Abstract
Background
Cronobacter sakazakii is an emerging opportunistic pathogen that is associated with rare but life-threatening cases of severe diseases: meningitis, necrotizing enterocolitis, and sepsis in premature and full-term infants. However, the pathogenesis mechanism of this pathogen remains largely unknown. To determine its pathogenesis at the genomic level, the genome of C. sakazakii ATCC 29544T was completely sequenced and analyzed.
Results
The genomic DNA, containing a circular chromosome and three plasmids, is composed of 4,511,265 bp with a GC content of 56.71%, containing 4380 predicted open reading frames (ORFs), 22 rRNA genes, and 83 tRNA genes. The plasmids, designated pCSK29544_p1, pCSK29544_p2, and pCSK29544_p3, were 93,905-bp, 4938-bp, and 53,457-bp with GC contents of 57.02, 54.88, and 50.07%, respectively. They were also predicted to have 72, 6, and 57 ORFs without RNA genes.
Conclusions
The strain ATCC 29544T genome has ompA and ibeB-homologous cusC genes, probably associated with the invasion of human brain microvascular endothelial cells (BMECs). In addition, gene clusters for siderophore production (iucABCD/iutA) and the related transport system (eitCBAD) were detected in pCSK29544_p1 plasmid, indicating better iron uptake ability for survival. Furthermore, to survive under extremely dry condition like milk powder, this genome has gene clusters for biosynthesis of capsular proteins (CSK29544_00281-00284) and cellulose (CSK29544_01124-01127) for biofilm formation and a gene cluster for utilization of sialic acid in the milk (nanKTAR). The genome information of C. sakazakii ATCC 29544T would provide further understanding of its pathogenesis at the molecular level for the regulation of pathogenicity and the development of a rapid detection method using biomarkers.
Electronic supplementary material
The online version of this article (doi:10.1186/s13099-016-0150-0) contains supplementary material, which is available to authorized users.
Keywords: Cronobacter sakazakii, Complete genome sequence, Pathogenesis, Infant milk formula, Virulence factor
Background
Enterobacter sakazakii has been reclassified into seven species in the genus Cronobacter according to biochemical and genetic evaluations [1, 2]. Among them, Cronobacter sakazakii is a well-known opportunistic food-borne pathogen causing bacteremia, meningitis and necrotizing enterocolitis, particularly in low-birth-weight neonatal infants. This species is a Gram-negative, rod-shaped, peritrichous and yellow-pigmented facultative anaerobe belonging to the Enterobacteriaceae family [3]. C. sakazakii has been often found in human and infant gut microbiota [4, 5]. Although C. sakazakii food-borne outbreaks are quite low, the fatality to infants generally ranges from 40 to 80% [6]. Interestingly, C. sakazakii was reported to produce capsular or biofilm materials for its own protection from extremely dry conditions, as in formula milk powder, substantiating the high survival ability of C. sakazakii in the milk powder [7]. After human infection, C. sakazakii can invade the intestinal epithelial cells and even the brain microvascular endothelial cells (BMECs), demonstrating its potentials to cause meningitis [8]. Therefore, the biocontrol and regulation of C. sakazakii are urgently required. However, C. sakazakii is resistant to some antibiotics, indicating a problem with antibiotic therapies against C. sakazakii [9, 10], and its pathogenicity mechanism remains unknown. Recently, to unveil the knowledge about the Cronobacter ecology, multilocus sequence typing (MLST) analysis using seven housekeeping genes has been established to identify the diversity of the Cronobacter genus from various sources, and its application has facilitated understanding of the evolutionary relationships and environmental fitness of Cronobacter species [2]. In this study, to understand its infection and pathogenesis at the molecular level, the genome of a representative C. sakazakii type strain, ATCC 29544T, was completely sequenced and analyzed in this study using bioinformatics. This genome information would provide the researchers with genomic insights into the virulence and pathogenicity mechanisms of this species for the further development of a rapid detection method and a novel biocontrol strategy.
Methods
Growth conditions and DNA isolation
Cronobacter sakazakii ATCC 29544T was routinely cultivated using Luria-Bertani (LB) medium at 37 °C with shaking at 220 rpm. Bacterial cells were harvested in the mid-exponential growth phase using centrifugation at 16,000×g for 1 min and its genomic DNA was isolated using G-spin™ Genomic DNA Extraction Kit for Bacteria (iNtRON Biotechnology, Seongnam, South Korea). The concentration and purity of extracted DNA were determined by NanoVue (GE healthcare, Little Chalfont, United Kingdom).
Genome sequencing and assembly
The complete genome of C. sakazakii ATCC 29544T was sequenced at Macrogen, Seoul, South Korea, using a hybrid of PacBio RS II (Pacific Biosciences, Menlo Park, CA, USA) and Illumina HiSeq 2500 (San Diego, CA, USA). The sequence reads from PacBio RS II and Illumina HiSeq 2500 platforms were assembled using HGAP (version 2.0) and ALLPATHS-LG (version r47449), respectively. The final genome coverages were on average 1321 X Illumina and 73 X PacBio, respectively.
Genome annotation
The ORFs were predicted using Glimmer3 [11] and GeneMark.hmm [12]. The gene prediction results were confirmed by manual curation. The genes of rRNA and tRNA were predicted using RNAmmer 1.2 [13] and tRNAscan-SE [14], respectively. The genome annotation was conducted using NCBI BLASTP [15] and a predicted protein analysis using InterProScan 5 [16] for the prediction of protein functions.
The analysis of comparative genomes and phage-associated regions
Phage-associated gene clusters in the genome sequences of C. sakazakii ATCC 29544 were searched using PHAST server [17].
Quality assurance
Cronobacter sakazakii ATCC 29544T was obtained from American Type Culture Collection (ATCC) and its morphological observation using a transmission electron microscopy (TEM) showed a traditional shape of C. sakazakii as a short rod (Additional file 1: Figure S1). In addition, this strain was confirmed to C. sakazakii using 16S rRNA gene sequencing. For genome sequencing, the raw read sequences were selected and assembled when their quality scores were more than 40 as cutoffs. The complete genome sequence after genome assembly was also used for confirmation using ANI analysis with previously reported complete genome sequences of C. sakazakii.
Initial findings
General genome properties
The complete genome of C. sakazakii ATCC 29544T is composed of a circular chromosome and three plasmids (Fig. 1). The chromosome is 4,511,265 bp in DNA length with a GC content of 56.71%, 4380 ORFs, 22 rRNA genes, and 83 tRNA genes. The plasmids, designated pCSK29544_p1, pCSK29544_p2 and pCSK29544_p3, are 93,905, 4938 and 53,457 bp in DNA length with GC contents of 57.02, 54.88 and 50.07%, respectively. The plasmids have predicted ORFs of 72, 6, and 57.
Fig. 1.
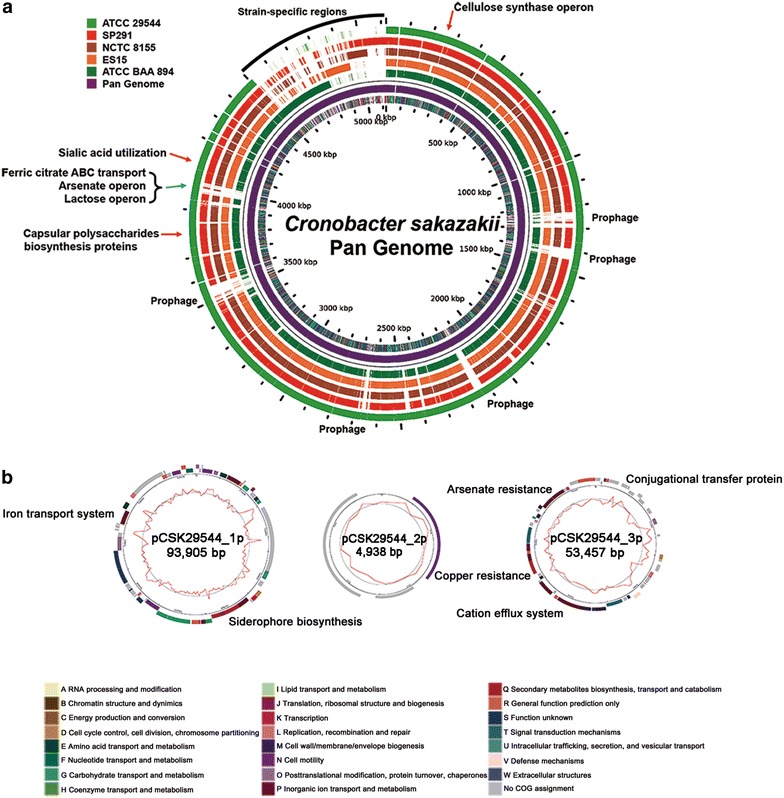
a Pan genome map of five C. sakazakii strains using GView [34]. The cut off value of BLASTN homology was 80%. The inner circle represents COG functional categories of C. sakazakii ATCC 29544T. Red arrows indicate gene cluster conserved in all C. sakazakii strains and blue arrows indicate ATCC 29544T—specific gene clusters. Black curved bar indicates other C. sakazakii strain-specific regions. b Plasmid maps from C. sakazakii ATCC 29544T. The curved bars in the outer circle indicate the predicted ORFs by strand, and colored by COG categories. The inner circle with red peaks indicates the G + C content
Pathogenesis and virulence factors
Cronobacter sakazakii infects human neonates and infants via mostly contaminated reconstituted infant formula milk, causing serious human diseases, including bacteremia, necrotizing enterocolitis, and even meningitis [6]. The high survival rate of C. sakazakii, even under extremely dried conditions, as in milk formula powder, has not been investigated at the molecular level. To understand this extraordinary property of C. sakazakii, molecular studies have been recently performed. Interestingly, the gene clusters associated with biosynthesis of capsular polysaccharides (CSK29544_00281-00284) and cellulose (CSK29544_01124-01127) were detected. Recently, the biofilm formation and cellulose (as a component of biofilm) production of C. sakazakii were experimentally confirmed [18, 19], suggesting that these gene clusters may be involved in the biofilm formation. This biofilm formation of C. sakazakii may contribute to the survival in the infant formula conditions [19]. However, exopolysaccharide (EPS) production was not observed in C. sakazakii ATCC 29544T [20], suggesting that capsular polysaccharide gene cluster may be associated with biofilm formation, not with EPS production. In particular, C. sakazakii is the only Cronobacter species that has the nanKTAR gene cluster to utilize sialic acid [21]. Interestingly, C. sakazakii ATCC 29544T has this gene cluster (CSK29544_00587-590), indicating its ability to use it. This unique ability may be involved in its adaptation to the milk conditions because sialic acid is one of the components of milk [22].
Upon human infection, C. sakazakii invades brain microvascular endothelial cells (BMECs), barriers to protect the brain from infection of meningitic pathogens, including Escherichia coli K1 and Neisseria meningitides, which cause meningitis in neonates and infants [23, 24]. Outer membrane protein A (OmpA) of C. sakazakii is a critical determinant, contributing in vitro invasion of human BMECs by enhancing cell adhesion [24]. In addition, a few genes (ibeA, ibeB, and yijP) in meningitic E. coli were also suggested to be associated with the invasion of human BMECs [25–27]. Interestingly, two genes, ompA (CSK29544_03699) for BMEC adhesion and the ibeB-homologous cusC (pCSK29544_3p0028) for the penetration of BMECs, were detected in the genome of C. sakazakii ATCC 29544T−, suggesting this strain may invade human BMECs. However, ibeA and yijP were not detected in the genome. It is noteworthy that this ibeB-homologous cusC is located in the gene cluster of the complete copper- and silver-resistance cation efflux system as encoded by cusABCF (CSK29544_3p0026–CSK29544_3p0031) in the plasmid pCSK29544_3p, suggesting this BMEC invasion ability may be transferrable to other C. sakazakii or that the strain ATCC 29544T may have been acquired from other C. sakazakii for human BMEC invasion. The presence of a conjugational transfer protein encoded by traX (CSK29544_3p0007) supports this hypothesis.
Cronobacter sakazakii ATCC 29544T harbors three plasmids, pCSK29544_1p (1p), pCSK29544_2p (2p), and pCSK29544_3p (3p). Interestingly, two large plasmids (1p and 3p) encode virulence-associated genes, related to iron uptake and BMEC invasion. Therefore, these two plasmids may contribute to host dominance and pathogenesis, respectively.
Iron is an essential element for dominant survival and colonization via bacterial competition for iron uptake because it plays an important role in the electron transport system for energy production [28]. To accomplish this dominant survival and colonization against other bacteria, C. sakazakii has an iron acquisition system, including siderophore production (iucABCD/iutA) and an ABC-type transport system (eitCBAD) in the plasmid pESA3 [29]. The strain ATCC 29544T also has this privileged iron acquisition system, including siderophore biosynthesis (iucABCD/iutA; CSK29544_1p0024–CSK29544_1p0028) and an ABC-type iron transport system (eitCBAD; CSK29544_1p0056–CSK29544_1p0059). However, this iron acquisition system is present in the plasmid 1p, similar to pESA3 of C. sakazakii BAA-894 [29]. Interestingly, C. sakazakii BAA-894 has multiple copies of pESA3 plasmids in a host, which is highly homologous to the plasmid 1p, suggesting the plasmid 1p may be multi copied, too [30]. These results indicate that the host strain may take advantage of better iron uptake ability in the given environments, probably due to the presence of multiple copies of this iron acquisition system encoded in the plasmid 1p.
Recent studies have revealed that milk formula contains an at least six-times-higher arsenic concentration than that of breast milk, suggesting bacterial survival in milk formula may require arsenic resistance [31]. In addition to the previously mentioned BMEC invasion ability of C. sakazakii ATCC 29544T via the ibeB-homologous cusC gene in the cation efflux system, the plasmid 3p has an arsenic resistance system as encoded by arsRDABC, suggesting the strain ATCC 29544T may have arsenic resistance activity for survival in formula milk powder and that this ability may have been acquired via plasmid conjugational transfer.
Comparative genome analysis
To date, five complete genome sequences of C. sakazakii and their annotation data are available in the GenBank database. To compare these genomes, whole genomic DNA sequence-based average nucleotide index (ANI) analysis and Roary matrix-based protein sequence analysis were performed. ANI analysis showed that C. sakazakii NCTC 8155 and SP291 are the most closely related, forming a taxonomical group, and the strains ATCC 29544T, ATCC BAA894, and ES15 are also similar to this group (Fig. 2a). Subsequent Roary matrix-based protein sequence analysis of these five genomes supports this relationship among them (Fig. 2b). However, the strain ATCC 29544T is more closely related to the group in the Roary matrix-based tree.
Fig. 2.
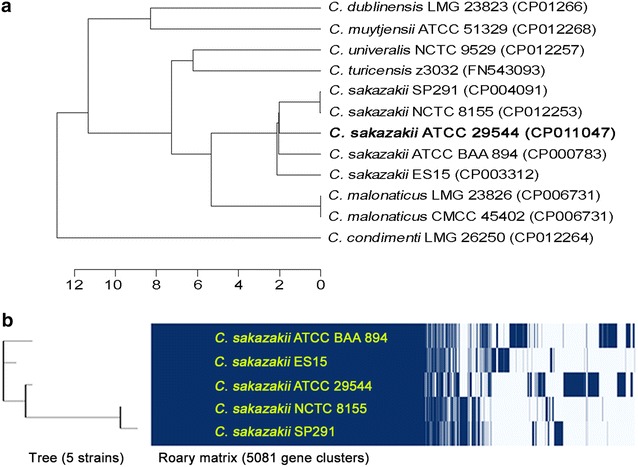
a Phylogenetic tree analysis of C. sakazakii ATCC 29544T and other Cronobacter species strains by average nucleotide identity (ANI) using JSpecies. b Roary matrix-based protein sequence comparison and the associated taxonomical tree of five C. sakazakii strains [35]
Moreover, pan genome analysis of these five genomes was conducted and their strain-specific regions were detected (Fig. 1). Interestingly, three functional gene clusters and seven prophage regions are ATCC 29544T-specific. In addition to the iron acquisition system consisting of siderophore production and ABC-type transport system in the plasmid 1p, ATCC 29544T-specific iron uptake-associated gene cluster containing fecABDCEI for dominant uptake of ferric dicitrate is present in the chromosome, probably related to survival in the iron-limited condition [32]. The second arsenic resistance gene cluster consisting of arsRCB for arsenate sensing, reduction, and efflux pumping was also detected, which is similar to the gene cluster in the plasmid 3p, probably for dominant survival in the human gut containing low concentration of arsenic after human infection [33] (Additional file 1: Fig. S2A). Comparative analysis of their protein sequences showed that they are >94% identical to each other, probably for synergistic arsenic resistance activity. Unlike other two gene clusters shared by the chromosome and plasmid, two copies of lactose operons are present in the chromosomes of ATCC 29544T, NCTC 8155, and SP291, probably related to the Roary matrix-based tree (Fig. 2b). While one of these operons (CSK29544_04271–04273) are shared by all C. sakazakii genomes, the second lactose operon (CSK29544_00439–00442) is ATCC 29544T-specific. This second lactose operon is similar to other lactose operon in Klebsiella and Enterobacter (data not shown), suggesting that this operon may be originated from other genus bacterium. The presence of multiple mobile elements containing transposases and relatively low protein sequence identity support this hypothesis (Additional file 1: Fig. S2B). Furthermore, pan genome analysis showed strain-specific region (Fig. 1). Most of genes in this region belong to prophages and they have very low homology among them, indicating that these prophages are also strain-specific. Interestingly, the strain ATCC 29544T contains seven strain-specific prophage regions with five intact prophages and two incomplete prophages (Table 1). However, most of genes in the prophages encode hypothetical proteins.
Table 1.
Genome statistics
| Attribute | Value | % of Totala |
|---|---|---|
| Genome size (bp) | 4,663,565 | 100 |
| DNA coding (bp) | 4,116,350 | 88.27 |
| DNA G + C (bp) | 2,641,511 | 56.64 |
| Total genes | 4620 | 100 |
| Protein coding genes | 4515 | 97.72 |
| RNA genes | 105 | 2.27 |
| rRNA operons | 22 | 0.48 |
| tRNA genes | 83 | 1.8 |
| Genes with function prediction | 3212 | 69.52 |
| Predicted prophages | 7 | 6.18 |
| CRISPR repeats | 2 | 0.04 |
aThe total is based on either the size of the genome (chromosome and 3 plasmids) in base pairs or the total number of total genes in the annotated genome
Future directions
This genome information of C. sakazakii ATCC 29544T provides genomic insights into the presence of various virulence factors and their associated pathogenesis mechanisms at molecular level. In addition, this genome information showed that the strain ATCC 29544T has several distinct features probably contributing to dominant survival under the given habitats and even various stress conditions like extremely dry or nutrient-limited conditions. Therefore, this genome information would be useful for further development of a novel strategy for control of C. sakazakii and of rapid detection method using strain-specific DNA markers in genomic level.
Authors’ contributions
SK, JHL, and SR initiated and supervised the study. SK and JHL drafted the manuscript. SK conducted laboratory experiments, and SK and YTK worked on the genome sequencing and annotated the genome. SK, YTK, HY, JHL, and SR discussed and analyzed the data and revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The genome sequences of C. sakazakii ATCC 29544T were deposited in the NCBI GenBank server under the accession numbers CP011047, CP011048, CP011049, and CP011050 for a chromosome and three plasmids, respectively.
Funding
This research was supported by a grant (14162MFDS972) from the Ministry of Food and Drug Safety, Korea, in 2016 and by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (NRF-2015R1C1A1A01053815).
Abbreviations
- BMECs
brain microvascular endothelial cells
- ANI
average nucleotide identity
- LB
Luria–Bertani medium
- MLST
multilocus sequence typing
- TEM
transmission electron microscopy
- COG
clusters of orthologous groups
Additional file
Additional file 1: Figure S1. The morphology of C. sakazakii ATCC 29544 imaged by energy-filtering transmission electron microscopy (EF-TEM). EF-TEM photograph was obtained by 2% uranyl acetate on copper grids and examined with ET-TEM at a voltage of 120 kV (LIBRA 120, Zeiss, Oberkochen, Germany). Figure S2: Comparative analysis of two ATCC 29544-specific gene clusters: (A) lac operon and (B) arsenic resistance. The amino acid sequence identities between associated genes are indicated as percentages.
Footnotes
Ju-Hoon Lee and Sangryeol Ryu contributed equally to this work
Contributor Information
Seongok Kim, Email: seongok@snu.ac.kr.
You-Tae Kim, Email: youeutae@khu.ac.kr.
Hyunjin Yoon, Email: yoonh@ajou.ac.kr.
Ju-Hoon Lee, Email: juhlee@khu.ac.kr.
Sangryeol Ryu, Email: sangryu@snu.ac.kr.
References
- 1.Iversen C, Lehner A, Mullane N, Bidlas E, Cleenwerck I, Marugg J, et al. The taxonomy of Enterobacter sakazakii: proposal of a new genus Cronobacter gen. nov. and descriptions of Cronobacter sakazakii comb. nov. Cronobacter sakazakii subsp. sakazakii, comb. nov., Cronobacter sakazakii subsp. malonaticus subsp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov. and Cronobacter genomospecies 1. BMC Evol Biol. 2007;7:64. doi: 10.1186/1471-2148-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Joseph S, Forsythe SJ. Insights into the emergent bacterial pathogen Cronobacter spp., generated by multilocus sequence typing and analysis. Front Microbiol. 2012;3:397. doi: 10.3389/fmicb.2012.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farmer JJ, Asbury MA, Hickman FW, Brenner DJ. Enterobacter sakazakii—a new species of Enterobacteriaceae isolated from clinical specimens. Int J Syst Bacteriol. 1980;30:569–584. doi: 10.1099/00207713-30-3-569. [DOI] [Google Scholar]
- 4.Hallstrom M, Eerola E, Vuento R, Janas M, Tammela O. Effects of mode of delivery and necrotising enterocolitis on the intestinal microflora in preterm infants. Eur J Clin Microbiol Infect Dis. 2004;23:463–470. doi: 10.1007/s10096-004-1146-0. [DOI] [PubMed] [Google Scholar]
- 5.Mao B, Li D, Zhao J, Liu X, Gu Z, Chen YQ, et al. In vitro fermentation of fructooligosaccharides with human gut bacteria. Food Funct. 2015;6:947–954. doi: 10.1039/C4FO01082E. [DOI] [PubMed] [Google Scholar]
- 6.Healy B, Cooney S, O’Brien S, Iversen C, Whyte P, Nally J, et al. Cronobacter (Enterobacter sakazakii): an opportunistic foodborne pathogen. Foodborne Pathog Dis. 2010;7:339–350. doi: 10.1089/fpd.2009.0379. [DOI] [PubMed] [Google Scholar]
- 7.Hurrell E, Kucerova E, Loughlin M, Caubilla-Barron J, Forsythe SJ. Biofilm formation on enteral feeding tubes by Cronobacter sakazakii, Salmonella serovars and other Enterobacteriaceae. Int J Food Microbiol. 2009;136:227–231. doi: 10.1016/j.ijfoodmicro.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 8.Giri CP, Shima K, Tall BD, Curtis S, Sathyamoorthy V, Hanisch B, et al. Cronobacter spp. (previously Enterobacter sakazakii) invade and translocate across both cultured human intestinal epithelial cells and human brain microvascular endothelial cells. Microb Pathog. 2012;52:140–147. doi: 10.1016/j.micpath.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Kilonzo-Nthenge A, Rotich E, Godwin S, Nahashon S, Chen F. Prevalence and antimicrobial resistance of Cronobacter sakazakii isolated from domestic kitchens in middle Tennessee, United States. J Food Prot. 2012;75:1512–1517. doi: 10.4315/0362-028X.JFP-11-442. [DOI] [PubMed] [Google Scholar]
- 10.Xu X, Li C, Wu Q, Zhang J, Huang J, Yang G. Prevalence, molecular characterization, and antibiotic susceptibility of Cronobacter spp. in Chinese ready-to-eat foods. Int J Food Microbiol. 2015;204:17–23. doi: 10.1016/j.ijfoodmicro.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 11.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lukashin AV, Borodovsky M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 16.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi Y, Kim S, Hwang H, Kim KP, Kang DH, Ryu S. Plasmid-encoded MCP is involved in virulence, motility, and biofilm formation of Cronobacter sakazakii ATCC 29544. Infect Immun. 2015;83:197–204. doi: 10.1128/IAI.02633-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu L, Grim CJ, Franco AA, Jarvis KG, Sathyamoorthy V, Kothary MH, et al. Analysis of the cellulose synthase operon genes, bcsA, bcsB, and bcsC in Cronobacter species: prevalence among species and their roles in biofilm formation and cell-cell aggregation. Food Microbiol. 2015;52:97–105. doi: 10.1016/j.fm.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Lehner A, Riedel K, Eberl L, Breeuwer P, Diep B, Stephan R. Biofilm formation, extracellular polysaccharide production, and cell-to-cell signaling in various Enterobacter sakazakii strains: aspects promoting environmental persistence. J Food Prot. 2005;68:2287–2294. doi: 10.4315/0362-028X-68.11.2287. [DOI] [PubMed] [Google Scholar]
- 21.Joseph S, Hariri S, Masood N, Forsythe S. Sialic acid utilization by Cronobacter sakazakii. Microb Inform Exp. 2013;3:3. doi: 10.1186/2042-5783-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang B, Brand-Miller J, McVeagh P, Petocz P. Concentration and distribution of sialic acid in human milk and infant formulas. Am J Clin Nutr. 2001;74:510–515. doi: 10.1093/ajcn/74.4.510. [DOI] [PubMed] [Google Scholar]
- 23.Nassif X, Bourdoulous S, Eugene E, Couraud PO. How do extracellular pathogens cross the blood-brain barrier? Trends Microbiol. 2002;10:227–232. doi: 10.1016/S0966-842X(02)02349-1. [DOI] [PubMed] [Google Scholar]
- 24.Nair MK, Venkitanarayanan K, Silbart LK, Kim KS. Outer membrane protein A (OmpA) of Cronobacter sakazakii binds fibronectin and contributes to invasion of human brain microvascular endothelial cells. Foodborne Pathog Dis. 2009;6:495–501. doi: 10.1089/fpd.2008.0228. [DOI] [PubMed] [Google Scholar]
- 25.Huang SH, Wan ZS, Chen YH, Jong AY, Kim KS. Further characterization of Escherichia coli brain microvascular endothelial cell invasion gene ibeA by deletion, complementation, and protein expression. J Infect Dis. 2001;183:1071–1078. doi: 10.1086/319290. [DOI] [PubMed] [Google Scholar]
- 26.Huang SH, Chen YH, Fu Q, Stins M, Wang Y, Wass C, et al. Identification and characterization of an Escherichia coli invasion gene locus, ibeB, required for penetration of brain microvascular endothelial cells. Infect Immun. 1999;67:2103–2109. doi: 10.1128/iai.67.5.2103-2109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Huang SH, Wass CA, Stins MF, Kim KS. The gene locus yijP contributes to Escherichia coli K1 invasion of brain microvascular endothelial cells. Infect Immun. 1999;67:4751–4756. doi: 10.1128/iai.67.9.4751-4756.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mietzner TA, Morse SA. The role of iron-binding proteins in the survival of pathogenic bacteria. Annu Rev Nutr. 1994;14:471–493. doi: 10.1146/annurev.nu.14.070194.002351. [DOI] [PubMed] [Google Scholar]
- 29.Franco AA, Hu L, Grim CJ, Gopinath G, Sathyamoorthy V, Jarvis KG, et al. Characterization of putative virulence genes on the related RepFIB plasmids harbored by Cronobacter spp. Appl Environ Microbiol. 2011;77:3255–3267. doi: 10.1128/AEM.03023-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kucerova E, Clifton SW, Xia XQ, Long F, Porwollik S, Fulton L, et al. Genome sequence of Cronobacter sakazakii BAA-894 and comparative genomic hybridization analysis with other Cronobacter species. PLoS ONE. 2010;5:e9556. doi: 10.1371/journal.pone.0009556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carignan CC, Cottingham KL, Jackson BP, Farzan SF, Gandolfi AJ, Punshon T, et al. Estimated exposure to arsenic in breastfed and formula-fed infants in a United States cohort. Environ Health Perspect. 2015;123:500–506. doi: 10.1289/ehp.1408789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harvie DR, Ellar DJ. A ferric dicitrate uptake system is required for the full virulence of Bacillus cereus. Curr Microbiol. 2005;50:246–250. doi: 10.1007/s00284-004-4442-0. [DOI] [PubMed] [Google Scholar]
- 33.Ratnaike RN. Acute and chronic arsenic toxicity. Postgrad Med J. 2003;79:391–396. doi: 10.1136/pmj.79.933.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petkau A, Stuart-Edwards M, Stothard P, Van Domselaar G. Interactive microbial genome visualization with GView. Bioinformatics. 2010;26:3125–3126. doi: 10.1093/bioinformatics/btq588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequences of C. sakazakii ATCC 29544T were deposited in the NCBI GenBank server under the accession numbers CP011047, CP011048, CP011049, and CP011050 for a chromosome and three plasmids, respectively.


