Abstract
Nitric oxide (NO) is a widespread signaling molecule, and numerous targets of its action exist in plants. Whereas the activity of NO in erythrocytes, microorganisms, and invertebrates has been shown to be regulated by several hemoglobins, the function of plant hemoglobins in NO detoxification has not yet been elucidated. Here, we show that Arabidopsis thaliana nonsymbiotic hemoglobin AHb1 scavenges NO through production of S-nitrosohemoglobin and reduces NO emission under hypoxic stress, indicating its role in NO detoxification. However, AHb1 does not affect NO-mediated hypersensitive cell death in response to avirulent Pseudomonas syringae, suggesting that it is not involved in the removal of NO bursts originated from acute responses when NO mediates crucial defense signaling functions.
INTRODUCTION
The broad chemistry of nitric oxide (NO) involves an array of interrelated redox forms with different chemical reactivities. It may have beneficial effects, for example, as a messenger in immune responses, but unregulated NO production can lead to either apoptotic or necrotic cell death, depending on the severity of the damage (Murphy, 1999). In plants, NO production is involved in diverse physiological processes, including disease resistance, growth, and development (Wendehenne et al., 2001). This wide variety of effects reflects the basic signaling mechanisms that are used by virtually all mammalian and plant cells in addition to many lower organisms. Hemoglobins are well known regulators of NO homeostasis. In humans, hemoglobins regulate the activity of NO through either detoxification (Joshi et al., 2002) or delivery through transnitrosylation reactions (Gow et al., 1999). In bacteria, flavohemoglobins consume NO enzymatically (Gardner et al., 1998; Hausladen et al., 1998), catalyzing the reaction of nitroxyl equivalent with molecular oxygen (Hausladen et al., 2001). Yeast flavohemoglobin possesses the same NO reductase/denitrosylase activity as bacteria (Liu et al., 1999). Ascaris hemoglobin has a slow NO consumption activity that involves the intermediacy of S-nitrosylated hemoglobin and subserves oxygen removal from the perienteric cavity (Minning et al., 1999). Thus, the primordial function of hemoglobins present not only in erythrocytes but also in microorganisms, invertebrates, and plants may well be to protect against nitrosative stress and to modulate NO signaling functions (Durner et al., 1999).
In plants, there are at least three distinct types of hemoglobins, which have been categorized as symbiotic, nonsymbiotic, and truncated hemoglobins. Symbiotic hemoglobins are found specifically in symbiotic legume root nodules where they scavenge and transport molecular oxygen to protect Rhizobium nitrogenase from inactivation (Appleby, 1984). Nonsymbiotic hemoglobins occur at much lower abundance but appear ubiquitous in the plant kingdom (Dordas et al., 2003a). The most recently discovered plant hemoglobins, the truncated hemoglobins, also appear to be present throughout the plant kingdom and share some characteristics with nonsymbiotic hemoglobin (Watts et al., 2001).
There are two classes of nonsymbiotic hemoglobins. Class-1 proteins have an extremely high affinity for oxygen and are induced in plants during hypoxic stress, whereas class-2 hemoglobins have lower affinity for oxygen and are inducible by low temperature. Although the presence of stress-induced hemoglobins is widespread in the plant kingdom, their function has not been elucidated. Nonsymbiotic hemoglobins exhibit a so-called hexacoordination in which the ligand binding site on the heme prosthetic group is occupied by a His residue, such that ligand binding involves competitive displacement of this intramolecular coordination (Goodman and Hargrove, 2001). Many hexacoordinate hemoglobins are upregulated during hypoxia or similar stresses, and their expression is directly associated with protection against hypoxic challenge (Hunt et al., 2002; Dordas et al., 2003b), a stress condition generating copious amount of NO (Dordas et al., 2003a).
AHb1, which is one of two nonsymbiotic hemoglobins encoded by the Arabidopsis thaliana genome, has been shown to have a high affinity for oxygen, with a dissociation constant for oxyhemoglobin of 0.12 s−1 (Trevaskis et al., 1997). This would result in conditions whereby the protein will remain oxygenated at oxygen concentrations far below those at which anaerobic processes are activated. Thus, it is unlikely that this hemoglobin would function as either a facilitator of oxygen diffusion or as an oxygen sensor.
In this report, we provide evidence suggesting that hexacoordinate AHb1 functions as an NO dioxygenase, metabolizing NO to nitrate using NADPH as electron donor. We also show that S-nitrosohemoglobin is endogenously produced in plants, indicating that Cys residue(s) play a conserved role in processing of NO and S-nitrosothiols (SNO). AHb1 reduces NO emission under hypoxic stress, confirming its role in NO detoxification. However, the kinetics of NO detoxification reaction is low; thus, it does not interfere with the NO bursts originated by an acute response such as that occurring during the hypersensitive resistance response to pathogen attack, when NO-mediated signaling functions are needed for the execution of the cell death program and the activation of disease resistance genes.
RESULTS
Characteristics of Recombinant AHb1 and Reaction with NO
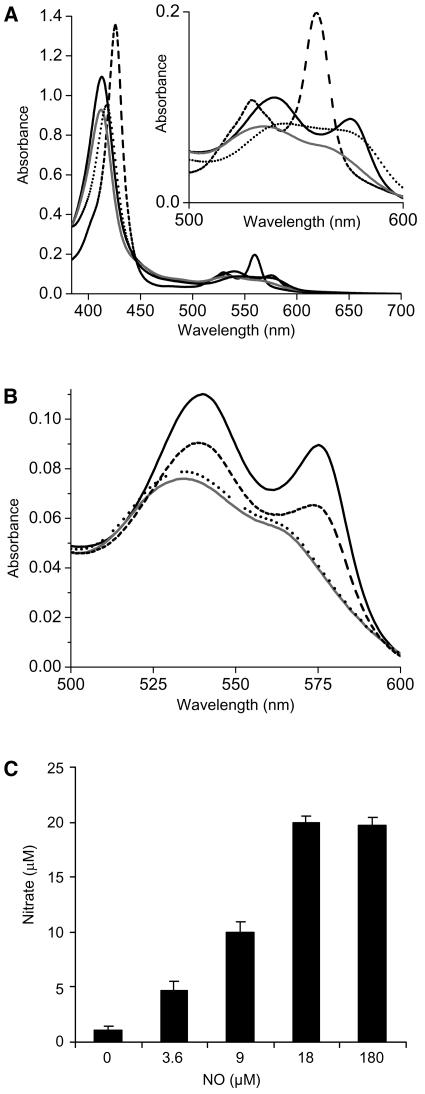
A recombinant form of AHb1 was expressed in Escherichia coli and purified to homogeneity to study its possible interactions with NO. Its molecular mass, as assessed by size exclusion chromatography on Superose 12, was 36 kD, suggesting that native AHb1 is a homodimer. Reduced, deoxygenated AHb1 exhibited a split peak in the visible region that is characteristic of a hexacoordinate heme iron and hexacoordinate hemoglobins (Figure 1A). The positions of the absorbance peaks are nearly identical to those of rice (Oryza sativa) Hb1 (Goodman and Hargrove, 2001), further suggesting bis-histidyl coordination in both the ferric and deoxy ferrous forms. Treatment of AHb1(FeII) with excess NO resulted in a new absorption spectrum with a maximum at 418 nm, typical of nitrosyl-hemoglobin (Gow et al., 1999) (Figure 1A), and the rapid NO-dependent oxidation of hemoglobin to methemoglobin was accompanied by nitrate accumulation (Figures 1B and 1C). This reaction occurred even with substoichiometric amounts of NO, and the accumulation of both metAHb1 and nitrate was dependent on the concentration of NO (Figures 1B and 1C), implying a direct oxidation to AHb1(FeIII). However, even when NO was provided in excess, nitrate only accumulated to stoichiometric equivalence with metAHb1 (Figure 1C), indicating that metAHb1 cannot further metabolize NO.
Figure 1.
Spectral Features of AHb1 and Its Interaction with NO.
(A) Absorbance spectra. AHb1 is purified in an oxygenated state (solid line). AHb1(FeII) was produced by adding an excess of solid sodium dithionite (dashed line): the two resolved bands at 530 and 560 nm (see inset) and the sharp Soret peak at 425 nm are reminiscent of low-spin hexacoordinated species. AHb1(FeIII) was obtained by adding an excess of potassium ferricyanide (gray line). Addition of 150 μM NO to 15 μM AHb1 reduced in the presence of an excess of dithionite led to the appearance of AHb1(FeII)NO (dotted line).
(B) Interaction of oxygenated AHb1 with NO. Conversion of 15 μM oxygenated AHb1 (solid line) to metAHb1 (gray line) was recorded 3 min after incubation with either 5 μM NO (dashed line) or 150 μM NO (dotted line).
(C) Nitrate production from 18 μM oxygenated AHb1 incubated with the indicated concentrations of NO. Experiments were repeated three times with similar results.
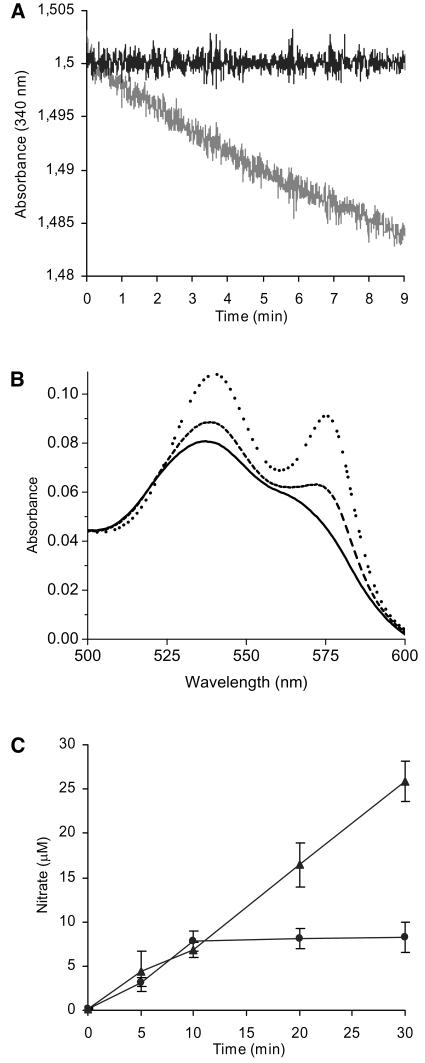
This reaction has biological relevance only if AHb1(FeII) can be regenerated. Other hemoglobins require a reductase activity for ferric heme iron reduction. For example, bacterial flavohemoglobin possesses a reductase domain (Poole and Hughes, 2000), whereas human myoglobin interacts with a flavoprotein, namely metmyoglobin-reductase, which uses an electron from NADPH (Flogel et al., 2001). It was found that metAHb1 can be directly reduced by NADPH, as for hemoglobin from Ascaris (Minning et al., 1999), and whereas AHb1(FeIII) consumed NADPH, AHb1(FeII)O2 did not (Figure 2A). Myoglobin was used as control and showed a rate of NADPH oxidation that was at least fourfold lower in the same experimental conditions (data not shown). Spectroscopic analysis showed that the addition of NADPH converted oxidized AHb1 to an oxygenated state (Figure 2B). To confirm this enzymatic cycle for NO metabolism, we showed that in the presence of excess NO and NADPH, nitrate accumulated linearly with time to levels severalfold beyond stoichiometric equivalence with AHb1 (Figure 2C). In the presence of saturating concentrations of NO, the specific activity of AHb1 was 24 nmol/min/mg. Because methemoglobin reductase activity was found in alfalfa (Medicago sativa) root cultures (Igamberdiev et al., 2004), it is possible that AHb1 activity may be augmented in vivo by specific interaction with a reductase system, as suggested for barley (Hordeum vulgare) hemoglobin (Igamberdiev et al., 2004) and demonstrated for human myoglobin (Flogel et al., 2001).
Figure 2.
NADPH-Dependent Catalysis of Nitrate Production by AHb1.
(A) NADPH consumption by oxygenated (black line) and oxidized (gray line) 6 μM AHb1.
(B) Conversion of 20 μM metAHb1 (black line) to the oxygenated state 3 min (dashed line) and 10 min (dotted line) after addition of 1 mM NADPH.
(C) Nitrate production by 8 μM AHb1 plus 80 μM NO in presence (triangle) or absence (circle) of 300 μM NADPH. Experiments were repeated three times with similar results.
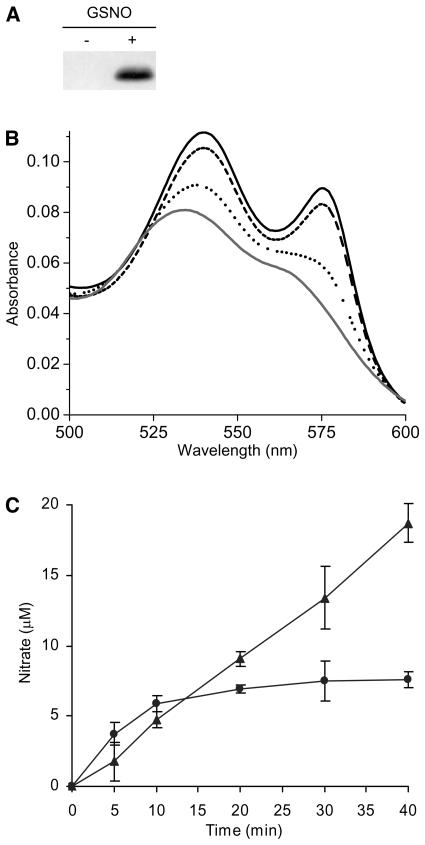
A similar catalytic cycle was also observed using S-nitrosoglutathione (GSNO) as a substrate. This nitrosylating compound represents a major transport form of NO in biological systems (Becker et al., 1995). GSNO transfers NO to human hemoglobin (Jia et al., 1996) through S-nitrosylation of a proximal Cys residue that is conserved in β-globins of all birds and mammals to yield S-nitrosohemoglobin (Gross and Lane, 1999). Incubation of AHb1 with GSNO under conditions that favored transnitrosylation of Cys residues (Jia et al., 1996) produced S-nitroso AHb1 (Figure 3A). FeII was oxidized (Figure 3B), and nitrate was produced in equimolar amounts with respect to heme concentration (Figure 3C). Furthermore, NADPH stimulated continuous production of nitrate severalfold beyond stoichiometric levels of AHb1 heme, indicating that AHb1 has the capacity to catalyze the metabolism of NO bound to thiol groups (Figure 3C).
Figure 3.
Interaction of AHb1 with GSNO.
(A) Protein gel blot analysis of S-nitroso AHb1 subjected to the biotin-switch assay. metAHb1 (20 μM) was incubated in the absence (−) or in the presence (+) of 200 μM GSNO.
(B) Conversion of 17 μM oxygenated AHb1 (solid line) to metAHb1 (gray line) 3 min (dashed line) and 10 min (dotted line) after incubation with 170 μM GSNO. The amount of NO released by 200 μM GSNO after 10 min of incubation in 10 mM phosphate buffer, pH 7.4, was <0.2 μM, as revealed by 4-amino-5-methylamino-2′,7′-difluorofluorescein (DAF-FM) fluorescence analysis. The absorbance maxima of S-nitroso AHb1 are masked by the absorption spectra of heme.
(C) Nitrate production by 8 μM oxygenated AHb1 plus 80 μM GSNO in the presence (triangle) or in the absence (circle) of 300 μM NADPH. Experiments were repeated three times with similar results.
Functional Effects of AHb1 in Transgenic Plants
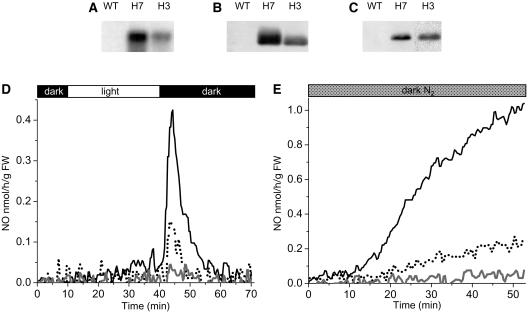
To investigate whether AHb1 can metabolize NO in vivo, we generated transgenic Arabidopsis ecotype Columbia-0 (Col-0) plants overexpressing AHb1. By RNA gel blot analysis of homozygous T3 plants, we selected two lines, referred to as H3 and H7, which were characterized by moderate and strong overexpression of sense AHb1 transcripts, respectively (Figure 4A). These lines showed correspondingly elevated levels of AHb1 protein (Figure 4B). The nitrate reductase–mediated emission of NO, resulting from the accumulation of nitrite on transfer of light-adapted plants to darkness (Kaiser et al., 2002), was significantly lower in the AHb1-overexpressing plants compared with the wild type (Figure 4D). To study the in vivo interaction of AHb1 with NO, S-nitrosylated proteins extracts were isolated by the biotin-switch method from Arabidopsis plants grown under normal conditions and then immunoprecipitated with antibiotin antibodies. Protein gel blot analysis of S-nitrosylated proteins using an anti-AHb1 antibody revealed the presence of S-nitrosylated AHb1 in overexpressing plants (Figure 4C). The absence of an AHb1 signal in wild-type plants is most likely attributable to insufficient levels of this protein in the total cellular extract. Thus, examination of S-nitrosylation of AHb1 in transgenic plants indicates that AHb1 scavenges endogenous NO in vivo through production of S-nitrosohemoglobin.
Figure 4.
Characterization of Arabidopsis Plants Overexpressing AHb1.
(A) RNA gel blot hybridization analysis of AHb1 expression in the wild type and in transgenic lines H7 and H3.
(B) Protein gel blot analysis of total proteins from leaves of wild-type, H7, and H3 plants probed with antibody to AHb1.
(C) Nitrosylation of AHb1 in vivo. S-Nitrosylated proteins from leaves of wild-type, H7, and H3 plants were biotinylated, immunoprecipitated with antibiotin antibody, and immunoblotted with antibodies to AHb1.
(D) Gas phase NO emission pattern of the wild-type (solid line), H7 (gray line), and H3 (dotted line) plants measured during a dark-light-dark treatment. The dark and light exposure is expressed on the bar at top. FW, fresh weight.
(E) Gas phase NO emission pattern of the wild-type (solid line), H7 (gray line), and H3 (dotted line) transgenic plants subjected to hypoxic stress by flushing the chamber with N2 in the dark. Experiments were repeated three times with similar results.
AHb1 is strongly induced during hypoxia, and as demonstrated for alfalfa hemoglobin (Dordas et al., 2003b), its overexpression in transgenic plants enhances tolerance to hypoxic stress (Hunt et al., 2002). Several mechanisms have been proposed to explain this effect, including the maintenance of the energy status of the cell in the absence of mitochondrial respiration (Sowa et al., 1998; Dordas et al., 2003b). However, it has recently emerged that anoxia activates nitrate reductase leading to elevated NO emission from leaves (Rockel et al., 2002). As predicted from the properties of AHb1 in vitro, hypoxia-stimulated NO emission was dramatically suppressed in the AHb1-overexpressing plants (Figure 4E).
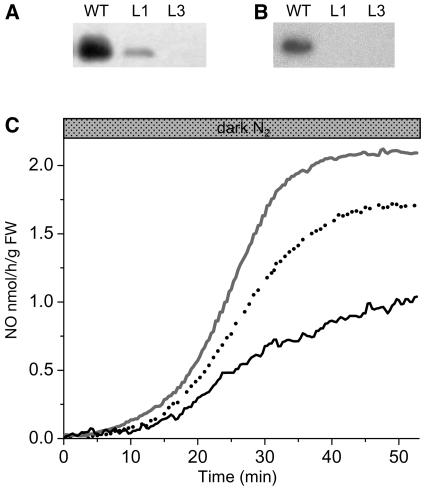
To further confirm the function of AHb1 in NO detoxification under hypoxia, we generated transgenic Arabidopsis plants expressing AHb1 in antisense orientation. By protein gel blot analysis of T3 homozygous plants, we selected two lines, referred to as L1 and L3, characterized by moderate and strong reduction of AHb1 accumulation under hypoxic stress, respectively (Figure 5A). Protein gel blot analysis of S-nitrosylated proteins using the anti-AHb1 antibody revealed the presence of S-nitrosylated AHb1 only in the wild type (Figure 5B), most likely because of insufficient levels of this protein in the total cellular extract from antisense lines. Analysis of NO emission under hypoxia revealed a dramatic accumulation of NO in the antisense lines compared with the wild type (Figure 5C).
Figure 5.
Characterization of AHb1 Antisense Arabidopsis Plants.
(A) Protein gel blot analysis of total proteins from the wild type and from antisense lines L1 and L3 exposed for 24 h to hypoxic stress.
(B) Nitrosylation of AHb1 in vivo. S-Nitrosylated proteins from the wild type and from antisense lines L1 and L3 exposed for 24 h to hypoxic stress were biotinylated, immunoprecipitated with antibiotin antibody, and immunoblotted with antibodies to AHb1.
(C) Gas phase NO emission pattern of wild-type (solid line), L1 (dotted line), and L3 (gray line) antisense lines subjected to hypoxic stress by flushing the chamber with N2 in the dark. Experiments were repeated three times with similar results.
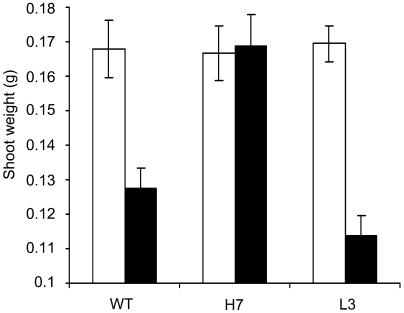
These observations, corroborated by the direct correlation between weight of shoots and AHb1 level observed after exposure of wild-type and transgenic Arabidopsis plants to hypoxic stress (Figure 6) and by the large accumulation of NO observed in hypoxic root cultures of hemoglobin-deficient alfalfa (Dordas et al., 2003b) and maize (Zea mays) mutants (Dordas et al., 2004), support the hypothesis that stress-induced hemoglobins may function as NO dioxygenases (Dordas et al., 2003a; Igamberdiev et al., 2004) protecting against nitrosative stress associated with hypoxia and confirm that AHb1 metabolizes NO in vivo.
Figure 6.
Growth after Hypoxic Stress of Arabidopsis Plants.
Shoot weight of 10 plants from the wild type and transgenic H7 and L3 lines either exposed to 24 h of hypoxic stress (closed bars) or aligned on the recovery plates without the stress (open bars). Data are mean of five replicates. The experiment was repeated two times with similar results.
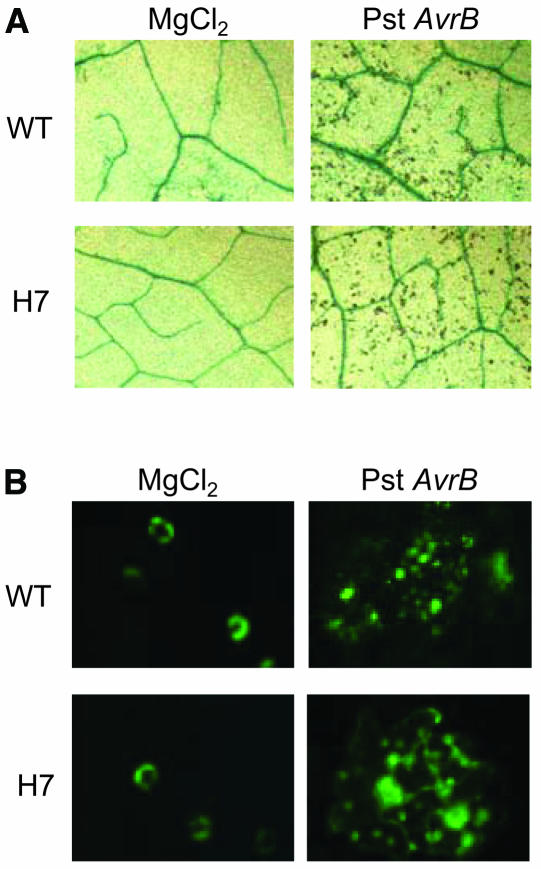
However, the accumulation of NO does not always have negative effects. Initial investigations suggested that plants use NO as a signaling molecule via pathways remarkably similar to those found in mammals (Wendehenne et al., 2001). For example, NO plays a key role in plant defense against pathogens by contributing with reactive oxygen species to trigger hypersensitive cell death and by inducing genes involved in the synthesis of defense products (Delledonne et al., 1998). Infiltration of leaves from Arabidopsis Col-0, which contain the RPM1 resistance gene, with Pseudomonas syringae pv tomato carrying the avrB avirulence gene, leads to rapid accumulation of NO and subsequent activation of the hypersensitive cell death program, causing localized lesion formation at the site of infection first apparent ∼1 d after inoculation (Grant et al., 1995). Infiltration of leaves of AHb1-overexpressing Arabidopsis plants did not affect NO accumulation and did not attenuate hypersensitive cell death (Figure 7), indicating that AHb1 does not interfere with NO bursts originated by acute responses, when NO mediates signaling functions through the resistance response(s).
Figure 7.
Analysis of the Hypersensitive Disease-Resistance Response in Arabidopsis.
(A) Trypan blue staining of dead cells in leaves from wild-type and H7 plants 24 h after infiltration with 108 colony-forming units (cfu) mL−1 avirulent P. syringae. Negative controls were infiltrated with 10 mM MgCl2.
(B) Visualization of NO by fluorescence microscopy using DAF-FM diacetate in epidermis of wild-type and H7 leaves 2 h after infiltration with 108 cfu mL−1 avirulent P. syringae. Negative controls were infiltrated with 10 mM MgCl2. Experiments were repeated three times with similar results.
DISCUSSION
Most if not all plants possess different nonsymbiotic hemoglobins whose expression patterns vary from tissue to tissue as well as in response to different types of stress (Hunt et al., 2001). The physiological role of this class of hemoglobins has remained elusive, although many functions, including roles in oxygen storage and sensing and detoxification of NO and/or other reactive species, have been proposed (Dordas et al., 2003a). The most unusual feature of nonsymbiotic hemoglobins is hexacoordination of the heme group (Hargrove et al., 2000), which differs markedly from the open binding site for exogenous ligands of pentacoordinate hemoglobins. The heme group in nonsymbiotic hemoglobins is coordinated by two His residues in a manner resembling cytochrome b5. Unlike the latter, however, nonsymbiotic hemoglobins bind exogenous ligands rapidly and with high affinity (Trent et al., 2001a; Kundu et al., 2003b). Such hexacoordination has also been observed in truncated hemoglobins from the photosynthetic microorganisms Synechocystis (Hvitved et al., 2001) and Clamydomonas (Couture et al., 1999). The identification of hexacoordinate neuroglobin and histoglobin in humans and other animals (Trent et al., 2001b; Burmester et al., 2002), accompanied with the existence of hexacoordination in Drosophila hemoglobin (Hankeln et al., 2002), suggests that hexacoordinate hemoglobins are common to a diverse group of organisms. The finding that hexacoordinate hemoglobins are upregulated by similar conditions in both plants and animals supports the intriguing hypothesis that these proteins share a common function.
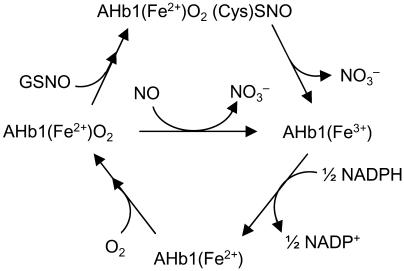
We found that the nonsymbiotic hexacoordinate hemoglobin AHb1 from Arabidopsis metabolizes both NO and GSNO. The nitrate forming reactions involve an Fe(III) intermediate, which is efficiently reduced by NADPH, and the reaction of AHb1 with GSNO involves S-nitrosylation of the protein (Figure 8). Moreover, S-nitrosylated AHb1 is endogenously produced in plants, thus indicating that AHb1 scavenges NO in vivo through production of S-nitrosohemoglobin. This article reports S-nitrosylation of a plant hemoglobin, suggesting a conserved role of hemoglobin S-nitrosothiols in processing NO and NO-donating SNO across humans, nematodes, and plants.
Figure 8.
Proposed Reaction Cycle for NO Metabolism by AHb1.
Oxygenated AHb1 is oxidized by either NO or GSNO with nitrate formation. The recovery of oxygenated AHb1 and its availability for another NO degradation cycle is ensured by regeneration of metAHb1 by NADPH to AHb1 and subsequent association with O2.
Microbial flavohemoglobins are well known to function in NO/SNO detoxification. However, they are dual-domain hemoglobins that use a flavin domain to reduce the ferric heme iron. They bind NO very tightly at the heme site and then react it with oxygen, behaving essentially as denitrosylases (Hausladen et al., 2001). Hexacoordinate AHb1 appears to operate more like Ascaris hemoglobin, which has evolved a different, Cys-dependent mechanism to achieve NO/O2 detoxification. Both AHb1 and Ascaris hemoglobin lack the proximal Cys residues involved in NO-related functions of human hemoglobin (Jia et al., 1996). However, AHb1 contains two Cys residues, E15 and E16 in the distal pocket, one of which is homologous to the distal Cys E15 of Ascaris.
It has been speculated that the ancestral hemoglobin is an enzyme that used redox chemistry to consume NO and resist nitrosative stress (Durner et al., 1999). The evolution of hemoglobin has led to loss of the reductase domain and the incorporation of thiols for NO-related functions. Plant hemoglobins diverged from worm hemoglobins ∼1500 million years ago, although there are little remaining structural similarities. Both, however, evolved Cys residues in the distal pocket that are not present in microbial flavohemoglobins. The function of these Cys residues remains a major unresolved issue. The perienteric hemoglobin of Ascaris is an NO-primed deoxygenase that employs NO to detoxify O2. In this case, NO is transferred oxidatively from the heme to a critical Cys within the heme pocket where it is ready to react with O2. NADPH then supports the transformation of NO/O2 into nitrate (Minning et al., 1999). The evidence reported here suggests that although AHb1 is structurally very different from the massive worm hemoglobin, it has retained the same chemistry, at least in its capacity to metabolize NO and NO-donating SNO. Similar to Ascaris hemoglobin, AHb1 retains the primitive function in NO/SNO detoxification by positioning Cys residues in the distal heme pocket, whereas NO delivery function in mammalian hemoglobin is accomplished through the use of proximal thiols. We therefore suggest that distal Cys residue(s) is a signature for the presence of NO detoxifying/SNO processing activities in hemoglobin. Although these data indicate that AHb1 scavenges NO via S-nitrosylation, future work in which AHb1 Cys are removed via mutagenesis will be necessary to demonstrate this unequivocally.
Systems for NO scavenging have not hitherto been experimentally identified in either plants or in other animal tissues lacking myoglobin. Flavohemoglobins scavenge NO in some bacteria and yeast. However, no examples of a hexacoordinate flavohemoglobin have been found, and no organism appears to posses both hexacoordinate hemoglobin and flavohemoglobin genes (Kundu et al., 2003a). Our data suggest that in those organisms lacking NO-scavenging flavohemoglobins, hexacoordinate hemoglobins have a complementary function. The pattern of regulation of nonsymbiotic hemoglobins by hypoxia and the phenotypic effects of altered expression indicate that one important physiological function for this novel NO scavenging activity is protection against nitrosative stress associated with hypoxia and related stresses. We observed a direct correlation between weight of shoots and AHb1 level in Arabidopsis plants exposed to hypoxic stress. This observation is corroborated by previous reports. Plants overexpressing AHb1 were found to survive to hypoxic treatment that killed wild-type plants, and the increased survival was parallel with increases in the weights of roots and shoots (Hunt et al., 2002). Furthermore, transgenic alfalfa root cultures expressing barley nonsymbiotic hemoglobin maintained growth when placed under hypoxia, whereas wild-type cultures and cultures underexpressing hemoglobin experienced 30 to 70% decline under the same condition (Dordas et al., 2003b). ATP levels and ATP/ADP ratios were significantly reduced during hypoxia in antisense lines, and overexpressing lines had significantly reduced cell breakdown, thus confirming the role of nonsymbiotic hemoglobin in protection during hypoxia, a stress condition generating copious amount of NO (Dordas et al., 2003b). However, NO also accumulates under normal growth conditions as it is produced from nitrite either through light-mediated nonenzymatic conversion by carotenoids or by the action of NADPH nitrate reductases (Klepper, 1990). AHb1 is induced by nitrate (Wang et al., 2000), and AHb1 may well protect against the NO generated in response to nitrogen fertilization (Klepper, 1990).
The effect of NO depends on many complex conditions (Tamir et al., 1993). Unregulated NO accumulation perturbs normal metabolism and irremediably damages plant cells, but its controlled production mediates several important biological actions implicated in plant growth, development, and defense (Beligni and Lamattina, 2001). Therefore, it is of great interest to determine whether the NO scavenging activity of nonsymbiotic hemoglobins contributes to the modulation of the wide array of NO-mediated signal systems. The key features of plant resistance to pathogens have been shown to depend on the production of NO, such as defense gene expression and activation of a hypersensitive reaction in cooperation with reactive oxygen species (Delledonne et al., 2001). Inhibitors of NO synthase or scavengers of NO are able to block the hypersensitive disease resistance response and cause increased disease susceptibility (Delledonne et al., 2003). However, infiltration of leaves of Arabidopsis plants overexpressing AHb1 with an avirulent bacterial pathogen did not reduce NO accumulation, did not attenuate hypersensitive cell death, and did not promote the spreading of chlorosis observed when NO levels were kept low by coinfiltration of either NO synthase inhibitors or NO scavengers (Delledonne et al., 1998). NO is a critical signal in plants required for activation of the immune response. The lack of interference during this pathogenic response and the slow kinetics of the NO detoxification reaction suggest that although AHb1 scavenges NO, it is not involved in the removal of NO bursts originated by acute responses occurring during the hypersensitive response, when NO-mediated signaling functions are needed for the execution of a proper defense reaction.
METHODS
Recombinant AHb1
The complete cDNA sequence of AHb1 (accession number U94998) was obtained by RT-PCR using the following primers: forward 5′-CATATGGAGAGTGAAGGAAAG-3′ and reverse 5′-GGATCCTTAGTTGGAAAGATTCAT-3′. The amplified fragment was cloned in pGEM-T vector (Promega, Madison, WI), sequenced, introduced into the expression vector pET11a (Novagen, Madison, WI), and then expressed in Escherichia coli BL21 (Novagen). AHb1 was purified as described (Trevaskis et al., 1997) except that the crude extract was first passed over a Phenyl Sepharose Fast Flow column (Amersham Pharmacia Biotech, Uppsala) and then on a Q-Sepharose Fast Flow ion-exchange column (Amersham Pharmacia Biotech). SDS-PAGE followed by silver staining of the recombinant protein revealed a single band; AHb1 purity was further confirmed by ratio of absorbance at 412 and 280 nm (data not shown). Spectra were recorded in an SLM UV/VIS Spectrometer Aminco DW2000 (SLM/Aminco, Urbana, IL). Spectroscopic analyses were performed in 10 mM phosphate buffer, pH 7.4. The heme content was determined by the pyridine hemochromagen method (Riggs, 1981).
Reaction of AHb1 with NO
Treatments of AHb1 with NO were performed by adding NOR1 (Calbiochem, San Diego), a synthetic NO donor releasing indicated amounts of NO. Nitrate produced from oxygenated AHb1 was quantified by vanadium(III) reduction of nitrate combined with detection by the acidic Griess reaction (Miranda et al., 2001). AHb1 denatured by boiling was used as negative control. NADPH consumption was followed at 340 nm in 10 mM phosphate buffer, pH 7.4. S-nitrosylated hemoglobin was detected with the biotin-switch method (Jaffrey et al., 2001).
Immunoprecipitation of S-Nitrosylated Proteins
The immunoprecipitation of S-nitrosylated proteins was performed with antibiotin antibody on samples prepared with a slight modification of the biotin-switch method (Jaffrey et al., 2001), which is specific for S-nitrosylated proteins (Foster and Stamler, 2004). In brief, Arabidopsis thaliana leaves were homogenized in MAE buffer (25 mM Hepes, 1 mM EDTA, and 0.2% Triton X-100, pH 7.7) containing complete protease inhibitor cocktail (Sigma, St. Louis). The extract was centrifuged at 4°C for 20 min, and the protein concentration in the supernatant was measured by Bradford assay (Bio-Rad, Hercules, CA). For the biotin switch, 1 mg of protein extracts was incubated with 20 mM methyl-methanthiosulfate and 2.5% SDS at 50°C for 30 min with frequent vortexing to block free Cys. Methyl-methanthiosulfate was then removed by protein precipitation with two volumes of cold acetone, and proteins were resuspended in 0.1 mL of RB buffer (25 mM Hepes, 1 mM EDTA, and 1% SDS, pH 7.7). After the addition of 1 mM HPDP-biotin (Pierce, Rockford, IL) and 1 mM ascorbic acid, the mixture was incubated for 1.5 h at room temperature in the dark with intermittent vortexing.
Biotinylated proteins were purified by immunoprecipitation for 2 h at 4°C with 20 μL antibiotin-IPA (Pierce) previously equilibrated in PBS following the manufacturer's instructions. Beads were washed three times with PBS, resuspended in SDS-PAGE solubilization buffer, incubated 5 min at 95°C, and then centrifuged for 5 min at 20,000g. Supernatant was then loaded in 12% SDS-PAGE and subjected to protein gel blot analysis using an anti-AHb1 antibody. As negative controls, a few samples were immunoprecipitated without being previously subjected to the biotin switch. The system was optimized using purified recombinant AHb1 exposed (positive control) and unexposed (negative control) to GSNO.
Constructs and Transgenic Plants
The complete cDNA sequence of AHb1 was introduced into the binary vector pBI121 (Clontech, Palo Alto, CA) in either sense (pBI-AHb1) or antisense (pMD-antiAHb1) orientation under the control of the 35S promoter of Cauliflower mosaic virus. The two constructs were then mobilized in Agrobacterium tumefaciens GV3101 and used for transformation of Arabidopsis ecotype Col-0 plants by floral dipping (Clough and Bent, 1998). Progeny from self-pollination of the transformants (T1) giving a 3:1 (resistant:susceptible) segregation of the kanamycin marker were selfed, and homozygous T3 plants were analyzed for AHb1 expression. Where indicated, 3-week-old plants were subjected to hypoxic stress by incubation for 24 h in anaerobic jars (Oxoid, Basingstoke, UK) containing Anaerocult A (Merck, Rahway, NJ).
RNA Gel Blot Hybridization
Total RNA was isolated from leaves using total RNA isolation reagent (TRIzol; Invitrogen, Carlsbad, CA). RNA samples (5 μg) were separated by electrophoresis through formaldehyde-agarose gels and blotted on nylon membranes (Hybond-N; Amersham). Arabidopsis cDNA clone AHb1 was labeled using [32P-α]dCTP by random priming (random primer kit; Amersham). Hybridizations and washes were performed as described (Church and Gilbert, 1984). RNA loading and transfer were checked by hybridization with an rRNA probe (Pepper et al., 1994).
Protein Gel Blot Analysis
Immunoblotting was performed using standard protocols (Sambrook and Russell, 2001). Protein samples were subjected to SDS-PAGE on 12% w/v polyacrylamide and transferred onto a polyvinylidene difluoride membrane. The membrane was stained with Ponceau red to check for equivalency in protein loading. Probing and detection of immunocomplexes were performed as described in the SuperSignal West Pico detection system (Pierce). Polyclonal AHb1 antibodies were generated by injecting purified recombinant AHb1 into rabbits at 3-week intervals and were used at 1:1000 dilution.
NO Emission
NO emission from whole leaves was measured by chemiluminescence detection (Rockel et al., 2002). Leaves from three plants were placed in a transparent lid container with 2 liters of air volume. A constant flow of NO-free air of 1.5 liters/min was pulled through the container and subsequently through a chemiluminescence detector CLD 770 AL ppt (Eco-Physics, Munich, Germany) by a vacuum pump connected to an ozone destroyer. Light provided by a 400-W HQi-lamp (Schreder) was adjusted to a quantum flux density of 100 μmol m−2 s−1 photosynthetically active radiation.
Hypoxic Stress Response Assay
Arabidopsis plants were grown in vitro on MS media-agar. Plates with 3-week-old plants were placed in jars designed for growing anaerobic bacteria (Oxoid) containing Anaerocult A (Merck). The jars were then sealed and kept at room temperature in the dark for 24 h. After treatment, plants were transferred to a new plate containing MS media-agar. The recovery plates were incubated in a culture room at 26°C under diffuse light for 2 weeks, after which plant shoot weight was measured to assess plant response to hypoxia.
Hypersensitive Response
Arabidopsis leaves were infiltrated with 6 μL of a bacterial suspension containing 1 × 108 cfu mL−1 avirulent Pseudomonas syringae pv tomato carrying avrB avirulence gene in 10 mM MgCl2. One day after infiltration, dead cells were visualized by Trypan blue staining (Koch and Slusarenko, 1990). The NO burst was revealed 2 h after bacterial challenge by transferring abaxial epidermal peels in a dish containing 3 mL of loading buffer (10 mM Tris/HCl, pH 7.2) supplemented with 20 μM DAF-FM diacetate and maintained in the dark for 10 min. Epidermal sections were washed twice with loading buffer for 20 min. Images were taken using an Olympus IX70 fluorescence inverted microscope (Tokyo) (470 to 490 nm excitation filter, 515-nm long-pass emission filter). Data acquired were analyzed using Image Pro Plus software (Media Cybernetics, Silver Spring, MD).
Acknowledgments
This work was supported by grants from Ministero dell'lstruzione, dell'Università e della Ricerca (Cofin 2002, Area 07). M.D. is supported by the EMBO Young Investigators Program.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Massimo Delledonne (massimo.delledonne@univr.it).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.025379.
References
- Appleby, C.A. (1984). Leghemoglobins and Rhizobium respiration. Annu. Rev. Plant Physiol. 35, 443–478. [Google Scholar]
- Becker, K., Gui, M., and Schirmer, R.H. (1995). Inhibition of human glutathione reductase by S-nitrosoglutathione. Eur. J. Biochem. 234, 472–478. [DOI] [PubMed] [Google Scholar]
- Beligni, M.V., and Lamattina, L. (2001). Nitric oxide in plants: The history is just beginning. Plant Cell Environ. 24, 267–278. [Google Scholar]
- Burmester, T., Ebner, B., Weich, B., and Hankeln, T. (2002). Cytoglobin: A novel globin type ubiquitously expressed in vertebrate tissues. Mol. Biol. Evol. 19, 416–421. [DOI] [PubMed] [Google Scholar]
- Church, G.M., and Gilbert, W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated tranformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Couture, M., Das, T.K., Lee, H.C., Peisach, J., Rousseau, D.L., Wittenberg, B.A., Wittenberg, J.B., and Guertin, M. (1999). Chlamydomonas chloroplast ferrous hemoglobin. Heme pocket structure and reactions with ligands. J. Biol. Chem. 274, 6898–6910. [DOI] [PubMed] [Google Scholar]
- Delledonne, M., Polverari, A., and Murgia, I. (2003). The functions of nitric oxide-mediated signaling and changes in gene expression during the hypersensitive response. Antioxid. Redox Signal. 5, 33–41. [DOI] [PubMed] [Google Scholar]
- Delledonne, M., Xia, Y., Dixon, R.A., and Lamb, C. (1998). Nitric oxide functions as a signal in plant disease resistance. Nature 394, 585–588. [DOI] [PubMed] [Google Scholar]
- Delledonne, M., Zeier, J., Marocco, A., and Lamb, C. (2001). Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc. Natl. Acad. Sci. USA 98, 13454–13459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dordas, C., Hasinoff, B.B., Igamberdiev, A.U., Manac'h, N., Rivoal, J., and Hill, R.D. (2003. b). Expression of a stress-induced hemoglobin affects NO levels produced by alfalfa root cultures under hypoxic stress. Plant J. 35, 763–770. [DOI] [PubMed] [Google Scholar]
- Dordas, C., Hasinoff, B.B., Rivoal, J., and Hill, R.D. (2004). Class-1 hemoglobins, nitrate and NO levels in anoxic maize cell-suspension cultures. Planta 219, 66–72. [DOI] [PubMed] [Google Scholar]
- Dordas, C., Rivoal, J., and Hill, R.D. (2003. a). Plant haemoglobins, nitric oxide and hypoxic stress. Ann. Bot. 91, 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durner, J., Gow, A.J., Stamler, J.S., and Glazebrook, J. (1999). Ancient origins of nitric oxide signaling in biological systems. Proc. Natl. Acad. Sci. USA 96, 14206–14207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flogel, U., Merx, M.W., Godecke, A., Decking, U.K., and Schrader, J. (2001). Myoglobin: A scavenger of bioactive NO. Proc. Natl. Acad. Sci. USA 98, 735–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, M.W., and Stamler, J.S. (2004). New insights into protein S-nitrosylation. Mitochondria as a model system. J. Biol. Chem. 279, 25891–25897. [DOI] [PubMed] [Google Scholar]
- Gardner, P.R., Gardner, A.M., Martin, L.A., and Salzman, A.L. (1998). Nitric oxide dioxygenase: An enzymic function for flavohemoglobin. Proc. Natl. Acad. Sci. USA 95, 10378–10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman, M.D., and Hargrove, M.S. (2001). Quaternary structure of rice nonsymbiotic hemoglobin. J. Biol. Chem. 276, 6834–6839. [DOI] [PubMed] [Google Scholar]
- Gow, A.J., Luchsinger, B.P., Pawloski, J.R., Singel, D.J., and Stamler, J.S. (1999). The oxyhemoglobin reaction of nitric oxide. Proc. Natl. Acad. Sci. USA 96, 9027–9032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, M.R., Godiard, L., Straube, E., Ashfield, T., Lewald, J., Sattler, A., Innes, R.W., and Dangl, J.L. (1995). Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269, 843–846. [DOI] [PubMed] [Google Scholar]
- Gross, S.S., and Lane, P. (1999). Physiological reactions of nitric oxide and hemoglobin: A radical rethink. Proc. Natl. Acad. Sci. USA 96, 9967–9969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankeln, T., Jaenicke, V., Kiger, L., Dewilde, S., Ungerechts, G., Schmidt, M., Urban, J., Marden, M.C., Moens, L., and Burmester, T. (2002). Characterization of Drosophila hemoglobin. Evidence for hemoglobin-mediated respiration in insects. J. Biol. Chem. 277, 29012–29017. [DOI] [PubMed] [Google Scholar]
- Hargrove, M.S., Brucker, E.A., Stec, B., Sarath, G., Arredondo-Peter, R., Klucas, R.V., Olson, J.S., and Phillips, G.N., Jr. (2000). Crystal structure of a nonsymbiotic plant hemoglobin. Structure Fold. Des. 8, 1005–1014. [DOI] [PubMed] [Google Scholar]
- Hausladen, A., Gow, A., and Stamler, J.S. (2001). Flavohemoglobin denitrosylase catalyzes the reaction of a nitroxyl equivalent with molecular oxygen. Proc. Natl. Acad. Sci. USA 98, 10108–10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausladen, A., Gow, A.J., and Stamler, J.S. (1998). Nitrosative stress: Metabolic pathway involving the flavohemoglobin. Proc. Natl. Acad. Sci. USA 95, 14100–14105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, P.W., Klok, E.J., Trevaskis, B., Watts, R.A., Ellis, M.H., Peacock, W.J., and Dennis, E.S. (2002). Increased level of hemoglobin 1 enhances survival of hypoxic stress and promotes early growth in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 99, 17197–17202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, P.W., Watts, R.A., Trevaskis, B., Llewelyn, D.J., Burnell, J., Dennis, E.S., and Peacock, W.J. (2001). Expression and evolution of functionally distinct haemoglobin genes in plants. Plant Mol. Biol. 47, 677–692. [DOI] [PubMed] [Google Scholar]
- Hvitved, A.N., Trent III, J.T., Premer, S.A., and Hargrove, M.S. (2001). Ligand binding and hexacoordination in synechocystis hemoglobin. J. Biol. Chem. 276, 34714–34721. [DOI] [PubMed] [Google Scholar]
- Igamberdiev, A.U., Seregelyes, C., Manac'h, N., and Hill, R.D. (2004). NADH-dependent metabolism of nitric oxide in alfalfa root cultures expressing barley hemoglobin. Planta 219, 95–102. [DOI] [PubMed] [Google Scholar]
- Jaffrey, S.R., Erdjument-Bromage, H., Ferris, C.D., Tempst, P., and Snyder, S.H. (2001). Protein S-nitrosylation: A physiological signal for neuronal nitric oxide. Nat. Cell Biol. 3, 193–197. [DOI] [PubMed] [Google Scholar]
- Jia, L., Bonaventura, C., Bonaventura, J., and Stamler, J.S. (1996). S-nitrosohaemoglobin: A dynamic activity of blood involved in vascular control. Nature 380, 221–226. [DOI] [PubMed] [Google Scholar]
- Joshi, M.S., Ferguson, T.B.J., Han, T.H., Hyduke, D.R., Liao, J.C., Rassaf, T., Bryan, N., Feelisch, M., and Lancaster, J.R.J. (2002). Nitric oxide is consumed, rather than conserved, by reaction with oxyhemoglobin under physiological conditions. Proc. Natl. Acad. Sci. USA 99, 10341–10346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser, W.M., Weiner, H., Kandlbimder, A., Tsai, C.B., Rockel, P., Sonoda, M., and Planchet, E. (2002). Modulation of nitrate reductase: Some new insights, an unusual case and a potentially important side reaction. J. Exp. Bot. 53, 875–882. [DOI] [PubMed] [Google Scholar]
- Klepper, L. (1990). Comparison between nox evolution mechanisms of wild-type and nr1 mutant soybean leaves. Plant Physiol. 93, 26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, E., and Slusarenko, A. (1990). Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu, S., Premer, S.A., Hoy, J.A., Trent III, J.T., and Hargrove, M.S. (2003. b). Direct measurement of equilibrium constants for high-affinity hemoglobins. Biophys. J. 84, 3931–3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu, S., Trent III, J.T., and Hargrove, M.S. (2003. a). Plants, humans and hemoglobins. Trends Plant Sci. 8, 387–393. [DOI] [PubMed] [Google Scholar]
- Liu, L., Zeng, M., and Stamler, J.S. (1999). Hemoglobin induction in mouse macrophages. Proc. Natl. Acad. Sci. USA 96, 6643–6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minning, D.M., Gow, A.J., Bonaventura, J., Braun, R., Dewhirst, M., Goldberg, D.E., and Stamler, J.S. (1999). Ascaris haemoglobin is a nitric oxide-activated ‘deoxygenase’. Nature 401, 497–502. [DOI] [PubMed] [Google Scholar]
- Miranda, K.M., Espey, M.G., and Wink, D.A. (2001). A rapid, simple spectrophotometric method for simultaneous detection of nitrate and nitrite. Nitric Oxide 5, 67–71. [DOI] [PubMed] [Google Scholar]
- Murphy, M.P. (1999). Nitric oxide and cell death. Biochim. Biophys. Acta 1411, 401–414. [DOI] [PubMed] [Google Scholar]
- Pepper, A., Delaney, T., Washburn, T., Poole, D., and Chory, J. (1994). DET1, a negative regulator of light-mediated development and gene expression in Arabidopsis, encodes a novel nuclear-localized protein. Cell 78, 109–116. [DOI] [PubMed] [Google Scholar]
- Poole, R.K., and Hughes, M.N. (2000). New functions for the ancient globin family: Bacterial responses to nitric oxide and nitrosative stress. Mol. Microbiol. 36, 775–783. [DOI] [PubMed] [Google Scholar]
- Riggs, A. (1981). Hemoglobin quantification: The pyridine hemochromogen method. Methods Enzymol. 76, 5–28.7329272 [Google Scholar]
- Rockel, P., Strube, F., Rockel, A., Wildt, J., and Kaiser, W.M. (2002). Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J. Exp. Bot. 53, 103–110. [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D.W. (2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sowa, A.W., Duff, S.M.G., Guy, P.A., and Hill, R.D. (1998). Altering hemoglobin levels changes energy status in maize cells under hypoxia. Proc. Natl. Acad. Sci. USA 95, 10317–10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamir, S., Lewis, R.S., de Rojas Walker, T., Deen, W.M., Wishnok, J.S., and Tannenbaum, S.R. (1993). The influence of delivery rate on the chemistry and biological effects of nitric oxide. Chem. Res. Toxicol. 6, 895–899. [DOI] [PubMed] [Google Scholar]
- Trent III, J.T., Hvitved, A.N., and Hargrove, M.S. (2001. a). A model for ligand binding to hexacoordinate hemoglobins. Biochemistry 40, 6155–6163. [DOI] [PubMed] [Google Scholar]
- Trent III, J.T., Watts, R.A., and Hargrove, M.S. (2001. b). Human neuroglobin, a hexacoordinate hemoglobin that reversibly binds oxygen. J. Biol. Chem. 276, 30106–30110. [DOI] [PubMed] [Google Scholar]
- Trevaskis, B., Watts, R.A., Andersson, C.R., Llewellyn, D.J., Hargrove, M.S., Olson, J.S., Dennis, E.S., and Peacock, W.J. (1997). Two hemoglobin genes in Arabidopsis thaliana: The evolutionary origins of leghemoglobins. Proc. Natl. Acad. Sci. USA 94, 12230–12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R., Guegler, K., LaBrie, S.T., and Crawford, N.M. (2000). Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell 12, 1491–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts, R.A., Hunt, P.W., Hvitved, A.N., Hargrove, M.S., Peacock, W.J., and Dennis, E.S. (2001). A hemoglobin from plants homologous to truncated hemoglobins of microorganisms. Proc. Natl. Acad. Sci. USA 98, 10119–10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendehenne, D., Pugin, A., Klessig, D.F., and Durner, J. (2001). Nitric oxide: Comparative synthesis and signaling in animal and plant cells. Trends Plant Sci. 6, 177–183. [DOI] [PubMed] [Google Scholar]