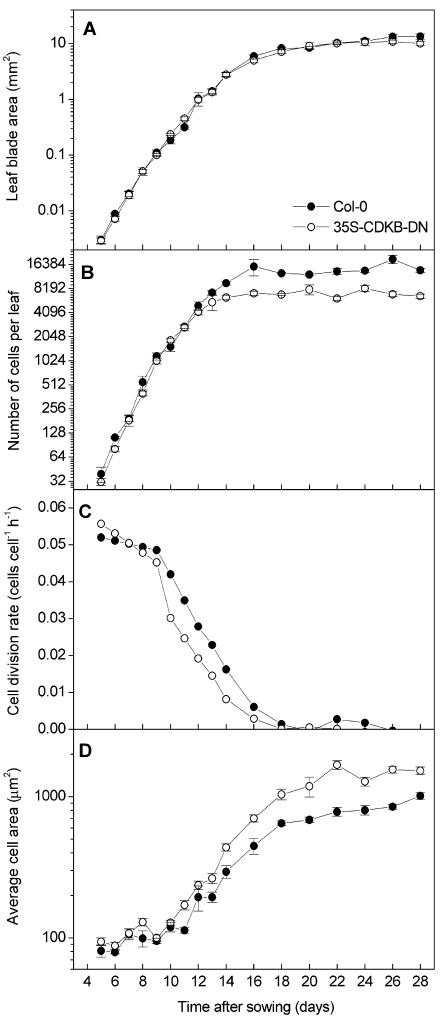
The effects of overexpression of the wild-type and dominant negative CDKB1;1 genes on leaf development and cell cycle duration were studied by a kinematic analysis. From day 5 until day 28 after sowing, the first leaf pairs of transgenic and wild-type plants grown side by side under the same conditions were harvested, and the leaf blade area was measured by image analysis (De Veylder et al., 2001). Subsequently, the average cell area of abaxial epidermal cells was determined from drawing-tube images, and total cell number was extrapolated as the ratio of leaf blade and average cell areas. Plants overproducing the native CDKB1;1 gene did not differ from wild-type plants (data not shown). However, when wild-type and CDKB1;1.N161-overexpressing plants were compared, significant differences were noticed (Figure 1). Leaf blade area expansion was similar in the wild-type and CDKB1;1.N161 transgenic plants: first-leaf size expanded exponentially until day 12, after which expansion rates steadily decreased to zero at day 20, when the leaves reached their mature size (Figure 1A). CDKB1;1.N161-overexpressing leaves contained only half the number of epidermal cells at maturity (Figure 1B). The divergence between wild-type and transgenic lines occurred between 10 and 16 d after sowing. Supporting this observation, average cell division rates for the whole leaf blade, calculated as exponential increase of the cell number, were approximately constant and comparable in wild-type and CDKB1;1.N161-overexpressing leaves until 9 d after sowing. However, from day 10 to day 16, which mark the exit from cell division, cell division rates were constantly lower in the transgenic plants (Figure 1C). Simultaneously, the average cell size, which initially was ∼100 μm in both lines, increased significantly faster in the transgenic lines, with cells reaching nearly twice the size of that of wild-type cells (Figure 1D). At day 14 after sowing, the abaxial epidermal pavement cells of the first leaf of CDKB1;1.N161-overexpressing plants had the characteristically jigsaw puzzle–shaped appearance. By contrast, wild-type pavement cells of the same age were less lobed, suggesting an advanced differentiation of CDKB1;1.N161-overexpressing cells (Figures 2A and 2B). Taken together, these data indicate that cell cycle exit occurred more rapidly in the transgenic plants.
Figure 1.
Kinematic Analysis of Leaf Growth of the First Leaf Pair of Wild-Type Columbia-0 and CDKB1;1.N161-Overproducing Plants.
(A) Leaf blade area. Col-0, Columbia-0.
(B) Epidermal cell number on the abaxial side of the leaf.
(C) Average cell division rates of the epidermal cells on the abaxial side of the leaf.
(D) Epidermal cell size on the abaxial side of the leaf. Error bars denote standard errors (n = 4 to 10).
Symbols in (B), (C), and (D) as in (A).
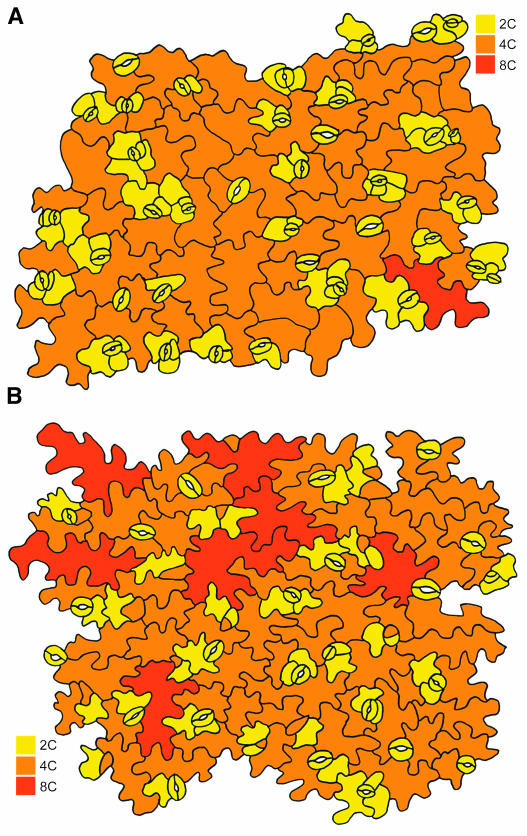
Figure 2.
Abaxial Epidermal Peel of the First Leaves 14 d after Germination.
(A) Wild-type plants.
(B) Plants overexpressing CDKB1;1.N161.
The ploidy level of the cells is indicated by color according to the legend.
CDKB1;1 Activity Is Required to Suppress Endoreduplication
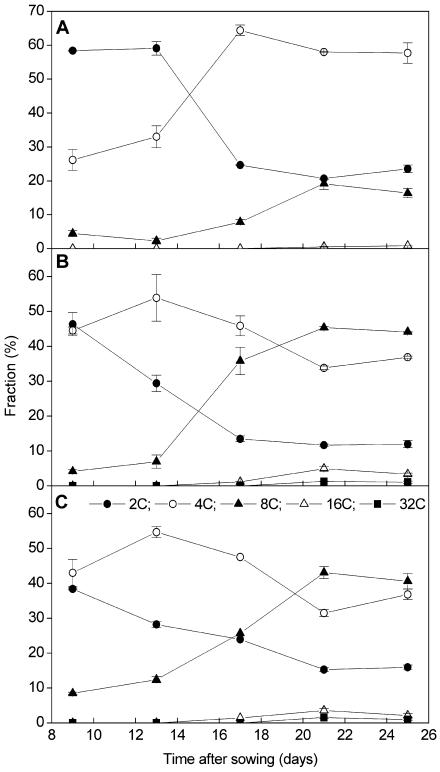
Previously, we have shown that the exit of the mitotic cell cycle in Arabidopsis is associated with the start of endoreduplication (Jacqmard et al., 1999). To study the onset of endoreduplication in the plants overexpressing CDKB1;1 and CDKB1;1.N161, the ploidy level of the first pair of true leaves was measured by flow cytometry from 9 to 25 d after sowing, harvested at 4-d intervals. Ploidy levels in wild-type and CDKB1;1-overexpressing leaves were similar at any stage of leaf development (Figure 3A; data not shown). The 2C and 4C levels remained fairly constant from days 9 to 13. From day 13 onward, the amount of cells with a 2C content decreased steeply, correlated with an increase in the number of cells with a 4C and 8C content until day 21. Afterwards, the ploidy levels remained essentially constant. In two independent CDKB1;1.N161 lines, the 4C/2C ratio was significantly higher at day 9 than that in wild-type leaves (Figures 3B and 3C). The increase in the number of cells with a 4C DNA content reflects the need for CDKB1;1 activity to progress through mitosis (Porceddu et al., 2001; Boudolf et al., 2004). In contrast with wild-type leaves, the 2C levels in CDKB1;1.N161 leaves already decreased between day 9 and day 13, which was accompanied by a further increase in 4C and 8C levels (Figures 3B and 3C). These data illustrate that the CDKB1;1.N161 leaves entered the endocycle earlier than wild-type plants. An advance entry into the endocycle can also clearly been seen at the cellular level. When the ploidy levels were compared of individual abaxial pavement cells in the first leaves 14 d after germination, a higher number of 8C cells could be observed in the transgenic plants (Figures 2A and 2B). In mature leaves, the ploidy levels were overall higher in the transgenic than in the control lines with 8C levels, reaching up to 45.3% ± 4.4% (35S promoter of Cauliflower mosaic virus [CaMV35S]:CDKB1;1.N161 line 1.2) and 43.1% ± 1.7% (CaMV35S:CDKB1;1.N161 line 9.2), compared with 19.2% ± 1.6% in control leaves. In addition, a small but reproducible 32C peak was detected in the transgenic lines but not in the pavement cells of control leaves. This endoreduplication phenotype could not be reverted by the co-overexpression with CDKA;1 (data not shown), indicating that the effect on the endocycle is specifically because of an inhibition of the B-type CDK activity.
Figure 3.
DNA Ploidy Level Distribution of the First Leaves of Wild-Type (Col-0) and CDKB1;1.N161-Overproducing Plants during Development.
(A) Wild type (Col-0).
(B) CaMV35S:CDKB1;1.N161 line 1.2.
(C) CaMV35S:CDKB1;1.N161 line 9.2. Leaves were harvested at the indicated time points. Data represent average ± sd.
Symbols in (A) and (B) as in (C).
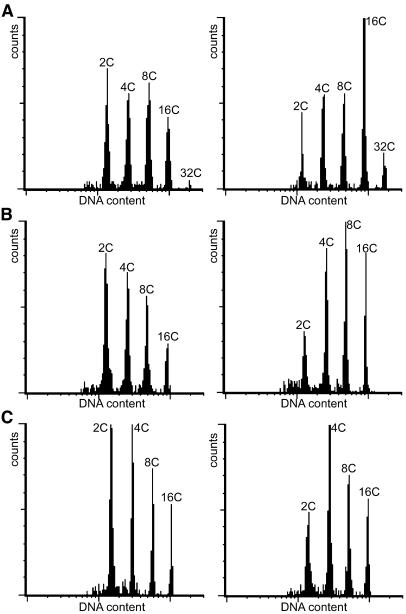
Reduced CDKB1;1 activity also stimulated endoreduplication in other tissues than leaves. In cotyledons, hypocotyls, and roots, the relative abundance of cells with a high ploidy level increased at the cost of cells with a low DNA content (Figure 4). No significant effect on the ploidy level of trichomes was observed, which might be because CaMV35S promoter activity in these cells is too low to counteract the endogeneous CDKB1;1 protein.
Figure 4.
DNA Ploidy Level Distribution of Wild-Type Plants (Col-0; Left) and CDKB1;1.N161-Overproducing Plants (Right) in Different Tissues.
(A) Cotyledons.
(B) Hypocotyls.
(C) Roots.
CDKB1;1 Is Only Expressed during the Mitotic Phase of Leaf Development
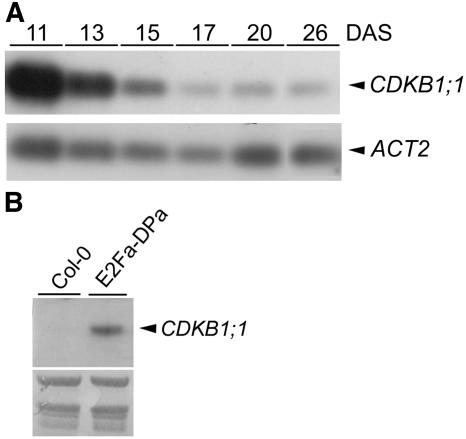
The temporal expression pattern of CDKB1;1 during leaf development was studied by RT-PCR with RNA prepared from the first leaves harvested between 11 and 26 d after sowing. CDKB1;1 transcript levels were high at day 11 and thereafter decreased to low basal levels (Figure 5A), which coincided with the onset of endoreduplication. By contrast, CDKA;1 was expressed at an almost constant level during leaf development (data not shown).
Figure 5.
Expression Analysis of the CDKB1;1 Gene.
(A) Transcript levels of CDKB1;1 during leaf development. cDNA prepared from the first leaf pair harvested at the indicated time points was subjected to semiquantitative RT-PCR analysis with gene-specific primers. The actin 2 (ACT2) gene was used as loading control. DAS, days after sowing.
(B) RNA gel blot analysis of CDKB1;1. RNA was extracted from 12-d-old control (Col-0) and E2Fa-DPa seedlings. Equal loading of the gel was confirmed by methylene blue staining of the membrane (bottom panel).
CDKB1;1.N161 Suppresses Ectopic Divisions Triggered by E2Fa-DPa
Previously, we have postulated that the decision of cells to undergo a mitotic cell cycle or to endoreduplicate depends on the presence of an MIF. Because of its role in the G2-M transition and the observed increase of endoreduplication upon overexpression of its dominant negative allele, CDKB1;1 is a good candidate to be part of this MIF. To test this hypothesis, E2Fa-DPa–overexpressing plants were crossed with a CDKB1;1.N161–overexpressing line. As control, CaMV35:CDKB1;1.N161 and CaMV35S:E2Fa-DPa lines were crossed separately with wild-type plants. The fidelity of the crosses was confirmed by PCR analysis and RNA gel blots (data not shown). In a first series of experiments, 6-d-old seedlings were analyzed by flow cytometry. As described above for the developing leaves, overexpression of CDKB1;1.N161 resulted in an increase of the 8C and 16C populations, corresponding with a decrease in the number of cells with a 2C DNA content (Table 1). As reported previously, overexpression of E2Fa-DPa resulted in the appearance of two additional endocycles (Table 1; De Veylder et al., 2002). When CDKB1;1.N161 and E2Fa-DPa were co-overexpressed, an additive phenotype was seen, resulting in an even stronger reduction in the amount of 2C cells, whereas the population of 16C and 32C cells increased.
Table 1.
DNA Ploidy Levels in 6-d-Old Wild-Type and Transgenic Seedlings
| Line | 2C (%) | 4C (%) | 8C (%) | 16C (%) | 32C (%) | 64C (%) |
|---|---|---|---|---|---|---|
| Col-0 | 26.6 ± 3.3 | 34.6 ± 1.4 | 30.2 ± 1.4 | 6.6 ± 1.6 | ||
| CDKB1;1.N161 × Col-0 | 14.7 ± 1.1 | 29.1 ± 4.2 | 35.9 ± 2.7 | 14.7 ± 0.9 | ||
| Col-0 × E2Fa-DPa | 18.6 ± 2.3 | 32.6 ± 1.6 | 23.5 ± 0.7 | 16.0 ± 1.6 | 5.4 ± 0.8 | 0.9 ± 0.2 |
| CDKB1;1.N161 × E2Fa-DPa | 13.1 ± 1.2 | 27.6 ± 1.9 | 25.4 ± 1.7 | 23.7 ± 2.1 | 8.0 ± 0.8 | 1.1 ± 0.2 |
Data represent average ± SD (n = 4 to 6).
While CDKB1;1.N161 overexpression stimulated the endocycle phenotype of E2Fa-DPa transgenics, it simultaneously suppressed the occurrence of ectopic cell divisions, as can been seen for abaxial epidermal pavement cells of 6-d-old cotyledons (Figure 6, Table 2). Similarly as described above for leaves, overexpression of CDKB1;1.N161 in cotyledons led to an increase in pavement cell size (Figures 6A and 6B). By contrast, the cotyledon size of E2Fa-DPa transgenic plants was considerably smaller, but many more epidermal cells were observed because of ectopic cell division (Figure 6C). However, when CDKB1;1.N161 was expressed in an E2Fa-DPa transgenic background, the ectopic cell division phenotype was repressed (Figure 6D), resulting in an average cell size being only slightly smaller than that of wild-type cells.
Figure 6.
Drawing-Tube Images of Wild-Type and Transgenic Plants.
(A) Wild type (Col-0).
(B) CaMV35S:CDKB1;1N161 × Col-0.
(C) Col-0 × CaMV35S:E2Fa-DPa.
(D) CaMV35S:CDKB1;1.N161 × CaMV35S:E2Fa-DPa.
Table 2.
Abaxial Pavement Cell Size and Cell Number in Cotyledons of CDKB1;1.N161 and E2Fa-DPa Transgenic Lines
| Abaxial Pavement Cells
|
|||
|---|---|---|---|
| Line | Cotyledon Size (mm2) | Size (μm2) | Estimated Number |
| Col-0 | 2.6 ± 0.2 | 2082 ± 110 | 1284 ± 170 |
| CDKB1.1.N161 × Col-0 | 2.6 ± 0.1 | 3925 ± 209 | 664 ± 40 |
| Col-0 × E2Fa-DPa | 1.6 ± 0.2 | 232 ± 33 | 7563 ± 1115 |
| CDKB1;1.N161 × E2Fa-DPa | 1.8 ± 0.1 | 2010 ± 110 | 910 ± 63 |
All measurements were performed on cotyledons harvested 6 d after sowing. The indicated values are means ± SE (n = 5 to 10).
CDKB1;1 Transcription Is Regulated by E2Fa-DPa
Previously, microarray analysis has revealed that CDKB1;1 transcripts are upregulated in E2Fa-DPa–overproducing plants (Vlieghe et al., 2003). To validate this result, CDKB1;1 expression levels were determined in wild-type versus E2Fa-DPa transgenic plants via RNA gel blotting. In wild-type control plants, no CDKB1;1 transcripts were detected, reflecting low expression levels (Figure 5B). By contrast, in the E2Fa-DPa transgenic plants a strong CDKB1;1 hybridization signal was observed, confirming the previous microarray result.
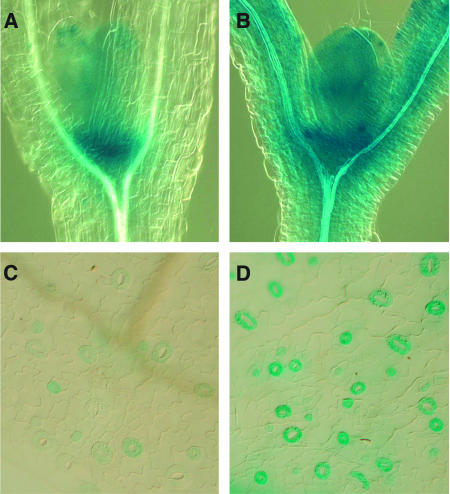
To test the tissue specificity of the CDKB1;1 induction in E2Fa-DPa seedlings, a CDKB1;1 promoter β-glucuronidase (GUS) construct was crossed into the E2Fa-DPa transgenic lines, and the spatial GUS expression pattern was compared with that of control plants. In the vegetative part of control plants, CDKB1;1 promoter activity could be observed in the shoot apical meristem, in the young leaf primordia, and in stomata (Figures 7A and 7C; Boudolf et al., 2004). In the E2Fa-DPa transgenic plants, GUS staining was significantly stronger (Figures 7B and 7D). GUS activity was present not only in a much broader region surrounding the meristem, probably in correlation with the occurrence of ectopic cell divisions, but was also stronger at the cellular level, as seen in leaf primordia and stomatal cells. These data suggest that the increased CDKB1;1 transcript levels observed in the E2Fa-DPa transgenic lines are not only the result of the ectopic cell division but also originate by a direct stimulation of CDKB1;1 promoter activity.
Figure 7.
Activity of the CDKB1;1 Promoter Fused to the GUS Reporter (pCDKB;1:GUS) in Wild-Type (Col-0) and E2Fa-DPa–Overexpressing Plants Visualized by Histochemical Staining.
(A) GUS activity in the shoot apex of untransformed plants.
(B) GUS activity in the shoot apex of E2Fa-DPa transgenic plants.
(C) GUS activity in stomata of untransformed plants.
(D) GUS activity in stomata of E2Fa-DPa transgenic plants.
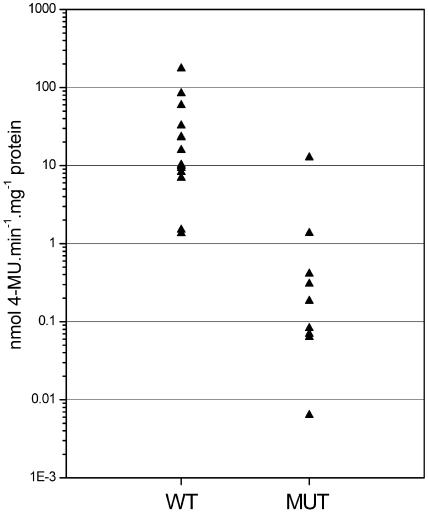
In the promoter of the CDKB1;1 gene, an E2F-like binding site (TTTCCCGC) can be detected, located 151 bp downstream of the start codon. To investigate how this promoter element contributes to the level of CDKB1;1 transcription, the E2F binding site was mutated into TTTCCAAC. This mutation has previously been demonstrated to impair E2F-DP binding (Kosugi and Ohashi, 2002). Both wild-type and mutant promoters were transcriptionally fused to the GUS reporter gene, and these reporter constructs were used to transform tobacco (Nicotiana tabacum) Bright Yellow-2 (BY-2) cells. For each construct, 14 independent calli were isolated, in which the level of GUS protein was quantified by fluorometry. On the average, GUS activity was 29-fold higher in the calli containing the wild-type promoter construct than in those harboring the mutant promoter (Figure 8). These data illustrate that the E2F cis-acting element is a major factor in regulating CDKB1;1 promoter activity.
Figure 8.
Quantification of Wild-Type and Mutant CDKB1;1 Promoter Activity.
GUS activity was measured fluorometrically in protein extracts prepared from transgenic BY-2 cells transformed with the wild type (n = 14) or mutant (MUT; n = 14) CDKB1;1 promoter fused to the GUS reporter gene. 4-MU, 4-methylumbelliferone.
DISCUSSION
In yeast and Drosophila, downregulation of M-phase–associated CDK activity is sufficient to drive cells into the endoreduplication cycle (Hayles et al., 1994; Sauer et al., 1995). A similar mechanism is probably operational in plants because the onset of endoreduplication in maize endosperm, during tomato fruit development, and Arabidopsis leaf development is correlated with the inhibition of mitotic CDK activity (Grafi and Larkins, 1995; Joubès et al., 1999). Here, we have demonstrated that CDKB1;1 controls the balance between mitotic cell division and endoreduplication in both wild-type and transgenic plants overproducing both E2Fa and DPa. Such a role for CDKB1;1 was hinted by its temporal expression pattern during leaf development. CDKB1;1 is highly expressed in dividing cells and is downregulated at the onset of endoreduplication. This result is in agreement with previously published data that demonstrate that CDKB1;1 transcription occurs only in the mitotically dividing cells of the shoot apex (Segers et al., 1996). Additionally, we found that by decreasing the CDKB1;1 activity, cells exit the mitotic cell cycle earlier, accompanied with an early onset of the endocycle. This untimely exit of the mitotic cell cycle results in leaves with fewer but larger cells than those in wild-type leaves, which have a higher ploidy level. An increase in cell size in response to an inhibition of cell division has been observed previously and can be attributed to an uncoupling of cell division and cell expansion (Hemerly et al., 1995; De Veylder et al., 2001). Because of the observed correlation between ploidy level and cell dimension, an increase in ploidy level in response to a perturbation in the cell cycle is often seen as a mechanism to accommodate the increase in cell size. As such, the extra endoreduplication observed in the CDKB1;1.N161 plants might be triggered by the need for cell growth. However, the amplified ploidy level is more readily a primary phenotype triggered by CDKB1;1.N161 rather than a secondary effect induced by cell expansion because an increase in the endoreduplication level could already be observed when cell number and size were indistinguishable between wild-type and transgenic lines. Because both wild-type and CDKB1;1.N161-overexpressing plants exit the endocycle at the same stage during leaf development (leaf 1 at 21 d after sowing), the higher ploidy levels observed in the CDKB1;1.N161 plants can be largely attributed to the earlier start of the endocycle program.
Whereas overproduction of the dominant negative CDKB1;1.N161 stimulates endoreduplication, overexpression of the wild-type CDKB1;1 gene had no effect on the ploidy distribution of wild-type plants. This discrepancy can be explained by the lack of a regulatory factor (such as a cyclin) for CDKB1;1 activity. Indeed, overexpression of CDKB1;1 in a wild-type background does not significantly alter the amount of extractable CDK activity (Boudolf et al., 2004). A good candidate to act in concert with CDKB1;1 is the CYCA3;2 cyclin, whose overproduction has been demonstrated to strongly inhibit the endocycle (Yu et al., 2003).
CDKB1;1 transcription is regulated through an E2F cis-acting element in its promoter, which is surprising because E2F-DP transcription factors are presumed to operate at the G1-S transition, whereas CDKB1;1 kinase activity peaks at G2-M. However, in synchronized cultures, CDKB1;1 transcript levels increase from the G1-S transition onwards, followed by an increase in CDKB1;1 protein and associated activity during S-phase (Porceddu et al., 2001; Sorrell et al., 2001; Breyne et al., 2002; Menges et al., 2002). In all eukaryotes, DNA replication is initiated by the stepwise binding of proteins to the origins of replication (Larkins et al., 2001; Nishitani and Lygerou, 2002). After binding of the DNA replication-licensing proteins ORC, CDC6, CDC18, and CDT1 to these origins, which form the prereplication complex, replication is initiated by the recruitment of MCM proteins. As soon as replication starts, the MCM proteins dissemble from the replication complex, and reformation of the complex is inhibited until sister chromatids are separated in mitosis. In fission yeast, the mitotic cyclin B/cdc2 complex has been demonstrated to prevent the relicensing of replicated DNA by association with the replication complex during chromosome duplication, preventing the MCM proteins to reinitiate DNA synthesis (Wuarin et al., 2002). Similarly, CDKB1;1 activity might repress the endocycle in plants by association with the prereplication complex. This hypothesis suggests that besides its anticipated role at the G2-M transition, CDKB1;1 also plays a role earlier in the cell cycle.
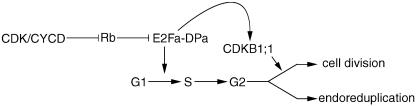
Previously, we have demonstrated that E2Fa-DPa regulates both the mitotic and endoreduplication cycles of Arabidopsis. Cells in which this heterodimeric transcription factor are overexpressed were shown to either undergo extra mitotic divisions or to endoreduplicate. The decision to divide mitotically or to endoreduplicate was postulated to depend on the presence of an MIF (De Veylder et al., 2002). Upon the coexpression of CDKB1;1.N161, the ectopic cell division phenotype of the E2Fa-DPa transgenic plants was repressed, whereas the endoreduplication phenotype is enhanced. These data illustrate that factors important for mitosis, which are stimulated by E2Fa-DPa overexpression are titrated out by CDKB1;1.N161. Therefore, CDKB1;1 is the most probable candidate to be part of the MIF. Cells that possess CDKB1;1 activity undergo mitotic division, whereas those that lack it are stimulated by E2Fa-DPa to enter the endocycle (Figure 9).
Figure 9.
Model Illustrating the Interaction between E2Fa-DPa and CDKB1;1 Activity to Drive the Mitotic Cell Cycle.
For details, see text. Rb, retinoblastoma.
Although the switch between mitosis and endocycle almost certainly occurs at the protein level by the specific destruction of M-phase–specific regulators by the CCS52 complex, the absence of CDKB1;1 transcripts in polyploid tissues suggests a mechanism that is operational to suppress the transcriptional activation of CDKB1;1 in endoreduplicating cells. The Arabidopsis E2F factors can be functionally subdivided into activators (E2Fa and E2Fb) and a repressor (E2Fc) (De Veylder et al., 2003). It is possible that the CDKB1;1 promoter is associated sequentially with different E2F proteins, with repressing E2Fs replacing an activating E2F complex when cells shift from the mitotic cell cycle into the endocycle. However, in contrast with the proliferating cell nuclear antigen promoter of tobacco, in which the E2F binding sites function as negative regulatory elements that repress transcription in mature leaves (Egelkrout et al., 2001, 2002), deletion of the E2F binding site in the CDKB1;1 promoter does not result in increased transcriptional activity in maturing leaves (data not shown), strongly suggesting that CDKB1;1 transcription is solely regulated by activating E2Fs. Therefore, the difference in CDKB1;1 promoter activity in mitotically dividing versus endoreduplicating cells might rather be regulated through the interaction of E2F with other transcriptional activators and repressors bound to the promoter. The combinatorial nature of E2F transcription factors has been demonstrated in mammals, in which activating E2Fs are recruited to a promoter via synergistic interactions with adjacent transcription factors (Schlisio et al., 2002; Giangrande et al., 2003, 2004). At the moment, the nature of the transcription factor that would distinguish an endoreduplicating cell from a mitotically dividing cell remains unknown.
Although our understanding of how cell cycle transitions are regulated has gradually improved, we hardly comprehend how the different transitions communicate with each other. The transcriptional induction of CDKB1;1 by E2F-DP transcription factors suggests a mechanism by which the G1-S and G2-M cell cycle checkpoints communicate. The existence of such linkage has already been hinted by the observation that ectopic expression of the D-type cyclin CYCD3;1 not only induces DNA replication but also cell division in Arabidopsis trichomes (Schnittger et al., 2002). In budding yeast, the combination of chromatin immunoprecipitation assays with DNA microarray analysis has revealed that proteins that operate as activators during one stage of the cell cycle can contribute to the transcriptional activation of proteins that function during the next stage, forming a fully connected regulatory circuit (Simon et al., 2001). The scarce data available suggest that a similar circuit might be operational in plants. However, it is clear that to fully understand cell cycle functioning, experiments aimed at unraveling connections between different checkpoints will be crucial.
METHODS
Generation of Transgenic Plants
Plants harboring the wild-type CDKB1;1 or mutant CDKB1;1.N161 gene under the control of CaMV35S (Boudolf et al., 2004) and plants coexpressing the DPa and E2Fa genes (De Veylder et al., 2002) were constructed as described previously. Triple CaMV35S:CDKB1;1.N161-E2Fa-DPa plants were obtained by crossing double homozygous CaMV35S:E2Fa-DPa plants with homozygous CaMV35S:CDKB1;1.N161 plants.
Kinematic Analysis of Leaf Growth
Leaf growth was analyzed kinematically as described (De Veylder et al., 2001). Briefly, wild-type and CDKB1;1.N161-overproducing plants were germinated and grown in round 12-cm Petri dishes filled with 100 mL of 1× MS medium (Duchefa, Haarlem, The Netherlands) and 0.6% plant tissue culture agar (LabM, Bury, UK) at 22°C and 65 μE m−2 s−1 radiation in a 16-h-light/8-h-dark photoperiod. From day 5 until day 28 after sowing, plants were harvested, cleared overnight in methanol, and subsequently stored in lactic acid for microscopy. The youngest plants were mounted on a slide and covered. The leaf primordia were observed under a microscope fitted with differential interference contrast optics (DMLB; Leica, Wetzlar, Germany). The total (blade) area of leaves 1 and 2 of each seedling were first determined from drawing-tube images with the public domain image analysis program ImageJ (version 1.30; http://rsb.info.nih.gov/ij/). At older stages, the primordia were digitized directly with a charge-coupled device camera mounted on a binocular (Stemi SV11; Zeiss, Jena, Germany). Cell density was determined from scanned drawing-tube images of outlines of at least 20 cells of the abaxial epidermis located 25 and 75% from the distance between the tip and the base of the leaf primordium (or blade once the petiole was present), halfway between the midrib and the leaf margin. In the youngest primordia (up to day 6), a single group of cells was drawn. The following parameters were determined: total area of all cells in the drawing, total number of cells, and number of guard cells. From these data, we calculated the average cell area and estimated the total number of cells per leaf by dividing the leaf area by the average cell area (averaged between the apical and basal positions). Finally, average cell division rates for the whole leaf were determined as the slope of the log2-transformed number of cells per leaf, which was done using five-point differentiation formulas (Erickson, 1976).
Flow Cytometric Analysis of Leaves
Leaves were chopped with a razor blade in 300 μL of buffer (45 mM MgCl2, 30 mM sodium citrate, 20 mM 3-[N-morpholino]propanesulfonic acid, pH 7, and 1% Triton X-100) (Galbraith et al., 1991). To the supernatants, 1 μL of 4′,6-diamidino-2-phenylindole from a stock of 1 mg/mL was added, which was filtered over a 30-μm mesh. The nuclei were analyzed with the BRYTE HS or CyFlow flow cytometer, using Win-Bryte software (Bio-Rad, Hercules, CA) or FloMax software (Partec, Münster, Germany), respectively.
Epidermal Peels and Measurements of Nuclear DNA Quantity
The protocol for making epidermal peels was adapted from Melaragno et al. (1993). Three-week-old plants were fixed in a solution of three-quarters 95% ethanol and one-quarter glacial acetic acid for 2 h at room temperature and stored in 70% ethanol at 4°C. Fixed tissue was soaked first in water and then in 0.5 M EDTA, pH 8. Leaves were placed with the abaxial epidermis side down on a glass slide and held in place with forceps, while tissue was removed from the abaxial epidermis with another pair of fine forceps. A drop of 4′,6-diamidino-2-phenylindole at a concentration of 0.005 mg/mL in McIlvaines's buffer, pH 4.1 (60 mL 0.1 mol/L citric acid + 40 mL 0.2 mol/L Na2HPO4), was placed on the epidermal peel. Peels were mounted in Vectashield mounting medium (Vector Laboratories, Burlingame, CA) and observed under a 20× objective on a Zeiss Axioskop equipped with an Axiocam CCD camera (Zeiss). Images were obtained with the Axiovision software and were analyzed in gray scale with the public domain image analysis program ImageJ (version 1.28; http://rsb.info.nih.gov/ij/). Relative fluorescence units were measured as integrated density, which is the product of the area and the average fluorescence of the selected nucleus.
Promoter Analysis
The intergenic region between the CDKB1;1 (At3g54180) open reading frame and the upstream open reading frame (486 bp) was isolated from Col-0 genomic DNA by PCR using the 5′-AAAAAGCAGGCTGAAGATGCAAAAGAAGAAATTGCTGGTGG-3′ and 5′-AGAAAGCTGGGTTTCTGAGAGGTTTCGTAAAATTGAACTGTG-3′ primers, and the resulting PCR fragment was cloned into the GATEWAY pDONR 207 vector (Invitrogen, Carlsbad, CA) by attB × attP recombination reaction. The resulting plasmid was used to mutate the E2F binding site by PCR with the outward-oriented 5′-AAGGAAAAACGAAGTGACGTGGAAG-3′ and 5′-CTCCTTTGGATTTTGAATTCGTTCG-3′ primers. The PCR fragment was phosphorylated with the T4 polynucleotide kinase and self-ligated. The introduced mutations were confirmed by sequencing. The wild-type and mutant promoter fragments were subsequently recombined into the pKGWFS7 vector by attL × attR recombination reaction at the GATEWAY recombination site located in front of the gene coding for the enhanced green fluorescent protein-GUS (Karimi et al., 2002), resulting in the pCDKBWT and pCDKBmut vectors, respectively. The vectors were used to transform tobacco (Nicotiana tabacum) BY-2 cells as described (Joubès et al., 2004). For both constructs, 14 calli were selected whose GUS activity was measured as described (Zambre et al., 2003).
RT-PCR Analysis
RNA was extracted from leaves of Arabidopsis thaliana Heynh. (ecotype Col-0) with TRIzol reagent (Invitrogen). First-strand cDNA synthesis was performed on 3 μg of total RNA with the Superscript RT II kit (Invitrogen) and oligo(dT)18 according to the manufacturer's instructions. A 1-μL aliquot of the total reverse transcription reaction volume (20 μL) was used as template in the semiquantitative RT-PCR amplification, ensuring that the amount of amplified product remained in linear proportion to the initial template present in the reaction. Ten microliters from the PCR reaction was separated on a 0.8% agarose gel and transferred onto Hybond N+ membranes (Amersham Biosciences, Little Chalfont, UK). The membranes were hybridized at 65°C with fluorescein-labeled probes (Gene Images random prime module; Amersham Biosciences). The hybridized bands were detected with the CDP Star detection module (Amersham Biosciences). Primers used were as follows: 5′-GGTGGTGACATGTGGTCTGTTGG-3′ and 5′-CGCAGTGTGGAAACACCCGG-3′ for CDKB1;1 and 5′-GGCTCCTCTTAACCCAAAGGC-3′ and 5′-CACACCATCACCAGAATCCAGC-3′ for ACT2 (At3g18780).
Acknowledgments
The authors thank the members of the cell cycle group for fruitful discussions and suggestions and Martine De Cock for help in preparing the manuscript. This research was supported by grants from the Interuniversity Poles of Attraction Programme-Belgian Science Policy (P5/13), the European Union (European Cell Cycle Consortium QLG2-CT1999-00454), and the Bilateral Scientific and Technological Cooperation Programme Flanders-Hungary (BIL 03/09). V.B. and K.V. are indebted to the Instituut voor de aanmoediging van Innovatie door Wetenschap en Technologie in Vlaanderen for a predoctoral fellowship. J.A.T.A. was a doctorate fellow from the Consejo Nacional de Ciencia y Tecnologia, Mexico (Grant 120198), and L.D.V. is a postdoctoral fellow of the Fund for Scientific Research (Flanders).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Lieven De Veylder (lieven.deveylder@psb.ugent.be).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.024398.
References
- Boudolf, V., Barrôco, R., de Almeida Engler, J., Verkest, A., Beeckman, T., Naudt, M., Inzé, D., and De Veylder, L. (2004). B1-type cyclin-dependent kinases are essential for the formation of stomatal complexes in Arabidopsis thaliana. Plant Cell 16, 945–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breyne, P., Dreesen, R., Vandepoele, K., De Veylder, L., Van Breusegem, F., Callewaert, L., Rombauts, S., Raes, J., Cannoot, B., Engler, G., Inzé, D., and Zabeau, M. (2002). Transcriptome analysis during cell division in plants. Proc. Natl. Acad. Sci. USA 99, 14825–14830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky, V.Y., and Uryvaeva, I.V. (1977). Cell polyploidy: Its relation to tissue growth and function. Int. Rev. Cytol. 50, 275–332. [DOI] [PubMed] [Google Scholar]
- Cebolla, A., Vinardell, J.M., Kiss, E., Oláh, B., Roudier, F., Kondorosi, A., and Kondorosi, E. (1999). The mitotic inhibitor ccs52 is required for endoreduplication and ploidy-dependent cell enlargement in plants. EMBO J. 18, 4476–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amato, F. (1964). Endopolyploidy as a factor in plant tissue development. Caryologia 17, 41–52. [Google Scholar]
- De Veylder, L., Beeckman, T., Beemster, G.T.S., de Almeida Engler, J., Ormenese, S., Maes, S., Naudts, M., Van Der Schueren, E., Jacqmard, A., Engler, G., and Inzé, D. (2002). Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa/DPa transcription factor. EMBO J. 21, 1360–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder, L., Beeckman, T., Beemster, G.T.S., Krols, L., Terras, F., Landrieu, I., Van Der Schueren, E., Maes, S., Naudts, M., and Inzé, D. (2001). Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13, 1653–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder, L., Joubès, J., and Inzé, D. (2003). Plant cell cycle transitions. Curr. Opin. Plant Biol. 6, 536–543. [DOI] [PubMed] [Google Scholar]
- Egelkrout, E.M., Mariconti, L., Settlage, S.B., Cella, R., Robertson, D., and Hanley-Bowdoin, L. (2002). Two E2F elements regulate the proliferating cell nuclear antigen promoter differently during leaf development. Plant Cell 14, 3225–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelkrout, E.M., Robertson, D., and Hanley-Bowdoin, L. (2001). Proliferating cell nuclear antigen transcription is repressed through an E2F consensus element and activated by geminivirus infection in mature leaves. Plant Cell 13, 1437–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson, R.O. (1976). Modeling of plant growth. Annu. Rev. Plant Physiol. 27, 407–434. [Google Scholar]
- Folkers, U., Berger, J., and Hülskamp, M. (1997). Cell morphogenesis of trichomes in Arabidopsis: Differential control of primary and secondary branching by branch initiation regulators and cell growth. Development 124, 3779–3786. [DOI] [PubMed] [Google Scholar]
- Galbraith, D.W., Harkins, K.R., and Knapp, S. (1991). Systemic endopolyploidy in Arabidopsis thaliana. Plant Physiol. 96, 985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giangrande, P.H., Hallstrom, T.C., Tunyaplin, C., Calame, K., and Nevins, J.R. (2003). Identification of E-box factor TFE3 as a functional partner for the E2F3 transcription factor. Mol. Cell. Biol. 23, 3707–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giangrande, P.H., Zhu, W., Rempel, R.E., Laakso, N., and Nevins, J.R. (2004). Combinatorial gene control involving E2F and E Box family members. EMBO J. 23, 1336–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafi, G., and Larkins, B.A. (1995). Endoreduplication in maize endosperm: Involvement of M phase-promoting factor inhibition and induction of S phase-related kinases. Science 269, 1262–1264. [DOI] [PubMed] [Google Scholar]
- Hayles, J., Fisher, D., Woollard, A., and Nurse, P. (1994). Temporal order of S phase and mitosis in fission yeast is determined by the state of the p34cdc2-mitotic B cyclin complex. Cell 78, 813–822. [DOI] [PubMed] [Google Scholar]
- Hemerly, A., de Almeida Engler, J., Bergounioux, C., Van Montagu, M., Engler, G., Inzé, D., and Ferreira, P. (1995). Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO J. 14, 3925–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacqmard, A., De Veylder, L., Segers, G., de Almeida Engler, J., Bernier, G., Van Montagu, M., and Inzé, D. (1999). Expression of CKS1At in Arabidopsis thaliana indicates a role for the protein in both the mitotic and the endoreduplication cycle. Planta 207, 496–504. [DOI] [PubMed] [Google Scholar]
- Joubès, J., and Chevalier, C. (2000). Endoreduplication in higher plants. Plant Mol. Biol. 43, 735–745. [DOI] [PubMed] [Google Scholar]
- Joubès, J., De Schutter, K., Verkest, A., Inzé, D., and De Veylder, L. (2004). Conditional, recombinase-mediated, expression of genes in plant cell cultures. Plant J. 37, 889–896. [DOI] [PubMed] [Google Scholar]
- Joubès, J., Phan, T.-H., Just, D., Rothan, C., Bergounioux, C., Raymond, P., and Chevalier, C. (1999). Molecular and biochemical characterization of the involvement of cyclin-dependent kinase A during the early development of tomato fruit. Plant Physiol. 121, 857–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi, M., Inzé, D., and Depicker, A. (2002). GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Kondorosi, E., Roudier, F., and Gendreau, E. (2000). Plant cell-size control: Growing by ploidy? Curr. Opin. Plant Biol. 3, 488–492. [DOI] [PubMed] [Google Scholar]
- Kosugi, S., and Ohashi, Y. (2002). E2F sites that can interact with E2F proteins cloned from rice are required for meristematic tissue-specific expression of rice and tobacco proliferating cell nuclear antigen promoters. Plant J. 29, 45–59. [DOI] [PubMed] [Google Scholar]
- Kosugi, S., and Ohashi, Y. (2003). Constitutive E2F expression in tobacco plants exhibits altered cell cycle control and morphological change in a cell type-specific manner. Plant Physiol. 132, 2012–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib, K., Craven, R.A., Crawford, K., and Nurse, P. (1995). Dominant mutants identify new roles for p34cdc2 in mitosis. EMBO J. 14, 2155–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkins, B.A., Dilkes, B.P., Dante, R.A., Coelho, C.M., Woo, Y.-m., and Liu, Y. (2001). Investigating the hows and whys of DNA endoreduplication. J. Exp. Bot. 52, 183–192. [PubMed] [Google Scholar]
- Melaragno, J.E., Mehrotra, B., and Coleman, A.W. (1993). Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5, 1661–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges, M., Hennig, L., Gruissem, W., and Murray, J.A.H. (2002). Cell cycle-regulated gene expression in Arabidopsis. J. Biol. Chem. 277, 41987–42002. [DOI] [PubMed] [Google Scholar]
- Nishitani, H., and Lygerou, Z. (2002). Control of DNA replication licensing in a cell cycle. Genes Cells 7, 523–534. [DOI] [PubMed] [Google Scholar]
- Porceddu, A., Stals, H., Reichheld, J.-P., Segers, G., De Veylder, L., De Pinho Barrôco, R., Casteels, P., Van Montagu, M., Inzé, D., and Mironov, V. (2001). A plant-specific cyclin-dependent kinase is involved in the control of G2/M progression in plants. J. Biol. Chem. 276, 36354–36360. [DOI] [PubMed] [Google Scholar]
- Sauer, K., Knoblich, J.A., Richardson, H., and Lehner, C.F. (1995). Distinct modes of cyclin E/cdc2c kinase regulation and S-phase control in mitotic and endoreduplication cycles of Drosophila embryogenesis. Genes Dev. 9, 1327–1339. [DOI] [PubMed] [Google Scholar]
- Schlisio, S., Halperin, T., Vidal, M., and Nevins, J.R. (2002). Interaction of YY1 and E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J. 21, 5775–5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger, A., Schöbinger, U., Bouyer, D., Weinl, C., Stierhof, Y.-D., and Hülskamp, M. (2002). Ectopic D-type cyclin expression induces not only DNA replication but also cell division in Arabidopsis trichomes. Proc. Natl. Acad. Sci. USA 99, 6410–6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segers, G., Gadisseur, I., Bergounioux, C., de Almeida Engler, J., Jacqmard, A., Van Montagu, M., and Inzé, D. (1996). The Arabidopsis cyclin-dependent kinase gene cdc2bAt is preferentially expressed during S and G2 phases of the cell cycle. Plant J. 10, 601–612. [DOI] [PubMed] [Google Scholar]
- Simon, I., Barnett, J., Hannett, N., Harbison, C.T., Rinaldi, N.J., Volkert, T.L., Wyrick, J.J., Zeitlinger, J., Gifford, D.K., Jaakkola, T.S., and Young, R.A. (2001). Serial regulation of transcriptional regulators in the yeast cell cycle. Cell 106, 697–708. [DOI] [PubMed] [Google Scholar]
- Sorrell, D.A., Menges, M., Healy, J.M.S., Deveaux, Y., Amano, X., Su, Y., Nakagami, H., Shinmyo, A., Doonan, J.H., Sekine, M., and Murray, J.A.H. (2001). Cell cycle regulation of cyclin-dependent kinases in tobacco cultivar Bright Yellow-2 cells. Plant Physiol. 126, 1214–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto-Shirasu, K., and Roberts, K. (2003). “Big it up”: Endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 6, 1–10. [DOI] [PubMed] [Google Scholar]
- Sun, Y., Dilkes, B.P., Zhang, C., Dante, R.A., Carneiro, N.P., Lowe, K.S., Jung, R., Gordon-Kamm, W.J., and Larkins, B.A. (1999). Characterization of maize (Zea mays L.) Wee1 and its activity in developing endosperm. Proc. Natl. Acad. Sci. USA 96, 4180–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traas, J., Hülskamp, M., Gendreau, E., and Höfte, H. (1998). Endoreduplication and development: Rule without dividing? Curr. Opin. Plant Biol. 1, 498–503. [DOI] [PubMed] [Google Scholar]
- Trimarchi, J.M., and Lees, J.A. (2002). Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 3, 11–20. [DOI] [PubMed] [Google Scholar]
- van den Heuvel, S., and Harlow, E. (1993). Distinct roles for cyclin-dependent kinases in cell cycle control. Science 262, 2050–2054. [DOI] [PubMed]
- Vandepoele, K., Raes, J., De Veylder, L., Rouzé, P., Rombauts, S., and Inzé, D. (2002). Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14, 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieghe, K., Florquin, K., Vuylsteke, M., Rombauts, S., Maes, S., Ormenese, S., Van Hummelen, P., Van de Peer, Y., Inzé, D., and De Veylder, L. (2003). Microarray analysis of E2Fa-DPa-overexpressing plants reveals changes in the expression levels of genes involved in DNA replication, cell wall biosynthesis, and nitrogen assimilation. J. Cell Sci. 116, 4249–4259. [DOI] [PubMed] [Google Scholar]
- Wuarin, J., Buck, V., Nurse, P., and Millar, J.B.A. (2002). Stable association of mitotic cyclin B/Cdc2 to replication origins prevents endoreduplication. Cell 111, 419–431. [DOI] [PubMed] [Google Scholar]
- Yoshizumi, T., Nagata, N., Shimada, H., and Matsui, M. (1999). An Arabidopsis cell cycle-dependent kinase-related gene, CDC2b, plays a role in regulating seedling growth in darkness. Plant Cell 11, 1883–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y., Steinmetz, A., Meyer, D., Brown, S., and Shen, W.-H. (2003). The tobacco A-type cyclin, Nicta;CYCA3;2, at the nexus of cell division and differentiation. Plant Cell 15, 2763–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambre, M., Terryn, N., De Clercq, J., De Buck, S., Dillen, W., Van Montagu, M., Van Der Straeten, D., and Angenon, G. (2003). Light strongly promotes gene transfer from Agrobacterium tumefaciens to plant cells. Planta 216, 580–586. [DOI] [PubMed] [Google Scholar]