Abstract
The rice (Oryza sativa) spotted leaf11 (spl11) mutant was identified from an ethyl methanesulfonate–mutagenized indica cultivar IR68 population and was previously shown to display a spontaneous cell death phenotype and enhanced resistance to rice fungal and bacterial pathogens. Here, we have isolated Spl11 via a map-based cloning strategy. The isolation of the Spl11 gene was facilitated by the identification of three additional spl11 alleles from an IR64 mutant collection. The predicted SPL11 protein contains both a U-box domain and an armadillo (ARM) repeat domain, which were demonstrated in yeast and mammalian systems to be involved in ubiquitination and protein–protein interactions, respectively. Amino acid sequence comparison indicated that the similarity between SPL11 and other plant U-box-ARM proteins is mostly restricted to the U-box and ARM repeat regions. A single base substitution was detected in spl11, which results in a premature stop codon in the SPL11 protein. Expression analysis indicated that Spl11 is induced in both incompatible and compatible rice–blast interactions. In vitro ubiquitination assay indicated that the SPL11 protein possesses E3 ubiquitin ligase activity that is dependent on an intact U-box domain, suggesting a role of the ubiquitination system in the control of plant cell death and defense.
INTRODUCTION
In multicellular organisms, cell death can occur as either a physiological cell death or a nonphysiological cell death (Vaux and Korsmeyer, 1999). Physiological cell death, also coined as programmed cell death (PCD), refers to a process programmed by the organism to kill its own cells in purpose. The most predominant form of PCD in animals is apoptosis, which is morphologically characterized by membrane blebbing, cell volume loss, nuclear condensation, and DNA fragmentation (Kerr et al., 1972). In plants, PCD occurs during both normal development and in response to pathogen infection. Prominent examples of developmentally PCD include the degeneration of cereal aleurone cells, the development of treacheary elements in xylogenesis, leaf senescence, and cell death in plant reproduction (Kuriyama and Fukuda, 2002). In plant–microbe interactions, PCD occurs during both plant hypersensitive response (HR) to avirulent pathogen infection and plant disease susceptibility under virulent pathogen attack (Greenberg, 1997).
HR cell death is characterized by the rapid localized cell death that occurs at the site of infection caused by avirulent pathogens. This response appears to be triggered through the recognition of an avirulent factor by a corresponding resistance (R) protein in the plant. A large number of mutants characterized by misregulated cell death phenotypes mimicking the HR have been identified in maize (Zea mays) (Walbot et al., 1983), Arabidopsis thaliana (Lorrain et al., 2003), barley (Hordeum vulgare) (Wolter et al., 1993), and rice (Oryza sativa) (Yin et al., 2000). The constitutive activation of cell death and defense pathways in some of the mutants suggests that these mutations might define genes involved in the regulation of HR in wild-type plants. These mutants are collectively called lesion mimics based on their spontaneous lesion formation in the absence of pathogen infection. More than a dozen genes controlling lesion mimics have been isolated to date. The proteins encoded by these genes fall into various functional groups, including membrane associated protein (Büschges et al., 1997), ion channel (Balagué et al., 2003), zinc-finger protein (Dietrich et al., 1997), heat stress transcription factor (Yamanouchi et al., 2002), and components involved in the biosynthesis/metabolic pathways of fatty acid/lipids (Kachroo et al., 2001), porphyrin (Hu et al., 1998), and phenolics (Gray et al., 1997). Studies of these lesion mimic mutants have begun to shed light on the control of PCD and its connections to disease resistance in plants. For example, analyses of Arabidopsis double mutants between the lesion stimulating disease mutant lsd1 (Dietrich et al., 1997) and mutants for two positive regulators for R gene function, enhanced disease susceptibility1 (EDS1) and phytoalexin deficient4 (PAD4), have indicated that both EDS1 and PAD4 are required for runaway cell death in the lsd1 mutant (Rusterucci et al., 2001). It was suggested that EDS1 and PAD4, two signaling genes that mediate some but not all R responses in Arabidopsis, regulate a reactive oxygen intermediates/salicylic acid–dependent defense signal amplification loop that is modulated by LSD1 (Rusterucci et al., 2001).
The ubiquitin/proteasome pathway is the major selective protein degradation system in eukaryotes. It is initiated by the formation of a thiol-ester linkage between the ubiquitin molecule and the Cys residue at the active site of the ubiquitin-activating enzyme (E1) in an ATP-dependent manner. The activated ubiquitin is then transferred to the active site of the ubiquitin-conjugating enzyme (E2). Finally, a ubiquitin-ligase (E3) binds E2 and catalyzes the formation of an isopeptide linkage between the activated ubiquitin and the substrate protein. In the last decade, ubiquitination has emerged as one of the key regulatory mechanisms of apoptosis in mammalian systems (Lee and Peter, 2003). In plants, ubiquitination-mediated protein degradation has been shown to play a significant role in multiple cellular processes, such as photomorphogenesis and regulation of hormone signaling (Sullivan et al., 2003). Recent data suggest that ubiquitination may also play an important role in plant defense against pathogens. The identification of two F-box proteins and several RING-type E3 ubiquitin ligases in the regulation of plant defense as well as the finding of a possible SGT1-mediated link between ubiquitination and R gene–mediated resistance have suggested a possible role for ubiquitination in plant disease resistance signaling (Devoto et al., 2003). Nevertheless, direct evidence for the involvement of the ubiquitination/proteolysis pathway in signaling and regulating plant PCD and disease resistance has not been established.
Many lesion mimic mutants have been identified in rice, and some of these mutants display altered early defense signaling or disease resistance (Takahashi et al., 1999; Yin et al., 2000). Disruption of a heat stress transcription factor was found recently to be responsible for the phenotype of the stress-inducible rice lesion mimic mutant spl7 (Yamanouchi et al., 2002). The rice lesion mimic mutation spotted leaf11 (spl11) was identified from an ethyl methanesulfonate–mutagenized indica cultivar IR68 population and was shown to be inherited in a recessive monogenic fashion (Singh et al., 1995). Phenotypic characterization showed that spl11 confers enhanced, nonrace-specific resistance to both Magnaporthe grisea and Xanthomonas oryzae pv oryzae, the pathogens that cause rice blast and bacterial blight diseases, respectively (Yin et al., 2000). In addition, correlation between the lesion development on leaves and the activation of several defense-related genes and enhanced resistance of the spl11 mutant to pathogens was also observed. To understand the molecular basis by which Spl11 suppresses cell death and the relationship between the spontaneous cell death and the activation of defenses in spl11, we have isolated the Spl11 gene by a map-based cloning strategy. The isolation of the Spl11 gene was facilitated by the identification of three additional spl11 alleles from an IR64 mutant collection (Leung et al., 2001). The Spl11 gene encodes a novel protein with both a U-box domain and six armadillo (ARM) repeats. A point mutation was identified in spl11 that resulted in a premature stop codon in the SPL11 protein. We also showed that SPL11 possesses an E3 ubiquitin ligase activity in vitro, and the intact SPL11 U-box domain is essential for this activity, suggesting an involvement of ubiquitination in the control of plant PCD and pathogen defense.
RESULTS
Genetic and Physical Mapping of the Spl11 Locus
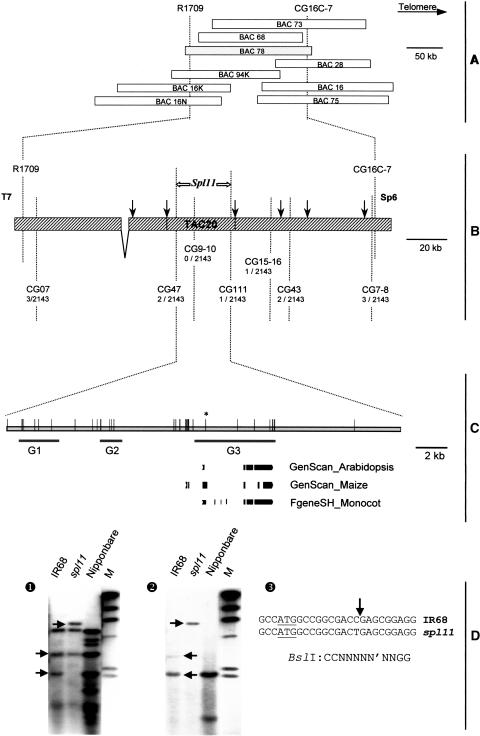
Spl11 was previously physically delimited to a 160-kb DNA segment by markers R1709 and CG16C-7 in the BAC clone BAC78 (Figure 1A) (Zeng et al., 2002). BAC78 was partially sequenced using a shotgun sequencing approach. Assembly of the sequences produced two continuous sequence contigs that cover 95.7% of BAC78 (Figure 1B). To narrow down the Spl11 gene to a smaller region, two new mapping populations were generated by crossing the spl11 mutant with two japonica cultivars, TP309 and Nipponbare. The spl11 mutation was first backcrossed into TP309. Homozygous progenies derived from the cross showing the same lesion mimic phenotype as spl11 (designated as TP309spl11/spl11) were then used as the pollen donor to cross with Nipponbare. Plants from the F2 and F3 generation of the cross were used for subsequent mapping analysis. Seven cleaved amplified polymorphism sequence markers (Weining and Langridge, 1991) were developed from the sequences available in the 160-kb DNA region. Recombination analysis of 297 F2 lesion mimic plants and 1846 F3 individuals indicated that the Spl11 locus was localized within a 27-kb DNA region bracketed by CG47 and CG111 (Figure 1B).
Figure 1.
Physical Delineation of the spl11 Mutation.
(A) Schematic representation of the BAC contig spanning the Spl11 locus. The overlaps between BAC inserts are displayed to scale as open bars. The dotted vertical lines mark the positions of DNA markers. The BAC insert containing the Spl11 locus is highlighted in light gray. Orientation of rice chromosome 12 is indicated in the top right corner.
(B) Fine physical mapping of Spl11 in BAC78. The two cross-hatched gray bars denote the sequenced regions in BAC78. The vertical dotted lines denote the positions of the respective cleaved amplified polymorphism sequence markers. The number of recombinants/number of segregants tested is indicated for each marker. Arrows above the bars mark the NotI cutting sites of the BAC78 insert. T7 and Sp6 indicate the orientation of the insert cloned into the BAC vector pBeloBAC11. The position of the subclone TAC20 insert that contains the Spl11 gene is displayed.
(C) Prediction of potential coding sequences in the 27-kb region of TAC20 where the Spl11 gene was physically delimited. The gray bar depicts the sequenced area. The three solid gray lines designated as G1, G2, and G3 indicate the regions with high coding probability. The vertical lines mark the BslI cutting sites. The asterisk denotes the putative mutation site in spl11. Exons predicted in G3 by the programs GENSCAN and Fgenesh using different matrixes are displayed in dark gray.
(D) RFLP fingerprinting of IR68, spl11, and Nipponbare genomic DNA at the Spl11 locus and detection of a point mutation detected in the spl11 gene. Nineteen restriction enzymes were analyzed but only the results of BslI are shown. (1) Genomic DNA was digested with BslI and then separated on a 1.0% agarose gel. A TAC20 insert digested with HindIII was used as the probe for hybridization. (2) The same blot hybridized with a TAC20 insert in (1) was used. DNA spanning the putative mutated BslI site was amplified from TAC20 and used as the probe. Gray arrows denote the polymorphic bands. M, λ/HindIII DNA marker (New England Biolabs, Beverly, MA). (3) DNA sequence in the vicinity of the spl11 mutation. The C-to-T point mutation in spl11 is denoted by an arrow. This point mutation causes a premature stop codon as marked by the underline. The asterisk marks the start codon for the SPL11 protein.
The gene prediction programs GENSCAN (Burge and Karlin, 1997) and Fgenesh (Solovyev et al., 1995) were then used to identify possible genes in the 27-kb DNA region. Three DNA intervals with high probability of containing a coding region were detected. We designated the putative genes encoded by these DNA intervals G1, G2, and G3 (Figure 1C). BAC78 was subcloned into a modified transformation-competent bacterial artificial chromosome (TAC) vector using NotI as the cloning restriction enzyme (Qu et al., 2003). The subclone that covers the 27-kb region, named TAC20, was then used as a probe to fingerprint spl11 and IR68 genomic DNA by restiction fragment length polymorphism (RFLP) analysis (Figure 1C). Among the 19 restriction enzymes analyzed, TAC20 showed polymorphism between spl11 and IR68 only with restriction enzyme BslI (Figure 1D, 1). Based on these results, we postulated that a mutation is most likely located within the Spl11 gene, which resulted in the absence of a BslI restriction site normally present in wild-type IR68 genomic DNA (Figure 1C). To test this hypothesis, DNA spanning the putative mutated BslI cutting site was then amplified from the TAC20 plasmid DNA and used as a probe to hybridize with the blot previously used in the fingerprinting experiment. The second hybridization gave exactly the same polymorphism pattern between spl11 and IR68 (Figure 1D, 2), indicating that a mutation did occur in the vicinity of the BslI restriction site.
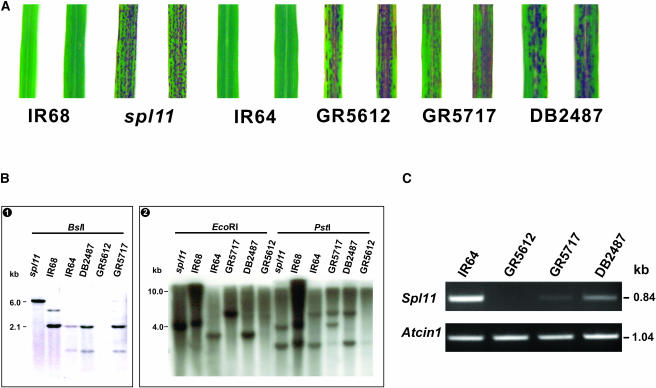
Figure 2.
Analysis of IR64 Lesion Mimic Mutants Allelic to spl11.
(A) Lesion phenotype of spl11 and IR64 background lesion mimic mutants. Picture was taken of leaves from 2-month-old plants.
(B) DNA gel blot analysis of the Spl11 locus in wild-type plants and different mutant lines. (1) Genomic DNA was restricted by BslI and then separated on a 1.0% agarose gel. A 2.5-kb genomic DNA fragment at the 5′ end of the Spl11 gene was used as the probe. (2) Genomic DNA was digested by EcoRI and PstI, respectively. The same probe in (1) was used for the hybridization. Changes detected at the Spl11 locus in the IR64 mutants are described in the text.
(C) Transcript analysis of Spl11 in IR64 lesion mimic mutants by RT-PCR. Spl11-specific primers were used to amplify a 0.84-kb Spl11 cDNA fragment from total RNA. The rice Actin1-specific primers were used in the RT-PCR to quantify the cDNA template. The experiment was repeated three times.
Sequence alignment between the mutated BslI region and the 27-kb DNA stretch indicated that the putative deleted BslI restriction site (designated as ΔBslI) in spl11 is located in the first predicted exon of G3 shortly downstream of the start codon (Figure 1C). A search for ESTs in the 27-kb DNA sequence using the BLAST2 algorithm and available databases identified rice ESTs matching the 3′ end of G3 only, which suggests that G3 was most probably the candidate Spl11 gene.
Identification of spl11 Alleles in an IR64 Mutant Collection
To facilitate the cloning of the Spl11 gene, we searched for lesion mimic mutants with a similar phenotype to that of the spl11 mutant from an IR64 mutant collection (Figure 2A). Three mutants were identified from either diepoxybutane-treated (DB2487) or radiation (γ-ray)-treated (GR5612 and GR5717) IR64 populations (Leung et al., 2001). Allelism tests indicated that the mutations that occurred in all three IR64 lesion mimic mutants are recessive and allelic to spl11 (Table 1). Moreover, alterations at the Spl11 locus were detected in two of the three mutants by RFLP analysis (Figure 2B). PCR analysis was also performed on these mutants using several Spl11-specific primer pairs. A combination of the PCR and DNA gel blot analysis data suggested a 2.5-kb genomic DNA deletion at the 5′ end of the Spl11 locus in mutant GR5612 and a 1.4-kb deletion near the Spl11 start codon in mutant GR5717 (data not shown). However, no visible change at the Spl11 locus was detected in mutant DB2487 when 20 enzymes were used in the RFLP analysis. This could reflect the fact that these enzymes could not detect the small deletion or point mutation in DB2487. Therefore, we evaluated ∼2000 F2 plants from the two reciprocal crosses between spl11 and DB2487. All the F2 plants were lesion mimics, suggesting that the mutation in DB2487 was allelic to the spl11 mutation. The segregation ratio in the F2 generation of the cross between IR64 and DB2487 fitted 3 to 1 (85 wild type to 23 lesion mimic, χ2 = 0.79, P = 0.37), further indicating that the mutation in mutant DB2487 was controlled by a single recessive locus.
Table 1.
Allelism Tests between spl11 and Three IR64 Lesion Mimic Mutantsa
| Cultivar/Mutant Line | IR64 | GR5717 | GR5612 | DB2487 | spl11 |
|---|---|---|---|---|---|
| IR64 | –b | 11c WT/12d | 13 WT/14 | 12 WT/13 | – |
| GR5717 | – | 71 LM/71 | 65 LM/65 | 33 LM/33 | |
| GR5612 | – | 35 LM/35 | 56 LM/56 | ||
| DB2487 | – | 58 LM/58 |
When crossed to IR64, the lesion mimic mutants were used as the female parent. WT, wild type; LM, lesion mimic.
Corresponding cross was not made.
The number of F1 plants with corresponding phenotype.
Total number of F1 plants of the corresponding cross.
To test whether the mutation at the Spl11 locus affected the transcript accumulation of the gene in these mutants, RT-PCR analysis using Spl11-specific primers was performed. As shown in Figure 2C, the expression of the candidate Spl11 gene in mutant GR5612 was completely disrupted, corroborated by DNA gel blot analysis data showing deletion of a 2.5-kb fragment (Figure 2B). There was only a trace expression of Spl11 in mutant GR5717. The expression of Spl11 was reduced in mutant DB2487 compared with that of wild-type IR64. The perfect association between genomic changes at the Spl11 locus and the lesion mimic phenotype of the three IR64 spl11 alleles, along with changes observed in candidate gene expression of the mutants, provided strong evidence that the candidate gene within the G3 fragment encodes Spl11.
Functional Complementation of spl11
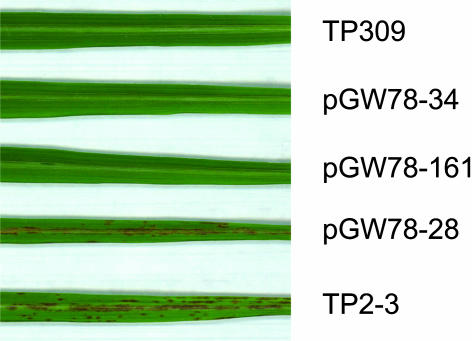
To get final confirmation that the candidate gene encoded within the G3 DNA fragment was Spl11, we made a pCAMBIA1301-derived binary plasmid, pGW78, that contains the full-length Spl11 genomic DNA and a 2.55-kb fragment of the upstream sequence to complement the spl11 mutation. To improve the efficiency of transformation, the spl11 mutation was first introgressed into japonica cultivar TP309, the most commonly used cultivar in rice transformation. The seeds produced by the lesion mimic plant TP309spl11/spl11 were then used for the complementation test. Plasmid pGW78 was transferred into the spl11 mutant via the Agrobacterium tumefaciens–mediated transformation system (Qu et al., 2003). In total, 44 independently transformed transgenic lines were generated, among which 40 lines were successfully complemented. DNA gel blot analysis of the transgenic plants revealed that all the plants carry the mutation originally introgressed from the spl11 plant and zero to two copies of the transgene (data not shown). None of the transgenic plants containing the completely integrated transgene showed any development of lesions within 2 months after the regeneration (Figure 3). These results confirmed that the gene encoded within the G3 DNA interval was responsible for the phenotype of the spl11 mutant.
Figure 3.
Functional Complementation Test of the Spl11 Candidate Gene.
The leaves of 2-month-old plants are shown. Wild-type TP309 and mutant TP2-3 (TP309spl11/spl11) are used as nontransgenic controls. Lines pGW78-34 and pGW78-161 are Spl11 transgenic plants: spl11 mutant TP2-3 (TP309spl11/spl11) with the 8.06-kb XbaI-PacI fragment of the wild-type gene. Line pGW78-28 indicated as an example of failure in transformation.
The Spl11 Gene Encodes a U-Box/ARM Protein with Homology to Drosophila melanogaster ARM
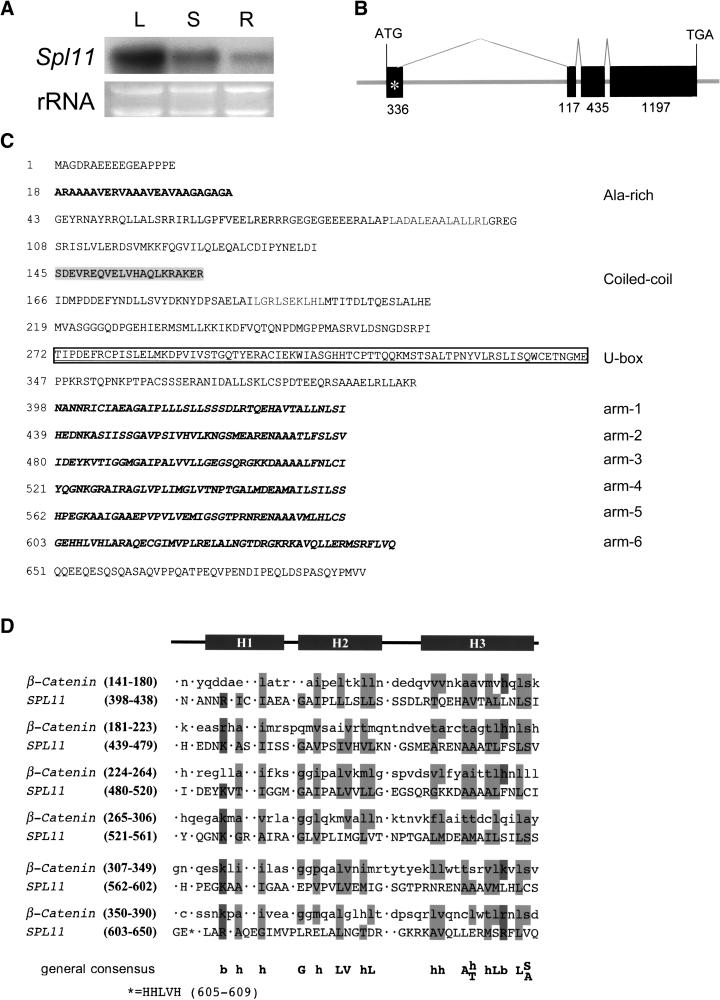
The successful complementation of the spl11 mutation prompted us to obtain the cDNA in the G3 DNA interval. RNA gel blot analysis using G3 genomic DNA as a probe revealed an ∼2.6-kb mRNA in the leaf, stem, and root of IR68 (Figure 4A). Spl11 shows highest expression in the leaf, and lowest in the root. A 2106-bp cDNA fragment that covers the central region and 3′ end of the G3 gene was identified from a Nipponbare leaf cDNA library. The 2106-bp cDNA sequence completely matches the corresponding predicted exons in G3. A modified rapid amplification of cDNA ends (RACE) amplification method using primers derived from the predicted Spl11 gene sequence was then used to obtain the 5′ Spl11 cDNA. A full-length cDNA sequence of 2518 bp for the G3 DNA interval was generated when the RACE result and the cDNA clone sequence were combined (Figure 4B). An open reading frame of 2085 bp starting at position 81 was detected in this full-length cDNA. The deduced protein of the complete open reading frame had 694 amino acids and a molecular mass of 75.3 kD, with a predicted isoelectric point of pH 5.2 (http://us.expasy.org/tools/#primary).
Figure 4.
Transcript Abundance of Spl11 in Different Tissues, Gene Structure, and Deduced Amino Acid Sequence of Spl11.
(A) RNA gel blot analysis of the Spl11 transcript accumulation in different tissues. Total RNA from 3-week-old leaves (L), stems (S), and roots (R) of IR68 were probed with a 0.84-kb cDNA fragment at the 3′ portion of Spl11. Ethidium bromide staining of rRNA was used as a loading control.
(B) Spl11 gene structure. Exons are denoted as black boxes. The number below each exon indicates the length of the exon in base pairs.
(C) Deduced amino acid sequence of Spl11. The N-terminal Ala-rich region is in bold. Amino acids of the coiled-coil domain (145 to 165) are highlighted. The U-box domain (272 to 346) is boxed. Amino acids of the ARM repeat motifs are displayed in italics and boldface.
(D) Sequence alignments of the ARM repeat of β-catenin and SPL11. Numbers of the ranges of amino acids composing each repeat are shown on the left. The repeats are structurally similar, with each repeat containing three helices, H1, H2, and H3, as indicated. The chemically conserved hydrophobic and polar residues are highlighted in dark and light gray, respectively.
A database search with the deduced SPL11 amino acid sequence showed that the 75 amino acids at positions 272 to 346 of SPL11 share high similarity to the consensus U-box domain sequence that was first identified in the yeast protein UFD2 (Koegl et al., 1999) (Figure 4C). The U-box domain contains ∼70 amino acids and is conserved among fungi, plants, and animals (Aravind and Koonin, 2000). It has been shown that the U-box domain is indispensable for E3 ubiquitin ligase activity of several U-box proteins (Jiang et al., 2001). The presence of the U-box domain in the SPL11 protein therefore suggests a probable E3 ubiquitin ligase activity for SPL11.
The database search also showed that the central and C-terminal regions of the SPL11 protein share similarity with the ARM repeats of β-catenin, the vertebrate homolog of Drosophila segment polarity protein ARM (Riggleman et al., 1989). The ARM repeats are tandemly repeated copies of the ARM motif, each containing 38 to 45 amino acid residues (Peifer et al., 1994). Structural characteristics of the ARM motif suggest its involvement in protein–protein interactions, which has been demonstrated in several cases (Huber et al., 1997). In total, six ARM repeat motifs were detected in SPL11 (Figure 4C). Alignment of the ARM repeats in SPL11 with β-catenin repeats 1 to 6 is shown in Figure 4D. Despite the significant variability in sequence among individual motifs, the chemical nature of the residues within each motif is generally conserved. Homologous modeling between the corresponding SPL11 and β-catenin ARM repeat region indicated that their structure matches well (data not shown) (Guex and Peitsch, 1997). This suggests that the ARM repeats of SPL11, like that of β-catenin, might be in physical contact with its interactor(s).
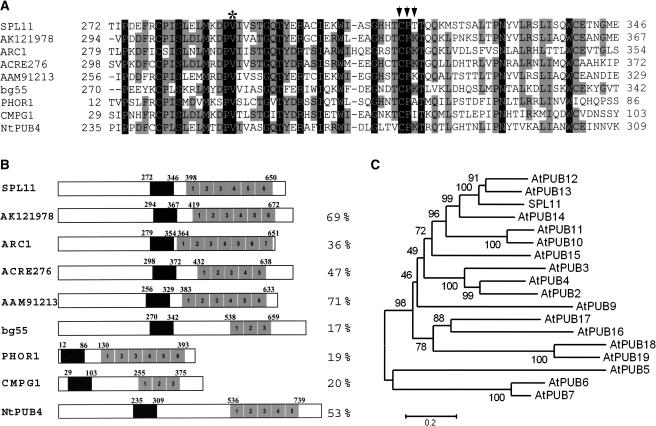
Only two plant proteins bearing both U-box and ARM repeat domains similar to those of SPL11 have been functionally characterized so far. One of them, ARC1, was isolated in a yeast two-hybrid screen for S receptor kinase–interacting proteins and was shown to possess an E3 ubiquitin ligase activity that positively regulates self-incompatibility of Brassica (Stone et al., 2003). The other one is PHOR1, which is a photoperiod-responsive protein involved in gibberellin signaling (Amador et al., 2001). In the Arabidopsis genome, more than 40 U-box-ARM proteins were identified using sophisticated data-mining approaches (Mudgil et al., 2004). In addition to Arabidopsis and rice U-box/ARM repeat proteins homologous to SPL11, BLAST2 algorithm search of the National Center for Biotechnology Information database identified several expressed U-box-ARM proteins from other plant species as well. Two of them, ACRE276 and NtPUB4, were isolated from tobacco (Nicotiana tabacum) and were speculated to be involved in Cf9/Avr9 elicited defense and tobacco development signaling, respectively (Durrant et al., 2000; Kim et al., 2003). The parsley (Petroselinum crispum) CMPG1 responded immediately after pathogen infection and the mangrove (Bruguiera gymnorrhiza) bg55 was induced in high salinity stress (Kirsch et al., 2001; Banzai et al., 2002). SPL11 is related to these proteins in amino acid sequence and overall structure. The sequence similarity between SPL11 and these proteins is mostly restricted to the U-box and ARM repeats. The sequence identity between SPL11 and these proteins in the U-box domain ranges from 75 to 47%, with those amino acid residues key to the U-box function highly conserved (Ohi et al., 2003) (Figure 5A). The overall distribution and position of the ARM repeats in these proteins are shown in Figure 5B. The number of ARM repeats in these proteins is different, varying from 3 to 7. Sequence comparison between the ARM repeats of these proteins and SPL11's ARM repeats indicated a sequence identity ranging from 19 to 71% in the ARM domain (Figure 5B; see Supplemental Figure 1 online). Phylogenetic analysis between Arabidopsis U-box/ARM repeat proteins (Azevedo et al., 2001) and SPL11 indicated that SPL11 is evolutionally most close to AtPUB13 (Figure 5C; see Supplemental Figure 2 online). Compared with other rice U-box/ARM repeat proteins, of which full-length cDNAs are available in the public database, SPL11 is most highly related to the protein deduced from the cDNA AK121978, with overall 57% sequence identity (data not shown). No significant SPL11 homolog was identified in human, animals, and yeast, suggesting SPL11 might be unique to plants.
Figure 5.
Amino Acid Sequence Alignments between SPL11 and U-Box-ARM Proteins from Other Plant Species.
(A) Sequence alignments in the highly conserved U-box domain of SPL11 and those of other U-box-ARM proteins. The numbers on the left or right indicate the amino acid residues. Gaps, which were introduced to maximize alignment, are indicated by dashes. The residues conserved among the compared sequences are boxed in black or light gray based on the degree of conservation. AK121978 from O. sativa (GI:37991601), ARC1 from B. napus (GI:2558938), ACRE276 from N. tabacum (GI:30013679), AAM91213 from A. thaliana (GI:22136270), bg55 from B. gymnorrhiza (GI:14149112), PHOR1 from Solanum tuberosum (GI:13539578), CMPG1 from P. crispum (GI:14582202), and NtPUB4 from N. tabacum (GI:28974687) are shown. Only the one most highly related to SPL11 from rice and Arabidopsis is included in the alignment because of the large number of U-box-ARM proteins present in the rice and Arabidopsis genomes. The asterisk and arrows marked the amino acids that were mutated in the SPL11 E3 ligase activity assay.
(B) Schematic representation of SPL11 and other U-box-ARM proteins of plants. The black box indicates the U-box domain, and the individual ARM repeat of the ARM domain is indicated by a numbered, shaded box. The percentage of sequence identity of the ARM repeats from plant U-box-ARM proteins to their most homologous ARM repeats in SPL11 is indicated. Detail sequence alignment in the ARM domain of SPL11 with those of other U-box-ARM proteins is indicated in Supplemental Figure 1 online.
(C) Phylogenetic relationship between SPL11 and Arabidopsis U-box/ARM repeat proteins. The phylogenies were generated with neighbor joining with 400 bootstrap replicates and were rooted at midpoint. The bootstrap values are shown as percentages. AtPUB8 (locus At4g21350) was not included in the tree because no EST, SAGE tag, or cDNA was identified for the corresponding predicted gene.
Molecular Properties of the Spl11 Gene and Spl11 Analogs in the Rice Genome
To determine the exact mutation site of the Spl11 gene in the spl11 mutant, the genomic DNA fragments that span the ΔBslI restriction site were identified from both spl11 and IR68. DNA sequencing revealed a unique nucleotide substitution of T for C in the spl11 gene, a substitution that eliminates the BslI restriction site originally present in the wild-type IR68 genome (Figure 1D, 3). This point mutation occurs in the first exon of the Spl11 gene, resulting in a premature stop codon in the spl11 transcript.
DNA gel blot analysis of nine rice japonica or indica cultivars indicated that the rice genome contains a single copy of the Spl11 gene (data not shown). Nevertheless, a whole genome scale sequence analysis revealed 83 annotated U-box proteins in rice, among which 32 showed a U-box-ARM overall structure (L.-R. Zeng, unpublished data). A search for SPL11-like proteins in the rice full-length cDNA database KOME (http://cdna01.dna.affrc.go.jp/cDNA/) identified 11 U-box-ARM proteins in addition to a partial cDNA of Spl11 (see Supplemental Table 1 online). The existence of a large number of U-box-ARM proteins in the rice genome suggests their probable involvement in a wide range of cellular processes.
SPL11 Possesses E3 Ubiquitin Ligase Activity in Vitro, and the U-Box Domain Is Essential for the E3 Ligase Activity
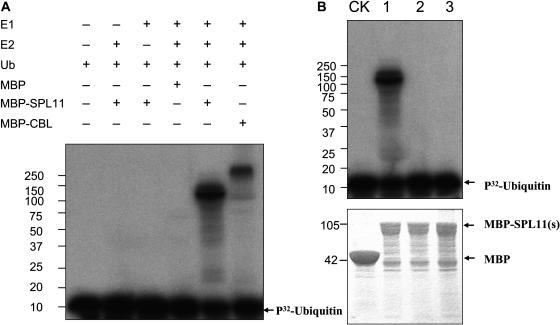
Because one of the important features of U-box containing proteins is to function as E3 ubiquitin ligases (Hatakeyama et al., 2001), we wanted to determine whether SPL11 also possesses E3 ligase activity. SPL11 (residues 112 to 694) was expressed in Escherichia coli as a fusion with maltose binding protein (MBP) and was purified by affinity chromatography. Mouse E3 ubiquitin ligase CBL (GI:38605691) was included in the experiment as a positive control.
In the presence of wheat (Triticum aestivum) E1 and an Arabidopsis E2 AtUBC9, ubiquitination activity was observed for the purified MBP-SPL11 and MBP-CBL proteins (Figure 6A, lanes 5 and 6 from the left, respectively), whereas in the absence of any of the E1, E2, or E3 (lanes 1 to 4 from the left), no signal was detected (Figure 6A). These results indicated that SPL11 possesses E3 ligase activity.
Figure 6.
E3 Ubiquitin Ligase Activity of SPL11.
(A) MBP-SPL11 and MBP-CBL fusion proteins were assayed for E3 activity in the presence of E1 (from wheat, GI:136632), E2 (AtUBC9, GI:20136191), and 32P-labeled ubiquitins. The numbers on the left denote the molecular mass of marker proteins in kilodaltons. Mouse E3 ubiquitin ligase CBL (GI:38605691) was used as a positive control. 32P-ubiquitin is indicated by an arrow. MBP itself was used as a negative control.
(B) E3 ligase activity of SPL11 and its mutants. CK, MBP; lane 1, wild-type SPL11; lane 2, SPL11 (V290R); lane 3, SPL11ΔC314P315T316. 32P-ubiquitin is indicated by arrow. The bottom panel shows two times of the corresponding amount of MBP and MBP fusion proteins used in the E3 activity assay.
It has been shown that the U-box is essential for the E3 ligase activity of U-box proteins (Hatakeyama et al., 2001). To test if an intact U-box domain is required for SPL11 E3 ligase activity as well, two versions of SPL11 bearing mutation in the U-box domain were tested for E3 activity. The first version carried a single mutation that results in a Val290 to Arg290 single amino acid residue change (Figure 5A, asterisk). This Val is highly conserved among different U-box proteins and a Val to Ile point mutation in the yeast protein Prp19p U-box domain leads to pre-mRNA splicing deficiency in vivo (Ohi and Gould, 2002). The second version carried a three–amino acid residue deletion (ΔC314P315T316) in the U-box domain (Figure 5A, arrows). The C314 and P315 also are highly conserved and were reported to be essential for correct folding of the U-box domain to form an appropriate interface interacting with E2 ubiquitin conjugase (Ohi et al., 2003). In vitro ubiquitination analysis indicated that the E3 ligase activity was completely abolished in both versions of SPL11 (Figure 6B), indicating that an intact U-box domain is required for SPL11's E3 ligase activity.
Expression Pattern of Spl11 in Blast-Infected Rice Plants
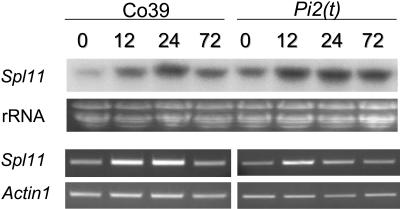
It is assumed that Spl11 is involved in rice defense signaling based on the enhanced resistance of the spl11 mutant to rice pathogen attack (Yin et al., 2000). To investigate the role of Spl11 in defense against rice pathogens, the expression of Spl11 in rice blast-infected resistant [carrying the Pi2(t) R gene] and susceptible [without the Pi2(t) gene] plants was monitored by RNA gel blot and RT-PCR analyses (Liu et al., 2002). RNA was isolated from both resistant and susceptible plants 0, 12, 24, and 72 h after inoculation with isolate PO6-6 avirulent to Pi2(t). Both RNA gel blot hybridization and RT-PCR results indicated that Spl11 was induced at 12 and 24 h after blast inoculation in resistant and susceptible interactions (Figure 7). No difference was detected in the pattern and level of Spl11 expression between the resistant and susceptible plants. Inoculation with PO6-6 on IR68 plants (resistant reaction) confirmed the induction of Spl11 by rice blast at early infection stages (data not shown). These results suggest that Spl11 is not R gene dependent and might be involved in the basal defense signaling against rice blast.
Figure 7.
Expression Patterns of Spl11 in Rice–Blast Interaction.
Total RNA was isolated from infected leaves at the indicated hours after rice blast inoculation. Approximately 10 μg of total RNA was loaded in each lane in RNA gel blotting. 32P-labeled Spl11 cDNA fragment (0.84 kb) was used as the probe in the RNA hybridization. A pair of Spl11-specific primers was used in the RT-PCR analysis. The rRNA gel shows the loading quantification of the RNAs in the RNA gel blot analysis. The amplification of the rice Actin1 gene was used as a control for equal amount of total RNAs in the RT-PCR analysis. The numbers denote the hours after rice blast inoculation.
DISCUSSION
To expand our understanding of cell death in plant disease resistance, we previously characterized nine rice lesion mimic mutants for their resistance to fungal and bacterial pathogens (Yin et al., 2000). The clear cell death phenotype and the intimate association of cell death with defense activation in the spl11 mutant suggested that the activation of a cell death pathway that is coordinated with the induction of defense responses leads to the spl11 phenotype. In plant–pathogen systems, coordination of PCD and defense responses occurs in both incompatible and compatible interactions. The activation of cell death and defense responses in these two types of disease reactions is dissimilar in terms of the time, location, and magnitude of their occurrence as demonstrated in Arabidopsis (Tao et al., 2003; Wu et al., 2003). In spl11, the two processes are difficult to distinguish because of the constitutive activation of both. The induction of Spl11 in both incompatible and compatible rice–blast interactions suggests that it might be involved in basal resistance instead of R gene–mediated race-specific defense. The specific role of Spl11 in fighting against rice blast invasion remains to be elucidated. The enhanced resistance of spl11 to rice blast and bacterial blight diseases supports the argument that the cell death pathway activated in spl11 might be interconnected with the defense pathways to the two rice pathogens.
A link between U-box–mediated ubiquitination and cell death has not been clearly established in plants. Our finding that Spl11 encodes a U-box protein endowed with E3 ubquitin ligase activity and the U-box domain is essential for its E3 activity is significant for our understanding of PCD and disease resistance in plants. The ubiquitination related to the spontaneous cell death phenotype of spl11 is analogous to the wide involvement of ubiquitination in the regulation of apoptosis in mammals (Lee and Peter, 2003). Mechanistically, regulation of apoptosis by ubiquitination always occurs via the ubiquitination of key pro- and anti-apoptotic regulators. In animals, a large number of components forming a complicated signaling network involved in the regulation and execution of apoptosis have been identified in the last decade. The HECT domain protein family and RING-finger domain–containing protein family, including the Skp1-Cdc53/Cullin1-F-box (SCF) multisubunit E3 complexes, have been implicated in targeting these apoptotic regulators for degradation (Wilson et al., 2002; Wing et al., 2002; Miyazaki et al., 2003). Although it is unclear at present how SPL11-mediated ubiquitination is regulated and how it functionally contributes to PCD and defense activation in the spl11 mutant, the indication of E3 activity for SPL11 suggested an involvement of a new family of E3 ubiquitin ligases in plant PCD and defense.
Non-U-box protein–mediated ubiquitination recently has been shown to be associated with plant disease resistance in several cases. For example, several RING-finger–type E3 ubiquitin ligases were induced after elicitor or pathogen treatments (Durrant et al., 2000; Takai et al., 2002). Recently, the plant SGT1 protein, which interacts with a convergence component of multiple R gene–mediated signaling pathways, RAR1, was found to interact with the SCF ubiquitin ligase complex as well as the COP9 signalosome (Azevedo et al., 2002). Despite all these findings, key questions remain to be addressed, such as (1) determining which substrates are targeted by the ubiquitination in plant defenses, and (2) determining when and where (i.e., at what level) ubiquitination operates in regulating the defense reactions. It is possible that one or more substrates targeted by SPL11 are functionally related in PCD and defense signaling pathways. In animals, the inhibitors of apoptosis proteins (IAPs) contain at least one baculoviral IAP repeat (BIR) domain at the N terminus and often a RING domain at the C terminus. The combination of BIR-mediated binding, and hence inactivation of proteins and RING-mediated proteolysis of proteins, has been shown to be central to the role of IAPs in regulating apoptosis (Lee and Peter, 2003). In this regard, it would be interesting to determine whether the ARM repeat domain in SPL11 functions with the U-box domain in a way similar to that between the BIR domain and the RING domain in animal IAPs. The identification and characterization of the substrates in SPL11-mediated ubiquitination will be essential to answer such questions and to obtain an in-depth understanding of the role of the SPL11-mediated ubiquitination in PCD and defense.
The Spl11 mutant phenotype could be caused by the disruption of negative regulation of PCD and defense activation or simply be a reflection of perturbed cellular homeostasis. Although it is difficult to distinguish these two possibilities with certainty, two lines of evidence support the first explanation. First, most of the genes differentially expressed in spl11 in our microarray hybridization experiment of spl11 are related to cell death and defense (L.-R. Zeng, T. Zhu, and G.-L. Wang, unpublished data). By contrast, few genes involved in other well-defined cellular processes could be identified, suggesting that the spl11 mutant, like the Arabidopsis mutant acd11, is not excessively pleiotropic (Brodersen et al., 2002). Second, we have identified several putative spl11 suppressors in the screening for mutants with alleviated spl11 phenotype from a diepoxybutane-treated spl11 population (H. Leung and G.-L. Wang, unpublished data). Multiple genes might suppress the lesion formation because there was a wide range of lesion phenotypes in terms of lesion numbers among these mutants. The identification of spl11 suppressors suggests that the spl11 phenotype is genetically programmed.
Only a few U-box proteins can be identified in the genomes of human, Caenorhabditis elegans, and Drosophila (Azevedo et al., 2001). By contrast, dozens of U-box proteins were identified in Arabidopsis and the rice genome (Mudgil et al., 2004; L.-R. Zeng and G.-L. Wang, unpublished data). Such proteins likely exist widely in other plants as well. The existence of a large number of such proteins in plants suggests that they may play diverse roles in multiple processes. So far, SPL11 represents the first case of a U-box/ARM repeat structure protein related to both PCD and defense. Our results indicated that ubiquitination is involved in PCD and defense in plants. This is consistent with the emerging plant disease defense model suggesting that the signaling components/regulators in the plant PCD and defense pathways need to be inactivated and degraded in a temporally and spatially ordered manner similar to what was observed in animals. This strongly suggests that the ubiquitination pathway could play a significant role in regulating plant PCD and defense as well. Therefore, the cloning and characterization of the Spl11 gene hereby opens a way to dissect the mechanism by which the ubiquitination system contributes to the control of PCD and disease resistance in plants.
METHODS
Plant Growth
The F2 population derived from the cross between TP309spl11/spl11 and Nipponbare used for the mapping analysis was grown in the greenhouse in the winter season at Columbus, OH, at 28 to 18°C day and night temperatures. The F3 population of the same cross was grown in the greenhouse in the summer at Columbus, OH, at 32 to 26°C day and night temperatures. Genomic DNA was prepared from young leaves of plants around 45 d old.
Identification of spl11 Allelic IR64 Mutants
A large deletion mutant bank was established at the International Rice Research Institute from chemical- and irradiation-treated IR64 populations as described (Leung et al., 2001). Among the morphological mutants collected in the bank, more than 30 were lesion mimic mutants (C. Wu and H. Leung, unpublished data). Mutants with phenotypes similar to that of spl11 were then crossed with IR64 and spl11 or were crossed with each other for genetic analysis. Those mutant lines allelic to spl11 were then subjected to molecular analysis.
DNA Gel Blot Analysis
Genomic DNA from young leaves was extracted and purified according to the method described (Dellaporta et al., 1984) with extraction buffer modification. The extraction buffer included 100 mM Tris-HCl buffer, pH 8.0, 25 mM EDTA, pH 8.0, 2% (w/v) sorbitol, 0.25% (w/v) hexadecyl trimethyl ammonium bromide, 0.25% (w/v) polyvinyl polyprolidone, 1% N-lauroyl sarcosine, and 1.4 M sodium chloride. Approximately 2 μg of rice (Oryza sativa) genomic DNA was digested with an appropriate enzyme and fractionated on a 1.0% agarose gel by electrophoresis. For DNA gel blotting analysis, the gel was first soaked in 0.25 M HCl for 10 to 20 min, rinsed with distilled water, and then soaked in 0.4 M NaOH for 10 min. The fractionated DNA was then transferred to a Hybond N+ nylon filter under alkaline conditions (0.4 M NaOH). The prehybridization, hybridization, and washing of the filter were conducted using standard procedures (Sambrook and Russell, 2001).
RNA Gel Blot Analysis
Total RNA was isolated with the RNAwiz RNA isolation reagent (Ambion, Austin, TX) from 3- to 4-week-old rice leaves according to the protocol provided by the manufacturer. Approximately 10 μg of total RNA from each sample was mixed with an equal volume of northernMax gel loading solution (Ambion), heated at 50°C for 30 min, then cooled on ice to denature the RNA. The denatured samples were then separated on a 1.4% agarose gel in 1× BPTE electrophoresis buffer (10 mM Pipes, 30 mM Bis-Tris, and 1 mM EDTA) and blotted to a Hybond-N+ nylon membrane (Amersham Biosciences, Piscataway, NJ) with 20× SSC solution (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate). The prehybridization and hybridization were performed using standard procedures (Sambrook and Russell, 2001). After hybridization, the blot was washed twice in 1× SSC and 0.5% SDS solution at 65°C for 5 min, followed by washing in 0.5× SSC and 0.5% SDS solution at 65°C for 10 min.
RT-PCR Amplification
To detect changes in Spl11 expression in IR64 mutants, 1 μg of total RNA per sample was used to synthesize the first-strand cDNA using the AMV reverse transcriptase system (Promega, Madison, WI). The synthesis was conducted according to the protocol provided by the manufacturer. The 20-μL first-strand cDNA product was diluted to 120 μL final volume with 1× TE buffer. For PCR amplification of the Spl11 cDNA fragment, 1.5 μL of the diluted first strand cDNA was used in a 25-μL reaction volume with the Taq enzyme from New England Biolabs. Spl11-specific primers Uc-3 (5′-GATGCTTGCCTTATTGTCCTCA-3′) and Uc-4 (5′-ACGGATTGATATGCCTGACGAT-3′) were used for the amplification. The reaction mixture was cycled through the following temperature profiles: 94°C for 210 s for one cycle, followed by 94°C for 40 s, 62°C for 40 s, and 72°C for 60 s for 22 cycles, and a final incubation at 72°C for 5 min. For amplification of the rice Actin1 gene, primer pair Actin-F (5′-CGTCTGCGATAATGGAACTGG-3′) and Actin-R (5′-CTGCTGGAATGTGCTGAGAGAT-3′) were used.
For amplification of the 5′ end of the Spl11 cDNA, ∼1.6 μg mRNA was first purified from total RNA using the Oligotex mRNA mini kit (Qiagen, Valencia, CA). Approximately 12.5% DMSO (final concentration) was added to the first-strand cDNA synthesis reaction to break the secondary structures of the RNA. The oligo(dT) primer was replaced by the Spl11-specific primer Uc-3 (5′-GATGCTTGCCTTATTGTCCTCA-3′) in the synthesis reaction. The reaction was performed at 38.5°C for 1 h using the AMV reverse transcriptase system and was then denatured at 95°C for 5 min. The reverse transcription was followed by PCR that was performed with primers RACE1 (5′-CGTCAGGCATATCAATCCGTTCTTT-3′) and URACE3 (5′-CCCCACTATTTACCATTCTGCCACT-3′) using approximately one-tenth of the reverse transcription products. Two percent DMSO and 0.25 M betaine (trimethylglycine) were added to the reaction mixture to overcome the difficulty in the amplification of the high GC content region. The reaction mixture was cycled through the following temperature profiles using the ThermalACE Taq enzyme (Invitrogen, Carlsbad, CA): 98°C for 180 s for one cycle, followed by 98°C for 30 s, 54°C for 40 s, and 72°C for 45 s for 32 cycles, and a final incubation at 72°C for 10 min.
E3 Ubiquitin Ligase Activity Assay
DNA fragments of Spl11 containing sequence for both the U-box domain and the ARM domain (1749 bp) and mouse E3 ubiquitin ligase gene CBL (GI:38605691) were cloned into the pMAL-c2 vector (New England Biolabs) and expressed in Escherichia coli. The fusion proteins were prepared according to the manufacturer's instructions. For the E3 ubiquitin ligase activity assay of the fusion proteins, wheat (Triticum aestivum) E1 (GI:136632) and Arabidopsis thaliana E2 AtUBC9 (GI:20136191) were used for the assay. Both wheat E1 and AtUBC9 were cloned in frame into vector pET28a (Novagen, Madison, WI, now part of EMD Biosciences, San Diego) and expressed in E. coli strain BL21. Protein from the E1- or E2-expressing E. coli was used in the E3 ubiquitin ligase assay in which ∼50 ng of E1, 50 ng of E2, and 1 μg of E3 were added. The two SPL11 mutants that contain mutation in the U-box domain are prepared using the Quickchange site-directed mutatgenesis kit (Stratagene, La Jolla, CA) according to the protocol provided by the manufacturer. The sequence of the primer pair used for the preparation of the Val290 to Arg290 mutant is as follows: M1F (5′-CTTGAGCTGATGAAGGATCCTAGAATAGTGTCTACAGGGCAGACA-3′) and M1R (5′-TGTCTGCCCTGTAGACACTATTCTAGGATCCTTCATCAGCTCAAG-3′). The primers for the preparation of the mutant containing a small deletion (ΔC314P315T316) in the U-box are as follows: M2F (5′-ATAGCATCAGGCCATCATACCACGCAACAGAAGATG-3′) and M2R (5′-CATCTTCTGTTGCGTGGTATGATGGCCTGATGCTAT-3′). The in vitro E3 ligase assays were performed as described (Xie et al., 2002).
Complementation
The BAC78 that contains the Spl11 gene was first subcloned into the modified transformation-competent BAC vector pTAC8 (Qu et al., 2003) using NotI as the restriction enzyme. The insert of different subclones was determined by pulse-field gel eletrophoresis and PCR amplification using primer pairs specific to each NotI-digested fragment of BAC78. The subclone containing the Spl11 gene, TAC20, was then digested with PacI and separated on a 0.8% agarose gel. The 9.4-kb fragment from the PacI-digested TAC20 that contains the Spl11 gene was recovered from the gel and then digested with XbaI to remove the 1.4-kb PacI-XbaI fragment. A 365-bp nos terminator DNA was amplified from vector pBI221 (Clontech, Palo Alto, CA) with primers that contain adapter sequences harboring the PacI and HindIII restriction sites, respectively. The 8.0-kb XbaI-PacI genomic DNA fragment containing the entire Spl11 gene and sufficient cis element (a 2.6-kb DNA fragment upstream of the start codon) was then ligated together with the nos terminator DNA fragment into the binary vector pCAMBIA1301 (CAMBIA, Canberra, Australia). This final binary construct (pGW78) was used for the complementation of the spl11 mutation. The pGW78 was mobilized into Agrobacterium tumefaciens strain LBA4404 (Hoekema et al., 1983) by electroporation and was used to transform spl11 plants (TP309spl11/spl11) (Qu et al., 2003). The phenotype of the T1 transformants was scored under standard plant growth conditions as described above.
Protein Sequence Alignments and Phylogenetic Analysis
All the protein sequence alignments were conducted using the program ClustalX (Thompson et al., 1997). The aligned sequence data was then inputted into the MEGA2 program (Kumar et al., 2001) to construct the phylogenetic tree.
Sequence data for the mRNA and genomic DNA of the Spl11 gene have been deposited with the EMBL/GenBank data libraries under accession numbers AY652589 and AY652590, respectively.
Supplementary Material
Acknowledgments
The authors thank Bo Zhou for the kind gift of the total RNA from rice blast–treated Pi2(t)-resistant and susceptible plants, Maria Bellizzi for technical assistance in the rice transformation, and Bill Hardy, Marietta Ryba-White, and Jean Leach for editorial help. This project was in part supported by the start-up fund from the Ohio Agricultural Research and Development Center and the Ohio State University to G.-L.W. and by the Swiss Agency for Development and Cooperation to H.L. at the International Rice Research Institute. B.H.N. is partially supported by the BK21 program of the Ministry of Education and the Crop Functional Genomics Center of the Ministry of Science and Technology.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Guo-Liang Wang (wang.620@osu.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.025171.
References
- Amador, V., Monte, E., Garcia-Martinez, J.L., and Prat, S. (2001). Gibberellins signal nuclear import of PHOR1, a photoperiod-responsive protein with homology to Drosophila armadillo. Cell 106, 343–354. [DOI] [PubMed] [Google Scholar]
- Aravind, L., and Koonin, E.V. (2000). The U box is a modified RING finger: A common domain in ubiquitination. Curr. Biol. 10, R132–R134. [DOI] [PubMed] [Google Scholar]
- Azevedo, C., Sadanandom, A., Kitagawa, K., Freialdenhoven, A., Shirasu, K., and Schulze-Lefert, P. (2002). The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295, 2073–2076. [DOI] [PubMed] [Google Scholar]
- Azevedo, C., Santos-Rosa, M.J., and Shirasu, K. (2001). The U-box protein family in plants. Trends Plant Sci. 6, 354–358. [DOI] [PubMed] [Google Scholar]
- Balagué, C., Lin, B., Alcon, C., Flottes, G., Malmstrom, S., Kohler, C., Neuhaus, G., Pelletier, G., Gaymard, F., and Roby, D. (2003). HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide-gated channel ion channel family. Plant Cell 15, 365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banzai, T., Hershkovits, G., Katcoff, D.J., Hanagata, N., Dubinsky, Z., and Karube, I. (2002). Identification and characterization of mRNA transcripts differentially expressed in response to high salinity by means of differential display in the mangrove, Bruguiera gymnorrhiza. Plant Sci. 162, 499–505. [Google Scholar]
- Brodersen, P., Petersen, M., Pike, H.M., Olszak, B., Skov, S., Odum, N., Jorgensen, L.B., Brown, R.E., and Mundy, J. (2002). Knockout of Arabidopsis accelerated-cell-death11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev. 16, 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge, C., and Karlin, S. (1997). Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268, 78–94. [DOI] [PubMed] [Google Scholar]
- Büschges, R., et al. (1997). The barley Mlo gene: A novel control element of plant pathogen resistance. Cell 88, 695–705. [DOI] [PubMed] [Google Scholar]
- Dellaporta, S.L., Wood, J., and Hicks, J.B. (1984). Maize DNA miniprep. In Molecular Biology of Plants: A Laboratory Course Manual, M. Russell, ed (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 36–37.
- Devoto, A., Muskett, P.R., and Shirasu, K. (2003). Role of ubiquitination in the regulation of plant defence against pathogens. Curr. Opin. Plant Biol. 6, 307–311. [DOI] [PubMed] [Google Scholar]
- Dietrich, R.A., Richberg, M.H., Schmidt, R., Dean, C., and Dangl, J.L. (1997). A novel zinc finger protein is encoded by the Arabidopsis LSD1 gene and functions as a negative regulator of plant cell death. Cell 88, 685–694. [DOI] [PubMed] [Google Scholar]
- Durrant, W.E., Rowland, O., Piedras, P., Hammond-Kosack, K.E., and Jones, J.D.G. (2000). cDNA-AFLP reveals a striking overlap in race-specific resistance and wound response gene expression profiles. Plant Cell 12, 963–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, J., Close, P.S., Briggs, S.P., and Johal, G.S. (1997). A novel suppressor of cell death in plants encoded by the Lls1 gene of maize. Cell 89, 25–31. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T. (1997). Programmed cell death in plant-pathogen interactions. In Annual Review of Plant Physiology and Plant Molecular Biology, R.L. Jones, C.R. Somerville, and V. Walbot, eds (Palo Alto, CA: Annual Reviews), pp. 525–545. [DOI] [PubMed]
- Guex, N., and Peitsch, M.C. (1997). SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- Hatakeyama, S., Yada, M., Matsumoto, M., Ishida, N., and Nakayama, K.I. (2001). U box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 276, 33111–33120. [DOI] [PubMed] [Google Scholar]
- Hoekema, A., Hirsch, P.R., Hooykaas, P.J.J., and Schilperoort, R.A. (1983). A binary plant vector strategy based on separation of vir and Tregion of the Agrobacterium tumefaciens Ti-plasmid. Nature 303, 179–180. [Google Scholar]
- Hu, G., Yamada, K., Briggs, S.P., and Johal, G.S. (1998). A porphyrin pathway impairment is responsible for the phenotype of a dominant disease lesion mimic mutant of maize. Plant Cell 10, 1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber, A.H., Nelson, W.J., and Weis, W.I. (1997). Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell 90, 871–882. [DOI] [PubMed] [Google Scholar]
- Jiang, J., Ballinger, C.A., Wu, Y., Dai, Q., Cyr, D.M., Hohfeld, J., and Patterson, C. (2001). CHIP is a U-box-dependent E3 ubiquitin ligase: Identification of Hsc70 as a target for ubiquitylation. J. Biol. Chem. 276, 42938–42944. [DOI] [PubMed] [Google Scholar]
- Kachroo, P., Shanklin, J., Shah, J., Whittle, E.J., and Klessig, D.F. (2001). A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc. Natl. Acad. Sci. USA 98, 9448–9453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr, J.F.R., Wyllie, A.H., and Currie, A.R. (1972). Apoptosis: A basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 26, 239–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M., Cho, H.S., Kim do, M., Lee, J.H., and Pai, H.S. (2003). CHRK1, a chitinase-related receptor-like kinase, interacts with NtPUB4, an armadillo repeat protein, in tobacco. Biochim. Biophys. Acta 1651, 50–59. [DOI] [PubMed] [Google Scholar]
- Kirsch, C., Logemann, E., Lippok, B., Schmelzer, E., and Hahlbrock, K. (2001). A highly specific pathogen-responsive promoter element from the immediate-early activated CMPG1 gene in Petroselinum crispum. Plant J. 26, 217–227. [DOI] [PubMed] [Google Scholar]
- Koegl, M., Hoppe, T., Schlenker, S., Ulrich, H.D., Mayer, T.U., and Jentsch, S. (1999). A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96, 635–644. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., Jakobsen, I.B., and Nei, M. (2001). MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics 17, 1244–1245. [DOI] [PubMed] [Google Scholar]
- Kuriyama, H., and Fukuda, H. (2002). Developmental programmed cell death in plants. Curr. Opin. Plant Biol. 5, 568–573. [DOI] [PubMed] [Google Scholar]
- Lee, J.C., and Peter, M.E. (2003). Regulation of apoptosis by ubiquitination. Immunol. Rev. 193, 39–47. [DOI] [PubMed] [Google Scholar]
- Leung, H., et al. (2001). Deletion mutants for functional genomics: Progress in phenotyping, sequence assignment, and database development. In Rice Genetics IV, G.S. Khush, D.S. Brar, and B. Hardy, eds (New Delhi, India: Science Publishers), pp. 239–251.
- Liu, G., Lu, G., Zeng, L., and Wang, G.L. (2002). Two broad-spectrum blast resistance genes, Pi9(t) and Pi2(t), are physically linked on rice chromosome 6. Mol. Genet. Genomics 267, 472–480. [DOI] [PubMed] [Google Scholar]
- Lorrain, S., Vailleau, F., Balague, C., and Roby, D. (2003). Lesion mimic mutants: Keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 8, 263–271. [DOI] [PubMed] [Google Scholar]
- Miyazaki, K., Ozaki, T., Kato, C., Hanamoto, T., Fujita, T., Irino, S., Watanabe, K., Nakagawa, T., and Nakagawara, A. (2003). A novel HECT-type E3 ubiquitin ligase, NEDL2, stabilizes p73 and enhances its transcriptional activity. Biochem. Biophys. Res. Commun. 308, 106–113. [DOI] [PubMed] [Google Scholar]
- Mudgil, Y., Shiu, S.H., Stone, S.L., Salt, J.N., and Goring, D.R. (2004). A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol. 134. [DOI] [PMC free article] [PubMed]
- Ohi, M.D., and Gould, K.L. (2002). Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. RNA 8, 798–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi, M.D., Vander Kooi, C.W., Rosenberg, J.A., Chazin, W.J., and Gould, K.L. (2003). Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat. Struct. Biol. 10, 250–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer, M., Berg, S., and Reynolds, A.B. (1994). A repeating amino acid motif shared by proteins with diverse cellular roles. Cell 76, 789–791. [DOI] [PubMed] [Google Scholar]
- Qu, S., Coaker, G., Francis, D., Zhou, B., and Wang, G.-L. (2003). Development of a new transformation-competent artificial chromosome (TAC) vector and construction of tomato and rice TAC libraries. Mol. Breed. 12, 297–308. [Google Scholar]
- Riggleman, B., Wieschaus, E., and Schedl, P. (1989). Molecular analysis of the armadillo locus: Uniformly distributed transcripts and a protein with novel internal repeats are associated with a Drosophila segment polarity gene. Genes Dev. 3, 96–113. [DOI] [PubMed] [Google Scholar]
- Rusterucci, C., Aviv, D.H., Holt III, B.F., Dangl, J.L., and Parker, J.E. (2001). The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell 13, 2211–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D.W. (2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Singh, K., Multani, D.S., and Khush, G.S. (1995). A new spotted leaf mutant in rice. Rice Genet. Newsl. 12, 192–193. [Google Scholar]
- Solovyev, V.V., Salamov, A.A., and Lawrence, C.B. (1995). Identification of human gene structure using linear discriminant functions and dynamic programming. Proc. Int. Conf. Intell. Syst. Mol. Biol. 3, 367–375. [PubMed] [Google Scholar]
- Stone, S.L., Anderson, E.M., Mullen, R.T., and Goring, D.R. (2003). ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell 15, 885–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, J.A., Shirasu, K., and Deng, X.W. (2003). The diverse roles of ubiquitin and the 26S proteasome in the life of plants. Nat. Rev. Genet. 4, 948–958. [DOI] [PubMed] [Google Scholar]
- Takahashi, A., Kawasaki, T., Henmi, K., Shil, K., Kodama, O., Satoh, H., and Shimamoto, K. (1999). Lesion mimic mutants of rice with alterations in early signaling events of defense. Plant J. 17, 535–545. [DOI] [PubMed] [Google Scholar]
- Takai, R., Matsuda, N., Nakano, A., Hasegawa, K., Akimoto, C., Shibuya, N., and Minami, E. (2002). EL5, a rice N-acetylchitooligosaccharide elicitor-responsive RING-H2 finger protein, is a ubiquitin ligase which functions in vitro in co-operation with an elicitor-responsive ubiquitin-conjugating enzyme, OsUBC5b. Plant J. 30, 447–455. [DOI] [PubMed] [Google Scholar]
- Tao, Y., Xie, Z., Chen, W., Glazebrook, J., Chang, H.S., Han, B., Zhu, T., Zou, G., and Katagiri, F. (2003). Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaux, D.L., and Korsmeyer, S.J. (1999). Cell death in development. Cell 96, 245–254. [DOI] [PubMed] [Google Scholar]
- Walbot, V., Hoisington, D.A., and Neuffer, M.G. (1983). Disease lesion mimic mutations. In Genetic Engineering of Plants, T. Kosuge, C.P. Meredith, and A. Hollaender, eds (New York: Plenum Publishing), pp. 431–442.
- Weining, S., and Langridge, P. (1991). Identification and mapping of polymorphism in cereal based on the polymerase chain reaction. Theor. Appl. Genet. 82, 209–216. [DOI] [PubMed] [Google Scholar]
- Wilson, R., Goyal, L., Ditzel, M., Zachariou, A., Baker, D.A., Agapite, J., Steller, H., and Meier, P. (2002). The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nat. Cell Biol. 4, 445–450. [DOI] [PubMed] [Google Scholar]
- Wing, J.P., Schreader, B.A., Yokokura, T., Wang, Y., Andrews, P.S., Huseinovic, N., Dong, C.K., Ogdahl, J.L., Schwartz, L.M., White, K., and Nambu, J.R. (2002). Drosophila Morgue is an F box/ubiquitin conjugase domain protein important for grim-reaper mediated apoptosis. Nat. Cell Biol. 4, 451–456. [DOI] [PubMed] [Google Scholar]
- Wolter, M., Hollricher, K., Salamini, F., and Schulze-Lefert, P. (1993). The mlo resistance alleles to powdery mildew infection in barley trigger a developmentally controlled defence mimic phenotype. Mol. Gen. Genet. 239, 122–128. [DOI] [PubMed] [Google Scholar]
- Wu, Y., Wood, M.D., Tao, Y., and Katagiri, F. (2003). Direct delivery of bacterial avirulence proteins into resistant Arabidopsis protoplasts leads to hypersensitive cell death. Plant J. 33, 131–137. [DOI] [PubMed] [Google Scholar]
- Xie, Q., Guo, H.S., Dallman, G., Fang, S., Weissman, A.M., and Chua, N.H. (2002). SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419, 167–170. [DOI] [PubMed] [Google Scholar]
- Yamanouchi, U., Yano, M., Lin, H., Ashikari, M., and Yamada, K. (2002). A rice spotted leaf gene, Spl7, encodes a heat stress transcription factor protein. Proc. Natl. Acad. Sci. USA 99, 7530–7535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Z., Chen, J., Zeng, L., Goh, M., Leung, H., Khush, G.S., and Wang, G.L. (2000). Characterizing rice lesion mimic mutants and identifying a mutant with broad-spectrum resistance to rice blast and bacterial blight. Mol. Plant-Microbe Interact. 13, 869–876. [DOI] [PubMed] [Google Scholar]
- Zeng, L., Yin, Z., Chen, J., Leung, H., and Wang, G.L. (2002). Fine genetic mapping and physical delimitation of the lesion mimic gene Spl11 to a 160-kb DNA segment of the rice genome. Mol. Genet. Genomics 268, 253–261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.