Abstract
pH (Low) Insertion Peptides (pHLIP peptides) find application in studies of membrane-associated folding because spontaneous insertion of these peptides is conveniently triggered by varying pH. Here, we employed small-angle X-ray scattering (SAXS) to investigate a wild-type (WT) pHLIP peptide oligomeric state in solution at high concentrations and monitor changes in the liposome structure upon peptide insertion into the bilayer. We established that even at high concentrations (up to 300 µM) the WT pHLIP peptide at pH 8.0 does not form oligomers larger than tetramers (which exhibit concentration-dependent transfer to the monomeric state, as was shown previously). This finding has significance for medical applications when high concentration of the peptide is injected into blood and diluted in blood circulation. The interaction of WT pHLIP peptide with liposomes does not alter the unilamellar vesicle structure upon peptide adsorption by the lipid bilayer at high pH or upon insertion across the bilayer at low pH. At the same time, SAXS data clearly demonstrate the insertion of the peptide into the membrane at low pH, which opens the possibility of investigating the kinetic process of polypeptide insertion and exit from the membrane in real time by time-resolved SAXS.
Graphical Abstract
INTRODUCTION
Folding and insertion into a membrane of constitutive membrane proteins are facilitated by complex molecular machines in vivo, including the translocon that places most transmembrane (TM) helices across the bilayer.1–3 Although they are assisted in their pathways by the Get proteins,4 nonconstitutive membrane proteins (such as tail-anchored proteins) can spontaneously insert and fold themselves across the lipid bilayer of a membrane.5,6 They may do so when released by the Get complex in vivo. The molecular mechanisms of spontaneous polypeptide folding/insertion and exit/unfolding are of interest in several contexts, including the action of antimicrobial peptides, the folding and degradation of membrane proteins, and biotechnological/medical applications based on these processes.
Stability and folding of all membrane proteins, irrespective of the mechanism of their insertion into the membrane, are governed by the formation of polypeptide secondary structures in the lipid bilayer environment, driven by the hydrophobic interactions and by hydrogen bonding. A convenient experimental approach is to trigger a coil–helix transition and peptide insertion into a bilayer by a pH jump. The protonation enhances the hydrophobicity of the peptide and, therefore, its affinity to the nonpolar environment of a membrane. One such case of a synthetic peptide with pH-dependent membrane-insertion properties has been investigated by White and Ladokhin.7 Another example is the pH Low Insertion Peptide (pHLIP peptides) family, which is the subject of this study.
The wild-type (WT) pHLIP peptide was derived from the Chelix of bacteriorhodopsin.8 Later, many different pHLIP variants were introduced.9–15 All peptides share the same property: pH-dependent insertion into the lipid bilayer of a membrane, which is accompanied by a coil–helix transition and formation of a transmembrane (TM) helix. Importantly, the main principle of membrane-associated folding was employed to introduce a novel class of rationally designed pHLIP delivery agents. The pHLIP peptides have medical utility, as they can target acidic diseased tissues, such as tumors, and deliver therapeutic cargo molecules across membrane or target nanoparticles to cancer cells. The wide medical efficacy of pHLIP peptides is shown in imaging16–21 and therapeutic22–27 applications.
The peptides of the pHLIP family exist in three major states: state I is attributed to the peptides in a coil conformation in aqueous solution at >pH 7.4; if a membrane is introduced into the system, then the peptides stay in equilibrium between aqueous free and membrane-bound states, which is called state II. Finally, when pH is lowered (<pH 6.0), the Asp/Glu residues in pHLIP peptides are protonated, which enhances peptide hydrophobicity and promotes peptide partition into a bilayer, which is accompanied by folding. The system reaches the minimum of free energy when peptides adopt the TM helical conformation, which is assigned to state III. Thermodynamics and kinetics studies were carried out to establish the molecular mechanism of peptide’s adsorption by the bilayer (transition from state I to state II) and insertion into the membrane (transition from state II to state III).10,12,28,29 Our ultimate goal is to introduce a physical model that describes spontaneous insertion of a polypeptide into the anisotropic environment of a lipid bilayer and formation of a polypeptide helical structure. We have already taken the first step and proposed a mathematical formalism for describing the coil–helix transition of a polypeptide adsorbed at the membrane.30 To introduce thermodynamics and kinetics models, we have to have full knowledge about the conformational changes within both the polypeptide and membrane during peptide insertion and folding. In all previous studies, the main emphasis was on monitoring the conformational changes that occur in the pHLIP peptides. Here, we used small-angle X-ray scattering (SAXS) to monitor changes that might occur within the lipid bilayer of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) liposomes when the WT pHLIP peptide interacts with liposomes at high and low pH values. We selected to work with the WT pHLIP peptide and POPC liposome system because the system has been very intensively characterized previously, and changes in the SAXS signal could be attributed to particular events of peptide–membrane interactions.
X-ray and neutron scattering methods have been widely used to elucidate the nanostructure of vesicles and oriented membranes.31–33 These techniques are sensitive to both membrane thickness perturbations and lateral inhomogeneities.31,34 In the case of unilamellar vesicles, the mean radius, size polydispersity, average membrane thickness, and internal membrane structure can be derived by means of the separated form factor model.35,36 Here, we have used SAXS over an extended size scale to monitor changes in the unilamellar vesicle form factor and membrane nanostructure.
EXPERIMENTAL PROCEDURES
Peptide Preparation
WT pHLIP peptide (AEQNPIYWARYADWLFTTPLLLLDLALLVDADEGT) was synthesized and purified at the W.M. KECK Biotechnology center at Yale. The synthesized peptide was dissolved directly in buffer and then centrifuged to remove large aggregates. The concentration of the peptide was calculated spectrophotometrically by measuring the absorbance at 280 nm using the extinction coefficient ε280 = 13 940 M−1 cm−1.
Liposome Preparation
Large unilamellar and multilamellar vesicles were prepared by extrusion. POPC (Avanti Polar Lipids) was dissolved in chloroform, desolvated in a rotary evaporator, and dried under high vacuum for several hours. The phospholipid film was then rehydrated in 10 mM phosphate buffer, pH 8.0, vortexed until the lipid bilayer was completely dissolved, and extruded 51 times through the membranes with 50 nm pore size.
Steady-State Fluorescence and Circular Dichroism (CD)
Steady-state fluorescence, CD, and OCD measurements were carried out at 25 °C by on a PC1 spectrofluorometer (ISS Inc.) and a MOS 450 spectropolarimeter (Bio-Logic Inc.), respectively. The concentrations of the peptide and POPC lipids were 150 µM and 4.4 mM, respectively. Tryptophan fluorescence of the peptide was excited at 295 nm and recorded with the excitation and emission slits set at 1 nm. The polarizers in the excitation and emission paths were set at the “magic” angle (54.7° from the vertical orientation) and vertically (0°), respectively. Peptide CD spectra were recorded from 190 to 260 nm (where no PMT saturation was observed) with 0.5 nm increment using a cuvette with an optical path length of 0.5 cm.
SAXS
Synchrotron SAXS measurements were carried out at beamline ID02 of the ESRF in Grenoble, France.33 The incident X-ray wavelength (λ) was 0.995 Å, and the sample-to-detector distances were 3 and 1 m, covering a scattering vector range of 0.03–6 nm−1. The magnitude of the scattering vector, q, is defined as q = (4π/λ) sin(θ/2), with θ being the scattering angle. Samples were contained in a temperature-controlled (25 °C) flow-through quartz capillary cell with a diameter of 1.8 mm. The measured two-dimensional SAXS patterns were normalized to an absolute intensity scale using the standard procedure and azimuthally averaged to obtain the intensity profile as a function of q.33 The typical exposure time was 0.1 s (with an incident flux of 1013 photons/s), and 3–4 frames were acquired for each sample, which were subsequently averaged after excluding any possible radiation damage. The background buffer was also measured by the same procedure with 10 frames in each case. The averaged buffer background was subtracted from each averaged sample intensity profile to obtain the presented I(q) data. In different experiments, the concentration of the peptide was 150 or 300 µM, and that of POPC liposomes of nominal size of 50 nm was 4.4 or 8.8 mM. SAXS intensities were measured for original POPC liposomes and those incubated with a peptide at pH 8.0 and 5.0, as well as peptide alone in buffer. In addition, we carried out measurements with 100 and 200 nm liposomes: the overall trend was the same; however, their analysis is more complicated because of the presence of the multilamellar structure.
Analysis of SAXS Data
The background-subtracted scattered intensity from a suspension of particulate system can be expressed as
| (1) |
where N is the number of particles per unit volume, F(q)2 is the single particle scattering function whose amplitude is given by the Fourier transform of the radial electron density profile, and S(q) is the structure factor describing the interaction between particles. In this study, the suspensions are relatively dilute (volume fraction less than 0.01) and the interaction between particles can be neglected corresponding to S(q) ≈ 1. In the case of WT peptide in solution, F(q)2 is described by a geometric shape (cylinder) with a homogeneous scattering contrast.33 For unilamellar vesicles, the radius R, polydispersity, and the membrane internal structure can be simultaneously evaluated using the separated form factor model.31,32,35,36 The scattering contrast essentially comes from the bilayer region, and within the separated form factor model, F(q, R) is described by
| (2) |
where t is the bilayer thickness with the origin at the midplane (r = 0), ρ(r) is the radial electron density of the membrane and ρm that of the solvent, and re is the classical electron radius. By this definition, R is the distance from the center of the vesicle to the midplane of the bilayer. ρ(r) of the phospholipid membrane is approximately described by three Gaussian functions corresponding to the head group regions on both sides and middle hydrophobic chains.35,36
| (3) |
where ρ1 = ρ3 and ρ2 are the electron densities of the lipid head group and chain regions, respectively, Ri is the distance from the center of the group to the bilayer midplane, and σi is the width of the corresponding Gaussian function. The excess electron densities of the head group and the hydrophobic chains from the buffer, ρ1 − ρm and ρ2 − ρm, are denoted by ΔρH and ΔρC, respectively. The corresponding widths of the Gaussian function are represented by DH and DC, and the distance from the midplane of the bilayer to the center of the head group is denoted by XH.31,36
F(q, R) is given by the separated form factor within the thinshell assumption.35,36 However, real vesicles have a finite size distribution, and eq 1 needs to be weighted over the size distribution, f(R), which in this case is assumed to be the Schulz size distribution.33,36 In addition, S(q) ≈ 1 because the volume fraction of the unilamellar vesicles is of the order of 0.01 and POPC is not charged.
| (4) |
Model fits using eq 4 yield the mean radius (RV) and polydispersity (pV) of the unilamellar vesicles, size of the head group and tail regions, and the corresponding electron densities. It is assumed that the same geometric shape of the vesicle is maintained, but the height and widths of the three Gaussian functions in eq 3 change when the peptide interacts with the lipid bilayer of the membrane.
RESULTS
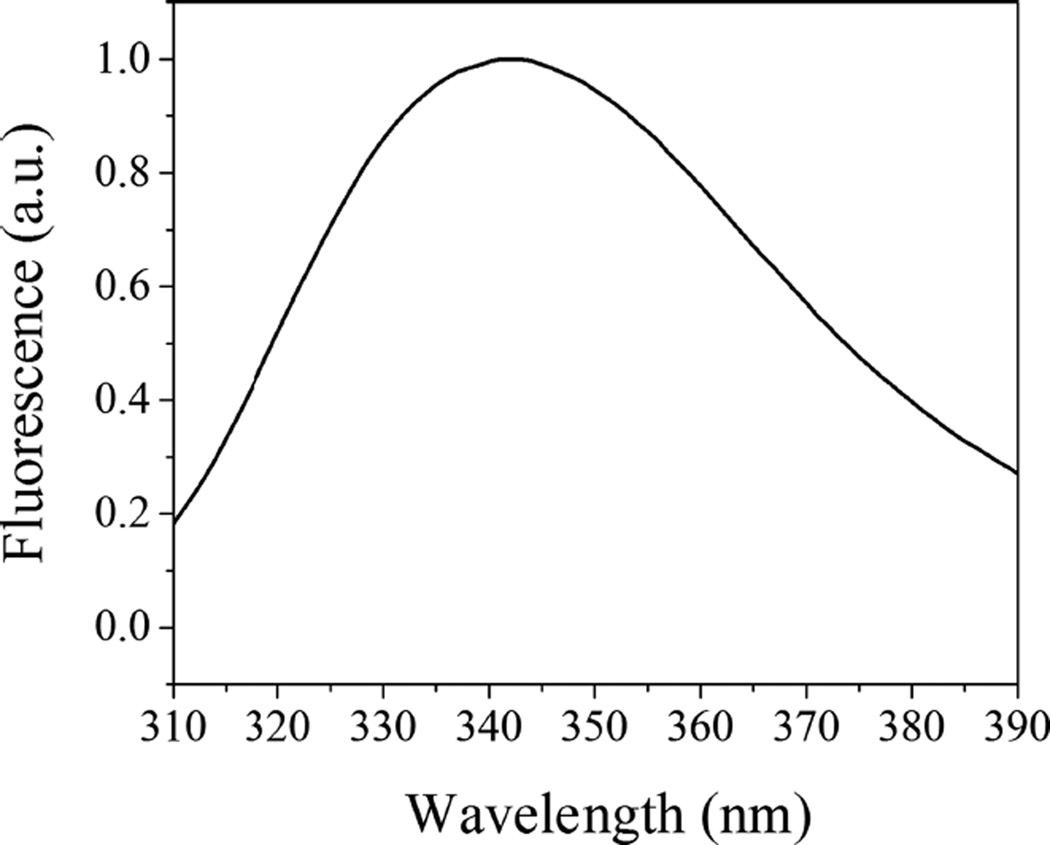
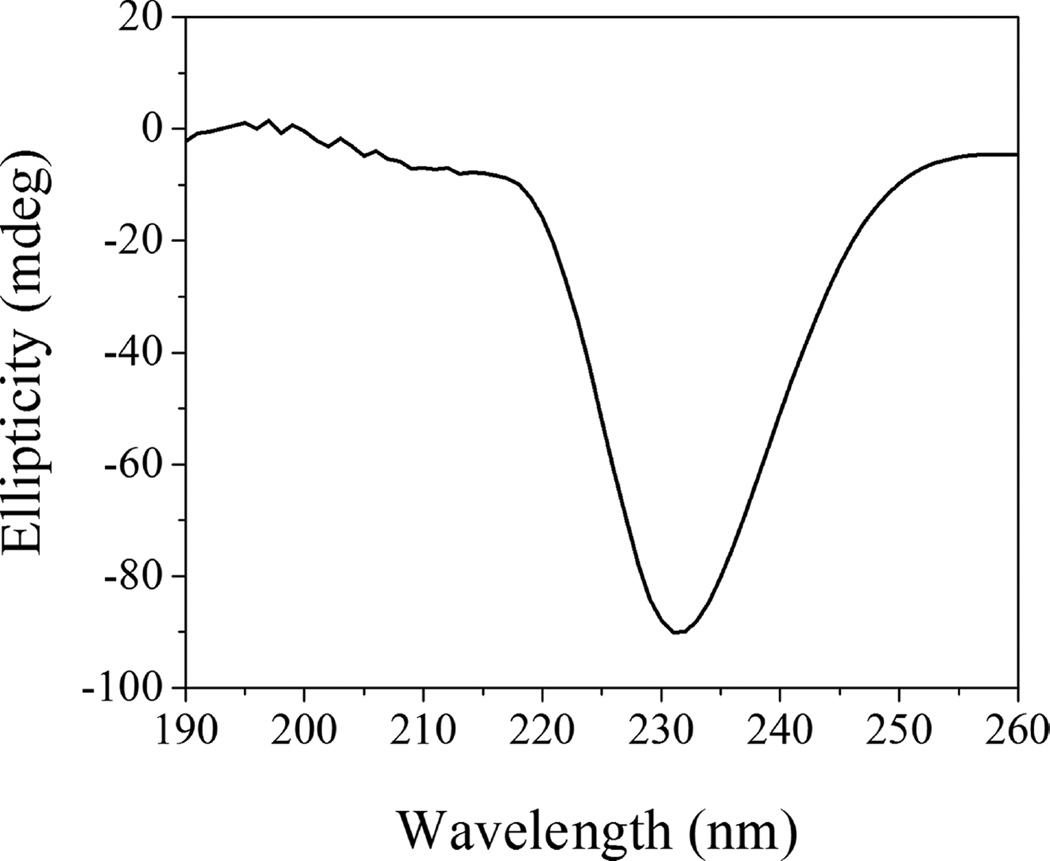
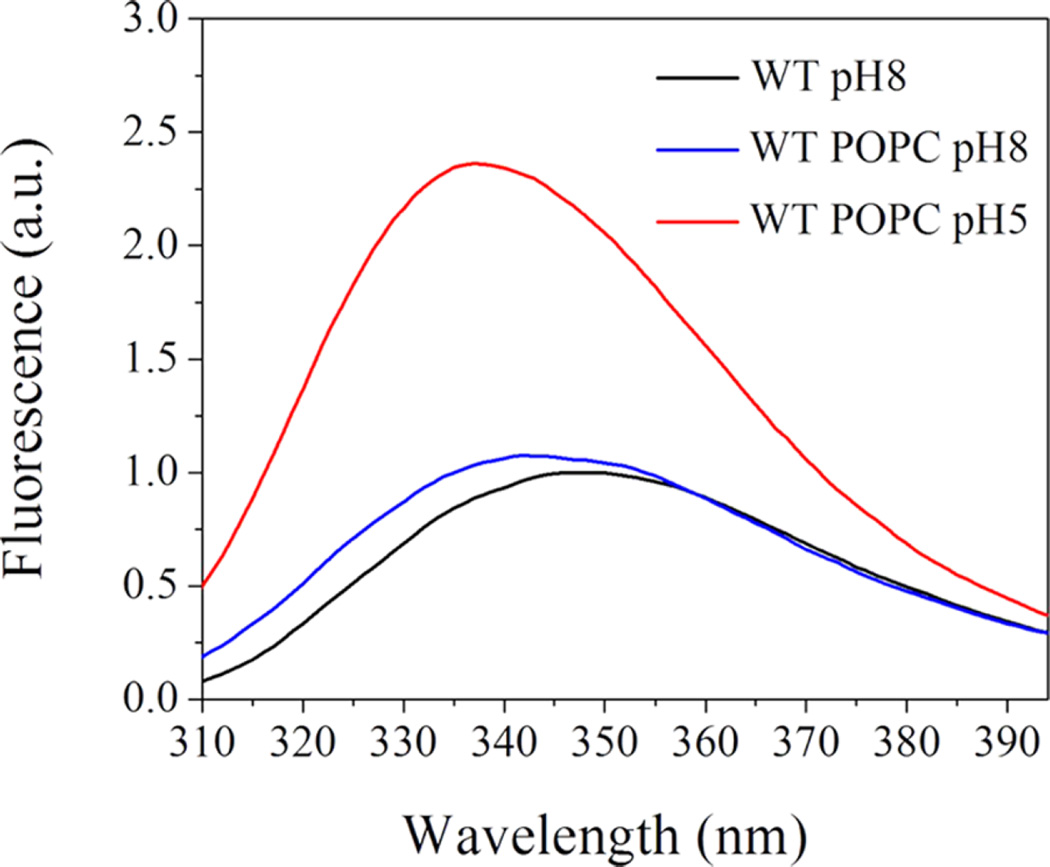
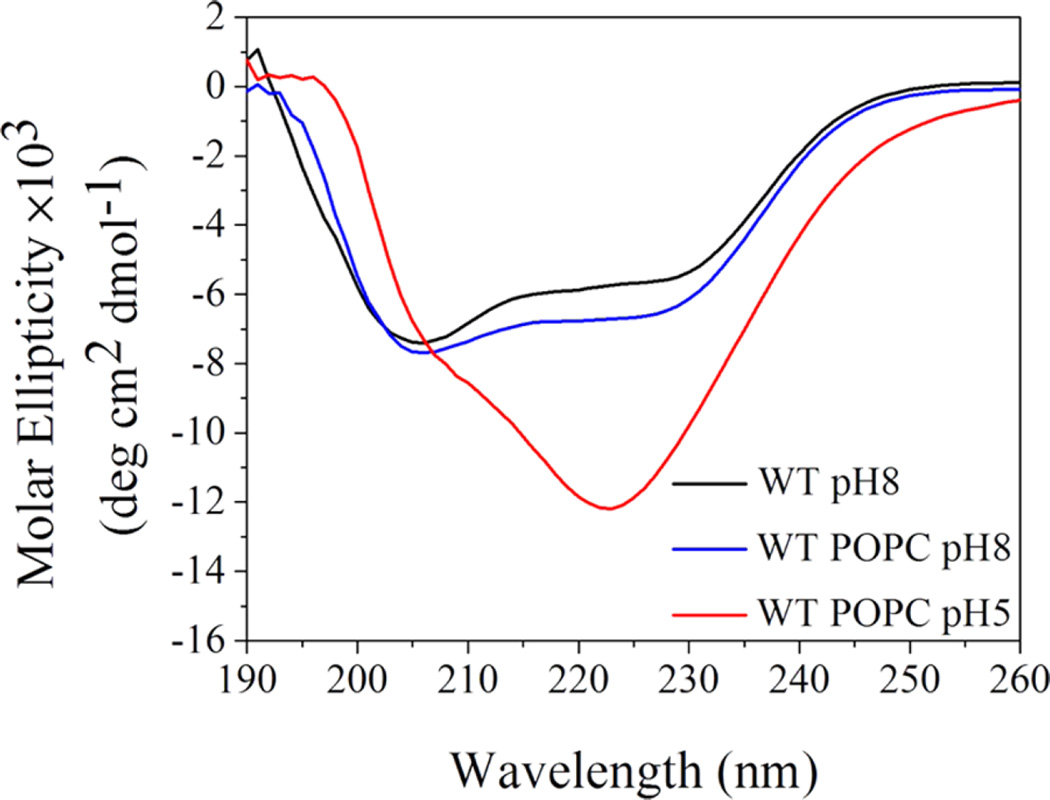
We employed SAXS to follow changes in the lipid bilayer of POPC liposomes induced by WT pHLIP peptide adsorption at pH 8.0 and insertion at pH 5.0. These experiments were carried out at much higher peptide and lipid concentrations (150 and 300 µM for peptides and 4.4 and 8.8 mM for POPC lipids) as compared to concentrations of peptides and lipids used before in biophysical studies. Previously, we have shown that the WT pHLIP peptide is monomeric in aqueous solution at pH 8.0 at concentrations of 7 µM and less; however, it oligomerizes and forms tetramers at a concentration of 50 µM.37 Here, we first measured CD, fluorescence, and scattering profile of 150 and 300 µM of the WT peptide at pH 8.0. The position of the maximum of the fluorescence spectrum of the WT peptide is 341 nm (Figure 1), and it is very similar to the values established previously for 50 µM of the peptide.37 The CD spectrum shows a negative signal at 232 nm (Figure 2) characteristic of exciton formation. The exciton (sharing of electronic density) is observed, when rings of several aromatic residues form stacks.38 The exciton formation was monitored previously for 50 µM of the peptide.37 At the same time, we do not observe the appearance of any elements of the secondary structure for the WT peptide at 150 µM at pH 8.0.
Figure 1.
Tryptophan fluorescence excited at 295 nm of 150 µM of WT pHLIP peptide in phosphate buffer at pH 8.0.
Figure 2.
Circular dichroism signal of 150 µM of WT pHLIP peptide in phosphate buffer at pH 8.0.
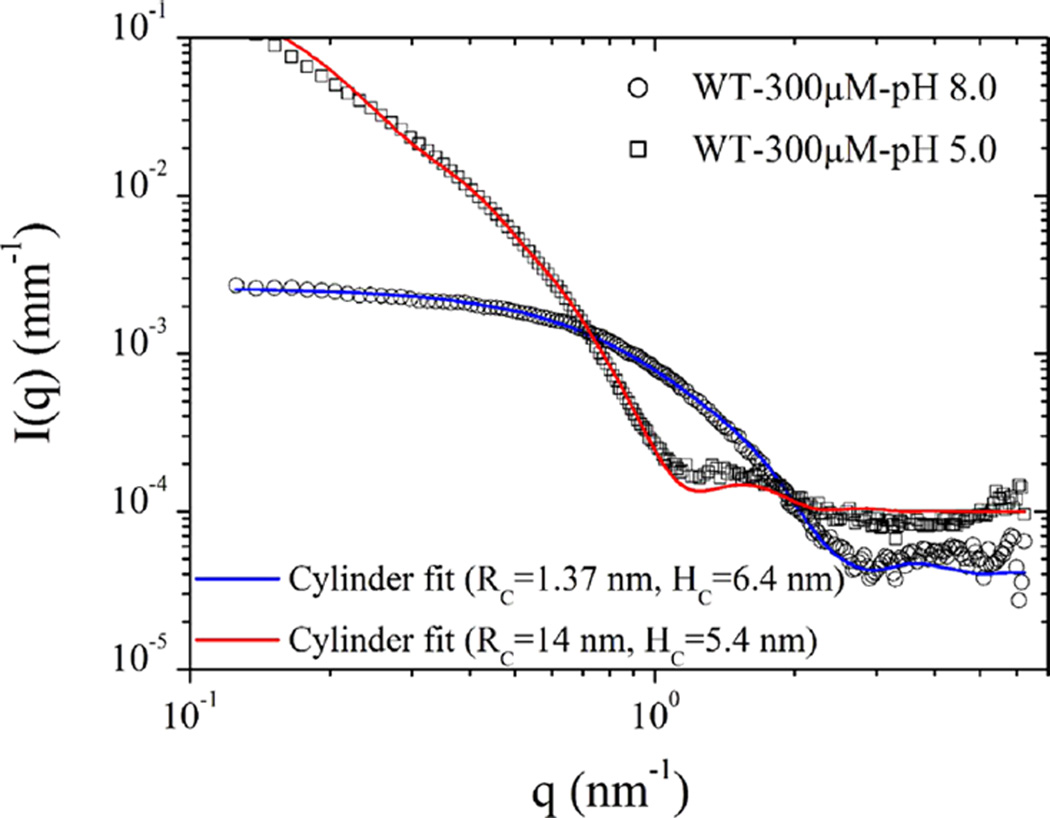
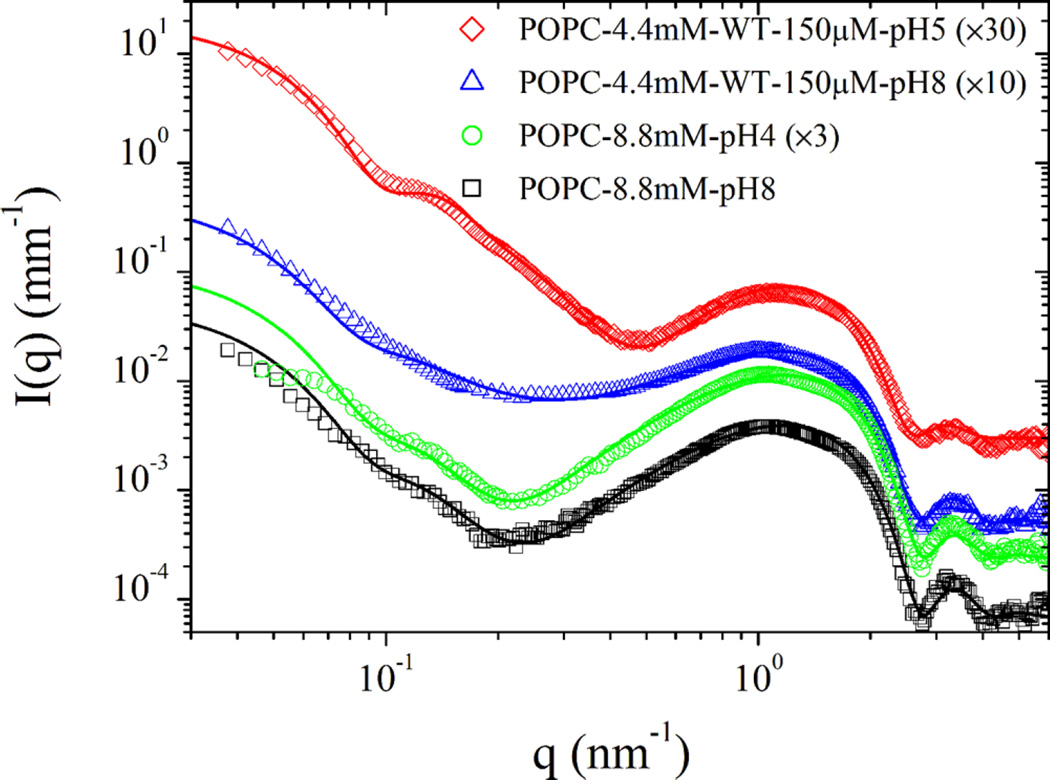
SAXS measurements provided very similar results for 150 and 300 µM WT pHLIP peptide at pH 8.0. Figure 3 shows the scattering of 300 µM WT pHLIP peptide in solutions of pH 8.0 and pH 5.0. The scattering curves can be described well by eq 1 with F(q)2 being the scattering function of polydisperse cylinders33 and S(q) ≈ 1. Although a solid cylinder or disk may not be the exact geometrical form the peptide oligomers adopt in solution, the model serves to indicate the approximate lateral dimensions and association states. The fit parameters including an additive constant background (IB) are summarized in Table 1.
Figure 3.
Normalized SAXS intensities obtained from 300 µM solutions of WT pHLIP peptide at pH 8.0 and pH 5.0. At pH 8.0, the scattering profile is described by a cylinder-like function corresponding to a tetrameric form of the peptide. At pH 5.0 in the absence of the membrane, the WT pHLIP peptide forms large flat aggregates. Table 1 summarizes the results of data analysis.
Table 1.
Analysis of SAXS Data Obtained from 300 µM Solutions of the WT pHLIP Peptide in Phosphate Buffer Saline at pH 8.0 and pH 5.0a
| pH | RC (nm) | HC (nm) | pC | IB (mm−1) |
|---|---|---|---|---|
| 8.0 | 1.37 | 6.4 | 0.1 | 4 × 10−5 |
| 5.0 | 14.0 | 5.4 | 0.1 | 1 × 10−4 |
Normalized SAXS intensities can be described by a polydisperse cylinder scattering function with parameters, mean radius (RC), height (HC), polydispersity in radius (pC), and a constant background (IB).
Using the solid cylinder model, we found that the mean radius (RC) and the height (HC) of the cylinder describing the scattering profile of the WT pHLIP peptide in solution at pH 8.0 are about 1.37 and 6.4 nm, respectively, which correspond to a cylinder volume of about 38 nm3. If we assume a coil configuration (which is supported by CD data) for the 35-residue-long WT pHLIP peptide and take into account that the contour length per residue (established previously), L, is 0.43 nm,39–41 then the average end-to-end length of the coil can be estimated to be d = L√N = 2.5 nm. Then for spherical shape, the volume occupied by a single WT peptide in a random coil conformation is about 8.2 nm3. Thus, about four peptides in a random coil configuration can occupy a volume of about 33–44 nm3, depending on the packing parameter. It is an interesting and important finding that the WT pHLIP peptide transforms from its monomeric form at <10 µM at pH 8.0 to a tetrameric oligomer at 50 µM and remains in the terameric form at high concentrations (at least up to 300 µM). At lower pH, the WT pHLIP peptide formed much bigger aggregates, which can also be approximately described by a cylinder scattering function but with a very different aspect ratio. The model curve for pH 4 in Figure 3 corresponds to a mean radius of 14 nm with 10% polydispersity and height of 5.4 nm. These parameters suggest that the pHLIP chains have further coiled and aggregated laterally to form a flat disklike object at low pH.
Next, we proceed to experiments on liposomes. As spectral signals (especially CD) are destroyed by scattering from high concentrations of liposomes (more than 4 mM of lipids), we prepared samples of 150 µM WT pHLIP peptide in state I (peptide at pH 8.0, no liposomes), state II (peptide at pH 8.0 and 4 mM POPC liposomes), and state III (peptide at pH 5.0 with 4 mM POPC liposomes). Then, each sample was diluted 7 times, and CD and fluorescence spectra were measured immediately. The changes in the fluorescence signal (Figure 4) is typical for pHLIP peptides,12,37 resulting in an increase in the fluorescence intensity and shift of the position of the maximum of emission to shorter wavelengths, which is indicative of pH-dependent insertion into the lipid bilayer of the membrane. CD demonstrates the presence of exciton structures in state I, indicative of oligomerization (Figure 5). Moreover, the exciton signal was observed for the peptide in the presence of the membrane at pH 8.0. We need to outline that at a low lipid/peptide ratio (27 in our case), we expect to have significant population of membrane-unbound peptides because at pH 8.0, one peptide interacts with about 50 lipid head groups on average.28 At the same time, at low pH, the affinity of the peptide to the membrane is higher and one peptide interacts with about 10–15 lipids on average.28 As a result, the main population of peptides is in a membraneinserted, helical conformation, which is supported by the CD spectrum with minimum at In at 225 nm.
Figure 4.
Three states of the WT pHLIP peptide were monitored by changes in tryptophan fluorescence. State I (black line) represents a peptide in solution at pH 8.0. State II (blue line) is a peptide in solution in the presence of POPC liposomes at pH 8.0. State III (red line) is a peptide in solution in the presence of POPC liposomes at pH 5.0.
Figure 5.
Three states of the WT pHLIP peptide were monitored by changes in the CD signal. State I (black line) represents a peptide in solution at pH 8.0. State II (blue line) is a peptide in solution in the presence of POPC liposomes at pH 8.0. State III (red line) is a peptide in solution in the presence of POPC liposomes at pH 5.0.
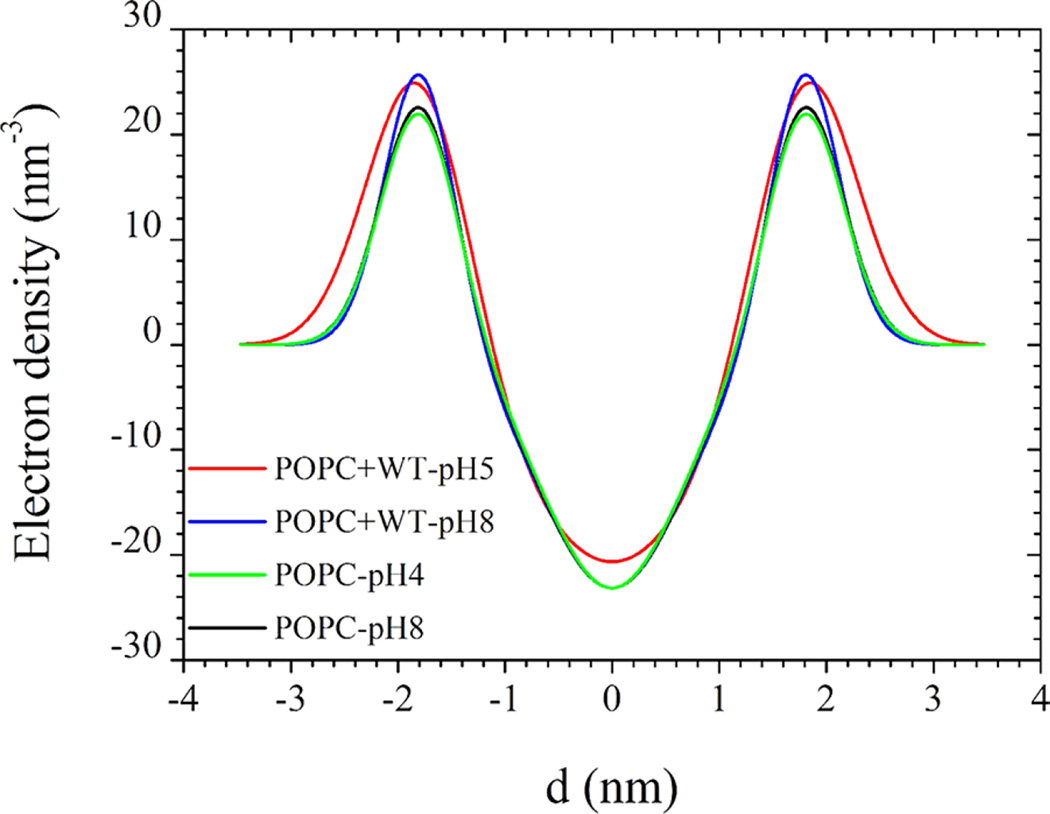
In Figure 6, we present the SAXS from POPC vesicles in the absence and presence of the WT pHLIP peptide. Because of the cancelling effect of positive and negative contrasts from the lipid head group and hydrophobic chains, respectively, the net low q forward scattering from pure POPC vesicles is rather weak and barely measurable over the buffer and instrument background. The main feature in SAXS is the bilayer form factor, which can be described by eq 4, confirming the unilamellar nature of the liposomes with a mean radius of about 28.0 nm. There is no significant variation in the bilayer form factor with pH as indicated by the similar shape of the curves. The best fit excess electron density profiles using eqs 3 and 4 are displayed in Figure 7. As expected, the membrane electron density profiles of pure POPC liposomes perfectly superimpose at higher and lower pH values. The same scattering features are observed at lower POPC concentrations with factor 2 and 4 dilutions, but the intensity statistics at high q values become poorer. The obtained electron density profile of POPC unilamellar liposomes is comparable to that reported for oriented lipid membranes32,42 though less structured (for a similar model to that in eq 3), presumably due to shape fluctuations.
Figure 6.
SAXS intensities from the POPC liposomes of nominal diameter 50 nm without and with the WT pHLIP peptide. The low-q region shows the unilamellar vesicle features, whereas the high-q region depicts the prototypical bilayer form factor. Data were analyzed using the polydisperse unilamellar vesicle model with three Gaussian electron density profiles for the lipid bilayer using eqs 3 and 4. Main parameters of the model are tabulated in Table 2. For clarity, successive scattering curves have been multiplied by the factor indicated in parenthesis.
Figure 7.
Excess electron density profiles (above the buffer level) presented as a sum of three Gaussian functions obtained from the analysis of SAXS intensities presented in Figure 6.
With the addition of the WT pHLIP peptide, the low-q scattering features of POPC vesicles became more pronounced as a result of an increase in the positive contrast of the phospholipid membrane. Indeed, these low-q scattering features are different from the pure-peptide scattering profiles presented in Figure 3. At pH 8.0, a fraction of the peptides remained free in the solution. In eq 4, it was necessary to include either a constant background or a fraction of free peptide (up to 30%) intensity to obtain a complete description of the data. However, at low pH in the presence of the membrane, larger peptide aggregates as those depicted in Figure 3 could not be observed, as most of the peptides were inserted into the membrane. The SAXS data are in agreement with CD measurements. The main parameters of the SAXS model are tabulated in Table 2.
Table 2.
Main Parameters Derived from SAXS Modeling of Data in Figure 6 by Means of the Separated Form Factor Approacha
| sample | RV (nm) | pV | ΔρH (nm−3) | Δ ρC (nm−3) | DH (nm) | DC (nm) | XH (nm) | IB (mm−1) |
|---|---|---|---|---|---|---|---|---|
| POPC— 8.8 mM, pH 8.0 | 28.0 | 0.28 | 23.1 | −23.2 | 0.36 | 0.66 | 1.8 | 7 × 10−5 |
| POPC—8.8 mM, pH 5.0 | 28.0 | 0.28 | 22.5 | −23.2 | 0.36 | 0.65 | 1.8 | 8 × 10−5 |
| POPC—4.4 mM; WT—150 µm, pH 8.0 | 28.6 | 0.30 | 26.3 | −23.2 | 0.33 | 0.67 | 1.82 | 5 × 10−5 |
| POPC—4.4 mM; WT—150 µm, pH 5.0 | 28.3 | 0.23 | 28.1 | −20.7 | 0.48 | 0.94 | 1.8 | 8 × 10−5 |
Parameters RV, pV, ΔρH, ΔρC, DH, DC, XH, and IB represent the mean vesicle radius, vesicle size polydispersity, excess electron densities of the head group and hydrophobic chains, Gaussian widths of the head group and hydrophobic chains, distance to the center of the head group from the bilayer midplane, and a constant background, respectively.
At the low pH, the typical unilamellar liposome scattering signature became more evident, confirming the insertion of the WT pHLIP peptide into the membrane, thereby significantly enhancing the positive contrast of the membranes. Scattering curves with the peptide were analyzed using the same parameters of the unilamellar liposomes, but the membrane electron density profile was allowed to vary. The corresponding excess electron density profiles are shown in Figure 7. At a higher pH (pH 8.0), the inner hydrophobic part has a similar shape as that in pure POPC liposomes, indicating that the peptide molecules were residing at the outer leaflets of the membranes. This has led to an increase in the electron density of the head group region without significant broadening of the peak, implying that the peptide chains have penetrated into the head group region. However, at lower pH (pH 5.0) both the inner and outer electron densities have increased, meaning that the peptides have been inserted across the bilayer of the membrane. In addition, the peak at the outer hydrophilic part has significantly broadened, implying that both ends of the peptide impose steric constraints on the lipid head groups. The resulting enhanced electron density led to a stronger low-q scattering feature of the unilamellar liposomes.
DISCUSSION
The pHLIP peptides find a wide range of applications in biomedical sciences16,20–22,43,44 and, also, they prove to be a very convenient model system for the investigation of polypeptide insertion into the lipid bilayer of a membrane triggered by pH.12,28 Whereas various spectroscopic methods applied previously probed changes in the conformation of peptides, SAXS employed in this study allowed us, for the first time, to monitor the changes in the liposome and lipid bilayer structure. Until now, most experiments were carried out at low peptide concentrations (about 2–10 µM) and high lipid/peptide ratios (about 100–300). In this study, we used high peptide concentrations (about 150–300 µM) and low lipid/peptide ratios (about 30). First, we investigated WT pHLIP peptide conformational states in solution in the absence of liposomes at high and low pH. Interestingly, in the range of concentrations from 50 µM up to 300 µM, the peptide remains in the tetrameric configuration and does not form larger aggregates at pH 8. This is a very important finding related to potential medical applications in which a high dosage of pHLIP peptides may be required for human administration. At low pH, when Asp and Glu residues are protonated, the peptide in solution in the absence of liposomes forms higher order of aggregates. However, in the presence of the membrane, these larger aggregates are not observed at low pH. The interaction of the WT pHLIP peptide with liposomes has not altered the unilamellar vesicle structure upon peptide adsorption by the lipid bilayer at high pH or upon insertion across the bilayer at low pH. From the analysis of SAXS data, an average change in the electron density profile of the membrane upon insertion of the WT pHLIP peptide was derived. It is clearly shown that the insertion of the peptide into the membrane at lower pH corresponding to a significant increase in the electron density and low-q scattering. The membrane electron density profiles of POPC unilamellar vesicles without peptides are comparable to those reported in the literature for oriented POPC membranes.32,42 Although the model could be improved further with additional Gaussian terms in eq 3, it was sufficient to capture the main structural features of the POPC–WT pHLIP liposomes.
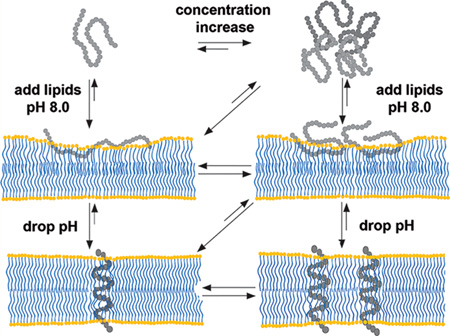
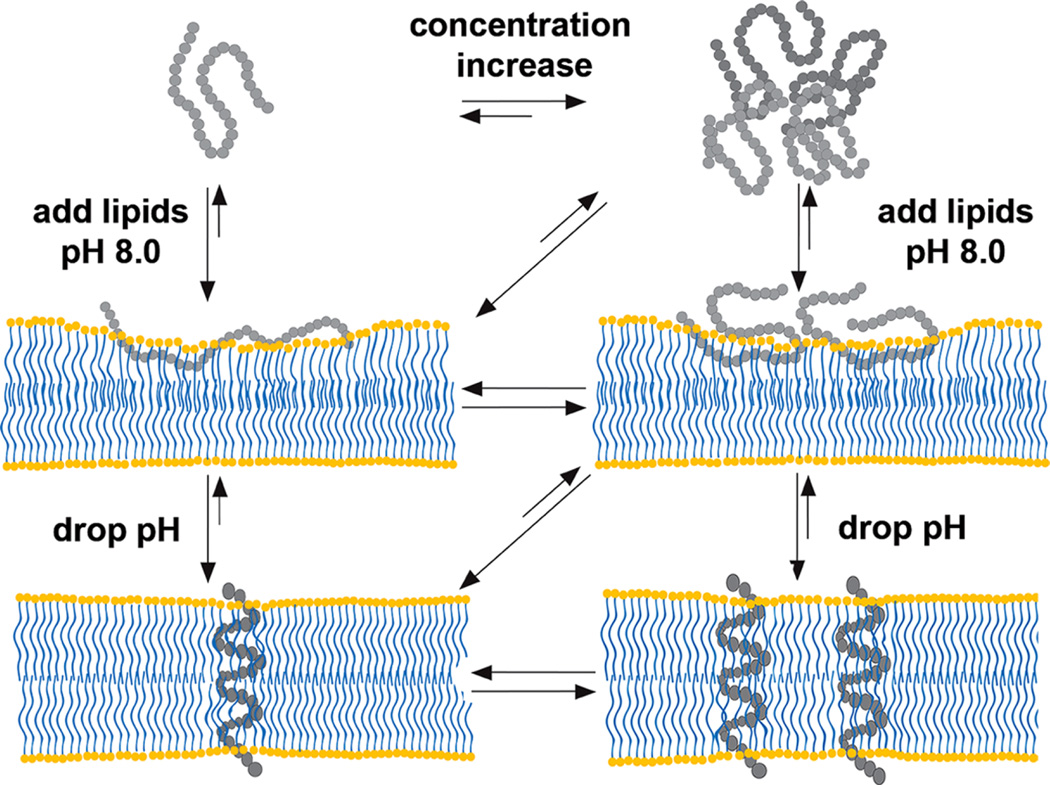
Figure 8 schematically illustrates the behavior of the peptide in aqueous solution at low and high concentrations, and interactions with the membrane at low and high pH values derived from numerous previous and current spectroscopic and SAXS studies. At high/neutral pH and at low concentrations (<7 µM), the peptide is monomeric in aqueous solution.37 With increase in peptide concentration (up to 30 µM), the WT pHLIP peptide associates into tetrameric oligomers and stays as a tetramer with concentration increase by about 10-fold (and potentially even higher concentrations). When an excess of membrane is present, the equilibrium is shifted toward the membrane-adsorbed state of the peptide (called state II). Depending on the lipid/peptide ratio, the polypeptide can adopt different conformations within a membrane. At low lipid/peptide ratios, the “parking problem” exists, which results in partial adsorption of the WT pHLIP peptide by the membrane, opposite to the extended peptide configuration at the surface of the lipid bilayer at high lipid/peptide ratios.28 Also, depending on the hydrophobicity of the polypeptide sequence and the presence of charged residues, various pHLIP peptides exhibit different penetration depths within the bilayer.10,45 Our data, obtained in this study, indicate that the WT pHLIP peptide is adsorbed within the polar head group region of the bilayer. A drop in the pH results in the protonation of Asp/Glu residues and an increase in the overall polypeptide hydrophobicity, which trigger polypeptide partitioning into membrane and folding. Most of the pHLIP peptides adopt very similar TM helical orientation (called state III).10 We have shown that at low peptide concentrations, TM helices do not oligomerize.37 Our scattering data obtained on liposomes with WT pHLIP peptide at low pH suggest uniform distribution of helices at high peptide concentrations and low lipid/peptide ratios. However, further investigation is required to confirm this feature.
Figure 8.
Schematic presentation of WT pHLIP peptide interaction with the lipid bilayer of the membrane; see description in the text.
The obtained results clearly indicate that SAXS could be used to monitor polypeptide interactions with the lipid bilayer of a membrane in real time upon pH change. Our next step will be a kinetic study directed toward monitoring changes in SAXS intensities upon peptide insertion into a membrane and exit from the membrane with pH jumps in correlation with changes in the tryptophan fluorescence signal of the peptide.
Acknowledgments
This work was supported by the National Institute of Health grant number GM073857 to OAA and YKR. ESRF is acknowledged for provision of synchrotron beam time.
ABBREVIATIONS
- CD
circular dichroism
- ED
electron density
- PBS
phosphate buffer saline
- pHLIP
pH Low Insertion Peptide
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- SAXS
small angle X-ray scattering
- TM
transmembrane
- WT
wild type
Footnotes
Author Contributions
T.N., O.A.A., and Y.K.R. designed research; D.W., A.G.K., M.A., and T.N. performed SAXS measurements and D.W. performed spectral measurements; T.N. analyzed SAXS data; D.W., O.A.A., Y.K.R., and T.N. wrote paper.
Notes
The authors declare the following competing financial interest(s): OA Andreev and YK Reshetnyak have founded and have a financial interest in a company, pHLIP, Inc., with the aim of bringing pHLIP technology to the clinic. The company has had no involvement in funding the studies reported here.
REFERENCES
- 1.Van den Berg B, Clemons WM, Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 2.Osborne AR, Rapoport TA, van den Berg B. Protein translocation by the Sec61/SecY channel. Annu. Rev. Cell. Dev. Biol. 2005;21:529–550. doi: 10.1146/annurev.cellbio.21.012704.133214. [DOI] [PubMed] [Google Scholar]
- 3.White SH, von Heijne G. The machinery of membrane protein assembly. Curr. Opin. Struct. Biol. 2004;14:397–404. doi: 10.1016/j.sbi.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Simpson PJ, Schwappach B, Dohlman HG, Isaacson RL. Structures of Get3, Get4, and Get5 provide new models for TA membrane protein targeting. Structure. 2010;18:897–902. doi: 10.1016/j.str.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brambillasca S, Yabal M, Makarow M, Borgese N. Unassisted translocation of large polypeptide domains across phospholipid bilayers. J. Cell Biol. 2006;175:767–777. doi: 10.1083/jcb.200608101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Renthal R. Helix insertion into bilayers and the evolution of membrane proteins. Cell. Mol. Life Sci. 2010;67:1077–1088. doi: 10.1007/s00018-009-0234-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ladokhin AS, White SH. Interfacial folding and membrane insertion of a designed helical peptide. Biochemistry. 2004;43:5782–5791. doi: 10.1021/bi0361259. [DOI] [PubMed] [Google Scholar]
- 8.Hunt JF, Earnest TN, Bousche O, Kalghatgi K, Reilly K, Horvath C, Rothschild KJ, Engelman DM. A biophysical study of integral membrane protein folding. Biochemistry. 1997;36:15156–15176. doi: 10.1021/bi970146j. [DOI] [PubMed] [Google Scholar]
- 9.Reshetnyak YK, Andreev OA, Lehnert U, Engelman DM. Translocation of molecules into cells by pH-dependent insertion of a transmembrane helix. Proc. Natl. Acad. Sci. U.S.A. 2006;103:6460–6465. doi: 10.1073/pnas.0601463103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weerakkody D, Moshnikova A, Thakur MS, Moshnikova V, Daniels J, Engelman DM, Andreev OA, Reshetnyak YK. Family of pH (low) insertion peptides for tumor targeting. Proc. Natl. Acad. Sci. U.S.A. 2013;110:5834–5839. doi: 10.1073/pnas.1303708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Musial-Siwek M, Karabadzhak A, Andreev OA, Reshetnyak YK, Engelman DM. Tuning the insertion properties of pHLIP. Biochim. Biophys. Acta, Biomembr. 2010;1798:1041–1046. doi: 10.1016/j.bbamem.2009.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karabadzhak AG, Weerakkody D, Wijesinghe D, Thakur MS, Engelman DM, Andreev OA, Markin VS, Reshetnyak YK. Modulation of the pHLIP transmembrane helix insertion pathway. Biophys. J. 2012;102:1846–1855. doi: 10.1016/j.bpj.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrera FN, Weerakkody D, Anderson M, Andreev OA, Reshetnyak YK, Engelman DM. Roles of carboxyl groups in the transmembrane insertion of peptides. J. Mol. Biol. 2011;413:359–371. doi: 10.1016/j.jmb.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Onyango JO, Chung MS, Eng CH, Klees LM, Langenbacher R, Yao L, An M. Noncanonical amino acids to improve the pH response of pHLIP insertion at tumor acidity. Angew. Chem., Int. Ed. 2015;54:3658–3663. doi: 10.1002/anie.201409770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen VP, Alves DS, Scott HL, Davis FL, Barrera FN. A Novel Soluble Peptide with pH-Responsive Membrane Insertion. Biochemistry. 2015;54:6567–6575. doi: 10.1021/acs.biochem.5b00856. [DOI] [PubMed] [Google Scholar]
- 16.Tapmeier TT, Moshnikova A, Beech J, Allen D, Kinchesh P, Smart S, Harris A, McIntyre A, Engelman DM, Andreev OA, et al. The pH low insertion peptide pHLIP Variant 3 as a novel marker of acidic malignant lesions. Proc. Natl. Acad. Sci. U.S.A. 2015;112:9710–9715. doi: 10.1073/pnas.1509488112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Viola-Villegas NT, Carlin SD, Ackerstaff E, Sevak KK, Divilov V, Serganova I, Kruchevsky N, Anderson M, Blasberg RG, Andreev OA, et al. Understanding the pharmacological properties of a metabolic PET tracer in prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 2014;111:7254–7259. doi: 10.1073/pnas.1405240111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cruz-Monserrate Z, Roland CL, Deng D, Arumugam T, Moshnikova A, Andreev OA, Reshetnyak YK, Logsdon CD. Targeting pancreatic ductal adenocarcinoma acidic microenvironment. Sci. Rep. 2014;4(4410) doi: 10.1038/srep04410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sosunov EA, Anyukhovsky EP, Sosunov AA, Moshnikova A, Wijesinghe D, Engelman DM, Reshetnyak YK, Andreev OA. pH (low) insertion peptide (pHLIP) targets ischemic myocardium. Proc. Natl. Acad. Sci. U.S.A. 2013;110:82–86. doi: 10.1073/pnas.1220038110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Demoin DW, Wyatt LC, Edwards KJ, Abdel-Atti D, Sarparanta M, Pourat J, Longo VA, Carlin SD, Engelman DM, Andreev OA, Reshetnyak YK, Viola-Villegas N, Lewis JS. PET Imaging of Extracellular pH in Tumors with (64)Cu- and (18)F-Labeled pHLIP Peptides: A Structure-Activity Optimization Study. Bioconjugate Chem. 2016;27:2014–2023. doi: 10.1021/acs.bioconjchem.6b00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reshetnyak YK, Yao L, Zheng S, Kuznetsov S, Engelman DM, Andreev OA. Measuring tumor aggressiveness and targeting metastatic lesions with fluorescent pHLIP. Mol. Imaging Biol. 2011;13:1146–1156. doi: 10.1007/s11307-010-0457-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antosh MP, Wijesinghe DD, Shrestha S, Lanou R, Huang YH, Hasselbacher T, Fox D, Neretti N, Sun S, Katenka N, et al. Enhancement of radiation effect on cancer cells by gold-pHLIP. Proc. Natl. Acad. Sci. U.S.A. 2015;112:5372–5376. doi: 10.1073/pnas.1501628112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.An M, Wijesinghe D, Andreev OA, Reshetnyak YK, Engelman DM. pH-(low)-insertion-peptide (pHLIP) translocation of membrane impermeable phalloidin toxin inhibits cancer cell proliferation. Proc. Natl. Acad. Sci. U.S.A. 2010;107:20246–20250. doi: 10.1073/pnas.1014403107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moshnikova A, Moshnikova V, Andreev OA, Reshetnyak YK. Antiproliferative effect of pHLIP-amanitin. Biochemistry. 2013;52:1171–1178. doi: 10.1021/bi301647y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng CJ, Bahal R, Babar IA, Pincus Z, Barrera F, Liu C, Svoronos A, Braddock DT, Glazer PM, Engelman DM, et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2014;518:107–110. doi: 10.1038/nature13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burns KE, Thevenin D. Down-regulation of PAR1 activity with a pHLIP-based allosteric antagonist induces cancer cell death. Biochem. J. 2015;472:287–295. doi: 10.1042/BJ20150876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burns KE, Robinson MK, Thevenin D. Inhibition of cancer cell proliferation and breast tumor targeting of pHLIP-monomethyl auristatin E conjugates. Mol. Pharmaceutics. 2015;12:1250–1258. doi: 10.1021/mp500779k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reshetnyak YK, Andreev OA, Segala M, Markin VS, Engelman DM. Energetics of peptide (pHLIP) binding to and folding across a lipid bilayer membrane. Proc. Natl. Acad. Sci. U.S.A. 2008;105:15340–15345. doi: 10.1073/pnas.0804746105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andreev OA, Karabadzhak AG, Weerakkody D, Andreev GO, Engelman DM, Reshetnyak YK. pH (low) insertion peptide (pHLIP) inserts across a lipid bilayer as a helix and exits by a different path. Proc. Natl. Acad. Sci. U.S.A. 2010;107:4081–4086. doi: 10.1073/pnas.0914330107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma GP, Reshetnyak YK, Andreev OA, Karbach M, Muller G. Coil-helix transition of polypeptide at water-lipid interface. J. Stat. Mech. 2015;2015(P01034) doi: 10.1088/1742-5468/2015/01/P01034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kučerka N, Nieh M-P, Katsaras J. Small-Angle Scattering from Homogenous and Heterogeneous Lipid Bilayers. In: Iglic A, Tien HT, editors. Advances in Planar Lipid Bilayers and Liposomes. Vol. 12. Burlington: Academic Press; 2010. pp. 201–235. [Google Scholar]
- 32.Kučerka N, Liu Y, Chu N, Petrache HI, Tristram-Nagle S, Nagle JF. Structure of fully hydrated fluid phase DMPC and DLPC lipid bilayers using X-ray scattering from oriented multilamellar arrays and from unilamellar vesicles. Biophys. J. 2005;88:2626–2637. doi: 10.1529/biophysj.104.056606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narayanan T, Gummel J, Gradzielski M. Probing the Self-Assembly of Unilamellar Vesicles Using Time-Resolved SAXS. In: Iglic A, Kulkarni CV, editors. Advances in Planar Lipid Bilayers and Liposomes. Vol. 20. Burlington: Academic Press; 2014. pp. 171–196. [Google Scholar]
- 34.Marquardt D, Heberle FA, Nickels JD, Pabst G, Katsaras J. On scattered waves and lipid domains: detecting membrane rafts with X-rays and neutrons. Soft Matter. 2015;11:9055–9072. doi: 10.1039/c5sm01807b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kiselev MA, Zemlyanaya EV, Aswal VK, Neubert RH. What can we learn about the lipid vesicle structure from the smallangle neutron scattering experiment? Eur. Biophys. J. 2006;35:477–493. doi: 10.1007/s00249-006-0055-9. [DOI] [PubMed] [Google Scholar]
- 36.Pencer J, Krueger S, Adams CP, Katsaras J. Method of separated form factors for polydisperse vesicles. J. Appl. Crystallogr. 2006;39:293. [Google Scholar]
- 37.Reshetnyak YK, Segala M, Andreev OA, Engelman DM. A monomeric membrane peptide that lives in three worlds: in solution, attached to, and inserted across lipid bilayers. Biophys. J. 2007;93:2363–2372. doi: 10.1529/biophysj.107.109967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy RS, Gopi HN, Raghothama S, Gilardi RD, Karle IL, Balaram P. Peptide hairpins with strand segments containing alpha- and beta-amino acid residues: cross-strand aromatic interactions of facing Phe residues. Biopolymers. 2005;80:787–799. doi: 10.1002/bip.20294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dietz H, Rief M. Protein structure by mechanical triangulation. Proc. Natl. Acad. Sci. U.S.A. 2006;103:1244–1247. doi: 10.1073/pnas.0509217103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carrion-Vazquez M, Li H, Lu H, Marszalek PE, Oberhauser AF, Fernandez JM. The mechanical stability of ubiquitin is linkage dependent. Nat. Struct. Biol. 2003;10:738–743. doi: 10.1038/nsb965. [DOI] [PubMed] [Google Scholar]
- 41.Oesterhelt F, Oesterhelt D, Pfeiffer M, Engel A, Gaub HE, Muller DJ. Unfolding pathways of individual bacteriorhodopsins. Science. 2000;288:143–146. doi: 10.1126/science.288.5463.143. [DOI] [PubMed] [Google Scholar]
- 42.Pabst G, Rappolt M, Amenitsch H, Laggner P. Structural information from multilamellar liposomes at full hydration: Full qrange fitting with high quality x-ray data. Phys. Rev. E. 2000;62:4000–4009. doi: 10.1103/physreve.62.4000. [DOI] [PubMed] [Google Scholar]
- 43.Adochite RC, Moshnikova A, Golijanin J, Andreev OA, Katenka N, Reshetnyak YK. Comparative study of tumor targeting and biodistribution of pH (Low) Insertion Peptides (pHLIP® peptides) conjugated with different fluorescent dyes. Mol. Imaging Biol. 2016;18:686–696. doi: 10.1007/s11307-016-0949-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng CJ, Bahal R, Babar IA, Pincus Z, Barrera F, Liu C, Svoronos A, Braddock DT, Glazer PM, Engelman DM, et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015;518:107–110. doi: 10.1038/nature13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zoonens M, Reshetnyak YK, Engelman DM. Bilayer interactions of pHLIP, a peptide that can deliver drugs and target tumors. Biophys. J. 2008;95:225–235. doi: 10.1529/biophysj.107.124156. [DOI] [PMC free article] [PubMed] [Google Scholar]