Abstract
Background
Expanded malaria control efforts in Sénégal have resulted in increased use of rapid diagnostic tests (RDT) to identify the primary disease-causing Plasmodium species, Plasmodium falciparum. However, the type of RDT utilized in Sénégal does not detect other malaria-causing species such as Plasmodium ovale spp., Plasmodium malariae, or Plasmodium vivax. Consequently, there is a lack of information about the frequency and types of malaria infections occurring in Sénégal. This study set out to better determine whether species other than P. falciparum were evident among patients evaluated for possible malaria infection in Kédougou, Sénégal.
Methods
Real-time polymerase chain reaction speciation assays for P. vivax, P. ovale spp., and P. malariae were developed and validated by sequencing and DNA extracted from 475 Plasmodium falciparum-specific HRP2-based RDT collected between 2013 and 2014 from a facility-based sample of symptomatic patients from two health clinics in Kédougou, a hyper-endemic region in southeastern Sénégal, were analysed.
Results
Plasmodium malariae (n = 3) and P. ovale wallikeri (n = 2) were observed as co-infections with P. falciparum among patients with positive RDT results (n = 187), including one patient positive for all three species. Among 288 negative RDT samples, samples positive for P. falciparum (n = 24), P. ovale curtisi (n = 3), P. ovale wallikeri (n = 1), and P. malariae (n = 3) were identified, corresponding to a non-falciparum positivity rate of 2.5%.
Conclusions
These findings emphasize the limitations of the RDT used for malaria diagnosis and demonstrate that non-P. falciparum malaria infections occur in Sénégal. Current RDT used for routine clinical diagnosis do not necessarily provide an accurate reflection of malaria transmission in Kédougou, Sénégal, and more sensitive and specific methods are required for diagnosis and patient care, as well as surveillance and elimination activities. These findings have implications for other malaria endemic settings where species besides P. falciparum may be transmitted and overlooked by control or elimination activities.
Electronic supplementary material
The online version of this article (doi:10.1186/s12936-016-1661-3) contains supplementary material, which is available to authorized users.
Keywords: Plasmodium falciparum, Plasmodium ovale curtisi, Plasmodium ovale wallikeri, Plasmodium malariae, Rapid diagnostic test
Background
Among Plasmodium species that infect humans, Plasmodium falciparum and Plasmodium vivax are responsible for most of the global mortality and morbidity. However, with intensified efforts to control the incidence of these primary species, there is rising concern over the prevalence of additional malaria species including Plasmodium ovale spp. and Plasmodium malariae into the niches previously occupied by P. falciparum and P. vivax [1–3]. Should these other species be undetected by current diagnostic methods, there is the potential that they may make substantive and possibly increasing contributions to overall malaria burden.
Globally, there have been sporadic reports of changing trends in the prevalence of malaria species other than P. falciparum. For example, there are increasing reports of P. ovale spp. in various countries across the African continent, including Ethiopia, Uganda, Equatorial Guinea, and Kenya [1–3]. Historically, and recently, there have been ad hoc reports of malaria infections caused by species other than P. falciparum in Sénégal. While previous reports in Sénégal have indicated low or reduced incidence of these species [4, 5], there are limited data from other regions of the country, and it remains unknown if these observations can be generalized nationwide. Furthermore, P. malariae is relatively under-reported in these regions [6]. Recently, multiple clinical cases of P. vivax infection were detected in Sénégal [7]. However, it remains unclear if the paucity of data describing other malaria species is due to a lack of inadequate diagnostics or a consequence of relative lack of transmission in these regions. Overall there is insufficient information about the prevalence or dynamics of these malaria species, especially in the context of recent efforts to reduce or eliminate malaria. Moreover, no recent attempt has been made to systematically determine the prevalence of non-falciparum malaria parasites in Sénégal.
The region of Kédougou in southeastern Sénégal is an area of high malaria transmission that borders the countries of Mali and Guinea. It is characterized by relatively high humidity and an ecology that supports seasonal malaria transmission, with entomological inoculation rates between 100 and 200 and incidence rates of greater than 25 per 1000 individuals [8–10]. Human migration to and from the region is encouraged by ongoing mining activities in Kédougou that have expanded in the most recent decade.
In addition to using previously-reported methods, this study also developed and validated a set of real-time PCR speciation assays to detect P. ovale spp., P. malariae, and P. vivax. These assays were applied to analyse nearly 500 rapid diagnostic tests (RDT) from two health centers in Kédougou, Sénégal in order to assess the prevalence of non-falciparum species in this region.
Methods
Sample collection site
Kédougou (12°56′00″ N, 12º21,00″W) is situated in the extreme southeastern part of Sénégal, in a tropical savanna located 710 km from Dakar, the nation’s capital (Fig. 1). Its 151,715 inhabitants comprise around 5% of the national population. The passive case detection in this study was based on patients aged 6 months or older reporting to health clinics in Tomboronkonto and Dindefelo for suspected malaria with fever or history of fever in the previous 48 h. Samples were collected from the first patient over 5 years of age as well as the first patient under 5 years of age reporting daily, for 4 days of the week over the course of a single year, a sampling scheme considered roughly representative of the symptomatic population reporting to health care facilities. These patients were diagnosed by RDT (First Response Malaria Ag [HRP2], Premier Medical Corp., USA), and parasite material from these discarded RDT was extracted for species identification. Institutional Review Board (IRB) review of these discarded diagnostic materials that were anonymous and de-identified were deemed non-human subjects material and used accordingly.
Fig. 1.
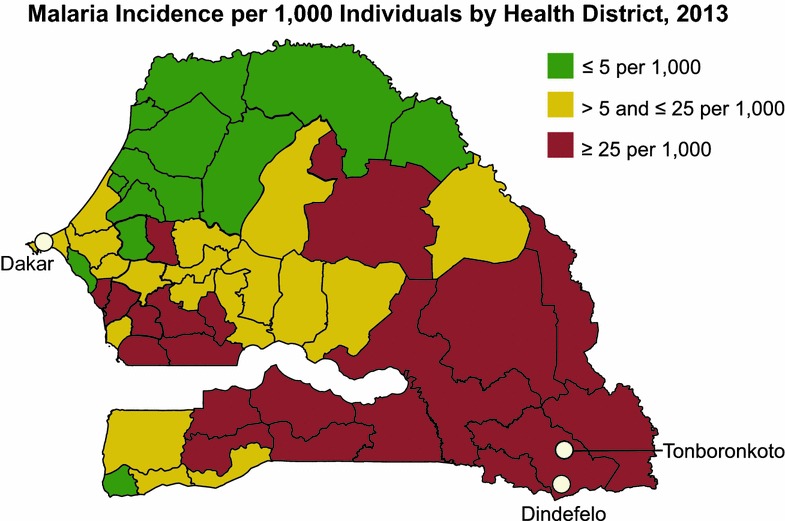
Study area. Map of Sénégal showing the location of health clinics in Kédougou relative to Dakar. Shading indicates malaria incidence levels. Figure adapted from the National Strategic Framework 2014–2018 [10]
Sample type and extraction
Parasite DNA was isolated from the filter pad of the RDT using the Promega DNA IQ Casework Pro Kit for Maxwell 16 (Promega Corp., Madison, WI, USA) as directed by the manufacturer.
Speciation assays
First, species-specific TaqMan real-time assays were developed to target the Plasmodium plasmepsin 4 gene based on previously-reported sequence data [11] (Table 1). Each DNA sample was run in duplicate with plasmid-based standard curves from 50,000 to 0.5 copies. The control plasmids contained species-specific plasmepsin 4 genes (Genbank accession numbers AF001208.1, AF001209.1, and 144 AF001210.1 for P. vivax, P. ovale spp., and P. malariae, respectively).
Table 1.
Primer and probe sequences
| Gene | Primer name | Primer sequence (5′– >3′) | Amplicon size (bp) | Specificity | Reference |
|---|---|---|---|---|---|
| Porbp2 | Porbp2TMfwd | TTGCAAACAAAAGTGCTCC | 120 | P. ovale | [2] |
| Porbp2TMrev | CCTAATTCTCTTTGT(G/A)CCC | ||||
| Potra | PoTRA fwd3 | GCACAAAAATGGTGCTAACC | 787 | P. ovale | [2] |
| PoTRA rev3 | ATCCATTTACCTTCCATTGC | ||||
| PoTRA fwd5 | ACGGCAAACCCGATAAACAC | 245–355 | |||
| PoTRA rev5 | GTGTTTGTAGTATTTACAGG | ||||
| Plasmepsin 4 | AIRSAMA_Fwd | ACTGACACTGATGATTTAGAACCCATTT | 1353 | P. vivax | |
| AIRSAMA_Rev | TGGAGAGATCTTTCCATCCTAAACCT | ||||
| AIRSAMA_Probe | CAGCAGCGTCGAGTTT | ||||
| Plasmepsin 4 | Ovale_CCRR9V5_Fwd | ACTCTTGGTTATTTGTCTGCACCTT | 1353 | P. ovale | |
| Ovale_CCRR9V5_Rev | CTATGTTACCATAAACAGGTTCTAAATCATCTGT | ||||
| Ovale_CCRR9V5_Probe | TCAGTTGCTTCAACAAATTT | ||||
| PF3D7_0718800 | pf07_0076_Fwd | CGACCCTGATGTTGTTGTTGGA | 79 | P. falciparum | [11] |
| pf07_0076_Rev | GGCTTTTTTCCATTTCTGTAGTTAAGATTCA | ||||
| pf07_0076_Probe | CAACAGCTCCAAAATAT | ||||
| Plasmepsin 4 | Malariae_CCWR2K1_Fwd | TTCAGTCAGGATATGTAAAACAAAATTATTTAGGTAGT | 1356 | P. malariae | |
| Malariae_CCWR2K1_Rev | CCTACTTCCCCTTCACCATAAAACA | ||||
| Malariae_CCWR2K1_Probe | TCGTCTAGTTCTATTACGTCATTTTC |
Next, each extracted RDT sample was pre-amplified [12] with species-specific primers (Table 1). The P. falciparum primers targeted the PF3D7_0718800 gene, as described previously [13].
Briefly, each reaction was performed with 3 µL of pre-amplified parasite DNA in a 15 µL total reaction volume (2× qPCR Master Mix, 20× primer-probe mix, and PCR-grade water). Amplification and detection were performed under the following conditions: 15-min incubation at 37 °C prior to amplification, initial denaturation for 15 min at 95 °C, then 40 cycles of denaturation for 10 s at 95 °C, and 1 min at 60 °C with data collection.
The specificities of the P. malariae, P. vivax, and P. ovale spp. real-time assays were validated by Sanger sequencing (Macrogen Corp., Rockville, MD) of the amplicon products, both from the synthesized plasmids and the amplification-positive samples identified in the current study.
Plasmodium ovale follow-up
Plasmodium ovale curtisi and Plasmodium ovale wallikeri were identified among P. ovale–positive samples using PoTRA3 and PoTRA5 primers in a nested PCR amplification of the tryptophan rich repeat region as described previously [10]. The PCR mixture consisted of 5 µL of DNA in a final volume of 20 µL (6 µL Phusion high-fidelity master mix [New England Biolabs, Ipswich, MA, USA], 2 µL of 10 pmol each forward and reverse primer, and 5 µL PCR-grade water). Infections with P. ovale wallikeri yielded a product of 245 bp, and P. ovale curtisi a 317 bp product.
DNA sequences at loci of interest were determined by direct sequencing of the final PCR product using PoTRA5 primers (Table 1). Sequences are available as GenBank accession numbers KX417699–KX417705.
High-resolution melting assays
Samples positive for P. ovale were additionally tested by high-resolution melting to differentiate sub-species using the porbp2 primers, as described previously [2].
Results
Validation of plasmepsin 4 P. ovale spp., P. malariae, and P. vivax assays
Amplicon products of these assays using control DNA templates and plasmid controls were confirmed to match sequences deposited to GenBank with 100% sequence concordance.
Overall RDT performance
A total of 475 HRP2-based RDT collected in Kédougou by the Programme National de Lutte contre le Paludisme (National Malaria Control Programme or PNLP) were evaluated in this study (Fig. 2). Of these, 187 were positive for P. falciparum (RDT positivity rate: 39.3%, 187 of 475 samples). Furthermore, among the 288 samples negative for P. falciparum by RDT, 24 were positive for amplification of the P. falciparum PF3D7_0718800 gene (Additional file 1: Figure S1), indicating the presence of this species even in RDT-negative samples (PCR positivity rate: 44.4%, 211 of 475 samples). The detection of Plasmodium showed similar trends between the two clinic settings (Tomboronkonto and Dindefelo, Additional file 2: Figure S2).
Fig. 2.
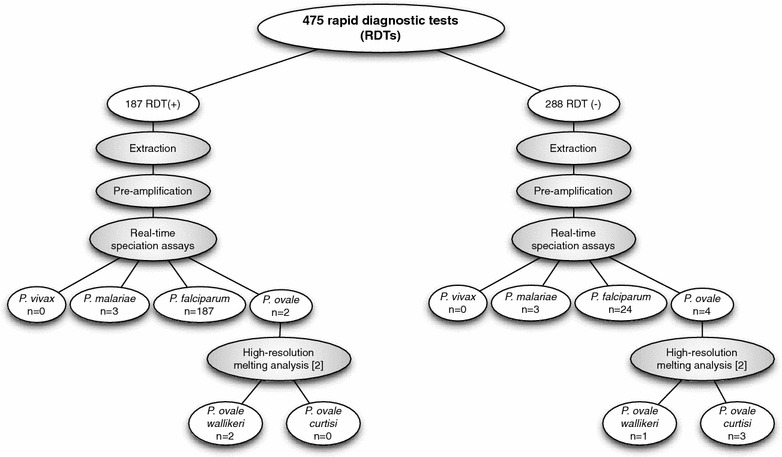
Flow chart of patient samples and species analysis
Evidence of malaria species other than Plasmodium falciparum among samples collected by passive case detection in Kédougou, Sénégal
Analysis of the RDT-positive samples revealed co-infection of P. falciparum with P. malariae (n = 3) and P. ovale (n = 2); with one sample positive for all three species. Both P. ovale (n = 4) and P. malariae (n = 3) were identified from among the 288 RDT-negative samples (Table 1). Although previous reports [7] indicate P. vivax infections in this region, in this study population there were no samples positive for P. vivax.
Sympatric circulation of Plasmodium ovale wallikeri and Plasmodium ovale curtisi
To further distinguish between the known subtypes of P. ovale infection, we employed a high-resolution melting analysis approach and confirmed that two of the RDT-positive samples were positive for P. ovale wallikeri; of four RDT-negative samples, three and one were positive for P. ovale curtisi and P. ovale wallikeri, respectively (Figs. 3, 4). These data indicate that both subtypes of P. ovale are evident in the southeastern part of Sénégal.
Fig. 3.
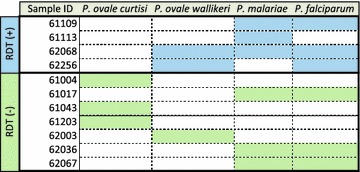
Summary of sample species positivity by rapid diagnostic test status. Shaded boxes indicate samples positive for the respective Plasmodium species based upon the PCR-based testing described
Fig. 4.
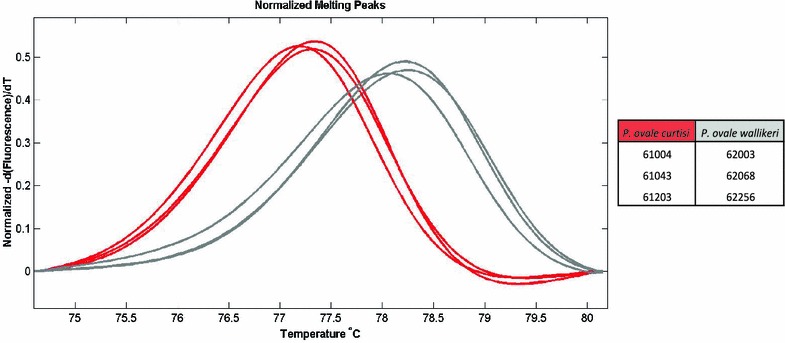
High-resolution melting differentiation of P. ovale curtisi and P. ovale wallikeri
To confirm the sensitivity and specificity of the genotyping results for P. malariae and P. ovale, we used a sequencing approach. Sanger sequencing of the positive samples was used to confirm the presence of P. malariae and P. ovale among the samples, and a BLAST search of the resultant sequences showed high identity with previously reported sequences. Sequencing was also able to distinguish between the P. ovale curtisi and P. ovale wallikeri identified in these samples; the sequencing results were concordant with those of the high-resolution melt findings.
Discussion
After significant restructuring in 2005, the Programme National de Lutte contre le Paludisme (PNLP) in Sénégal developed a control strategy implemented between 2006 and 2010 that involved distribution of RDT to all health facilities (2007), nationwide access to artemisinin combination therapies (2007 and 2008), and distribution of insecticide-treated bed nets (2007–2009). While the initial results of this strategy were encouraging based on the nationwide reported incidence of P. falciparum, several regions have experienced malaria rebound since 2009 [14]. Of concern is that HRP2-based surveillance may miss both non-P. falciparum malaria species as well as low-density P. falciparum infections and infections composed exclusively of P. falciparum parasites that lack HRP2 and HRP3 expression [15].
The PNLP has also conducted nationwide cross-sectional surveys for disease surveillance. Kédougou, in southeastern Sénégal, is a region that borders Mali and Guinea with relatively high transmission, as indicated by entomological inoculation rates (EIRs) of 100–200 [9, 10]. Recent reports have identified P. vivax malaria in Kédougou [7]. Although P. vivax was not observed in the population assessed in the current study, P. ovale wallikeri, P. ovale curtisi, and P. malariae were identified, none of which are tested for by the current RDT. It remains to be determined if this incidence is changing, particularly in comparison with reduced P. falciparum transmission in response to increased eradication efforts that target P. falciparum vectors and their characteristic behaviors, which might not be common in other Plasmodium species. Indeed, there is evidence of significant biological differences between P. ovale spp. and P. falciparum, with increased latency times reported in patients infected with the former and differences in clinical features between P. ovale curtisi and P. ovale wallikeri [16]. Additional studies are necessary to more fully characterize the nationwide prevalence of these non-falciparum species; to identify their source as importation or local transmission; their primary transmission vectors; and to differentiate between relapse, reinfection, or recrudescence events where applicable.
The results of this study underscore the potential limitations of the routine use of standard RDT for malaria detection in Sénégal both in terms of missing P. falciparum infections as well as missing non-P. falciparum malaria infections in Kédougou, Sénégal. The current RDT likely detects most of the infections in this region, particularly those without co-infections; however, while the relative proportion of non-falciparum infections remains low, it may increase with improved patient access to diagnosis and treatment. Furthermore, the findings of the current study, including the presence of P. falciparum genetic material in RDT-negative samples and subsequent differences in positivity rates (39.3 vs. 44.4%), are similar to those of previous reports [17] that PCR-based detection of parasites genomic material is more sensitive than standard RDT. This suggests the need for more sensitive methods to detect infections, particularly in asymptomatic patients and in the context of declining transmission and malaria incidence rates. This study developed tools that allow for the sensitive identification of these infections for surveillance of the incidences of infection and their changes in response to control efforts.
The lack of detection of P. falciparum may be due to either low parasite densities, below the level of detection of the RDT or lack of specific HRP2 loci in the parasite population that results in a negative diagnostic test. Reports of non-falciparum malaria are of concern with control and elimination efforts that increasingly rely on HRP2-based and P. falciparum-specific RDT, as these may miss infections that contribute to continued malaria transmission, and thus may undermine elimination efforts and impact morbidity and mortality due to malaria. Failure to detect malaria infections other than P. falciparum is most likely is due to the specificity of the RDT, which was not designed to detect these malaria species. Regardless of the specific reasons why the RDT failed to detect these infections, these findings point to the need for appropriate diagnosis and surveillance of malaria infection to provide both adequate clinical treatment and to direct elimination strategies to address the nature of ongoing malaria infection in a given endemic setting. Future studies are necessary to assess and track the prevalence of HRP2 deletions in endemic populations in this region.
This study has several limitations. First, the pre-amplification step, like nested and multi-step PCR reactions, offers increased sensitivity to detect parasites at low concentrations, but prevents us from determining the threshold cycles (Ct) values of the original samples; furthermore, RDT were administered by regular health care providers rather than staff specifically trained for malaria testing. Blood smears were not obtained from the patients, and trained microscopists were not available; therefore, parasite densities in these patients were not determined. Finally, the sampling scheme was not a cross-sectional study, so there is a potential bias in the study population. Future cross-sectional studies including larger sample sizes and asymptomatic patients are necessary to validate these findings.
Despite these limitations, the findings of this study suggest the importance of assessing the true prevalence of specific malaria infections using appropriate diagnostic methods to evaluate the requirements for health care services and develop appropriate strategies for malaria elimination. Such quantification is increasingly important in order for malaria elimination programmes to implement species-appropriate detection, prevention, and treatment strategies.
Authors’ contributions
RFD, ABD, DFW, SKV, and DN conceived of the research, participated in the analysis, and helped to write the manuscript. ABD, JFG, BD, JIT, FBD, MB, MN, ASB, YDN, and DN participated in the study design and sample collection. RFD, ABD, and KD extracted samples and performed PCR reactions. DFW, SKV, and DN offered critical review of the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We would like to thank Younous Diedhiou for collecting the RDT, the Sénégal PNLP for providing the RDT, the Region Medical Kédougou for permission to use these discarded diagnostic materials, and the people of Kédougou for their support and participation.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets generated and analysed during the current study are available in the GenBank repository as accession numbers KX417699–KX417705.
Ethics approval and consent to participate
Institutional Review Board (IRB) review of these anonymous and de-identified discarded diagnostic materials were deemed non-human subjects material and used accordingly.
Funding
This work was made possible through support provided by the United States President’s Malaria Initiative, and the U.S. Agency for International Development, under the terms of an Interagency Agreement with the Centers for Disease Control and Prevention (CDC). The opinions expressed herein are those of the authors and do not necessarily reflect the views of the U.S. Agency for International Development or the Centers for Disease Control and Prevention.
In addition, this work was also supported by a grant from the Bill and Melinda Gates Foundation (Grant # OPP1053604).
Additional files
Additional file 1: Figure S1. Plasmodium spp. detection in Kédougou, 2013–2014 (n = 475 samples).
Additional file 2: Figure S2. Plasmodium falciparum detection in Kédougou by health center, 2013–2014 (n = 475).
Footnotes
Rachel F. Daniels and Awa Bineta Deme contributed equally to this work
Contributor Information
Rachel F. Daniels, Email: rdaniels@hsph.harvard.edu
Awa Bineta Deme, Email: deme.awa@gmail.com.
Jules F. Gomis, Email: jules.gomis@gmail.com
Baba Dieye, Email: dieyebaba2004@yahoo.fr.
Katelyn Durfee, Email: kdurfee@hsph.harvard.edu.
Julie I. Thwing, Email: jthwing@cdc.gov
Fatou B. Fall, Email: fall1fatou@yahoo.fr
Mady Ba, Email: mady.ba@pnlp.sn.
Medoune Ndiop, Email: mnzop5@yahoo.fr.
Aida S. Badiane, Email: asbadiane@gmail.com
Yaye Die Ndiaye, Email: ydndiaye@gmail.com.
Dyann F. Wirth, Email: dfwirth@hsph.harvard.edu
Sarah K. Volkman, Email: svolkman@hsph.harvard.edu
Daouda Ndiaye, Email: daouda.ndiaye@ucad.edu.sn.
References
- 1.Alemu A, Fuehrer H-P, Getnet G, Tessema B, Noedl H. Plasmodium ovale curtisi and Plasmodium ovale wallikeri in North-West Ethiopia. Malar J. 2013;12:346. doi: 10.1186/1475-2875-12-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oguike MC, Betson M, Burke M, Nolder D, Stothard JR, Kleinschmidt I, et al. Plasmodium ovale curtisi and Plasmodium ovale wallikeri circulate simultaneously in African communities. Int J Parasitol. 2011;41:677–683. doi: 10.1016/j.ijpara.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller RH, Obuya CO, Wanja EW, Ogutu B, Waitumbi J, Luckhart S, et al. Characterization of Plasmodium ovale curtisi and P. ovale wallikeri in Western Kenya utilizing a novel species-specific real-time PCR assay. PLoS Negl Trop Dis. 2015;9:e0003469. doi: 10.1371/journal.pntd.0003469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roucher C, Rogier C, Sokhna C, Tall A, Trape J-F. A 20-year longitudinal study of Plasmodium ovale and Plasmodium malariae prevalence and morbidity in a West African population. PLoS ONE. 2014;9:e87169. doi: 10.1371/journal.pone.0087169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diallo MA, Badiane AS, Diongue K, Deme A, Lucchi NW, Gaye M, et al. Non-falciparum malaria in Dakar: a confirmed case of Plasmodium ovale wallikeri infection. Malar J. 2016;15:429. doi: 10.1186/s12936-016-1485-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Badiane AS, Diongue K, Diallo S, Ndongo AA, Diedhiou CK, Deme AB, et al. Acute kidney injury associated with Plasmodium malariae infection. Malar J. 2014;13:226. doi: 10.1186/1475-2875-13-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niang M, Thiam LG, Sow A, Loucoubar C, Bob NS, Diop F, et al. A molecular survey of acute febrile illnesses reveals Plasmodium vivax infections in Kedougou, southeastern Sénégal. Malar J. 2015;14:281. doi: 10.1186/s12936-015-0808-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Continuous Demographic and Health Survey in Sénégal (Continuous DHS) 2012–2013. Agence Nationale de la Statistique et de la Démographie (ANSD) [Sénégal]. ICF International: Calverton; 2013.
- 9.MAP: malaria atlas project [Internet]. map.ox.ac.uk. http://www.map.ox.ac.uk/explore/countries/sen/. Accessed 11 Oct 2016.
- 10.National Strategic Framework 2014–2018 [Internet]. www.pnlp.sn. http://www.pnlp.sn/telechargements/Documents-strategiques/PNLP_CADRE_STRATEGIQUE.pdf. Accessed 16 Sep 2016.
- 11.Dame JB, Yowell CA, Omara-Opyene L, Carlton JM, Cooper RA, Li T. Plasmepsin 4, the food vacuole aspartic proteinase found in all Plasmodium spp. infecting man. Mol Biochem Parasitol. 2003;130:1–12. doi: 10.1016/S0166-6851(03)00137-3. [DOI] [PubMed] [Google Scholar]
- 12.Mharakurwa S, Daniels R, Scott A, Wirth DF, Thuma P, Volkman SK. Pre-amplification methods for tracking low-grade Plasmodium falciparum populations during scaled-up interventions in Southern Zambia. Malar J. 2014;13:89. doi: 10.1186/1475-2875-13-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daniels R, Volkman SK, Milner DA, Mahesh N, Neafsey DE, Park DJ, et al. A general SNP-based molecular barcode for Plasmodium falciparum identification and tracking. Malar J. 2008;7:223. doi: 10.1186/1475-2875-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daniels RF, Schaffner SF, Wenger EA, Proctor JL, Chang H-H, Wong W, et al. Modeling malaria genomics reveals transmission decline and rebound in Sénégal. Proc Natl Acad Sci USA. 2015;112:7067–7072. doi: 10.1073/pnas.1505691112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deme AB, Park DJ, Bei AK, Sarr O, Badiane AS, Gueye PEHO, et al. Analysis of pfhrp2 genetic diversity in Sénégal and implications for use of rapid diagnostic tests. Malar J. 2014;13:34. doi: 10.1186/1475-2875-13-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rojo-Marcos G, Rubio-Muñoz JM, Ramírez-Olivencia G, García-Bujalance S, Elcuaz-Romano R, Díaz-Menéndez M, et al. Comparison of imported Plasmodium ovale curtisi and P. ovale wallikeri infections among patients in Spain, 2005–2011. Emerg Infect Dis. 2014;20:409–416. doi: 10.3201/eid2003.130745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu L, van den Hoogen LL, Slater H, Walker PGT, Ghani AC, Drakeley CJ, et al. Comparison of diagnostics for the detection of asymptomatic Plasmodium falciparum infections to inform control and elimination strategies. Nature. 2015;528:S86–S93. doi: 10.1038/nature16039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analysed during the current study are available in the GenBank repository as accession numbers KX417699–KX417705.


