The crystal structure of (perchlorato-κO)(1,4,7,10-tetraazacyclododecane-κ4 N)copper(II) perchlorate is reported. The crystal was grown from a solution of methanol at ambient temperature which resulted in no co-crystallization of solvent.
Keywords: crystal structure, copper(II), cyclen, perchlorate
Abstract
The crystal structure of the title salt, [Cu(ClO4)(C8H20N4)]ClO4, is reported. The CuII ion exhibits a square-pyramidal geometry and is coordinated by the four N atoms of the neutral 1,4,7,10-tetraazacyclododecane (cyclen) ligand and an O atom from one perchlorate anion, with the second perchlorate ion hydrogen-bonded to one of the amine N atoms of the cyclen ligand. Additional N—H⋯O hydrogen bonds between the amine H atoms and the coordinating and non-coordinating perchlorate groups create a three-dimensional network structure. Crystals were grown from a concentrated methanol solution at ambient temperature, resulting in no co-crystallization of solvent.
Chemical context
Azamacrocycle ligands, including 1,4,7,10-tetraazacyclododecane (cyclen), are of significant importance in research due to their ability to form stable metal complexes, allowing for their use in a wide range of applications. Some of these complexes have been studied for their use as chemical sensors, contrast agents in MRI and PET, antimicrobial agents and as biomimetic catalysts (De León-Rodríguez et al., 2010 ▸; Yoo et al., 2005 ▸). Copper–cyclen complexes have been studied extensively for their ability to perform catalytic DNA cleavage and peptide hydrolysis (Zhang et al., 2016 ▸; Li et al. 2014 ▸; Hormann et al., 2015 ▸). Although the synthesis of a similar CuII complex has been reported previously, no crystal structure of the complex, [Cu(1,4,7,10-tetraazacyclododecane)](ClO4)2, has previously been published (Kruppa et al., 2006 ▸).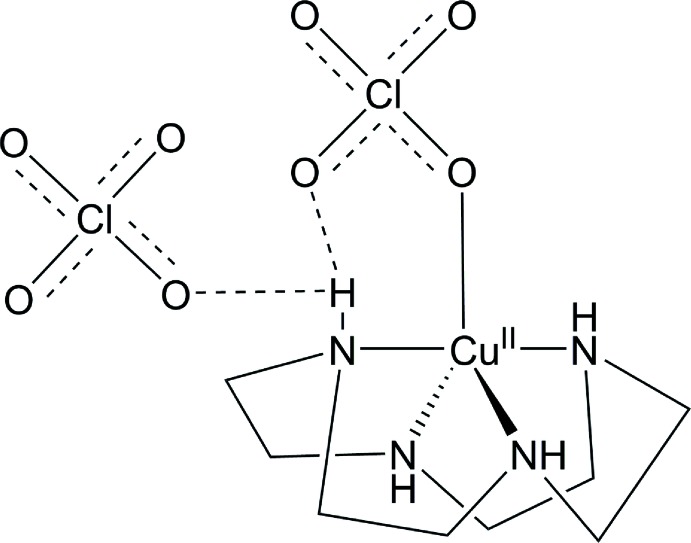
Structural commentary
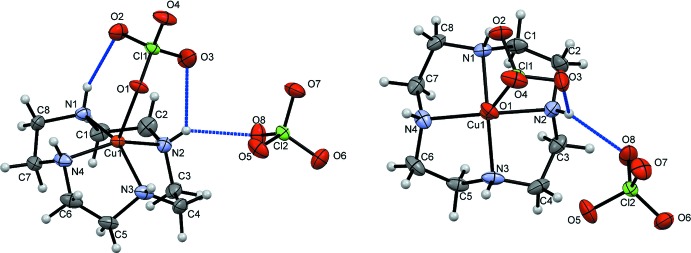
In the title complex (Fig. 1 ▸), the copper(II) ion coordinated by the four nitrogen atoms of the cyclen ligand and one oxygen atom of a perchlorate ligand. The five-coordinate cupric ion shows a nearly ideal square-pyramidal geometry (τ5 = 0.049; Addison et al., 1984 ▸). The Cu—N bond lengths range from 2.004 (1) to 2.015 (1) Å, which are typical values. The CuII ion exhibits a tetragonal distortion that leads to a longer apical bond with Cu1—O1 = 2.266 (1) Å, which is 0.12 Å longer than the average Cu—O distance (Clay et al., 1979 ▸; Rohde & Merzweiler, 2010 ▸). The average N—Cu—O bond angle is 103.8 (8)°. Three hydrogen bonds are present within the asymmetric unit, with two extending from O2 and O3 of the bound perchlorate anion to N1—H1 and N2—H2, respectively. The third hydrogen bond extends from N2—H2 to O8 of the unbound anion; the numerical details are given in Table 1 ▸.
Figure 1.
Side (left) and top (right) views, as defined by the cyclen ligand ring, of [Cu(cyclen)](ClO4)2 represented with ellipsoids at the 50% probability level. Hydrogen bonds are drawn in blue.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O6i | 0.86 (1) | 2.50 (2) | 3.171 (1) | 135 (1) |
| N1—H1⋯O2 | 0.86 (1) | 2.39 (1) | 3.093 (1) | 139 (1) |
| N2—H2⋯O8ii | 0.88 (2) | 2.31 (2) | 3.050 (1) | 142 (1) |
| N2—H2⋯O3 | 0.88 (2) | 2.44 (2) | 3.052 (2) | 127 (1) |
| N3—H3⋯O1ii | 0.86 (2) | 2.40 (1) | 3.245 (1) | 169 (2) |
| N3—H3⋯O4ii | 0.86 (2) | 2.55 (2) | 3.132 (1) | 126 (1) |
| N4—H4⋯O5 | 0.86 (2) | 2.36 (1) | 3.096 (1) | 143 (1) |
Symmetry codes: (i) 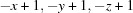 ; (ii)
; (ii) 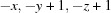 .
.
Supramolecular features
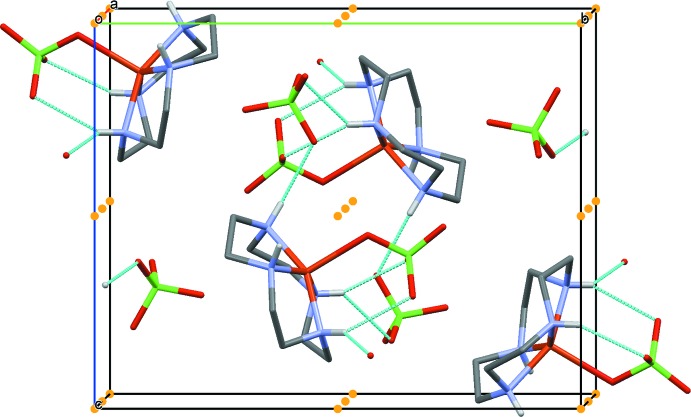
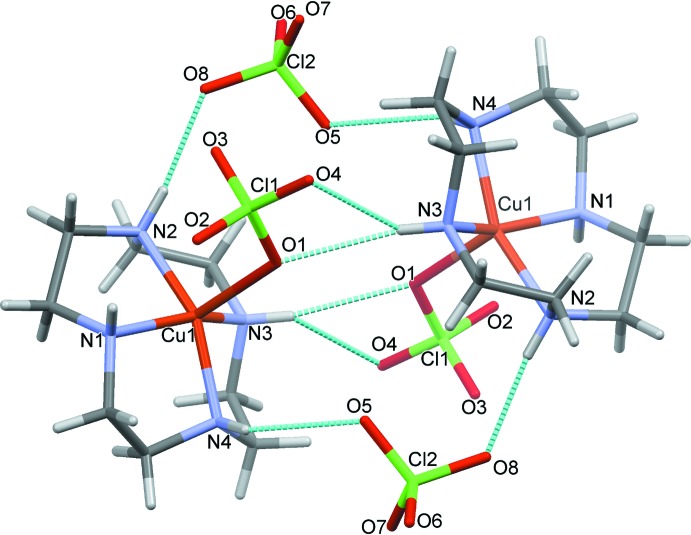
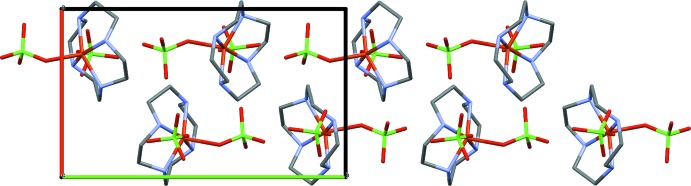
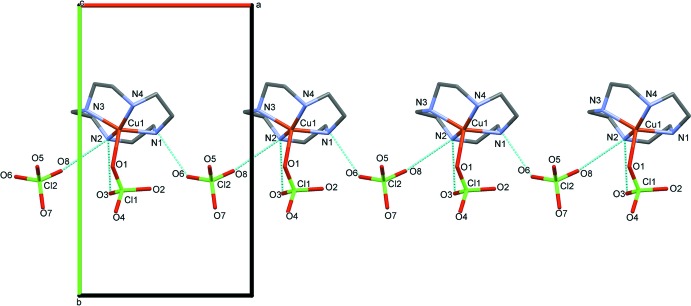
The crystal structure exhibits three unique symmetry elements: an inversion center, a twofold screw axis and a glide plane. The complex cations of two asymmetric units hydrogen-bond across an inversion center, which is clearly visible when viewed along the a axis (Fig. 2 ▸), creating a dimer. These hydrogen bonds (N3—H3⋯O1, N3—H3⋯O4, N4—H4⋯O5) have an average N⋯O distance of 3.16 Å (Fig. 3 ▸). The complexes assemble in rows parallel to the b axis (Fig. 4 ▸) due in part to weak electrostatic interactions between the bound perchlorate anion and a neighboring cyclen ligand. A hydrogen bond between the cyclen ligand and a neighboring perchlorate anion (N1—H1⋯O3) allows the building units to assemble parallel to the a axis (Fig. 5 ▸).
Figure 2.
View of the unit cell along the a axis. An inversion center (yellow dots) exists between two asymmetric units, creating the dimeric unit defined at the center of the unit cell. Hydrogen bonds are drawn in blue.
Figure 3.
A view of hydrogen bonding within a dimer pair. Hydrogen bonds are drawn in blue. Carbon and hydrogen atom labels have been omitted for clarity.
Figure 4.
Packing of the complex cations, as viewed along the c axis of the unit cell. The a axis is drawn in red and the b axis is drawn in green.
Figure 5.
Hydrogen bonding between complex cations and anions, as viewed along the c axis. Hydrogen bonds are drawn in blue. The a axis is drawn in red and the b axis is drawn in green.
Database survey
A database survey resulted in several similar Cu–cyclen complexes with five-coordinate copper(II). Four structures chosen for further analysis contained a copper(II) ion coordinated by either five nitrogen atoms or four nitrogen atoms and one oxygen atom (Rohde & Merzweiler, 2010 ▸; Sarma et al., 2010 ▸; Péréz-Toro et al., 2015 ▸; Guo et al., 2008 ▸). Where applicable, the complexes have similar Cu—O bond lengths to that of the title complex, with only slight deviations. The title complex and surveyed complexes have similar Cu—N distances with a standard deviation of 0.018 Å.
Synthesis and crystallization
The title complex was synthesized by a modified method as reported by Kruppa et al. (2006 ▸). Under a nitrogen atmosphere, 1,4,7,10-tetraazacyclododecane (247 mg, 1.4 mmol) and copper(II) perchlorate hexahydrate (527 mg, 1.4 mmol) were separately dissolved in 2.8 mL anhydrous methanol each and combined. The resulting purple solution formed a precipitate. The reaction mixture was heated to reflux for 30 min then filtered. The filtrate was evaporated to dryness to yield a purple amorphous solid. X-ray quality crystals were grown by dissolving the solid in a minimum amount of methanol followed by slow evaporation at ambient temperature. The title complex [Cu(cyclen)](ClO4)2 was isolated as purple crystals in 84% yield (1.2 mmol, 526 mg). IR [ATR, ν (cm−1)]: 3281, 2939, 1478, 1072, 617. MS (MALDI–TOF, MeOH): m/z = 334.2 [Cu(cyclen)2+ + ClO4 −]−.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. H atoms attached to carbon were positioned geometrically and constrained to ride on their parent atoms. The H atoms attached to nitrogen were located in a difference map and restrained to have comparable bond lengths. U iso(H) values were set to 1.2U eq(C/N).
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | [Cu(ClO4)(C8H20N4)]ClO4 |
| M r | 434.72 |
| Crystal system, space group | Monoclinic, P21/n |
| Temperature (K) | 173 |
| a, b, c (Å) | 8.9387 (2), 15.0607 (4), 11.9235 (3) |
| β (°) | 92.949 (1) |
| V (Å3) | 1603.05 (7) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 1.74 |
| Crystal size (mm) | 0.23 × 0.21 × 0.18 |
| Data collection | |
| Diffractometer | Bruker SMART APEXII CCD |
| Absorption correction | Multi-scan (SADABS; Sheldrick, 2014 ▸) |
| T min, T max | 0.667, 0.747 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 43306, 7519, 6655 |
| R int | 0.021 |
| (sin θ/λ)max (Å−1) | 0.830 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.025, 0.068, 1.02 |
| No. of reflections | 7519 |
| No. of parameters | 221 |
| No. of restraints | 6 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.60, −0.44 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989016019563/zl2687sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016019563/zl2687Isup2.hkl
CCDC reference: 1521075
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
Special thanks to The University of Alabama Department of Chemistry for funding and facilities. We also thank the Undergraduate Creativity and Research Academy (UCRA) at UA, the Research Grants Committee (RGC) at UA, and acknowledge the NSF EPSCoR Track 2 Seed Grant to ETP (PI N. Hammer, grant No. OIA-1539035) for generous financial support.
supplementary crystallographic information
Crystal data
| [Cu(ClO4)(C8H20N4)]ClO4 | F(000) = 892 |
| Mr = 434.72 | Dx = 1.801 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| a = 8.9387 (2) Å | Cell parameters from 9899 reflections |
| b = 15.0607 (4) Å | θ = 2.3–35.9° |
| c = 11.9235 (3) Å | µ = 1.74 mm−1 |
| β = 92.949 (1)° | T = 173 K |
| V = 1603.05 (7) Å3 | Block, purple |
| Z = 4 | 0.23 × 0.21 × 0.18 mm |
Data collection
| Bruker SMART APEXII CCD diffractometer | 6655 reflections with I > 2σ(I) |
| Radiation source: fine focus sealed tube | Rint = 0.021 |
| phi and ω scans | θmax = 36.2°, θmin = 2.2° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 2014) | h = −14→13 |
| Tmin = 0.667, Tmax = 0.747 | k = −24→24 |
| 43306 measured reflections | l = −19→12 |
| 7519 independent reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.025 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.068 | w = 1/[σ2(Fo2) + (0.0357P)2 + 0.4526P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.02 | (Δ/σ)max = 0.003 |
| 7519 reflections | Δρmax = 0.60 e Å−3 |
| 221 parameters | Δρmin = −0.44 e Å−3 |
| 6 restraints | Extinction correction: SHELXL2014 (Sheldrick, 2015), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0025 (3) |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.42855 (13) | 0.41595 (9) | 0.83806 (10) | 0.0315 (2) | |
| H1A | 0.5240 | 0.4277 | 0.8813 | 0.038* | |
| H1B | 0.4043 | 0.3521 | 0.8450 | 0.038* | |
| C2 | 0.30439 (15) | 0.47211 (10) | 0.88238 (9) | 0.0341 (2) | |
| H2A | 0.2850 | 0.4540 | 0.9601 | 0.041* | |
| H2B | 0.3344 | 0.5354 | 0.8835 | 0.041* | |
| C3 | 0.07219 (14) | 0.38402 (9) | 0.83922 (11) | 0.0330 (2) | |
| H3A | 0.0198 | 0.3977 | 0.9083 | 0.040* | |
| H3B | 0.1353 | 0.3308 | 0.8533 | 0.040* | |
| C4 | −0.04050 (12) | 0.36717 (8) | 0.74254 (13) | 0.0353 (3) | |
| H4A | −0.0945 | 0.3110 | 0.7554 | 0.042* | |
| H4B | −0.1148 | 0.4160 | 0.7378 | 0.042* | |
| C5 | 0.08786 (13) | 0.27094 (7) | 0.60632 (10) | 0.0297 (2) | |
| H5A | 0.0006 | 0.2334 | 0.5836 | 0.036* | |
| H5B | 0.1415 | 0.2431 | 0.6720 | 0.036* | |
| C6 | 0.19107 (16) | 0.27947 (8) | 0.51035 (10) | 0.0345 (2) | |
| H6A | 0.2356 | 0.2209 | 0.4943 | 0.041* | |
| H6B | 0.1335 | 0.2998 | 0.4420 | 0.041* | |
| C7 | 0.44486 (13) | 0.30541 (7) | 0.60226 (11) | 0.0302 (2) | |
| H7A | 0.5046 | 0.2707 | 0.5500 | 0.036* | |
| H7B | 0.4126 | 0.2651 | 0.6620 | 0.036* | |
| C8 | 0.53867 (12) | 0.38048 (8) | 0.65356 (12) | 0.0315 (2) | |
| H8A | 0.6201 | 0.3558 | 0.7037 | 0.038* | |
| H8B | 0.5847 | 0.4147 | 0.5934 | 0.038* | |
| H1 | 0.4734 (19) | 0.4937 (9) | 0.7172 (14) | 0.038* | |
| H2 | 0.1146 (18) | 0.5097 (10) | 0.8063 (14) | 0.038* | |
| H3 | −0.0155 (19) | 0.3802 (12) | 0.5781 (13) | 0.038* | |
| H4 | 0.3389 (19) | 0.3719 (11) | 0.4820 (12) | 0.038* | |
| N1 | 0.44205 (10) | 0.43993 (6) | 0.71871 (8) | 0.02310 (15) | |
| N2 | 0.16614 (10) | 0.46012 (7) | 0.80849 (8) | 0.02595 (16) | |
| N3 | 0.03746 (11) | 0.36146 (6) | 0.63546 (9) | 0.02866 (19) | |
| N4 | 0.31208 (12) | 0.34444 (6) | 0.54087 (8) | 0.02774 (18) | |
| O1 | 0.19181 (10) | 0.55583 (5) | 0.55695 (6) | 0.02513 (14) | |
| O2 | 0.40695 (9) | 0.63197 (6) | 0.62923 (8) | 0.03461 (19) | |
| O3 | 0.17927 (13) | 0.64843 (8) | 0.71488 (9) | 0.0461 (3) | |
| O4 | 0.21084 (11) | 0.71021 (6) | 0.53609 (9) | 0.0388 (2) | |
| O5 | 0.24547 (13) | 0.44353 (6) | 0.31635 (7) | 0.0375 (2) | |
| O6 | 0.36978 (11) | 0.40926 (8) | 0.15317 (8) | 0.0381 (2) | |
| O7 | 0.21333 (13) | 0.30342 (6) | 0.23229 (9) | 0.0411 (2) | |
| O8 | 0.10864 (11) | 0.42926 (7) | 0.14227 (8) | 0.0380 (2) | |
| Cl1 | 0.24776 (3) | 0.63777 (2) | 0.61025 (2) | 0.02082 (4) | |
| Cl2 | 0.23354 (3) | 0.39588 (2) | 0.21099 (2) | 0.02216 (5) | |
| Cu1 | 0.23119 (2) | 0.42802 (2) | 0.65438 (2) | 0.01773 (3) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0265 (5) | 0.0390 (6) | 0.0278 (5) | −0.0022 (4) | −0.0100 (4) | 0.0095 (4) |
| C2 | 0.0368 (6) | 0.0465 (7) | 0.0184 (4) | −0.0037 (5) | −0.0038 (4) | −0.0015 (4) |
| C3 | 0.0296 (5) | 0.0328 (5) | 0.0375 (6) | 0.0004 (4) | 0.0110 (4) | 0.0077 (4) |
| C4 | 0.0176 (4) | 0.0285 (5) | 0.0600 (8) | −0.0005 (4) | 0.0049 (5) | 0.0049 (5) |
| C5 | 0.0299 (5) | 0.0191 (4) | 0.0386 (5) | −0.0049 (3) | −0.0117 (4) | 0.0052 (4) |
| C6 | 0.0523 (7) | 0.0237 (5) | 0.0266 (5) | −0.0087 (5) | −0.0064 (5) | −0.0022 (4) |
| C7 | 0.0307 (5) | 0.0206 (4) | 0.0402 (6) | 0.0008 (4) | 0.0099 (4) | 0.0013 (4) |
| C8 | 0.0199 (4) | 0.0252 (5) | 0.0501 (7) | −0.0006 (3) | 0.0081 (4) | 0.0014 (4) |
| N1 | 0.0190 (3) | 0.0217 (3) | 0.0282 (4) | −0.0023 (3) | −0.0021 (3) | 0.0033 (3) |
| N2 | 0.0241 (4) | 0.0280 (4) | 0.0259 (4) | 0.0011 (3) | 0.0030 (3) | 0.0013 (3) |
| N3 | 0.0236 (4) | 0.0208 (4) | 0.0400 (5) | −0.0022 (3) | −0.0134 (3) | 0.0065 (3) |
| N4 | 0.0403 (5) | 0.0208 (4) | 0.0224 (4) | −0.0040 (3) | 0.0032 (3) | 0.0018 (3) |
| O1 | 0.0361 (4) | 0.0155 (3) | 0.0227 (3) | −0.0025 (3) | −0.0091 (3) | −0.0012 (2) |
| O2 | 0.0213 (4) | 0.0348 (4) | 0.0466 (5) | −0.0019 (3) | −0.0095 (3) | 0.0080 (4) |
| O3 | 0.0542 (6) | 0.0452 (6) | 0.0408 (5) | −0.0136 (5) | 0.0192 (4) | −0.0216 (4) |
| O4 | 0.0420 (5) | 0.0181 (3) | 0.0538 (5) | −0.0016 (3) | −0.0219 (4) | 0.0084 (3) |
| O5 | 0.0614 (6) | 0.0281 (4) | 0.0222 (3) | 0.0054 (4) | −0.0054 (4) | −0.0058 (3) |
| O6 | 0.0258 (4) | 0.0516 (6) | 0.0371 (4) | −0.0021 (4) | 0.0033 (3) | 0.0019 (4) |
| O7 | 0.0536 (6) | 0.0203 (4) | 0.0493 (5) | −0.0032 (4) | 0.0031 (5) | −0.0015 (4) |
| O8 | 0.0302 (4) | 0.0538 (6) | 0.0295 (4) | 0.0164 (4) | −0.0042 (3) | −0.0005 (4) |
| Cl1 | 0.02111 (9) | 0.01695 (8) | 0.02384 (9) | −0.00122 (7) | −0.00426 (7) | −0.00201 (7) |
| Cl2 | 0.02481 (10) | 0.02058 (9) | 0.02081 (9) | 0.00237 (7) | −0.00149 (7) | −0.00166 (7) |
| Cu1 | 0.01806 (5) | 0.01641 (5) | 0.01824 (5) | −0.00136 (3) | −0.00382 (4) | 0.00267 (3) |
Geometric parameters (Å, º)
| C1—N1 | 1.4791 (14) | C7—H7A | 0.9900 |
| C1—C2 | 1.5122 (19) | C7—H7B | 0.9900 |
| C1—H1A | 0.9900 | C8—N1 | 1.4901 (15) |
| C1—H1B | 0.9900 | C8—H8A | 0.9900 |
| C2—N2 | 1.4913 (15) | C8—H8B | 0.9900 |
| C2—H2A | 0.9900 | N1—Cu1 | 2.0061 (9) |
| C2—H2B | 0.9900 | N1—H1 | 0.858 (14) |
| C3—N2 | 1.4781 (16) | N2—Cu1 | 2.0145 (9) |
| C3—C4 | 1.513 (2) | N2—H2 | 0.876 (14) |
| C3—H3A | 0.9900 | N3—Cu1 | 2.0036 (9) |
| C3—H3B | 0.9900 | N3—H3 | 0.859 (13) |
| C4—N3 | 1.4881 (18) | N4—Cu1 | 2.0099 (10) |
| C4—H4A | 0.9900 | N4—H4 | 0.859 (13) |
| C4—H4B | 0.9900 | O1—Cl1 | 1.4644 (7) |
| C5—N3 | 1.4826 (15) | O1—Cu1 | 2.2664 (7) |
| C5—C6 | 1.5118 (19) | O2—Cl1 | 1.4320 (8) |
| C5—H5A | 0.9900 | O3—Cl1 | 1.4267 (10) |
| C5—H5B | 0.9900 | O4—Cl1 | 1.4321 (9) |
| C6—N4 | 1.4898 (15) | O5—Cl2 | 1.4459 (9) |
| C6—H6A | 0.9900 | O6—Cl2 | 1.4441 (10) |
| C6—H6B | 0.9900 | O7—Cl2 | 1.4285 (9) |
| C7—N4 | 1.4835 (16) | O8—Cl2 | 1.4409 (9) |
| C7—C8 | 1.5174 (17) | ||
| N1—C1—C2 | 107.30 (9) | C1—N1—C8 | 115.66 (9) |
| N1—C1—H1A | 110.3 | C1—N1—Cu1 | 102.97 (7) |
| C2—C1—H1A | 110.3 | C8—N1—Cu1 | 107.77 (7) |
| N1—C1—H1B | 110.3 | C1—N1—H1 | 107.1 (11) |
| C2—C1—H1B | 110.3 | C8—N1—H1 | 110.9 (12) |
| H1A—C1—H1B | 108.5 | Cu1—N1—H1 | 112.2 (11) |
| N2—C2—C1 | 109.06 (9) | C3—N2—C2 | 114.31 (10) |
| N2—C2—H2A | 109.9 | C3—N2—Cu1 | 103.55 (7) |
| C1—C2—H2A | 109.9 | C2—N2—Cu1 | 107.38 (7) |
| N2—C2—H2B | 109.9 | C3—N2—H2 | 111.3 (12) |
| C1—C2—H2B | 109.9 | C2—N2—H2 | 109.4 (12) |
| H2A—C2—H2B | 108.3 | Cu1—N2—H2 | 110.7 (11) |
| N2—C3—C4 | 107.81 (10) | C5—N3—C4 | 114.55 (9) |
| N2—C3—H3A | 110.1 | C5—N3—Cu1 | 102.42 (7) |
| C4—C3—H3A | 110.1 | C4—N3—Cu1 | 108.34 (7) |
| N2—C3—H3B | 110.1 | C5—N3—H3 | 106.1 (12) |
| C4—C3—H3B | 110.1 | C4—N3—H3 | 113.6 (12) |
| H3A—C3—H3B | 108.5 | Cu1—N3—H3 | 111.3 (12) |
| N3—C4—C3 | 109.92 (9) | C7—N4—C6 | 114.37 (9) |
| N3—C4—H4A | 109.7 | C7—N4—Cu1 | 102.84 (7) |
| C3—C4—H4A | 109.7 | C6—N4—Cu1 | 107.10 (8) |
| N3—C4—H4B | 109.7 | C7—N4—H4 | 110.1 (12) |
| C3—C4—H4B | 109.7 | C6—N4—H4 | 110.2 (12) |
| H4A—C4—H4B | 108.2 | Cu1—N4—H4 | 112.0 (12) |
| N3—C5—C6 | 107.69 (9) | Cl1—O1—Cu1 | 116.92 (4) |
| N3—C5—H5A | 110.2 | O3—Cl1—O2 | 109.65 (7) |
| C6—C5—H5A | 110.2 | O3—Cl1—O4 | 111.02 (7) |
| N3—C5—H5B | 110.2 | O2—Cl1—O4 | 109.85 (6) |
| C6—C5—H5B | 110.2 | O3—Cl1—O1 | 108.78 (6) |
| H5A—C5—H5B | 108.5 | O2—Cl1—O1 | 109.36 (5) |
| N4—C6—C5 | 109.55 (9) | O4—Cl1—O1 | 108.14 (5) |
| N4—C6—H6A | 109.8 | O7—Cl2—O8 | 109.87 (7) |
| C5—C6—H6A | 109.8 | O7—Cl2—O6 | 109.78 (7) |
| N4—C6—H6B | 109.8 | O8—Cl2—O6 | 109.12 (6) |
| C5—C6—H6B | 109.8 | O7—Cl2—O5 | 109.49 (6) |
| H6A—C6—H6B | 108.2 | O8—Cl2—O5 | 109.95 (6) |
| N4—C7—C8 | 108.37 (9) | O6—Cl2—O5 | 108.61 (6) |
| N4—C7—H7A | 110.0 | N3—Cu1—N1 | 151.33 (4) |
| C8—C7—H7A | 110.0 | N3—Cu1—N4 | 87.11 (4) |
| N4—C7—H7B | 110.0 | N1—Cu1—N4 | 87.14 (4) |
| C8—C7—H7B | 110.0 | N3—Cu1—N2 | 86.24 (4) |
| H7A—C7—H7B | 108.4 | N1—Cu1—N2 | 86.52 (4) |
| N1—C8—C7 | 109.56 (9) | N4—Cu1—N2 | 153.54 (4) |
| N1—C8—H8A | 109.8 | N3—Cu1—O1 | 104.87 (3) |
| C7—C8—H8A | 109.8 | N1—Cu1—O1 | 103.78 (3) |
| N1—C8—H8B | 109.8 | N4—Cu1—O1 | 103.79 (3) |
| C7—C8—H8B | 109.8 | N2—Cu1—O1 | 102.66 (3) |
| H8A—C8—H8B | 108.2 | ||
| N1—C1—C2—N2 | 54.05 (13) | C6—C5—N3—C4 | −168.48 (9) |
| N2—C3—C4—N3 | 50.52 (13) | C6—C5—N3—Cu1 | −51.42 (9) |
| N3—C5—C6—N4 | 53.24 (12) | C3—C4—N3—C5 | 89.53 (11) |
| N4—C7—C8—N1 | 51.48 (13) | C3—C4—N3—Cu1 | −24.09 (11) |
| C2—C1—N1—C8 | −169.09 (9) | C8—C7—N4—C6 | −165.51 (10) |
| C2—C1—N1—Cu1 | −51.82 (10) | C8—C7—N4—Cu1 | −49.77 (10) |
| C7—C8—N1—C1 | 89.34 (12) | C5—C6—N4—C7 | 87.24 (12) |
| C7—C8—N1—Cu1 | −25.21 (11) | C5—C6—N4—Cu1 | −26.00 (11) |
| C4—C3—N2—C2 | −166.49 (10) | Cu1—O1—Cl1—O3 | −58.89 (8) |
| C4—C3—N2—Cu1 | −50.00 (10) | Cu1—O1—Cl1—O2 | 60.85 (7) |
| C1—C2—N2—C3 | 87.01 (12) | Cu1—O1—Cl1—O4 | −179.55 (6) |
| C1—C2—N2—Cu1 | −27.24 (11) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O6i | 0.86 (1) | 2.50 (2) | 3.171 (1) | 135 (1) |
| N1—H1···O2 | 0.86 (1) | 2.39 (1) | 3.093 (1) | 139 (1) |
| N2—H2···O8ii | 0.88 (2) | 2.31 (2) | 3.050 (1) | 142 (1) |
| N2—H2···O3 | 0.88 (2) | 2.44 (2) | 3.052 (2) | 127 (1) |
| N3—H3···O1ii | 0.86 (2) | 2.40 (1) | 3.245 (1) | 169 (2) |
| N3—H3···O4ii | 0.86 (2) | 2.55 (2) | 3.132 (1) | 126 (1) |
| N4—H4···O5 | 0.86 (2) | 2.36 (1) | 3.096 (1) | 143 (1) |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x, −y+1, −z+1.
References
- Addison, A. W., Rao, N. T., Reedijk, J., van Rijn, J. & Verschoor, G. C. (1984). J. Chem. Soc. Dalton Trans. pp. 1349–1356.
- Bruker (2013). SAINT-Plus and APEX2. Bruker AXS Inc., Madison, Wisconsin, USA.
- Clay, R., Murray-Rust, P. & Murray-Rust, J. (1979). Acta Cryst. B35, 1894–1895.
- De León-Rodríguez, L. M., Viswanathan, S. & Sherry, A. D. (2010). Contrast Media Mol. Imaging, 5, 121–125. [DOI] [PMC free article] [PubMed]
- Guo, J.-F., Yeung, W.-F., Gao, S., Lee, G.-H., Peng, S.-M., Lam, M. H.-W. & Lau, T.-C. (2008). Eur. J. Inorg. Chem. pp. 158–163.
- Hormann, J., van der Meer, M., Sarkar, B. & Kulak, N. (2015). Eur. J. Inorg. Chem. pp. 4722–4730.
- Hübschle, C. B., Sheldrick, G. M. & Dittrich, B. (2011). J. Appl. Cryst. 44, 1281–1284. [DOI] [PMC free article] [PubMed]
- Kruppa, M., Frank, D., Leffler-Schuster, H. & König, B. (2006). Inorg. Chim. Acta, 359, 1159–1168.
- Li, S., Chen, J.-X., Xiang, Q.-X., Zhang, L.-Q., Zhou, C.-H., Xie, J.-Q., Yu, L. & Li, F.-Z. (2014). Eur. J. Med. Chem. 84, 677–686. [DOI] [PubMed]
- Pérez-Toro, I., Domínguez-Martín, A., Choquesillo-Lazarte, D., Vílchez-Rodríguez, E., González-Pérez, J. M., Castiñeiras, A. & Niclós-Gutiérrez, J. (2015). J. Inorg. Biochem. 148, 84–92. [DOI] [PubMed]
- Rohde, D. & Merzweiler, K. (2010). Acta Cryst. E66, m894. [DOI] [PMC free article] [PubMed]
- Sarma, M., Chatterjee, T. & Das, S. K. (2010). Inorg. Chem. Commun. 13, 1114–1117.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2014). SADABS. University of Göttingen, Germany.
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Yoo, S. H., Lee, B. J., Kim, H. & Suh, J. (2005). J. Am. Chem. Soc. 127, 9593–9602. [DOI] [PubMed]
- Zhang, X., Liu, X., Phillips, D. L. & Zhao, C. (2016). ACS Catal. 6, 248–257.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989016019563/zl2687sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016019563/zl2687Isup2.hkl
CCDC reference: 1521075
Additional supporting information: crystallographic information; 3D view; checkCIF report