Abstract
The first enzyme of the phenylpropanoid pathway, Phe ammonia-lyase (PAL), is encoded by four genes in Arabidopsis thaliana. Whereas PAL function is well established in various plants, an insight into the functional significance of individual gene family members is lacking. We show that in the absence of clear phenotypic alterations in the Arabidopsis pal1 and pal2 single mutants and with limited phenotypic alterations in the pal1 pal2 double mutant, significant modifications occur in the transcriptome and metabolome of the pal mutants. The disruption of PAL led to transcriptomic adaptation of components of the phenylpropanoid biosynthesis, carbohydrate metabolism, and amino acid metabolism, revealing complex interactions at the level of gene expression between these pathways. Corresponding biochemical changes included a decrease in the three major flavonol glycosides, glycosylated vanillic acid, scopolin, and two novel feruloyl malates coupled to coniferyl alcohol. Moreover, Phe overaccumulated in the double mutant, and the levels of many other amino acids were significantly imbalanced. The lignin content was significantly reduced, and the syringyl/guaiacyl ratio of lignin monomers had increased. Together, from the molecular phenotype, common and specific functions of PAL1 and PAL2 are delineated, and PAL1 is qualified as being more important for the generation of phenylpropanoids.
INTRODUCTION
The phenylpropanoid compounds derived from the shikimic acid pathway are precursors to diverse classes of phenolics, such as lignin, flavonoids, isoflavonoids, coumarins, and stilbenes. One of the intriguing characteristics of phenylpropanoids is the rapid modification in fluxes and rates of their synthesis and metabolism. This aspect is corroborated by the occurrence of specific phenolics in certain cell types, in response to specific stimuli (light, pathogens, and wounding) or during particular developmental stages.
The end products of the shikimate pathway, the aromatic amino acids Phe, Trp, and Tyr, are generated from the precursors phospho-enol-pyruvate and erythrose-4-phosphate. Phe ammonia-lyase (PAL; EC 4.3.1.5), the first enzyme of the phenylpropanoid pathway, removes Phe from aromatic amino acid biosynthesis to generate trans-cinnamic acid and NH3 in a nonoxidative deamination. Therefore, PAL commits the flux of primary metabolites into the phenylpropanoid pathway and becomes rate limiting when reduced below a threshold of 20 to 25% in tobacco (Nicotiana tabacum) (Bate et al., 1994; Sewalt et al., 1997). Many studies have revealed the regulatory mechanisms for various PAL isoforms in response to different stimuli. PAL activity is mainly regulated via de novo synthesis of the enzyme, but it can also be modulated by inactivation of the protein. PAL transcript levels increase when furnished with its substrate Phe as, among others, evidenced through feeding studies in lignifying Pinus taeda cells (Anterola et al., 2002). PAL activity and PAL gene transcription are inhibited by trans-cinnamic acid, its product (Bolwell et al., 1986; Mavandad et al., 1990; Blount et al., 2000). Moreover, PAL is subject to posttranslational phosphorylation by a specific calcium-dependent protein kinase (Cheng et al., 2001).
Given that alterations in the expression of monolignol biosynthesis genes can lead to differential accumulation of pathway intermediates that, in turn, can affect the transcription of other genes (Boerjan et al., 2003), we hypothesize that it would be possible to reveal the full spectrum of the PAL function and to dissect functional redundancy of PAL isoforms by means of thorough molecular phenotyping. We have recently identified the 34 Arabidopsis thaliana genes that encode the 10 known enzyme functions in monolignol biosynthesis (Raes et al., 2003). The Arabidopsis genome harbors four PAL genes that fall into two phylogenetic groups consisting of PAL1 and PAL2, and PAL3 and PAL4. On the basis of their strong expression in the inflorescence stem, phylogenetic relationship, and occurrence of promoter elements, PAL1 and PAL2 were selected as good candidates for monolignol production during developmental lignification of vascular tissues (Raes et al., 2003).
Here, we show that in the absence of phenotypic alterations in the pal1 and pal2 single mutants and with limited phenotypic alterations in the pal1 pal2 double mutant, significant modifications occur at the transcriptome and metabolome levels in these mutants. Transcript-profiling approaches reveal far-reaching consequences of the disruption of the PAL function on the transcription of genes that encode enzymes of the phenylpropanoid, carbohydrate, and amino acid metabolisms, establishing a clear link between primary and secondary metabolisms. Furthermore, many of these alterations at the transcriptional level are translated into the corresponding biochemical effects as witnessed by targeted metabolite profiling. Our data indicate common as well as discrete functions of PAL1 and PAL2 and demonstrate that a single mutation can profoundly affect the transcription of a plethora of genes, also outside the phenylpropanoid pathway, and the biosynthesis of a variety of metabolites.
RESULTS
Morphological Characterization of the pal1, pal2, and pal1 pal2 Mutants
T-DNA insertion mutants for PAL1 and PAL2 were generated by exon-trap mutagenesis in the Arabidopsis ecotype C24 (Babiychuk et al., 1997; see Methods). T-DNA insertions resided in the first intron of both genes (data not shown).
Although flavonoid deficiency is known to influence several aspects of plant development, such as lateral root formation, inflorescence growth, and seed viability, the pal1 and pal2 exon-trap mutants had no visible phenotypic alterations. The pal1 pal2 double mutant exhibited no gross morphological differences either, except that it had become sterile. As shown in Figure 1, the siliques remained small (3 to 4 mm versus ∼12 mm in the wild type). The empty ovule sacs had initially enlarged but shriveled later, suggesting male sterility. Reduced pollen viability was also observed in transgenic tobacco downregulated for PAL (Elkind et al., 1990). Because of the failure to produce viable seeds, the double mutant generated recurrent flushes of accessory paraclades that also failed to produce seeds. These flushes continued long after the wild type and single mutants had ceased growth.
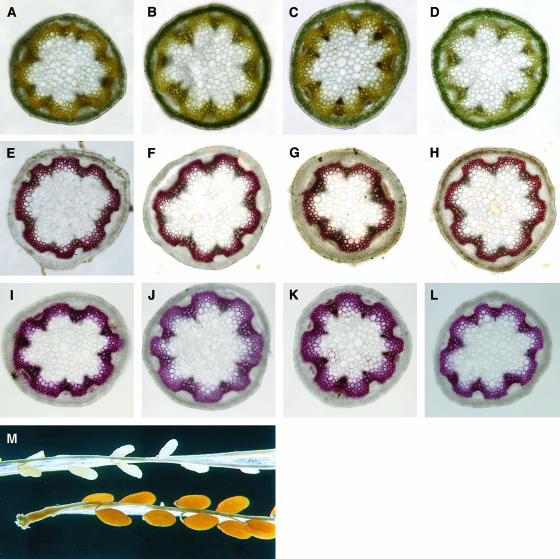
Figure 1.
Phenotypes of the Wild-Type C24 and pal1, pal2, and pal1 pal2 Mutants.
Lignin staining in the basal part of 3-month-old inflorescence stems ([A] to [L]) and silique phenotype (M).
(A) The wild type stained with aniline sulfate.
(B) pal1 mutant stained with aniline sulfate.
(C) pal2 mutant stained with aniline sulfate.
(D) pal1 pal2 mutant stained with aniline sulfate.
(E) The wild type stained with KMnO4:HCl:NH3.
(F) pal1 mutant stained with KMnO4:HCl:NH3.
(G) pal2 mutant stained with KMnO4:HCl:NH3.
(H) pal1 pal2 mutant stained with KMnO4:HCl:NH3.
(I) The wild type stained with phloroglucinol:HCl.
(J) pal1 mutant stained with phloroglucinol:HCl.
(K) pal2 mutant stained with phloroglucinol:HCl.
(L) pal1 pal2 mutant stained with phloroglucinol:HCl.
(M) Siliques of the wild type (bottom) and pal1 pal2 mutant (top) at seed maturity of the wild type.
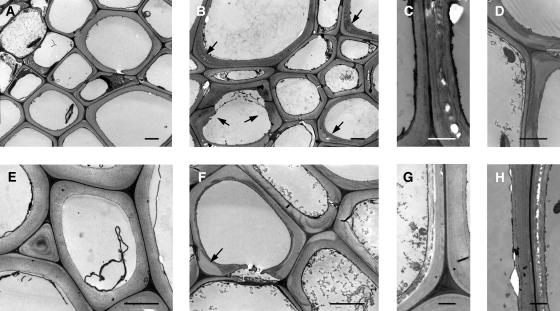
Although no growth differences could be seen with respect to time of bolting, height, or diameter of the inflorescence stem, slight differences in lignin accumulation were observed by lignin histochemistry in inflorescence stems of 2.5-month-old plants. As shown in Figure 1, a small reduction in lignin deposition was detected in the pal1 mutant and a more significant reduction in the pal1 pal2 double mutant (Figures 1A to 1L). Cell walls were further investigated at the ultrastructural level by transmission electron microscopy, as presented in Figure 2. The cell walls of xylem and interfascicular fibers were evenly thickened and uniformly stained in the wild type (Figures 2A and 2E). Double mutants were indistinguishable from the wild type in terms of cell size, cell number, or thickness of the cell wall. However, the cell wall of the interfascicular fibers and the large vessels appeared unevenly stained, and cell corners were often whiter (Figures 2B and 2F). In several cells, various signs of a reduced cell wall strength were observed: layers of the cell wall were loosened and had numerous little holes, or cellulose microfibrils became apparent (Figures 2C, 2D, 2G, and 2H). The latter phenomena were never observed in the wild type. At a low frequency, cell walls of the double mutant were aberrantly shaped and irregular (Figure 2B), similar to what has been observed in a more dramatic way in the lignin-deficient irregular xylem 4 Arabidopsis mutants (Jones et al., 2001).
Figure 2.
Cell Wall Ultrastructure in the Basal Part of 3-Month-Old Inflorescence Stems of the Wild Type and pal1 pal2 Mutants.
(A) Xylem region of the wild type.
(B) Xylem region of pal1 pal2 mutants. Arrows point to aberrantly shaped walls and to regions punctured with little holes.
(C) and (D) Cell walls in the xylem region of pal1 pal2 mutants. In (D), loosened cellulose fibrils become apparent.
(E) Interfascicular fiber region of the wild type.
(F) Interfascicular fiber region of pal1 pal2 mutants. Arrow points to whiter cell corners.
(G) and (H) Cell walls in the interfascicular fiber region of pal1 pal2 mutants.
Bar in (A), (B), (E), and (F) = 2.5 μm; bar in (C), (D), (G), and (H) = 1 μm.
In conclusion, pal1 and pal2 mutants have no obvious visible phenotype. Only the combination of both mutations in the double mutant leads to infertility and clear changes in lignin accumulation and secondary cell wall ultrastructure.
Expression of the Four Arabidopsis PAL Genes Is Altered in the Mutants
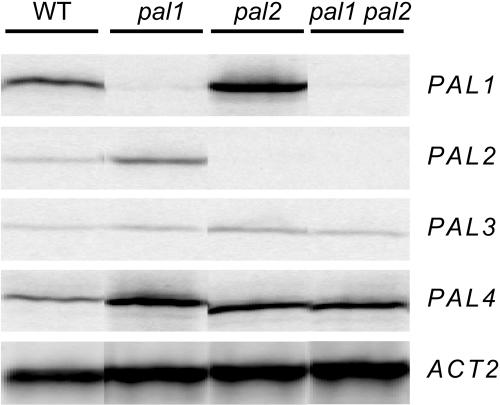
One possibility to explain the limited phenotypic differences in the pal single and double mutants is that the disruption of the PAL1 and PAL2 function is compensated for by other PAL isoforms. A comprehensive screen of the complete Arabidopsis genome revealed four PAL genes (Raes et al., 2003). The PAL genes are expressed in many tissues (including seed, seedling, leaf, root, flower, silique, and inflorescence stem), with PAL3 being generally expressed at a lower level (Wanner et al., 1995; Raes et al., 2003). Expression of PAL1 and PAL2 is detected most abundantly in roots and stems, where it increases during the later stages of inflorescence stem development. Because PAL1, PAL2, and PAL4 genes are most abundantly expressed during the later stages of inflorescence stem development (Raes et al., 2003), major effects of knock downs should manifest themselves in the inflorescence stem. Semiquantitative RT-PCR with specific primers was performed to clarify to what extent the expression levels of each of the four genes were affected in the mutants. As shown in Figure 3, T-DNA insertions in the PAL1 and PAL2 genes effectively disrupted the expression of these genes in the respective mutants. On the other hand, transcript levels of the PAL1 and PAL2 genes were increased in the pal2 and pal1 mutants, respectively, indicating a compensatory response (Figure 3). Also, the PAL4 gene was significantly induced in all mutants. The impact of PAL3, although altered in the pal2 mutant, was presumably weak because of its generally low expression level (Figure 3; Wanner et al., 1995). Therefore, the PAL transcripts are mainly derived from PAL2 and PAL4 in pal1 mutants, from PAL1 and PAL4 in pal2 mutants, and from PAL4 in the double mutant.
Figure 3.
Expression of PAL Genes in 10-Week-Old Inflorescence Stems of the Wild Type and pal1, pal2, and pal1 pal2 Mutants.
Semiquantitative RT-PCRs were performed on a pool of 10 individuals of each respective genotype (see Methods). The experiment was repeated with similar results.
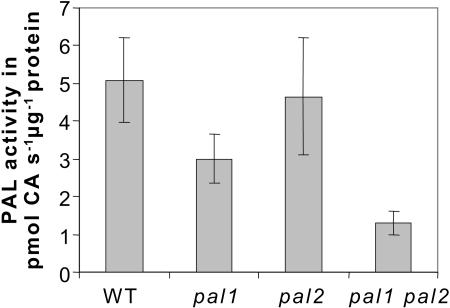
To estimate the consequences of upregulation of other isoforms at the enzyme level, PAL activity was determined in inflorescence stems of greenhouse-grown plants. As shown in Figure 4, PAL activity in the inflorescence stem was clearly reduced in the pal1 but not in the pal2 mutant. In the double mutant, the 25% residual PAL activity must be derived mainly from PAL4 transcripts (Figures 3 and 4). Additional activity measurements in siliques and leaves of these plants revealed none and a 50% reduced activity in all genotypes, respectively (data not shown). Thus, most clear-cut differences in activity were observed in the inflorescence stem, the tissue where PAL1 and PAL2 are most highly expressed. However, the lesion in PAL2 was almost completely compensated for at the enzyme level. Following the PAL activity during inflorescence growth, PAL activity was highest in 10-week-old, seed-setting but still green plants (Figure 4; data not shown). Assuming that at this age the differences in PAL activity between the wild type and mutants would also be highest, inflorescence stem material of 10-week-old plants was used to conduct all further experiments.
Figure 4.
PAL Activity in 10-Week-Old Inflorescence Stems of the Wild Type and pal1, pal2, and pal1 pal2 Mutants.
PAL activity of crude extracts was determined in inflorescence stems (n = 10) and is expressed as average PAL activity in pmol cinnamic acid (CA) s−1 μg−1 protein with standard deviation.
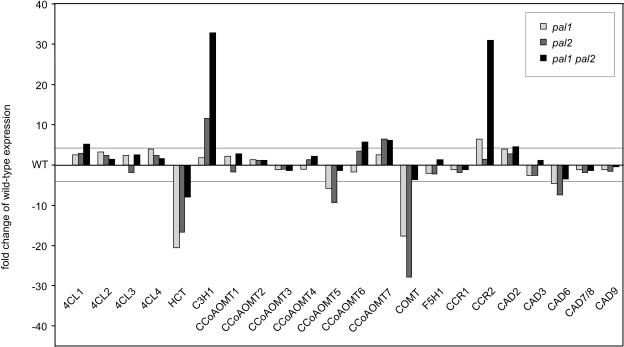
Expression of 10 Genes Encoding Enzymes of Monolignol Biosynthesis Is Strongly Altered in the pal Mutants
Our search was extended for consequences of PAL disruption on the transcription of the remaining 30 genes encoding the nine other enzymes of monolignol biosynthesis (Raes et al., 2003). Their expression was assayed by RT-PCR with gene-specific primers for each of the genes. Given the rapid alterations in expression levels known to occur for many phenylpropanoid genes and the semiquantitative nature of RT-PCR, a conservative approach of fourfold change was taken to delineate those genes with clear changes in expression. As shown in Figure 5, 23 of the 30 genes were expressed in 10-week-old inflorescence stems. Among the genes that were judged important for the developmental lignification of vascular tissues based on expression, phylogenetic relationship, and occurrence of promoter elements (Raes et al., 2003), pronounced changes in expression were found for 4-coumarate:CoA ligase (4CL1), para-coumarate 3-hydroxylase (C3H1), hydroxycinnamoyl-CoA:shikimate/quinate hydroxycinnamoyltransferase (HCT), caffeic acid O-methyltransferase (COMT), and cinnamyl alcohol dehydrogenase (CAD6). Interestingly, similar trends in alteration of gene expression were observed for most of them in both single pal mutants (Figure 5). Only the expression of C3H1 was higher in the mutants, whereas that of HCT, COMT, and CAD6 was lower (Figure 5). C3H1, hydroxylating the C3 position of p-coumaroyl shikimic/quinic acid, might be very prone to transcriptional regulation by the concentration of certain pathway intermediates. On the other hand, HCT activity is positively regulated by cinnamic acid (Lamb, 1977); hence, the decreased transcription probably correlated with the reduced synthesis of cinnamic acid in the pal mutants. Moreover, the HCT promoter contains both an H and a G box, known to be necessary for feed-forward induction by p-coumaric acid (Loake et al., 1992; Lindsay et al., 2002; Raes et al., 2003). Thus, our data underscore the responsiveness of the HCT gene to regulation by early pathway intermediates.
Figure 5.
Expression of the Monolignol Biosynthesis Genes in pal Mutants.
Genes (34) were annotated with sensitive annotation methods for the 10 enzymes currently known to participate in the generation of monolignols (Raes et al., 2003). Semiquantitative RT-PCRs were performed on two pools of 10 inflorescence stems of each respective genotype (see Methods). Expression levels were normalized to ACT2 gene expression before calculation of fold changes in expression levels taking the wild-type level as reference. Bars represent the average expression level of two biological repeats. Lines mark the level of fourfold upregulation or downregulation. Note that the following genes were not found to be expressed at this stage of inflorescence stem development (10-week-old plants): C4H, C3H2, C3H3, F5H2, CAD1, CAD4, and CAD5. For primers and gene nomenclature, see Raes et al. (2003).
Caffeoyl-CoA 3-O-methyltransferases CCoAOMT5, CCoAOMT6, and CCoAOMT7, CAD2, and cinnamoyl-CoA reductase (CCR2) with an altered expression in pal mutants had not been selected as candidates for a role in developmental lignification (Figure 5; Raes et al., 2003). CCR2, assigned a primary function in pathogen response (Lauvergeat et al., 2001), was also induced in the cellulose-deficient mutants de-etiolated 3, radially swollen 1, and ectopic lignification 1-1 (Caño-Delgado et al., 2003). Thus, CCR2 as well as the others might become subject to transcriptional regulation upon alterations in the concentration of pathway intermediates.
pal Mutants Are Affected in Transcription of Genes Encoding Enzymes of Amino Acid, Phenylpropanoid, and Carbohydrate Metabolisms
In view of the dramatic alterations in transcription of the most closely related genes, we investigated the consequence of the pal mutations on the transcriptome by a near genome-wide cDNA-amplified fragment length polymorphism (AFLP) transcript profiling of 10-week-old inflorescence stems. Transcript profiling was performed on two biologically independent sets of the wild type and the three mutants. Each single sample was a pool of 10 individual plants to normalize for the expected variability in gene expression of individual plants. In the expression profiles, differential transcript-derived fragments were identified by their increase/decrease in one or all mutants as compared with the wild type. Fragments were withheld only when a similar expression pattern was observed in both biological sets. For 63 differential sequence tags, selected in this manner and classified in Table 1 according to functional categories, the corresponding genes in Arabidopsis were identified. Among these 63 genes, three groups accounted for one-third of the sequences (or 43% of the genes with a known function): genes related to amino acid synthesis and metabolism, to the phenylpropanoid pathway, and to carbohydrate metabolism (Table 1).
Table 1.
Identities and Expression Patterns of the Sequence Tags Isolated for Their Differential Expression in the Wild Type and pal Mutants by cDNA-AFLP Transcript Profiling
| Expression in
|
Nearest Homolog (PEDANT at MIPS)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| AGI | WT | pal1 | pal2 | DM | Description (MIPS) | AGI/GI | Description | E Value |
| Amino Acid–Related | ||||||||
| At3g04940 | 0 | 0 | 1 | 0 | Cys synthase (AtcysD1) | |||
| At5g17920 | 0 | 0 | 1 | 0 | Met synthase | |||
| At1g69570 | 0 | 0 | 1 | 1 | H-protein promoter binding factor-2b, putative | At5g39660 | Promoter-binding protein like | 1 × 10−118 |
| At3g15450 | 0 | 0 | 1 | 1 | Unknown protein | At5g10240 | Asn synthetase ASN3 | 3 × 10−71 |
| At4g33010 | 0 | 1 | 0 | 0 | P-Protein-like protein | GI:20471026 | Gly dehydrogenase, Homo sapiens | 0 |
| At2g26080 | 0 | 1 | 1 | 1 | Putative Gly dehydrogenase | GI:20471026 | Gly dehydrogenase, H. sapiens | 0 |
| At2g27820 | 0 | 1 | 1 | 1 | Putative chorismate mutase/prephenate dehydratase | |||
| At5g54810 | 0 | 1 | 1 | 1 | Trp synthase β chain 1 precursor | |||
| Carbohydrate-Related | ||||||||
| At2g21170 | 0 | 0 | 1 | 0 | Putative triosephosphate isomerase | |||
| At1g32900 | 0 | 1 | 0 | 0 | Granule-bound starch synthase-like protein | |||
| At5g17420 | 0 | 1 | 0 | 1 | Cellulose synthase catalytic subunit (IRX3) | |||
| At1g35580 | 0 | 1 | 1 | 1 | Invertase, putative | |||
| At3g48530 | 0 | 1 | 1 | 1 | Unknown protein | GI:7300717 | SNF4Aγ gene product, Drosophila melanogaster | 5 × 10−64 |
| At5g58090 | 0 | 1 | 1 | 1 | β-1,3-glucanase 6 | |||
| At3g46970 | 1 | 0 | 1 | 1 | Starch phosphorylase H (cytosolic form)-like protein | |||
| Phenylpropanoid-Related | ||||||||
| At3g53260 | 0 | 1 | 0 | 0 | Phe ammonia-lyase (AtPAL2) | |||
| At3g02280 | 0 | 1 | 0 | 0 | Putative NADPH-ferrihemoprotein reductase ATR3 | At4g24520 | NADPH-ferrihemoprotein reductase ATR1 | 0 |
| At1g51680 | 0 | 1 | 0 | 1 | 4-Coumarate:CoA ligase (At4CL1) | At4g05160 | 4-Coumarate:CoA ligase-like protein | 1 × 10−147 |
| At1g43620 | 0 | 1 | 1 | 1 | Hypothetical protein | GI:6562348 | UDP-glucoronosyl/glucosyl transferase, Caenorhabditis elegans | 1 × 10−124 |
| At2g32520 | 0 | 1 | 1 | 1 | Putative carboxymethylenebutenolidase | |||
| At1g49140 | 1 | 0 | 0 | 0 | Similar to NADH-ubiquinone oxidoreductase precursor | |||
| Light-Related | ||||||||
| At2g43280 | 0 | 0 | 1 | 0 | Unknown protein | At5g28530 | Far-red-impaired response protein (FAR1)-like | 3 × 10−52 |
| At1g65070 | 0 | 1 | 1 | 1 | (No name given) | GI:33876770 | mutS protein homolog 5, H. sapiens | 1 × 10−150 |
| At1g53090 | 1 | 1 | 0 | 1 | Phytochrome A suppressor SPA1, putative | At4g11110 | COP1-like protein | 2 × 10−71 |
| At3g22170 | 1 | 1 | 0 | 1 | Far-red-impaired response protein, putative | At5g28530 | Far-red-impaired response protein (FAR1)-like | 1 × 10−141 |
| Stress Response | ||||||||
| At3g11170 | 0 | 1 | 1 | 1 | ω-3 Fatty acid desaturase, chloroplast precursor | |||
| At4g16070 | 0 | 1 | 1 | 1 | Unknown protein | At3g49050 | Calmodulin binding heat shock–like protein | 9 × 10−61 |
| At4g16860 | 0 | 1 | 1 | 1 | Disease resistance RPP5-like protein | |||
| At5g42650 | 0 | 1 | 1 | 1 | Allene oxide synthase | |||
| At5g62130 | 0 | 1 | 1 | 1 | Unknown protein | GI:83226 | PER1, Saccharomyces cerevisiae | 1 × 10−99 |
| Signal Transduction/Transcriptional Regulation | ||||||||
| At3g11500 | 0 | 0 | 1 | 0 | Putative small nuclear ribonucleoprotein polypeptide G | |||
| At1g07410 | 0 | 0 | 1 | 1 | Small G protein, putative | |||
| At5g57210 | 0 | 0 | 1 | 1 | Microtubule-associated protein-like | GI:12188746 | Rab6 GTPase-activating protein, H. sapiens | 4 × 10−50 |
| At1g04390 | 0 | 1 | 0 | 0 | Hypothetical protein | GI:6705975 | KIAA0740 protein, H. sapiens | 2 × 10−54 |
| At1g21920 | 0 | 1 | 0 | 1 | Phosphatidylinositol-4-phosphate 5-kinase-like protein | |||
| At1g23230 | 0 | 1 | 1 | 0 | Conserved hypothetical protein | GI:7291500 | CG3695 gene product, D. melanogaster | 0 |
| At1g23380 | 0 | 1 | 1 | 1 | Knotted-like homeobox protein, putative | At5g25220 | KNAT3 homeodomain protein | 1 × 10−44 |
| At3g28450 | 0 | 1 | 1 | 1 | Receptor kinase, putative | At5g16000 | Receptor protein kinase-like protein | 1 × 10−117 |
| At3g61570 | 0 | 1 | 1 | 1 | Putative protein | GI:29387238 | CDC42 binding protein kinase β, H. sapiens | 2 × 10−29 |
| At4g18010 | 0 | 1 | 1 | 1 | Putative inositol polyphosphate 5-phosphatase At5P2 | GI:21396493 | Inositol polyphosphate 5-phosphatase, H. sapiens | 1 × 10−101 |
| At5g14670 | 0 | 1 | 1 | 1 | ADP-ribosylation factor-like protein | |||
| At2g41090 | 1 | 0 | 0 | 0 | Calcium binding protein (CaBP-22) | At4g04710 | Putative calcium-dependent protein kinase | 5 × 10−34 |
| At1g73080 | 1 | 0 | 1 | 1 | Unknown protein | At5g07280 | Receptor-like protein kinase-like protein | 0 |
| At2g21660 | 1 | 0 | 1 | 1 | Gly-rich RNA binding protein | |||
| At3g12300 | 1 | 1 | 1 | 0 | Unknown protein | GI:7297748 | CG17118 gene product, D. melanogaster | 5 × 10−48 |
| Transport | ||||||||
| At5g03555 | 0 | 1 | 0 | 0 | Uracil transporter-like protein | |||
| At5g02270 | 0 | 1 | 0 | 1 | ABC transporter-like protein | |||
| At4g34950 | 0 | 1 | 1 | 1 | Putative protein | GI:16411641 | Strongly similar to transmembrane transporter | 2 × 10−34 |
| At3g08040 | 1 | 0 | 0 | 0 | Putative integral membrane protein | |||
| Unknown | ||||||||
| At1g28070 | 0 | 0 | 1 | 1 | Hypothetical protein | |||
| At1g28700 | 0 | 0 | 1 | 1 | Unknown protein | |||
| At1g75810 | 0 | 0 | 1 | 1 | Hypothetical protein | |||
| At2g40080 | 0 | 0 | 1 | 1 | Unknown protein | |||
| At3g26000 | 0 | 0 | 1 | 1 | Unknown protein | |||
| At4g32340 | 0 | 0 | 1 | 1 | Unknown protein | |||
| At4g12070 | 0 | 1 | 0 | 1 | Unknown protein | |||
| At1g67210 | 0 | 1 | 1 | 0 | Unknown protein | |||
| At1g73330 | 0 | 1 | 1 | 1 | Putative protein | |||
| At3g47833 | 0 | 1 | 1 | 1 | Unknown protein | |||
| At3g51250 | 0 | 1 | 1 | 1 | Unknown protein | |||
| At4g27080 | 1 | 0 | 0 | 0 | Unknown protein | |||
| At4g14260 | 1 | 0 | 1 | 1 | Hypothetical protein | |||
| At3g01860 | 1 | 1 | 0 | 1 | Unknown protein | |||
The description of the genes is retrieved from the Munich Information Center for Protein Sequences (MIPS); when the gene was annotated as unknown, the closest homolog is given as obtained from PEDANT at MIPS (Frishman et al., 2003). Expression was scored in the wild type and pal1, pal2 and pal1 pal2 double mutants (DM) with 0 for absence/decrease and 1 for presence/increase in expression, taking the wild type as reference. AGI, Arabidopsis Genome Initiative.
The transcriptomes of pal1 and pal2 mutants from the same biological material were also compared with the wild type with a 6K-cDNA microarray. As shown in Table 2, expression significantly changed (twofold at P = 0.001) for 36 genes, of which only three differential genes were common in both mutants. Convincingly, half of these genes (or 61% of the genes with a known function) fell into the same three functional categories (amino acid, phenylpropanoid, and carbohydrates) as in the cDNA-AFLP experiment. Only two genes were found with both methods: a homolog of Asn synthase (ASN3, At3g15450) and a putative protein (At1g73330). The fact that 27 of the 63 tags found in the cDNA-AFLP experiment are present on the microarray suggests that many of these tags were not detected because of cross-hybridization with transcripts of other gene family members or transcript abundance. Clearly, cDNA-AFLP detected more differentially expressed genes involved in signal transduction/gene regulation and unknowns than the microarray (15 versus 3 and 14 versus 5, respectively). This observation underscores the complementarity of both approaches and the higher sensitivity of the cDNA-AFLP method for low-abundant messengers and members of gene families.
Table 2.
Identities and Expression Patterns of the Sequence Tags Identified for Their Differential Expression in the Wild Type and pal1 and pal2 Mutants by Hybridization with a 6K Arabidopsis Microarray
| Expression in pal1
|
Expression in pal2
|
Nearest Homolog (PEDANT at MIPS)
|
||||||
|---|---|---|---|---|---|---|---|---|
| AGI | Fold Change | P Value | Fold Change | P Value | Description (MIPS) | AGI/GI | Description | E Value |
| Amino Acid–Related | ||||||||
| At3g15450 | 3.64 | 2.21 × 10−8 | Unknown protein | At5g10240 | Asn synthetase ASN3 | 3 × 10−71 | ||
| At4g27450 | 2.33 | 7.87 × 10−7 | Unknown protein | At5g10240 | Asn synthetase ASN3 | 2 × 10−59 | ||
| At4g28410 | 0.40 | 4.04 × 10−8 | Hypothetical protein | At4g28420 | Tyr transaminase-like protein | 1 × 10−46 | ||
| At3g25990 | 0.46 | 4.47 × 10−7 | Putative DNA binding protein, GT-1 | |||||
| Carbohydrate-Related | ||||||||
| At5g49360 | 2.19 | 1.91 × 10−4 | 3.23 | 6.30 × 10−6 | Xylosidase | |||
| At5g38410 | 2.01 | 9.54 × 10−8 | 2.05 | 7.55 × 10−8 | Rubisco small chain 3b precursor | |||
| At1g70290 | 2.79 | 2.29 × 10−6 | Putative trehalose-6-phosphate synthase | |||||
| At4g36670 | 2.40 | 1.41 × 10−5 | Sugar transporter-like protein | |||||
| At3g01500 | 2.16 | 1.19 × 10−4 | Carbonic anhydrase, chloroplast precursor | |||||
| At1g79550 | 2.07 | 1.02 × 10−8 | Phosphoglycerate kinase (EC 2.7.2.3) like protein | |||||
| At2g39730 | 2.03 | 5.47 × 10−7 | Rubisco activase | |||||
| At3g23490 | 0.49 | 2.40 × 10−6 | Cyanate lyase (CYN) | |||||
| Phenylpropanoid-Related | ||||||||
| At2g22990 | 2.29 | 9.35 × 10−5 | Putative Ser carboxypeptidase Ia | |||||
| At2g34660 | 0.42 | 1.15 × 10−5 | MRP-like ABC transporter | |||||
| At5g17220 | 0.41 | 6.20 × 10−4 | GST-like protein | |||||
| At5g42980 | 0.40 | 6.00 × 10−4 | Thioredoxin | |||||
| At2g37040 | 0.30 | 2.22 × 10−7 | Phe ammonia lyase (PAL1) | |||||
| At5g42800 | 0.26 | 4.96 × 10−5 | Dihydroflavonol 4-reductase | |||||
| At5g13930 | 0.15 | 2.11 × 10−6 | Chalcone synthase | |||||
| Light-Related | ||||||||
| At2g41460 | 2.98 | 6.95 × 10−6 | DNA-(apurinic or apyrimidinic site) lyase (ARP)b | |||||
| At1g68520 | 2.67 | 2.11 × 10−5 | Putative B-box zinc finger protein | At5g15850 | CONSTANS-like 1 | 4 × 10−26 | ||
| At1g29930 | 2.64 | 9.19 × 10−7 | Unknown protein | At3g61470 | Lhca2 protein | 1 × 10−76 | ||
| At1g55670 | 2.09 | 4.15 × 10−7 | Photosystem I subunit V precursor (PsaG) | |||||
| Stress Response | ||||||||
| At5g24490 | 2.57 | 1.08 × 10−4 | Unknown protein | GI:2636057 | Stress-induced protein, Bacillus subtilis | 2 × 10−78 | ||
| At3g52470 | 2.55 | 3.35 × 10−5 | Unknown protein | At5g22200 | NDR1/HIN1-like protein | 6 × 10−67 | ||
| At2g40000 | 2.18 | 1.60 × 10−5 | Putative nematode-resistance protein | |||||
| At1g75040 | 2.04 | 1.18 × 10−6 | Thaumatin-like protein | At5g38280 | Receptor Ser/Thr kinase PR5K | 7 × 10−97 | ||
| Signal Transduction/Transcriptional Regulation | ||||||||
| At1g80440 | 2.14 | 2.75 × 10−4 | 3.08 | 1.08 × 10−5 | Unknown protein | GI:28374149 | Kelch-like protein 3, H. sapiens | 9 × 10−61 |
| At1g13260 | 2.28 | 2.18 × 10−5 | Unknown protein | At5g20730 | Putative protein | 2 × 10−66 | ||
| At1g78830 | 2.02 | 2.16 × 10−4 | Unknown protein | At4g21370 | Receptor kinase-like protein | 1 × 10−102 | ||
| Miscellaneous | ||||||||
| At2g33830 | 2.23 | 2.48 × 10−6 | Putative auxin-regulated protein | |||||
| Unknown | ||||||||
| At5g21940 | 2.49 | 1.11 × 10−6 | Unknown protein | |||||
| At4g27380 | 2.47 | 7.59 × 10−10 | Unknown protein | |||||
| At2g15890 | 2.10 | 4.30 × 10−8 | Unknown protein | |||||
| At1g14880 | 2.03 | 4.15 × 10−4 | Unknown protein | |||||
| At1g73330 | 0.39 | 1.91 × 10−7 | Putative protein | |||||
Expression is given as fold change of wild-type expression. The description of the genes is retrieved from MIPS; when the gene was annotated as unknown, the closest homolog is given as obtained from PEDANT at MIPS (Frishman et al., 2003).
Note that this gene is annotated as Ser carboxypeptidase at MIPS but has been shown to correspond to the SNG1 locus, encoding sinapoylglucose:malate sinapoyltransferase (Lehfeldt et al., 2000).
It cannot be ruled out that the upregulation of ARP partly results from fragments of the ARP gene used in the promoter-trap construct.
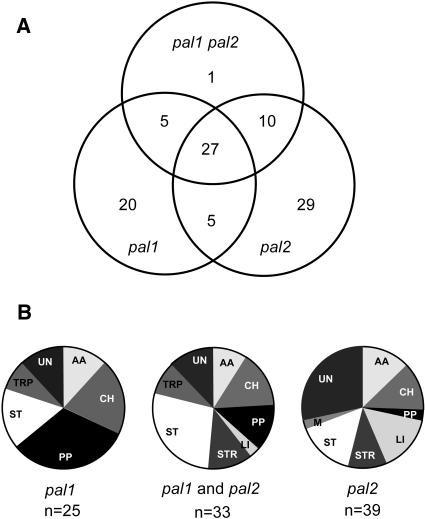
Figure 6 shows an interesting trend by clustering all 97 tags according to their expression in the mutants: the largest group of genes linked specifically to the pal1 mutation identifies genes of the phenylpropanoid pathway. Remarkably, ∼50% of the differentials found in pal1 and pal2 mutants were common to pal1 and pal2, and 27 of 32 of these common differentials were also found in the double mutant (Figure 6). Only one gene (At3g12300, unknown protein; Table 1) required both mutations for a change in expression, altogether corroborating that the differentially expressed genes were linked to the defect in PAL1 and PAL2, and not to stochastic gene expression.
Figure 6.
Number and Functional Categories of Genes with Altered Expression in pal1, pal2, and pal1 pal2 Mutants as Compared with the Wild Type.
(A) Summary from both cDNA-AFLP and microarray analyses (Tables 1 and 2) corrected for the genes that were revealed by both methods.
(B) Functional categories of the groups of genes controlled either by pal1, pal2, or both. The group pal1 comprises genes differentially expressed in only pal1 (n = 20) or pal1 and the double mutant (n = 5). The group pal2 comprises genes differentially expressed in only pal2 (n = 29) or pal2 and the double mutant (n = 10). The group pal1 and pal2 comprises genes differentially expressed in both single mutants but not in the double mutant (n = 5), genes expressed in pal1, pal2, and the double mutant (n = 27), and genes expressed exclusively in the double mutant (n = 1). AA, amino acid metabolism; CH, carbohydrate metabolism; LI, light related; M, miscellaneous; PP, phenylpropanoid related; ST, signal transduction; STR, stress response; TRP, transport; UN, unknown.
As summarized in Figure 7, 43 and 61% of the differentially expressed genes with a known function identified through cDNA-AFLP and microarray, respectively, were related to amino acid, phenylpropanoid, and carbohydrate metabolisms. With respect to amino acid metabolism, three tags recognized enzymes of the aromatic amino acid biosynthesis and metabolism: chorismate mutase (CM), the first committed step in Phe and Tyr biosynthesis; Trp synthase (TSB), performing the final step in biosynthesis of Trp; and Tyr transaminase (TAT), degrading Phe and Tyr (Tables 1 and 2, Figure 7). Two genes that are homologs of ASN3 are upregulated in pal2 mutants (Tables 1 and 2). Four other tags correspond to genes encoding components of the Gly cleavage complex, primarily involved in photorespiration: two different homologs of Gly dehydrogenase and two DNA binding proteins (At1g69570 and At3g25990) that recognize motifs present in the H protein promoter with the H protein itself being part of the Gly decarboxylase enzyme complex (Tables 1 and 2). In addition, Met synthase, upregulated in the pal2 mutant, represents an interesting tag because it catalyzes the methylation of homocysteine with 5-methyl-tetrahydrofolate as methyl donor. This reaction does not only occur during de novo synthesis but also during recycling of S-adenosyl-l-homocysteine after S-adenosyl-Met (SAM) has donated its methyl group. Besides the incorporation of Met into proteins, 80% of Met is converted into SAM for various transmethylations that are also very frequent in the phenylpropanoid pathway (Figure 7).
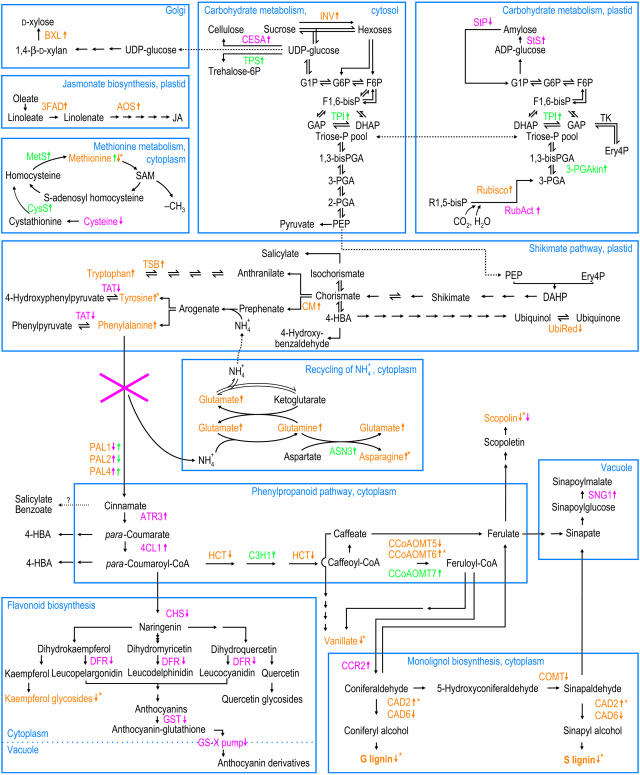
Figure 7.
Integration of the Metabolic Pathways Affected by a Decreased Carbon Flux into the Phenylpropanoid Pathway, as Evidenced by Molecular Alterations in pal Mutants.
Results of the transcriptome analyses (RT-PCR, cDNA-AFLP, and microarray) and the biochemical analyses (extractable phenolics, amino acids, and lignin) are incorporated. A dashed line indicates transport into another compartment; right and left harpoons mark a reversible enzyme reaction and arrows an irreversible enzyme reaction, respectively; question mark denotes a not fully established path. Arrows pointed up or down indicate the upregulation or downregulation of the gene or metabolite, respectively. The following color code is used: red, transcripts and metabolites linked with the pal1 but not the pal2 mutation; green, transcripts and metabolites linked with the pal2 but not the pal1 mutation; orange, transcripts and metabolites linked with the pal1 and pal2 mutations; orange with asterisk, transcripts and metabolites occurring only in the pal1 pal2 double mutant. Abbreviations are listed per pathway. Carbohydrate metabolism: 1,3-BisPGA, 1,3-bisphosphoglycerate; BXL, β-xylosidase; CESA, cellulose synthase; DHAP, dihydroxyacetone phosphate; Ery4P, erythrose 4-phosphate; F1,6-BisP, fructose 1,6-bisphosphate; F6P, fructose 6-phosphate; G1P, glucose 1-phosphate; G6P, glucose 6-phosphate; GAP, glyceraldehyde 3-phosphate; INV, invertase; PEP, phospho-enol-pyruvate; 2-PGA, 2-phosphoglycerate; 3-PGA, 3-phosphoglycerate; 3-PGAkin, 3-phosphoglycerate kinase; R1,5-BisP, ribulose 1,5-bisphosphate; RubAct, Rubisco activase; StP, starch phosphorylase; StS, starch synthase; TK, transketolase; TPI, triose-phosphate isomerase; TPS, trehalose 6-phosphate synthase. Jasmonate biosynthesis: AOS, allene oxide synthase; 3FAD, ω-3 fatty acid desaturase; JA, jasmonate. Methionine metabolism: CysS, cystathionine β-lyase (Cys synthase); MetS, Met synthase. Shikimate pathway: DAHP, 3-deoxy-d-arabino-heptulosonate 7-phosphate; Ery4P, erythrose 4-phosphate; 4-HBA, para-hydroxybenzoate; 4-HBald, para-hydroxybenzaldehyde; PEP, phospho-enol-pyruvate; UbiRed, NADH-ubiquinone oxidoreductase. Recycling of NH4+1: ASN3, Asn synthase 3. Phenylpropanoid pathway: ATR3, NADPH-ferrihemoprotein reductase 3; CCoAOMT, caffeoyl-CoA 3-O-methyltransferase; 4-HBA, para-hydroxybenzoate. Monolignol biosynthesis: G lignin, guaiacyl lignin monomers; S lignin, syringyl lignin monomers. Flavonoid biosynthesis: CHS, chalcone synthase.
Among the phenylpropanoid pathway-related transcripts, PAL1, PAL2, and 4CL1 were expressed in a manner consistent with the results from the RT-PCR experiment (Tables 1 and 2, Figures 3 and 5). The NADPH ferrihemoprotein reductase ATR3, found particularly induced in the pal1 mutant, is probably part of a microsomal P450 system that associates multiple P450 enzymes and NADPH:cytochrome P450 reductases. ATR homologs from poplar (Populus trichocarpa × P. deltoides) and Arabidopsis were shown to efficiently support the activity of the P450 enzyme cinnamate 4-hydroxylase (C4H), when coexpressed in yeast (Urban et al., 1997; Mizutani and Ohta, 1998; Ro et al., 2002). Differential tags from a carboxymethylenebutenolidase, dihydroflavonol 4-reductase (DFR), sinapoylglucose:malate sinapoyltransferase (SNG1), a UDP-glucosyl transferase, a glutathione S-transferase (GST), and a glutathione S-conjugate pump (GS-X pump) correspond to enzymes active toward the endpoints of biosynthetic pathways originating from the phenylpropanoid pathway (Figure 7). The UDP-glucosyl transferase belongs to the glycosyltransferase family 1; characterized members use phenylpropanoids, such as flavonol and sinapate, as substrates (http://afmb.cnrs-mrs.fr/CAZY/). The GSTs of the φ and τ subgroups, to which the downregulated GST also belongs (GSTF12; Dixon et al., 2002), have been implicated in cytoplasmic stabilization of flavonoids and flavonoid conjugation before uptake into the vacuole (Mueller et al., 2000; Dixon et al., 2002). The ABC transporter (AtMRP2) identified here encodes a functional GS-X pump of the plant vacuolar membrane with a high capacity for glutathionated anthocyanins (Lu et al., 1998; Liu et al., 2001; Sánchez-Fernández et al., 2001).
Perturbations in the PAL function seem to lead to adjustments in carbon fixation and carbohydrate metabolism (Tables 1 and 2, Figure 7). Ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) is upregulated in pal1 and pal2 mutants, and Rubisco activase is upregulated in the pal1 mutant. Both are central parts of CO2 fixation into organic carbon (Table 2, Figure 7). Enzymes, such as phosphoglycerate kinase and triose phosphate isomerase, that metabolize the phosphoglycerate generated by Rubisco, are also upregulated in pal2 mutants (Tables 1 and 2, Figure 7). SNF4 (Akin γ), upregulated in all pal mutants, plays important regulatory roles in carbohydrate metabolism in yeast and Arabidopsis (Table 1; Bouly et al., 1999). Trehalose 6-phosphate synthase, upregulated in pal2 mutants, generates trehalose 6-phosphate that, in turn, has an important function in controlling the flux into glycolysis in yeast and controls carbohydrate use in plants (Table 2; Eastmond and Graham, 2003; Schluepmann et al., 2003). Invertase, upregulated in all mutants, and starch phosphorylase, upregulated in the pal1 mutant, participate in the generation of hexose phosphates from sucrose and starch degradation, respectively. It is tempting to think that some of these tags identify components that are involved in the generation of substrates for cell wall biosynthesis because cellulose synthase was also induced in the pal1 and pal1 pal2 double mutants (Table 1). β-Xylosidase, upregulated in both pal1 and pal2 mutants (Table 2), hydrolyzes β-d-xylose residues of (most likely) glucuronoarabinoxylans. The glucuronoarabinoxylans in turn are able to covalently bind lignins and to attach to cellulose (Hatfield et al., 1999).
Two smaller groups of tags relate to light signal transduction and stress responses, two processes in which the importance of flavonoids is well documented. Light-related transcripts are found preferentially in the pal2 mutant. SPA1 and FAR are both specific signaling intermediates of phytochrome A signal transduction (Hoecker et al., 1999; Hudson et al., 1999). Two different FAR homologs, one upregulated and one downregulated in the pal2 mutant, are found (Table 1). Moreover, the pal2 mutant also contains a CONSTANS-related transcription factor that is upregulated (Table 2). Two other light-related tags are mutS and DNA (apurinic or apyrimidinic) lyase, both involved in DNA repair that is probably increased because of the lack of UV light-protecting flavonoids (see below). ω-3 Fatty acid desaturase and allene oxide synthase participate and rate limit, respectively, the biosynthesis of jasmonate and are upregulated in all pal mutants (Table 1, Figure 7).
pal1 pal2 Double Mutants Lack Three Major Flavonol Glycosides
The alterations in gene transcription related to particular pathways prompted us to conduct a more in-depth biochemical characterization of the mutants. In accordance with the transcriptome data, and despite the near wild-type visible appearance of the mutants, severe modifications in amino acids, phenolics, and lignin accumulation were detected.
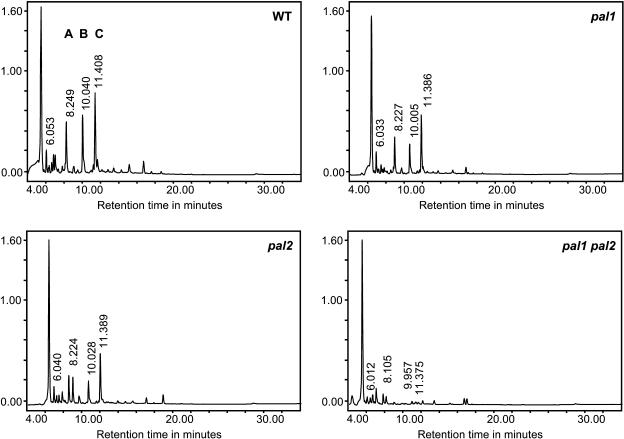
When the soluble phenolic compounds in the inflorescence stem of the three mutants and the wild type were compared by reverse-phase HPLC, significant changes in the profile were noticed. As shown in the maxplots presented in Figure 8, reduced PAL activity led to a decreased accumulation of several phenolics. Out of 89 peaks in the analyzed chromatograms, eight peaks differed significantly in abundance between the wild type and pal mutants. As shown in Table 3, total phenolics were significantly diminished to approximately one-third of the wild-type amount in the pal1 pal2 double mutants, slightly reduced to 75% in pal1 mutants, and not affected in the pal2 mutant. This decrease in total phenolics correlates with the levels of PAL activity (Figure 4) and is mainly attributable to the depletion of the three major peaks denoted as A, B, and C in Figure 8. As shown in Figure 9A, these peaks correspond, on the basis of their retention time (RT), photodiode array, and mass spectrometry (MS) spectra, to previously identified kaempferol glycosides (Veit and Pauli, 1999): kaempferol 3-O-β-[β-d-glucopyranosyl(1-6)-d-glucopyranoside]-7-O-α-l-rhamnopyranoside (Glc-Glc-Rha-kaempferol), kaempferol 3-O-β-d-glucopyranoside-7-O-α-l-rhamnopyranoside (Glc-Rha-kaempferol), and kaempferol 3-O-α-l-rhamnopyranoside-7-O-α-l-rhamnopyranoside (Rha-Rha-kaempferol), respectively. The alterations in the accumulation of these peaks in the single mutants were not statistically significant but showed a trend to lower levels in the pal1 mutant (Table 3). These glycosides belong to the flavonol glycosides that are the major UV light-absorbing metabolites in Arabidopsis (Graham, 1998). Interestingly, other UV light-protecting compounds in Arabidopsis, such as sinapoyl glucose and sinapoyl malate (Landry et al., 1995), were not altered in leaves or inflorescence stems (thin-layer chromatography; data not shown). To address whether the reduction of the three major peaks in the double mutant was a result of a reduced synthesis of trans-cinnamic acid, double mutants and the wild type were fed for 24 h with trans-cinnamic acid or para-coumaric acid. Kaempferol glycoside peaks could not be restored in the double mutant (data not shown), suggesting that the reduced PAL activity has not only simple effects on the amount of its reaction product but imbalances the phenylpropanoid pathway in a more sophisticated way.
Figure 8.
Profiles of Soluble Phenolic Metabolites in 3-Month-Old Inflorescence Stems of the Wild-Type C24, pal1, and pal2, pal1 pal2 Mutants.
Soluble phenolics are separated by reverse-phase HPLC. The maxplot chromatograms are given as absorbance units at 200 to 450 nm and are scaled down to show the differences in the most abundant peaks. A, B, and C denote the three major kaempferol glycosides decreased in the double mutant.
Table 3.
Soluble Phenolics That Accumulate Differentially in the Inflorescence Stems of the Wild Type and pal Mutants as Resolved by HPLC
| Peak Height in μV/μg Dry Weight (Average with se)
|
|||||
|---|---|---|---|---|---|
| RT (min)a | Identity | Wild Type | pal1 | pal2 | pal1 pal2 |
| Total | 99.87 ± 6.65 | 73.98 ± 8.43 | 106.01 ± 12.95 | 34.14 ± 5.73* | |
| 6.033 | Glycosylated vanillic acid | 5.89 ± 0.29 | 5.65 ± 0.19 | 6.08 ± 0.99 | 2.26 ± 0.34* |
| 7.331 | Scopolin | 0.67 ± 0.07 | 0.41 ± 0.02* | 0.64 ± 0.09 | 0.19 ± 0.04* |
| 8.227 | Glc-Glc-Rha-kaempferol | 19.12 ± 2.07 | 12.95 ± 2.21 | 17.49 ± 2.62 | 0.21 ± 0.08* |
| 8.350 | Not identified | ND | ND | ND | 0.03 ± 0.01 |
| 10.005 | Glc-Rha-kaempferol | 20.67 ± 2.59 | 12.61 ± 3.35 | 19.39 ± 3.31 | 0.31 ± 0.10* |
| 11.178 | FM(4–O–8)Gb | 1.21 ± 0.15 | 0.62 ± 0.06* | 0.83 ± 0.11 | 0.22 ± 0.05* |
| 11.386 | Rha-Rha-kaempferol | 25.89 ± 2.32 | 19.71 ± 3.02 | 27.66 ± 3.70 | 0.64 ± 0.23* |
| 15.119 | FM(5–8)Gc | 1.34 ± 0.13 | 0.74 ± 0.09* | 1.16 ± 1.27 | 0.42 ± 0.09* |
The 106 μV corresponds to an absorbance of 1 OD. One-way ANOVA and post-hoc least square difference tests for peak height were performed (see Methods) and are marked by an asterisk, if found significantly different from the wild type at P = 0.05. ND, not detected.
The RTs correspond to those in an individual pal1 chromatogram, shown in Figure 8.
Feruloyl malate coupled 4–O–8 to coniferyl alcohol.
Feruloyl malate coupled 5–8 to coniferyl alcohol.
Figure 9.
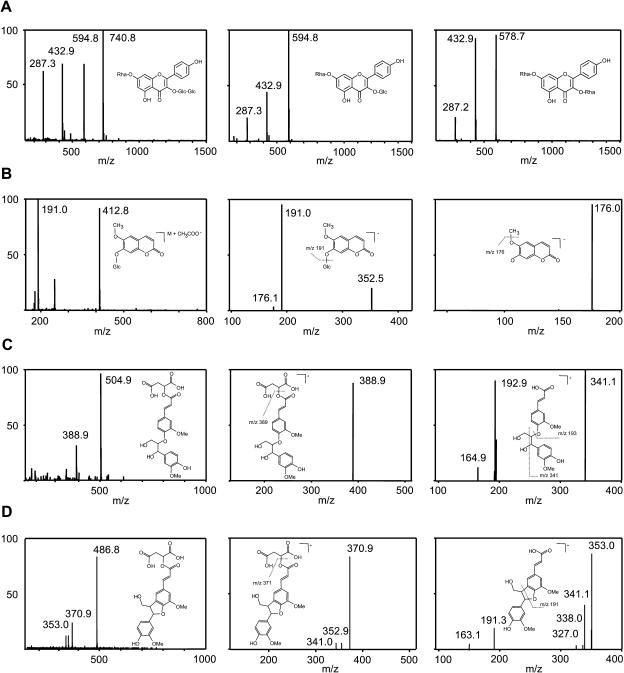
Identification by Mass Spectrometry of Five Soluble Phenolics Significantly Altered in pal Mutants.
All MS spectra are given as relative abundance of molecules with a scale to 100%.
(A) Full MS spectra obtained in the positive ionization mode for the three major peaks found to be altered significantly in pal mutants (indicated with A, B, and C in Figure 8). The mass-to-charge (m/z) values in the MS spectra for A, B, and C confirm that these compounds are hexose-conjugated kaempferols. The A in Figure 8 corresponds to kaempferol 3-O-β-[β-d-glucopyranosyl (1-6)-d-glucopyranoside]-7-O-α-l-rhamnopyranoside, B to kaempferol 3-O-β-d-glucopyranoside-7-O-α-l-rhamnopyranoside, and C to kaempferol 3-O-α-l-rhamnopyranoside-7-O-α-l-rhamnopyranoside.
(B) Identification of the HPLC peak at RT 7.331 min as scopolin by full MS in the negative ionization mode, MS2 of m/z 413, and MS3 of m/z 191.
(C) Identification of the HPLC peak at RT 11.178 min as feruloyl malate coupled 4–O–8 to coniferyl alcohol by full MS, MS2 of m/z 505, and MS3 of m/z 389. The MS4 of m/z 193, with the characteristic fragmentation pattern of ferulic acid, is not shown.
(D) Identification of the HPLC peak at RT 15.119 min as feruloyl malate coupled 5–8 to coniferyl alcohol by full MS, MS2 of m/z 487, and MS3 of m/z 371. Fragmentation of m/z 371 yielded peaks at m/z 353, 341, and 338, in abundances reminiscent of a phenylcoumaran (K. Morreel and W. Boerjan, unpublished data).
Feruloyl Malate Esters Coupled to Coniferyl Alcohol Are Less Abundant in pal1 and pal1 pal2 Double Mutants
Besides the three major UV light–absorbing kaempferol glycosides, five other peaks were significantly altered in the mutants, one of which occurred only in the double mutant (RT 8.350 min; not identified, Table 3). The compound at RT 6.033 min (Figure 8) was identified as glycosylated vanillic acid by RT, photodiode array spectrum, and comparison with an authentic standard (Table 3).
The remaining three peaks were significantly reduced in pal1 and the double mutant, but not in the pal2 mutant (RT 7.331 min, 11.178 min, and 15.119 min; Table 3). MS analysis of the compound eluting at 7.331 min indicated that this compound is scopolin or isoscopolin (Figure 9B). The peak was identified as scopolin because its aglycon released after hydrolysis consisted exclusively of scopoletin (RT 11.37 min) and not of isoscopoletin (RT 10.78 min). Scopolin had been detected previously in tobacco leaf extracts (Baumert et al., 2001) but not in Arabidopsis.
The compound eluting at 11.178 min has a UV/VIS spectrum characterized by absorption maxima at 235 and 329 nm. MS analysis suggested that this peak corresponds to the 4–O–8-cross-coupling product of feruloyl malate and coniferyl alcohol, FM(4–O–8)G, or 3-{4-[2-hydroxy-2-(4-hydroxy-3-methoxy-phenyl)-1-hydroxymethyl-ethoxy]-3-methoxy-phenyl}-acrylic acid (Figure 9C). The core structure FA(4–O–8)G (FA, ferulic acid) was authenticated using previously isolated FA(4–O–8)G from ryegrass (Lolium perenne) (Ralph et al., 1992). The UV/VIS spectrum (λmax at 242 and 332 nm) of the third compound (RT 15.119 min) was similar to that of FM(4–O–8)G. Based on MS analysis and its partial authentication with FA(5–8)G, this compound is suggested to be the related FM(5–8)G compound 2-{3-[2-(4-hydroxy-3-methoxy-phenyl)-3-hydroxymethyl-7-methoxy-2,3-dihydro-benzofuran-5-yl]-acryloyloxy}-succinic acid (Figure 9D). Thus, the two latter products resulted from coupling of coniferyl alcohol (at its favored 8-position) with feruloyl malate (at its 4–O- or 5-position) and had not been described before.
In summary, only downregulation of both PAL1 and PAL2 leads to significant changes in the accumulation of the major soluble phenolic compounds, the kaempferol glycosides (Table 3). On the other hand, the reduced accumulation of compounds produced by other side branches of phenylpropanoid metabolism, such as scopolin (hydroxycoumarin), and the feruloyl malates coupled to coniferyl alcohol were clearly linked with the mutation in PAL1 (Table 3, Figure 9). Sinapate esters were notably unaffected in all mutants, indicating that either this pathway is essential or that the enzymes producing sinapate esters are in metabolic complexes with PAL isoforms other than PAL1 and PAL2. This observation suggests that PAL1 and PAL2 are either associated with different cell types or with different metabolic complexes within the cell or that PAL1 has a higher activity than PAL2.
pal1 pal2 Mutants Overaccumulate Phe and Are Perturbed in Other Nonaromatic Amino Acids
Given the high number of differential transcripts related to amino acid metabolism, amino acid concentrations were determined in the wild type and the pal mutants. As shown in Table 4, the reduction in PAL activity had dramatic consequences for the accumulation of several amino acids, not only the aromatic ones. The content of most amino acids was increased in all mutants. As expected, Phe accumulated to >100-fold in the double mutant as compared with the wild type, whereas the other two aromatic amino acids, Tyr and Trp, accumulated to fourfold (Table 4). Accordingly, the CM gene, the first committed enzyme toward Phe and Tyr, as well as the TSB gene, were only expressed in the three mutants but were undetectable in the wild type in the transcript profiling experiment (Table 1).
Table 4.
Concentrations of Amino Acids in 3-Month-Old Inflorescence Stems of the Wild Type and pal1, pal2, and pal1 pal2 Mutants as Determined by HPLC
| Peak Height in μAU/μg Dry Weight (Average with se)
|
|||||
|---|---|---|---|---|---|
| Amino Acid | RT (min) | Wild Type | pal1 | pal2 | pal1 pal2 |
| Asp | 3.75 | 567.0 ± 65.6 | 741.3 ± 35.2 | 722.7 ± 38.7 | 930.7 ± 182.6 |
| Glu | 4.00 | 1185.8 ± 133.8 | 1744.0 ± 42.1 | 1745.2 ±83.3 | 2728.5 ± 527.3* |
| Asn | 6.05 | ND | ND | ND | 4.2 ± 4.2 |
| Ser | 6.20 | 117.0 ± 24.7 | 147.7 ± 5.7 | 184.3 ± 11.4 | 177.6 ± 25.8 |
| Gln | 6.48 | 78.3 ± 16.1 | 135.8 ± 4.6 | 211.2 ± 29.8 | 741.2 ± 149.0* |
| Gly | 6.65 | 1.1 ± 1.1 | ND | ND | ND |
| His | 7.20 | 8.3 ± 2.0 | 15.1 ± 3.4 | 15.9 ± 1.1 | 25.5 ± 6.1* |
| Arg | 8.10 | ND | ND | ND | ND |
| Thr | 8.30 | 101.7 ± 15.3 | 200.2 ± 5.5 | 181.8 ± 12.4 | 536.7 ± 98.3* |
| Ala | 8.65 | 97.0 ± 21.2 | 128.1 ± 2.6 | 191.9 ± 14.0 | 300.8 ± 40.1* |
| Pro | 8.95 | 23.3 ± 4.7 | 58.0 ± 3.3 | 47.0 ± 6.1 | 198.2 ± 58.8* |
| Tyr | 11.20 | 45.0 ± 13.5 | 58.4 ± 4.6 | 50.1 ± 6.1 | 166.0 ± 31.2* |
| Val | 11.85 | 112.1 ± 18.3 | 191.3 ± 5.0 | 237.1 ± 7.6 | 494.6 ± 78.3* |
| Met | 12.20 | 439.1 ± 41.8 | 522.7 ± 34.9 | 717.1 ± 117.2* | 26.3 ± 3.2* |
| Cys | 12.70 | 7.2 ± 1.2 | 1.1 ± 1.1* | 6.8 ± 0.7 | 8.2 ± 0.8 |
| Ile | 13.45 | 40.1 ± 7.2 | 77.1 ± 0.9 | 86.4 ± 6.7 | 161.9 ± 23.7* |
| Leu | 13.65 | 43.6 ± 5.9 | 91.8 ± 4.7* | 70.3 ± 3.8 | 103.9 ± 14.9* |
| Phe | 14.35 | 19.9 ± 4.1 | 72.8 ± 2.4 | 60.9 ± 4.9 | 2653.3 ± 740.7* |
| Trp | 14.62 | 27.3 ± 3.1 | 49.2 ± 3.3 | 67.4 ± 5.6 | 121.3 ± 31.1* |
| Lys | 15.30 | 11.8 ± 3.6 | 24.7 ± 0.8* | 18.2 ± 1.6 | 21.9 ± 2.6* |
Data are average of five individuals per genotype. One-way ANOVA and post-hoc Dunnett's tests were conducted (see Methods). Values significantly different from the wild type at P < 0.05 are indicated by asterisks. ND, not detected; AU, absorption units.
Another affected amino acid was Met that was significantly increased in the pal2 mutant but decreased to 6% of the wild-type level in the double mutant (Table 4). A transcript corresponding to Met synthase was exclusively upregulated in the pal2 mutant (Table 1). Finally, and noteworthy, Gly was only detected in the wild type (Table 4). Four transcripts related to Gly degradation were exclusively expressed or upregulated in the pal mutants (Tables 1 and 2). By contrast, Asn was only detected in the double mutant (Table 4). Correspondingly transcripts encoding putative proteins with strong similarity to ASN were upregulated in pal2 and pal1 pal2 mutants (Tables 1 and 2).
In conclusion, blocking PAL led to a strong overaccumulation of Phe and imbalanced the abundance of many other amino acids. Quite surprisingly, 15 out of 20 amino acids changed significantly in at least one genotype (Table 4). The fact that the concentration of these amino acids increases, except for Met in the double mutant and Cys in pal1, suggests an overarching mechanism for homeostasis of amino acid levels that acts across individual biosynthetic pathways. Similarly, plants overexpressing a Trp or Tyr decarboxylase, thus artificially creating a metabolic sink, were altered in the aromatic and three nonaromatic amino acids (Yao et al., 1995; Guillet et al., 2000).
pal1 pal2 Mutants Have Less Lignin with a Higher Syringyl/Guaiacyl Ratio
To determine lignification changes, lignin contents were measured via the acetyl bromide method, and monomer compositions of the wild type and pal mutants were determined by derivatization followed by reductive cleavage (DFRC) analysis (see Methods). As presented in Table 5, inflorescence stems of 3-month-old wild-type and mutant plants clearly differed in lignin content as well as monomer composition despite the fact that these differences were hardly apparent in histochemical analysis (Figure 1). Double mutants had 30% residual lignin (Table 5), and the molar ratio of S units in the noncondensed lignin fraction was nearly double that in the wild type. S-rich lignins have higher cleavable β-aryl ether levels and less condensed structures, involving 5–5-, 8–5-, and 4–O–5-units (Ralph et al., 2004), explaining the higher amount of G and S monomers from the noncondensed fraction per gram of lignin. A similar reduction in total lignin and increase in the S/G ratio was found in transgenic tobacco downregulated for PAL (Sewalt et al., 1997; Korth et al., 2001).
Table 5.
Lignin Content and Monomer Composition in Inflorescence Stems of 3-Month-Old Wild-Type, pal1, pal2, and pal1 pal2 Mutants
| Genotype | mg Lignin/g Cell Wall | μmol G/g Lignin | μmol S/g Lignin | Molar Ratio S/G |
|---|---|---|---|---|
| Wild type | 200 ± 8 | 761 ± 103 | 154 ± 9 | 0.21 ± 0.02 |
| pal1 | 139 ± 2 | 715 ± 56 | 197 ± 15 | 0.28 ± 0.003 |
| pal2 | 125 ± 3 | 755 ± 63 | 216 ± 11 | 0.30 ± 0.03 |
| pal1 pal2 | 69 ± 1 | 1020 ± 30 | 389 ± 33 | 0.38 ± 0.04 |
Lignin content was determined as acetyl bromide soluble lignin (ABSL). Monomer compositions were determined by DFRC (see Methods). Data were obtained with at least three individuals/genotype; each individual was measured twice. Data are expressed as averages with se.
The determination of lignin content by acetyl bromide leads to an underestimation of the amount of monomers linked by ester bonds. Therefore, isolated cell wall material was saponified and analyzed by HPLC. The HPLC profiles of the wild type and double mutant were virtually identical (data not shown), indicating that the amount of esters incorporated into the cell wall was not significantly changed.
DISCUSSION
Molecular Phenotyping Is Informative
Genome-wide studies of the transcriptome allow explaining a mutant phenotype at the molecular level at a depth that was not possible before the development of genomic tools. The extension of the phenotypic description to the molecular level increases the resolution so that functional redundancy of closely related gene family members can be separated. Thus, these tools are excellent to provide an insight into the functional significance of individual gene family members. Furthermore, our study underscores that the disruption of a single gene can have wide-ranging consequences on the transcription of other genes, revealing the intricacy of interactions among metabolic pathways, even in the absence of a visible phenotype (Tables 1 and 2). Elements of most of the pathways, altered in pal mutants, have been previously described for their responsiveness to pathogen attack or elicitation (Figure 7; Somssich and Hahlbrock, 1998). However, in contrast with transcriptome profiling after challenging the plant, typically designed to reconstruct a pathway response to a particular agent or environmental stress, our aim was to describe the adaptation of the transcriptome to genetic differences at a particular locus. In a similar study on the brevipedicellus mutant that is disrupted in a KNOX gene and displays premature lignin deposition, expression differed in 12 genes related to lignification or cell wall biosynthesis (Mele et al., 2003). Among these are 4CL1, PAL1, TAT, and cellulose synthase, genes that are also differentially expressed in the pal mutants (Tables 1 and 2). Moreover, ASN, differentially expressed in the brevipedicellus and pal2 mutant, is known to respond dramatically at the transcriptional level to metabolites, light, and development (Lam et al., 1998). Thus, our study might have identified genes that are under metabolic regulation, as opposed to genes involved in a pathway's response to environmental or developmental signals.
Altogether, the pal1 and pal2 mutations cause 58 and 72 genes, respectively, to change in expression within our experiment and degree of transcriptome resolution (20% with the microarray and 60% with the cDNA-AFLP; Figure 6). In the pal mutants, the expression of several differentially expressed genes depends probably on the level of one or more biochemical compounds generated or consumed by PAL, such as Phe and cinnamic acid or their upstream and downstream derivatives. However, among the differentially expressed genes, there will also be genes that are expressed as a result of secondary effects of altered cellular processes.
A Reduced Carbon Flux into the Phenylpropanoid Pathway Affects the Homeostasis of Amino Acid and Carbohydrate Metabolism
Under normal conditions, 20% of all fixed carbon is pumped into the shikimate pathway (Herrmann, 1995). One of the major drains to this pathway is PAL, consuming Phe. Because the pal1 pal2 double mutant has 30% residual total extractable phenolics and 30% residual lignin, carbon supply must be either adjusted or diverted to other pathways (Tables 3 and 5). Taking into account, moreover, that PAL1 expression occurs only in the vascular bundle (Leyva et al., 1995) and that PAL2 expression also preferentially correlates with lignified inflorescence stems (Raes et al., 2003), most of the observed molecular alterations in the mutants are expected to be manifest in a limited number of cell types in the stem (Figure 1).
Assuming that most transcriptomic changes concern the same few cell types, the three major downstream branches of the phenylpropanoid pathway (flavonoids, lignins, and sinapate esters) undergo a different adaptation to the reduced flux into the pathway in the mutants: the production of flavonoids and lignin is clearly reduced. Additionally, hydroxycoumarins, glycosylated vanillic acid, and FM(4–O–8)G and FM(5–8)G are reduced in abundance (Table 3), whereas sinapate esters remain unchanged. Because all flavonoids, sinapate esters, and hydroxycoumarins have UV light-protecting capacities, the sinapate esters remain probably produced because they are most effective in UV light shielding. Alternatively, sinapate esters could be derived from a metabolic complex involving a PAL isoform other than PAL1 or PAL2.
As summarized in Figure 7, signs of molecular adjustments are not only found in the phenylpropanoid pathway but also in the shikimate pathway, in the related cell wall biosynthesis, in components of central metabolism such as glycolysis, and even in distant stress-related pathways. In the transcriptome data (Figures 3 and 5, Tables 1 and 2), the complete sequence of the early phenylpropanoid biosynthesis is reconstructed, except for C4H (Figure 7). Similarly, in PAL-suppressed tobacco plants, C4H expression remains unchanged (Blount et al., 2000). However, a NADPH P450 reductase gene ATR3, shown to support C4H activity in vitro, was upregulated (Table 1; Urban et al., 1997; Mizutani and Ohta, 1998; Ro et al., 2002). Probably the reduced flux through PAL superinduces transcription of the consecutive genes: the respective other PAL gene(s), ATR3, 4CL1, and C3H1. To the contrary, genes encoding components in the flavonoid pathway (e.g., CHS, DFR, GST, and AtMRP2) and genes thought to be specific for lignification (e.g., COMT and CAD6) are downregulated.
With respect to the shikimate pathway generating the aromatic amino acids, transcripts for the enzyme at the branch point (CM) and two enzymes at the respective end points (TSB and TAT) were differentially expressed (Figure 7). All three hold a great potential for exerting control over the flux in this pathway. Phe, accumulating to 100-fold in the double mutant, and Trp, accumulating to fourfold in the double mutant, exert negative and positive feedback control on CM, respectively, and Trp also negatively regulates anthranilate synthase in its own biosynthesis (Bentley, 1990). On the other hand, Trp synthase is upregulated in pal mutants, pointing to a counterbalancing mechanism for decreased anthranilate synthase. TAT, transcriptionally downregulated in pal1 mutants, degrades Phe and Tyr (Figure 7). Its expression might be downregulated to limit the degradation of Phe to keep feeding substrate into the PAL reaction. Together, a severe imbalance in a part of the pathway might override classical feedback mechanisms. The accumulation of aromatic amino acids seems to cause further adjustments in the abundance of as many as 15 amino acids, indicating a general amino acid homeostasis (Table 4).
Clearly, many changes also occur in the primary metabolism of the pal mutants (Figure 7). Several studies reveal the reverse, namely, consequences of downregulation or disruption of enzyme functions in primary metabolism for secondary metabolism. For example, downregulation of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase (shikimate pathway) or plastid transketolase (TK, Figure 7) leads to a decrease in aromatic amino acids, phenylpropanoids, and lignin (Jones et al., 1995; Henkes et al., 2001). On the contrary, here, far-reaching consequences for primary metabolism caused by a disruption in an enzyme of secondary metabolism have been found at the transcriptional level (Figure 7). One striking example is the upregulation of the Rubisco-encoding gene in pal1 and pal2 mutants (Table 2). Only the opposite (namely, Rubisco downregulation and a corresponding decrease in the Phe pool) have been shown before in tobacco (Matt et al., 2002). These observations raise the possibility that primary and secondary metabolism are interlocked through Phe pools.
Reduction in Lignin Is Not Compensated for by Cellulose in pal Mutants
Lignin and polysaccharides are two principal components of the secondary thickened cell wall. A common assumption is that these cell wall components are coordinately regulated and can compensate for each other. In the pal mutants, having reduced lignin levels, several transcripts related to cell wall polysaccharides (most importantly, cellulose synthase and β-xylosidase) were identified as upregulated, but the corresponding changes in cell wall composition could not be detected. No clear differences between the wild type and mutants were found by cellulose histochemistry and analysis of the total polysaccharide composition (data not shown). Thus, the decrease in lignin does not seem to be compensated for by cellulose. Arabidopsis mutants with a reduced cellulose synthesis can exhibit ectopic lignification (Taylor et al., 1999; Caño-Delgado et al., 2000, 2003), whereas irregular xylem 4 mutants, disrupted in the CCR gene, have less lignin but not more cellulose (Jones et al., 2001). Thus, a reduction in cellulose synthesis, which precedes lignin deposition, is sensed to induce extra lignification (Ellis et al., 2002; Caño-Delgado et al., 2003), but a reduction in lignin deposition is not necessarily compensated for by cellulose biosynthesis. In a few cases, in which a reduced lignin accumulation was accompanied by an increased cellulose content, alterations in development have been observed (Hu et al., 1999; Li et al., 2003). An increased cellulose content in lignin-deficient plants might arise from pleiotropic effects of decreased lignification rather than from compensation for lignin by cellulose in each single cell. Alternatively, the lack of polysaccharide changes in the pal mutants has to be explained by the subtle nature of these changes or the failure of the methods applied; a complex mixture of different cell types present in the whole inflorescence stem is used for chemical analyses, whereas only the fraction of cells with secondary cell wall thickening is expected to display changes.
A Role in Cross-Linking for the Novel Coupling Products of Feruloyl Malate and Coniferyl Alcohol?
Although no gross differences were observed, various signs of decreased cell wall rigidity, particularly in the interfascicular fibers, were noticed in the double mutant (Figure 2). This phenotype is probably linked to the 30% residual lignin levels in the pal1 pal2 mutants but could possibly also arise from the decreased abundance of the novel compounds, FM(4–O–8)G and FM(5–8)G, in the pal1 and pal1 pal2 mutants (Table 3). The malate esters of FA coupled to coniferyl alcohol have not yet been identified before in plants, but ajugol, methyl, and glucose esters of FA(4–O–8)G and/or FA(5–8)G have been reported (Yamamoto et al., 1993; Wahl et al., 1995; Liu et al., 1999). The core structure FA(4–O–8)G esterified to arabinosyl units has been found in saponified grass and cereal cell walls (Jacquet et al., 1995; Grabber et al., 2002; Bunzel et al., 2004). Generally, FA is esterified to the polysaccharides during their biosynthesis in the endomembrane system, and the feruloyl esters can undergo oxidative coupling, thereby linking polysaccharides in the cell wall (Ralph et al., 1999; Fry, 2004). It is possible that the malate ester of FA (coupled with coniferyl alcohol) is an intermediate that can become transesterified to polysaccharides. If this were the case, the reduced rigidity of the cell wall observed in the double mutant could partly arise from weaker cross-coupling of polysaccharides with each other and with lignin.
The Molecular Phenotypes of pal1 and pal2 Mutants Reveal Distinct Roles for PAL1 and PAL2 in Phenylpropanoid Production
Isoforms of PAL have been shown to differ in posttranslational modification, metabolite sensitivity, localization, and association with metabolite channeling complexes (Sarma et al., 1998; Allwood et al., 1999; Rasmussen and Dixon, 1999; Kao et al., 2002). Only few studies have examined the contribution of isoforms to different branches of the phenylpropanoid pathway (Howles et al., 1996; Sarma et al., 1998; Rasmussen and Dixon, 1999). However, the specificity of the isoforms from other species cannot readily be transferred to those in Arabidopsis.
pal1 and pal2 mutants are indistinguishable at the phenotypic level, and only the double mutant has slight phenotypic changes together with a clear reduction in kaempferol glycosides and lignin (Figures 1 and 2, Tables 3 and 5). Considering the 34 genes that code for the enzymes of monolignol biosynthesis, similar trends in alteration of gene expression were observed for most of them in both single pal mutants (e.g., HCT, CCoAOMT5, COMT, and CAD6, Figure 5). Although this observation argues for a functional overlap of PAL1 and PAL2, the fact that expression levels of these four genes were closer to the wild-type level in the double mutant than in the single mutants corroborates a more complex metabolic interaction (Figure 5). On the other hand, expression of several genes was only significantly increased in the double mutant (4CL1 and CCoAOMT6; Figure 5), arguing for a synergistic action of PAL1 and PAL2 in the generation of particular phenylpropanoids. Still, the differential expression of several genes is correlated with only one of the two mutations: PAL3, C3H1, and CCoAOMT7 with pal2, and CCR2 with pal1 (Figures 3 and 5). Although it is too early to speculate on how this transcriptional regulation corresponds to the differential accumulation of a particular metabolite or with particular metabolic complexes, these transcriptional differences clearly indicate directions toward the specific roles of these two PAL isoforms.
When the altered transcripts of other pathways in either pal1 or pal2 (and the double mutant) are compared, more tags relate to light, UV light protection, and stress in pal2 than in pal1 mutants (Figures 5, 6, and 7). On the other hand, pal1 mutants are more altered in phenylpropanoid-related transcripts (PAL1, PAL2, ATR3, 4CL1, DFR, CHS, GST, GS-X pump, and SNG1, Figure 7). In accordance with these transcriptomic changes, the pal1, but not the pal2, mutant has reduced total extractable phenolics because of a decrease in sugar-conjugated kaempferols, scopolin, FM(4–O–8)G, and FM(5–8)G (Table 3).
The difference in aromatic carbon flux seems to translate in different ways to carbon flux in both mutants. The lack of C6-C3 carbon probably signals in both mutants a demand for more carbon; transcripts correlated with photosynthetic carbon fixation and with sucrose degradation are upregulated in both mutants (e.g., Rubisco and invertase; Figure 7). However, based on the transcriptomic changes that exclusively occur in pal1 mutants, the excess carbon is stored away as starch in pal1 mutants (starch synthase: up, starch phosphorylase; involved in starch degradation: down; Figure 7). On the other hand, in pal2 mutants, carbon is rather redirected toward other processes; transcripts related to glycolysis and gluconeogenesis (e.g., trehalose 6P-synthase, triose-P isomerase, phosphoglycerate kinase: up; Figure 7). Possibly part of this carbon is used to generate more amino acids because the levels of many amino acids are higher in pal2 than in pal1 (Table 4).
The understanding of the transcriptome and biochemistry provide novel insight into the functional differences of the two PAL isoforms. On the basis of these observations, a PAL1 knock down has more severe consequences for the phenylpropanoid pathway than a PAL2 knock down. Whereas both enzymes contribute to the production of lignin precursors, PAL1 is of higher importance for the generation of flavonoids in the inflorescence stem. To elaborate on the specific functions of the PAL isoforms in Arabidopsis, future experiments need to address the function of the two other PALs and the potentially different localization of all PALs at the cellular and tissue levels.
The application of a similar molecular phenotyping strategy to other mutants in the 34 genes that encode the 10 known enzymes in the monolignol pathway (Raes et al., 2003) will allow us to answer long-standing questions on the metabolic regulation within the phenylpropanoid pathway, between metabolic pathways, and on the functional divergence in the gene families. In addition, comparative transcriptome analysis of this set of mutants should allow the discrimination between genes regulated by metabolites and genes expressed as a result of pleiotropic effects of a particular mutation. The comparative approach will also help to elucidate the function of the class of unknown genes that still constitute 20% of all differential transcripts identified in the pal mutants. These unknown genes are expected to fall into the same broad functional categories as the other differential genes; hence, new candidate genes that play a role in phenylpropanoid biosynthesis will be identified.
METHODS
Plant Growth Conditions
For germination, seeds were surface sterilized and placed on MS medium (Duchefa, Haarlem, The Netherlands) supplemented with 10 g L−1 sucrose (and if required with 50 mg L−1 kanamycin). After the seeds had undergone a cold treatment for homogenous germination (overnight at 4°C), they were exposed to 20°C, 50 μmol m−2 s−1 light intensity, and 70% humidity under a 16-h-light/8-h-dark cycle. Plants were transferred to a greenhouse after 14 d. Conditions were as follows: 50 μmol m−2 s−1 light intensity at plant level (MBFR/U 400-W incandescent lamps; Philips, Eindhoven, The Netherlands), a 16-h-light/8-h-dark cycle, 40% relative humidity, 23°C, and without shielding from incident daylight. Plant material was always harvested between 11 am and 1 pm.
Generation of the pal Mutants and Double Mutant and Identification of the Genotypes
Primary transformants of SL12-17 (pal1) and of L1-138 (pal2) were obtained through transformation with the pSL1 and pSL4 constructs, respectively (Babiychuk et al., 1997). The exon-trap vector contains, starting from the right border of the T-DNA, the first intron and second exon of the apurinic endonuclease gene fused in frame to the neomycin phosphotransferase II (NPTII) gene lacking its translation initiation codon. The first intron of the apurinic endonuclease gene lacks its 5′ donor splice site. Together, kanamycin resistance after integration of this T-DNA is only achieved in those plants, where the host gene provides promoter activity, an ATG codon, and a 5′ donor splice site. After the intron is spliced out, a chimeric transcript of the host gene, apurinic endonuclease exon 2, and NPTII is made. Insertions in the PAL1 and PAL2 genes were revealed by 5′ rapid amplification of cDNA ends using a NPTII primer. The precise insertion place for both genes was delineated to the first intron through sequencing of a genomic PCR product spanning from the first PAL1 and PAL2 exon to the 5′ end of the NPTII gene (data not shown). Single locus, homozygous lines were obtained by self-fertilization and segregation analysis.
The pal1 pal2 double mutant was generated by crossing pal1 and pal2, using either as female and male parent. In the F2 progeny of both crosses, sterile double mutants were identified in the expected Mendelian ratios. Therefore, the double mutant had to be identified for each experiment in the progeny of plants homozygous for one but heterozygous for the other mutation. To identify the wild-type and mutated PAL1 and PAL2 genes, the following primers were used as sense and antisense primers, respectively: for pal1, 5′-ATGGAGATTAACGGGGCACAC-3′ and 5′-GCATCAGAGCAGCCGATTGTCTGTT-3′; for PAL1, 5′-ATGGAGATTAACGGGGCACAC-3′ and 5′-CATGGCGGCTCTTGTGGCGG-3′; for pal2, 5′-ATGGATCAAATCGAAGCAATG-3′ and 5′-GCATCAGAGCAGCCGATTGTCTGTT-3′; for PAL2, 5′-ATGGATCAAATCGAAGCAATG-3′ and 5′-CATGGCGGCTCTTGTGGCGG-3′. PCR reactions (reaction buffer as supplied with the Taq polymerase, deoxynucleotide triphosphate at 200 pmol, primers at 100 ng) were performed as follows: 30 cycles at 94°C for 30 s, at 50°C for 30 s, at 72°C for 2 min, and a final extension step at 72°C for 10 min. PCR products were separated in 1% agarose gels.
DNA and RNA Extractions
For the identification of double mutants in the progeny of plants homozygous for one but heterozygous for the other mutation, DNA was extracted for use in PCR reactions according to Edwards et al. (1991). Total RNA from stem tissues was extracted with the TRIzol method (Invitrogen, Carlsbad, CA). Total RNA (5 μg) was subsequently reverse transcribed into double-stranded cDNA.
RT-PCRs
All primers used for the amplification of monolignol biosynthesis genes were as described in Raes et al. (2003) and are available at http://www.psb.ugent.be/bioinformatics/. All these primers spanned over at least one intron, so that amplification from the contaminating genomic template could easily be recognized. The ACT2 gene (At3g18780), to which the RT-PCRs were normalized when expressed as relative expression levels, was amplified with the following sense and antisense primers, respectively: 5′-GTTGCACCACCTGAAAGGAAGT-3′ and 5′-CAATGGGACTAAAACGCAAAAC-3′ to generate a fragment of 364 bp on cDNA.
In RT-PCR experiments, reactions in 25 μL (reaction buffer as supplied with the Taq polymerase, 50 ng of each primer) contained a modified nucleotide mix: dCTP, dTTP, and dGTP were at 200 pmol, whereas dATP was reduced to 20 pmol. To each reaction, 0.1 μL of 33P-labeled dATP (10 mCi mL−1, 2500 Ci mmol−1) was added, resulting in a hot-to-cold dATP ratio of 1:2500. Products were separated on 4.5% polyacrylamide or 1% agarose gels and visualized on dried gels through autoradiography. To assure at least semiquantitative assays, the linear range of the PCR reaction was determined for each gene by testing at least two template concentrations (1 μL 1:10 diluted cDNA and 1 μL undiluted cDNA) and three different PCR reaction cycles (21, 24, and 30).
Transcript Profiling and Handling of Differentially Expressed cDNA Fragments
cDNA-AFLP starting with 5 μg of total RNA was performed as described by Breyne et al. (2002). Amplified fragments were considered interesting when their pattern of expression was the same in the two biological sets used. Fragments were cut out, reamplified, and directly sequenced. Sequences were subjected to a tBLASTX against GenBank and a BLASTN against all Arabidopsis thaliana accessions at EMBL. The best-hit sequence was aligned with the fragment and analyzed for the correct BstYI and MseI restriction sites in the respective part of the sequence, for the correct nucleotide extensions at the restriction sites, and for the correct size of the resulting BstYI/MseI fragment. Only when all criteria were fulfilled was the database hit considered correct.
Microarray Hybridization
The 6528 genes were spotted in duplicate on the microarray, of which 6008 were from a unigene clone collection from Incyte (Arabidopsis Gem I; Incyte, Palo Alto, CA) and 520 positive and negative controls from the Universal Score Card spike set (Amersham Biosciences, Little Chalfont, UK); for details, see http://www.microarrays.be/service.htm/currently available arrays. Total RNA (5 μg) of each sample was reverse transcribed and amplified according to a modified protocol for in vitro transcription and subsequently fluorescently labeled with Cy5 or Cy3 (Amersham Biosciences; http://www.microarrays.be/service.htm/protocols). Hybridization and washing were performed in an automated hybridization station (Amersham Biosciences) for a 16-h cycle (http://www.microarrays.be/service.htm/protocols). The arrays were scanned at 532 and 635 nm by a Generation III scanner (Amersham Biosciences), and images were analyzed with ArrayVision (Imaging Research, St. Catharines, Ontario, Canada). Spot intensities were measured as artifact-removed total intensities (ARVol) without correction for background.
Microarray Data Analysis
Slides were normalized by plotting for each single slide M = log2(R/G) against A = log2√R × G for each spot (Yang et al., 2002). Subsequent Lowess normalization (Lowess window width = 0.2) yielded adjusted log2R and log2G signal intensities. Normalized spots were processed by two sequential analyses of variance (ANOVAs; Wolfinger et al., 2001). Using this mixed model approach, as implemented in restricted maximum likelihood assay in Genstat (Payne and Arnold, 2002), the fixed and random effects were fitted simultaneously, and significance for the fixed genotype effects was calculated using the Wald statistics. The P value cutoff was set at 0.001. No further adjustments for multiple testing were performed.
Determination of PAL Activity
PAL activity was determined as previously described (Legrand et al., 1976). Homogenized samples were extracted with 0.1 M sodium borate, pH 8.8, and centrifuged for 10 min at 4°C and 14,000 rpm. The total protein content of the supernatant was determined according to the Bradford assay. Samples of 10 to 50 μg total protein were then allowed to digest a mixture of 100 nmol l-Phe and 0.01 nmol 14C-l-Phe (ratio 10,000:1 cold:hot) for 3 h at 37°C. The volume was increased by the addition of water and extracted with a double volume of cyclohexane/ether (1:1 [v/v]). The organic phase was measured in a scintillation counter for the 14C-trans-cinnamic acid. The partitioning coefficient of cinnamic acid between the water and the organic phase was determined to be ∼50% under our experimental conditions. Using 0.2 nmol 14C-l-Phe as a standard for the counts/minute, the pmol cinnamic acid s−1 μg−1 protein was calculated.
Light Microscopy and Lignin Histochemistry
Vibroslices (50 to 100 μm thick) of agarose-embedded fresh stem material of 3-month-old plants were collected in water or 92% ethanol and immediately stained. Lignin staining after Wiesner was performed in 1% phloroglucinol in 92% ethanol for 2 min. The specimens were subsequently incubated in 25% HCl and immediately observed. Specimens for lignin staining after Mäule were stained in 1% KMnO4 for 5 min, rinsed with water, incubated in 37% HCl:H2O (1:1 [v/v]) for 2 min, and observed after the addition of a drop of NH3. Lignin staining with concentrated aniline sulfate in water proceeded for 3 to 5 min and was stopped by the addition of diluted H2SO4:H2O (1:2 [v/v]). Specimens were observed in water (Spearing, 1953).
Transmission Electron Microscopy
Basal parts of 3-month-old inflorescence stems of all genotypes were harvested and trimmed to smaller pieces before fixation. These pieces were fixed with a mixture of 4% paraformaldehyde and 3% glutaraldehyde and postfixed with 1% OsO4 and 1.5% K3Fe(CN)6 in 0.1 M Na-cacodylate buffer, pH 7.2. Samples were dehydrated through a graded ethanol series, including a bulk staining with 2% uranyl acetate at the 50% ethanol step. Ethanol was subsequently replaced by LR white hard grade embedding medium (London Resin, Basingstoke, UK) and samples embedded in LR white. Ultrathin sections of gold interference color were cut using an ultramicrotome and collected on collodion-coated Cu grids of 200 mesh. The sections were post-stained in an ultrostainer (Leica, Herburgg, Switzerland) for 15 min in 2% uranyl acetate (Ultrostain 1; Leica) at 40°C and 4 min in lead citrate (Ultrostain 2; Leica) at 20°C. The sections were examined with a transmission electron microscope 1010 (JEOL, Tokyo, Japan).
Analysis of Soluble Phenolics and Amino Acids
Three-month-old, fully grown, still-green plants of the four genotypes (10 wild-type plants, 6 plants of pal1, 6 plants of pal2, and 7 plants of pal1 pal2) were harvested after flowering had ceased. Whole inflorescence stems (without leaves, siliques, or flowers) were immediately frozen and homogenized in liquid nitrogen. Soluble phenolics were analyzed as described by Meyermans et al. (2000). Briefly, the material was extracted with methanol and the supernatant dried and extracted with cyclohexane/water containing 0.1% trifluoroacetic acid (1:1 [v/v]). Phenolics in the aqueous phase were separated and analyzed by reverse-phase HPLC. Absorbance spectra were recorded with a 996 diode array detector (Waters, Milford, MA) scanning from 200 to 450 nm. The peak height was quantified at the maximum absorbance value between 250 and 450 nm. Data collection and integration were done using the Millenium software (Waters). Peak height/dry weight of 89 peaks (subtracting the major but indifferent peak at retention time of 5.46 in the beginning of the chromatogram) were analyzed with weighted one-way ANOVA, with P = 0.05 for the F test as well as for the least square difference post-hoc test.
Liquid chromatography–mass spectrometry analysis of the water phase was performed using a gradient separation from 83% solvent A (aqueous, 1% acetic acid) to 77% solvent B (acetonitrile, 1% acetic acid) in 21 min on a reverse-phase Luna C18 column (150 × 2.1 mm, 3 μm; Phenomenex, Torrance, CA) at a flow of 0.30 mL min−1 (SpectraSystem P1000XR HPLC pump; Thermo Separation Products, Riviera Beach, FL) and a column temperature of 40°C (SpectraSystem AS1000 autosampler; Thermo Separation Products). UV/VIS absorption spectra were taken between 200 and 450 nm on a SpectraSystem UV6000LP detector (Thermo Separation Products). Full MS spectra were obtained in the positive or negative ionization mode on a LCQ Classic MS instrument (ThermoQuest, San Jose, CA), using atmospheric pressure chemical ionization as ion source. Vaporizer temperature, capillary temperature, sheath gas, aux gas, and the source current were set at 450°C, 150°C, 64, 3, and 5 μA, or at 400°C, 150°C, 23, 6, and 5 μA in the positive or negative ionization mode, respectively. MSn spectra were obtained by single reaction monitoring using a collision energy of 35%.
The methanol extracts were also used for amino acid analysis. Of the extracts, 5 μL was directly applied to an amino acid analyzer (420A derivatizer with an online 130A separation system; Applied Biosystems, Foster City, CA). Amino acids were identified by cochromatography with authentic standards. Peak height/dry weight were analyzed with one-way ANOVA, with P = 0.05 for the F test and the Dunnett's post-hoc test.
The lyophilized scopolin peak and synthetic scopolin were subjected to acid hydrolysis (1 N HCl, 3 h at 90°C). The reaction products were resolved on HPLC, as described above, and authenticated with standards of scopoletin and isoscopoletin. Synthetic compounds for authentication were vanillate (Acros, Geel, Belgium), scopoletin (Sigma-Aldrich, St. Louis, MO), isoscopoletin (Fluka, Buchs, Switzerland), and FA(4-O-8)G (Ralph et al., 1992). FA(8-5)G was isolated from the mixture obtained by cross-coupling coniferyl alcohol with ethyl FA via MnO2 in acetone followed by saponification (H. Kim and J. Ralph, unpublished data). Scopolin was synthesized from scopoletin (Chaudhury et al., 1948).
Substrate Feeding Assays and Thin-Layer Chromatography of Sinapate Esters
Substrate feeding assays with 1 mM of cinnamic acid or para-coumaric acid were performed on 15 cm inflorescence stems. Stems were cut under water to avoid collapse of the vascular stream and placed into water with the respective substrate for 24 h. Stem pieces from above the part that was in contact with the substrate were harvested, immediately frozen, and analyzed as above. Thin-layer chromatography of sinapate esters were run as described by Lehfeldt et al. (2000) using leaves and inflorescence stems of 10-week-old greenhouse-grown plants.
Acetyl Bromide Soluble Lignin Determination and DFRC Analysis of Lignin
The inflorescence stems of five plants of each C24, pal1, pal2, and pal1 pal2 were harvested individually after 3 months of growth. Plants were green, and flowering had ceased. Stem pieces were immediately frozen in liquid nitrogen and freeze-dried. The ABSL assay was performed as detailed (Fukushima and Hatfield, 2001). ABSL was calculated using an extinction coefficient of 17.2. DFRC lignin analysis was performed essentially as described previously (Lu and Ralph, 1997; Ralph and Lu, 1998). Monomer determination was performed using response factors (coniferyl diacetate (G), 1.39; sinapyl diacetate (S), 1.44) derived from monomer standards against the internal standard (4,4′-ethylidenebisphenol; Aldrich, Milwaukee, WI).
Cell Wall Isolation and Saponification
Inflorescence stem material (1.5 g) of the wild type and double mutant was ground in liquid nitrogen and taken up in 30 mL 50 mM NaCl. This mixture was kept at 4°C overnight. Samples were centrifuged for 10 min at 3500 rpm, and the pellet was extracted with 40 mL 80% ethanol and sonicated for 20 min. This extraction was repeated twice. Centrifugation, extraction, and sonication were repeated exactly with the following extractives: acetone, chloroform:methanol (1:1), and acetone. Samples were allowed to air dry. Saponification of isolated cell walls was performed overnight in 2 M NaOH at room temperature under nitrogen. Extracts were neutralized with 2 M HCl under nitrogen and centrifuged. The supernatant was analyzed with liquid chromatography–mass spectrometry as above.
Carbohydrate Analysis
The xylem tissue from the stems was dried and ground (100 mesh). The powder was extracted successively with a mixture of toluene and ethanol (2:1 [v/v]), with ethanol, and with hot water, and then freeze-dried. The cell wall material was hydrolyzed by the 72% H2SO4 method and the sugars analyzed by gas chromatography of the alditol acetate derivatives as described (Chambat et al., 1997).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers ▪▪▪.
Acknowledgments
We thank Paul Van Hummelen for microarray hybridization, Hoon Kim (U.S. Dairy Forage Research Center) for synthesizing scopolin and FA(8–5)G, Debbie Bishop and Fachuang Lu (U.S. Dairy Forage Research Center) for the ABSL and DFRC analyses, and Martine De Cock for help in preparing the manuscript. This research was funded in part by the European Commission program EDEN (QLK5-CT-2001-00443) and the Department of Energy Biosciences program (DE-AI02-00ER15067 to J.R.). A.R. is a postdoctoral fellow of the Fund for Scientific Research, Flanders.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Antje Rohde (antje.rohde@psb.ugent.be) and Wout Boerjan (wout.boerjan@psb.ugent.be).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.023705.
References
- Allwood, E.G., Davies, D.R., Gerrish, C., Ellis, B.E., and Bolwell, G.P. (1999). Phosphorylation of phenylalanine ammonia-lyase: Evidence for a novel protein kinase and identification of the phosphorylated residue. FEBS Lett. 457, 47–52. [DOI] [PubMed] [Google Scholar]
- Anterola, A.M., Jeon, J.-H., Davin, L.B., and Lewis, N.G. (2002). Transcriptional control of monolignol biosynthesis in Pinus taeda: Factors affecting monolignol ratios and carbon allocation in phenylpropanoid metabolism. J. Biol. Chem. 277, 18272–18280. [DOI] [PubMed] [Google Scholar]
- Babiychuk, E., Fuangthong, M., Van Montagu, M., Inzé, D., and Kushnir, S. (1997). Efficient gene tagging in Arabidopsis thaliana using a gene trap approach. Proc. Natl. Acad. Sci. USA 94, 12722–12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bate, N.J., Orr, J., Ni, W., Meromi, A., Nadler-Hassar, T., Doerner, P.W., Dixon, R.A., Lamb, C.J., and Elkind, Y. (1994). Quantitative relationship between phenylalanine ammonia-lyase levels and phenylpropanoid accumulation in transgenic tobacco identifies a rate-determining step in natural product synthesis. Proc. Natl. Acad. Sci. USA 91, 7608–7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumert, A., Mock, H.-P., Schmidt, J., Herbers, K., Sonnewald, U., and Strack, D. (2001). Patterns of phenylpropanoids in non-inoculated and potato virus Y-inoculated leaves of transgenic tobacco plants expressing yeast-derived invertase. Phytochemistry 56, 535–541. [DOI] [PubMed] [Google Scholar]
- Bentley, R. (1990). The shikimate pathway–A metabolic tree with many branches. Crit. Rev. Biochem. Mol. Biol. 25, 307–384. [DOI] [PubMed] [Google Scholar]
- Blount, J.W., Korth, K.L., Masoud, S.A., Rasmussen, S., Lamb, C., and Dixon, R.A. (2000). Altering expression of cinnamic acid 4-hydroxylase in transgenic plants provides evidence for a feedback loop at the entry point into the phenylpropanoid pathway. Plant Physiol. 122, 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan, W., Ralph, J., and Baucher, M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546. [DOI] [PubMed] [Google Scholar]
- Bolwell, G.P., Cramer, C.L., Lamb, C.J., Schuch, W., and Dixon, R.A. (1986). l-Phenylalanine ammonia-lyase from Phaseolus vulgaris. Modulation of the levels of active enzyme by trans-cinnamic acid. Planta 169, 97–107. [DOI] [PubMed] [Google Scholar]
- Bouly, J.-P., Gissot, L., Lessard, P., Kreis, M., and Thomas, M. (1999). Arabidopsis thaliana proteins related to the yeast SIP and SNF4 interact with AKINα1, an SNF1-like protein kinase. Plant J. 18, 541–550. [DOI] [PubMed] [Google Scholar]
- Breyne, P., Dreesen, R., Vandepoele, K., De Veylder, L., Van Breusegem, F., Callewaert, L., Rombauts, S., Raes, J., Cannoot, B., Engler, G., Inzé, D., and Zabeau, M. (2002). Transcriptome analysis during cell division in plants. Proc. Natl. Acad. Sci. USA 99, 14825–14830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunzel, M., Ralph, J., Kim, H., Hatfield, R.D., and Steinhart, H. (2004). Are cereal grains lignified? J. Agric. Food Chem., in press. [DOI] [PubMed]
- Caño-Delgado, A., Penfield, S., Smith, C., Catley, M., and Bevan, M. (2003). Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J. 34, 351–362. [DOI] [PubMed] [Google Scholar]
- Caño-Delgado, A.I., Metzlaff, K., and Bevan, M.W. (2000). The eli1 mutation reveals a link between cell expansion and secondary cell wall formation in Arabidopsis thaliana. Development 127, 3395–3405. [DOI] [PubMed] [Google Scholar]
- Chambat, G., Cartier, N., Lefèbvre, A., Marais, M.-F., and Joseleau, J.-P. (1997). Changes in cell wall extracellular polysaccharides during the culture cycle of Rubus fruticosus cells in suspension culture. Plant Physiol. Biochem. 35, 655–664. [Google Scholar]
- Chaudhury, D.N., Holland, R.A., and Robertson, A. (1948). The synthesis of glycosides. Part XII. Fabiatrin. J. Chem. Soc. 1948, 1671–1672. [DOI] [PubMed]
- Cheng, S.-H., Sheen, J., Gerrish, C., and Bowlell, G.P. (2001). Molecular identification of phenylalanine ammonia-lyase as a substrate of a specific constitutively active Arabidopsis CDPK expressed in maize protoplasts. FEBS Lett. 503, 185–188. [DOI] [PubMed] [Google Scholar]
- Dixon, D.P., Lapthorn, A., and Edwards, R. (2002). Plant glutathione transferases. Genome Biol. 3, 2004.1–2004.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond, P.J., and Graham, I.A. (2003). Trehalose metabolism: A regulatory role for trehalose-6-phosphate? Curr. Opin. Plant Biol. 6, 231–235. [DOI] [PubMed] [Google Scholar]
- Edwards, K., Johnstone, C., and Thompson, C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19, 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkind, Y., Edwards, R., Mavandad, M., Hedrick, S.A., Ribak, O., Dixon, R.A., and Lamb, C.J. (1990). Abnormal plant development and down-regulation of phenylpropanoid biosynthesis in transgenic tobacco containing a heterologous phenylalanine ammonia-lyase gene. Proc. Natl. Acad. Sci. USA 87, 9057–9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, C., Karafyllidis, I., Wasternack, C., and Turner, J.G. (2002). The Arabidopsis mutant cev1 links well wall signaling to jasmonate and ethylene responses. Plant Cell 14, 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frishman, D., et al. (2003). The PEDANT genome database. Nucleic Acids Res. 31, 207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry, S.C. (2004). Primary cell wall metabolism: Tracking the careers of wall polymers in living plant cells. New Phytol. 161, 641–675. [DOI] [PubMed] [Google Scholar]
- Fukushima, R.S., and Hatfield, R.D. (2001). Extraction and isolation of lignin for utilization as a standard to determine lignin concentration using the acetyl bromide spectrophotometric method. J. Agric. Food Chem. 49, 3133–3139. [DOI] [PubMed] [Google Scholar]
- Grabber, J.H., Ralph, J., and Hatfield, R.D. (2002). Model studies of ferulate-coniferyl alcohol cross-product formation in primary maize walls: Implications for lignification in grasses. J. Agric. Food Chem. 50, 6008–6016. [DOI] [PubMed] [Google Scholar]
- Graham, T.L. (1998). Flavonoid and flavonol glycoside metabolism in Arabidopsis. Plant Physiol. Biochem. 36, 135–144. [Google Scholar]
- Guillet, G., Poupart, J., Basurco, J., and De Luca, V. (2000). Expression of tryptophan decarboxylase and tyrosine decarboxylase genes in tobacco results in altered biochemical and physiological phenotypes. Plant Physiol. 122, 933–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield, R.D., Ralph, J., and Grabber, J.H. (1999). Cell wall cross-linking by ferulates and diferulates in grasses. J. Sci. Food Agric. 79, 403–407. [Google Scholar]
- Henkes, S., Sonnewald, U., Badur, R., Flachmann, R., and Stitt, M. (2001). A small decrease of plastid transketolase activity in antisense tobacco transformants has dramatic effects on photosynthesis and phenylpropanoid metabolism. Plant Cell 13, 535–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann, K.M. (1995). The shikimate pathway: Early steps in the biosynthesis of aromatic compounds. Plant Cell 7, 907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker, U., Tepperman, J.M., and Quail, P.H. (1999). SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science 284, 496–499. [DOI] [PubMed] [Google Scholar]
- Howles, P.A., Sewalt, V.J.H., Paiva, N.L., Elkind, Y., Bate, N.J., Lamb, C., and Dixon, R.A. (1996). Overexpression of l-phenylalanine ammonia-lyase in transgenic tobacco plants reveals control points for flux into phenylpropanoid biosynthesis. Plant Physiol. 112, 1617–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, W.-J., Harding, S.A., Lung, J., Popko, J.L., Ralph, J., Stokke, D.D., Tsai, C.-J., and Chiang, V.L. (1999). Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat. Biotechnol. 17, 808–812. [DOI] [PubMed] [Google Scholar]
- Hudson, M., Ringli, C., Boylan, M.T., and Quail, P.H. (1999). The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev. 13, 2017–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet, G., Pollet, B., Lapierre, C., Mhamdi, F., and Rolando, C. (1995). New ether-linked ferulic acid-coniferyl alcohol dimers identified in grass straws. J. Agric. Food Chem. 43, 2746–2751. [Google Scholar]
- Jones, J.D., Henstrand, J.M., Handa, A.K., Herrmann, K.M., and Weller, S.C. (1995). Impaired wound induction of 3-deoxy-d-arabino-heptulosonate-7-phosphate (DAHP) synthase and altered stem development in transgenic potato plants expressing a DAHP synthase antisense construct. Plant Physiol. 108, 1413–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, L., Ennos, A.R., and Turner, S.R. (2001). Cloning and characterization of irregular xylem4 (irx4): A severely lignin-deficient mutant of Arabidopsis. Plant J. 26, 205–216. [DOI] [PubMed] [Google Scholar]
- Kao, Y.-Y., Harding, S.A., and Tsai, C.-J. (2002). Differential expression of two distinct phenylalanine ammonia-lyase genes in condensed tannin-accumulating and lignifiying cells of quakin aspen. Plant Physiol. 130, 796–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korth, K.L., Blount, J.W., Chen, F., Rasmussen, S., Lamb, C., and Dixon, R.A. (2001). Changes in phenylpropanoid metabolites associated with homology-dependent silencing of phenylalanine ammonia-lyase and its somatic reversion in tobacco. Physiol. Plant. 111, 137–143. [Google Scholar]
- Lam, H.-M., Hsieh, M.-H., and Coruzzi, G. (1998). Reciprocal regulation of distinct asparagine synthetase genes by light and metabolites in Arabidopsis thaliana. Plant J. 16, 345–353. [DOI] [PubMed] [Google Scholar]
- Lamb, C. (1977). trans-cinnamic acid as a mediator of the light-stimulated increase in hydroxycinnamoyl:CoA-quinate hydroxycinnamoyl transferase. FEBS Lett. 75, 37–40. [DOI] [PubMed] [Google Scholar]
- Landry, L.G., Chapple, C.C.S., and Last, R.L. (1995). Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiol. 109, 1159–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauvergeat, V., Lacomme, C., Lacombe, E., Lasserre, E., Roby, D., and Grima-Pettenati, J. (2001). Two cinnamoyl-CoA reductase (CCR) genes from Arabidopsis thaliana are differentially expressed during development and in response to infection with pathogenic bacteria. Phytochemistry 57, 1187–1195. [DOI] [PubMed] [Google Scholar]
- Legrand, M., Fritig, B., and Hirth, L. (1976). Enzymes of the phenylpropanoid pathway and the necrotic reaction of hypersensitive tobacco to tobacco mosaic virus. Phytochemistry 15, 1353–1359. [Google Scholar]
- Lehfeldt, C., Shirley, A.M., Meyer, K., Ruegger, M.O., Cusumano, J.C., Viitanen, P.V., Strack, D., and Chapple, C. (2000). Cloning of the SNG1 gene of Arabidopsis reveals a role for a serine carboxypeptidase-like protein as an acyltransferase in secondary metabolism. Plant Cell 12, 1295–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyva, A., Jarillo, J.A., Salinas, J., and Martinez-Zapater, J.M. (1995). Low temperature induces the accumulation of phenylalanine ammonia-lyase and chalcone synthase mRNAs of Arabidopsis thaliana in a light-dependent manner. Plant Physiol. 108, 39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Zhou, Y., Cheng, X., Sun, J., Marita, J.M., Ralph, J., and Chiang, V.L. (2003). Combinatorial modification of multiple lignin traits in trees through multigene cotransformation. Proc. Natl. Acad. Sci. USA 100, 4939–4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay, W.P., McAlister, F.M., Zhu, Q., He, X.-Z., Dröge-Laser, W., Hedrick, S., Doerner, P., Lamb, C., and Dixon, R.A. (2002). KAP-2, a protein that binds to the H-box in a bean chalcone synthase promoter, is a novel plant transcription factor with sequence identity to the large subunit of human Ku autoantigen. Plant Mol. Biol. 49, 503–514. [DOI] [PubMed] [Google Scholar]
- Liu, G., Sánchez-Fernández, R., Li, Z.-S., and Rea, P.A. (2001). Enhanced multispecificity of Arabidopsis vacuolar multidrug resistance-associated protein-type ATP-binding cassette transporter, AtMRP2. J. Biol. Chem. 276, 8648–8656. [DOI] [PubMed] [Google Scholar]
- Liu, H., Orjala, J., Sticher, O., and Rali, T. (1999). Acylated flavonol glycosides from leaves of Stenochlaena palustris. J. Nat. Prod. 62, 70–75. [DOI] [PubMed] [Google Scholar]
- Loake, G.J., Faktor, O., Lamb, C.J., and Dixon, R.A. (1992). Combination of H-box [CCTACC(N)7CT] and G-box [CACGTG] cis elements is necessary for feed-forward stimulation of a chalcone synthase promoter by the phenylpropanoid-pathway intermediate p-coumaric acid. Proc. Natl. Acad. Sci. USA 89, 9230–9234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, F., and Ralph, J. (1997). Derivatization followed by reductive cleavage (DFRC method), a new method for lignin analysis: Protocol for analysis of DFRC monomers. J. Agric. Food Chem. 45, 2590–2592. [Google Scholar]
- Lu, Y.-P., Li, Z.-S., Drozdowicz, Y.M., Hörtensteiner, S., Martinoia, E., and Rea, P.A. (1998). AtMRP2, an Arabidopsis ATP binding cassette transporter able to transport glutathione S-conjugates and chlorophyll catabolites: Functional comparisons with AtMRP1. Plant Cell 10, 267–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matt, P., Krapp, A., Haake, V., Mock, H.-P., and Stitt, M. (2002). Decreased Rubisco activity leads to dramatic changes of nitrate metabolism, amino acid metabolism and the levels of phenylpropanoids and nicotine in tobacco antisense RBCS transformants. Plant J. 30, 663–677. [DOI] [PubMed] [Google Scholar]
- Mavandad, M., Edwards, R., Liang, X., Lamb, C.J., and Dixon, R.A. (1990). Effects of trans-cinnamic acid on expression of the bean phenylalanine ammonia-lyase gene family. Plant Physiol. 94, 671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mele, G., Ori, N., Sato, Y., and Hake, S. (2003). The knotted1-like homeobox gene BREVIPEDICELLUS regulates cell differentiation by modulating metabolic pathways. Genes Dev. 17, 2088–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyermans, H., et al. (2000). Modification in lignin and accumulation of phenolic glucosides in poplar xylem upon down-regulation of caffeoyl-coenzyme A O-methyltransferase, an enzyme involved in lignin biosynthesis. J. Biol. Chem. 275, 36899–36909. [DOI] [PubMed] [Google Scholar]
- Mizutani, M., and Ohta, D. (1998). Two isoforms of NADPH:cytochrome P450 reductase in Arabidopsis thaliana. Gene structure, heterologous expression in insect cells, and differential regulation. Plant Physiol. 116, 357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, L.A., Goodman, C.D., Silday, R.A., and Walbot, V. (2000). AN9, a petunia glutathione S-transferase required for anthocyanin sequestration, is a flavonoid-binding protein. Plant Physiol. 123, 1561–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne, R.W., and Arnold, G.M. (2002). GenStat Release 6.1 Reference Manual Part 3: Procedure Library PL14. (Hemel Hempstead, UK: VSN International).
- Raes, J., Rohde, A., Christensen, J.H., Van de Peer, Y., and Boerjan, W. (2003). Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 133, 1051–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph, J., Hatfield, R.D., Grabber, J.H., Jung, H.-J. G., Quideau, S., and Helm, R.F. (1999). Cell wall cross-linking in grasses by ferulates and diferulates. In Lignin and Lignan Biosynthesis, ACS Symposium Series, Vol. 697, N.G. Lewis and S. Sarkanen, eds (Washington, D.C.: American Chemical Society), pp. 209–236.
- Ralph, J., Helm, R.F., Quideau, S., and Hatfield, R.D. (1992). Lignin–feruloyl ester cross-links in grasses. Part 1. Incorporation of feruloyl esters into coniferyl alcohol dehydrogenation polymers. J. Chem. Soc. Perkin Trans. 1 1, 2961–2969. [Google Scholar]
- Ralph, J., and Lu, F. (1998). The DFRC method for lignin analysis. 6. A simple modification for identifying natural acetates on lignins. J. Agric. Food Chem. 46, 4616–4619. [DOI] [PubMed] [Google Scholar]
- Ralph, J., Lundquist, K., Brunow, G., Lu, F., Kim, H., Schatz, P.F., Marita, J.M., Hatfield, R.D., Ralph, S.A., Christensen, J.H., and Boerjan, W. (2004). Lignins: Natural polymers from oxidative coupling of 4-hydroxyphenylpropanoids. Phytochem. Rev. 3, in press.
- Rasmussen, S., and Dixon, R.A. (1999). Transgene-mediated and elicitor-induced perturbation of metabolic channeling at the entry point into the phenylpropanoid pathway. Plant Cell 11, 1537–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro, D.-K., Ehlting, J., and Douglas, C.J. (2002). Cloning, functional expression, and subcellular localization of multiple NADPH-cytochrome P450 reductase from hybrid poplar. Plant Physiol. 130, 1837–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Fernández, R., Davies, T.G.E., Coleman, J.O.D., and Rea, P.A. (2001). The Arabidopsis thaliana ABC protein superfamily, a complete inventory. J. Biol. Chem. 276, 30231–30244. [DOI] [PubMed] [Google Scholar]
- Sarma, A.D., Sreelakshmi, Y., and Sharma, R. (1998). Differential expression and properties of phenylalanine ammonia-lyase isoforms in tomato leaves. Phytochemistry 49, 2233–2243. [DOI] [PubMed] [Google Scholar]
- Schluepmann, H., Pellny, T., van Dijken, A., Smeekens, S., and Paul, M. (2003). Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 100, 6849–6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewalt, V.J.H., Ni, W., Blount, J.W., Jung, H.G., Masoud, S.A., Howles, P.A., Lamb, C., and Dixon, R.A. (1997). Reduced lignin content and altered lignin composition in transgenic tobacco down-regulated in expression of l-phenylalanine ammonia-lyase or cinnamate 4-hydroxylase. Plant Physiol. 115, 41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somssich, I.E., and Hahlbrock, K. (1998). Pathogen defence in plants–A paradigm of biological complexity. Trends Plant Sci. 3, 86–90. [Google Scholar]
- Spearing, J.K. (1953). Simultaneous double-staining technique for histology of vascular plants. J. R. Microsc. Soc. 73, 162–163. [DOI] [PubMed] [Google Scholar]
- Taylor, N.G., Scheible, W.-R., Cutler, S., Somerville, C.R., and Turner, S.R. (1999). The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11, 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban, P., Mignotte, C., Kazmaier, M., Delorme, F., and Pompon, D. (1997). Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J. Biol. Chem. 272, 19176–19186. [DOI] [PubMed] [Google Scholar]
- Veit, M., and Pauli, G.F. (1999). Major flavonoids from Arabidopsis thaliana leaves. J. Nat. Prod. 62, 1301–1303. [DOI] [PubMed] [Google Scholar]
- Wahl, A., Roblot, F., and Cavè, A. (1995). Isolation and structure elucidation of xylobuxin, a new neolignan from Xylopia buxifolia. J. Nat. Prod. 58, 786–789. [Google Scholar]
- Wanner, L.A., Li, G., Ware, D., Somssich, I.E., and Davis, K.R. (1995). The phenylalanine ammonia-lyase gene family in Arabidopsis thaliana. Plant Mol. Biol. 27, 327–338. [DOI] [PubMed] [Google Scholar]
- Wolfinger, R.D., Gibson, G., Wolfinger, E.D., Bennett, L., Hamadeh, H., Bushel, P., Afshari, C., and Paules, R.S. (2001). Assessing gene significance from cDNA microarray expression data via mixed models. J. Comput. Biol. 8, 625–637. [DOI] [PubMed] [Google Scholar]
- Yamamoto, A., Nitta, S., Miyase, T., Ueno, A., and Wu, L.-J. (1993). Phenylethanoid and lignan-iridoid complex glycosides from roots of Buddleja davidii. Phytochemistry 32, 421–425. [DOI] [PubMed] [Google Scholar]
- Yang, Y.H., Dudoit, S., Luu, P., Lin, D.M., Peng, V., Ngai, J., and Speed, T.P. (2002). Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30, e15. [DOI] [PMC free article] [PubMed]
- Yao, K., De Luca, V., and Brisson, N. (1995). Creation of a metabolic sink of tryptophan alters the phenylpropanoid pathway and the susceptibility of potato to Phytophthora infestans. Plant Cell 7, 1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]