Abstract
Tomato (Lycopersicon esculentum) Pto kinase specifically recognizes the Pseudomonas effector proteins AvrPto and AvrPtoB, leading to induction of defense responses and hypersensitive cell death. Structural modeling of Pto combined with site-directed mutagenesis identified a patch of surface-exposed residues required for native regulation of signaling. Mutations in this area resulted in constitutive gain-of-function (CGF) forms of Pto that activated AvrPto-independent cell death via the cognate signaling pathway. The patch overlaps the peptide binding region of the kinase catalytic cleft and is part of a broader region required for interaction with bacterial effectors. We propose that the negative regulatory patch is normally occupied by a peptide that represses Pto signaling. Furthermore, we found that Pto kinase activity was required for Avr-dependent activation but dispensable for signaling by CGF forms of Pto. This suggests that Pto signals by a conformational change rather than phosphorylation of downstream substrates in the defense signaling pathway.
INTRODUCTION
Plants possess innate defense responses against attack by pathogenic organisms, including viruses, microbes, and invertebrates (Hammond-Kosack and Jones, 1997). Specific disease resistance, in which the host responds to a narrow subset of pathogens, is associated with precise recognition events described in genetic terms by the gene-for-gene hypothesis (Flor, 1971). In such cases, plant resistance is inherited as a single dominant or semidominant gene. For the plant resistance (R) gene to be functional the pathogen must carry a complementary, dominant avirulence (Avr) gene. Host susceptibility results if one or the other of the complementary partners is absent.
Ellingboe (1981) first proposed that direct interaction between cognate R and Avr gene products might underlie gene-for-gene resistance. Most R genes encode proteins containing a central conserved nucleotide binding site (NBS) domain (i.e., an NBS typical of Apaf-1, R proteins, and CED4) and C-terminal leucine-rich repeats (LRRs) (Dangl and Jones, 2001). Some data are consistent with a direct binding event between NBS-LRR–containing proteins and Avrs (Jia et al., 2000); however, the weight of evidence supports indirect perception (Dangl and Jones, 2001). Perception of AvrRpm1, AvrB, and AvrRpt2 was coordinated by the RIN4 protein (Mackey et al., 2002), which interacted with both cognate R determinants RPM1 and RPS2 (Axtell and Staskawicz, 2003; Mackey et al., 2002, 2003). Another example is Arabidopsis thaliana PBS1 kinase, which was cleaved by the avirulence determinant AvrPphB (Shao et al., 2003). This event was necessary and possibly sufficient for elicitation of resistance but was upstream of cognate recognition specified by the NBS-LRR protein RPS5. Thus, there is evidence for the involvement of additional proteins in the specific recognition of avirulence determinants. However, it is likely that NBS-LRR proteins play a key proximal role in recognition because they act as specificity determinants particularly through their LRR domains (Rathjen and Moffett, 2003). The role of the NBS-LRR protein is sometimes described in terms of the guard hypothesis, in which each R protein is proposed to recognize a putative complex between the avirulence protein and its intracellular target to initiate downstream signaling (Dangl and Jones, 2001). Downstream events associated with R signaling appear to be similar for all pathways and frequently include a cell death phenotype known as the hypersensitive response (HR) (Hammond-Kosack and Jones, 1996).
The Pto gene of tomato conditions race-specific resistance to Pseudomonas syringae pathovar tomato strains carrying avrPto or avrPtoB (Pedley and Martin, 2003). Pto is a Ser-Thr protein kinase with no apparent receptor domain (Martin et al., 1993). Pto appears to interact directly with AvrPto and AvrPtoB, presumably after their delivery into host cells via the type III protein secretion system (Pedley and Martin, 2003). Pto binds to AvrPto and AvrPtoB in yeast two-hybrid assays, and mutations in either Pto or avrPto that disrupt disease resistance in planta also abolish the interaction in yeast. However, no physical interaction in planta between Pto and either Avr has been reported. Downstream signaling by Pto requires the Prf gene, which encodes a large NBS-LRR protein (Salmeron et al., 1996). The functional role of Prf is unknown, although it acts coincident with or downstream of Pto in the signal transduction pathway (Rathjen et al., 1999). Further potential effectors of Pto signaling are PtiI, which encodes another protein kinase, and Pti 4/5/6, a family of transcription factor–like genes (Zhou et al., 1995, 1997). Both PtiI and Pti4 are substrates of Pto in vitro (Gu et al., 2000), although it is unknown whether these proteins interact with Pto in vivo.
Protein kinases are frequent points of control in diverse signaling pathways, and their structure and enzymology are well understood (Huse and Kuriyan, 2002). There are four kinase substructures that control ATP binding and orientation, binding of the peptide substrate, and catalytic phosphotransfer. These are folded within the bilobal kinase domain in precise molecular orientation to compose the functional enzyme. ATP binds to the smaller N-terminal lobe, whereas the substrate binding is controlled by the C-terminal lobe, with the catalytic loop resident in the interdomain cleft. Kinase activity can be repressed in various ways by altering the critical alignment of the key catalytic substructures or by preventing access of substrate molecules to their respective binding sites. Frequently, negative regulation is reversed by a phosphorylation event(s) in the activation segment, a loop region that lies within the catalytic cleft. The activation segment is composed of two recognizable regions: the T-loop, in which regulatory phosphorylation often occurs, and the P+1 loop, which forms the primary binding site for the substrate peptide. Regulatory phosphorylation causes the activation segment to flip out of the active site and assume a characteristic conformation, which may cause subdomain realignment and/or release of steric hindrance. The activated enzyme is then poised for catalysis.
Molecular analyses of the Pto protein showed that P+1 loop residues, rather than the activation segment per se, are important for the control of signal transduction. Firstly, Pto expressed and purified from Escherichia coli is active on several substrates (Sessa et al., 1998; Gu et al., 2000), which appears to exclude subdomain misalignment as an intrinsic mechanism of control. Secondly, structure–function studies revealed four residues within the P+1 loop necessary for AvrPto binding (Scofield et al., 1996); one of these (Thr-204) was sufficient to confer AvrPto binding to the Pto homolog Fen (Frederick et al., 1998). Scanning mutagenesis within the Pto T-loop region did not affect AvrPto binding, whereas mutation of the P+1 loop residues Thr-204 and Tyr-207 deleted the AvrPto interaction. Strikingly, Asp substitutions of Thr-204 and Tyr-207 conferred a CGF phenotype of AvrPto-independent HR to Pto (Rathjen et al., 1999). A functional Prf gene was required for this phenotype, indicating activation of the cognate signal transduction pathway. Therefore, residues required for AvrPto binding are also responsible for correct regulation of signaling activity.
In this study, a combination of site-directed mutagenesis and structural modeling revealed the involvement of most P+1 loop residues in Pto regulation. P+1 loop residues are part of a broader surface-exposed patch that mediates negative regulation of Pto signaling. This patch appears to be a shared docking site for both an unknown negative regulatory molecule and the specific Pto ligands AvrPto and AvrPtoB. The phosphorylation capability of Pto was dispensable for induction of the HR by Pto CGF mutants. Therefore, Pto does not phosphorylate the proximal protein in signal transduction and may act instead as an adaptor protein.
RESULTS
Mutations throughout the Pto P+1 Loop Confer CGF Activity
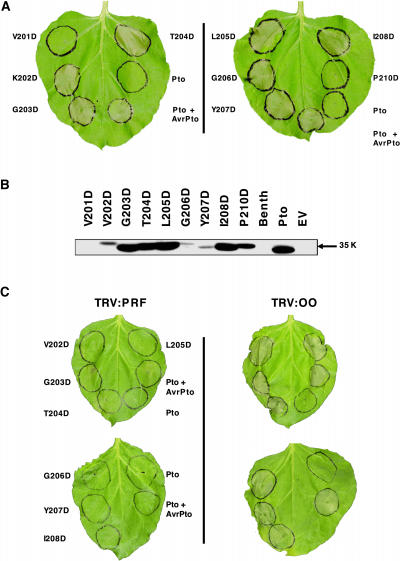
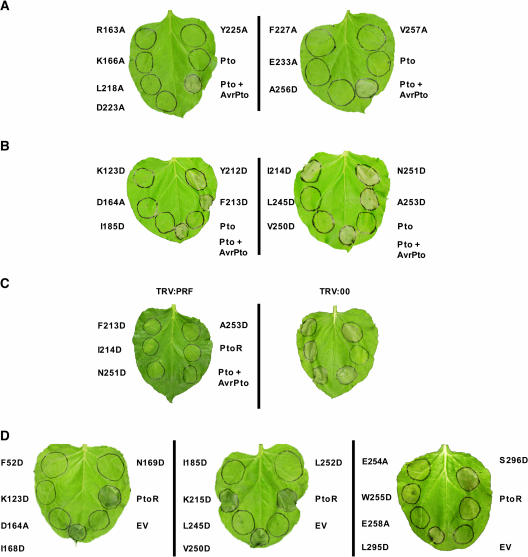
To explore if all P+1 loop residues are required for normal regulation of Pto, the amino acids between 201 and 210 (Rathjen et al., 1999; Moore et al., 2003) were mutated individually to Asp. Mutant genes were expressed transiently in Nicotiana benthamiana leaf tissue using Agrobacterium tumefaciens. Coexpression of wild-type Pto with avrPto resulted in the HR within 4 d, indicative of pathway activation, as did individual expression of ptoT204D or ptoY207D (Figure 1A). Transient expression of the mutants ptoV202D, ptoG203D, ptoL205D, ptoG206D, or ptoI208D also induced the HR, consistent with a role for these residues in control of Pto signaling. By contrast, expression of ptoV201D or ptoP210D, or wild-type Pto in the absence of avrPto, failed to elicit cell death. The requirement for Val-201 is unclear because the mutant protein did not accumulate (Figure 1B), whereas protein was detected from all other constructs using Pto-specific antisera. These data are consistent with involvement of most P+1 loop residues in control of Pto signaling with the further exception of Pro-210, which is the C-terminal residue in the sequence. Similarly, expression of ptoT199D (Rathjen et al., 1999) or ptoV200D (data not shown) did not induce the HR. Both of these residues are amino-proximal to the P+1 loop. Therefore, we conclude that the Pto P+1 loop is a negative regulator of Pto signaling leading to the HR.
Figure 1.
Acidic Mutations throughout the Pto P+1 Loop Confer CGF Activity.
(A) Expression of Pto P+1 loop mutants in N. benthamiana leaves. Leaves were infiltrated with A. tumefaciens containing each construct as indicated and the area of infiltration outlined with a pen. Leaves are shown 4 d after infiltration.
(B) Accumulation of mutant proteins in planta. Total protein was extracted from leaves and blotted against polyclonal Pto antisera. Equivalent loadings were confirmed by Coomassie blue staining of the membrane (data not shown). Benth, noninfiltrated leaf tissue; EV, empty vector control.
(C) Prf is required for cell death induced by P+1 loop mutants. Left, N. benthamiana leaves silenced for the Prf gene; right, leaves silenced with the empty VIGS vector. Top leaves were infiltrated with each Agrobacterium culture as indicated 4 weeks after inoculation with the tobacco rattle virus (TRV) silencing vector. Leaves were infiltrated identically on left and right.
We asked whether the cell death induced by P+1 loop mutants was attributable to specific elicitation of the cognate signal transduction pathway or to activation of a novel stress response. Signaling by Pto is dependent on the Prf gene, and virus-induced gene silencing (VIGS) of an N. benthamiana Prf homolog specifically compromises disease resistance to P. syringae pv tabaci expressing avrPto (Lu et al., 2003). Transient expression of the P+1 loop CGF mutants described above in N. benthamiana tissue silenced for Prf did not induce the HR (Figure 1C). Conversely, the HR induced by expression of these mutants was retained in control plants infected with the empty silencing vector. Thus, the loss of HR in Prf-silenced plants was attributable to the absence of Prf and not to secondary effects of VIGS. These data are consistent with specific activation of the Pto-Prf signal transduction pathway by the P+1 loop substitution mutants.
Surface-Exposed Amino Acids of the P+1 Loop Are Responsible for the CGF Phenotype
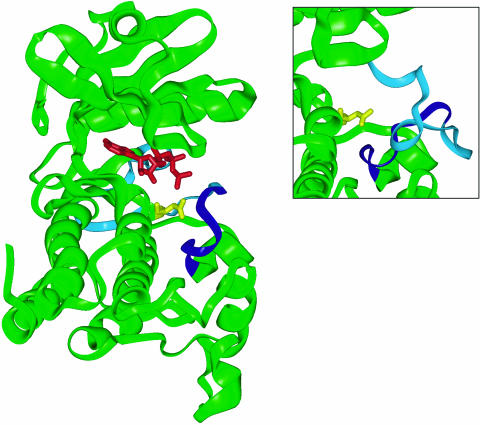
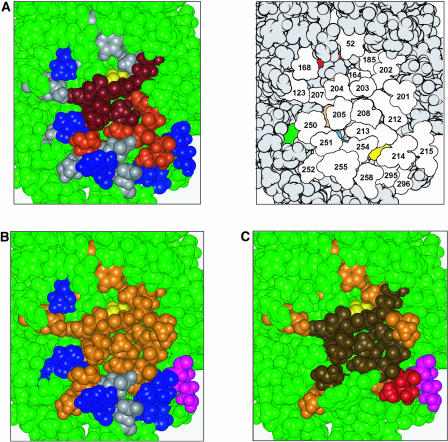
To explore the potential effect(s) of P+1 loop mutations on Pto kinase structure, a three-dimensional homology model was constructed using the crystal structures of the c-Abl and BTK Tyr kinases (Mao et al., 2001; Nagar et al., 2003). The BTK activation loop is in a noninhibitory conformation, whereas the c-Abl activation loop is oriented in essentially the opposite direction and obstructs the putative active site. We used these two structures to model alternative active and inactive structures for Pto. The models conform closely to the canonical bilobal kinase core but exclude the 20 N-terminal nonkinase amino acids for which a suitable template was not available (Figure 2). The junction of the two lobes forms the catalytic cleft, and the ATP molecule is shown docked onto the N-terminal lobe. The P+1 loop and the presumed catalytic residue Asp-164 are located in close proximity in the C-terminal lobe. P+1 loop residues were mapped onto the active Pto model. This revealed that the side chains of residues Val-201, Val-202, Gly-203, Thr-204, Leu-205, Gly-206, and Ile-208 are surface exposed, consistent with a role for these amino acids in substrate binding. The side chain of Tyr-207 is semi-buried; however, the partial exposure of the aromatic ring in our model is similar to crystal structures of cAPK, where it is thought to interact directly with substrate residues (Moore et al., 2003). Pro-210 is buried within the molecule. Thus, there is a correlation between the surface exposure of Pto P+1 loop residues and their ability to specify cell death when mutated to Asp. The data are consistent with a model in which the solvent-exposed residues mediate the regulation of Pto, possibly within the canonical role of P+1 loop residues in peptide binding.
Figure 2.
Homology Model of Pto.
Ribbon diagram of the Pto homology model in its active form, representing residues 20 to 321. The P+1 loop (amino acids 201 to 210) is shown in dark blue and the T-loop in cyan. The presumed catalytic residue Asp-164 is shown in yellow and bound ATP in red. Inset, the activation loop in its inactive conformation.
Kinase Activity Is Dispensable for Signaling by ptoL205D
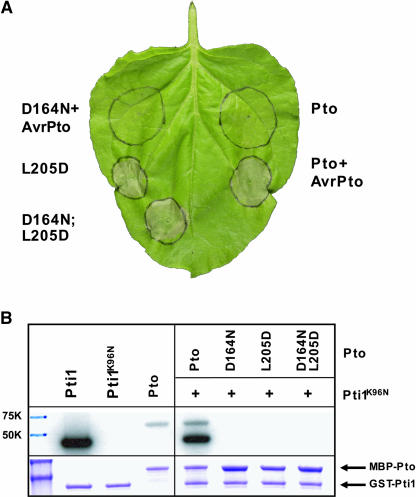
We showed previously that a pto double mutant containing a P+1 loop substitution and a kinase knockout mutation in Lys-69 (K69N) was unable to elicit cell death in planta (Rathjen et al., 1999). ptoK69N is unable to interact with AvrPto, whereas ptoD164N is kinase deficient but able to bind AvrPto in yeast, indicating minimal structural consequences of this mutation (Scofield et al., 1996; Tang et al., 1996). To test the role of kinase activity in CGF signaling, we introduced the mutation D164N to ptoL205D, chosen on the basis of its strong CGF phenotype. The AvrPto-dependent HR of Pto in N. benthamiana was abolished by the D164N mutation, consistent with previous data (Figure 3A; Rathjen et al., 1999). However, transient expression of ptoD164N:L205D in N. benthamiana tissue elicited a strong HR qualitatively similar to that of the single CGF mutant ptoL205D. To test the kinase activity of the CGF molecules, we established an in vitro assay based on phosphorylation of Pti1K96N by Pto (Figure 3B; Zhou et al., 1995). ptoL205D showed absent or greatly reduced autophosphorylation and transphosphorylation relative to wild-type Pto (cf. with Figure 6B), and these activities were completely absent in the double mutant ptoD164N:L205D. Staining of the radioactive gel confirmed equivalent protein loading in these assays. These data are consistent with the known role of the P+1 loop in kinase activity and dispensability of kinase activity for signaling once Pto has been made active by mutation. The data fundamentally contradict current models in which Pto acts at the apex of a phosphorylation cascade (Pedley and Martin, 2003) and dictate that P+1 loop mutations must act by some mechanism other than enhancement of kinase activity.
Figure 3.
Kinase Activity Is Dispensable for Signaling by pto L205D.
(A) In planta expression of Pto derivatives containing a substitution in the presumed catalytic residue (D164). Each mutant was expressed as indicated.
(B) Pto mutant proteins capable of inducing an HR in vivo lack kinase activity. Proteins were expressed as MBP fusions in E. coli, purified and tested for the ability to autophosphorylate (top band) or transphosphorylate a GST-PtiI substrate (bottom band). The MBP-Pto fusions correspond to ∼77 K and GST-PtiK96N to ∼59 K. Bottom panel, Coomassie blue–stained gel showing equivalent loading of fusion proteins in the autoradiograph.
Figure 6.
Kinase Activity Is Dispensable for Signaling by Pto CGF Mutants.
(A) Expression of Pto CGF mutants containing a second-site kinase knockout mutation in planta. Mutants were expressed as indicated.
(B) Diminished kinase activity of most Pto CGF mutants. Mutant proteins were expressed as GST fusions and assayed for the ability to autophosphorylate or transphosphorylate in vitro. Radiolabeled species were detected by autoradiography after SDS-PAGE. The position of GST-Pto fusion proteins and cleaved Pt1K96N fusion proteins are indicated. Bottom panel, Coomassie stain of the autoradiograph gel showing equivalent loading.
Identification of Non-P+1 Loop Residues That Control Pto Signaling
The data presented above require an alternative explanation for Pto signaling. We reasoned that the P+1 loop could represent a binding patch for a negative regulator, a dynamic loop that undergoes conformational change(s), or both. To investigate these possibilities, we used our active homology model to identify representative buried and surface-exposed residues within 2 Å of the P+1 loop region as candidates for further mutagenesis. Buried residues could hold the P+1 loop in an inactive form before stimulation by AvrPto, whereas surface-exposed residues might form part of the binding site for a putative regulatory molecule. Buried residues were changed to Ala to delete side chain contacts while maintaining hydrophobicity, whereas surface-exposed residues were mutated to Asp according to our original strategy.
Mutant genes encoding the buried residue variants ptoR163A, ptoK166A, ptoL218A, ptoD223A, ptoY225A, ptoF227A, ptoE233A, ptoA256D, and ptoV257A were expressed in N. benthamiana leaves and assessed for CGF activity. Cell death was not observed for any mutant that contained an Ala substitution in a putatively buried residue (Figure 4A). Mutants ptoL218A, ptoD223A, ptoA256D, and ptoV257A did not accumulate in planta, and so the expression phenotypes are noninformative (see Supplemental Figure 1A online). Similarly, ptoR163A, ptoK166A, ptoY225A, ptoF227A, and ptoE233A accumulated in vivo but did not respond to AvrPto coexpression and therefore are potentially misfolded. The lack of positive data from this experiment means that role(s) for buried residues in Pto regulation cannot be identified.
Figure 4.
Surface-Exposed Residues Are Responsible for Native Regulation of Pto.
(A) Expression of Ala substitution mutants in planta. Identity of each mutant is indicated.
(B) Expression of Asp substitution mutants in planta. Identity of each mutant is indicated.
(C) CGF mutants require Prf for signaling. Left, leaves silenced for Prf; right, leaves silenced for empty vector only. Pto mutants were expressed as indicated.
(D) In planta recognition of AvrPto by non-CGF pto mutants. Mutants were expressed as indicated in N. benthamiana leaves carrying a dexamethasone-inducible avrPto transgene.
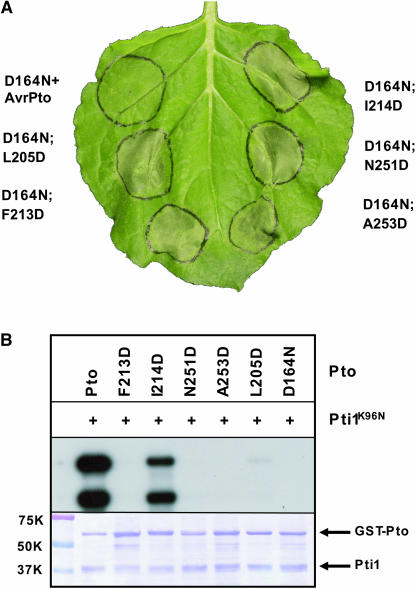
For mutant genes encoding substitutions in surface residues, expression of wild-type Pto or any of ptoK123D, ptoD164A, ptoI185D, ptoL245D, and ptoV250D did not induce the HR. Transient expression of each of ptoF213D, ptoI214D, ptoN251D, and ptoA253D elicited strong AvrPto-independent cell death in N. benthamiana leaves similar to PtoL205D (Figure 4B). ptoY212D displayed CGF activity in ∼50% of infiltrations. Signaling by ptoF213D, ptoI214D, ptoN251D, and ptoA253D was pathway dependent as judged by the inability to induce the HR in N. benthamiana silenced for Prf (Figure 4C). These CGF-conferring residues form a contiguous patch of surface-exposed residues in our three-dimensional model (Figure 5A) but are separated in the primary structure of Pto.
Figure 5.
Mapping a Negative Regulatory Patch on the Surface of Pto.
(A) A negative regulatory patch on Pto. Left, the P+1 loop residues 202 to 208 are dark red, catalytic residue D164 is yellow, and other CGF-conferring residues are red. Residues that retained the wild-type properties of Pto after mutation are shown in blue. Non-CGF mutants that did not respond to AvrPto in planta are colored gray. Right, a schematic of the same region defining the residues. Residues that are partially obscured in this projection are color coded as follows: 169, red; 206, orange; 210, yellow; 245, green; 253, blue.
(B) Residues required for interaction of Pto with AvrPto and AvrPtoB in yeast. The view is comparable to (A). Residues required for interaction with AvrPto and AvrPtoB are shown in orange; dispensable residues are colored blue. K215 (magenta) is a unique binding determinant for AvrPtoB. Residues with inconclusive binding function are in gray. The catalytic residue D164 is yellow.
(C) Overlap between regulatory and Avr-interaction patches. Overlay of (A) and (B). The negative regulatory patch directly overlaps the presumed Avr docking site (brown). Residues required solely for CGF activity (I214, red), Avr interaction (orange), or AvrPtoB binding (K215, magenta) are indicated.
Mapping of a Surface-Exposed Patch That Controls Pto Signaling
The preceding experiments identified surface-exposed residues that control Pto signaling. To further define this patch, we used our model to find additional surface-exposed residues contiguous with those responsible for CGF activity. These included Phe-52, Ile-168, Asn-169, Lys-215, Leu-252, Glu-254, Trp-255, Glu-258, Leu-295, and Ser-296. Substitution of any of these new residues to Asp did not confer CGF activity upon expression of each mutant gene in N. benthamiana leaves (data not shown). These data, and the negative CGF data from the previous series of surface-exposed mutants, could be significant because protein accumulated from the expression of all genes except for ptoL252D (see Supplemental Figure 1B online). These mutants could represent functionally wild-type molecules that respond to AvrPto coexpression and would thus identify residues that do not control Pto signaling. To test this, the non-CGF genes comprising ptoF52D, ptoK123D, ptoD164A, ptoI168D, ptoN169D, ptoI185D, ptoK215D, ptoL245D, ptoV250D, ptoL252D, ptoE254A, ptoW255D, ptoE258A, ptoL295D, and ptoS296D were coexpressed in planta with AvrPto. ptoI168D, ptoK215D, ptoV250D, ptoW255D, ptoL295D, and ptoS296D were able to induce AvrPto-dependent cell death (Figure 4D). Thus, these mutants behave as wild-type Pto in this assay and define a partial border of the proposed regulatory patch (Figure 5A). No conclusions regarding potential regulatory roles of residues Phe-52, Lys-123, Asp-164, Asn-169, Ile-185, Leu-245, Leu-252, Glu-254, and Glu-258 (Figures 4D and 5A) can be drawn because of the lack of an in planta phenotype.
Most Surface Patch Residues Show Impaired Interaction with AvrPto and AvrPtoB
The P+1 loop is an important AvrPto and AvrPtoB binding determinant (Rathjen et al., 1999; Kim et al., 2002). To test the role of non-P+1 loop residues in ligand binding activity, surface patch mutants were assayed for the ability to interact with AvrPto and AvrPtoB in yeast. Most mutants lacked the ability to interact with either avirulence protein (Table 1). These data are significant because all mutant fusion proteins accumulated in yeast, with the exception of ptoN169D, ptoE254A, and ptoE258A (see Supplemental Figures 2A and 2B online). Thus, most of the mutated residues were required for interaction with ligands and define a second patch that substantially overlaps the regulatory patch defined above (Figures 5B and 5C). Seven residues (Ile-168, Ile-214, Leu-245, Val-250, Trp-255, Leu-295, and Ser-296) were dispensable for interaction with both AvrPto and AvrPtoB (Figure 5B). Strikingly, the mutant ptoI214D elicited AvrPto-independent HR; therefore, CGF activity and the ability to interact with Avr proteins are not mutually exclusive. ptoL245D, and the buried residue mutant ptoF227A, retained the ability to interact with AvrPto and AvrPtoB in yeast but were not activated by AvrPto coexpression in planta (Table 1). These mutants are presumably deficient in kinase activity or interaction with the downstream effector of signaling. A further point of interest is Lys-215, located on the extreme right edge of the proposed binding patch (Figures 5B and 5C). ptoK215D was able to interact with AvrPto but not AvrPtoB. Therefore, the binding requirements for these ligands can be functionally separated.
Table 1.
Phenotypes of pto Mutants Generated in This Study
| In Planta Assays
|
Yeast Two-Hybrid
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Residue Class | pto Mutant | AA Class | CGF | AvrPto | Expression | AvrPto | AvrPtoB | Expression |
| P+1 loop residues | V201D | Hydrophobic | − | − | − | − | − | + |
| K202D | + | + | N/A | + | − | − | + | |
| G203D | Hydrophobic | + | N/A | + | − | − | + | |
| T204D | Polar | + | N/A | + | − | − | + | |
| L205D | Hydrophobic | + | N/A | + | − | − | + | |
| G206D | Hydrophobic | + | N/A | + | − | − | + | |
| Y207D | Polar | + | N/A | + | − | − | + | |
| I208D | Hydrophobic | + | N/A | + | − | − | + | |
| P210D | Hydrophobic | − | − | + | − | − | + | |
| Solvent-exposed residues | F52D | Hydrophobic | − | − | + | − | − | + |
| K123D | + | − | − | + | − | − | + | |
| D164A | − | − | − | + | − | − | + | |
| I168D | Hydrophobic | − | + | + | + | + | + | |
| N169D | Polar | − | − | + | − | − | + | |
| I185D | Hydrophobic | − | − | + | − | − | + | |
| Y212D | Polar | + | N/A | + | − | − | + | |
| F213D | Hydrophobic | + | N/A | + | − | − | + | |
| I214D | Hydrophobic | + | N/A | + | + | + | + | |
| K215D | + | − | + | + | + | − | + | |
| L245D | Hydrophobic | − | − | + | + | + | + | |
| V250D | Hydrophobic | − | + | + | + | + | + | |
| N251D | Polar | + | N/A | + | − | − | + | |
| L252D | Hydrophobic | − | − | − | − | − | + | |
| A253D | Hydrophobic | + | N/A | + | − | − | + | |
| E254A | − | − | − | + | − | − | − | |
| W255D | Polar | − | + | + | + | + | + | |
| E258A | − | − | − | + | − | − | − | |
| L295D | Hydrophobic | − | + | + | + | + | + | |
| S296D | Polar | − | + | + | + | + | + | |
| Buried residues | R163A | + | − | − | + | − | − | + |
| K166A | + | − | − | + | − | − | + | |
| L218A | Hydrophobic | − | − | − | + | − | + | |
| D223A | Polar | − | − | − | − | − | + | |
| Y225A | Polar | − | − | + | − | − | + | |
| F227A | Hydrophobic | − | − | + | + | + | + | |
| E233A | − | − | − | + | − | − | + | |
| A256D | Hydrophobic | − | − | − | − | − | + | |
| V257A | Hydrophobic | − | − | − | − | − | + | |
| D164N | − | − | − | + | + | + | + | |
| Pto | None | − | + | + | + | + | + | |
pto mutants are indicated using single letter amino acid (AA) codes. In planta assays: CGF, AvrPto-independent HR phenotype when expressed in planta, (+) indicates HR and (−) indicates no phenotype. AvrPto, AvrPto-dependent HR when coexpressed with each mutant in planta. N/A, not applicable because of CGF phenotype. Expression, accumulation of protein measured by protein gel blot analysis. Yeast two-hybrid interactions: AvrPto, interaction of each mutant with AvrPto in yeast, (+) represents a positive interaction and (−) designates a lack of interaction. AvrPtoB, interaction of each mutant with AvrPtoB in yeast.
Kinase Activity Is Not Required for HR Signaling of Non-P+1 Loop CGF Mutants
Kinase activity is required for AvrPto-dependent activation of Pto but dispensable for the CGF variant ptoL205D. Despite this, we reasoned that some of the non-P+1 loop CGF molecules described here might mimic intermediate forms of Pto activation and hence require phosphorylation ability. To test this, we constructed double mutants incorporating the kinase knockout mutation D164N to genetic constructs containing CGF substitutions. Transient expression of ptoD164N:F213D, ptoD164N:I214D, ptoD164N:N251D, or ptoD164N:A253D in N. benthamiana induced the HR, similar to the positive control ptoD164N:L205D (Figure 6A). The HR induced by the double mutants was dependent on Prf as assessed by VIGS experiments (see Supplemental Figure 3 online). Thus, novel surface patch CGF mutants appeared to be similar to ptoL205D in mimicking an apparently terminally activated form of Pto. To test the phosphorylation potential of these mutants, ptoF213D, ptoI214D, ptoN251D, and ptoA253D were expressed and purified as glutathione S-transferase (GST) fusion proteins (Figure 6B) and subjected to an in vitro kinase assay. Autophosphorylated Pto was visualized as a band of ∼62 K, and a transphosphorylation band using pti1K96N as substrate migrated at ∼40 K. ptoI214D showed strong phosphorylation activity similar to the wild type, whereas the kinase activity of ptoL205D was strongly diminished. ptoF213D, ptoN251D, and ptoA253D, and the negative control ptoD164N, did not have detectable kinase activity in this assay. These data are consistent with the dispensability of kinase activity for signaling of CGF forms of Pto and with previous data showing that ptoY207D lacked autophosphorylation activity (Rathjen et al., 1999).
DISCUSSION
The data presented here extend previous observations to firmly establish the P+1 loop as a key regulator of Pto signaling. Acidic substitutions of almost all P+1 loop residues conferred CGF activity to Pto. Most importantly, CGF-associated residues were predicted to be at least partially surface exposed, and we were able to use this criterion to identify further residues required for normal Pto regulation. The data suggest the existence of a surface patch that normally acts to repress signaling. We extended these observations to define a requirement for Pto kinase activity in Avr-dependent activation but not downstream signaling. Finally, integration of the preceding data with the Avr interaction phenotypes of surface patch mutants leads to an outline mechanism for activation of Pto by specific ligands.
The protein kinase P+1 loop was recognized originally for its role in stabilizing the substrate P+1 residue (i.e., the residue carboxy-proximal to the phosphorylated residue; Taylor et al., 1995). For the template kinase cAPK, the side chains of Leu-198, Pro-202, and Leu-205 form a hydrophobic pocket that interacts with the hydrophobic P+1 residue of the substrate. These residues correspond to Pto amino acids Val-201, Leu-205, and Ile-208; both L205D and I208D mutants induced cell death in this study, whereas V201D did not accumulate in planta. Subsequent modeling work has shown that other cAPK P+1 loop residues interact directly and indirectly with substrate residues (Moore et al., 2003). Thus, cAPK Gly-200 (equivalent to Pto Gly-203) forms a hydrogen bond with the backbone amide of the P+1 residue, and Glu-203 (Pto Gly-206) interacts with the P-6 Arg. Tyr-204 (Pto Tyr-207) has profound effects on substrate affinity, apparently because the aromatic ring is partially exposed and interacts with the P-2 Arg. Moore et al. (2003) have proposed that the term “peptide positioning” loop is a more accurate description of how this region of cAPK acts at several levels to secure the peptide substrate in the correct orientation before phosphorylation. Our results reveal that equivalent P+1 loop residues control either peptide binding in cAPK or CGF activity in Pto after mutation. Conservative extrapolation of these data suggests that the Pto P+1 loop functions in a canonical sense to bind a peptide, but the role of the proposed peptide is to repress Pto signaling. We propose that the effect of Asp substitutions is to delete the interaction with a negative regulator, leading to derepression of signaling. Several protein kinases, including cAPK, are known to be regulated in this manner by so-called pseudosubstrate molecules (Taylor and Radzio-Andzelm, 1997).
Certain P+1 loop residues are known to interact with distal residues during substrate recognition and catalysis (Smith et al., 1999). We reasoned that mutation of some of these residues might result in constitutively active molecules via indirect disruption of the P+1 loop. We analyzed our active model to identify residues linked to the P+1 loop in three dimensions as candidates for further mutagenesis. Experiments in which buried residues were substituted for Ala were inconclusive, either because of failure of the mutant protein to accumulate or the absence of phenotypes associated with these mutations. Such mutants may be misfolded or otherwise unstable. By contrast, acidic substitutions of several surface-exposed residues resulted in the CGF phenotype. We identified five residues that are not resident in the P+1 loop; Tyr-212, Phe-213, Ile-214, Asn-251, and Ala-253. These residues form an apparently contiguous surface patch with the P+1 loop residues but are separated from the P+1 loop in the primary sequence of Pto. Roles in substrate binding for the corresponding residues in other protein kinases have not been defined to our knowledge. We suggest that these residues represent additional sites of contact that mediate protein docking with the proposed inhibitory protein.
The CGF-responsible residues identified here appear to represent a concise patch. Several residues on the edge of the studied region did not alter the wild-type characteristics of Pto after substitution with Asp, including Ile-168, Lys-215, Val-250, Trp-255, Leu-295, and Ser-296. Such residues evidently do not mediate negative regulation of Pto or interaction with Avr proteins and so define a boundary of the regulatory region. The patch of amino acids that did confer CGF when substituted consists mostly of hydrophobic residues, including Gly-203, Leu-205, Gly-206, Ile-208, Phe-213, Ile-214, and Ala-253. Sites of protein–protein interaction are frequently hydrophobic in nature, although often include charged residues that form salt bridges between the binding partners. However, we did not identify any charged residues within the confines of the Pto surface patch. The Asp substitution strategy used here is a continuance of our earlier work and is slightly unorthodox in that the small residue Ala is usually used for scanning mutagenesis schemes. Incorporation of an acidic residue, such as Asp, should disrupt the presumed hydrophobic moment of the patch, potentially canceling a negative regulatory interaction leading to the observed CGF phenotype. By contrast, the effect of Ala substitutions is more subtle in that only the specific side chain interactions of the substituted amino acid would be deleted. Consistent with this reasoning, ptoY207D conferred a qualitatively stronger CGF HR then ptoY207A (Rathjen et al., 1999). Thus, the use of acidic substitutions may have been an important factor in our ability to detect the proposed surface patch.
In total, we identified 12 different CGF mutants that induced a Prf-dependent HR. The kinase dependence of these mutants provides insight into the possible effects of CGF mutations. We and others previously showed that the kinase activity of Pto is required for activation by AvrPto. This conclusion is supported most strongly by the mutant ptoD164N, which deletes kinase activity but is able to bind AvrPto and AvrPtoB in yeast (Rathjen et al., 1999; Table 1), whereas other kinase knockout mutations, such as K69N, destroy Avr interaction ability (Scofield et al., 1996; Tang et al., 1996). Lys-69 is an invariant residue that stabilizes ATP binding by interaction with the α and β phosphates through formation of a salt bridge with a conserved Glu in the αC helix (Huse and Kuriyan, 2002). Mutation of this residue would be expected to lead to significant conformational changes in the kinase structure. Surprisingly, we found that several CGF mutants containing the D164N mutation were able to induce the Prf-dependent HR in planta. Thus, the kinase activity of Pto is required only for activation and dispensable once the molecule is in the active form. Pto appears to act as a conformation-sensitive molecular adaptor in signaling, rather than transphosphorylating a substrate. Similar roles have been described for the kinase-like signaling molecules KSR and interleukin-1 receptor-associated kinases -M and -2 (Michaud et al., 1997; Janssens and Beyaert, 2003). Pto phosphorylates several substrates in vitro, including the protein kinase Pti1 and the transcription factor–like protein Pti4. Our results suggest that these proposed phosphorylation events are not necessary for either the Prf-dependent HR, or disease resistance per se, because HR and disease resistance have not been separated upstream of Prf.
The data suggest that the role of kinase activity in Pto activation is to remove the proposed negative regulator from the surface patch. This would correspond to the terminal stage of Pto activation mimicked by the CGF mutants. It follows that removal of the negative regulator is probably sufficient for activation because kinase activity is dispensable after this step. The putative regulatory molecule is likely to be a peptide following the argument above, but its identity is unknown. Theoretically, it could be part of the Pto molecule itself or an unidentified member of the signaling pathway. Overexpression of Pto does not induce the HR in N. benthamiana (although quantitative phenotypes are evident in transgenic tomato plants; Tang et al., 1999); therefore, it seems that the repression cannot be overcome by titration. This is consistent with Pto itself mediating repression. Our inactive model of Pto shows an extensive interaction between the P+1 and T-loops, which is essentially absent in the active form (Figure 2) and could provide a structural basis for Pto self-regulation. Conversely, Pto signaling could be controlled by another molecule. To explain why Pto overexpression does not lead to the HR, we suggest that the regulatory molecule could also play a positive role in signaling. In this scenario, the regulatory protein would be limiting such that only a small number of Pto-regulator complexes competent for signaling would form, whereas overexpressed Pto would accumulate as nonfunctional monomers. This is consistent with the suggested role of Pto as an adaptor in a signaling complex. Avr-dependent conformational activation of Pto would result in a concomitant change in the proposed regulatory protein, thus triggering downstream signaling. Identification of the proposed regulatory partner(s) is likely to provide considerable insight to control of signaling by Pto.
Many of the surface residues mutated here were required for interaction with both AvrPto and AvrPtoB in yeast. There was considerable overlap in the sequence requirements for binding each Avr protein, with the exception of Lys-215, which was required only for AvrPtoB. Like the CGF area, the region controlling Avr binding appeared to be a concise patch because some peripheral residues (Ile-168, Ile-214, Leu-245, Val-250, Leu-295, and Ser-296) were dispensable for interaction with ligands. Thus, we have partially mapped a border of a second surface area that overlaps the previous patch responsible for CGF activity (Figure 5C). Avr proteins could bind to this area directly, or alternatively, mutations in this region could cause a conformational change that disrupts a binding site in some other part of the Pto molecule. Generally speaking, there is an inverse correlation between the CGF activity of each mutant and the ability to bind either Avr. However, the mutant protein ptoI214D, which binds both Avr proteins but is constitutively active in vivo, shows that these activities are not mutually exclusive. This mutant appears to preclude a global conformational change in the Pto kinase domain upon activation because it retained both kinase activity and the ability to interact with ligands. If global rearrangement of Pto structure can indeed be ruled out, then the most likely explanation of our data is that Avr molecules bind to the same region as the proposed negative regulatory molecule. This hypothesis is attractive because it suggests a mechanism for kinase-dependent activation of Pto by Avr proteins.
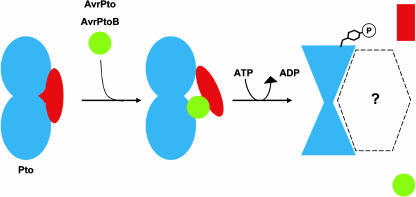
We suggest the following model for activation of Pto by specific ligands (Figure 7). Before stimulation, Pto is held in the inactive state by a regulatory peptide that binds to the P+1 loop in the kinase catalytic cleft. This peptide may repress kinase activity generally or simply suppress the specific phosphorylation event that leads to activation of signaling. The Avr protein binds to Pto, resulting in a phosphorylation event(s) that vacates the inhibitory peptide from the Pto peptide positioning loop. The overlap in negative regulatory and Avr interaction patches detected here suggests that these molecules might bind competitively to Pto. Thus, Avr binding might cause displacement of the inhibitory protein from the kinase catalytic cleft, allowing the regulatory phosphorylation event(s). The site of phosphorylation is unknown and could be on Pto itself or the proposed negative regulator. The outcome of phosphorylation is presumably a conformational change in Pto that is perceived by another protein. This scenario is reminiscent of the theoretical model proposed by Van der Biezen and Jones (1998), although the current data do not provide insight to the role of Prf in signaling by Pto.
Figure 7.
Model of Pto Activation by AvrPto and AvrPtoB.
Native Pto (blue) is regulated by an inhibitory peptide (red) that represses kinase activity. Upon secretion into the plant cell, AvrPto or AvrPtoB (green) interacts with Pto, displacing the regulatory peptide from the catalytic cleft and derepressing kinase activity. Activated Pto catalyzes a phosphorylation event(s) leading to a specific conformational change. An unknown effector protein recognizes the altered Pto conformation, leading to activation of the resistance pathway. Although the Pto-effector complex is shown dissociated from the Avr and inhibitor proteins, this remains speculative.
Another plant protein kinase that acts apparently as the primary receptor for a bacterial effector protein is Arabidopsis PBS1, which recognizes AvrPphB (Shao et al., 2003). However, this example appears to be different from Pto. AvrPphB is a protease that cleaves PBS1 in the T-loop region. This event is necessary for induction of resistance, but its consequence is unknown. However, it is known that the kinase activity of PBS1 is required for subsequent signaling. Thus, the next protein in the pathway could recognize a PBS1 phosphopeptide as suggested by Shao et al. (2003), or the cleavage event could stimulate the kinase activity of PBS1, resulting in phosphorylation of a substrate molecule. Either possibility is clearly different from our demonstration of kinase independence of Pto for downstream signaling. It may be necessary to define the roles of the respective NBS-LRR proteins of these pathways, Prf and RPS5, before these differences can be reconciled.
METHODS
Cloning and Expression of Pto Mutants
Standard techniques for manipulating plasmids in Escherichia coli strain DH5α were used (Sambrook et al., 1989). PCR-based site-directed mutagenesis was performed as described (Horton et al., 1989). All products contained 5′ BamHI and NcoI sites and 3′ XbaI and were cloned into pCRII (Invitrogen, Carlsbad, CA) before automated DNA sequencing (Applied Biosystems, Foster City, CA). For expression in planta, P+1 loop mutants were cloned exactly as described (Rathjen et al., 1999). Non-P+1 loop mutants were subcloned in the sense orientation into the vector pCB302-3 (Xiang et al., 1999) using BamHI and XbaI. All recombinant binary vectors were transferred to Agrobacterium tumefaciens strain GV2260 for expression in planta and the presence of the binary vector confirmed by PCR. For Y2H analysis, Pto and its mutant derivatives were cloned into the DNA binding domain vector pAS2-1 (Clontech, Palo Alto, CA) using NcoI and EcoRI. For expression in E. coli, Pto and its mutant derivatives were subcloned into pMalc2 (New England Biolabs, Beverly, MA) or pGEX-2TK (Amersham Biosciences, Buckinghamshire, UK).
Transient Agrobacterium-Mediated Expression
A. tumefaciens strain GV2260 containing the binary plasmid of interest was grown in LB media with appropriate antibiotics to stationary phase (∼2 d) at 28°C. Cultures were diluted 1:10 into fresh LB plus antibiotics and grown overnight at 28°C. Cells were pelleted and washed once in infiltration media (10 mM Mes, pH 5.6, 10 mM MgCl2, and 200 μM acetosyringone) and then resuspended to an OD600 of 1.0. Six-week-old Nicotiana benthamiana plants grown in the greenhouse were inoculated by pressure infiltration using a disposable syringe, and the approximate area of infiltration was outlined with a marker pen. Infiltrated plants were kept in laboratory conditions for 4 d to allow symptoms to develop.
VIGS
VIGS was performed by infiltrating Agrobacterium strains carrying either TRV empty vector (pTV00; Ratcliff et al., 2001) or TRV:PRF (Lu et al., 2003) into leaves of 2-week-old N. benthamiana seedlings. Systemic TRV infection developed 5 to 10 d postinoculation and systemic silencing after 3 to 4 weeks.
Construction of a Structural Model of Pto
A full description of the modeling procedure and PDB files containing coordinates of the active and inactive models can be found in the supplemental data online.
Yeast Two-Hybrid Analysis
Yeast two-hybrid analysis was performed using the MATCHMAKER GAL4 system (Clontech) following the manufacturer's protocol. AvrPto and AvrPtoB were expressed from the plasmid pACT2 and transformed into yeast strain Y187 (MATα). Pto and its mutant derivatives were expressed from plasmid pAS2-1 and transformed into yeast strain AH109 (MATa). Pairwise matings were set up between AH109:pto strains and either Y187:AvrPto or Y187:AvrPtoB. Yeast cotransformants were selected by growth on selective media. To assay for protein–protein interactions, cotransformants were replica plated onto separate selective media and growth was monitored based on activation of the ADE2 and HIS3 reporter genes. Growth on selective media was scored as a positive interaction between the fusion proteins, as presented in Table 1.
Protein Analysis
For the analysis of protein accumulation in planta, infiltrated leaf samples were extracted in 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 5 mM DTT, 1% Triton X-100, and 1.5% polyvinylpyrrolidone supplemented with protease inhibitors (Sigma, St. Louis, MO). Fifty micrograms of total protein was loaded onto a 10% SDS-PAGE gel, and after electrophoresis, the gels were electroblotted onto a polyvinylidene difluoride (PVDF) membrane. Immunological reactions were performed with a Pto polyclonal antisera before detection of immunocomplexes with a horseradish peroxidase–conjugated anti-rabbbit secondary antibody, using the ECL detection reagent (Amersham Biosciences). To determine protein accumulation in yeast, yeast cultures were grown in selective SD medium overnight. The culture were diluted to an OD600 = 0.15 to 0.2 and monitored until OD600 = 0.4 to 0.6 was reached. The cells were pelleted and proteins extracted by boiling in 50 mM sodium phosphate, pH 7.0, 25 mM Mes, pH 7.0, 3 M urea, 1% SDS, 10% β-mercaptoethanol, and 0.1% bromophenol blue, supplemented with protease inhibitors (Roche, Indianapolis, IN). OD units (0.5 total) per well were loaded onto an SDS-PAGE gel, and after electrophoresis, the gels were electroblotted onto PVDF membrane for Pto detection.
Protein Expression in E. coli
Pto and its mutant derivatives were expressed as fusion proteins with MBP (Rathjen et al., 1999) or GST in E. coli strain DH5α according to the manufacturer's instructions. For GST-Pto expression, cultures in LB media were incubated at 16°C until OD600 ∼0.4, then induced with 0.2 mM isopropylthio-β-galactoside and incubated at 16°C with shaking overnight. Cells were harvested and resuspended in Tris-buffered saline containing 1 mM DTT, 0.1% (v/v) Triton X-100, and bacterial protease inhibitor cocktail (Sigma). Cells were lysed by sonication and the extract clarified by centrifugation at 13,000g for 30 min at 4°C. The clear supernatants were filtered through 0.45-μm filters and GST-fusion proteins pulled down with glutathione-Sepharose beads (Amersham Biosciences) according to the standard protocol. GST-PtiIK96N was expressed as described (Rathjen et al., 1999) and purified over GSTrap FF columns (Amersham Biosciences) according to the manufacturer's instructions. GST-PtiIK96N was subsequently cleaved with thrombin at 22°C for 16 h. Thrombin was removed by benzamidine affinity chromatography. Cleaved PtiIK96N was concentrated by ultrafiltration (Centricon-10 concentrators; Amicon, Beverly, MA) and used for Pto kinase activity assays.
In Vitro Kinase Assay
Pto autophosphorylation and transphosphorylation activities (against PtiIK96N) were assayed in 50-μL reaction mixtures containing 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 0.5 mM MnCl2, 20 μM ATP, 5 μCi [γ-32P]-ATP, 5 μg GST-PtiIK96N, and immobilized GST-Pto equivalent to 5 μg of protein. For Figure 6B, PtiIK96N was cleaved from the GST moiety before the kinase assay reaction as described above. Reactions were held at 25°C for 30 min and terminated by adding SDS sample buffer and boiling for 5 min. Proteins were resolved on SDS-PAGE gels, then transferred onto PVDF membranes, visualized with Coomassie Brilliant Blue R 250, and subjected to autoradiography.
Supplementary Material
Acknowledgments
This work was initiated at the Center of Engineering Plants for Resistance to Pathogens (Davis, CA) under the supervision of R.W. Michelmore and supported by National Science Foundation Cooperative Agreement BIR-8920216. Part of this work was funded by the Biotechnology and Biological Science Research Council Grant 83/P15081 to J.P.R. The support of the Gatsby Charitable foundation is gratefully acknowledged.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with policy described in the Instructions for Authors (www.plantcell.org) is: John P. Rathjen (john.rathjen@sainsbury-laboratory.ac.uk).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.024141.
References
- Axtell, M.J., and Staskawicz, B. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112, 369–377. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D.G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Ellingboe, A.H. (1981). Changing concepts in host-pathogen genetics. Annu. Rev. Phytopathol. 19, 125–143. [Google Scholar]
- Flor, H.H. (1971). Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9, 275–298. [Google Scholar]
- Frederick, R.D., Thilmony, R.L., Sessa, G., and Martin, G.B. (1998). Recognition specificity for the bacterial avirulence protein AvrPto is determined by Thr-204 in the activation loop of the tomato Pto kinase. Mol. Cell 2, 241–245. [DOI] [PubMed] [Google Scholar]
- Gu, Y.-Q., Yang, C., Thara, V.K., Zhou, J., and Martin, G.B. (2000). Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell 12, 771–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond-Kosack, K.E., and Jones, J.D.G. (1996). Resistance gene-dependent plant defense responses. Plant Cell 8, 1773–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond-Kosack, K.E., and Jones, J.D.G. (1997). Plant disease resistance genes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 575–607. [DOI] [PubMed] [Google Scholar]
- Horton, R.M., Hunt, H.D., Ho, S.N., Pullen, J.K., and Pease, L.R. (1989). Engineering hybrid genes without the use of restriction enzymes: Gene splicing by overlap extension. Gene 77, 61–68. [DOI] [PubMed] [Google Scholar]
- Huse, M., and Kuriyan, J. (2002). The conformational plasticitiy of protein kinases. Cell 109, 275–282. [DOI] [PubMed] [Google Scholar]
- Janssens, S., and Beyaert, R. (2003). Functional diversity and regulation of different interleukin-1 receptor-associated kinase (IRAK) family members. Mol. Cell 11, 293–302. [DOI] [PubMed] [Google Scholar]
- Jia, Y., McAdams, S.A., Bryan, G.T., Hershey, H.P., and Valent, B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19, 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.J., Lin, N.-C., and Martin, G.B. (2002). Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell 109, 589–598. [DOI] [PubMed] [Google Scholar]
- Lu, R., Malcuit, I., Moffett, P., Ruiz, M.T., Peart, J., Wu, A.-J., Rathjen, J.P., Bendahmane, A., Day, L., and Baulcombe, D.C. (2003). High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 22, 5690–5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey, D., Belkhadir, Y., Alonso, J.M., Ecker, J.R., and Dangl, J. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112, 379–389. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Holt, B.F., Wiig, A., and Dangl, J.L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108, 743–754. [DOI] [PubMed] [Google Scholar]
- Mao, C., Zhou, M., and Uckun, F.M. (2001). Crystal structure of Bruton's tyrosine kinase domain suggests a novel pathway for activation and provides insights into the molecular basis of X-linked Agammaglobulinemia. J. Biol. Chem. 276, 41435–41443. [DOI] [PubMed] [Google Scholar]
- Martin, G.B., Brommonschenkel, S.H., Chunwongse, J., Frary, A., Ganal, M.W., Spivey, R., Wu, T., Earle, E.D., and Tanksley, S.D. (1993). Map-based cloning of protein kinase gene conferring disease resistance in tomato. Science 262, 1432–1436. [DOI] [PubMed] [Google Scholar]
- Michaud, N.R., Therrien, M., Cacace, A., Edsall, L.C., Spiegel, S., Rubin, G.M., and Morrison, D.K. (1997). KSR stimulates Raf-1 activity in a kinase-independent manner. Proc. Natl. Acad. Sci. USA 94, 12792–12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, M.J., Adams, J.A., and Taylor, S.S. (2003). Structural basis for peptide binding in protein kinase A. Role of glutamic acid 203 and tyrosine 204 in the peptide-positioning loop. J. Biol. Chem. 278, 10613–10618. [DOI] [PubMed] [Google Scholar]
- Nagar, B., Hantschel, O., Young, M., Scheffzek, K., Veach, D., Bornmann, W., Clarkson, B., Superti-Furga, G., and Kuriyan, J. (2003). Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell 112, 859–871. [DOI] [PubMed] [Google Scholar]
- Pedley, K.F., and Martin, G.B. (2003). Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Annu. Rev. Phytopathol. 41, 215–243. [DOI] [PubMed] [Google Scholar]
- Ratcliff, F., Martin-Hernandez, A., and Baulcombe, D. (2001). Technical advance. Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25, 237–245. [DOI] [PubMed] [Google Scholar]
- Rathjen, J.P., Chang, J.H., Staskawicz, B.J., and Michelmore, R.W. (1999). Constitutively active Pto induces a Prf-dependent hypersensitive response in the absence of avrPto. EMBO J. 18, 3232–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathjen, J.P., and Moffett, P. (2003). Early signal transduction events in specific plant disease resistance. Curr. Opin. Plant Biol. 6, 300–306. [DOI] [PubMed] [Google Scholar]
- Salmeron, J.M., Oldroyd, G.E.D., Rommens, C.M.T., Scofield, S.R., Kim, H.-S., Lavelle, D.T., Dahlbeck, D., and Staskawicz, B.J. (1996). Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell 86, 123–133. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Scofield, S.R., Tobias, C.M., Rathjen, J.P., Chang, J.H., Lavelle, D.T., Michelmore, R.W., and Staskawicz, B.J. (1996). Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science 274, 2063–2065. [DOI] [PubMed] [Google Scholar]
- Sessa, G., D'Ascenzo, M., Loh, Y.-T., and Martin, G.B. (1998). Biochemical properties of two protein kinases involved in disease resistance signaling in tomato. J. Biol. Chem. 273, 15860–15865. [DOI] [PubMed] [Google Scholar]
- Shao, F., Golstein, C., Ade, J., Stoutemyer, M., Dixon, J.E., and Innes, R.W. (2003). Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301, 1230–1233. [DOI] [PubMed] [Google Scholar]
- Smith, C.M., Radzio-Andzelm, E., Madhusudan, Akamine, P., and Taylor, S.S. (1999). The catayltic subunit of cAMP-dependent protein kinase: Prototype for an extended network of communication. Prog. Biophys. Mol. Biol. 71, 313–341. [DOI] [PubMed] [Google Scholar]
- Tang, X., Xie, M., Kim, Y.J., Zhou, J., Klessig, D.F., and Martin, G.B. (1999). Overexpression of Pto activates defense responses and confers broad resistance. Plant Cell 11, 15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, X.Y., Frederick, R.D., Zhou, J.M., Halterman, D.A., Jia, Y.L., and Martin, G.B. (1996). Initiation of plant disease resistance by physical interaction of AvrPto and Pto Kinase. Science 274, 2060–2063. [DOI] [PubMed] [Google Scholar]
- Taylor, S., Radzio-Andzelm, E., and Hunter, T. (1995). How do protein kinases discriminate between serine/threonine and tyrosine? Structural insights from the insulin receptor protein-tyrosine kinase. FASEB J. 9, 1255–1266. [DOI] [PubMed] [Google Scholar]
- Taylor, S.S., and Radzio-Andzelm, E. (1997). Protein kinase inhibition: Natural and synthetic variations on a theme. Curr. Opin. Chem. Biol. 1, 219–226. [DOI] [PubMed] [Google Scholar]
- Van der Biezen, E.A., and Jones, J.D.G. (1998). Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci. 23, 454–456. [DOI] [PubMed] [Google Scholar]
- Xiang, C., Han, P., Lutziger, I., Wang, K., and Oliver, D. (1999). A mini binary vector series for plant transformation. Plant Mol. Biol. 40, 711–717. [DOI] [PubMed] [Google Scholar]
- Zhou, J., Tang, X., and Martin, G. (1997). The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis-element of pathogeneis-related genes. EMBO J. 16, 3207–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J., Loh, Y.-T., Bressan, R.A., and Martin, G.B. (1995). The tomato gene Pti1 encodes a serine/threonine kinase that is phosphorylated by Pto and is involved in the hypersensitive response. Cell 83, 925–935. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.