Abstract
The internal transcribed spacer (ITS) as one part of nuclear ribosomal DNA is one of the most extensively sequenced molecular markers in plant systematics. The ITS repeats generally exhibit high-level within-individual homogeneity, while relatively small-scale polymorphism of ITS copies within individuals has often been reported in literature. Here, we identified large-scale polymorphism of ITS copies within individuals in the legume genus Lespedeza (Fabaceae). Divergent paralogs of ITS sequences, including putative pseudogenes, recombinants, and multiple functional ITS copies were sometimes detected in the same individual. Thirty-seven ITS pseudogenes could be easily detected according to nucleotide changes in conserved 5.8S motives, the significantly lower GC contents in at least one of three regions, and the lost ability of 5.8S rDNA sequence to fold into a conserved secondary structure. The distribution patterns of the putative functional clones were highly different between the traditionally recognized two subgenera, suggesting different rates of concerted evolution in two subgenera which could be attributable to their different extents/frequencies of hybridization, confirmed by our analysis of the single-copy nuclear gene PGK. These findings have significant implications in using ITS marker for reconstructing phylogeny and studying hybridization.
Concerted evolution is a form of multigene family evolution in which all the tendency of the different genes in a gene family or cluster are assumed to evolve as a unit in concert1,2. All the repeats are maintained in the genome with very similar sequences but differ between related species. The best known example of concerted evolution is among multicopy nrDNA genes3. Nuclear ribosomal DNA cistron is arranged as tandemly repeated units consisting of 18S, ITS1, 5.8S, ITS2, 26S in all land plants except bryophytes, and occurs with hundreds to thousands copies per haploid genome4. The internal transcribed spacer (ITS1 and ITS2) as one part of nrDNA is one of the most extensively sequenced molecular markers in plant systematics because of the rapid concerted evolution within and among component subunits, fast evolution rate, and short length and availability of universal primers5,6,7,8. The ITS region generally undergoes rapid concerted evolution via unequal crossing over, high-frequency gene conversion, and large deletion2,9. Hence, intra-individual polymorphism has generally been considered to be an exception. Nevertheless, under certain circumstances, polymorphic ITS copies within individuals have been identified in many different plant groups but in a relatively small scale10,11,12,13,14,15,16,17,18,19,20,21,22,23,24 which suggest concerted evolution remains incomplete across the repeats. Various reasons such as hybridization, polyploidization, recombination among copies, and pseudogenization of cistrons are considered to be the causes of polymorphic ITS copies5,17,24,25. Among ITS polymorphism, non-functional pseudogenes are prominent and can be distinguished by the GC content, secondary structure stability, substitution rates, the presence of conserved motifs, and phylogenetic positions6,19,21,22.
Speciation via hybridization is thought to be an important evolutionary mechanism in plants26,27. Natural hybridizations are well documented and have been widely recognized in angiosperm evolution26,27,28,29, with an estimated approximately 25% of vascular plants forming hybrids with other species30 and about 11% of plant species probably being derived from past hybridization events31. The phenomenon of hybridization is particularly likely to occur in plant species complexes in which species are evolutionarily young and sometimes not well differentiated, and the barriers between them are relatively weak32,33,34,35,36,37. Hybridization leads the evolutionary processes at the species level to a network rather than a bifurcating tree, confounding efforts to reconstruct the phylogeny of the complexes. In such cases, the DNA sequence information of the uniparentally inherited plastid and that of the biparentally inherited nrDNA which often causes parents homogenization due to concerted evolution, mask evidence of reticulation in hybridizing species38,39,40,41. However, single or low-copy nuclear genes are able to overcome this disadvantage32,42. They are biparental and independently inherited and do not often experience concerted evolution, and thus can provide evolutionary signatures of unique gene copies from ancestral genomes which allows tracing hybrid reticulate history within lineages42,43,44. In addition, the direction of hybridization can be inferred when information of single or low-copy nuclear genes is used together with plastid markers.
The legume genus Lespedeza Michx. (Fabaceae: Papilionoideae), well known for about 30 natural hybrids within the genus45,46,47, offers an ideal system for studying hybridization, introgression, and incomplete lineage sorting. Lespedeza belongs to tribe Desmodieae subtribe Lespedezinae. It comprises 45 herbaceous and shrubby species dispersed throughout East Asia and eastern North America (44 spp. in Ohashi and Nemoto48; one additional sp. in Xu et al.)49. Lespedeza is traditionally divided into two subgenera, i.e., subg. Lespedeza and subg. Macrolespedeza (Maxim.) H. Ohashi50,51,52. As traditionally defined, species of subg. Macrolespedeza are shrubs with only chasmogamous flowers while those of subg. Lespedeza are herbs or subshrubs with both cleistogamous and chasmogamous flowers. Geographically, species of subg. Lespedeza occur disjunctly in eastern North America and East Asia, while those of subg. Macrolespedeza are limited to East Asia. The two-subgenus classification based on macromorphology is supported by pollen features53. However, neither of the subgenera is recovered as monophyletic by a study based on one nuclear and five plastid markers; instead, the American species and Asian species of Lespedeza each were resolved as monophyletic and sister to each other51. Most recently, Ohashi and Nemoto48 proposed a new infrageneric system which confined subg. Lespedeza to North America, and subg. Macrolespedeza to Asia mainly based on the results of recent molecular work51.
Based on ITS and five plastid markers, Xu et al.51 found that the nuclear and plastid markers contained obviously incongruent phylogenetic signals at shallow-level phylogeny. Also, the resolution among species within the traditionally defined subg. Macrolespedeza was very poor. Surprisingly, multiple samples of the same species within subg. Macrolespedeza from different localities often did not group together based on either ITS or plastid markers. As we have known recently, polymorphic ITS copies within individuals have been identified in many different plant groups which may confound attempts to recover correct phylogenetic species relationships. Our preliminary study indicated that there is relatively high nrDNA ITS diversity within individuals of some species of Lespedeza, implying incomplete concerted evolution. Additionally, the uniparental inheritance of plastid gene tree reflects evolutionary processes other than those of phylogenetic descent which may mask evidence of reticulation in hybridizing species54,55. Either of the reasons may cause incongruence between the ITS and plastid phylogenies.
The objectives of this study included: (1) to thoroughly investigate the extent and pattern of intra- and inter-specific ITS diversity in Lespedeza; (2) to distinguish the presence and pattern of pseudogenes; (3) to reconstruct the reticulate evolution of species of Lespedeza; and (4) to compare newly obtained evolutionary relationships with complete distribution pattern of ITS clones to understand the impact of hybridization and introgression on the evolution of the ITS lineages and on phylogenetic reconstruction.
Results
A total of 555 sequences were newly generated for this study (Supplementary Information: Appendix).
Length variation and sequence diversity of ITS region
In total, 450 ITS clones were obtained from the 50 samples of Lespedeza. Two hundred ninety out of 450 clones were distinct clones. The entire length of ITS region (containing partial 18S and 26S) ranged from 587 (L. stuevei-3, including a 136-bp deletion in ITS1 and 5.8S) to 732 bp, and their alignment was 808 bp long, of which 546 (67.6%) were variable and 442 (54.7%) were potentially parsimony informative. The length of ITS1, 5.8S, ITS2 region varied from 127 (L. stuevei-3, including a 111-bp deletion) to 246 bp, 104 (L. patens-5, including a 59-bp deletion) to 163 bp, and 183 (L. caraganae-8, including a 30-bp deletion) to 218 bp, respectively.
GC content of the ITS region, secondary structure of 5.8S rDNA sequences and identification of pseudogenes and recombinants
As shown in Table S1 (Supplementary Information), the GC content of Lespedeza ranged from 46.55% to 67.06% in ITS1, 41.76% to 56.05% in 5.8S, and 46.43% to 69.48% in ITS2 region. Within the clones, 35 ITS sequences had significantly lower GC contents in at least one of three regions compared with that of others from the same sample. Most of the clones exhibited the predicted secondary structure for a functional 5.8S rRNA, with the presence of five conserved helices. However, 28 clones displayed non-compensatory substitutions and at least one helix could not be folded correctly. Thirty clones contained at least one substitution within one of the three conserved motifs. The nucleotide changes in three conserved motives of 5.8S rDNA accessions are summarized in Table 1. Based on the information of sequence length and substitution variation, GC content, presence of conserved motives in the 5.8S rDNA sequence, and ability of 5.8S rDNA sequence to fold into a conserved secondary structure, we identified 37 putative pseudogenes (Table 1). The sequence diversity across the ITS region of all putative pseudogenes, estimated either by the number of nucleotide differences (k) or by nucleotide diversity (Pi), was remarkably higher than the presumed functional sequences (Table 2).
Table 1. Sequence characteristics of putative ITS pseudogenes in Lespedeza.
| Accession name | GC content [%] | Position of nucleotide changes (nt-) in conserved 5.8S motives | Secondary structure of 5.8S | Note | ||||
|---|---|---|---|---|---|---|---|---|
| ITS1 | 5.8S | ITS2 | M1 | M2 | M3 | |||
| L. bicolor-1-3 | 62.30 | 55.41 | 67.14 | nt-16 | conserved | conserved | XXXXX | pseudogene |
| L. bicolor-1-4 | 63.52 | 50.96 | 63.85 | nt-16 | conserved | conserved | XXXXX | pseudogene |
| L. bicolor-1-5 | 61.89 | 53.50 | 62.19 | nt-12 | conserved | nt-6 | X-XX- | pseudogene |
| L. bicolor-1-6 | 62.30 | 50.96 | 63.38 | nt-16 | conserved | conserved | XXXXX | pseudogene |
| L. bicolor-2-8 | 64.75 | 55.41 | 64.73 | nt-4 | conserved | conserved | X-XXX | pseudogene |
| L. buergeri-3-4 | 65.16 | 52.87 | 67.61 | nt-16 | conserved | nt-8 | XXX-- | pseudogene |
| L. caraganae-8 | 52.23 | 45.86 | 52.46 | nt-12 | nt-7,12 | nt-7,9 | X-X-- | pseudogene |
| L. cuneata-2-2 | 65.59 | 54.78 | 64.79 | conserved | conserved | conserved | XXXXX | pseudogene |
| L. cuneata-2-5 | 61.18 | 52.87 | 61.03 | nt-6 | nt-8 | nt-8 | XXX-- | pseudogene |
| L. cyrtobotrya-3-5 | 63.34 | 55.41 | 68.54 | conserved | conserved | nt-8 | XXXXX | pseudogene |
| L. dunnii-2 | 63.22 | 55.41 | 62.39 | conserved | conserved | conserved | XXXXX | pseudogene |
| L. dunnii-6 | 59.49 | 55.41 | 64.79 | conserved | nt-6 | conserved | XXXXX | pseudogene |
| L. fordii-3-6 | 52.05 | 47.77 | 51.64 | nt-16 | nt-6,7 | nt-2,7 | ----- | pseudogene |
| L. fordii-3-8 | 46.55 | 41.76 | 46.43 | nt-1,12, 16 | nt-6,14 | nt-8,9,10 | X---- | pseudogene |
| L. fordii-3-9 | 51.64 | 47.77 | 51.64 | nt-16 | nt-6,7 | nt-2,7 | ----- | pseudogene |
| L. formosa-1-2 | 47.41 | 42.24 | 47.88 | nt-9,12, 16 | nt-6,14 | nt-8,9,10 | ----- | pseudogene |
| L. formosa-1-6 | 63.11 | 52.87 | 63.85 | conserved | nt-6 | conserved | XXXXX | pseudogene |
| L. formosa-1-10 | 60.25 | 50.96 | 61.03 | conserved | conserved | nt-9 | XX--- | pseudogene |
| L. formosa-4-3 | 63.11 | 53.50 | 65.10 | conserved | nt-7 | nt-1 | -XXXX | pseudogene |
| L. formosa-5-1 | 57.74 | 49.68 | 57.75 | conserved | nt-7 | nt-7 | XX--X | pseudogene |
| L. formosa-5-9 | 58.51 | 51.28 | 59.62 | nt-14 | conserved | nt-4 | XX-X- | pseudogene |
| L. formosa-5-10 | 58.51 | 51.28 | 59.62 | nt-14 | conserved | nt-4 | XX-X- | pseudogene |
| L. inschnica-2-2 | 61.48 | 52.23 | 63.85 | conserved | nt-7 | nt-8 | XXX-X | pseudogene |
| L. jiangxiensis-1-1 | 58.61 | 50.96 | 59.62 | nt-16 | conserved | conserved | XXXX- | pseudogene |
| L. jiangxiensis-1-6 | 58.61 | 54.03 | 66.67 | conserved | nt-1 | all missing | XXX-- | pseudogene |
| L. jiangxiensis-2-5 | 64.78 | 54.78 | 63.38 | conserved | conserved | conserved | XXXXX | pseudogene |
| L. lichiyuniae-3-5 | 60.08 | 51.59 | 58.69 | nt-16 | nt-7 | conserved | XXXXX | pseudogene |
| L. lichiyuniae-3-8 | 65.18 | 54.78 | 60.09 | conserved | conserved | conserved | XXXX- | pseudogene |
| L. patens-2 | 61.63 | 54.14 | 64.32 | conserved | conserved | conserved | XXXXX | pseudogene |
| L. patens-4 | 57.74 | 54.14 | 61.03 | conserved | conserved | conserved | XXXX- | pseudogene |
| L. patens-5 | 56.49 | 59.18 | 60.09 | missing 4-16 | all missing | nt-2 | ---XX | pseudogene |
| L. patens-6 | 58.09 | 49.68 | 57.28 | nt-16 | conserved | nt-7 | XX-XX | pseudogene |
| L. patens-10 | 54.20 | 42.31 | 47.02 | nt-9,12,16 | nt-6,14 | nt-8,9,10 | ----- | pseudogene |
| L. wilfordii-1-7 | 65.73 | 53.50 | 60.56 | conserved | conserved | conserved | XXXX- | pseudogene |
| L. wilfordii-2-1 | 61.89 | 51.92 | 60.09 | nt-14 | conserved | nt-4 | XXXX- | pseudogene |
| L. wilfordii-2-3 | 63.93 | 54.78 | 62.91 | conserved | conserved | conserved | XXXXX | pseudogene |
| L. wilfordii-2-7 | 59.43 | 51.92 | 59.62 | nt-14 | conserved | nt-4 | XXXX- | pseudogene |
| L. wilfordii-2-8 | 46.98 | 42.31 | 47.62 | nt-8,9,12, 16 | nt-6,14 | nt-8,9,10 | ----- | pseudogene |
| L. wilfordii-2-9 | 59.84 | 51.92 | 60.09 | nt-14 | conserved | nt-4 | XXXX- | pseudogene |
| L. wilfordii-3-3 | 54.51 | 51.59 | 59.62 | nt-2,11 | nt-9 | conserved | X-XXX | pseudogene |
Indicates the presence (x) or absence (−) of the ability to build up the helices (B4–B8), respectively.
Table 2. Nucleotide diversity of individual parts and the entire ITS region of Lespedeza analyzed.
| Region | ITS1 |
5.8S |
ITS2 |
ITS |
||||
|---|---|---|---|---|---|---|---|---|
| Sequence type | F | P | F | P | F | P | F | P |
| Pi | 0.067 | 0.139 | 0.005 | 0.154 | 0.061 | 0.179 | 0.038 | 0.146 |
| k | 7.357 | 23.827 | 0.473 | 12.765 | 10.614 | 27.376 | 18.825 | 57.927 |
F – presumed functional ITS paralogs; P – putative ITS pseudogenes; Pi – Nucleotide diversity; k – Average number of nucleotide differences.
Five putative pseudogenes (Lepedeza bicolor-1-6, L. jiangxiensis-2-5, L. lichiyuniae-3-8, L. patens-10, and L. wilfordii-1-7) were detected as recombinants with the RDP4 program. Another two recombinants (L. jiangxiensis-1-2, L. jiangxiensis-2-8) were identified by their sharp discontinuities in the patterns of sequence similarity and substitution pattern, though RDP4 did not identify these.
ITS phylogenetic analyses
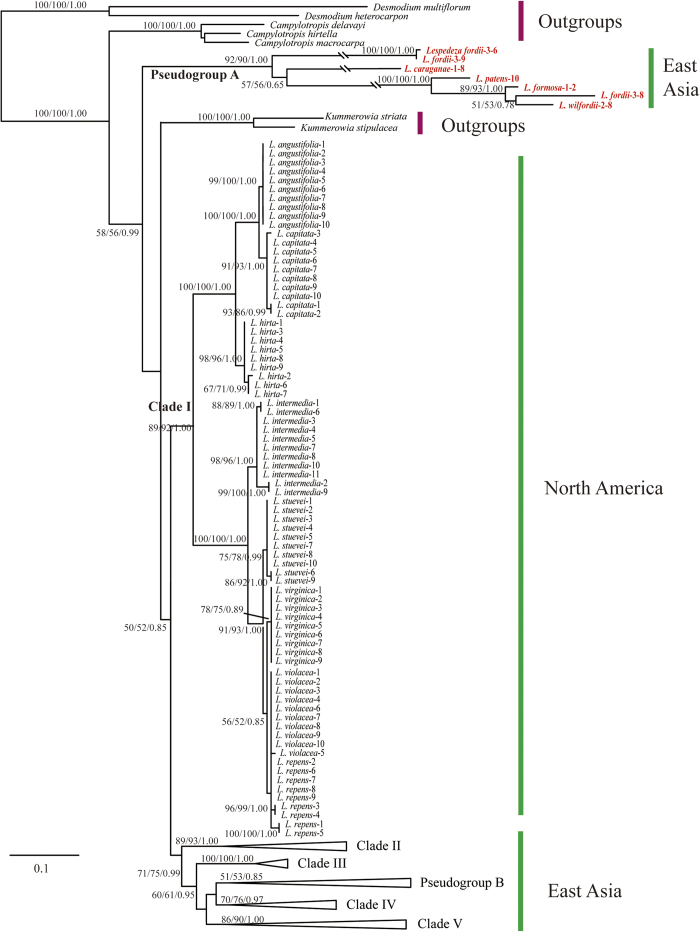
The ML analysis resulted in one optimal tree which is shown in Figs 1, 2, 3, 4, with ML bootstrap (LB), MP jackknife support values (PJ), and Bayesian posterior probabilities (PP). The MP and Bayesian tree topologies were similar (no well-supported conflicts) to the results from the ML analysis. Both Campylotropis and Kummerowia were strongly supported as monophyletic (LB = 100%, PJ = 100%, PP = 1.00), whereas Lespedeza was not resolved as monophyletic by influence of the putative pseudogenes. Within Lespedeza, seven major clades were identified (Fig. 1) which consisted of two pure pseudogene clades (Pseudogroups A, B) and five main functional gene clades (Clades I, II, III, IV, V). The relationships among Pseudogroup A, Kummerowia, and the rest of Lespedeza were unresolved. North American species of Lespedeza formed a well supported monophyletic group (Fig. 1: Clade I; LB = 89%, PJ = 92%, PP = 1.00) without any putative pseudogene, which was sister to the Asian species of Lespedeza except Pseudogroup A. Within the Asian species, six major clades were recovered and most of the relationships among them were unresolved (Fig. 1: Clades II, III, IV, V, Pseudogroups A, B).
Figure 1. Phylogram resulting from Maximum likelihood phylogenetic analysis of ITS cloning data.
There are seven main clades within Lespedeza (Clades I, II, III, IV, V, and Pseudogroups (A,B).They are expanded in Figs 2, 3, 4. Support values (maximum likelihood bootstrap support, maximum parsimony jackknife support and posterior probability) are shown along the branches. Dash shows that the bootstrap value or jackknife value is lower than 50. Pseudogenes are marked in red.
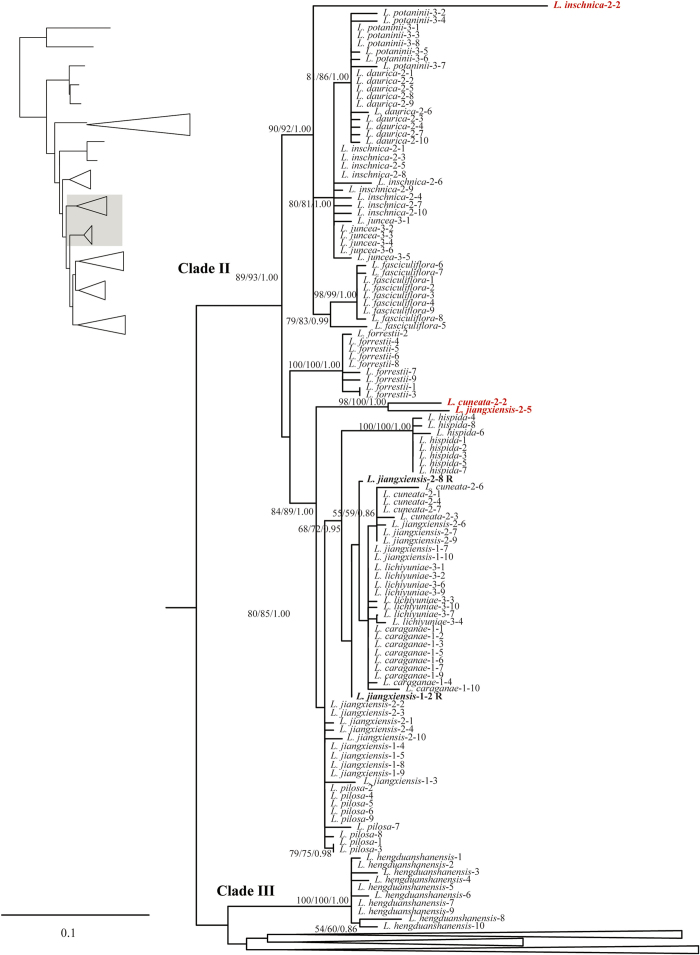
Figure 2. Detail of Clades II, III from Maximum likelihood phylogenetic analysis of ITS cloning data.
Support values (maximum likelihood bootstrap support, maximum parsimony jackknife support and posterior probability) are shown along the branches. Dash shows that the bootstrap value or jackknife value is lower than 50. Pseudogenes are marked in red and recombinants are labeled R.
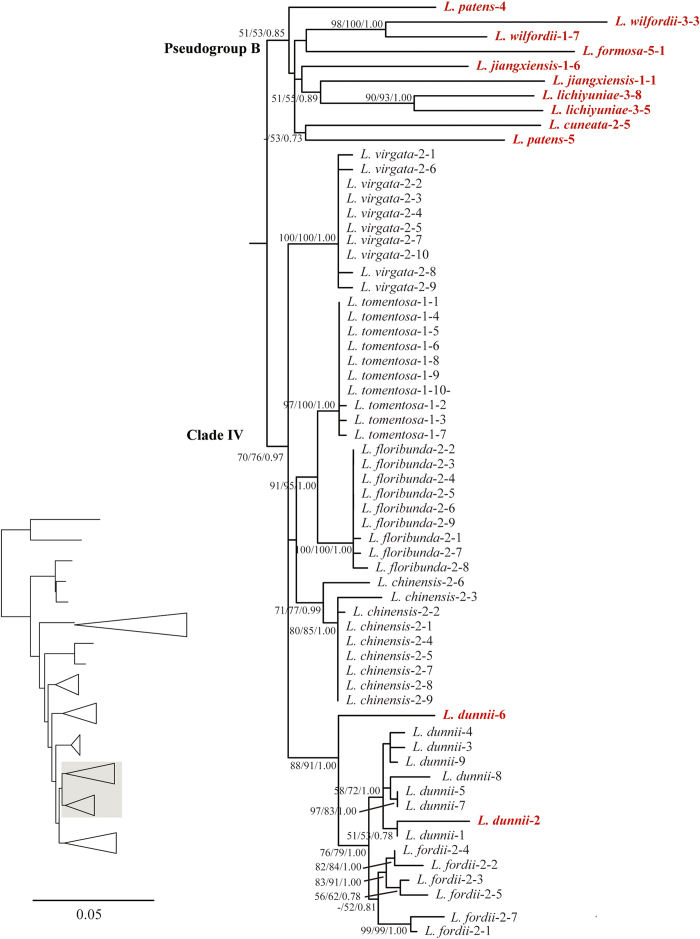
Figure 3. Detail of Clade IV, Pseudogroup B from Maximum likelihood phylogenetic analysis of ITS cloning data.
Support values (maximum likelihood bootstrap support, maximum parsimony jackknife support and posterior probability) are shown along the branches. Dash shows that the bootstrap value or jackknife value is lower than 50. Pseudogenes are marked in red.
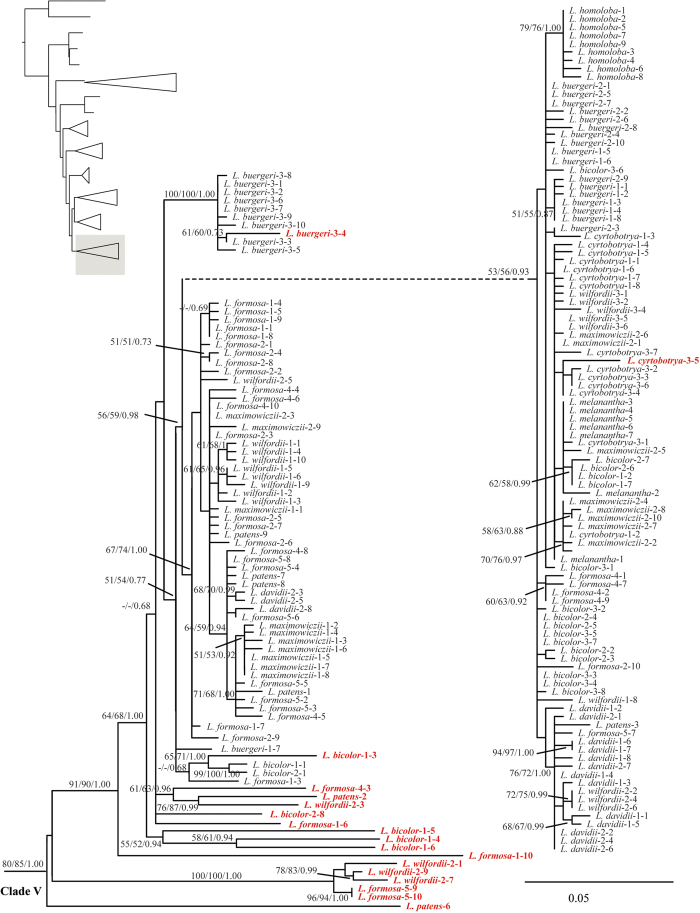
Figure 4. Detail of Clade V from Maximum likelihood phylogenetic analysis of ITS cloning data.
Support values (maximum likelihood bootstrap support, maximum parsimony jackknife support and posterior probability) are shown along the branches. Dash shows that the bootstrap value or jackknife value is lower than 50. Pseudogenes are marked in red.
PGK phylogenetic analyses
The final PGK dataset contained 105 accessions and 1,494 aligned nucleotides, of which 358 (24.0%) were variable and 198 (13.3%) were parsimony informative.
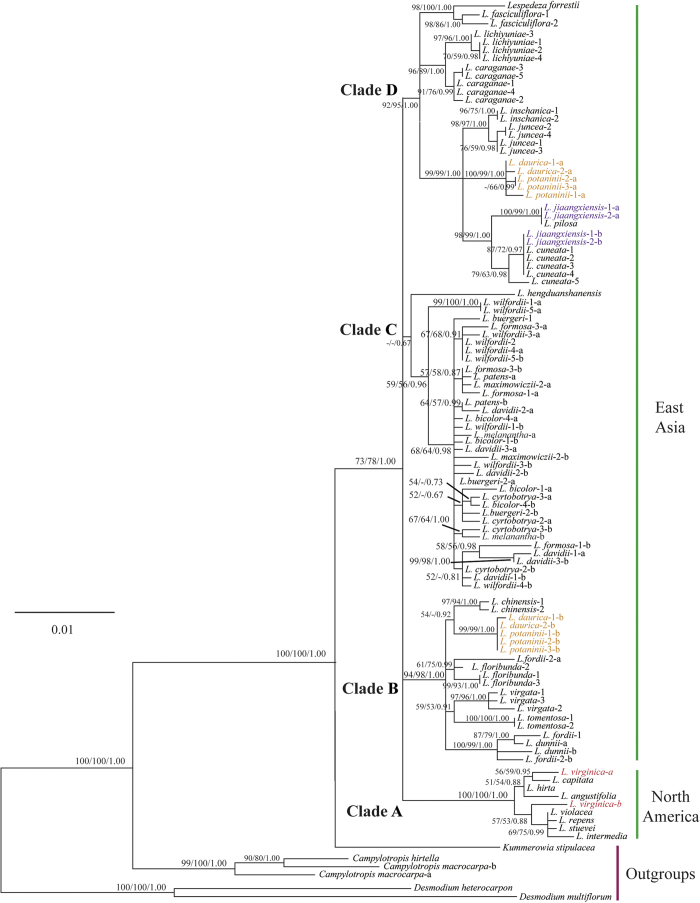
The phylogenetic analyses of the PGK dataset using MP, ML, and BI methods showed similar topologies. Lespedeza was resolved as a monophyletic group with moderate to strong support (LB = 73%, PJ = 78%, PP = 1.00) and is sister to Kummerowia and these two together are further sister to Campylotropis. Within Lespedeza, four major clades were identified (Fig. 5: Clades A, B, C, D). Three of the them were well supported (LB ≥ 92%, PJ = ≥ 95%, PP = 1.00), while one was weakly supported (PP = 0.67). North American species formed a well supported monophyletic group and further divided into two monophyletic subgroups with weak support (Fig. 5: Clade A). Within the Asian species, three major clades were recovered and their relationships were unresolved (Fig. 5: Clades B, C, D). Surprisingly, many samples show two divergent copies within an individual, e.g., L. daurica (Laxm.) Schindl., L. jiangxiensis Bo Xu, X.F.Gao & Li Bing Zhang, L. potaninii Vassiliev, and most samples of Clade C.
Figure 5. Phylogram resulting from Maximum likelihood phylogenetic analysis of PGK data.
There are four main clades within Lespedeza (Clades A,B,C,D). Support values (maximum likelihood bootstrap support, maximum parsimony jackknife support and posterior probability) are shown along the branches. Dash shows that the bootstrap value or jackknife value is lower than 50. Typical hybrid species are marked in multicolor.
Discussion
Characterization of ITS pseudogenes and recombinants in Lespedeza
In recent years, divergent ITS paralogs which may contain non-functional pseudogenes have been observed in more and more plant groups13,16,17,18,19,20,21,22,23. ITS pseudogenes can be identified with the GC content, secondary structure stability, substitution rates, the presence of conserved motifs, and phylogenetic positions6,19,21,22. In the present study, most ITS pseudogenes could be easily detected according to nucleotide changes in conserved 5.8S motives, the significantly lower GC contents in at least one of three regions, and the lost ability of 5.8S rDNA sequence to fold into a conserved secondary structure. As expected, all ITS pseudogenes in Lespedeza accumulated methylation related substitutions, which reduced the GC content and led to the loss of gene function. In agreement with other studies19, the putative pseudogenes in Lespedeza showed remarkably increased nucleotide substitutions and sequence diversity compared with presumed functional sequences.
Our phylogenetic results suggested that the putative pseudogenes in Lespedeza are mainly divided into three groups, i.e., Pseudogroup A (LB = 92%, PJ = 90%, PP = 1.00), Pseudogroup B (LB = 51%, PJ = 53%, PP = 0.85), and some others are mixed with functional copies. Pseudogroups A and B formed a large genetic differentiation with functional copies, suggesting that these pseudogenes originated with a long history. There are three different distribution patterns for the mixed pseudogenes: 1) The differentiation between pseudogenes and their functional copies is relatively small and they are still resolved in a monophyletic clade in the phylogenetic analysis, e.g., L. buergeri-3-4, L. cyrtobotrya-3-5 (Fig. 4), suggesting that their origins did not predate the divergence of the species; 2) The differentiation between pseudogenes and their functional copies is relatively large and the pseudogenes are resolved in a separate clade, e.g., L. inschnica-2-2, L. formosa-1-10 (Figs 2 and 4); and 3) The pseudogenes from different species form a well-supported clade probably due to the result of long-branch attraction, e.g., L. cuneata-2-2, L. jiangxiensis-2-5 (Fig. 2).
For the recombinants in Lespedeza detected by the RDP4 program, most of their putative parent sequences were also found to be pseudogenes. The putative recombination mainly occur between non-functional pseudogenes maybe due to their early origin and accumulation of high-level homoplastic mutations. The two recombinants (L. jiangxiensis-1-2, L. jiangxiensis-2-8; Fig. 2) identified by their sharp discontinuities are resolved in basal positions to either parental lineage, which indicates that they may have resulted in the loss of clade resolution in the phylogenetic tree and obscure the real phylogenetic relationships. However, we could not distinguish the recombinants by PCR-mediated recombination from those by natural recombination.
Incomplete concerted evolution of ITS in Lespedeza
Despite the concerted evolution of rDNA, polymorphic ITS copies within individuals have been identified in many different plant groups10,16,18,19,20,23. The high-level intra-individual polymorphism (290 out of 446) in the ITS region in Lespedeza suggests that incomplete concert evolution exists in this genus. Divergent paralogs of ITS sequences, including putative pseudogenes, multiple functional ITS copies, and probably also recombinants, were detected in the same individual in our study.
Interestingly, the resolution patterns of the putative functional clones were highly different between the two traditional subgenera. Within subg. Macrolespedeza, the clones from most species were mixed together, suggesting low rate of concerted evolution in this subgenus. In contrast, clones from each species of subg. Lespedeza each formed a monophyletic group or unresolved, implying a higher rate of concerted evolution in the subgenus. The distinct resolution patterns of the ITS clones in the two subgenera can be attributed to their different evolutionary histories. Several biological processes might retard or disrupt concert evolution, such as polyploidization56,57, agamospermy58, multiple nucleolar organizer regions (NORs;24,57,59,60, longer generation times61, and hybridization17,20,25,58. In Lespedeza, all taxa investigated so far except L. daurica and L. potaninii are diploid62,63,64 and no agamospermous taxa have been reported. The number of NORs in Lespedeza is currently unknown, but most diploid angiosperms have only one or two NORs per genome38,65,66. Thus, polyploidization, agamospermy, and a large number of NORs may not be the major reasons for the intra-individual polymorphism and great difference of resolution patterns of the ITS clones in the two subgenera. A long generation time has been suggested as a mechanism that might retard concerted evolution in Paeonia61. Within Lespedeza, members of subg. Lespedeza are herbs or subshrubs, while those of subg. Macrolespedeza are shrubs. Although we do not have any detailed information on the generation times of the two subgenera, normally shrubs have relatively longer generation times than herbs and subshrubs. Therefore, different generation time may have been one of the reasons responsible for the different resolution patterns of the ITS clones in the two subgenera. Hybridization could be accounted for the high-level intra-individual polymorphism of ITS found in many other plant groups, including Amelanchier Medik58, Iris L.67, Musaceae20, Paeonia L.61, Platanus L.17, Pyrus L.18, and Quercus L.25. Since interspecific hybridization is well-known within Lespedeza45,46,47, it appears to be plausible to assume that hybridization may have created the similar high-level intra-individual polymorphism of ITS in the genus. The great difference between the resolution patterns of the ITS clones in the two subgenera could be attributed to their different extents/frequencies of hybridization. This inference is further corroborated by the analysis of nuclear gene PGK which shows rampant hybridization may have played a key role in the evolution of subg. Macrolespedeza.
Phylogeny of Lespedeza
In our present phylogenetic analyses based on ITS paralogs, Lespedeza is not resolved as monophyletic (Fig. 1) because of the putative pseudogenes, whereas it is resolved as a monophyletic group with better support based on PGK gene data than it was in our previous study based on ITS and plastid loci51. North American species are strongly supported as monophyletic based on PGK gene data, in accordance with previous studies. In contrast, the monophyly of the Asian taxa is not recovered based on PGK gene data but resolved as paraphyletic in relation to the North American species and Kummerowia, different from previous studies which resolved the Asian species as monophyletic and sister to the North American species51,52. The PGK data do not support Ohashi & Nemoto48 classification which recognized the Asian and North American species as two subgenera. Notably, monophyletic groups are generally recognized, while some authors advocated for paraphyletic groups.
The major clades in Lespedeza identified with previous ITS and plastid data are recovered in the analyses of PGK data (Fig. 5: Clades A, B, C, D). However, PGK data are found to provide better resolution among the species complexes, especially within Clade D (Fig. 5). For example, the relationships among L. caraganae Bunge, L. cuneata (Dum. Cours.) G. Don, L. inschanica Schindl., L. juncea Pers., and L. lichiyuniae Nemoto & Ohashi were unresolved based on previous ITS and plastid data51, but are well resolved based on the PGK data. In the PGK gene tree (Fig. 5), multiple accessions contain two divergent copies within an individual, e.g., L. daurica, L. jiangxiensis, L. potaninii, and most accessions of Clade C, which is suggestive that frequent hybridization may have played an important role in the evolution of Lespedeza.
Hybridization within Lespedeza
Natural hybridization is recognized as an important creative force and evolutionary process in generating angiosperm species diversity26,27,68. The signature of hybridization can be revealed by phylogenetic incongruence between different markers, or the existence of two divergent alleles of single-copy nuclear gene within one individual. Our new data, as well as previous study51, suggest that several species or species complexes of Lespedeza may be of hybrid origin and hybridization may have played a critical role in the evolution of the genus. Hybrid speciation within Lespedeza occurs either at a homoploid level (between two species of the same ploidy) or via allopolyploidy (speciation via hybridization and genome doubling). Listed below are several examples of the hybridization within Lespedeza.
Lespedeza jiangxiensis was newly described from Jiangxi Province, China69. Our previous study51 based on ITS and five plastid markers identified L. pilosa Siebold & Zucc. as the closest relative of L. jiangxiensis. Interestingly, however, L. pilosa is remarkably different from L. jiangxiensis morphologically. In the present study, we have identified two divergent PGK copies in L. jiangxiensis (Fig. 5: Clade D). The two divergent copies grouped with L. cuneata and L. pilosa, respectively, indicating that L. jiangxiensis is a hybrid progeny formed between L. cuneata and L. pilosa. Furthermore, L. pilosa can be inferred as the female parent when analyzed in concert with plastid markers51.
Lespedeza virginica (L.) Britton is a North American species with purple flowers. In the earlier studies based on both ITS and plastid markers52,70, L. virginica formed a clade with L. intermedia (S. Watson ex A. Gray) Britton, L. procumbens Michx., L. repens (L.) W.P.C. Barton, L. stuevei Nutt., and L. violacea (L.) Pers., which all have purple flowers. In our previous study51, the position of L. virginica in the ITS tree was in accordance with that in earlier studies, but in our tree based on combined plastid data L. virginica grouped with L. angustifolia (Pursh) Elliott, L. capitata Michx., L. hirta (L.) Hornem., and L. leptostachya Engelm. ex A. Gray, which all have white flowers. This suggests that the plastids of our L. virginica sample are acquired from another taxon with white flowers (e.g., L. angustifolia, L. capitata, L. hirta, or L. leptostachya). This hypothesis is corroborated by our new nuclear PGK data in this study, which clearly indicate that our L. virginica sample is a hybrid progeny formed between L. virginica and L. capitata (Fig. 5: Clade A).
Lespedeza daurica and L. potaninii are only tetraploid species known so far in the genus62,63,71. The relationships of L. daurica and L. potaninii have been well discussed. Some taxonomists regarded L. potaninii as a subspecies of L. daurica or a species of its own50,72. However, Ohashi et al.73 considered L. potaninii being conspecific with L. daurica and treated L. potaninii as an ecological form growing in sunny dry areas. Our previous study51 showed that L. daurica and L. potaninii formed a monophyletic clade and were closely related with L. inschanica and L. juncea. In the present study (Fig. 5), all five accessions of L. daurica and L. potaninii each have two divergent copies and these two copies each formed a monophyletic clade (Fig. 5: Clades B, D), suggesting that L. daurica and L. potaninii originated from interbreeding between L. chinensis (weak support) and a species of Clade D (e.g., L. cuneata, L. inschanica, L. juncea). This hypothesis is reinforced by chromosome evidence. The chromosome number 2n = 22 is common throughout Clade B and 2n = 20 throughout Clade D, while the chromosome numbers of L. daurica and L. potaninii are 2n = 4262,63,71. Therefore, the chromosomal data well support the speciation of L. daurica and L. potaninii via hybridization between Clade B and Clade D resulting in genome doubling.
Subg. Macrolespedeza is characterized by having a woody habit and only chasmogamous flowers in compound racemes. In our previous study based on ITS and plastid data51, multiple accessions of the same species of subg. Macrolespedeza from different geographical localities often did not group together. It appears that multiple copies of some genes exist intensively in subg. Macrolespedeza. Surprisingly, this hypothesis is corroborated by our present study which shows that all the twenty samples of subg. Macrolespedeza except L. buergeri-1 and L. wilfordii-2 have two divergent PGK copies (Fig. 5: Clade C). Two reasons can be accounted for an individual having two copies of a single-copy nuclear gene: (1) It has undergone a gene duplication for this gene; and (2) It is a hybrid progeny while the two copies are different alleles from different parents. In the first case, if there is a gene duplication of PGK gene in the evolution of subg. Macrolespedeza, the divergent copies of the subgenus should be grouped into two distinct clades. However, in our study, the divergent copies from most species are mixed together on the tree (Fig. 5). Thus, gene duplication can not be the major factor for the remarkable presence of such divergent PGK gene copies in subg. Macrolespedeza. Notably, species of subg. Macrolespedeza are often geographically distributed sympatrically and have not yet been clearly distinguished from one another due to their lack of significant morphological characters and the presence of interspecific hybrids46,51,64. The two divergent copies from most species were mixed together on the tree, implying that widespread and frequent hybridization may have played a significant role in the evolution of subg. Macrolespedeza. This is consistent with the findings of previous studies on other groups of plants, which showed that hybridization is particularly likely to occur in plant species complexes in which species are evolutionarily young and sometimes not well differentiated32,33,34,35,36,37.
Parents homogenization of ITS
Our putative hybrid samples of L. daurica, L. potaninii, and L. virginica, based on PGK gene and plastid loci which only showed one parental repeat type in the analysis of ITS clones, indicating that the alternative repeat type may be lost due to the impact of concerted evolution which may have caused parents homogenization. In L. virginica, homogenization of polymorphism is biased towards paternal type copies. By contrast, in allopolyploid species of L. daurica and L. potaninii, all the samples show a biased homogenization towards maternal type repeats. Different directions, degrees, and patterns of concerted evolution have been reported in different plant groups especially among allopolyploids6,38,41,74,75. For example, in Cardamine L. (Brassicaceae), homogenization towards maternal alleles were uniform across multiple accessions of allopolyploid species74. Similarly, in most populations of both tetraploids, Tragopogon L. (Poaceae) showed a reduction in paternal rDNA homologs76. In contrast, in Nicotiana tabacum L. (Solanaceae), after hybridization of the parental diploid species, the paternal dominated its rDNA and the maternal rDNA repeat was then eliminated from the allopolyploid genome77. Meanwhile, bidirectional interlocus concerted evolution, in which repeats become homogenized to alternative progenitor diploids in different allopolyploid species, was also found in Gossypium L.38,75 and Oryza L.38,75. Despite the underlying driven mechanisms responsible for different unidirectional homogenization are not well understood in our study, the parents-biased homogenization of ITS following hybridization and polyploidization has very significant implications for phylogeny reconstruction, especially when based on rDNA sequences. If not well recognized, such biased homogenization following hybridization or polyploidization could lead to incorrect tree inference and erroneous estimates of the evolutionary relationships.
The impact of ITS evolution on phylogeny
The ITS region, as a spacer diverging at relatively high rate, is one of the most widely used molecular makers in phylogenetic inference of plants at generic and species levels. Since intra-individual polymorphisms and pseudogenes of ITS were discovered in Zea L.11, they have been increasingly detected in many plant groups15,16,17,18,19,20,21,23,25,58. Many of these studies show and discuss the phylogenetic consequences of the existence of pseudogenes. Zheng et al.18 found that functional ITS copies led to poorly resolved phylogeny as a result of low sequence divergence, while certain types of pseudogenes and some relict pseudogenes offered more credible clues for the evolutionary history of species of Pyrus (Rosaceae). Similar results have been obtained by Razafimandimbison et al.15 in Naucleeae (Rubiaceae), which clearly showed that divergent putative pseudogenes could be useful for phylogenetic analyses, especially when no sequences of their functional counterparts were available. Although the two examples above demonstrate that pseudogenes can be helpful to resolve phylogenetic relationships of closely related species to a certain extent, in most cases, non-concerted ITS evolution due to the existence of pseudogenes brings many difficulties, and even leads to misunderstanding of evolutionary relationships among taxa10,16,17,19,23,25,78,79. In our present study, divergent pseudogenes in Lespedeza accumulated high-level homoplastic mutations leading to random relationships among species partially due to long-branch attractions. Similar results were found in Brassicaceae and Cycas L.13,19. Recombination of divergent sequences following hybridization leads to chimeric DNA sequences which may also cause phylogenetic errors. Two recombinations of ITS (might have been resulted from PCR process though) are discovered in our putative hybrid species L. jiangxiensis and are resolved as basal to the parental lineages. Alvarez and Wendel6 reviewed the recombination of divergent sequences following hybridization in several different plants and found that this process may mask the real phylogenetic signals. As mentioned above, the parents-biased homogenization of ITS following hybridization and polyploidization can also result in incorrect tree inference and erroneous estimates of the evolutionary relationships. Therefore, when using ITS marker for reconstructing phylogeny and studying evolution of hybridization and polyploidization, one must be aware of parents homogenization and the impact of pseudogenes and recombinations.
Methods
Taxon sampling
In total, 50 samples representing 37 species of Lespedeza were cloned for nrDNA ITS sequence analyses and 71 samples representing 36 species of Lespedeza were included for PGK gene analyses. We also included two species of Kummerowia Schindl. [K. stipulacea (Maximowicz) Makino and K. striata (Thunberg) Schindl.], three species of Campylotropis Bunge [C. delavayi (Franch.) Schindl., C. hirtella (Franch.) Schindl., and C. macrocarpa (Bunge) Rehd.], and two species of Desmodium Desv. [D. microphyllum (Thunb.) DC. and D. heterocarpon (Linn.) DC.] as outgroups, following Xu et al.51. Most of our samples were collected in the field and in a few cases leaf material was directly taken from herbarium specimens. Voucher information and GenBank accession numbers for each accession are listed in Appendix.
DNA extraction, amplification, and sequencing
Total genomic DNA was extracted from silica-dried material or sometimes from herbarium fragments using the TIANGEN plant genomic DNA extraction kit (TIANGEN Biotech., Beijing, China) following the manufacturers’ protocols.
The internal transcribed spacer (ITS) and a single-copy nuclear gene coding for phosphoglycerate kinase (PGK) were used in this study. The ITS sequences were amplified with primers ITS4 and ITSA80. Those of PGK region were amplified with newly designed primers xb_pgk1F (GACAGTATT GGTCCAGAAGTAG) and xb_pgk1R (CAGAATCTCCTCCTCCAATAAT).
All PCR conditions followed Xu et al.51. Amplified fragments were purified with the TIANquick Mini Purification Kit (TIANGEN). Purified PCR products were sequenced by Invitrogen (Shanghai, China). If sequences had more than one heterozygous peak or could not be directly sequenced in PGK, cloning was performed. All the accessions of ITS within Lespedeza were cloned. Cloning was carried out using the pEASY-T3 Cloning Kit (TransGen Biotech, Beijing, China), following the manufacturer’s protocols. Finally, six to twelve positive clones were sequenced.
Sequence treatment, alignment and gap coding
Sequencher 4.1 (Gene Codes Corp., Ann Arbor, MI, USA) was used to assemble and edit complementary strands. PCR errors were corrected by comparing the cloned sequences with the original sequences from direct sequencing. Sequences obtained for each fragment were initially aligned using Clustal X 1.8181 and then manually adjusted in BioEdit82. Gap characters were coded following the modified complex indel coding83,84 for maximum parsimony (MP) and simple coding84 for the Bayesian Inference (BI) using SeqState85. Single-base repeats as well as ambiguously aligned indels were excluded.
ITS sequence analysis, secondary structure reconstruction, and recombination test
The boundaries of the ITS1, 5.8S and ITS2 regions were identified according to previously published ITS sequences from GenBank. GC content calculation and haplotype identification were performed using MEGA486. DnaSP87 was used to calculate the average number of nucleotide difference (K), the nucleotide diversity (p). Secondary structures of the 5.8S region were predicted under specific settings for base pairing (Hribova et al.20, for helix B4, F 45 105 3; helix B5, F 48 61 3; helix B6, F 69 96 3; helix B7, F 110 118 3; and for helix B8, F119 142 4 and F 126 135 3) using Mfold version 2.3 on a web server (http://unafold.rna.albany.edu/? q=mfold/RNA-Folding-Form2.3)88. For the 5.8S rDNA sequences, we inspected for the presence of three angiosperm conserved motifs: M1(5′-CGATGAAGAACGTAGC), M2 (5′-GAATTGCAGAATCC-3′), and M3 (5′-TTTGAACGCA-3′) following Harpke and Peterson89. Recombination Detection Program (RDP4)90 was used to detect putative recombinants.
Phylogenetic analysis
Unweighted maximum parsimony (MP) analyses were conducted for each locus in PAUP* ver. 4.0b1091 using 1000 tree-bisection-reconnection (TBR) searches with MAXTREES set to increase without limit. Parsimony jackknife (JK) analyses92 with heuristic search were conducted using PAUP* with the removal probability set to approximately 37%, and “jac” resampling emulated. One thousand replicates were performed with 10 TBR searches per replicate.
jModelTest v2.1.193 was used to select the best-fitting likelihood model for each dataset. The best-fitting models and parameter values selected using the Akaike Information Criterion94.
Maximum likelihood (ML)95 tree searches and ML bootstrapping were performed using RAxML-HPC2 on XSEDE v8.0.2496 on the web server Cipres Science Gateway97, with 5000 rapid bootstrap analyses followed by a search for the best-scoring tree in a single run.
Bayesian inference (BI) was conducted with mixed models using MrBayes v3.2.198. Two runs of four Markov chain Monte Carlo chains were conducted, each beginning with a random tree and sampling one tree every 1000 generations of 10,000,000 generations. Convergence among chains was checked using Tracer 1.599 and the first 25% was discarded as burn-in. The remaining trees were used to calculate a 50% majority-rule consensus topology and posterior probabilities (PP).
Additional Information
How to cite this article: Xu, B. et al. ITS non-concerted evolution and rampant hybridization in the legume genus Lespedeza (Fabaceae). Sci. Rep. 7, 40057; doi: 10.1038/srep40057 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We are grateful to Xue-Li Zhao, Zhang-Ming Zhu, Meng Li, and Li-Na Guo for their help with fieldwork. Special thanks go to Heng-Ning Deng for his assistance in the lab. This research was supported by the National Natural Science Foundation of China (Grant No. 31200169, 31570196). the China Postdoctoral Science Foundation (Grant No. 2012M511946), Project of Platform Construction for Plant Resources of Sichuan Province (2016TJPT0001-3), and a grant from the Fondation Franklinia to the Flora of China project.
Footnotes
Author Contributions B.X., X.-M.Z., X.-F.G. and L.-B.Z. designed the study; B.X., X.-F.G., D.-P.J. and L.-B.Z. collected the samples; B.X. performed all the experiments and statistical analyses; B.X. wrote the paper; X.-F.G., X.-M.Z. and L.-B.Z. edited the manuscript. All authors reviewed the manuscript and approved the manuscript for publication.
References
- Elder J. F. & Turner B. J. Concerted Evolution of Repetitive DNA-Sequences in Eukaryotes. Q. Rev. Biol. 70, 297–320 (1995). [DOI] [PubMed] [Google Scholar]
- Liao D. Q. Concerted evolution: Molecular mechanism and biological implications. Am. J. Hum. Genet. 64, 24–30, doi: 10.1086/302221 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley A. R. D. & Kobayashi T. Highly efficient concerted evolution in the ribosomal DNA repeats: Total rDNA repeat variation revealed by whole-genome shotgun sequence data. Genome Res. 17, 184–191, doi: 10.1101/Gr.5457707 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone T. et al. Bryophyte 5S rDNA was inserted into 45S rDNA repeat units after the divergence from higher land plants. Plant Mol. Biol. 41, 679–685, doi: 10.1023/A:1006398419556 (1999). [DOI] [PubMed] [Google Scholar]
- Baldwin B. et al. The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Ann. Mo. Bot. Gard. 82, 247–277 (1995). [Google Scholar]
- Alvarez I. & Wendel J. F. Ribosomal ITS sequences and plant phylogenetic inference. Mol. Phylogenet. Evol. 29, 417–434, doi: 10.1016/S1055-7903(03)00208-2 (2003). [DOI] [PubMed] [Google Scholar]
- Schoch C. L. et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 109, 6241–6246, doi: 10.1073/pnas.1117018109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D. Z. et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Natl. Acad. Sci. USA 108, 19641–19646, doi: 10.1073/pnas.1104551108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley A. R. D. & Kobayashi T. Monitoring the Rate and Dynamics of Concerted Evolution in the Ribosomal DNA Repeats of Saccharomyces cerevisiae Using Experimental Evolution. Mol. Biol. Evol. 28, 2883–2891, doi: 10.1093/molbev/msr117 (2011). [DOI] [PubMed] [Google Scholar]
- Mayol M. & Rossello J. A. Why nuclear ribosomal DNA spacers (ITS) tell different stories in Quercus. Mol. Phylogenet. Evol. 19, 167–176, doi: 10.1006/mpev.2001.0934 (2001). [DOI] [PubMed] [Google Scholar]
- Buckler E. S. & Holtsford T. P. Zea ribosomal repeat evolution and substitution patterns. Mol. Biol. Evol. 13, 623–632 (1996). [DOI] [PubMed] [Google Scholar]
- Hartmann S., Nason J. D. & Bhattacharya D. Extensive ribosomal DNA genic variation in the columnar cactus Lophocereus. J. Mol. Evol. 53, 124–134 (2001). [DOI] [PubMed] [Google Scholar]
- Bailey C. D., Carr T. G., Harris S. A. & Hughes C. E. Characterization of angiosperm nrDNA polymorphism, paralogy, and pseudogenes. Mol. Phylogenet. Evol. 29, 435–455, doi: 10.1016/j.ympev.2003.08.021 (2003). [DOI] [PubMed] [Google Scholar]
- Popp M. & Oxelman B. Evolution of a RNA polymerase gene family in Silene (Caryophyllaceae) - Incomplete concerted evolution and topological congruence among paralogues. Syst. Biol. 53, 914–932, doi: 10.1080/10635150490888840 (2004). [DOI] [PubMed] [Google Scholar]
- Razafimandimbison S., Kellogg E. & Bremer B. Recent Origin and Phylogenetic Utility of Divergent ITS Putative Pseudogenes: A Case Study from Naucleeae (Rubiaceae). Syst. Biol. 53, 177–192, doi: 10.1080/10635150490423278 (2004). [DOI] [PubMed] [Google Scholar]
- Harpke D. & Peterson A. Non-concerted ITS evolution in Mammillaria (Cactaceae). Mol. Phylogenet. Evol. 41, 579–593, doi: 10.1016/j.ympev.2006.05.036 (2006). [DOI] [PubMed] [Google Scholar]
- Grimm G. W. & Denk T. ITS evolution in Platanus (Platanaceae): Homoeologues, pseudogenes and ancient hybridization. Ann. Bot. 101, 403–419, doi: 10.1093/Aob/Mcm305 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X. Y., Cai D. Y., Yao L. H. & Teng Y. W. Non-concerted ITS evolution, early origin and phylogenetic utility of ITS pseudogenes in Pyrus. Mol. Phylogenet. Evol. 48, 892–903 (2008). [DOI] [PubMed] [Google Scholar]
- Xiao L. Q., Moller M. & Zhu H. High nrDNA ITS polymorphism in the ancient extant seed plant Cycas: Incomplete concerted evolution and the origin of pseudogenes. Mol. Phylogenet. Evol. 55, 168–177, doi: 10.1016/j.ympev.2009.11.020 (2010). [DOI] [PubMed] [Google Scholar]
- Hribova E. et al. The ITS1-5.8S-ITS2 Sequence Region in the Musaceae: Structure, Diversity and Use in Molecular Phylogeny. Plos One 6, doi: ARTN e17863 10.1371/journal.pone.0017863 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queiroz C. D., Batista F. R. D. & de Oliveira L. O. Evolution of the 5.8S nrDNA gene and internal transcribed spacers in Carapichea ipecacuanha (Rubiaceae) within a phylogeographic context. Mol. Phylogenet. Evol. 59, 293–302, doi: 10.1016/j.ympev.2011.01.013 (2011). [DOI] [PubMed] [Google Scholar]
- Li Y., Jiao L. & Yao Y. J. Non-concerted ITS evolution in fungi, as revealed from the important medicinal fungus Ophiocordyceps sinensis. Mol. Phylogenet. Evol. 68, 373–379, doi: 10.1016/j.ympev.2013.04.010 (2013). [DOI] [PubMed] [Google Scholar]
- Xu J. et al. Polymorphism and evolution of ribosomal DNA in tea (Camellia sinensis, Theaceae). Mol. Phylogenet. Evol. 89, 63–72, doi: 10.1016/j.ympev.2015.03.020 (2015). [DOI] [PubMed] [Google Scholar]
- Won H. & Renner S. S. The internal transcribed spacer of nuclear ribosomal DNA in the gymnosperm Gnetum. Mol. Phylogenet. Evol. 36, 581–597, doi: 10.1016/j.ympev.2005.03.011 (2005). [DOI] [PubMed] [Google Scholar]
- Muir G., Fleming C. C. & Schlotterer C. Three divergent rDNA clusters predate the species divergence in Quercus petraea (matt.) liebl. and Quercus robur L. Mol. Biol. Evol. 18, 112–119 (2001). [DOI] [PubMed] [Google Scholar]
- Yakimowski S. B. & Rieseberg L. H. The Role of Homoploid Hybridization in Evolution: A Century of Studies Synthesizing Genetics and Ecology. Am. J. Bot. 101, 1247–1258, doi: 10.3732/Ajb.1400201 (2014). [DOI] [PubMed] [Google Scholar]
- Soltis P. S. & Soltis D. E. The Role of Hybridization in Plant Speciation. Annu. Rev. Plant Biol. 60, 561–588, doi: 10.1146/annurev.arplant.043008.092039 (2009). [DOI] [PubMed] [Google Scholar]
- Wissemann V. Plant evolution by means of hybridization. Syst. Biodivers. 5, 243–253, doi: 10.1017/S1477200007002381 (2007). [DOI] [Google Scholar]
- Vriesendorp B. & Bakker F. T. Reconstructing patterns of reticulate evolution in angiosperms: what can we do? Taxon 54, 593–604 (2005). [Google Scholar]
- Mallet J. Hybridization as an invasion of the genome. Trends Ecol. Evol. 20, 229–237, doi: 10.1016/j.tree.2005.02.010 (2005). [DOI] [PubMed] [Google Scholar]
- Ellstrand N. C., Whitkus R. & Rieseberg L. H. Distribution of spontaneous plant hybrids. Proc. Natl. Acad. Sci. USA 93, 5090–5093, doi: 10.1073/pnas.93.10.5090 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy T., Cole L. W., Chang T. H. & Lindqvist C. Untangling reticulate evolutionary relationships among New World and Hawaiian mints (Stachydeae, Lamiaceae). Mol. Phylogenet. Evol. 89, 46–62, doi: 10.1016/j.ympev.2015.03.023 (2015). [DOI] [PubMed] [Google Scholar]
- Guggisberg A., Mansion G. & Conti E. Disentangling Reticulate Evolution in an Arctic-Alpine Polyploid Complex. Syst. Biol. 58, 55–73, doi: 10.1093/sysbio/syp010 (2009). [DOI] [PubMed] [Google Scholar]
- Frajman B., Eggens F. & Oxelman B. Hybrid Origins and Homoploid Reticulate Evolution within Heliosperma (Sileneae, Caryophyllaceae)—A Multigene Phylogenetic Approach with Relative Dating. Syst. Biol. 58, 328–345, doi: 10.1093/sysbio/syp030 (2009). [DOI] [PubMed] [Google Scholar]
- Chang Y. F., Li J., Lu S. G. & Sehneider H. Species diversity and reticulate evolution in the Asplenium normale complex (Aspleniaceae) in China and adjacent areas. Taxon 62, 673–687 (2013). [Google Scholar]
- Russell A. et al. Reticulate evolution in diploid and tetraploid species of Polystachya (Orchidaceae) as shown by plastid DNA sequences and low-copy nuclear genes. Ann. Bot. 106, 37–56, doi: 10.1093/aob/mcq092 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta J. H., Ebihara A. & Ito M. Reticulate evolution in the Crepidomanes minutum species complex (Hymenophyllaceae). Am. J. Bot. 98, 1782–1800, doi: 10.3732/ajb.1000484 (2011). [DOI] [PubMed] [Google Scholar]
- Wendel J. F., Schnabel A. & Seelanan T. Bidirectional Interlocus Concerted Evolution Following Allopolyploid Speciation in Cotton (Gossypium). Proc. Natl. Acad. Sci. USA 92, 280–284, doi: 10.1073/pnas.92.1.280 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase M. W. et al. Molecular systematics, GISH and the origin of hybrid taxa in Nicotiana (Solanaceae). Ann. Bot. 92, 107–127, doi: 10.1093/Aob/Mcg087 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauscher J. T., Doyle J. J. & Brown A. H. D. Internal transcribed spacer repeat-specific primers and the analysis of hybridization in the Glycine tomentella (Leguminosae) polyploid complex. Mol. Ecol. 11, 2691–2702, doi: 10.1046/j.1365-294X.2002.01640.x (2002). [DOI] [PubMed] [Google Scholar]
- Aguilar J. F., Rossello J. A. & Feliner G. N. Nuclear ribosomal DNA (nrDNA) concerted evolution in natural and artificial hybrids of Armeria (Plumbaginaceae). Mol. Ecol. 8, 1341–1346 (1999). [DOI] [PubMed] [Google Scholar]
- Sang T. Utility of low-copy nuclear gene sequences in plant phylogenetics. Crit. Rev. Biochem. Mol. Biol. 37, 121–147, doi: 10.1080/10409230290771474 (2002). [DOI] [PubMed] [Google Scholar]
- Duarte J. M. et al. Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels. BMC Evol. Biol. 10, doi: 10.1186/1471-2148-10-61 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele P. R., Guisinger-Bellian M., Linder C. R. & Jansen R. K. Phylogenetic utility of 141 low-copy nuclear regions in taxa at different taxonomic levels in two distantly related families of rosids. Mol. Phylogenet. Evol. 48, 1013–1026, doi: 10.1016/j.ympev.2008.05.017 (2008). [DOI] [PubMed] [Google Scholar]
- Clewell A. F. Native North American species of Lespedeza (Leguminosae). Rhodora 68, 359–405 (1966). [Google Scholar]
- Akiyama S. & Ohba H. Studies on hybrids in the genus Lespedeza sect. Macrolespedeza (1) A putative hybrid between L. Buergeri Miq. and L. cyrtobotrya Miq. J. Jpn. Bot. 57, 232–240 (1982). [Google Scholar]
- Ohashi H. Desmodieae Tribe in Legumes of the World (ed. Lewis G., Schrire B., Mackinder B. & Lock M.) 433–453 (Royal Botanic Gardens, Kew, 2005). [Google Scholar]
- Ohashi H. & Nemoto T. A New System of Lespedeza (Leguminosae Tribe Desmodieae). J. Jpn. Bot. 89, 1–11 (2014). [Google Scholar]
- Xu B., Gao X.-F. & Zhang L.-B. Lespedeza hengduanshanensis comb. & stat. nov.(Leguminosae: Papilionoideae: Desmodieae) from the Hengduan Mountains of SW China. Phytotaxa 202, 165–168 (2015). [Google Scholar]
- Huang P.-H., Ohashi H. & Nemoto T. Lespedeza Michaux in Flora of China Vol. 10 (ed. Wu C.Y., Raven P.H. & Hong D. Y.) 302–311 (Science Press, 2010). [Google Scholar]
- Xu B., Wu N., Gao X.-F. & Zhang L.-B. Analysis of DNA sequences of six chloroplast and nuclear genes suggests incongruence, introgression, and incomplete lineage sorting in the evolution of Lespedeza (Fabaceae). Mol. Phylogenet. Evol. 62, 346–358, doi: 10.1016/j.ympev.2011.10.007 (2012). [DOI] [PubMed] [Google Scholar]
- Han J. E., Chung K.-H., Nemoto T. & Choi B.-H. Phylogenetic analysis of eastern Asian and eastern North American disjunct Lespedeza (Fabaceae) inferred from nuclear ribosomal ITS and plastid region sequences. Bot. J. Linn. Soc. 164, 221–235 (2010). [Google Scholar]
- Xu B., Gao X.-F., Wu N. & Zhang L.-B. Pollen diversity and its systematic implications in Lespedeza (Fabaceae). Syst. Bot. 36, 352–361 (2011). [Google Scholar]
- Soltis D. E. & Kuzoff R. K. Discordance between nuclear and chloroplast phylogenies in the Heuchera group (Saxifragaceae). Evolution 49, 727–742 (1995). [DOI] [PubMed] [Google Scholar]
- Okuyama Y. et al. Nonuniform concerted evolution and chloroplast capture: heterogeneity of observed introgression patterns in three molecular data partition phylogenies of Asian Mitella (Saxifragaceae). Mol. Biol. Evol. 22, 285–296 (2005). [DOI] [PubMed] [Google Scholar]
- Suh Y., Thien L. B., Reeve H. E. & Zimmer E. A. Molecular evolution and phylogenetic implications of internal transcribed spacer sequences of ribosomal DNA in Winteraceae. Am. J. Bot. 80, 1042–1055 (1993). [Google Scholar]
- Karvonen P. & Savolainen O. Variation and inheritance of ribosomal DNA in Pinus sylvestris L.(Scots pine). Heredity 71, 614–622 (1993). [Google Scholar]
- Campbell C. S., Wojciechowski M. F., Baldwin B. G., Alice L. A. & Donoghue M. J. Persistent nuclear ribosomal DNA sequence polymorphism in the Amelanchier agamic complex (Rosaceae). Mol. Biol. Evol. 14, 81–90 (1997). [DOI] [PubMed] [Google Scholar]
- Lubaretz O., Fuchs J., Ahne R., Meister A. & Schubert I. Karyotyping of three Pinaceae species via fluorescent in situ hybridization and computer-aided chromosome analysis. Theor. Appl. Genet. 92, 411–416 (1996). [DOI] [PubMed] [Google Scholar]
- Quijada A., Liston A., Delgado P., Vázquez-Lobo A. & Alvarez-Buylla E. Variation in the nuclear ribosomal DNA internal transcribed spacer (ITS) region of Pinus rzedowskii revealed by PCR-RFLP. Theor. Appl. Genet. 96, 539–544 (1998). [DOI] [PubMed] [Google Scholar]
- Sang T., Crawford D. J. & Stuessy T. F. Documentation of Reticulate Evolution in Peonies (Peonia) Using Internal Transcribed Spacer Sequences of Nuclear Ribosomal DNA - Implications for Biogeography and Concerted Evolution. Proc. Natl. Acad. Sci. USA 92, 6813–6817, doi: 10.1073/pnas.92.15.6813 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce W. P. Cytology of the genus Lespedeza. Am. J. Bot. 29, 736–744 (1939). [Google Scholar]
- Xia Y.-Q., Su J.-K. & Xiong D.-S. The investigation on the karyotypes of ten species of Lespedeza. Grassl. China 2, 27–34 (1989). [Google Scholar]
- Akiyama S. A revision of the genus Lespedeza section Macrolespedeza (Leguminosae). Bull. Univ. Mus. Univ. Tokyo 33, 1–170 (1988). [Google Scholar]
- Long E. O. & Dawid I. B. Repeated genes in eukaryotes. Annu. Rev. Biochem. 49, 727–764 (1980). [DOI] [PubMed] [Google Scholar]
- Rogers S. O. & Bendich A. J. Ribosomal RNA genes in plants: variability in copy number and in the intergenic spacer. Plant Mol. Biol. 9, 509–520 (1987). [DOI] [PubMed] [Google Scholar]
- Arnold M. L., Bennett B. D. & Zimmer E. A. Natural hybridization between Iris fulva and Iris hexagona: pattern of ribosomal DNA variation. Evolution 40, 1512–1521 (1990). [DOI] [PubMed] [Google Scholar]
- Rieseberg L. H. The Role of Hybridization in Evolution - Old Wine in New Skins. Am. J. Bot. 82, 944–953 (1995). [Google Scholar]
- Xu B., Gao X.-F. & Zhang L.-B. Lespedeza jiangxiensis, sp. nov.(Fabaceae) from China based on Molecular and Morphological Data. Syst. Bot. 38, 118–126 (2013). [Google Scholar]
- Nemoto T., Yokoyama J., Fukuda T., Iokawa Y. & Ohashi H. Phylogeny of Lespedeza (Leguminosae) Based on Chloroplast trnL-trnF Sequences. J. Jpn. Bot. 85, 213–229 (2010). [Google Scholar]
- Yan G., Zhang S., Yan J., Fu X. & Wang L. Chromosome numbers and geographical distribution of 68 species of forage plants. Grassl. China 1989, 53–60 (1989). [Google Scholar]
- Chen C.-J. Lespedeza Michaux in Vascular plants of the Hengduan Mountains Vol. 1 (ed. Wang W.-T. & Wu S.-G.) 986–989 (Science Press, 1993). [Google Scholar]
- Ohashi H., Nemoto T. & Ohashi K. A revision of Lespedeza subgenus Lespedeza (Leguminosae) of China. J. Jpn. Bot. 84, 143–166 (2009). [Google Scholar]
- Zozomová-Lihová J. et al. When fathers are instant losers: homogenization of rDNA loci in recently formed Cardamine× schulzii trigenomic allopolyploid. New Phytol. 203, 1096–1108 (2014). [DOI] [PubMed] [Google Scholar]
- Bao Y., Wendel J. F. & Ge S. Multiple patterns of rDNA evolution following polyploidy in Oryza. Mol. Phylogenet. Evol. 55, 136–142 (2010). [DOI] [PubMed] [Google Scholar]
- Kovarik A. et al. Rapid concerted evolution of nuclear ribosomal DNA in two Tragopogon allopolyploids of recent and recurrent origin. Genetics 169, 931–944 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov R. A., Borisjuk N. V., Panchuk I. I., Schweizer D. & Hemleben V. Elimination and rearrangement of parental rDNA in the allotetraploid Nicotiana tabacum. Mol. Biol. Evol. 16, 311–320 (1999). [DOI] [PubMed] [Google Scholar]
- Buckler E. S., Ippolito A. & Holtsford T. P. The evolution of ribosomal DNA: Divergent paralogues and phylogenetic implications. Genetics 145, 821–832 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner D. L. & Banik M. T. Intragenomic variation in the ITS rDNA region obscures phylogenetic relationships and inflates estimates of operational taxonomic units in genus Laetiporus. Mycologia 103, 731–740, doi: 10.3852/10-331 (2011). [DOI] [PubMed] [Google Scholar]
- White T. J., Bruns T., Lee S. & Taylor J. In PCR protocols: a guide to methods and applications (ed. Innis M. A., Gefland D. H., Sninsky J. J. & White T. J.) 315–322 (Academic Press Inc., 1990). [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F. & Higgins D. G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic acids symposium series 95–98 (1999).10780396 [Google Scholar]
- Müller K. Incorporating information from length-mutational events into phylogenetic analysis. Mol. Phylogenet. Evol. 38, 667–676 (2006). [DOI] [PubMed] [Google Scholar]
- Simmons M. P. & Ochoterena H. Gaps as characters in sequence-based phylogenetic analyses. Syst. Biol. 49, 369–381 (2000). [PubMed] [Google Scholar]
- Müller K. SeqState. Appl. Bioinformatics 4, 65–69 (2005). [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M. & Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 (2007). [DOI] [PubMed] [Google Scholar]
- Librado P. & Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (2009). [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpke D. & Peterson A. 5.8 S motifs for the identification of pseudogenic ITS regions. Botany 86, 300–305 (2008). [Google Scholar]
- Martin D. P., Murrell B., Golden M., Khoosal A. & Muhire B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evolution 1, vev003, doi: 10.1093/ve/vev003 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D. PAUP 4.0 b10: Phylogenetic analysis using parsimony (Sinauer Associates, 2002). [Google Scholar]
- Farris J. S., Albert V. A., Källersjö M., Lipscomb D. & Kluge A. G. Parsimony jackknifing outperforms neighbor-joining. Cladistics 12, 99–124 (1996). [DOI] [PubMed] [Google Scholar]
- Darriba D., Taboada G. L., Doallo R. & Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Meth. 9, 772–772 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akaike H. A new look at the statistical model identification. IEEE Trans. Automat. Contr. 19, 716–723 (1974). [Google Scholar]
- Felsenstein J. Maximum likelihood and minimum-steps methods for estimating evolutionary trees from data on discrete characters. Syst. Biol. 22, 240–249 (1973). [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. A., Pfeiffer W. & Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE), 1–8, doi: 10.1109/GCE.2010.5676129 (2010). [DOI] [Google Scholar]
- Ronquist F. et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. & Drummond A. Tracer v1. 5. http://tree.bio.ed.ac.uk/software/ tracer/, doi: http://tree.bio.ed.ac.uk/software/tracer/ (2009).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.