Abstract
Oncogenic microRNAs (miRs) have emerged as diagnostic biomarkers and novel molecular targets for anti-cancer drug therapies. Real-time quantitative PCR (qPCR) is one of the most powerful techniques for analyzing miRs; however, the use of unsuitable normalizers might bias the results. Tumour heterogeneity makes even more difficult the selection of an adequate endogenous normalizer control. Here, we have evaluated five potential referenced small RNAs (U6, rRNA5s, SNORD44, SNORD24 and hsa-miR-24c-3p) using RedFinder algorisms to perform a stability expression analysis in i) normal colon cells, ii) colon and breast cancer cell lines and iii) cancer stem-like cell subpopulations. We identified SNORD44 as a suitable housekeeping gene for qPCR analysis comparing normal and cancer cells. However, this small nucleolar RNA was not a useful normalizer for cancer stem-like cell subpopulations versus subpopulations without stemness properties. In addition, we show for the first time that hsa-miR-24c-3p is the most stable normalizer for comparing these two subpopulations. Also, we have identified by bioinformatic and qPCR analysis, different miR expression patterns in colon cancer versus non tumour cells using the previously selected suitable normalizers. Our results emphasize the importance of select suitable normalizers to ensure the robustness and reliability of qPCR data for analyzing miR expression.
MicroRNAs (miRs) are involved in the regulation of many physiological processes, including development, apoptosis and cell growth; as well as in pathological processes such as cancerogenesis1. Aberrant expression of miRs has been associated with tumour initiation, progression and patient outcome2,3. Since miRs are stable in tissues and blood plasma4, oncogenic miRs have emerged as diagnostic biomarkers and novel molecular targets for anti-cancer drug therapies5. Real-time quantitative PCR (qPCR) is one of the most powerful techniques for analyzing miR expression because of its sensitivity and specificity6,7,8. qPCR analysis to test miR expression is based in the use of endogenous controls for results normalization, reliability and reproducibility. However, there are several factors, such as RNA quality and purity, manipulability errors, differential stability and heterogeneity of the sample, among others, that may introduce variations in qPCR results. Moreover, for human samples it should be taken in account the endogenous variations of the biological individuals to avoid an erroneous interpretation of the data9. In addition, tumour heterogeneity makes even more difficult the selection of an adequate endogenous control. Housekeeping genes, ribosomal, small nuclear or nucleolar RNAs are frequently used as internal controls. However, according to experimental data, expression levels of these genes may differ in neoplastic and normal tissues10, and these variations may introduce bias to experimental results.
Tumour heterogeneity is characterized, in part, by different cell subpopulations including a small subset of cancer cells that act as initiating tumour cells or cancer stem cells (CSCs)11. This subpopulation maintains self-renewal, promotes cancer growth and it is responsible of drug/treatment resistance, tumour recurrence and metastasis12. Drug resistance and tumour recurrence in CSCs are mainly explained by the overexpression of multidrug resistance membrane proteins and the aldehyde dehydrogenase (ALDH) enzyme among others13. On the other hand, metastasis is one of the most crucial steps in cancer progression and the main cause of cancer-related mortality. Metastatic cancer cells are characterized for undergoing an epithelial-to-mesenchymal transition (EMT), losing their attachment to the epithelial niche and acquiring a mesenchymal phenotype14. ALDH activity has been extensively used for CSCs identification and isolation15. Recently, we have developed a non-aggressive, easy, inexpensive and reproducible methodology to isolate prospectively cancer stem-like cells based on their differential sensibility to trypsin exposure16. Trypsin-sensitive (TS) cancer cell subpopulation shows increased CSC properties when compared with the total population (TP) and/or the trypsin-resistant (TR) subpopulation16. Aberrant miR expression and the implication of these miRs on the biological complexities of CSCs, makes miR determination a powerful tool with great clinical potential, encouraging further studies with this approach. It has been well established that the use of a single or invalidated reference gene is not suitable to obtain reliable qPCR data6,7,10. The most well-known reference genes used in cancer are among others, U6, rRNA5s, SNORD44 and SNORD24; however, their use has resulted in substantial discrepancies10 as we discuss in the next section. The aim of this study was to validate small RNAs as suitable normalizers comparing the expression of selected miRs between several human cancer cell lines, a normal (non-tumour) epithelial human cell line and enriched CSCs subpopulations isolated by ALDH activity and differential trypsinization methodologies. For this purpose, key points to obtaining reliable qPCR data were evaluated by Bestkeeper, NormFinder and comparative ΔCt stability methods. Moreover, we used miRanda-mirSVR, TargetScan 6.0 and Pictar bioinformatic analysis to predict miRs involved in self-renewal pathways and others cancer related pathways differently expressed in cancer cells versus non tumour cells.
Results
Determination of the best normalizer for miR expression in tumour cells and non-tumour cells
We evaluated the expression pattern of the most commonly used small RNAs normalizers in HT-29 and HCT-116 colon cancer cell lines and in the CCD-18Co normal colon established cell line. We selected U6, rRNA5s, SNORD44 and SNORD24. We also used the hsa-miR-24c-3p since in some cases it has shown stability between several cell subpopulations17.
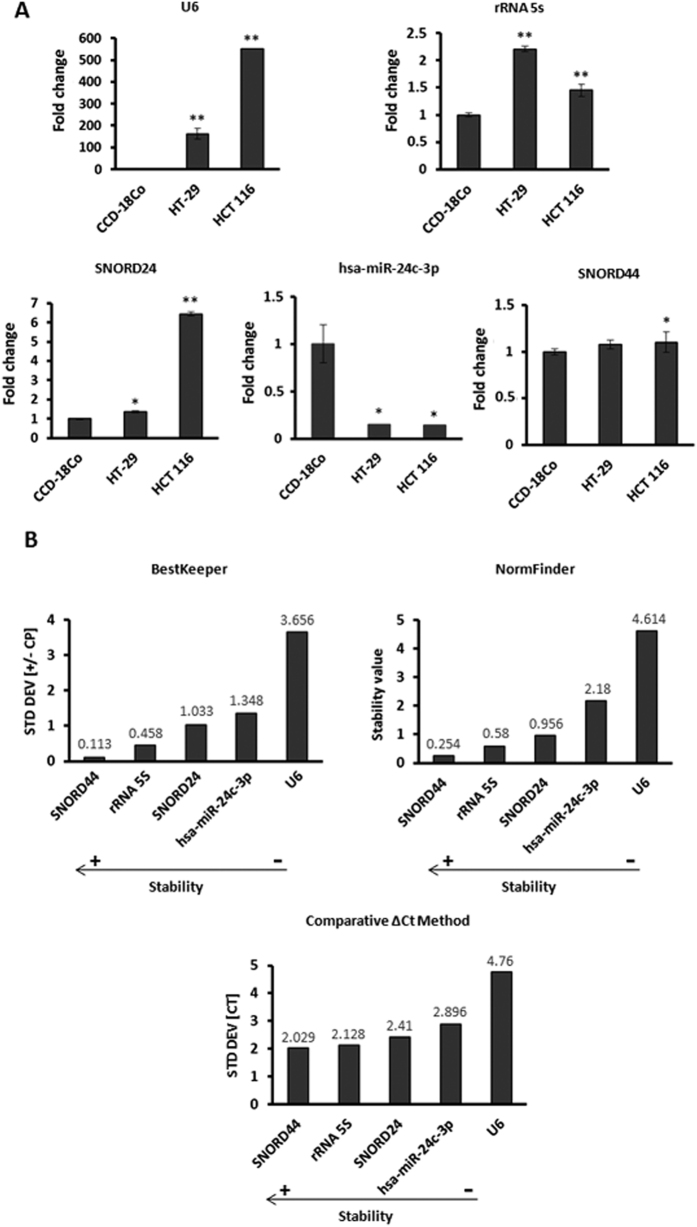
We compared miR expression levels relatively normalized to the artificial UniSp6 RNA spike from C. elegans. As shown in Fig. 1A, U6 was significantly overexpressed up to 500-fold (p < 0.01) in HCT-116 and up to 100-fold (p < 0.01) for HT-29 colon cancer cell lines in comparison with normal colon cell line. The rRNA5s also displayed increased expression levels in both HT-29 and HCT-116 colon cancer cell lines up to 2.3-fold (p < 0.01) and 1.48-fold (p < 0.01), respectively. Moreover, SNORD24 was significantly overexpressed in HT-29 (1.4-fold, p < 0.05) and HCT-116 (6.1-fold, p < 0.01) colon cancer cell lines in comparison with the CCD-18Co normal colon cell line. In contrast, the hsa-miR-24c-5p was significantly downregulated in both HT-29 and HCT-116 respect to the CCD-18Co normal colon cell line. Interestingly, SNORD44 did not show significant differences in colon cancer cell lines in comparison with epithelial colon normal cells (Fig. 1A). Next, we evaluated the expression stability of the endogenous candidate normalizers using the values generated by Bestkeeper, NormFinder and comparative ΔCt methods and comparing normal cells versus cancer cells (Fig. 1B). Whereas U6, rRNA5s, SNORD24 and hsa-miR-24c-3p were not suitable normalizers for colon cell lines, SNORD44 showed high expression stability between tumour and normal colon cell lines (Fig. 1B).
Figure 1. Relative expression and stability of five non-coding RNAs candidate reference genes for miR qPCR analysis in HCT-116 and HT-29 colon cancer cell lines versus CCD-18Co normal colon cell line.
(A) Fold change of U6, rRNA5s, SNORD24. hsa-miR-24c-3p and SNORD44. Values were normalized using the UniSp6 RNA Spike-in control primer set. Data are mean values ± SEM of three independent experiments. Significance was calculated using Student’s t-test. *p < 0.05; **p < 0.01. (B) The stability of U6, rRNA5s, SNORD24, hsa-miR-24c-3p and SNORD44 was determined by BestKeeper, Normfinder and comparative ΔCt method algorithms.
Determination of a suitable endogenous normalizer for cancer stem-like cell subpopulations
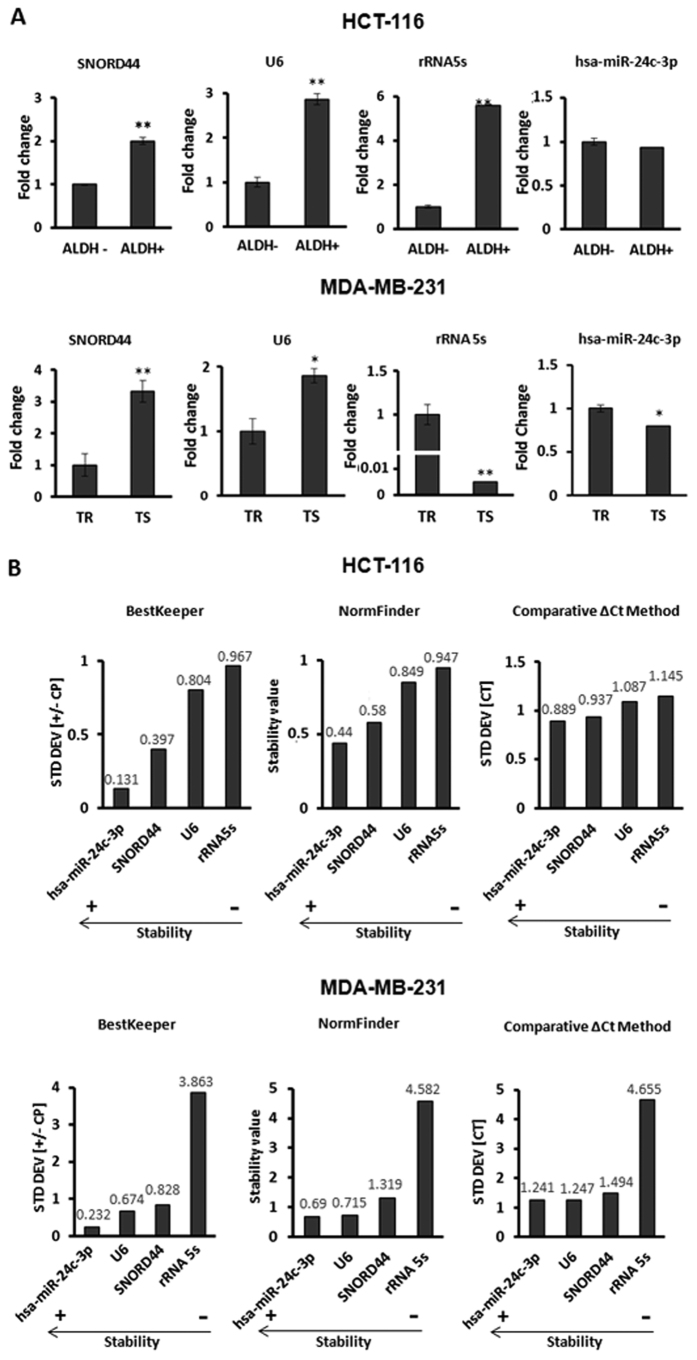
For the selection of small RNAs that could be used as normalizers in qPCR analysis of miR expression for cancer stem-like cell subpopulations, we analyzed two cell subpopulations with stemness properties: i) ALDH+ sorted cells by FACS and ii) low adherent cells isolated by differential trypsinization, as we have previously described in Morata-Tarifa et al. 201616. We compared the expression of selected miRs between those two enriched CSC subpopulations, HCT-116 ALDH+ human colon cancer cells and MDA-MB-231 trypsin sensitive (TS) human breast cancer cells, with their corresponding ALDH− and trypsin resistant (TR) subpopulations with not stemness properties. The artificial UniSp6 RNA spike from C. elegands was used as control. Since SNORD24 showed a weak expression in these cell subpopulations, it was discarded as a potential housekeeping candidate.
Firstly, SNORD44 was analyzed since it was the one showing greater stability between tumour and non tumour cell lines (Fig. 1). However, SNORD44 was significantly over-expressed in the ALDH+ subpopulation in comparison to ALDH− subpopulation obtained from HCT-116 cell line (Fig. 2A). Similarly, SNORD44 also showed a significantly higher expression in the TS subpopulation in comparison to the TR subpopulation selected from MDA-MB-231 cell line (Fig. 2B). Therefore, we decided to analyze other commonly used small RNA normalizers such as U6. However, this small RNA also showed a significantly higher expression in the ALDH+ and TS subpopulations regarding their respective non CSC ALDH− and TR subpopulations (Fig. 2A). As in the two previous cases, rRNA5s significantly varied in the subpopulations analyzed showing higher expression in the ALDH+ subpopulation regarding the ALDH− subpopulation in HCT-116 colon cancer cells (Fig. 2A). However, rRNA5s was downregulated in the TS breast cancer cell subpopulation in comparison to the TR subpopulation (Fig. 2A).
Figure 2. Relative gene expression and stability of the selected non-coding RNAs candidate reference genes for miR qPCR analysis in colon and breast cancer stem-like cell subpopulations versus the respective subpopulations without stemness properties.
(A) Fold changes for SNORD44, U6, rRNA5s and hsa-miR-24c-3p in ALDH + versus ALDH- cells subpopulations from HCT-116 colon cancer cell line and in trypsin sensible (TS) versus trypsin resistant (TR) subpopulations from MDA-MB-231 breast cancer cell line. Values were normalized using the UniSp6 RNA Spike-in control primer set. Data are mean values ± SEM of three independent experiments. Significance was calculated using Student’s t-test. *p < 0.05; **p < 0.01. (B) Stability of SNORD44, U6, rRNA5s and hsa-miR-24c-3p was determined by both BestKeeper and Normfinder computational programs and comparative ΔCt method algorithms in HCT-116 and MDA-MB-231 subpopulations.
Finally, we found that hsa-miR-24c-3p did not show significant differences between ALDH+ CSC subpopulation when compared with ALDH− non CSC subpopulation for colon cancer cells (Fig. 2A). Although a minimal statistical difference was found between TS CSC subpopulation and TR non CSC subpopulation from breast cancer cells (Fig. 2A), hsa-miR-24c-3p showed more stability than SNORD44, U6 and rRNA5s, when were analyzed by Bestkeeper, NormFinder and comparative ΔCt methods, contrasting ALDH+ with ALDH− subpopulations and TS with TR subpopulations, in both colon and breast cancer cells respectively (Fig. 2B).
Expression patterns of miRs in several colon cell lines
Once identified the most stable normalizer for miR expression analysis for each cell line or each CSC subpopulation, we decided to undertake a bioinformatic study for the identification of miRs targeting mRNAs involved in cancer pathways. We used miRanda-mirSVR (http://www.microrna.org/microrna/getGeneForm.do), TargetScan 6.0 (www.targetscan.org) and Pictar (http://pictar.mdc-berlin.de/) to identify predicted miRs targeting mRNAs involved in self-renewal pathways (Notch, Wnt and Hedgehog) and other CSC related pathways as shown in Supplementary Material and Methods section. We focused on 10 miRs identified by at least two of these applications and with better prediction algorithms (Table 1). The selected miR sequences are shown in Supplementary Table 1. These miRs were analyzed in colon cancer cell lines HCT-116 and HT-29 and in the non-tumor cells CCD-18Co (Fig. 3) using SNORD44 as normalizer based on our previously described results. As shown in Fig. 3, tumour cell lines showed a significantly higher expression of miR-142-3p. However, miR-590-5p and miR-15b-5p were overexpressed in HT-29 cells, while in HCT-116 did not display significant changes. In addition, a dramatic decrease in miR-199a-5p and miR-370 expression levels for the two tumour cell lines was observed when compared to the CCD18-Co non-tumour cell line. The analysis of selected miRs in cancer stem-like subpopulation using the hsa-miR-24c-3p normalizer was previously published in Morata-Tarifa C et al. 201616.
Table 1. List of miRs with the best prediction algorithm values.
| Targets | miRs | TargetScan 6.0 (Context + score) | miRanda-mirSVR (mirSVR score) | PicTar (probability) |
|---|---|---|---|---|
| NOTCH1 | hsa-miR-34a | −0.34 | −1.2401 | 0.94 |
| hsa-miR-34c | −0.33 | −1.2426 | 0.94 | |
| NOTCH2 | hsa-miR-15b | −0.24 | ||
| hsa-miR-34a | −0.13 | 0.87 | ||
| hsa-miR-34c | −0.12 | 0.89 | ||
| APH1B | hsa-miR-590 | −0.7131 | ||
| MAML1 | hsa-miR-93 | 0.82 | ||
| RBPJ | hsa-miR-590 | −0.22 | −0.9176 | |
| hsa-miR-15b | −0.9352 | |||
| FZD4 | hsa-miR-199a | −0.34 | −0.4930 | 0.85 |
| FZD5 | hsa-miR-100 | −0.30 | ||
| FZD6 | hsa-miR-199a | −0.43 | −1.0331 | 0.97 |
| FZD8 | hsa-miR-100 | −0.34 | −1.1406 | 0.92 |
| FZD10 | hsa-miR-15b | −0.51 | −1.3160 | 0.95 |
| AXIN | hsa-miR-15b | −0.23 | −0.8583 | 0.98 |
| GSK3B | hsa-miR-199a | −0.20/−0.09 | 0.99 | |
| APC | hsa-miR-142 | −0.40 | −1.2461 | |
| PTCH1 | hsa-miR-15b | −0.24 | −0.5850 | |
| SMO | hsa-miR-370 | −0.33/−0.26 | −0.2491 | |
| CDKN1A | hsa-miR-93 | −0.6463 | 0.93 | |
| ABCB5 | hsa-miR-15b | −1.2476 | ||
| MYC | hsa-miR-34c | 0.99 |
Figure 3. Relative gene expression of bioinformatic selected miRs differentially expressed by qPCR analysis in HCT-116 and HT-29 colon cancer cell lines versus CCD-18Co normal colon cell line.
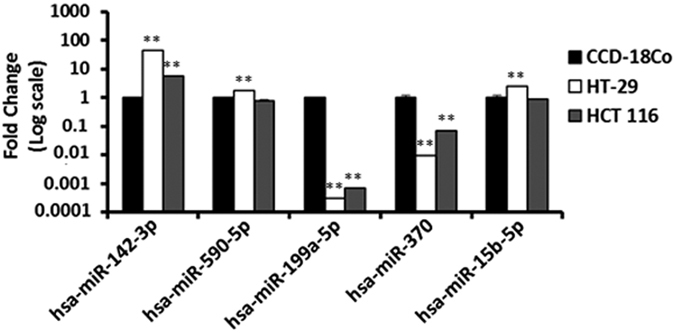
Values were normalized using the previously determined SNORD44 normalizer. Data are mean values ± SEM of three independent experiments. Significance was calculated using Student’s t-test. *p < 0.05; **p < 0.01.
Discussion
Several studies based on miR expression profile have been performed in cell lines, normal and neoplastic tissues or in liquid biopsies to find diagnostic and therapeutic biomarkers for cancer18. Since small changes in the expression of a single miR may affect multiple genes, accurate measurement of miR expression is a critical prerequisite. However, some of the miR expression studies showed differing results suggesting that these discrepancies could be attributed to the different platforms and methods employed and the diversity of samples and control groups. Furthermore, the use of unsuitable reference genes seems to be one of the main reasons for the differences among the results obtained by qPCR studies. This fact highlights the importance of a correct standardization strategy in the qPCR analysis of miR profiles10,19. Currently, the majority of the studies investigating miR expression in cancer have used reference genes without a systematic validation of their stability in every model of cancer analysis. In fact, to the best of our knowledge, this is the first report detailing the identification and validation of suitable reference genes for miR qPCR assays, comparing colon cancer cells with the normal (non-tumour) colon cells. Moreover, we have also analyzed for the first time cancer stem-like cell subpopulations isolated by different methods in comparison with cancer cells without stemness properties. To do so, we performed a bioinformatic stability analysis to determine the most reliable normalizer for accurate miR profile studies in every experimental condition, evaluating a panel of five referenced small RNAs candidates such as U6, rRNA5s, SNORD44, SNORD24 and hsa-miR-24c-3p.
Although U6 and rRNA5s are commonly used in the normalization of miRs in different tumour samples20,21,22; however, in our study both miRs were upregulated in colon cancer cells respect to normal colon cells and in colon and breast cancer stem-like cells in comparison to cells without stemness properties. Therefore, these results suggest that both miRs could not be valid normalizers for studies comparing miR expression levels between non-malignant samples and cancer samples. For example, it has previously been shown that U6 and rRNA5s were overexpressed in serum from breast cancer patients’ respect to healthy women’ serum23. Furthermore, U6 expression levels in liver and breast cancer tissues were higher than those expressed in adjacent normal tissues24. These studies support the notion that the expression and distribution of U6 and rRNA5s exhibits a high degree of variability among several types of human cells.
Although SNORD24 has been recommended as an endogenous reference exhibiting the most stable expression levels between tissues and cells25 however, in our analysis SNORD24 expression was significantly variable. In contrast, SNORD44 expression level was similar in cancer and normal colon cell lines, although it was not suitable as normalizer for cancer stem-like subpopulations. In agreement with our results SNORD44 has proved to be a stable normalizer in cancer and normal endometrial tissues26, although evidences that it is overexpressed in cancer respect to normal tissues have emerged27.
We also evaluated the expression stabilities of referenced genes using Bestkeeper, NormFinder and comparative ΔCt methods algorithms. These analyses showed that SNORD44 was the most stable normalizer for studies with both cancer and normal colon cell lines. However, in qPCR analysis of cancer stem-like subpopulations, SNORD44 was not the most stable normalizer. In fact U6, rRNA5s and SNORD44 were overexpressed in the ALDH+ and TS subpopulations respect to the ALDH− and TR subpopulations. Conversely, hsa-miR-24c-3p was the most stable miR among the several cancer stem cell subpopulations analyzed using both qPCR analysis and the three algorithms described. In fact, it has been previously reported that hsa-miR-24c-3p shows stability in CD44+ and CD44− prostate cancer cell subpopulations17. In addition, it has been validated in bone marrow and peripheral blood samples from patients with neuroblastoma compared to healthy patients28, and used as normalizer for pancreatic ductal tumour assays29. Although hsa-miR-24c-3p was a valid normalizer for comparing cancer stem-like subpopulations, as we previously published16, however it did not show stability in tumour cells compared to normal cells. Therefore, our results encourage the need to set standards miR normalizers considering normal or cancer cell lines, different cell sub-populations or normal or cancer tissue samples.
Once identified the most stable normalizer we performed qPCR determinations of miRs identified through the bioinformatic analysis to validate its potential as candidate oncogenic miRs in cancer. Using SNORD44 as normalizer in both HCT116 and HT-29 colon cancer cell lines, and in CCD-18Co normal colon cell line, we found that miR-142-3p was overexpressed in the colon tumour cells in comparison to normal cell line. miR-142-3p mediates Wnt signalling pathway activation in breast cancer through the inhibition of APC protein30, a protein that is also involved in intercellular adhesion and that is mutated in a high percentage of colorectal tumours31. Furthermore, miR-590-5p and miR-15b-5p were slightly but significantly elevated in the HT-29 cell line respect to non- tumour cells. Whereas it is widely known that miR-15b is elevated in samples from patients with colon cancer32,33, to our knowledge, this is the first implication of miR-590-5p overexpression in colon cancer cells. Moreover, it has been demonstrated that miR-590-5p is overexpressed in renal carcinoma cell lines34, hepatocellular35 and cervical cancer tissue36. On the other hand, we have identified a downregulation of miR-199-5p and miR-370 in colon cancer cells, as it has been showed in other cancer types such as renal cancer, ovarian cancer, hepatocellular carcinoma, bladder cancer, and larynx cancer37,38,39,40. In addition, in endometrial ovarian cancer cells, miR-370 suppressed cell viability and colony formation and increased chemosensitivity41, being also downregulated in chemoresistant breast cancer cells42. The analysis of selected miRs in cancer stem-like subpopulations using the hsa-miR-24c-3p normalizer was previously published in Morata-Tarifa C et al. 201616.
In summary, our results encourage deeper analysis in miR patterns comparing cancer to healthy tissues, emphasizing the importance of a previous selection of suitable normalizers for each cancer, normal or cancer stem-like subpopulations or every tumour model of analysis. The selection of the best housekeeping will ensure the robustness and reliability of the results in precision or personalized medicine for cancer.
Materials and Methods
Cell lines and cell culture
Human colon cancer cell lines HCT-116 (ATCC CCL-247) and HT-29 (ATCC HTB-38), the triple negative human breast cancer cell line MDA-MB-231 (ATCC HTB-26), and the normal human colon cell line CCD-18Co (ATCC CRL1459) were obtained from American Type Culture Collection (ATCC) and cultured according to the ATCC indications.
Differential trypsinization
Selection of cancer cells with CSC-like properties or with a more differentiated phenotype was performed using the differential trypsinization method as we previously described by Morata-Tarifa C et al. 201616. Briefly, CSC-like cells were obtained based on their lower attachment capability to the cell culture surface by incubating cell cultures at 60–80% of confluence with 0.05% trypsin for 2 minutes at 37 °C. More differentiated cancer cells were isolated based on their resistance to detach from the cell culture surface by enzymatic digestion with diluted trypsin at 0.05%. These two different subpopulations, with differential phenotypic and functional properties, were named as Trypsin-Sensitive (TS) and Trypsin-Resistant (TR), respectively; its CSC properties were characterized as indicated by Morata-Tarifa et al. 201616.
Aldefluor assay
Aldehyde dehydrogenase (ALDH) enzyme activity in viable cells was assayed using Aldefluor® kit assay (Stem Cell Technologies, Grenoble, France) according to the manufacturer’s instructions. Analyses were performed in a FACS CANTO II (BD Biosciences) cytometer using the FACS DIVA software.
Bioinformatic miRs prediction
Predictions were realized looking for miRs targeting mRNA of proteins that enhance or inhibit CSC-related pathways (Supplementary information). miRs were selected based on three bioinformatics online tools: TargetScan 6.0., Pictar and miRanda-mirSVR. Predictions were selected based on the most favorable (lowest) context + score, higher probabilities and lowest sum of mirSVR scores, respectively43.
RNA isolation and Quantitaitve Real Time Polimerase Chain Reaction (qPCR)
Total RNA, including miRs, was obtained from the different cell lines and subpopulations using the miRNeasy Mini Kit (Qiagen, Limburgo, Netherland) following manufacturer´s instructions. Reverse transcription from miRs was performed using the miRCURY LNATM Synthesis kit II (Exiqon, Vedbaek, Denmark) following manufacturer’s protocol. qPCR was performed using miRCURY LNA TM EXILENT SYBR Green (Exiqon) in a CFX96 real-time PCR detection system (Bio-Rad). Each reaction was performed in triplicate and the comparative threshold cycle (Ct) method was used to calculate the amplification factor. UniSp6 RNA Spike-in control primer set is used to amplify UniSp6 RNA Spike-in template for the control of the cDNA synthesis step. The standard curve was constructed by 5-fold serial dilutions of cDNA.
Determination of stable normalizer RNAs
The stability of candidate reference miRs was evaluated using RefFinder (see Supplementary Material and Methods information), a user-friendly web-based comprehensive tool for evaluating and screening reference genes from extensive experimental datasets. RefFinder integrates the computational programs Normfinder, BestKeeper and comparative ΔCt method algorithms, to separately and comprehensively compare and rank the tested candidate reference genes with the data obtained for all subpopulations or cell types studied.
Statistical analysis
All graphs present mean ± SEM from ≥ 3 assays. Student’s t-test was used for experiments with two groups. Comparisons of > 2 groups used one way analysis of variance (ANOVA) followed by Dunnett’s or Tukey’s post hoc analysis. Some experiments used two way analysis of variance followed by Tukey´s post hoc tests. P-values < 0.05 were considered statistically significant in all cases.
Additional Information
How to cite this article: Morata-Tarifa, C. et al. Validation of suitable normalizers for miR expression patterns analysis covering tumour heterogeneity. Sci. Rep. 7, 39782; doi: 10.1038/srep39782 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported in part by grants from the Fundación Pública Andaluza Progreso y Salud, Consejería de Salud Junta de Andalucía in collaboration with JANNSSEN CILAG,S.A. (project numbers PI-0533-2014, PI-0441-2014), from the Ministerio de Economía y Competitividad (MINECO, FEDER funds, grant number MAT2015-62644-C2-2-R) from the Ministry of Economy and Competitiveness, Instituto de Salud Carlos III (FEDER funds, project DTS15/00174) and from the Chair “Doctors Galera-Requena in cancer stem cell research”.
Footnotes
Author Contributions C.M.-T. Conception and design, collection and/or assembly of data, data analysis and interpretation; M.P.-R. Collection and/or assembly of data, data analysis and interpretation. C.G.-L. Assembly of data, data analysis and interpretation; H.B. Data analysis and interpretation, manuscript writing; M.P. Data analysis and interpretation, manuscript writing; M.A.G. Conception and design, assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript and financial support. J.A.M. Conception and design, financial support, administrative support, provision of study material, data analysis and interpretation, manuscript writing and final approval of manuscript.
References
- Su Z. et al. MicroRNAs in apoptosis, autophagy and necroptosis. Oncotarget 6, 8474–8490, doi: 10.18632/oncotarget.3523 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B. P., Burge C. B. & Bartel D. P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20, doi: 10.1016/j.cell.2004.12.035 (2004). [DOI] [PubMed] [Google Scholar]
- Volinia S. et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 103, 2257–2261, doi: 10.1073/pnas.0510565103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J. A. et al. The microRNA spectrum in 12 body fluids. Clin Chem 56, 1733–41, doi: 10.1373/clinchem.2010.147405 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiske S., Suetani R. J., Neilsen P. M. & Callen D. F. The oncogenic role of miR-155 in breast cancer. Cancer Epidemiol Biomarkers Prev 21, 1236–43, doi: 10.1158/1055-9965.EPI-12-0173 (2012). [DOI] [PubMed] [Google Scholar]
- Vandesompele J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S. A. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol 29, 23–39, doi: 10.1677/jme.0.0290023 (2002). [DOI] [PubMed] [Google Scholar]
- Bustin S. A., Benes V., Nolan T. & Pfaffl M. W. Quantitative real-time RTPCR–a perspective. J Mol Endocrinol 34, 597–601, doi: 10.1677/jme.1.01755 (2005). [DOI] [PubMed] [Google Scholar]
- Dheda K. et al. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal Biochem 344, 141–143, doi: 10.1016/j.ab.2005.05.022 (2005). [DOI] [PubMed] [Google Scholar]
- Peltier H. J. & Latham G. J. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA 14, 844–852, doi: 10.1261/rna.939908 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Neerven S. M., Tieken M., Vermeulen L. & Bijlsma M. F. Bidirectional interconversion of stem and non-stem cancer cell populations: A reassessment of theoretical models for tumor heterogeneity. Mol Cell Oncol 3, doi: 10.1080/23723556.2015.1098791 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirino V. et al. Cancer stem cells in solid tumors: an overview and new approaches for their isolation and characterization. FASEB J 27, 13–24, doi: 10.1096/fj.12-218222 (2013). [DOI] [PubMed] [Google Scholar]
- Raha D. et al. The cancer stem cell marker aldehyde dehydrogenase is required to maintain a drug-tolerant tumor cell subpopulation. Cancer Res 74, 3579–3590, doi: 10.1158/0008-5472.CAN-13-3456. (2014). [DOI] [PubMed] [Google Scholar]
- Ombrato L. & Malanchi I. The EMT universe: space between cancer cell dissemination and metastasis initiation. Crit Rev Oncog 19, 349–361, doi: 10.1615/CritRevOncog.2014011802. (2014). [DOI] [PubMed] [Google Scholar]
- Ginestier C. et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 1, 555–567, doi: 10.1016/j.stem.2007.08.014 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morata Tarifa C. et al. Low adherent cancer cell subpopulations are enriched in tumorigenic and metastatic epithelial-to-mesenchymal transition-induced cancer stem-like cells. Sci Rep 6, doi: 10.1038/srep18772 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med 17, 211–5, doi: 10.1038/nm.2284 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saumet A. & Lecellier C. H. microRNAs and Personalized Medicine: Evaluating Their Potential as Cancer Biomarkers. Adv Exp Med Biol 888, 5–15, doi: 10.1007/978-3-319-22671-2_2 (2015). [DOI] [PubMed] [Google Scholar]
- Dijkstra J. R., van Kempen L. C., Nagtegaal I. D. & Bustin S. A. Critical appraisal of quantitative PCR results in colorectal cancer research: Can we rely onpublished qPCR results? Mol Oncol 8, 813–818, doi: 10.1016/j.molonc.2013.12.016 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. et al. MiRNA-615-5p Functions as a Tumor Suppressor in Pancreatic Ductal Adenocarcinoma by Targeting AKT2. PLoS One 10, e0119783, doi: 10.1371/journal.pone.0119783 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G. J., Li J. S., Zhou H., Xiao H. X., Li Y. & Zhou T. MicroRNA-106b promotes colorectal cancer cell migration and invasion by directly targeting DL C1. J Exp Clin Cancer Res 34, doi: 10.1186/s13046-015-0189-7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng et al. MiR-106b induces cell radioresistance via the PTEN/PI3K/AKT pathways and p21 in colorectal cancer. J Transl Med 13, 252, doi: 10.1186/s12967-015-0592-z (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appaiah H. N. et al. Persistent upregulation of U6:SNORD44 small RNA ratio in the serum of breast cancer patients. Breast Cancer Res 13, R86, doi: 10.1186/bcr2943 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou G. et al. Differential distribution of U6 (RNU6-1) expression in human carcinoma tissues demonstrates the requirement for caution in the internal control gene selection for microRNA quantification. Int J Mol Med 36, 1400–8, doi: 10.3892/ijmm.2015.2338 (2015). [DOI] [PubMed] [Google Scholar]
- Sauer E., Babion I., Madea B. & Courts C. An evidence based strategy for normalization of quantitative PCR data from miRNA expression analysis in forensic organ tissue identification. Forensic Sci Int Genet 13, 217–23, doi: 10.1016/j.fsigen.2014.08.005 (2014). [DOI] [PubMed] [Google Scholar]
- Torres A., Torres K., Wdowiak P., Paszkowski T. & Maciejewski R. Selection and validation of endogenous controls for microRNA expression studies in endometrioid endometrial cancer tissues. Gynecol Oncol 130, 588–94, doi: 10.1016/j.ygyno.2013.06.026 (2013). [DOI] [PubMed] [Google Scholar]
- Martens-Uzunova E. S. et al. C/D-box snoRNA-derived RNA production is associated with malignant transformation and metastatic progression in prostate cancer. Oncotarget 6, 17430–44, doi: 10.18632/oncotarget.4172 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viprey V. F., Corrias M. V. & Burchill S. A. Identification of reference microRNAs and suitability of archived hemopoietic samples for robust microRNA expression profiling. Anal Biochem 421, 566–72, doi: 10.1016/j.ab.2011.10.022 (2012). [DOI] [PubMed] [Google Scholar]
- Munding J. B. et al. Global microRNA expression profiling of microdissected tissues identifies miR-135b as a novel biomarker for pancreatic ductal adenocarcinoma. Int J Cancer 131, E86–95, doi: 10.1002/ijc.26466.27 (2012). [DOI] [PubMed] [Google Scholar]
- Isobe T. et al. miR-142 regulates the tumorigenicity of human breast cancer stem cells through the canonical WNT signaling pathway. Elife 3, doi: 10.7554/eLife.0197 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig D. O. & Tsikitis V. L. Molecular markers for colon diagnosis, prognosis and targeted therapy. Surg Oncol 111, 96–102, doi: 10.1002/jso.23806 (2015). [DOI] [PubMed] [Google Scholar]
- Li j. et al. Inhibition of miR-15b decreases cell migration and metastasis in colorectal cancer. Tumour Biol 1–9, doi: 10.1007/s13277-015-4396-9 (2016). [DOI] [PubMed] [Google Scholar]
- Giraldez M. D. et al. Circulating microRNAs as biomarkers of colorectal cancer: results from a genome-wide profiling and validation study. Clin Gastroenterol Hepatol 11, 681–8. e3, doi: 10.1016/j.cgh.2012.12.009 (2013). [DOI] [PubMed] [Google Scholar]
- Xiao X., Tang C., Xiao S., Fu C. & Yu P. Enhancement of proliferation and invasion by MicroRNA-590-5p via targeting PBRM1 in clear cell renal carcinoma cells. Oncol Res 20, 537–44, doi: 10.3727/096504013X13775486749335 (2013). [DOI] [PubMed] [Google Scholar]
- Jiang X. et al. MicroRNA-590-5p regulates proliferation and invasion in human hepatocellular carcinoma cells by targeting TGF-β RII. Mol Cells 33, 545–51, doi: 10.1007/s10059-012-2267-4 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y. et al. MicroRNA-590 promotes cervical cancer cell growth and invasion by targeting CHL1. Cell Biochem 115, 847–53, doi: 10.1002/jcb.24726 (2014). [DOI] [PubMed] [Google Scholar]
- Sun Z., Zhang Z., Liu Z., Qiu B., Liu K. & Dong G. MicroRNA-335 inhibits invasion and metastasis of colorectal cancer by targeting ZEB2. Med Oncol 31, 982, doi: 10.1007/s12032-014-0982-8 (2014). [DOI] [PubMed] [Google Scholar]
- Kinose Y., Sawada K., Nakamura K. & Kimura T. The role of microRNAs in ovarian cancer. Biomed Res Int 249393, doi: 10.1155/2014/249393 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. et al. Up-regulation of p21(WAF1/CIP1) by miRNAs and its implications in bladder cancer cells. FEBS Lett 588, 4654–64. doi: 10.1016/j.febslet.2014.10.037 (2014). [DOI] [PubMed] [Google Scholar]
- Yungang W., Xiaoyu L., Pang T., Wenming L. & Pan X. miR-370 targeted FoxM1 functions as a tumor suppressor in laryngeal squamous cell carcinoma (LSCC). Biomed Pharmacother 68, 149–54, doi: 10.1016/j.biopha.2013.08.008 (2014). [DOI] [PubMed] [Google Scholar]
- Chen X. P., Chen Y. G., Lan J. Y. & Shen Z. J. MicroRNA-370 suppresses proliferation and promotes endometrioid ovarian cancer chemosensitivity to cDDP by negatively regulating ENG. Cancer Lett 353, 201–10, doi: 10.1016/j.canlet.2014.07.026 (2014). [DOI] [PubMed] [Google Scholar]
- Lv J. et al. miRNA expression patterns in chemoresistant breast cancer tissues. Biomed Pharmacother 68, 935–42, doi: 10.1016/j.biopha.2014.09.011 (2014). [DOI] [PubMed] [Google Scholar]
- Garcia D. M. et al. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol 18(10), 1139–46, doi: 10.1038/nsmb.2115 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.