Abstract
Checkpoint blockade with antibodies against CTLA-4 or PD-1 elicits durable tumor regressions in metastatic cancer, but these dramatic responses are confined to a minority of patients1–3. This suboptimal outcome is likely due in part to the complex network of immunosuppressive pathways present in advanced tumors, which are unlikely to be overcome by intervention at a single signaling checkpoint4–8. Here we demonstrate a combination immunotherapy that recruits a variety of innate and adaptive immune cells to eliminate large tumor burdens in syngeneic tumor models and a genetically engineered mouse melanoma model; to our knowledge tumors of this size have not previously been curable by treatments relying on endogenous immunity. Maximal anti-tumor efficacy required four components: a tumor antigen targeting antibody, an extended half-life recombinant IL-29, anti-PD-1, and a powerful T-cell vaccine10. Depletion experiments revealed that CD8+ T-cells, cross-presenting DCs, and several other innate immune cell subsets were required for tumor regression. Effective treatment induced infiltration of immune cells and production of inflammatory cytokines in the tumor, enhanced antibody-mediated tumor antigen uptake, and promoted antigen spreading. These results demonstrate the capacity of an elicited endogenous immune response to destroy large, established tumors and elucidate essential characteristics of combination immunotherapies capable of curing a majority of tumors in experimental settings typically viewed as intractable.
Checkpoint blockade therapies prove that an endogenous immune response can regress human tumors, but significant responses remain restricted to a minority of patients6,11. Combining checkpoint blockade with other agents may improve response rates12–14, and immunotherapies implementing three or more agents in tandem are already in clinical testing15–18. However, even in preclinical models, complete tumor rejection is usually only achieved by treating very small tumors and/or treating at very early times preceding the establishment of a fully developed tumor microenvironment, unless adoptive transfer of ex vivo-expanded T-cells is employed19,20. Autochthonous tumors induced in genetically-engineered animals, which may generate immunosuppressive networks more akin to human tumors, are also generally refractory to immunotherapy21,22. As such, the general characteristics of an endogenous immune response capable of reliably eradicating large immunosuppressive tumor burdens remain unclear.
We recently reported that therapies combining anti-tumor antibodies and extended half-life IL-2 (IL-2 fused with mouse serum albumin (MSA-IL-2) or Fc) exhibited synergistic therapeutic effects in mouse models of cancer9. This combination was curative when administered with adoptive T-cell therapy. We hypothesized that similar therapeutic results might be achieved in the setting of a wholly endogenous immune response by replacing T-cell transfer with a powerful vaccine. To this end, we employed a potent lymph node-targeted vaccine composed of peptide antigens and CpG DNA conjugated to albumin-binding lipids. These amphiphile-vaccines reversibly bind to albumin present in interstitial fluid and efficiently traffic to lymph nodes, leading to robust T-cell responses10. We also included systemic anti-PD-1 administration due to its capacity to enhance vaccine responses and further modulate the tumor microenvironment by blocking suppressive signals that dampen anti-tumor immunity7,11. In multiple syngeneic tumor models as well as the BRaf/Pten genetically engineered mouse model of melanoma, this quaternary combination immunotherapy cured a majority of mice with established tumors and elicited long-lived protective T cell memory responses.
RESULTS
Tumor regression by combination therapy
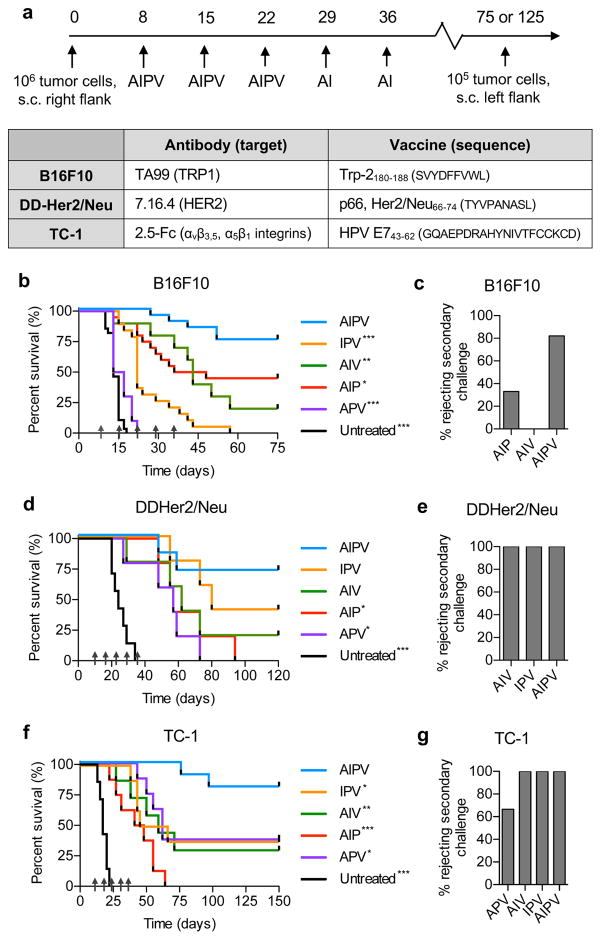
Motivated by the synergy observed between extended half-life IL-2 and anti-tumor antibodies9 and our recent discovery of a strategy to efficiently target peptide vaccines to lymph nodes10, we explored combination treatments bringing these reagents together with checkpoint blockade. To simplify reference to this four-component therapy, we designate combinations of these components by concatenated single initials A (anti-tumor antibody), I (MSA-IL-2), P (anti-PD-1), and V (amphiphile-vaccine). We first evaluated this combination treatment in three subcutaneous (s.c.) syngeneic tumor models: B16F10 melanoma, DD-Her2/neu (a rat Her2/neu-expressing breast cancer tumor), and TC-1 tumors expressing the HPV oncoantigens E6 and E7. For each tumor, appropriate A and V components were selected (Fig. 1a). Antibodies against Trp1 (TA99) and Her2/neu (7.16.4) were used to treat B16F10 and DD-Her2/neu, respectively, while an antibody surrogate 2.5F-Fc (Fc fused with an engineered binding protein that recognizes tumor-expressed αvβ3, αvβ5, and α5β1 integrins23) was used to treat TC-1 tumors (Supplementary Fig. 1a–c). Amphiphile-peptide vaccines encoding the well characterized immunodominant Trp2, Her2/neu, and HPV E7 epitopes were synthesized (Supplementary Fig. 1d–g, and ref10). Tumor cells (106) were injected s.c. and allowed to grow for 8 days prior to initiation of therapy, resulting in tumor sizes of ~40–60 mm2 at initial treatment, depending on the tumor model. The protein components were injected i.p. and the vaccine was administered s.c. at the base of the tail. Therapies were given once per week, with P and V administered 3 times and A and I given 5 times (Fig. 1a).
Figure 1. AIPV immunotherapy cures large established tumors and establishes protective memory in multiple tumor models.
a, Components of AIPV therapy and timeline of treatment. b–g, Groups of mice were inoculated with 106 tumor cells s.c. in the flank: B16F10 (b–c, n = 10/group for APV, 20/group for all other groups) and TC-1 (f–g, n = 8/group) tumor cells were injected in C57Bl/6 mice while DD-Her2/neu tumor cells (d–e, n = 7 for untreated and AIPV, n = 5 for all other groups) were injected in balb/c mice. On day 8 post implantation, AIPV treatment or ternary combinations were initiated following the timeline in a. Shown are survival (b, d, f) over time and the fraction of long term survivors that rejected a rechallenge with 105 tumor cells on day 75 for B16F10 and day 125 for TC-1 and DD-Her2/Neu (c, e, g). Data compiled from 2–3 independent experiments. Arrows indicate treatment time points. * p<0.05, ** p<0.01, *** p<0.001 versus AIPV by Log-rank (Mantel-Cox) test.
This AIPV regimen induced robust tumor regression and durable cures in 75% of mice bearing B16F10 melanoma tumors (Fig. 1b), while combinations of 3 or fewer of the AIPV components elicited weaker therapeutic responses ranging from modestly reduced efficacy (AIP) to outcomes barely different from untreated tumors (APV) (Fig. 1b, Supplementary Fig. 2a–b and g). Greater than 80% of AIPV-treated long-term survivors rejected a rechallenge with 105 B16F10 cells two months after cessation of therapy, suggesting formation of effective immunological memory (Fig. 1d). Similarly, in the DD-Her2/neu and TC-1 tumor models greater than 70% of mice achieved complete tumor rejection when treated with AIPV, and 100% of long term survivors rejected tumors on rechallenge (Fig. 1d–g, Supplementary Fig. 2c–f and h–i). Triple combinations of AIPV components demonstrated significantly lower efficacy in primary tumor rejection, though each of the 3 tumor models showed a different hierarchy of efficacy among the ternary combinations. A limited exploration of alternate 4-agent combinations revealed that anti-PD-1 could be replaced by anti-CTLA-4, but several other combinations failed to replicate AIPV in eliminating B16F10 tumors (Supplementary Fig. 3). Importantly, despite high rates of response, AIPV therapy was associated with minimal systemic toxicity, as mice did not show weight loss or significant elevation of liver enzymes. (Supplementary Fig. 4a–c).
Combination therapy impacts tumor immune infiltration
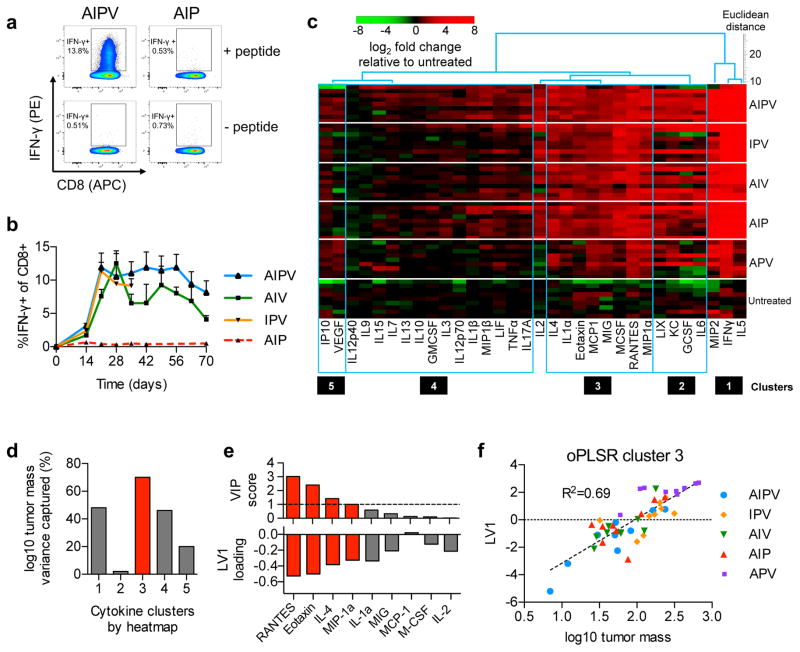
To understand the cellular and molecular mechanisms underlying the observed therapeutic effect of the quaternary combination therapy, we further analyzed responses to AIPV therapy in the B16F10 model as representative of an immunosuppressive, poorly immunogenic tumor. AIPV treatment induced expansion of an interferon-γ-producing Trp2-specific CD8+ T-cell population in the blood of mice (Fig. 2a–b); abundance of these cells correlated with vitiligo in treated animals, which is associated with favorable responses to immunotherapy in the clinic24 (Supplementary Fig. 4d–f). However, comparable Trp2-specific T-cell responses were elicited by AIV and IPV treatments, which failed to induce comparable therapeutic outcomes (Fig. 2b). We thus looked for changes within tumors that correlated more closely with survival. Luminex quantification of intratumoral cytokines and chemokines revealed five clusters of co-regulated proteins, with many pro-inflammatory factors increased in tumors treated with effective therapies (Fig. 2c, Supplementary Fig. 5). We used partial least squares regression (PLSR) to construct models using cytokines from each cluster to predict log10 tumor mass. Analysis of the variance in tumor mass explained by each model revealed that cluster 3 cytokines captured the most variance (Fig. 2d), and therefore provided the most predictive model. Variable importance in projection (VIP) scores identified 4 predictive biomarkers from cluster 3– MIP-1α, RANTES, eotaxin, and IL-4– whose levels were inversely correlated with tumor size and predicted tumor mass across all 5 treatment combinations (Fig. 2e–f, R2=0.69).
Figure 2. AIPV therapy primes sustained vaccine-specific T-cell responses and remodels the microenvironment of established tumors.
B16F10 tumors in C57Bl/6 mice were treated with AIPV or indicated ternary combinations of the therapy components as in Fig. 1a. a–b, Peripheral blood cells were stimulated with Trp2 peptide for 6 hours in the presence of brefeldin A and then analyzed by intracellular cytokine staining. (a) Representative intracellular cytokine staining to detect Trp2-specific CD8+ T-cells in peripheral blood on day 21. (b) Mean±s.e.m. percentage IFN-γ+ among CD8+ T-cells over indicated time points (n = 5/group, b). c–f, Mice bearing B16F10 tumors were treated with the regimens indicated on the right. Seventeen days later, tumors were isolated and cytokine and chemokine levels were measured by Luminex (n = 9/group). (c) Five co-regulated clusters of cytokines and chemokines were identified by hierarchical cluster analysis and then pruning clusters by applying a threshold to linkage distance (Supplementary Fig. 5). (d) These clusters were independently tested for their ability to predict log-transformed tumor size in treated mice on day 17 using a single latent variable (LV) in an orthogonalized partial least squares regression (oPLSR) model. (e) For the best performing cluster 3 (red bar), VIP scores (a measure of each cytokine’s contribution to the LV score and overall regression performance) were used to assess the contribution of individual proteins from the cluster to the oPLSR model; VIP scores >1 (dotted line) were considered significant (top, red bars) and the loading factors for LV1 of the oPLSR model are shown (bottom, red bars). (f) A scatter plot shows the oPLSR model performance for individual mice from all treatment groups and the best fit linear regression (dotted line) (f).
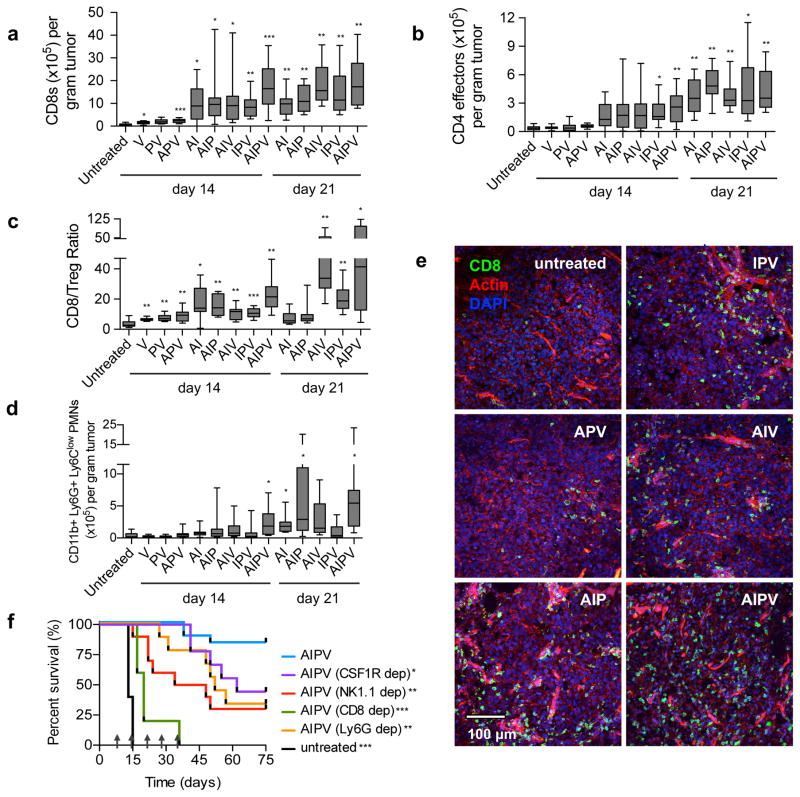
Motivated by this chemoattractant-rich protective signature, we analyzed cellular infiltrates in treated tumors. AIPV therapy induced tumor infiltration by effector CD4+ and CD8+ T-cells, CD11b+Ly6G+Ly6Clow neutrophils25,26, NK cells, and other myeloid cells (Fig. 3a–d, Supplementary Fig. 6a–d). PD-1+TIM-3+CD8+ cells were infrequent in all treatment groups, but AIPV induced a large increase in the CD8+ T-cell/Treg ratio (Fig. 3c, Supplementary Fig. 6g–h), an interesting outcome given the potential for IL-2 treatment to increase Treg numbers. Notably however, no single leukocyte population examined individually correlated with therapeutic outcome, as several treatment subcombinations elicited infiltration of individual cell populations equivalent to AIPV. Immunohistochemical analysis of treated tumors revealed CD8+ T-cells were present at high density throughout AIPV-treated tumors, with less effective triple-combination therapies eliciting progressively fewer CD8+ cells in the bulk tumor mass (Fig. 3e, Supplementary Fig. 6e–f). Antibody depletion experiments showed that CD8+ T-cells were critical to tumor rejection (Fig. 3f, Supplementary Fig. 7a). Macrophage, NK cell, or neutrophil depletion also led to significant reductions in overall survival rates, while IL-5 blockade or CD4+ T-cell depletion did not impact efficacy (Fig. 3f, Supplementary Fig. 7a–b). Thus, AIPV treatment elicited substantial remodeling of the tumor microenvironment with contributions from diverse effector cells.
Figure 3. AIPV therapy induces pronounced immune infiltration of tumors with efficacy dependent on innate and adaptive immune cells.
a–d, B16F10 tumors treated as indicated were isolated on days 14 and 21 and quantified by flow cytometry (n = 15 animals/group for day 14, 9 animals/group for day 21, at least 2 independent experiments). Shown are boxplots (whiskers 5–95%) for tumor-infiltrating CD8+ T-cells (a), CD4+FoxP3− T-cells (b), the ratio of CD8+ T-cells to CD4+CD25hiFoxp3+ Tregs (c), CD11b+LyG+LyClow polymorphonuclear (PMN) cells (d), and representative immunofluorescence images from tumors on day 14; scale bar is 100μm (e). f, depleting antibodies against the indicated surface markers were administered i.p. beginning one day prior to initiation of AIPV therapy to deplete macrophages (CSF1R), NK cells (NK1.1), CD8+ T-cells (CD8), or neutrophils (Ly6G). Shown is survival over time (n = 5 animals/group for CD8 depletion, n = 10 animals/group for NK1.1, CSF1R, and Ly6G depletion, 2 independent experiments). Arrows indicate treatment time points. *P < 0.05, **P < 0.01, ***P < 0.001 by Welch’s t test versus untreated with Bonferroni correction for a–d. * P < 0.05, ** P < 0.01, *** P < 0.001 versus AIPV by Log-rank (Mantel-Cox) test for f.
Therapy promotes cross-presentation to generate de novo T cell responses
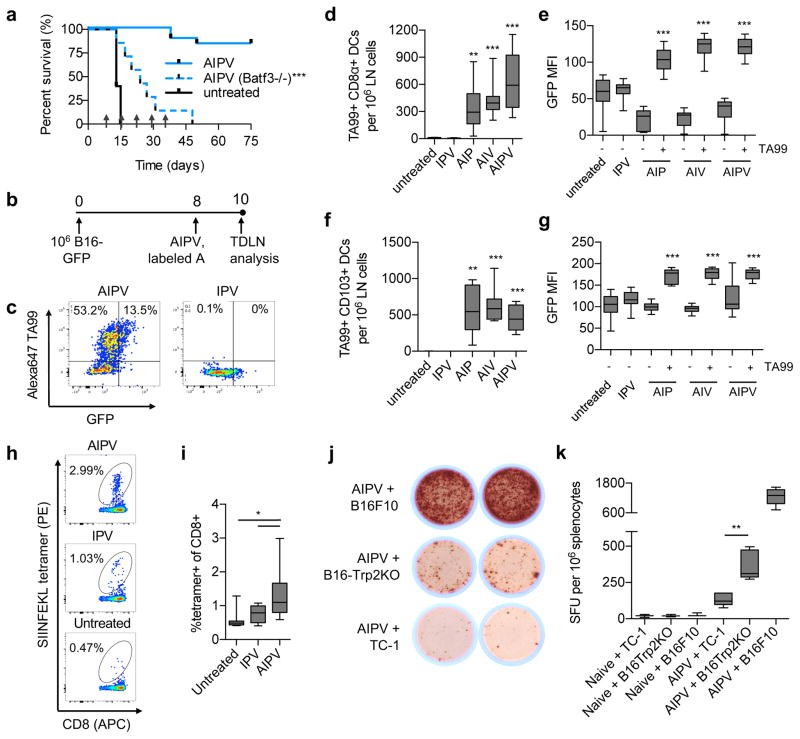
We next assessed whether de novo adaptive immune responses specific for antigens not encoded by the vaccine were primed by AIPV treatment. Given the critical role of CD8+ T-cells in the therapeutic effect, we first analyzed the impact of AIPV on antigen presentation in the tumor-draining lymph nodes (TDLNs). Batf3 is a transcription factor required for development of cross presenting DCs in mice,27 and AIPV-treated Batf3−/− mice failed to reject tumors, indicating a requirement for cross-presenting DCs for response to therapy (Fig. 4a). Anti-tumor antibodies promote cross-presentation of tumor antigens28. To further explore the role of anti-tumor antibody in this process, we treated mice bearing GFP-expressing B16F10 tumors with AIPV or subcombinations and analyzed the uptake of GFP and fluorescently labeled TA99 in dendritic cells in TDLNs (Fig. 4b–c). We examined two Batf3-dependent cross-presenting DC populations that may have distinct roles in tumors, CD8α+ DCs and CD103+ DCs29–31. Labeled TA99 accumulated in these DC populations, and treatment combinations including TA99 significantly increased GFP uptake by both CD8α+ and CD103+ DCs over untreated tumors (Fig. 4d–g). Further, ovalbumin (OVA)-expressing B16F10 tumors treated with AIPV (vaccinating against Trp2) induced T cell responses specific for the SIINFEKL OVA peptide that were substantially greater than responses observed in untreated or IPV-treated tumors, indicating a role for the antibody in priming T-cell responses to new tumor antigens (Fig. 4h–i). Further supporting the conclusion that AIPV induced a broad T cell response to multiple tumor antigens, splenic T cells from B16F10-tumor bearing mice treated with AIPV produced IFN-γ upon restimulation with either parental B16F10 cells or a Trp2-deleted B16F10 cell line (Supplementary Fig. 8a–b) (Fig. 4j–k). In addition, 50% of AIPV-treated mice cured of primary tumors also rejected a rechallenge with Trp2-deleted B16F10 cells on day 125 (Supplementary Fig. 8c). These data suggest the antibody component of AIPV promoted epitope spreading, leading to functional de novo T-cell responses against tumor antigens not encoded by the T cell vaccine.
Figure 4. Combination therapy elicits antibody-enhanced antigen spreading and de novo T-cell responses.
a, AIPV treatment was carried out in wild type or Batf3−/− mice (n = 7 animals/group) bearing established B16F10 tumors as in Fig. 1a; mice were euthanized when tumor area exceeded 100 mm2. Shown is survival over time. Arrows indicate treatment time points. b–g, B16-GFP cells (106) were injected s.c. (n = 10 animals/group) and AIPV or indicated ternary combination treatment was applied with AlexaFluor647-labeled TA99 antibody, followed by tumor-draining LN isolation, digestion, and analysis by flow cytometry (b). Shown are representative scatter plots of CD11c+ DCs (c), enumeration of TA99+ CD11c+CD8α+CD11b− and CD11c+CD103+ DCs (d and f, respectively) and MFI of GFP within CD11c+CD8α+CD11b− and CD11c+CD103+ DCs (e and g, respectively) gated on either TA99+ or TA99− cells. Shown are boxplots (whiskers 5–95%) for d–g. h–i, B16-OVA cells (106) were injected s.c. and tumors were treated with AIPV as in Fig. 1a (with vaccine against Trp2); staining with SIINFEKL/H-2Kb tetramers on peripheral blood mononuclear cells was performed on day 15. Shown are representative flow plots (h) and boxplots (whiskers 5–95%) (i) from 1 of 2 independent experiments (n = 10 animals/group). j–k, AIPV treated or naïve mice were challenged with 105 B16F10 on day 75, and 6 days later splenocytes were isolated and tested for reactivity against a Trp2-KO B16F10 line generated using CRISPR/Cas9 (Supplementary Fig. 8), parental B16F10 cells, or control TC-1 cells via ELISPOT. Shown are representative wells, j, and enumeration of counts by boxplot (whiskers 5–95%), k (n = 6 animals/group). *P < 0.05, **P < 0.01, ***P < 0.001 by Welch’s t test versus untreated with Bonferroni correction for d and f, by Welch’s t test versus the TA99- fraction within the same treatment group for e and g, by ANOVA with Bonferroni post-test for i, and by t test for k.
Therapy-induced endogenous antibody responses
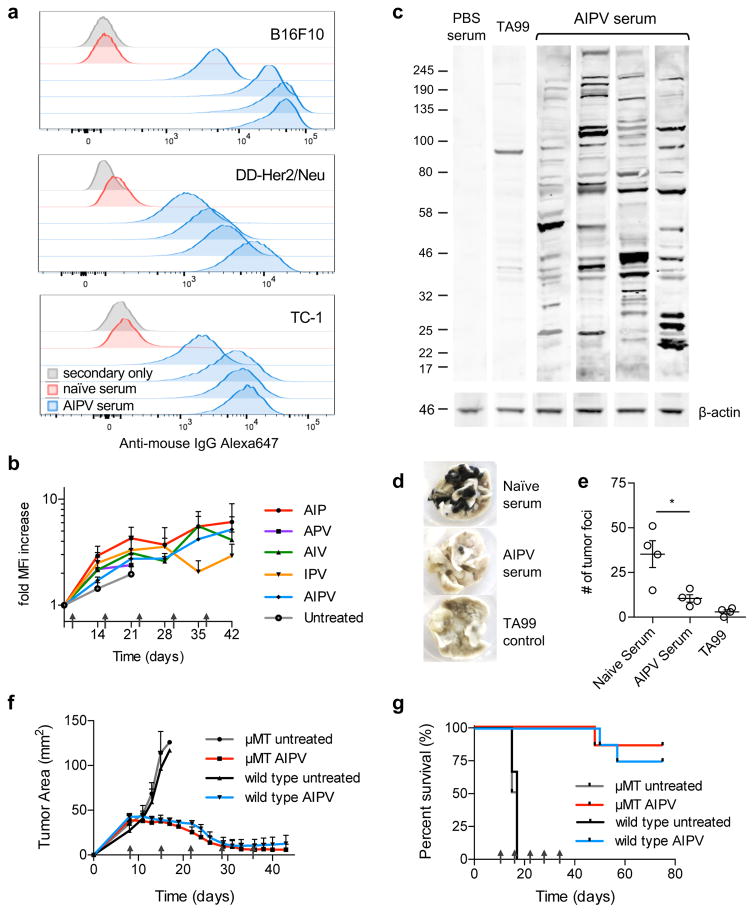
In parallel we evaluated endogenous antibody responses in AIPV-treated animals. In all 3 transplanted tumor models, AIPV therapy elicited antibodies that bound to the tumor cells (Fig. 5a). We developed an approach to deplete the injected antibody from sera recovered from mice undergoing treatment, in order to analyze the endogenous IgG response over time (Supplementary Fig. 9a–f). Using this approach, we found that both AIPV and the less effective triple subcombinations elicited anti-tumor antibodies, which were detectable as early as 7 days following start of treatment and increased in binding signal over 30 days for combinations that promoted better survival (Fig. 5b and Supplementary Fig. 9f). An immunoblot of sera from B16F10 tumor-bearing mice treated with AIPV against B16F10 lysate revealed that these endogenous antibodies recognized numerous antigens (Fig. 5c). These therapy-induced antibodies were functional, as serum transferred from AIPV-treated mice protected naive mice against intravenous B16F10 challenge (Fig. 5d–e). However, administration of AIPV therapy to B-cell-deficient μMT bearing established B16F10 tumors led to a similar frequency of tumor regressions as observed in wild type mice, suggesting that the endogenous antibody response was not essential for the therapeutic effect of the AIPV regimen (Fig. 5f–g). Together, these experiments indicate that AIPV broadly promoted both antibody and T-cell responses against epitopes not directly targeted by the therapy components.
Figure 5. AIPV therapy induces de novo endogenous anti-tumor antibody responses.
a, indicated tumor cells were incubated with 5% serum collected on day 150 from AIPV-treated mice that had previously rejected B16F10 (top), DD-Her2/neu (middle), or TC-1 (bottom) tumors, or age-matched naïve mice. The cells were then washed and stained with Alexa647 anti-mouse IgG secondary antibody then analyzed by flow cytometry (shown are results for sera from individual treated animals, representative of 2 independent experiments of n = at least 4). b, C57Bl/6 mice bearing B16F10 tumors were treated with the indicated combination therapies as in Fig. 1a using biotinylated TA99. Mice were bled weekly and serum was depleted of TA99-biotin using streptavidin resin (Supplementary Fig. 9), then binding of remaining endogenous IgG to B16F10 cells was measured by flow cytometry as in a. Plotted is the fold change in MFI of anti-mouse IgG over time±s.e.m. (n = 5/group). c, Western blot of B16F10 tumor cell lysate with serum from AIPV-treated mice, PBS-treated mice, or TA99 with detection using an anti-mouse-IgG-IRDye800 secondary antibody. Shown is 1 representative of 2 independent experiments. d–e, 350 μL serum from AIPV treated mice (day 150) or age-matched naïve mice (n = 4 animals/group) was incubated at 57°C for 1h to inactivate complement and transferred into naïve recipients, followed by challenge with 2.5x105 B16F10 cells intravenously. Lungs were isolated 17 days later and nodules were counted in a blinded fashion. Shown are representative lungs (d) and mean counts±s.e.m. (e). Data shown is 1 representative of 2 independent experiments. f–g, B cell-deficient μMT or wild type C57Bl/6 mice were inoculated with B16F10 tumors and left untreated or received AIPV therapy as in Fig. 1a. Shown are mean±s.e.m tumor growth curves (f) and overall survival (g) (n = 8/group for AIPV treated groups, n = 4/group for untreated). Arrows indicate treatment time points. *P < 0.05, **P < 0.01, ***P < 0.001 by one-way ANOVA with Bonferroni post-test.
Efficacy in an autochthonous genetically engineered mouse model
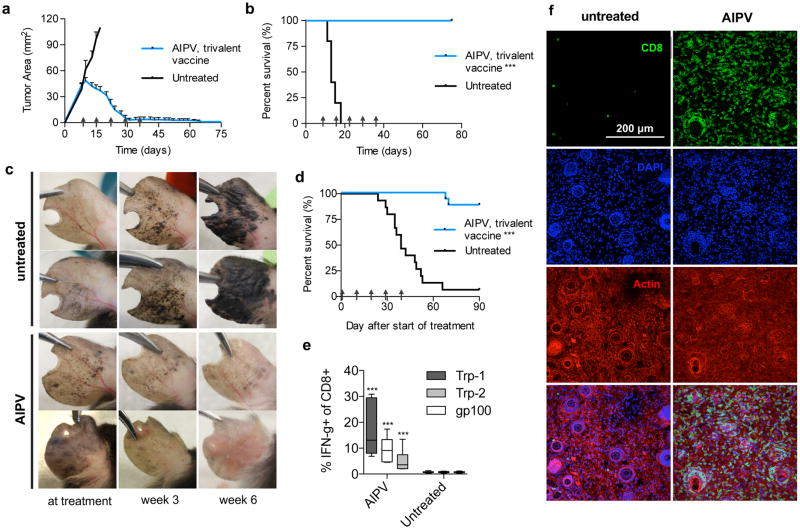
Transplanted tumors do not closely mimic the histology of human cancers, and often have a high mutational burden that constitutes potential neoantigens. To assess the efficacy of AIPV in a model more closely mimicking the pathophysiology of human disease and simultaneously determine the impact of a low frequency of neoantigens, we used the BRafCA PtenloxP Tyr::CreERT2 (BRaf/Pten) inducible melanoma model32. We first enhanced the potency of AIPV by increasing the number of antigens targeted by the vaccine component33. AIPV therapy using a trivalent amphiphile-vaccine targeting gp100, Trp1, and Trp2 cured 100% of B16F10 tumor-bearing mice with mean initial tumor sizes of 50 mm2 (Fig. 6a–b). To test this combination in genetically engineered mice, tumors were induced by painting tamoxifen on the ears of BRaf/Pten mice, and when visible lesions were present (~4 weeks), treatment was initiated. Melanomas grew progressively and by week 10 completely covered the ears of untreated mice (Fig. 6c). By contrast, trivalent-vaccine AIPV induced regression of pigmented lesions, leading to complete clearance in the majority of animals and a significant improvement in overall survival compared to untreated mice (Fig. 6c–d). This result was coincident with expansion of T-cells recognizing the three vaccine-encoded antigens in peripheral blood (Fig. 6e) and with infiltration of CD8+ T-cells throughout AIPV-treated Braf/Pten tumors (Fig. 6f). To our knowledge this is the first immunotherapy to regress a majority of induced lesions in this autochthonous tumor model.
Figure 6. AIPV with a trivalent vaccine is curative for established B16F10 tumors and induces regression in BRaf/Pten autochthonous melanoma.
a–b, Established B16F10 tumors were treated with AIPV as outlined in Fig. 1a using a trivalent vaccine targeting Trp2, Trp1, and gp 100. Shown are tumor area measurements (a, mean±s.e.m.) and survival (b) (n = 10 animals/group). c–e, BRafCAPtenloxPTyr::CreERT2 mice were painted with 4-hydroxytamoxifen on one ear and treated with AIPV (including trivalent vaccine) beginning 28 days later when visible lesions were apparent. Shown are representative photographs of AIPV treated or untreated ears at treatment, 3 weeks after treatment initiation, and 6 weeks after treatment initiation (c), and survival over time (d) (n = 16 animals/group, compiled from 4 independent experiments). Mice were euthanized when tumor coverage exceeded 90% of the ear. e, On day 48, peripheral blood was stimulated with peptide antigens for 6 hours in the presence of golgi inhibitor and intracellular cytokine staining was performed to assess CD8+ T-cell responses against each vaccine antigen. Shown are boxplots (whiskers 5–95%) (quantified as a fraction of total CD8+ T-cells, n = 7 animals/group). f, BRafCAPtenloxPTyr::CreERT2 mice were painted with 4-hydroxytamoxifen on the left flank and treated with AIPV (including trivalent vaccine) beginning on day 36. Shown are images of representative immunofluorescence from tumors taken on day 70 (n = 8 tumors per condition); scale bar is 200μm. Arrows indicate treatment time points. ***P<0.0001 by Welch’s t test for d, or by Log-rank (Mantel-Cox) test for b and e.
DISCUSSION
To date, complete elimination of large, poorly immunogenic tumors in immunocompetent animal models has only been reliably achieved by combination therapies employing large numbers of adoptively transferred T-cells14,19,34. Thus, it has remained an outstanding question whether an endogenous immune response can be stimulated to overcome advanced tumors in a majority of animals. Here we demonstrate in several transplanted tumor models and a genetically-engineered mouse model of melanoma the capacity of endogenous immunity, elicited by a safe four-component immunotherapy, to regress large established tumors in the majority of mice. All four components incorporated in AIPV were required for treatment of several difficult tumor models, and other treatment combinations were often less effective or very toxic, for example, utilizing anti-CD40 in place of the anti-tumor antibody caused inflammatory toxicity resulting in death of treated mice (unpublished data). Notably, the models studied here were selected in part due to their resistance to immunotherapy treatment; other commonly used immunogenic tumors (e.g., EG7.OVA, CT26, and MC-38) could be cured by double- or triple-component subcombinations of AIPV (unpublished data).
Many immunotherapy studies focus on CD8+ T-cell responses to tumors, but important roles for innate immune effector cells are also well described. Especially in the presence of tumor-opsonizing antibodies, NK cells, neutrophils and macrophages can all contribute to direct antibody-dependent phagocytosis, reactive oxygen-mediated cytotoxicity against tumors, and secretion of inflammatory cytokines and chemokines35–37. This innate attack creates intertwined positive feedback loops: opsonized tumor antigen is released that is efficiently cross presented by DCs13,38,39; inflammatory cytokines and immune complexes are released that activate DCs, stimulate their migration to draining lymph nodes, and promote adaptive effector functions9,40,41; and additional innate effectors and T-cells are recruited to the tumor9,40,42. Consistent with the importance of the innate immune response, maximal efficacy of AIPV therapy was dependent not only on CD8+ T-cells but also neutrophils, macrophages, NK cells, and cross-presenting DCs. AIPV therapy completely failed in Batf3-deficient mice, and was in fact even less effective than AIP treatment, suggesting that the loss in efficacy in these knockout animals reflected more than loss of responsiveness to the vaccine component (unpublished data). Although models of infectious disease and systemic immunization have implicated CD8α+ dendritic cells as key cross-presenting cells for T-cell immunity43,44, recent data has suggested that in growing tumors, a second Batf3-dependent DC lineage, CD103+ DCs, plays a critical role in tumor antigen presentation29,30. Consistent with these reports, we found both CD8α+ and CD103+ DCs acquired tumor antigen in a manner enhanced by the antibody component of AIPV.
Treatment of multiple tumor models with AIPV therapy revealed distinct hierarchies of importance for the four components. For example, the monoclonal antibody component was a critical contributor to efficacy in the B16F10 model, but the least important component in the DD-Her2/neu model. A key aspect of AIPV treatment is that this set of 4 agents collectively mounts an integrated response overcoming tumor resistance mechanisms in all of the models evaluated here– suggesting that the appropriate combination of immune effectors can overcome a range obstacles presented in tumor microenvironments. Notably, therapy excluding the extended-PK IL-2 (APV treatment) showed greatly reduced efficacy in all 3 transplanted tumor models, correlating with reduced abundance of inflammatory cytokines and chemokines in tumors and fewer infiltrating CD4+ and CD8+ T-cells. Which cell types IL-2 must act on for therapeutic efficacy remains to be defined, but besides supporting the proliferation and effector functions of T-cells, IL-2 is known to also drive expansion and enhance the sensitivity of NK cells45, promote neutrophilia and eosinophilia, and induce the production of myriad additional cytokines and chemokines that may support a combined innate and adaptive immune attack on tumors46. Despite the potential of IL-2 to expand Tregs, we found that intratumoral CD8:Treg ratios were amplified relative to untreated tumors during AIPV therapy.
Although it remains an understudied area, prior work has demonstrated that the number of antigens targeted by cancer vaccines can be an important variable, with vaccines targeting more antigens leading to greater anti-tumor efficacy33. In agreement with these data, we found that the efficacy of AIPV therapy was sensitive to the valency of the vaccine; targeting a single epitope in the B16F10 model led to a 75% cure rate (Fig. 1c), while a modest increase in vaccine valency to target 3 melanocyte antigens led to cures of 100% of treated mice (Fig. 6a–b). There is currently much enthusiasm for targeting neoantigens formed by mutations in tumors, but our data demonstrate that combination immunotherapy targeting only tumor-associated self antigens can elicit pronounced tumor regressions, even in the setting of a genetically-induced mouse model bearing an intrinsically low burden of mutations. However, AIPV also promoted responses to additional antigens not present in the vaccine, which could protect against challenge with tumor cells lacking the vaccine-targeted antigens. We attempted to evaluate whether AIPV could be protective in the absence of antigen spreading by treating pmel-1 transgenic mice19 that lack T-cells capable of responding to peptides other than gp100, but when treated with AIPV incorporating a gp100 peptide as the vaccine component, these animals developed severe inflammatory toxicity (unpublished observation). Antigen spreading promoted by immunotherapy has also observed in recent clinical studies melanoma patients47, and may be important for dealing with tumor heterogeneity and preventing escape by tumor antigen loss variants48.
Endogenous antibody responses were primed by AIPV and other combinations of the four monotherapies, and AIPV treatment induced antibodies that were protective when transferred to naïve recipients. Despite these findings, B-cells were not required for AIPV treatment efficacy. We speculate that because the therapy itself includes a potent monoclonal tumor-opsonizing antibody, the endogenous response may be dispensable. An interesting question for future work will be whether the monoclonal antibody component could be removed from the treatment regimen once the endogenous antibody response develops, i.e. by the second or third treatment.
AIPV incorporates two tumor-specific reagents, the lymph node-targeted peptide vaccine and a monoclonal opsonizing antibody. While still a challenge in some diseases, identification of T-cell target antigens shared by a proportion of tumors of a given type has been well delineated for many cancers over the past 20 years by the cancer vaccine field49 and a number of pipelines for finding unique mutant neoantigens have been recently developed50,51. In contrast, identifying an effective opsonizing monoclonal antibody presents a development hurdle to apply AIPV to an arbitrary given tumor. However, the demonstration that the integrin binding protein 2.5F-Fc, which contains an Fc domain for antibody effector functions and recognizes αvβ3, αvβ5, and α5β1 integrins over-expressed in a broad range of cancers52, suggests that the 2.5F-Fc protein may be applicable to many tumor types. Careful consideration of potential side effects is needed in the choice of target antigens for translation of this approach. Targeting tumor-associated antigens carries the risk of autoimmunity, for example immunotherapy targeting melanocyte self antigens is known to induce vitiligo, uveitis, and hearing loss in some patients due to loss of normal melanocytes.53,54 Sequencing of patient tumors for neoantigen identification is an attractive alternative, and the peptide vaccine platform used here is well suited to such an approach. Similarly, while antigen spreading as observed here may promote tumor rejection, an important issue for future work is understanding whether these de novo T-cell responses are focused on tumor-specific mutations or self antigens.
In summary, we have demonstrated that a rational combination of four complementary immunomodulatory agents, each of which have modest anti-tumor efficacy individually, can lead to robust complete responses in the setting of large, immunosuppressive tumors. Rejection of established tumors relied on an orchestrated response invoking diverse effector mechanisms of the immune system. Although a quaternary treatment regimen will be challenging for direct clinical translation due to the complexity of identifying optimal dosing levels and schedules for each component, insights from this successful protocol provide a framework for devising simpler regimens and extensions to other treatment modalities.
METHODS
Mice
B6 mice (C57BL/6NTac) were purchased from Taconic. Balb/c mice (BALB/cJ), Batf3−/− mice (B6.129S(C)-Batf3tm1Kmm/J), BRaf/Pten mice (B6.Cg-Braftm1Mmcm Ptentm1Hwu Tg(Tyr-cre/ERT2)13Bos/BosJ), and mT/mG mice (B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J) were purchased from The Jackson Laboratory. Mice used in the inducible cancer model (BRaf/Pten-TG) were crosses of BRaf/Pten and mT/mG bred in-house and having the following genotype: Braftm1Mmcm +/−, Ptentm1Hwu +/+, Tg(Tyr-cre/ERT2)13Bos +, Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo +/+, where “+” indicates presence of the mutant/transgenic allele. All BRaf/Pten mice were genotyped via Transnetyx. Mice were used in studies when 6–8 weeks old. All animal work was conducted under the approval of the Massachusetts Institute of Technology (MIT) Division of Comparative Medicine in accordance with federal, state, and local guidelines.
Cells
B16F10 were purchased from ATCC. TC-1 cells were kindly provided by Dr. T. C. Wu at John Hopkins University. B16-OVA and DD-Her2/neu cells were kindly provided by Dr. Glenn Dranoff at Dana-Farber. B16-GFP-Luc cells were generated as previously described55. DD-Her-2/neu breast tumor cells were derived from a spontaneous breast tumor in a Balb/c Her-2/neu (Balb-NeuT) transgenic mouse, in which MMTV promoter-driven breast epithelium-specific expression of the activated oncogenic form of rat Her-2/neu drives progressive development of breast tumors in transgenic Her-2/neu female mice56. The DD cell line was found to lose Her-2/neu expression in vitro, and thus cells were infected with pMFG-Her-2/neu virus to achieve stable expression of high levels of oncogenic rat Her-2/neu both in vitro and in vivo.HEK293 cells were purchased from Life Technologies.
Tumor cell lines were cultured in complete DMEM (DMEM supplemented with 10% FBS, 100 units/mL penicillin, 100 μg/mL streptomycin, and 4 mM L-alanyl-L-glutamine) with the exception of TC-1 cells, which were cultured in complete RPMI. T-cells and splenocytes were cultured in RPMI with 10% heat inactivated FBS, 20 mM HEPES, 1 mM sodium pyruvate, 0.05 mM beta-mercaptoethanol, 100 units/mL penicillin, 100 μg/mL streptomycin, 2 mM L-alanyl-L-glutamine, and 1X MEM non-essential amino acids. Hybridomas were cultured in CD Hybridoma AGT Medium (Life Technologies). HEK293 cells were cultured in Freestyle Media (Life Technologies). All cell lines and assay cultures were maintained at 37°C and 5% CO2. All cells were tested regularly for mycoplasma contamination and for rodent pathogens and none used tested positive at any point.
Vaccine and protein production
Amphiphile-CpG (amph-CpG) was produced as previously described, by solid phase synthesis of a Class B CpG 1826 sequence with G2 spacer (5′ diacyl lipid -*G*G*T*C*C*A*T*G*A* C*G*T*T*C*C*T*G*A*C*G*T*T- 3′) conjugated via the 5′ end to an 18-carbon diacyl tail10. Amphiphile-peptides (amph-peptides) were produced as previously described10, via conjugation of cys residues of peptide antigens to 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000] (Avanti Polar Lipids). Vaccine sequences are as follows: Trp2180-188 (CSVYDFFVWL); Her2/Neu66-74 (p66, CTYVPANASL), HPV E743-62 (GQAEPDRAHYNIVTFCCKCD), Tyrp1opt455-463 (optimized A463M57, CTAPDNLGYM), gp100opt20-39 (optimized S27P, EGP long58, CAVGALEGPRNQDWLGVPRQL). TA99 and MSA-IL2 were produced in HEK293 cells as previously described9. 2.5-Fc was generated by fusing an integrin (αVβ3, αVβ5, and α5β1) binding domain to a murine IgG2a Fc region to create an antibody-like tumor targeting molecule23. 2.5-Fc was produced by transient transfection of HEK293 cells and purified by Protein A. The 7.16.4 monoclonal antibody to rat Her2/neu was generated from hybridoma 7.16.4 purchased from ATCC, and was purified using Protein A. IFN-α was expressed as a SUMO fusion using the pE-SUMOpro vector (LifeSensors) in Rosetta-gami 2 (DE3) competent cells (Novagen). After IMAC purification using TALON metal affinity resin according to the manufacturer’s instructions (Clontech Laboratories), the SUMO tag was removed by incubation with SUMO protease and reapplication on TALON resin as detailed previously59. Finally, the protein was passed through Detoxi-Gel endotoxin removal resin (Thermo Scientific) until endotoxin levels were below 0.1 total EU/dose as measured by the QCL-1000 chromogenic LAL assay (Lonza). The IL-15 superagonist60 was produced by transiently transfecting HEK293 cells with a plasmid encoding His-tagged IL-15 superagonist and purified using Ni-NTA agarose (Qiagen) on a PD-10 column (Bio-Rad) followed by further purification using a His-Spin Protein Miniprep (Zymo Research). Anti-TGF-β, a homodimeric fusion between murine IgG2a Fc and the extracellular domain of the type II TGF-b receptor, was produced by transient transfection of HEK293 cells and purified by Protein A.
Tumor inoculation and subcutaneous tumor therapy
An inoculum of 106 tumor cells was injected s.c. on the flank of mice in 50 μL sterile PBS. Eight days following injection, treatment was initiated as indicated (Fig. 1a). The tumor-targeting antibody (A) was administered at 100 μg per dose i.p. for B16F10 and DD-Her2/Neu models. 2.5-Fc was was administered at 500 μg per dose (i.p.). MSA-IL2 (I) was administered i.p. at 30 μg (6 μg molar equivalent IL-2) per dose. Anti-PD-1 (P) (clone RMP1-14, BioXCell) was administered at 200 μg i.p. The vaccine, composed of 1.24 nmol amph-CpG and 20 μg of amph-peptide, was administered s.c. at base of the tail, half the dose given on each side. For experiments exploring other quaternary combos, the following doses were used as indicated in Supplementary Fig. 3: anti-CTLA-4 (clone 9D9, BioXCell) was administered at 200 μg per dose i.p., IFN-α was administered at 50 μg per dose i.p., IL-15 superagonist at 4 μg per dose i.p., and anti-TGF-β at 100 μg per dose i.p. Mice were randomized into treatment groups on day 8 following tumor inoculation, immediately prior to treatment. Tumor size was measured as an area (longest dimension x perpendicular dimension) three times weekly, and mice were euthanized when tumor area exceeded 100 mm2. Mice that rejected tumors were rechallenged as indicated with 105 tumor cells s.c. on the opposite flank.
Flow cytometry
Antibodies to CD8α (53–6.7), IFN-γ (XMG1.2), TNF-α (MP6-XT22), CD3ε (500A2), IgM (RMM-1), CD19 (6D5), NK1.1 (PK136), Ly-6G (1A8), F4/80 (BM8), CD11b (M1/70), CD25 (PC61), PD-1 (29F.1A12), Tim-3 (B8.2C12), Ly-6C (HK1.4), and CD64 (X54-5/7.1) were purchased from BioLegend. Antibodies to CD4 (RM4-5) and Foxp3 (FJK-16s) were purchased from eBioscience. Trp2 tetramer (iTAg Tetramer/PE H-2Kb TRP2) and OVA tetramer (iTAg Tetramer/PE H-2Kb OVA) were purchased from MBL. Tetramer staining was performed in buffer containing 50 nM dasatinib to enhance staining. Viability was assessed by LIVE/DEAD Fixable Aqua purchased from Life Technologies. TA99 was labeled with Alexa Fluor 647 NHS Ester (Life Technologies) to generate TA99-647. Foxp3 was stained using the Transcription Factor Buffer Set from BD.
Intracellular Cytokine Staining (ICS) was performed as described previously10. Peptides used for restimulation were 10 μg/mL of the relevant antigen: Trp2180-188 (SVYDFFVWL), Her2/Neu66-74 (TYVPANASL), Tyrp-1455-463 native (TAPDNLGYA), or gp10025-33 native (EGSRNQDWL). Immune cell infiltrates of tumors were analyzed as previously described9, briefly: tumors were resected from mice on day 14 or 21, weighed, and mechanically disrupted to generate a single cell suspension. Cells were stained to identify infiltrating immune and quantified using flow cytometry. Shown are cell counts normalized by tumor mass. For antibody and GFP uptake experiments, lymph nodes were resected from tumor bearing mice on day 10, digested with collagenase/dispase in media at 37°C for 20 minutes and mechanical disruption was used to generate a single cell suspension. Cells were stained to identify DC populations and run on the cytometer. Cells were analyzed using BD FACS LSR II, BD FACS LSR Fortessa, and BD FACSCanto flow cytometers, and data was analyzed using FlowJo.
Depletions
Cellular subsets were depleted by administering 400 μg of depleting antibody i.p. twice weekly beginning one day prior to therapy as indicated: CD8 T-cells with anti-CD8α (clone 2.43, BioXCell), CD4 T-cells with anti-CD4 (clone GK1.5, BioXCell), NK cells with anti-NK1.1 (clone PK136, BioXCell), neutrophils with anti-Ly-6G (clone 1A8, BioXCell), with the exception of CSF1R and IL-5, which were depleted using 300 μg (clone AFS98, BioXCell) every other day and 1 mg (clone TRFK5, BioXCell) weekly respectively.9 Cellular depletions of CD8 T cells, CD4 T cells, neutrophils, and NK cells were confirmed by flow cytometry of PBMC (Supplementary Fig. 7).
Autochthonous tumor model induction and therapy
BRafCA PtenloxP Tyr::CreERT2 mice27 on the C57BL/6 background61 were crossed with mT/mG mice62 to generate BRaf/Pten-TG mice. To induce tumors, 2 μL of 5 mg/mL tamoxifen was administered to the left ear on three consecutive days. Tumors were allowed to develop for 24–26 days, at which time visible pigmented tumors were present. Treatment schedule and doses were the same as B16F10 s.c. model, except that vaccine used was a combination of three amph-peptides (15 μg amph-gp100, 15 μg amph-Trp1, and 15 μg amph-Trp2) and 1.24 nmol amph-CpG. Mice were euthanized when pigmented lesions covered greater than 90% of the ear.
Liver enzyme measurements
Serum was isolated from mice 36 hours after treatment and liver enzymes AST and ALT were quantified using a colorimetric Aspartate Aminotransferase Activity Assay Kit (Sigma Aldrich) and Alanine Aminotransferase Activity Assay Kit (Sigma Aldrich) respectively according to the manufacturer’s protocol.
Endogenous antibody detection and serum transfer
For endogenous antibody binding measurements following therapy, B16F10 cells, TC-1 cells, or DD-Her2/Neu cells were incubated with 5% serum from AIPV-treated mice (day 150) or age-matched naïve mice in PBS for 30 minutes at 4°C, washed twice with PBS, 1% BSA, 5mM EDTA, then incubated with AlexaFluor647 labeled anti-mouse IgG for 20 minutes at 4°C. Cells were washed in PBS, 1% BSA, 5mM EDTA with DAPI included to exclude dead cells and were subsequently run on the BD FACSCanto for analysis. To measure endogenous antibody responses during therapy in the B16F10 model, TA99 antibody used as part of the therapy needed to be depleted from serum samples. To this end, biotinylated TA99 was used for therapy, which could be removed from recovered sera using streptavidin resin. TA99 was biotinylated using an N-hydroxy succinimide-activated biotin according to the manufacturer’s protocol (Thermo Fisher Scientific) and excess biotin was removed using desalting columns (Zeba). Biotin labeling was quantified using the colorimetric Biotin Quantitation Kit (Pierce), and a degree of labeling of 5.1 biotins per TA99 molecule was achieved for the experiments shown. For serum antibody measurement during therapy, serum was isolated from mice weekly and incubated with an excess of streptavidin resin (GenScript) for 1h at 25°C. Serum was frozen and stored at −80°C and thawed on the day of the binding assay, which was performed as described above.
For serum transfer studies, serum was obtained from AIPV treated mice (day 150) or age-matched naïve mice and complement was inactivated by incubating for 30 minutes at 57°C. Serum (350 μL) or TA99 (200 μg) was injected i.p. into naïve recipients six hours prior to injection of 250,000 B16F10 cells i.v. through the tail vein. Lungs were isolated 17 days later and lung nodules were counted in a blinded fashion.
Generation of Trp2 KO cell line
B16-Trp2-KO cells were generated using CRISPR-Cas9. B16-GFP-Cas9 cells were generated by transduction of B16F10 cells with lentivirus expressing humanized SpCas9-P2A-EGFP from an EFS promoter (unpublished, kindly provided by Tim Wang in Dr. David Sabatini’s Lab at the Whitehead Institute). Clones were isolated by single cell flow cytometry for GFP positive cells. A clone stably expressing Cas9 was designated B16-GFP-Cas9. Guide RNA expression vectors were created by cloning a human U6 promoter and sgRNA sequences into a minimal vector with an ampicillin-selectable marker and a ColE1 replication origin. The optimized sgRNA sequence described previously63 were used. Sequences for the sgRNAs (Supplementary Table 1) were designed using tools provided by the Zhang Lab at the Broad Institute (available at http://crispr.mit.edu/). B16-GFP-Cas9 cells were co-transfected with 2 sgRNAs expression vectors targeting Trp2 and a plasmid expressing EGFP and a puromycin selectable marker from a CAG promoter using Xfect (Clontech) according to manufacturer’s instructions. Stable cells were selected for with 2 μg/mL puromycin followed by single cell FACS. Clones were grown for 3 weeks before being analyzed for knockout by PCR amplification of the Trp2 gene (Primers shown in Supplementary Table 2). The final clone was then selected and designated B16-Trp2-KO. PCR amplification of genomic DNA showed a smaller fragment (Supplementary Fig. 8) and sequencing confirmed deletion of the a 283 bp fragment containing the Trp2 peptide sequence (data not shown).
ELISPOT assay
Target B16F10, TC-1, or B16-Trp2-KO cells were treated with 500 U/mL mIFN-γ (Peprotech) for 12 hr, then irradiated (120 Gy). Effector cells were splenocytes isolated from AIPV-treated mice that had rejected tumors 6 days after rechallenge with 106 B16F10 cells. A Mouse IFN-γ ELISPOT Kit (BD) was used. Targets cells were seeded at 25,000 cells per well. Effector cells were seeded at 106 cells per well. Plates were wrapped in foil and cultured for 24 hours then developed according to manufacturer’s protocol. Plates were scanned using a CTL-ImmunoSpot Plate Reader and data was analyzed using CTL ImmunoSpot Software.
Immunoblot
B16F10 cell lysate was run on an SDS-PAGE gel, transferred to nitrocellulose membranes, blocked for 1 hr at 25°C, and then stained with serum from treated mice and rabbit anti-beta-actin overnight at 4°C. Membranes were then washed and stained with goat anti-mouse-IRDye800 and anti-rabbit-IRDye680 for 1 hr at 25°C. Imaging was performed on an Odyssey scanner (Licor). As a control, TA99 was used in place of mouse serum in the protocol above to show the presence of Trp1 in the B16F10 lysate.
Tumor cytokine Luminex assay
Tumors were harvested at day 17 (2 days after 2nd treatment). All tumors from a treatment group were collected up to a maximum of ~250 mg for processing in a bead beater tube. Tissue samples were lysed in lysis buffer (5 μL/mg tumor): 50 mM tris-HCl (pH 7.5), 10% glycerol, 150 mM NaCl, 1% NP-40, and freshly supplemented with Halt Protease & Phosphatase Inhibitor Cocktail (Life Technologies). Lysates were aliquoted and stored at −80°C until analysis. Samples diluted 1:1 with Assay Buffer and assayed using a Luminex bead-based ELISA (MILLIPLEX MAP Mouse Cytokine/Chemokine Magnetic Bead Panel, Millipore) following manufacturer’s instructions with the following modifications: magnetic capture beads and detection antibodies were both used at 1/5x recommended amounts. Concentrations were normalized to total protein input. All samples and analytes were above the limit of detection.
Histology
Tumor inoculation and treatment were performed following the timeline in Fig. 1a. For hematoxylin and eosin staining, mice were sacrificed 3 days after the second treatment, and tumors were fixed in 10% formalin, embedded in paraffin, and stained with hematoxylin and eosin. For immunofluorescence, tumors were isolated on day 14 and processed as previously detailed64. Flank tumors were used to analyze infiltrate by immunofluorescence in the BRafCA PtenloxP Tyr::CreERT2 model to allow for higher quality sectioning. For staining, anti-CD8 (CT-CD8a, Cedar Lane), DAPI, and phalloidin were used. The images were acquired using an Olympus Fluoview FV1200 microscope equipped with 10X (NA 0.40) and 30X (NA 1.05) objectives and optimum lasers and filter sets. The images were acquired under identical acquisition setting and subsequently processed using Fiji image analysis software.
Multivariate Analysis of Cytokine Data
Cytokine levels for each tumor were normalized to the median of “Untreated” controls and log2 transformed. Heatmaps and clustering trees were generated using the clustergram function in Matlab (Mathworks). Euclidean distance was used for hierarchical clustering of cytokines only. The resulting dendrogram was pruned by collapsing leaves with Euclidean distance <95% confidence interval for all 32 potential leaves.
Partial least squares regression was applied using raw cytokine concentrations (pg/μg total protein) regressed against a vector of total tumor mass using PLS_Toolbox R7 (Eigenvector Research). Orthogonalization was used to maximize tumor mass variance captured in the first latent variable; model performance was assessed by random subsets cross-validation with maximum splits and iterations performed.
Statistical Analysis
Statistical methods were not used to predetermine necessary sample size, but sample sizes were chosen based on estimates from pilot experiments and previously published results such that appropriate statistical tests could yield significant results. Experiments were not performed in a blinded fashion. Intratumoral infiltrates, serum transfer foci quantitation, tetramer stain, AST, and ALT, were analyzed using one-way ANOVA with Bonferroni post-test using GraphPad Prism software. Where ANOVA was used, variance between groups was found to be similar by Bartlett’s test. Survival curves were analyzed using the Log-rank (Mantel-Cox) test, and two-tailed student’s t test was used for the antigen spreading ELISPOT. For the tumor infiltrate data and DC uptake data, Welch’s t test (with Bonferroni correction) was used for each condition versus untreated due to unequal variance between groups. No samples were excluded from analysis.
Supplementary Material
Acknowledgments
This work was supported in part by the Koch Institute Support (core) grant P30-CA14051 from the National Cancer Institute, the NIH (CA174795), the Bridge Project partnership between the Koch Institute for Integrative Cancer Research and the Dana Farber/Harvard Cancer Center (DF/HCC), the V Foundation, and the Ragon Institute. K.D.M. and J.M.E. are supported by the Fannie & John Hertz Foundation Fellowship. K.D.M., C.F.O., J.M.E., B.H.K., and E.F.Z. are supported by NSF Graduate Research Fellowships, A.M.R. is supported by the NIGMS/NIH Interdepartmental Biotechnology Training Program (NIH #T32GM008334), and A.T. is supported by the Siebel Scholarship. G.L.S. was supported by the NIH under Ruth L. Kirschstein National Research Service Award (CA180586). We thank T. C. Wu for kindly providing the TC-1 tumor cells and Glenn Dranoff for providing the DD-Her2/neu and B16-OVA cells. We thank Musie Ghebremichael (Ragon Institute of MGH, MIT, and Harvard) for helpful statistical advice. We thank the Koch Institute Swanson Biotechnology Center for technical support, specifically the applied therapeutics and whole animal imaging core facility, histology, and flow cytometry core facility. D.J.I. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Author Contributions
K.D.M., C.F.O., D.J.I., and K.D.W. designed the studies and wrote the manuscript. K.D.M. and C.F.O carried out experiments. E.F.Z, A.T., B.H.K., M.J.K, and H.S. assisted with protein production. J.M.E assisted in generating the Trp2-KO line using Crispr/Cas9. K.R., A.T., E.F.Z., W.A., R.T.W., A.M.R., R.L.K., N.K.M., and K.H. assisted with experiments. G.L.S. designed and analyzed intratumoral Luminex, and assisted in flow cytometry design and writing the manuscript. D.A.L. aided with multivariate analysis of intratumoral Luminex data. G.L.S. and M.H.Z. conducted intratumoral Luminex and assisted with BRaf/Pten experiments. S.K. assisted with immunofluorescence microscopy. J.R.C. supplied 2.5F-Fc reagents.
References
- 1.Hodi FS, et al. Improved Survival with Ipilimumab in Patients with Metastatic Melanoma. New England Journal of Medicine. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Topalian SL, et al. Safety, Activity, and Immune Correlates of Anti–PD-1 Antibody in Cancer. New England Journal of Medicine. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Larkin J, et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. The New England journal of medicine. 2015;373:23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mellman I, Coukos G, Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480:480–489. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drake CG. Combination immunotherapy approaches. Annals of Oncology. 2012;23:viii41–viii46. doi: 10.1093/annonc/mds262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gajewski TF. The Next Hurdle in Cancer Immunotherapy: Overcoming the Non-T-Cell-Inflamed Tumor Microenvironment. Semin Oncol. 2015;42:663–671. doi: 10.1053/j.seminoncol.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 8.Smyth MJ, Ngiow SF, Ribas A, Teng MWL. Combination cancer immunotherapies tailored to the tumour microenvironment. Nat Rev Clin Oncol. 2015 doi: 10.1038/nrclinonc.2015.209. advance online publication. [DOI] [PubMed] [Google Scholar]
- 9.Zhu EF, et al. Synergistic innate and adaptive immune response to combination immunotherapy with anti-tumor antigen antibodies and extended serum half-life IL-2. Cancer cell. 2015;27:489–501. doi: 10.1016/j.ccell.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu H, et al. Structure-based programming of lymph-node targeting in molecular vaccines. Nature. 2014;507:519–522. doi: 10.1038/nature12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zou W, Wolchok JD, Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci Transl Med. 2016;8:328rv324. doi: 10.1126/scitranslmed.aad7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curran MA, Kim M, Montalvo W, Al-Shamkhani A, Allison JP. Combination CTLA-4 blockade and 4-1BB activation enhances tumor rejection by increasing T-cell infiltration, proliferation, and cytokine production. PloS one. 2011;6:e19499. doi: 10.1371/journal.pone.0019499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carmi Y, et al. Allogeneic IgG combined with dendritic cell stimuli induce antitumour T-cell immunity. Nature. 2015;521:99–104. doi: 10.1038/nature14424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu-Lieskovan S, et al. Improved antitumor activity of immunotherapy with BRAF and MEK inhibitors in BRAFV600E melanoma. Science Translational Medicine. 2015;7:279ra241–279ra241. doi: 10.1126/scitranslmed.aaa4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rapoport AP, et al. Combination immunotherapy after ASCT for multiple myeloma using MAGE-A3/Poly-ICLC immunizations followed by adoptive transfer of vaccine-primed and costimulated autologous T cells. Clin Cancer Res. 2014;20:1355–1365. doi: 10.1158/1078-0432.CCR-13-2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lum LG, et al. Targeted T-cell Therapy in Stage IV Breast Cancer: A Phase I Clinical Trial. Clin Cancer Res. 2015;21:2305–2314. doi: 10.1158/1078-0432.CCR-14-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ribas A, et al. Phase I study combining anti-PD-L1 (MEDI4736) with BRAF (dabrafenib) and/or MEK (trametinib) inhibitors in advanced melanoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2015;33(suppl) abstr 3003. [Google Scholar]
- 18.Chen G, et al. A Feasibility Study of Cyclophosphamide, Trastuzumab, and an Allogeneic GM-CSF–Secreting Breast Tumor Vaccine for HER2+ Metastatic Breast Cancer. Cancer Immunology Research. 2014 doi: 10.1158/2326-6066.cir-14-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Overwijk WW, et al. Tumor regression and autoimmunity after reversal of a functionally tolerant state of self-reactive CD8+ T cells. The Journal of experimental medicine. 2003;198:569–580. doi: 10.1084/jem.20030590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klebanoff CA, et al. Determinants of successful CD8+ T-cell adoptive immunotherapy for large established tumors in mice. Clin Cancer Res. 2011;17:5343–5352. doi: 10.1158/1078-0432.CCR-11-0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DuPage M, et al. Endogenous T cell responses to antigens expressed in lung adenocarcinomas delay malignant tumor progression. Cancer cell. 2011;19:72–85. doi: 10.1016/j.ccr.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stromnes IM, et al. T Cells Engineered against a Native Antigen Can Surmount Immunologic and Physical Barriers to Treat Pancreatic Ductal Adenocarcinoma. Cancer cell. 2015;28:638–652. doi: 10.1016/j.ccell.2015.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore SJ, et al. Engineered knottin peptide enables noninvasive optical imaging of intracranial medulloblastoma. Proceedings of the National Academy of Sciences. 2013;110:14598–14603. doi: 10.1073/pnas.1311333110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phan GQ, Attia P, Steinberg SM, White DE, Rosenberg SA. Factors Associated With Response to High-Dose Interleukin-2 in Patients With Metastatic Melanoma. Journal of Clinical Oncology. 2001;19:3477–3482. doi: 10.1200/JCO.2001.19.15.3477. [DOI] [PubMed] [Google Scholar]
- 25.Rose S, Misharin A, Perlman H. A novel Ly6C/Ly6G-based strategy to analyze the mouse splenic myeloid compartment. Cytometry A. 2012;81:343–350. doi: 10.1002/cyto.a.22012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ostrand-Rosenberg S, Sinha P. Myeloid-derived suppressor cells: linking inflammation and cancer. J Immunol. 2009;182:4499–4506. doi: 10.4049/jimmunol.0802740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hildner K, et al. Batf3 Deficiency Reveals a Critical Role for CD8α Dendritic Cells in Cytotoxic T Cell Immunity. Science. 2008;322:1097. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rafiq K, Bergtold A, Clynes R. Immune complex – mediated antigen presentation induces tumor immunity. The Journal of clinical investigation. 2002;110:71–79. doi: 10.1172/JCI200215640.Introduction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Broz ML, et al. Dissecting the tumor myeloid compartment reveals rare activating antigen-presenting cells critical for T cell immunity. Cancer cell. 2014;26:638–652. doi: 10.1016/j.ccell.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts EW, et al. Critical Role for CD103+/CD141+ Dendritic Cells Bearing CCR7 for Tumor Antigen Trafficking and Priming of T Cell Immunity in Melanoma. Cancer cell. 2016 doi: 10.1016/j.ccell.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salmon H, et al. Expansion and Activation of CD103(+) Dendritic Cell Progenitors at the Tumor Site Enhances Tumor Responses to Therapeutic PD-L1 and BRAF Inhibition. Immunity. 2016;44:924–938. doi: 10.1016/j.immuni.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dankort D, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41:544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mendiratta SK, Thai G, Eslahi NK, Thull NM. Therapeutic tumor immunity induced by polyimmunization with melanoma antigens gp100 and TRP-2. Cancer research. 2001:859–863. [PubMed] [Google Scholar]
- 34.Nelson MH, et al. Toll-like receptor agonist therapy can profoundly augment the antitumor activity of adoptively transferred CD8(+) T cells without host preconditioning. J Immunother Cancer. 2016;4:6. doi: 10.1186/s40425-016-0110-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woo SR, Corrales L, Gajewski TF. Innate immune recognition of cancer. Annu Rev Immunol. 2015;33:445–474. doi: 10.1146/annurev-immunol-032414-112043. [DOI] [PubMed] [Google Scholar]
- 36.van Egmond M, Bakema JE. Neutrophils as effector cells for antibody-based immunotherapy of cancer. Semin Cancer Biol. 2013;23:190–199. doi: 10.1016/j.semcancer.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 37.Weiskopf K, Weissman IL. Macrophages are critical effectors of antibody therapies for cancer. MAbs. 2015;7:303–310. doi: 10.1080/19420862.2015.1011450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DiLillo DJ, Ravetch JV. Differential Fc-Receptor Engagement Drives an Anti-tumor Vaccinal Effect. Cell. 2015;161:1035–1045. doi: 10.1016/j.cell.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Montfoort N, et al. Antigen storage compartments in mature dendritic cells facilitate prolonged cytotoxic T lymphocyte cross-priming capacity. Proc Natl Acad Sci U S A. 2009;106:6730–6735. doi: 10.1073/pnas.0900969106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guilliams M, Bruhns P, Saeys Y, Hammad H, Lambrecht BN. The function of Fcgamma receptors in dendritic cells and macrophages. Nat Rev Immunol. 2014;14:94–108. doi: 10.1038/nri3582. [DOI] [PubMed] [Google Scholar]
- 41.Clatworthy MR, et al. Immune complexes stimulate CCR7-dependent dendritic cell migration to lymph nodes. Nat Med. 2014;20:1458–1463. doi: 10.1038/nm.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Albanesi M, et al. Neutrophils mediate antibody-induced antitumor effects in mice. Blood. 2013;122:3160–3164. doi: 10.1182/blood-2013-04-497446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allan RS, et al. Migratory dendritic cells transfer antigen to a lymph node-resident dendritic cell population for efficient CTL priming. Immunity. 2006;25:153–162. doi: 10.1016/j.immuni.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 44.den Haan JM, Lehar SM, Bevan MJ. CD8(+) but not CD8(−) dendritic cells cross-prime cytotoxic T cells in vivo. The Journal of experimental medicine. 2000;192:1685–1696. doi: 10.1084/jem.192.12.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gasteiger G, et al. IL-2-dependent tuning of NK cell sensitivity for target cells is controlled by regulatory T cells. The Journal of experimental medicine. 2013;210:1167–1178. doi: 10.1084/jem.20122462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.List J, et al. Cytokine responses to intraventricular injection of interleukin 2 into patients with leptomeningeal carcinomatosis: rapid induction of tumor necrosis factor alpha, interleukin 1 beta, interleukin 6, gamma-interferon, and soluble interleukin 2 receptor (Mr 55,000 protein) Cancer Res. 1992;52:1123–1128. [PubMed] [Google Scholar]
- 47.Carreno BM, et al. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science. 2015;348:803–808. doi: 10.1126/science.aaa3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DuPage M, Mazumdar C, Schmidt LM, Cheung AF, Jacks T. Expression of tumour-specific antigens underlies cancer immunoediting. Nature. 2012;482:405–409. doi: 10.1038/nature10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vigneron N, Stroobant V, Van den Eynde BJ, van der Bruggen P. Database of T cell-defined human tumor antigens: the 2013 update. Cancer Immunity Archive. 2013;13 [PMC free article] [PubMed] [Google Scholar]
- 50.Yadav M, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2014;515:572–576. doi: 10.1038/nature14001. [DOI] [PubMed] [Google Scholar]
- 51.Gubin MM, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515:577–581. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nature Reviews Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morgan RA, et al. Cancer Regression in Patients After Transfer of Genetically Engineered Lymphocytes. Science. 2006;314:126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson LA, et al. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood. 2009;114:535–546. doi: 10.1182/blood-2009-03-211714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tzeng A, Kwan BH, Opel CF, Navaratna T, Wittrup KD. Antigen specificity can be irrelevant to immunocytokine efficacy and biodistribution. Proc Natl Acad Sci U S A. 2015;112:3320–3325. doi: 10.1073/pnas.1416159112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boggio K, et al. Interleukin 12–mediated Prevention of Spontaneous Mammary Adenocarcinomas in Two Lines of Her-2/neu Transgenic Mice. The Journal of Experimental Medicine. 1998;188:589–596. doi: 10.1084/jem.188.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guevara P, et al. Optimization of a self antigen for presentation of multiple epitopes in cancer immunity. The Journal of Clinical Investigation. 116:1382–1390. doi: 10.1172/JCI25591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Stipdonk MJB, et al. Design of Agonistic Altered Peptides for the Robust Induction of CTL Directed towards H-2Db in Complex with the Melanoma-Associated Epitope gp100. Cancer Research. 2009;69:7784–7792. doi: 10.1158/0008-5472.can-09-1724. [DOI] [PubMed] [Google Scholar]
- 59.Malakhov MP, et al. SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. Journal of Structural and Functional Genomics. 5:75–86. doi: 10.1023/b:jsfg.0000029237.70316.52. [DOI] [PubMed] [Google Scholar]
- 60.Rubinstein MP, et al. Converting IL-15 to a superagonist by binding to soluble IL-15Rα. Proceedings of the National Academy of Sciences. 2006;103:9166–9171. doi: 10.1073/pnas.0600240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hooijkaas AI, Gadiot J, van der Valk M, Mooi WJ, Blank CU. Targeting BRAFV600E in an inducible murine model of melanoma. Am J Pathol. 2012;181:785–794. doi: 10.1016/j.ajpath.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 62.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 63.Chen B, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Joshi Nikhil S, et al. Regulatory T Cells in Tumor-Associated Tertiary Lymphoid Structures Suppress Anti-tumor T Cell Responses. Immunity. 43:579–590. doi: 10.1016/j.immuni.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.