Abstract
Alterations in expression of the DFF40 gene have been reported in some cancers. This study is an in vitro study of the therapeutic effects of gene transfer that lead to elevation in DFF40 expression within T-47D cells in the presence of sulfonamide drugs. In this study, we have constructed a eukaryotic expression vector for DFF40 and transfected it into T-47D cancer cells. We used real time RT-PCR to detect the expression of DFF40 and the MTT assay to determine effects of the sulfonamide drugs acetazolamide, sulfabenzamide, sulfathiazole and sulfacetamide on cell viability in the presence of increased and normal DFF40 levels. Cell cycle distribution was assessed by propidium iodide (PI) staining and the rates of apoptosis by annexin V/PI staining. The DNA laddering analysis was employed to evaluate apoptosis. We observed that overexpression of DFF40 was only effective in decreasing viability in cells incubated with acetazolamide and sulfabenzamide. There was enhanced apoptosis in these groups, particularly with acetazolamide. The cell cycle distribution analysis showed that in the presence of sulfonamide drugs there were no substantial changes in empty-vector or DFF40-transfected cells, except for those cells treated with sulfabenzamide or sulfathiazole. There was no DNA laddering in cells that expressed the empty vector when incubated with sulfonamide drugs. In contrast, we observed DNA laddering in cells that expressed DFF40 in the presence of acetazolamide. Our results have demonstrated that combinatorial use of some sulfonamides such as acetazolamide along with increased expression of DFF40 can potently kill tumor cells via apoptosis and may be beneficial for treatment of some chemoresistant cancers.
Keywords: DFF40, Sulfonamide drugs, Apoptosis, Cancer
Introduction
The majority of chemotherapy drugs act by induction of apoptosis in cancer cells. However, in numerous malignancies the cancer cells can escape from apoptosis by the up-regulation of anti-apoptotic proteins and down-regulation of pro-apoptotic proteins [1]. Thus, interference with the apoptosis pathways may provide effective method for cancer therapy [2]. DNA laddering is a late event during apoptosis. DFF40, the main enzyme involved in this process, cleaves DNA into 30–50 kbp and 200 bp during early and late apoptosis, respectively [3]. In non-apoptotic cells, DFF40 is inhibited by DFF45, which also has a chaperone role allowing for appropriate folding of DFF40 into an active nuclease [4]. Activated caspase-3 in apoptotic cells degrades DFF45 and activates DFF40. The deletion or aberrantly spliced transcripts of the DFF40 gene is observed in some cancers [5–7].
Sulfonamides are a large family of drugs that have various therapeutic potential uses which include anti-bacterial [8], anti-carbonic anhydrase [9], anti-metalloprotease [10], diuretic [11], anti-diabetic [12], anti-thyroid [13], antiviral, and anti-inflammatory [10] activities. Other studies have shown that these drugs can inhibit the proliferation and growth of cancer cells, which opens a new perspective in cancer treatment. Despite the common chemical domain in the sulfonamide family, different mechanisms are used to inhibit cancer cells and include carbonic anhydrase inhibition [14], disruptions to the cell cycle during G1/S and G2/M transitions by disruption of microtubule assembly [15–17], functional suppression of the transcriptional activator NF-Y [18], and angiogenesis inhibition [19, 20].
Here we evaluated the viability, cell cycle arrest, and induction of apoptosis in T-47D cells in the presence of four sulfa drugs: acetazolamide, sulfabenzamide, sulfathiazole and sulfacetamide in order to determine their anti-proliferative mechanisms of action. Acetazolamide as a potent carbonic anhydrase inhibitor is a good choice for treatment of glaucoma, benign intracranial hypertension, altitude sickness, and seizures [21–25]. Acetazolamide can also inhibit cancer cell growth and invasion by inhibition of carbonic anhydrases and decreased expression of aquaporin-1 mRNA and protein. Other mechanisms of action of this drug have not been clarified [14, 26]. Sulfabenzamide, sulfathiazole, and sulfacetamide are also used to treat specific vaginal infections. In this study we have overexpressed DFF40 as a proapoptotic protein and followed its effects on T-47D cell response to sulfonamide drugs.
Materials and methods
Reagents
Roswell Park Memorial Institute Medium 1640 (RPMI 1640), fetal bovine serum (FBS), streptomycin and penicillin were purchased from Gibco (UK). TRIzol reagent was obtained from the Invitrogen Corporation (Carlsbad, CA, USA). 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide (MTT) was purchased from Sigma (USA). The High Pure PCR Purification Kit, DNA Laddering and Annexin-V-FLOUS Staining Kits were obtained from Roche (Germany). The Plasmid Purification Kit was purchased from Qiagen. We obtained the Power SYBR® Green PCR Master Mix (Applied Biosystems, UK), cDNA Synthesis Kit, T4 DNA ligase and restriction enzymes from Fermentas Company (Germany). The pIRES2-EGFP and G418 were obtained from Clontech. The sodium salt of acetazolamide and other sulfonamides were purchased from Aventis Pharma (Canada) and Sina Darou (Iran), respectively.
Construction of the pIRES2-EGFP-DFF40 expression vector
The DFF40 sequence was prepared by PCR amplification from fibroblast cDNA by forward 5′-aatctcgaggcaat gctccagaagcccaag-3′ and reverse 5′-aatccgcggtcactggcg tttccgcacagg-3′ primers with restriction sites for XhoI and SacII (bold–underlined sequences). The PCR product and pIRES2-EGFP vector were digested with XhoI and SacII. Ligation of pIRES2-EGFP vector and PCR product of DFF40 was performed by mixing the vector and inserted DNA at a 1:5 ratio with 1 U of ligase (T4 DNA ligase) in 1× ligase buffer. Plasmid DNA was then transformed into DH5α Escherichia coli (Invitrogen) according to the manufacturer’s protocol. Selected colonies were amplified overnight using a 4 ml broth culture, purified using the plasmid purification kit, and sequenced for accuracy prior to use in transfection experiments. For stable transfection, the pIRES2-EGFP-DFF40 and pIRES2-EGFP vectors (empty vector) were linearized by AseI restriction enzyme and purified by the High Pure PCR Purification Kit.
Cell culture, stable transfection, and detection of the DFF40 mRNA in transfected cells
The human breast cancer cell line (T-47D) was obtained from the Cell Bank of Pasteur Institute, Tehran, Iran. T-47D cells were grown in RPMI 1640 supplemented with 10 % FBS, penicillin (100 unit/ml) and streptomycin (100 μg/ml). Cells were maintained in a humidified atmosphere with 5 % CO2 at 37 °C. The culture medium was changed every other day and the cells were passaged when they reached 80–90 % confluency. For transfection, 5 × 106 cells were resuspended in 0.5 ml of PBS, mixed with 20 μg plasmid DNA and electroporated (350 V, 500 μF). The transfection mixture was added to 14 ml of RPMI medium that contained 10 % FBS and seeded into a 75 cm2 flask. After a 2-day incubation period, the medium was replaced with medium that contained G418 (600 μg/ml). T-47D cells were transfected with the empty vector as the control.
Cellular DFF40 mRNA level was determined by real time RT-PCR. Total RNA was prepared from cultured cells using TRIzol reagent as recommended by the manufacturer’s single-step chloroform extraction protocol. cDNA was generated by reverse transcription of 1 μg of total RNA using random hexamer primers (100 μM) and RevertAid™ M-MuLV Reverse Transcriptase working at 25 °C for 5 min and 42 °C for 1 h in a total reaction volume of 20 μl. The cDNA (25 ng) was amplified by specific DFF40 primers (forward: 5′-ttggagtcccgatttcagag-3′, reverse: 5′-ctgtcgaagtagctgccattg-3′) and Power SYBR® Green PCR Master Mix in an ABI device (Applied Biosystems). Reaction parameters were: 95 °C for 10 min, followed by 95 °C for 10 s and 60 °C for 1 min for 30 cycles. Relative gene expression of DFF40 was calculated with the 2−(ΔΔCT) method using GAPDH as the reference gene. To confirm PCR specificity, we subjected the PCR products to a melting-curve analysis. The expression level of DFF45 was determined with DFF45 specific primers (forward: 5′-ttctgtgtctaccttccaatacta-3′, reverse: 5′-ctgtctg tttcatctac atcaaag-3′).
Incubation of cells with sulfonamide drugs
The sulfonamide drugs (acetazolamide, sulfabenzamide, sulfathiazole, and sulfacetamide) were dissolved at their LC50 concentrations (determined from the MTT assays) in RPMI supplemented with 10 % FBS, penicillin (100 unit/ml), and streptomycin (100 μg/ml). The cells in two groups (cells transfected with empty vector or DFF40) were seeded 24 h before treatment. At 50 % confluency, cells were incubated with freshly prepared drugs at respective LC50 concentrations. The cells were incubated for 48 h and then tested for viability, cell cycle distribution, and apoptosis.
Cell viability assay
The viability of cells that expressed the empty vector or DFF40 was determined in the presence of sulfonamide drugs by the MTT assay. The viable cells with an active respiratory chain and other electron transport systems can reduce MTT and other tetrazolium salts, and thereby form violet formazan crystals within the cells. In brief, after incubation with drugs, the medium was replaced with a 5:1 ratio of medium and MTT solution (5 mg/ml in PBS). The cells were incubated for 2 h at 37 °C until purple formazan crystals were formed. Finally, the MTT-containing medium was removed, the formazan crystals were dissolved in dimethyl sulfoxide (DMSO) and absorbance was read at 570 nm. Cell viability was calculated as percent value relative to the blank group that was cultured in RPMI alone.
Cell cycle phase distribution
Following incubation under various treatments, cells were collected, centrifuged and washed with PBS, then fixed with ice-cold 70 % ethanol for 2 h. The cell suspension was centrifuged, resuspended in PBS that contained propidium iodide (10 μg/ml) and RNase A (100 μg/ml), and stored at room temperature for 30 min before flow cytometry analysis. A total of 10,000 events were acquired using a FACS Calibur flow cytometer (BD Biosciences). The two-dimensional plots of DNA content (x-axis, PI fluorescence) versu count (y-axis) were drawn for analysis.
Induction of apoptosis and necrosis in the presence of sulfonamide drugs
Apoptotic and necrotic cells were detected by the Annexin-V-FLOUS Staining Kit. The cells that expressed empty or DFF40 vector were incubated with sulfonamide drugs for 48 h. Following incubation, both attached and floating cells were collected and stained with annexin V-FITC/PI. The percentages of early apoptotic, late apoptotic, and necrotic cells were determined by flow cytometry. The occurrence of apoptosis in each group was assayed three times.
DNA laddering
Cells that expressed the empty vector or DFF40 were cultured in 25 cm2 flasks and incubated with the sulfonamide drugs for 48 h. Following incubation, the cells were lysed in 200 μl lysis/binding buffer that contained 6 M guanidine-HCl, 10 mM urea, 10 mM Tris–HCl, and 20 % Triton X-100 for 10 min at 25 °C. After the 10 min incubation, 100 μl isopropanol was added to the samples and transferred into the upper reservoir of the filter tubes (Roche, Germany). The filter tubes were centrifuged for 1 min at 8,000 rpm. After washing, the bound DNA was separated from the filter tube by 200 μl pre-warmed elution buffer (70 °C). The amount of DNA was quantified spectrophotometrically at 260 nm. Equal amounts of DNA (3 μg) were loaded on a 1 % agarose gel and subjected to electrophoresis at 75 V for 90 min. DNA bands were visualized with UV light after ethidium bromide staining.
Results
Expression of exogenous DFF40 in T-47D cells
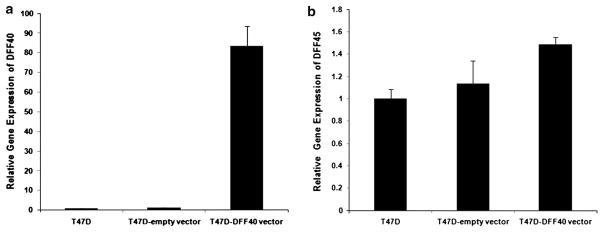
The eukaryotic expression vector pIRES2-EGFP-DFF40 was prepared and used to transfect T-47D cells by electroporation. After 3 days, the un-transfected cells were removed by G418 selection. We determined transfection efficiency by fluorescence microscopy of the GFP protein. The cells were then evaluated for expression of DFF40 by real time RT-PCR. The results showed that the DFF40 transfected cells expressed approximately 80-fold more DFF40 mRNA compared with cells transfected with the empty vector or non-transfected cells (Fig. 1a). Interestingly, the expression of DFF45 did not significantly change among the transfected cells (Fig. 1b).
Fig. 1.
Cellular DFF40 and DFF45 mRNA expression detected by real time RT-PCR in transfected T-47D cells. a DFF40 mRNA increased ~80-fold in DFF40 transfected cells compared with control cells. b Overexpression of DFF40 had no significant effect on expression of DFF45 mRNA. Relative gene expression was calculated with the 2−(ΔΔCT) method using GAPDH as the reference gene. Numerical results are mean ± SD from at least three replicated experiments
Cell viability assessment by MTT assay
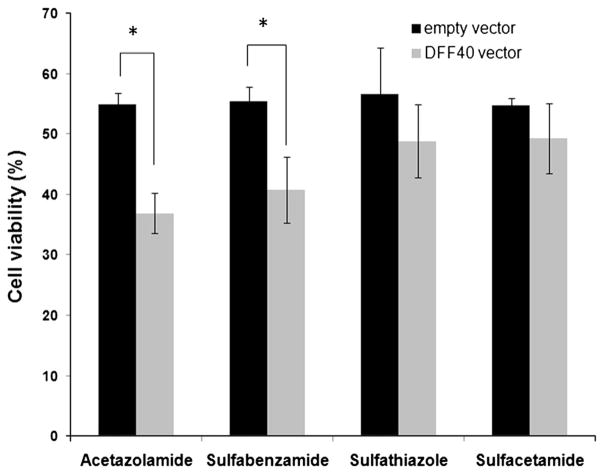
The two groups of cells (those transfected with DFF40 or empty vector) were incubated with sulfonamide drugs at their LC50. We performed the MTT assay for all groups after 48 h. Overexpression of DFF40 sensitized the T-47D cells to the cytotoxic effects of the sulfonamide drugs. However, a significant effect was only detected in cells incubated with acetazolamide and sulfabenzamide (Fig. 2). We assessed the cell cycle distribution and apoptosis in order to determine the mechanisms that contributed to reduced survival in these cells.
Fig. 2.
Impact of DFF40 expression and sulfa drug treatments on cell viability was evaluated by the MTT assay. Cell viability was calculated as percent value relative to the blank (untreated) group. Data are shown as mean ± SD from three independent experiments
Cell cycle analysis
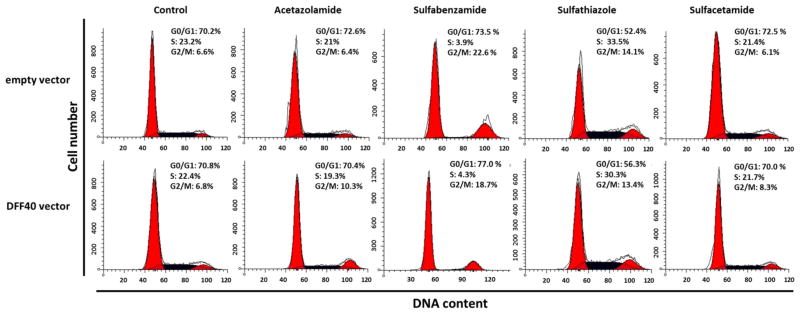
To determine the proportion of cells in different phases of the cell cycle, we used PI and flow cytometry to detect the DNA content in DFF40 and control cells (including the empty vector). We compared each phase between the two groups of cells (DFF40 and empty vector transfected cells) and concluded that the cell cycle arrest was not the main cause for reduction of cell viability in cells that overexpressed DFF40 (Fig. 3). We considered the effect of sulfonamide drugs on the cell cycle in order to study the mechanisms for cytotoxicity. There were no significant differences in the cell cycle distributions when the cells were incubated with sulfonamide drugs, with the exception of sulfabenzamide and sulfathiazole. In both groups (DFF40 and control group), incubation with sulfabenzamide resulted in partial G2/M arrest whereas incubation with sulfathiazole resulted in partial S+G2/M arrest (Fig. 3).
Fig. 3.
Cells were incubated with the LC50 concentration of each drug for 48 h. Cell cycle analysis was performed as described in Materials and Methods. Upper row shows cell cycle dissipation for empty vector transfected cells and lower row indicates the same dissipation for DFF40 transfected cells. Data are representative of at least three independent experiments
Apoptosis/cell death assay
We used annexin V-FITC/PI double staining to determine whether reduced cell viability is due to decreased cell proliferation or increased cell death. In the early stages of apoptosis, phosphatidylserine (PS) transfers from the inner part of the plasma membrane to the outer layer, by which PS becomes exposed at the external surface of the cell. Annexin V is a phospholipid binding protein with a high affinity for PS. In addition, the membrane of a necrotic cell is permeable to PI, causing its accumulation in the cell nucleus.
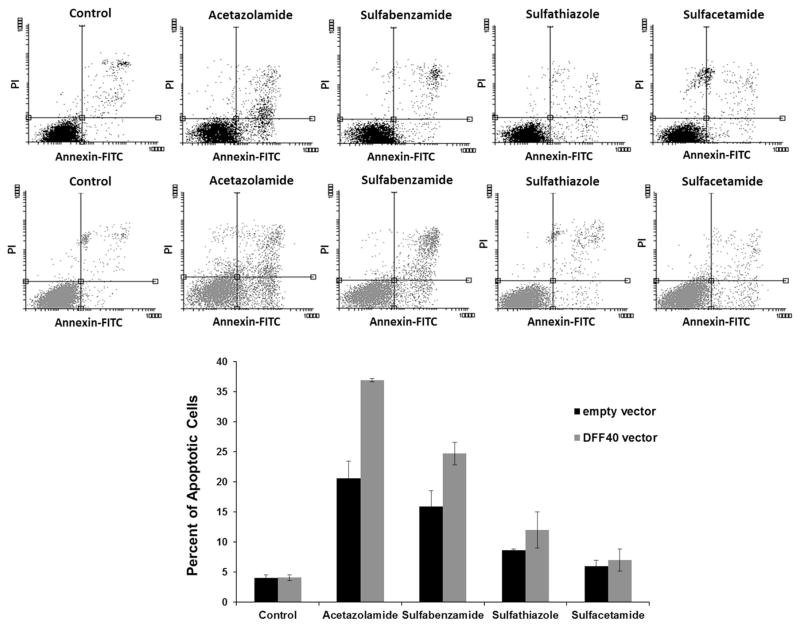
Analysis of data was performed using the Flow software 2 and two-dimensional plots were divided into four square regions, Annexin V-FITC− and PI+ (upper left; necrotic cells), Annexin V-FITC+ and PI+ (upper right; late apoptotic cells), Annexin V-FITC− and PI− (lower left; alive cells), and Annexin V-FITC+ and PI− (lower right; early apoptotic cells). Based on these results, acetazolamide and sulfabenzamide induced apoptosis. We observed 20 % apoptotic cells with acetazolamide treatment and 15 % apoptotic cells with sulfabenzamide treatment. These amounts increased to 35 and 25 % in cells that overexpressed DFF40. The changes in apoptosis were negligible with other drugs (Fig. 4).
Fig. 4.
Incubation with acetazolamide and sulfabenzamide induced apoptosis in DFF40 transfected cells. The cells transfected with empty or DFF40 vector were incubated with sulfonamides at LC50 concentrations for 48 h. The percentage of apoptotic cells (upper right and lower right) is shown. The percentages of apoptotic cells are shown as mean ± SD (n = 3)
DNA laddering
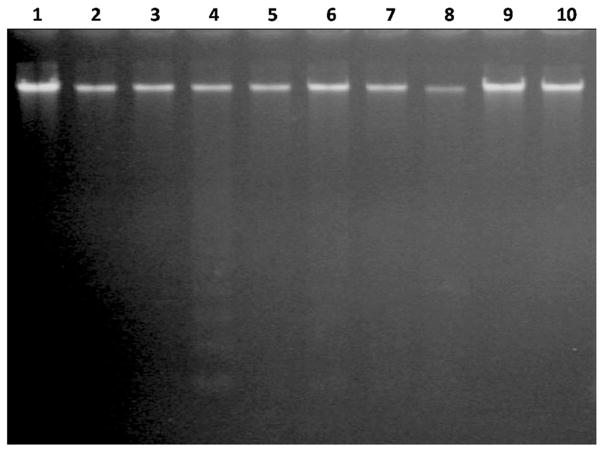
DNA laddering is a hallmark of late apoptosis where DNA becomes cleaved at inter-nucleosomal chromatin regions [27]. Our results showed that internucleosomal DNA fragmentation did not occur in cells incubated with LC50 concentrations of the sulfonamide drugs. However, overexpression of DFF40 in combination with acetazolamide resulted in nucleosomal cleavage of DNA (Fig. 5).
Fig. 5.
DNA gel electrophoresis. Cells transfected with empty or DFF40 vector were incubated with sulfonamide drugs. After 48 h, genomic DNA was extracted and subjected to gel electrophoresis. DNA ladder formation was observed in DFF40 transfected cells incubated with acetazolamide. Lane 1: Control (−DFF40); lane 2: Control (+DFF40); lane 3: Acetazolamide (−DFF40); lane 4: Acetazolamide (+DFF40); lane 5: Sulfabenzamide (−DFF40); lane 6: Sulfabenzamide (+DFF40); lane 7: Sulfathiazole (−DFF40); lane 8: Sulfathiazole (+DFF40); lane 9: Sulfacetamide (−DFF40); lane 10: Sulfacetamide (+DFF40)
Discussion
A major obstacle in chemotherapy treatment is the resistance of cancer cells to programmed cell death or apoptosis. Morphological and biochemical events in apoptotic cells are remarkably similar across cell types and species. These alterations include activation of caspases, rounding up and shrinkage of cells, externalization of PS, chromatin condensation, membrane blebbing and DNA fragmentation [28]. DNA fragmentation is a late process in apoptosis and DFF40 is a key factor in this process. Since DFF40 is a downstream element in apoptosis its activation causes irreparable DNA damage leading to assured cell death. Thus, DFF40 can be considered as a preferable target for inducing apoptosis in cancer cells.
In this study, our first priority was to enhance the anti-cancer potency of some chemical drugs via augmentation of cellular tendency toward apoptosis. This was achieved by overexpression of DFF40, the main DNA nuclease at the late stage of the apoptosis pathway and cell death. We stably overexpressed DFF40 and unlike the Yuichi Kimura et al. study who showed that transient expression of DFF40 induced cell death [29], we observed no cell death in the absence of apoptosis. DFF40 needs DFF45 for correct folding, however DFF45 can also inhibit it [27, 30, 31]. In fact, both the existence of DFF45 and also its elimination via apoptosis induction are compulsory for DFF40 to fragment DNA.
Here we have examined the effect of DFF40 overexpression in T-47D cells incubated with drugs from the sulfonamide family. We chose these drugs because our previous work showed that these cells were resistant to apoptosis in the presence of sulfa drugs [32]. Thus, if the sulfa drugs could induce apoptosis in the DFF40-overex-pressed cells, we would be able to confirm our hypothesis that increased expression of DFF40 results in enhanced apoptosis.
Sulfonamides have a classical role in treatment of infections; however, some have anti-cancer effects. Different mechanisms are reported to explain the anticancer effects of these drugs. Some have anticancer effects by inducing cell cycle arrest and apoptosis. J30 (a novel oral indoline-sulfonamide agent) prevents assembly of tubulin by strongly binding to the colchicine-binding site resulting in accumulation of cancer cells in the G2/M phase of the cell cycle. J30 also causes apoptosis by Bcl-2 phosphorylation and activation of the caspase-9 and caspase-3 cascades [17]. Others have shown the anticancer effects of sulfa drugs via induction of autophagy [32, 33]. Additional strategies regarding anticancer effects of sulfonamide drugs include inhibition of carbonic anhydrases, matrix metalloproteinases, and interference in gene expression. [14, 18, 19].
We initially studied the cell cycle arrest and apoptosis induction at LC50 values of four sulfonamide drugs, acetazolamide (26 mM), sulfabenzamide (10.8 mM), sulfathiazole (6.5 mM) and sulfacetamide (41 mM) in the T-47D cells that expressed the empty vector. There was no change in cell cycle distribution except in cells incubated with sulfabenzamide and sulfathiazole, in which we observed cell cycle arrest in G2/M and S+G2/M (16 and 18 % increase relative to the non-treated cells), respectively (Fig. 3). Our data illustrated that cell cycle arrest was not the main mechanism for the anticancer effects of the four sulfa drugs in decreasing cell viability (Fig. 2).
In order to determine whether apoptosis was the main reason for the anticancer effects of these sulfa drugs, we measured the apoptosis induction. There were 20 % apoptotic cells in the cells treated with acetazolamide and 15 % apoptotic cells in those treated with sulfabenzamide. There were negligible apoptotic cells observed in those treated with sulfathiazole and sulfacetamide (Fig. 4). However, the increased apoptosis observed under the various treatments were not enough to account for the 50 % reduction in viability observed in the MTT assays. Thus, cell cycle arrest and apoptosis induction were not the main mechanisms for the observed reduction in cell viability. However, apoptosis can be a subsidiary mechanism for the anticancer effects following acetazolamide and sulfabenzamide treatments.
Our results indicated that DFF40-overexpressing cells exhibited enhanced apoptosis in response to acetazolamide and sulfabenzamide. The net enhancements in apoptosis were 15 (acetazolamide) and 10 % (sulfabenzamide; Fig. 4). This supported the MTT results where the viability of these cells significantly decreased to 35 (acetazolamide) and 40 % (sulfabenzamide; Fig. 2). The equality between the intensified apoptosis and diminished viability of the cells has shown that the cause for further cell death in DFF40-overexpressed cells is apoptosis. This notion supports our hypothesis regarding augmentation of the potency of drugs for apoptosis in DFF40-over expressing cells. Thus, enhancement of DFF40 is able to increase cell sensitivity to apoptosis and could be envisaged as a therapeutic means for cancer treatment.
An additional tool for detection of cells undergoing apoptosis is DNA gel electrophoresis, which discerns DNA cleavage. Internucleosomal DNA fragmentation was only observed in DFF40-overexpressed cells incubated with acetazolamide (Fig. 5). Thus, enhanced apoptosis in DFF40-over expressing cells is the main cause for DNA laddering in acetazolamide treatment. A conceivable explanation for this observation is the enhancement of caspase-3 activity in the presence of sulfa drugs to eliminate DFF45 inhibiting effects on DFF40. Increased caspase-3 activity, in the presence of sulfa drugs, was reported in our recent studies [32, 33]. Upon the induction of apoptosis, DFF45 is cleaved by caspase-3 and released from DFF40. The activated DFF40 breaks DNA in the inter-nucleosomal chromatin regions [34].
Our previous studies showed that in the presence of sulfa drugs, despite increased caspase-3 activity, there was no DNA fragmentation in T-47D cells. In these cells the expression ratio of DFF45/DFF40 increased, which led to stronger DFF45 inhibitory effects on DFF40 in order to halt DNA fragmentation. In acetazolamide treatment, the increased DFF45/DFF40 ratio originated from reduction of DFF40 and unchanged expression level of DFF45 [35]. Thus, overexpression of DFF40 (Fig. 1a) with minimal effect on expression of DFF45 (Fig. 1b) results in a decline in the DFF45/DFF40 ratio, which causes DNA fragmentation in the presence of acetazolamide (Fig. 5).
In summary, the present results indicated that overexpression of DFF40 was effective for apoptosis amplification, particularly for those drugs that had the potential for induction of apoptosis. Among the four sulfonamide drugs, only acetazolamide acted in this manner and decreased the DFF45/DFF40 ratio which triggered apoptosis in the target cells. Thus, overexpression of DFF40 in the presence of some apoptosis-inducing drugs might be targeted as a therapeutic treatment of breast and perhaps other cancers.
Acknowledgments
The authors would like to thank Iran National Science Foundation (INSF) and Royan Institute of Stem Cell Biology and Technology for their financial support. The authors express their appreciation to the Research Council of the University of Tehran for their valuable patronages.
Contributor Information
Fatemeh Bagheri, Department of Cell and Molecular Biology, School of Biology, College of Science, University of Tehran, Tehran, Iran. Department of Stem Cells and Developmental Biology at Cell Sciences Research Center, Royan Institute for Stem Cell Biology and Technology, ACECR, Tehran, Iran.
Shahrokh Safarian, Department of Cell and Molecular Biology, School of Biology, College of Science, University of Tehran, Tehran, Iran.
Mohamadreza Baghaban Eslaminejad, Department of Stem Cells and Developmental Biology at Cell Sciences Research Center, Royan Institute for Stem Cell Biology and Technology, ACECR, Tehran, Iran.
Nader Sheibani, Department of Ophthalmology and Visual Sciences, University of Wisconsin School of Medicine and Public Health, Madison, WI, USA.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Igney FH, Krammer PH. Death and anti-death: tumour resistance to apoptosis. Nat Rev Cancer. 2002;2(4):277–288. doi: 10.1038/nrc776. [DOI] [PubMed] [Google Scholar]
- 3.Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391(6662):43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- 4.Otomo T, Sakahira H, Uegaki K, Nagata S, Yamazaki T. Structure of the heterodimeric complex between CAD domains of CAD and ICAD. Nat Struct Biol. 2000;7(8):658–662. doi: 10.1038/77957. [DOI] [PubMed] [Google Scholar]
- 5.Hsieh SY, Liaw SF, Lee SN, Hsieh PS, Lin KH, Chu CM, Liaw YF. Aberrant caspase-activated DNase (CAD) transcripts in human hepatoma cells. Br J Cancer. 2003;88(2):210–216. doi: 10.1038/sj.bjc.6600695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsieh SY, Chen WY, Yeh TS, Sheen IS, Huang SF. High-frequency Alu-mediated genomic recombination/deletion within the caspase-activated DNase gene in human hepatoma. Oncogene. 2005;24(43):6584–6589. doi: 10.1038/sj.onc.1208803. [DOI] [PubMed] [Google Scholar]
- 7.Yamaguchi K, Uzzo R, Dulin N, Finke JH, Kolenko V. Renal carcinoma cells undergo apoptosis without oligonucleosomal DNA fragmentation. Biochem Biophys Res Commun. 2004;318(3):710–713. doi: 10.1016/j.bbrc.2004.04.086. [DOI] [PubMed] [Google Scholar]
- 8.Seydel JK. Sulfonamides, structure-activity relationship, and mode of action. Structural problems of the antibacterial action of 4-aminobenzoic acid (PABA) antagonists. J Pharm Sci. 1968;57(9):1455–1478. doi: 10.1002/jps.2600570902. [DOI] [PubMed] [Google Scholar]
- 9.Lindskog S, Thorslund A. On the interaction of bovine cobalt carbonic anhydrase with sulfonamides. Eur J Biochem. 1968;3(4):453–460. doi: 10.1111/j.1432-1033.1967.tb19552.x. [DOI] [PubMed] [Google Scholar]
- 10.Supuran CT, Casini A, Scozzafava A. Protease inhibitors of the sulfonamide type: anticancer, antiinflammatory, and antiviral agents. Med Res Rev. 2003;23(5):535–558. doi: 10.1002/med.10047. [DOI] [PubMed] [Google Scholar]
- 11.Maren TH. Relatons between structure and biological activity of sulfonamides. Annu Rev Pharmacol Toxicol. 1976;16:309–327. doi: 10.1146/annurev.pa.16.040176.001521. [DOI] [PubMed] [Google Scholar]
- 12.Loubatieres-Mariani MM. The discovery of hypoglycemic sulfonamides. J Soc Biol. 2007;201(2):121–125. doi: 10.1051/jbio:2007014. [DOI] [PubMed] [Google Scholar]
- 13.Comby F, Lagorce JF, Moulard T, Buxeraud J, Raby C. Antibacterial sulfonamides, antiparasitic and antifungal derivatives of imidazole: evaluation of their antithyroid effects in the rat. Vet Res. 1993;24(4):316–326. [PubMed] [Google Scholar]
- 14.Parkkila S, Rajaniemi H, Parkkila AK, Kivela J, Waheed A, Pastorekova S, Pastorek J, Sly WS. Carbonic anhydrase inhibitor suppresses invasion of renal cancer cells in vitro. Proc Natl Acad Sci U S A. 2000;97(5):2220–2224. doi: 10.1073/pnas.040554897040554897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Owa T, Yoshino H, Okauchi T, Yoshimatsu K, Ozawa Y, Sugi NH, Nagasu T, Koyanagi N, Kitoh K. Discovery of novel antitumor sulfonamides targeting G1 phase of the cell cycle. J Med Chem. 1999;42(19):3789–3799. doi: 10.1021/jm9902638. jm9902638. [DOI] [PubMed] [Google Scholar]
- 16.Fukuoka K, Usuda J, Iwamoto Y, Fukumoto H, Nakamura T, Yoneda T, Narita N, Saijo N, Nishio K. Mechanisms of action of the novel sulfonamide anticancer agent E7070 on cell cycle progression in human non-small cell lung cancer cells. Invest New Drugs. 2001;19(3):219–227. doi: 10.1023/a:1010608317361. [DOI] [PubMed] [Google Scholar]
- 17.Liou JP, Hsu KS, Kuo CC, Chang CY, Chang JY. A novel oral indoline-sulfonamide agent, N-[1-(4-methoxybenzenesulfonyl)-2,3-dihydro-1H-indol-7-yl]-isonicotinamide (J30), exhibits potent activity against human cancer cells in vitro and in vivo through the disruption of microtubule. J Pharmacol Exp Ther. 2007;323(1):398–405. doi: 10.1124/jpet.107.126680. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka H, Ohshima N, Ikenoya M, Komori K, Katoh F, Hidaka H. HMN-176, an active metabolite of the synthetic antitumor agent HMN-214, restores chemosensitivity to multidrug-resistant cells by targeting the transcription factor NF-Y. Cancer Res. 2003;63(20):6942–6947. [PubMed] [Google Scholar]
- 19.Cheng XC, Wang Q, Fang H, Xu WF. Role of sulfonamide group in matrix metalloproteinase inhibitors. Curr Med Chem. 2008;15(4):368–373. doi: 10.2174/092986708783497300. [DOI] [PubMed] [Google Scholar]
- 20.Funahashi Y, Sugi NH, Semba T, Yamamoto Y, Hamaoka S, Tsukahara-Tamai N, Ozawa Y, Tsuruoka A, Nara K, Takahashi K, Okabe T, Kamata J, Owa T, Ueda N, Haneda T, Yonaga M, Yoshimatsu K, Wakabayashi T. Sulfonamide derivative, E7820, is a unique angiogenesis inhibitor suppressing an expression of integrin alpha2 subunit on endothelium. Cancer Res. 2002;62(21):6116–6123. [PubMed] [Google Scholar]
- 21.Reiss WG, Oles KS. Acetazolamide in the treatment of seizures. Ann Pharmacother. 1996;30(5):514–519. doi: 10.1177/106002809603000515. [DOI] [PubMed] [Google Scholar]
- 22.Kaur IP, Smitha R, Aggarwal D, Kapil M. Acetazolamide: future perspective in topical glaucoma therapeutics. Int J Pharm. 2002;248(1–2):1–14. doi: 10.1016/s0378-5173(02)00438-6. [DOI] [PubMed] [Google Scholar]
- 23.Imray C. Acetazolamide for the prophylaxis of acute mountain sickness. BMJ. 345:e7077. doi: 10.1136/bmj.e7077. [DOI] [PubMed] [Google Scholar]
- 24.Leaf DE, Goldfarb DS. Mechanisms of action of acetazolamide in the prophylaxis and treatment of acute mountain sickness. J Appl Physiol. 2007;102(4):1313–1322. doi: 10.1152/japplphysiol.01572.2005. [DOI] [PubMed] [Google Scholar]
- 25.Celebisoy N, Gokcay F, Sirin H, Akyurekli O. Treatment of idiopathic intracranial hypertension: topiramate vs acetazolamide, an open-label study. Acta Neurol Scand. 2007;116(5):322–327. doi: 10.1111/j.1600-0404.2007.00905.x. [DOI] [PubMed] [Google Scholar]
- 26.Xiang Y, Ma B, Li T, Yu HM, Li XJ. Acetazolamide suppresses tumor metastasis and related protein expression in mice bearing Lewis lung carcinoma. Acta Pharmacol Sin. 2002;23(8):745–751. [PubMed] [Google Scholar]
- 27.Liu X, Zou H, Slaughter C, Wang X. DFF, a heterodimeric protein that functions downstream of caspase-3 to trigger DNA fragmentation during apoptosis. Cell. 1997;89(2):175–184. doi: 10.1016/s0092-8674(00)80197-x. [DOI] [PubMed] [Google Scholar]
- 28.Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, Blagosklonny MV, El-Deiry WS, Golstein P, Green DR, Hengartner M, Knight RA, Kumar S, Lipton SA, Malorni W, Nunez G, Peter ME, Tschopp J, Yuan J, Piacentini M, Zhivotovsky B, Melino G. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16(1):3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimura Y, Sugimoto C, Matsukawa S, Sunaga H, Igawa H, Yamamoto H, Ito T, Saito H, Fujieda S. Combined treatment of cisplatin and overexpression of caspase-activated deoxyribonuclease (CAD) promotes apoptosis in vitro and in vivo. Oral Oncol. 2004;40(4):390–399. doi: 10.1016/j.oraloncology.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 30.Zhou P, Lugovskoy AA, McCarty JS, Li P, Wagner G. Solution structure of DFF40 and DFF45 N-terminal domain complex and mutual chaperone activity of DFF40 and DFF45. Proc Natl Acad Sci U S A. 2001;98(11):6051–6055. doi: 10.1073/pnas.111145098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X, Zou H, Widlak P, Garrard W, Wang X. Activation of the apoptotic endonuclease DFF40 (caspase-activated DNase or nuclease). Oligomerization and direct interaction with histone H1. J Biol Chem. 1999;274(20):13836–13840. doi: 10.1074/jbc.274.20.13836. [DOI] [PubMed] [Google Scholar]
- 32.Mohammadpour R, Safarian S, Sheibani N, Norouzi S, Razazan A. Death inducing and cytoprotective autophagy in T-47D cells by two common antibacterial drugs: sulphathiazole and sulphacetamide. Cell Biol Int. 2013;37(4):348–358. doi: 10.1002/cbin.10047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohammadpour R, Safarian S, Farahnak S, Hasheminasl S, Sheibani N. Sulfabenzamide promotes autophagic cell death in T-47D breast cancer cells through p53/DRAM pathway. J Cell Mol Biol. 2012;10(1):41–54. [Google Scholar]
- 34.Sakahira H, Enari M, Nagata S. Cleavage of CAD inhibitor in CAD activation and DNA degradation during apoptosis. Nature. 1998;391(6662):96–99. doi: 10.1038/34214. [DOI] [PubMed] [Google Scholar]
- 35.Mohammadpour R, Safarian S, Ejeian F, Sheikholya-Lavasani Z, Abdolmohammadi MH, Sheinabi N. Acetazolamide triggers death inducing autophagy in T-47D breast cancer cells. Cell Biol Int. 2013 doi: 10.1002/cbin.10197. [DOI] [PubMed] [Google Scholar]