Abstract
Background
Malaria is still a major public health issue worldwide, and one of the best approaches to fight the disease remains vector control. The current methods for mosquito identification include morphological methods that are generally time-consuming and require expertise, and molecular methods that require laboratory facilities with relatively expensive running costs. Matrix-Assisted Laser Desorption Ionization Time-Of-Flight Mass Spectrometry (MALDI-TOF MS) technology, routinely used for bacterial identification, has recently emerged in the field of entomology. The aim of the present study was to assess whether MALDI-TOF MS could successfully distinguish Anopheles stephensi mosquitoes according to their Plasmodium infection status.
Methods
C57BL/6 mice experimentally infected with Plasmodium berghei were exposed to An. stephensi bites. For the determination of An. stephensi infection status, mosquito cephalothoraxes were dissected and submitted to mass spectrometry analyses and DNA amplification for molecular analysis. Spectra were grouped according to mosquitoes’ infection status and spectra quality was validated based on intensity and reproducibility within each group. The in-lab MALDI-TOF MS arthropod reference spectra database, upgraded with representative spectra from both groups (infected/non-infected), was subsequently queried blindly with cephalothorax spectra from specimens of both groups.
Results
The MALDI TOF MS profiles generated from protein extracts prepared from the cephalothorax of An. stephensi allowed distinction between infected and uninfected mosquitoes. Correct classification was obtained in blind test analysis for (79/80) 98.75% of all mosquitoes tested. Only one of 80 specimens, an infected mosquito, was misclassified in the blind test analysis.
Conclusions
Matrix-Assisted Laser Desorption Ionization Time-Of-Flight Mass Spectrometry appears to be a promising, rapid and reliable tool for the epidemiological surveillance of Anopheles vectors, including their identification and their infection status.
Background
Malaria is caused by parasites of the Plasmodium genus. These pathogens are transmitted to humans during the blood meal of an infected female Anopheles mosquito [1]. Five species of Plasmodium are known to infect humans [1] and Plasmodium falciparum is the most virulent species for humans, causing cerebral malaria and death in the worst cases [1, 2]. A major weapon against malaria remains vector control, which involves monitoring of Anopheles mosquito populations and knowledge of their infection status with regards to Plasmodium species.
Currently, the identification of mosquitoes is mainly done by morphological or molecular methods [3]. Morphological identification is a reliable method and yet may be be limited by the requirement of identification keys, specific documentation and entomological expertise. Using these methods can be lengthy if a large number of samples must be identified. Moreover, morphological tools cannot differentiate mosquitoes belonging to a species complex, such as the Gambiae complex, which includes 8 species that are not all Plasmodium vectors [6]. Molecular techniques are an alternative. However, they require specific laboratory facilities. They may also be time-consuming and relatively expensive, especially when sequencing is required [4]. The infection status of arthropods can be determined in diverse ways, but three major methods are used for detection of Plasmodium in mosquitoes. The microscopic approach, which is routinely used in malaria-endemic countries, entails the observation of live or stained parasites in dissected or crushed mosquitoes [5]. Although it is an affordable and relevant method, it is time-consuming, and conclusions can be operator-dependent [6]. The second approach relies on immunological methods, such as the enzyme-linked immunosorbent assay (ELISA) or direct immunohistochemistry, both methods targeting Plasmodium antigens such as the circumsporozoite protein (CSP). Antibodies are frequently used to distinguish Plasmodium-infected from parasite-free Anopheles. However, this method presents several limitations, such as the diversity of antibodies required for the specific detection of Plasmodium species, the risk of cross-reaction with close parasite species by antibodies, or the difficulty of data interpretation when the signal-to-noise ratio is low [7]. Molecular methods such as standard PCR, nested-PCR and qPCR are adequate and sensitive for Plasmodium detection on whole and pooled mosquitoes [10, 11], but the preparation of the samples and cost of the reagents may limit their use. Thus, the development of a consensual alternative tool for rapid and inexpensive identification of mosquitoes, and also for the determination of their infection status, appears important for the development of malaria epidemiological surveillance.
In recent years, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF–MS) has been used for the identification and classification of microorganisms [15, 16], and it has also been applied in the identification of arthropods [7], including mosquitoes [17–19]. It requires the creation of a reference spectra database obtained from organisms unambiguously identified by morphology and molecular biology reference methods. More recently, MALDI-TOF has been described as a promising alternative for the detection of microorganisms in arthropods. Indeed, it was reported that this innovative tool could differentiate rickettsiae-infected and non-infected ticks, using only tick legs [8] or tick haemolymph [9]. The dual identification of arthropod species and infection status at the same time by MALDI-TOF could be revolutionary for vector monitoring and entomological diagnosis.
Based on these promising results, the goal of the present study was to assess whether MALDI-TOF could be used to distinguish Plasmodium-infected Anopheles from uninfected mosquitoes. To do so, mosquitoes were infected by feeding on C57BL/6 mice experimentally infected with Plasmodium berghei (ANKA strain). MS spectra from molecularly validated infected and non-infected mosquitoes were then compared to evaluate the ability of mass spectrometry to distinguish the two categories.
Methods
Mice and ethics statement
Female C57BL/6 J mice from the Charles River Laboratories (Saint-Germain-Nuelles, France) (8–10 week old, 20–25 g body weight) were maintained at 23–25 °C with a 12 h light/12 h dark cycle, with free access to food and water. Animal studies were conducted in agreement with the directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 and in agreement with decree No. 2013-118 of 1 February 2013 from the French Government. The study was approved by the local Ethics Committee (Comité d’Ethique en Expérimentation Animale Marseille 14) of Marseille, France.
Murine model of experimental malaria with Plasmodium berghei
Six female mice were infected with the same batch of P. berghei (ANKA strain) parasites by intraperitoneal injection of 2 × 106 parasitized red blood cells per mouse, after validation of the cell titer on KOVA-slides (Fisher Scientific, Illkirch, France). Parasitaemia, expressed as the percentage of parasitized red blood cells, was determined daily from day 3 post-infection by Giemsa-stained tail-blood smears, as previously described [10]. Gametocytaemia, expressed as the percentage of parasites in gametocyte form, was determined as well. Animals were monitored daily for clinical signs of cerebral malaria until mouse sacrifice. The main focus was on weight loss, which was the main clinical manifestation detectable during animal follow-up. Four mice of the same age and same breeding conditions were used as controls.
Anopheles rearing
Only Anopheles stephensi mosquitoes were used in this study, and all were from the Institut Pasteur laboratory rearing facility. They were reared in the laboratory (Aix-Marseille University, Marseille, France) using standard methods, with temperature of 26 ± 1 °C, relative humidity of 80 ± 10% and a 12 h light/12 h dark cycle in incubators (Panasonic cooled incubator). Larvae were reared until the pupal stage in trays containing 1 litre of distilled water supplemented with fish food (TetraMin Baby, Tetra GmbH, Herrenteich, Germany). Pupae were collected daily and transferred to a mosquito cage (Bug Dorm 1, BioQuip products). Adults were fed with a 10% glucose solution. For egg production, blood meals were given through a Parafilm-membrane and an artificial feeding device (Hemotek membrane feeding systems, Discovery Workshops UK), using fresh human blood [11]. Female An. stephensi planned to be used for this work were isolated and fed only with a 10% sugar solution from their emergence until their mouse blood meal 4 days later. After the blood meal on mice, Plasmodium exposed mosquitoes were reared at temperature of 21 ± 1 °C, relative humidity of 80 ± 10% and a 12 h light/12 h dark cycle in incubators.
Anopheles feeding and sampling
Based on parasitaemia greater than 1% and the appearance of gametocytes in blood, mice were anesthetized by intraperitoneal injection of ketamine (90 mg/kg) and xylazine (10 mg/kg). Immediately after complete anesthesia, mice abdomens were thoroughly shaved and disinfected with ethanol. Mice were then placed directly onto mosquito cages to allow engorgement for 1 h. Mosquitoes were killed 18 days after their infective blood meal by storage at −20 °C until further utilization.
Sample preparation for MALDI-TOF MS
Mosquitoes’ cephalothoraxes (head and thorax) were selected for MALDI-TOF MS. Eighteen days post mouse-blood feeding, each mosquito was briefly washed in 70% ethanol, rinsed in sterile water, dried and dissected. Glass powder and 50 µl of HPLC-grade water were added to mosquito cephalothoraxes (CTs) before crushing them using the TissueLyser instrument (Qiagen, Hilden, Germany) at a frequency of 30 movements per second for 60 s. This cycle was repeated twice and followed by a single 3 min cycle. The crushed CTs were centrifuged at 10,000 rpm for 30 s and 1 µL of the supernatant of each sample was carefully dropped on a MALDI-TOF target plate in quadruplicate (Bruker Daltonics, Wissembourg, France) [12]. Each spot was then recovered with 1 µL CHCA matrix solution composed of saturated α-cyano-4-hydroxycynnamic acid (CHCA) (Sigma, Lyon, France), 50% acetonitrile (v/v), 2.5% trifluoroacetic acid (v/v) (Aldrich, Dorset, UK) and HPLC-grade water. After drying for several minutes at room temperature, the target plate was introduced into the Microflex LT MALDI-TOF Mass Spectrometer device (Bruker Daltonics, Germany) for analysis. To control loading on mass spectra steel, matrix quality and MALDI-TOF apparatus performance, matrix solution only was also loaded in duplicate onto each MALDI-TOF plate with or without a E. coli bacterial test standard (Bruker protein Calibration Standard I).
DNA extraction and molecular analysis
Infection by P. berghei was simultaneously assessed in all the samples using MALDI-TOF and molecular biology. To do so, the remaining supernatant and debris of crushed CTs after MS plate loading were used for molecular determination of the infection status of the collected mosquitoes. Crushed CTs were vortexed with 200 µL of G2 buffer solution containing 20 µL of proteinase K (15 mg/mL) (Qiagen, Hilden, Germany) and incubated at 56 °C overnight prior to DNA extraction. DNA extractions from individual mosquito samples were performed with the Qiagen EZ1 Advanced XL Robot with respective Qiagen kits (Qiagen, Hilden, Germany), according to the manufacturer’s recommendations. DNA from each sample was eluted in 50 µL of Tris EDTA (TE) buffer (Qiagen, Hilden, Germany) and either immediately used or stored at −20 °C. Primers and probe targeting a 189 bp region of the 18S gene of all Plasmodium species were used to detect P. berghei DNA by real-time quantitative PCR (Table 1). Real-time quantitative PCR was carried out according to the manufacturer’s protocol using a CFX Connect™ Real-Time PCR Detection System (Bio-Rad) with the Eurogentec Takyon qPCR kit. DNA extracted from P. berghei-infected mouse blood was used as positive control. DNA extracted from mosquitoes of the non-infected group and DNA-free PCR mix were used as PCR negative controls. Standard PCR targeting a 710 bp region of the invertebrate cytochrome oxidase I (COI) gene [13] was performed on a randomly selected batch of 21 exposed but PCR negative mosquitoes.
Table 1.
Sequences of primers and probe used for the quantitative PCR detection of P. berghei in mosquitoes
| Target | Primer name | Sequence |
|---|---|---|
|
18S gene All Plasmodium species |
18S_F | 5′-AGGCAACAACAGGTCTGTGA-3′ |
| 18S_R | 5′-GCAATAATCTATCCCCATCACG-3′ | |
| 18S_P | 5′-6FAM- GAACTAGGCTGCACGCGTGCTACA-3′ |
Primers and probe targeting a 189 bp sequence of all Plasmodium species 18S gene used for the detection of Plasmodium berghei ANKA in mosquitoes
MALDI-TOF–MS parameters
Protein mass profiles were obtained using a Microflex LT MALDI-TOF Mass Spectrometer (Bruker Daltonics, Germany), with detection in the linear positive-ion mode at a laser frequency of 50 Hz within a mass range of 2–20 kDa. The acceleration voltage was 20 kV, and the extraction delay time was 200 ns. Each spectrum corresponds to ions obtained from 240 laser shots performed in six regions of the same spot and automatically acquired using the AutoXecute of the Flex Control v.2.4 software (Bruker Daltonics). The average spectrum profiles obtained were visualized with Flex analysis v.3.3 software and exported to ClinProTools software v.2.2 and MALDI-Biotyper v.3.0. (Bruker Daltonics, Germany) for data processing (smoothing, baseline subtraction, and peak picking) as previously described [14].
MALDI-TOF analysis and database creation
Spectrum quality was validated by assessing its general intensity, the smoothness of the peaks, the flatness of baseline and its reproducibility compared to other spectra of the same categories. Poor quality spectra were excluded from the analysis. Spectra reproducibility of CTs from An. stephensi specimens molecularly defined as P. berghei-infected or non-infected was evaluated by comparing the average spectra of each sample within its respective group using ClinProTools 2.2 and Flex analysis v.3.3 software (Bruker Daltonics, Germany). Reference spectra (MSP, Main Spectrum Profile) were generated by the automated function of the MALDI-Biotyper software v3.0 (Bruker Daltonics, Germany) by combining the results of the spectra of at least two specimens per condition in order to create a database. MSP were produced based on an unbiased algorithm, taking into consideration the peak position, intensity and frequency. Spectra from thirty control mosquitoes were selected based on their reproducibility and intensity for all further MS analyses. Random spectra of good intensity and good reproducibility of specimens from each batch (infected/non-infected) were loaded in MALDI-Biotyper 3.0 software to create a reference spectra database. Twelve spectra from control An. stephensi were added to the custom database, as well as six spectra of infected specimens and two spectra of exposed but PCR-negative specimens.
MALDI-TOF MS biomarker mass set
To determine the CT differential peaks between An. stephensi, infected or not with P. berghei, the MS spectra from both groups were loaded into ClinProTools 2.2 software. The software was used to generate a peak list for each group in the 2–20 kDa mass range and to identify discriminating peaks. The parameter settings in ClinProTools 2.2 software for spectra preparation were as follows: a resolution of 800 ppm; a noise threshold of 3.00 (intensity arbitrary unit); a maximum peak shift of 800 ppm and a match to calibration peaks of 20%. For the peak calculation, peak peaking was performed on single spectra with a signal-to-noise threshold of 3.00 and an aggregation of 800 ppm. The spectra were then analysed with the genetic algorithm (GA) model, which displayed a list of discriminating peaks [6]. Manual inspection and validation of the selected peaks by the operator gave a “recognition capability” (RC) value together with the highest “cross-validation” (CV) value. These values reflect the ability to distinguish the different spectra groups based on the chosen discriminatory peaks. Genetic Algorithm settings are optimized to provide the highest RC and CV values. All reference spectra from infected and non-infected An. stephensi were imported into ClinProTools software, in order to detect the presence of discriminating peaks between the spectra of infected and non-infected CTs.
Blind tests
The remaining CT spectra of each group after exclusion of those entered in the database were submitted to a blind test analysis. This blind test was performed against our in-lab database, composed of the present CT spectra from P. berghei-infected and non-infected An. stephensi, and also spectra from other arthropod species listed in Table 2 [12, 15–18]. The identification level of significance was determined using the log score values (LSVs) given by the MALDI-Biotyper software v.3.3., corresponding to a matched degree of signal intensities of mass spectra of the query and the reference spectra. LSVs ranged from 0 to 3. LSVs allow good evaluation of the reproducibility between a queried spectrum and a reference spectrum as it is the result of thorough comparison of peaks position and intensity between those two spectra. A LSV was obtained for each spectrum of the samples tested blindly.
Table 2.
List of the arthropod species present in our in-lab MALDI-TOF MS database
| Mosquitoes | Anopheles gambiae, A. coluzzi, An. funestus, An. ziemanni, An. arabiensis, An. wellcomei, An. rufipes, An. pharoensis, An. coustani, An. claviger, An. hyrcanus, An. maculipennis, Culex quinquefasciatus, Cx. pipiens, Cx. modestus, Cx. insignis, Cx. neavei, Aedes albopictus, Ae. excrucians, Ae. vexans, Ae. rusticus, Ae. dufouri, Ae. cinereus, Ae. fowleri, Ae. aegypti, Ae. caspius, Mansonia uniformis, Orthopodomyia reunionensis, Coquillettidia richiardii and Lutzia tigripes |
| Ticks | Amblyomma. variegatum infected or not by Rickettsia africae, Rh. sanguineus, Rh. bursa, Rh. sulcatus, Hyalomma m. rufipes, I. ricinus, I. hexagonus, D. marginatus infected or not by R. slovaca, D. reticulates, Haemaphysalis concinna |
| In ethanol: Am. cohaerens, Am. gemma, Am. variegatum, Ha. leachi, Hy. m.rufipes. Hy. truncatum, Hy. detritum, Rh. decoloratus, Rh. bergeoni, Rh e. evertsi, Rh. praetextatus, Rh. pulchellus, Rh. sanguineus, Rh. microplus, Rh. annulatus, Rh. turanicus, Rh.bursa | |
| Triatomines | Triatoma infestans, Rhodnius prolixus, Rh. pictipes, Rh. robustus, Eratyrus mucronatus, Panstrongylus geniculatus |
| Bedbugs | Cimex lectularius |
| Fleas | Ctenocephalides felis, Ct. canis, Archaeopsylla erinacei, Xenopsylla cheopis and Stenoponia tripectinata |
| Mites | Leptotrombidium chiangraiensis, L. imphalum, L. deliense |
| Sandflies | Phlebotomus papatasi, P. (Larrousius) longicuspis), P. (Larrousius) perfiliewi, P. (Larrousius) perniciosus, P. (Paraphlebotomus) sergenti, Sergentomyia minuta |
| Lice | Damalinia bovis, D. caprae, D. ovis, Haematopinus eurysternus, Linognatus vituli, L. africanus, Pediculus humanus |
Mosquito, tick, triatomine, bedbug reference spectra were obtained from legs protein extracts. Flea reference spectra were obtained from the whole body without abdomen protein extracts. Sandfly reference spectra were obtained from thorax, wings and legs protein extracts. Louse reference spectra were obtained from half of the body protein extracts. These species include field specimens or from insectary breeding, but also specimens collected from patients
Results
Assessment of Plasmodium berghei mouse infection and Anopheles feeding
All six mice were successfully infected with P. berghei-parasitized red blood cells as demonstrated by parasitaemia on follow-up and loss of body weight. While the blood smears performed at day 3 post-infection did not reveal any parasitized red blood cells, parasitaemia was detected in all mice and estimated at approximately 1.5% (mean + SD:1.5 ± 0.35) on day 4 after P. berghei inoculation (90 h post-infection). Approximately 5% (mean + SD: 5.08 ± 1.4) of the parasites detected were in gametocyte form (Fig. 1). This time point was selected for immediate mosquito engorgement, because a rapid degradation of gametocytes was noticed.
Fig. 1.
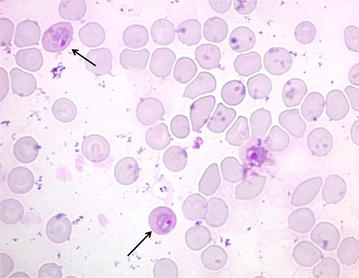
Giemsa-stained mouse tail-blood smear performed 4 days post-infection showing gametocytes. Gametocyte-infected cells are indicated by black arrows
One hundred and seventy female Anopheles mosquitoes were then allowed to engorge on the Plasmodium-infected anesthetized mice, while 145 mosquitoes fed on 4 healthy mice. After 1 h of feeding, 156 mosquitoes were engorged from the infected mice and 111 were engorged from parasite-free mice.
Molecular analysis
Eighteen days after the infective blood meal, 50 control Anopheles (fed on healthy mice) and 60 Plasmodium-exposed mosquitoes were collected. DNAs were extracted from the crushed CTs, and 20 of 60 parasite-exposed mosquito DNAs were positive in real-time PCR targeting of a region of the 18S gene of the Plasmodium species with cycle threshold (Ct) values ranging from 24.99 to 34.68. All control mosquitoes tested were negative for Plasmodium detection.
All mosquitoes tested by COI-standard PCR for validation of DNA extraction were positive.
MALDI-TOF analysis
To appraise water-crushing reproducibility, CT MS spectra of non-infected An. stephensi were submitted to MALDI-TOF MS analysis. Afterwards, the comparison of the MS spectra of infected and non-infected An. stephensi with Flex Analysis and ClinProTools software indicated correct reproducibility of the protein profiles of CTs from mosquitoes of the same group (Fig. 2). Nevertheless, mild variations were still observed between spectra of the same group. For this reason, spectra selection for the database was supported by the creation of a dendrogram. Dendrograms are based on the results of Composite Correlation Index matrix (CCI). CCIs are calculated by dividing spectra into intervals and comparing these intervals across a data set. The composition of correlations of all intervals provides the CCI, which is used a parameter that defines the distance between spectra. A CCI match value of 1 represents complete correlation, whereas a CCI match value of 0 represents an absence of correlation. Spectra chosen for the database upgrading were from different clusters, therefore representing the whole group, despite its relative heterogeneity.
Fig. 2.

Representative MS spectra of non-infected and Plasmodium berghei-infected Anopheles stephensi cephalothoraxes using Flex analysis 3.3 software. a.u., arbitrary units; m/z, mass-to-charge ratio. Vertical bar highlights of distinct spectral regions between infected and uninfected mosquitoes
Visual comparison of MS profiles between the two groups revealed a few specific distinct peaks. Precise determination of discriminatory peaks between the two categories was then assessed using the ClinProTools software. In order to distinguish mosquitoes according to their infection status, 20 spectra from P. berghei-infected mosquitoes were compared to 30 spectra from non-infected mosquitoes using the Genetic Algorithm (GA) tool. After verification of the peak report in the average spectrum, 13 discriminatory masses were determined (Table 3; Fig. 3). The selected peaks of the Genetic Algorithm displayed recognition capability and cross validation values of 100 and 98.72%, respectively.
Table 3.
Comparison of MALDI-TOF profiles of uninfected and P. berghei-infected An. stephensi mosquitoes and determination of distinguishing peak masses
| Mass m/z (Da) | An. stephensi | An. stephensi infected with Plasmodium berghei ANKA |
|---|---|---|
| 3368 | No | Yes |
| 3441 | No | Yes |
| 4430 | Yes | No |
| 5084 | Yes | No |
| 5187 | Yes | No |
| 5384 | No | Yes |
| 5644 | No | Yes |
| 5803 | No | Yes |
| 6351 | Yes | No |
| 6790 | Yes | No |
| 7521 | No | Yes |
| 8448 | Yes | No |
| 12,264 | No | Yes |
Determination of the CT differential peaks between An. stephensi infected or not with P. berghei obtained with the Genetic Algorithm tool of ClinProTools 2.2 software. All mosquitoes are from the same laboratory rearing
Fig. 3.
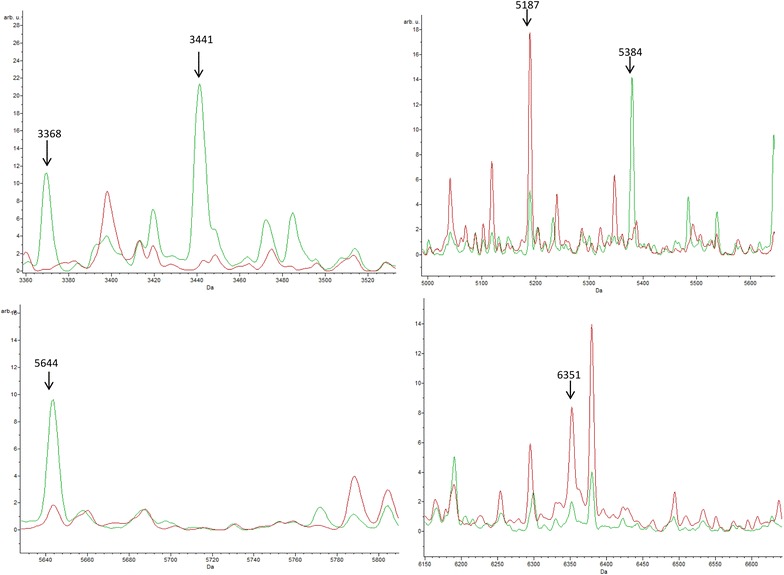
Comparison of MALDI-TOF MS profiles of mosquitoes infected or not by Plasmodium berghei using ClinProTools software. Red and green profiles correspond to average spectrum of control and infected mosquitoes respectively. Peak masses (Da) are indicated with an arrow above the referred peak. arb.u., arbitrary units; m/z, mass-to-charge ratio
The upgraded MS reference database containing the 20 new spectra added in this study was queried blindly, resulting in 100% correct species-level identification and 98.75% correct identification regarding the infection status for all tested samples, with LSVs ranging from 1.347 to 2.908. A total of 13/14 specimens considered as infected based on molecular analysis matched with reference spectra of P. berghei-infected An. stephensi. All control specimens were identified as non-infected by MALDI TOF MS. Also, mosquitoes exposed to parasitized mice but Plasmodium PCR-negative were identified as non-infected by MALDI TOF MS. Indeed, 100% of these spectra matched with the reference spectra of non-infected An. stephensi (Table 4).
Table 4.
Comparison of molecular biology and mass spectrometry performances for the detection of Plasmodium berghei in An. stephensi mosquitoes
| Controls | PCR+ | PCR- | TOTAL | |
|---|---|---|---|---|
| Total number of spectra | 42 | 20 | 38 | 100 |
| Number of spectra introduced in DB | 12 | 6 | 2 | 20 |
| Number of spectra tested blinded against DB | 30 | 14 | 36 | 80 |
| Percentage of MALDI-TOF consistency with PCR | 30/30 (100%) | 13/14 (92.8%) | 36/36 (100%) | 79/80 (98.75%) |
Results of MALDI-TOF blinded tests were based on the highest log score values and obtained with the MALDI BioTyper software. Controls: Anopheles fed on non-infected mice, PCR + : mosquitoes fed on infected mice and for which P. berghei-PCR was positive, PCR − : mosquitoes fed on infected mice and for which P. berghei-PCR was negative. DB database
Discussion
The present work shows preliminary evidence that MALDI-TOF technology can be used for Plasmodium detection. Anopheles stephensi and Plasmodium berghei ANKA is one of the most frequently used pairs for experimental models involving this murine parasite [26].
The blood smears performed on day 4 post-infection showed a low parasitaemia, as the average of the six blood smears was approximately 1% of infected red blood cells. However, approximately 5% of the parasites were in gametocyte form, some of them starting to deteriorate, highlighting the very short time window during which mosquitoes could be fed on the mice. Published literature suggests that peak gametocytaemias for P. berghei ANKA takes place around day 4 post-infection [27], with possible variations depending on the mouse strain and environmental parameters. However, the gametocytaemias described were always much lower than those observed in this work [19].
Plasmodium detection was performed on samples collected 18 days after the infective blood meal. The 18S gene-qPCR positivity cut-off was chosen according to the Ct value of the positive control, which was formerly validated by microscopy. The blood used as positive control was previously checked by microscopy for the presence of parasites in fresh blood and Giemsa-stained blood smears. As this cut-off was established at 35.6 Ct value empirically, positivity of samples with Ct values slightly superior to 35 could not be excluded with certainty. However, for the proof of concept of this work, we decided to use mosquitoes for which the infection status was certain. Mosquito sampling was done 18 days after the infective blood meal to ensure that the parasite had already undergone several multiplications and had completed its migration towards the salivary glands [29].
The in-lab MALDI TOF mosquito database was initially built using mosquito legs crushed manually with pestles [12]. However, for determination of infection status in this study, analyses were performed on CTs, where parasites are located when the mosquito is infected [5]. Moreover, standardization of the protocol required elimination of the manual sample preparation method and development of an automated crushing technique. The TissueLyser crushing device is traditionally used for mechanical cell disruption [20]. It was recently adapted for the standardization of MALDI-TOF preparation of arthropod samples, including An. gambiae and An. stephensi CTs (unpublished). This automated crushing technique provides reproducible MS spectra for An. stephensi CTs.
Molecular biology using a whole mosquito is a standard and validated technique for the detection of Plasmodium parasites [21]. Mosquitoes were considered as infected when parasites were detected in the CT by qPCR with Ct values inferior to 35, and DNA extractions were thoroughly validated by detection of the mosquito COI gene by standard PCR. Because PCR and mass spectrometry had to be performed on the same mosquito compartment, protein extraction was performed in high performance liquid chromatography-grade water instead of the standard solution of formic acid and acetonitrile, responsible for cell wall disruption and protein extraction, respectively [22]. Sample crushing in water already showed MALDI-TOF results comparable to crushing in formic acid in previous entomological studies [23]. However, the absence of acid impairs protein extraction and could explain the mild spectra variability that was observed within the same group, but also the poor quality of the spectra that were excluded from this study. Comparison of spectra from CTs crushed in water with acid/acetonitrile-derived spectra revealed high similarity of the profiles, with much higher quality and reproducibility for the samples crushed in acid solution. (unpublished). Here, spectra used to upgrade the database were thoroughly selected to be representative of the whole group, despite its variability. The single misidentification observed in this study could be attributable to the water crushing and its resulting variability. Comparison between the average profiles of the infected mosquitoes and non-infected mosquitoes revealed several peaks distinguishing the two categories. These modifications in the mosquito’s global proteome could be attributable to additional proteins linked to the parasite presence but also to the mosquito’s immune response to infection. Indeed, while it was admitted for a long time that infections are neutral for the arthropod vector, it has been very well described that Plasmodium infection triggers a significant immune response in Anopheles mosquitoes. During this response, several genes coding for proteins involved in innate immunity are upregulated, but some of them are also downregulated, which could explain why some peaks are disappearing on the average profile of infected mosquitoes [24, 25].
Besides the two main limitations of MALDI-TOF technology, which are the cost of the device and the comprehensiveness of the database, this high-throughput technique allows analysis of a large number of samples at limited cost compared to current methods.
This technique is already established in Africa, since bacteriology laboratories in Dakar, Senegal are now equipped with MALDI-TOF devices for clinical microbiology purposes. Productivity has consequently been increased there, and it has contributed to an intense development of these laboratories in the field of microbiology. With a running cost lower than $1 per sample, this technology remains less expensive than traditional methods, despite the high initial cost of the devices [26, 27]. Also, as previously mentioned by Yssouf et al. [23], the spectra files are available on request and transferable to any Bruker MALDI-TOF device, thus excluding any additional cost associated with the construction of this database (e.g., molecular biology). Since the application of MS to entomological studies does not lead to additional cost, the protocol described here remains entirely affordable. In fact, entomological applications have already been developed in Dakar, since MS identification of Culicoides was finalized there [28]. This preliminary work remains to be validated with field samples. Adjustments and upgrading of our database will be necessary in order to include other reference spectra, such as Anopheles gambiae infected or not by P. falciparum. It would be interesting to assess the consistency of MS spectra from diverse mosquito populations, and also the performance of the identification associated with plasmodial detection for species belonging to the same complex. The detection of the same plasmodial infection in different mosquito species was not evaluated here, nor the detection of different Plasmodium species in the same Anopheles species. It would be interesting to do so in further studies, along with an assessment of the ability of MALDI-TOF to identify Plasmodium mixed infections. If proved relevant, this technique will be a rapid and convenient tool for the monitoring of the mosquito population in these Plasmodium-endemic areas, which is crucial for determination of at-risk populations and for subsequent care. This tool will allow analysis of large numbers of mosquito specimens at limited cost, quickly providing all the necessary information, such as mosquito identification and Plasmodium infection status.
Conclusion
Matrix-Assisted Laser Desorption Ionization Time-Of-Flight appears to be a promising tool for the detection of Plasmodium spp. in mosquitoes, in addition to its ability to identify mosquitoes at the species level. This approach does not require knowledge of entomology or molecular biology once the database is comprehensive. However, any upgrade of the database will always require precise identification, initially provided by morphology and molecular biology. This high-throughput, fast and low-cost method makes it an appropriate technique for epidemiological studies and monitoring of Anopheles populations in Plasmodium-endemic areas, with the concomitant identification of mosquito species and their pathogens. The rapid identification of at-risk populations can provide a valuable advantage for vector control, which may include targeted installations of collective and individual protection against mosquitoes.
Authors’ contributions
PP, AV and ML designed and developed the experiments. ML, EP, AV and YB performed the experiments. ML, AV, LA and PP analysed the data. ML and PP wrote the paper. All authors read and approved the final manuscript.
Acknowledgements
We thank George Grau (Bosch Institute, Sydney) for providing the parasite strain and the Pasteur Institute of Paris, France for providing the strain of An. stephensi. We also thank Marielle Bedotto-Buffet for designing the 18S gene primers and probe used in the present study for the molecular detection of Plasmodium parasites, and Constentin Dieme and Adama Diarra for their help with the mosquito rearing.
Competing interests
The authors declare that they have no competing interests.
Ethics approval
Animal studies were conducted in agreement with the directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 and with the decree No. 2013-118 of 1 February 2013 from the French Government, and were approved by the local Ethics Committee (Comité d’Ethique en Expérimentation Animale Marseille 14) of Marseille, France.
Funding
This work has been carried out thanks to the support of the A*MIDEX project (No. ANR-11-IDEX-0001-02) funded by the Investissements d’Avenir French Government program, managed by the French National Research Agency (ANR).
Contributor Information
Maureen Laroche, Email: maureen.laroche972@gmail.com.
Lionel Almeras, Email: almeras.lionel@gmail.com.
Emilie Pecchi, Email: emilie.pecchi@univ-amu.com.
Yassina Bechah, Email: yassina.bechah@univ-amu.fr.
Didier Raoult, Email: didier.raoult@gmail.com.
Angèle Viola, Email: angele.viola@univ-amu.fr.
Philippe Parola, Phone: + 33 (0) 4 91 32 43 75, Email: philippe.parola@univ-amu.fr.
References
- 1.White NJ, Pukrittayakamee S, Hien TT, Faiz MA, Mokuolu OA, Dondorp AM. Malaria. Lancet. 2014;383:723–735. doi: 10.1016/S0140-6736(13)60024-0. [DOI] [PubMed] [Google Scholar]
- 2.WHO: World malaria report 2014. Geneva: World Health Organization; 2015.
- 3.Rajavel AR, Kumar NP, Natarajan R, Vanamail P, Rathinakumar A, Jambulingam P. Morphological and molecular characterization of the ecological, biological and behavioural variants of the JE vector Culex tritaeniorhynchus: an assessment of its taxonomic status. J Vector Borne Dis. 2015;52:40–51. [PubMed] [Google Scholar]
- 4.Yssouf A, Almeras L, Raoult D, Parola P. Emerging tools for identification of arthropod vectors. Future Microbiol. 2016;11:549–566. doi: 10.2217/fmb.16.5. [DOI] [PubMed] [Google Scholar]
- 5.Bassene H, Kengne P, Ndiath MO, Sokhna C, Dupressoir T, Fontenille D, et al. Comparison of PCR, ELISA-CSP and direct microscopic observation methods for the detection of Plasmodium falciparum sporozoites in Anopheles gambiae M in Senegal. Bull Soc Pathol Exot. 2009;102:233–237. [PubMed] [Google Scholar]
- 6.Wirtz RA, Burkot TR. Detection of malarial parasites in mosquitoes. Adv Dis Vector Res. 1991;8:77–106. doi: 10.1007/978-1-4612-3110-3_4. [DOI] [Google Scholar]
- 7.Durnez L, Van Bortel W, Denis L, Roelants P, Veracx A, Trung HD, et al. False positive circumsporozoite protein ELISA: a challenge for the estimation of the entomological inoculation rate of malaria and for vector incrimination. Malar J. 2011;10:195. doi: 10.1186/1475-2875-10-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yssouf A, Almeras L, Terras J, Socolovschi C, Raoult D, Parola P. Detection of Rickettsia spp in ticks by MALDI-TOF MS. PLoS Negl Trop Dis. 2015;9:e0003473. doi: 10.1371/journal.pntd.0003473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yssouf A, Almeras L, Berenger J-M, Laroche M, Raoult D, Parola P. Identification of tick species and disseminate pathogen using hemolymph by MALDI-TOF MS. Ticks Tick Borne Dis. 2015;6:579–586. doi: 10.1016/j.ttbdis.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 10.Grau GE, Piguet PF, Engers HD, Louis JA, Vassalli P, Lambert PH. L3T4 + T lymphocytes play a major role in the pathogenesis of murine cerebral malaria. J Immunol. 1986;137:2348–2354. [PubMed] [Google Scholar]
- 11.Dieme C, Bechah Y, Socolovschi C, Audoly G, Berenger J-M, Faye O, et al. Transmission potential of Rickettsia felis infection by Anopheles gambiae mosquitoes. Proc Natl Acad Sci USA. 2015;112:8088–8093. doi: 10.1073/pnas.1413835112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yssouf A, Socolovschi C, Flaudrops C, Ndiath MO, Sougoufara S, Dehecq J-S, et al. Matrix-assisted laser desorption ionization–time of flight mass spectrometry: an emerging tool for the rapid identification of mosquito vectors. PLoS ONE. 2013;8:e72380. doi: 10.1371/journal.pone.0072380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Marine Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 14.Dieme C, Yssouf A, Vega-Rúa A, Berenger JM. Accurate identification of Culicidae at aquatic developmental stages by MALDI-TOF MS profiling. Parasit Vectors. 2014;7:544. doi: 10.1186/s13071-014-0544-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yssouf A, Parola P, Lindström A, Lilja T, L’Ambert G, Bondesson U, et al. Identification of European mosquito species by MALDI-TOF MS. Parasitol Res. 2014;113:2375–2378. doi: 10.1007/s00436-014-3876-y. [DOI] [PubMed] [Google Scholar]
- 16.Yssouf A, Socolovschi C, Leulmi H, Kernif T, Bitam I, Audoly G, et al. Identification of flea species using MALDI-TOF/MS. Comp Immunol Microbiol Infect Dis. 2014;37:153–157. doi: 10.1016/j.cimid.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Yssouf A, Flaudrops C, Drali R, Kernif T, Socolovschi C, Berenger J-M, et al. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for rapid identification of tick vectors. J Clin Microbiol. 2013;51:522–528. doi: 10.1128/JCM.02665-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lafri I, Almeras L, Bitam I, Caputo A, Yssouf A, Forestier C-L, et al. Identification of algerian field-caught phlebotomine sand fly vectors by MALDI-TOF MS. PLoS Negl Trop Dis. 2016;10:e0004351. doi: 10.1371/journal.pntd.0004351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shinzawa N, Ishino T, Tachibana M, Tsuboi T, Torii M. Phenotypic dissection of a Plasmodium-refractory strain of malaria vector Anopheles stephensi: the reduced susceptibility to P. berghei and P. yoelii. PLoS ONE. 2013;8:e63753. doi: 10.1371/journal.pone.0063753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith B, Li N, Andersen AS, Slotved HC, Krogfelt KA. Optimising bacterial DNA extraction from faecal samples: comparison of three methods. Open Microbiol J. 2011;5:14–17. doi: 10.2174/1874285801105010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bass C, Nikou D, Blagborough AM, Vontas J, Sinden RE, Williamson MS, et al. PCR-based detection of Plasmodium in Anopheles mosquitoes: a comparison of a new high-throughput assay with existing methods. Malar J. 2008;7:177. doi: 10.1186/1475-2875-7-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sauer S, Freiwald A, Maier T, Kube M, Reinhardt R, Kostrzewa M, et al. Classification and identification of bacteria by mass spectrometry and computational analysis. PLoS ONE. 2008;3:e2843. doi: 10.1371/journal.pone.0002843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaufmann C, Ziegler D, Schaffner F, Carpenter S, Pflüger V, Mathis A. Evaluation of matrix-assisted laser desorption/ionization time of flight mass spectrometry for characterization of Culicoides nubeculosus biting midges. Med Vet Entomol. 2011;25:32–38. doi: 10.1111/j.1365-2915.2010.00927.x. [DOI] [PubMed] [Google Scholar]
- 24.Dimopoulos G, Müller HM, Levashina EA, Kafatos FC. Innate immune defense against malaria infection in the mosquito. Curr Opin Immunol. 2001;13:79–88. doi: 10.1016/S0952-7915(00)00186-2. [DOI] [PubMed] [Google Scholar]
- 25.Dimopoulos G, Christophides GK, Meister S, Schultz J, White KP, Barillas-Mury C, et al. Genome expression analysis of Anopheles gambiae: responses to injury, bacterial challenge, and malaria infection. Proc Natl Acad Sci USA. 2002;99:8814–8819. doi: 10.1073/pnas.092274999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan KE, Ellis BC, Lee R, Stamper PD, Zhang SX, Carroll KC. Prospective evaluation of a matrix-assisted laser desorption ionization-time of flight mass spectrometry system in a hospital clinical microbiology laboratory for identification of bacteria and yeasts: a bench-by-bench study for assessing the impact on time to identification and cost-effectiveness. J Clin Microbiol. 2012;50:3301–3308. doi: 10.1128/JCM.01405-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lo CI, Fall B, Sambe-Ba B, Diawara S, Gueye MW, Mediannikov O, et al. MALDI-TOF mass spectrometry: a powerful tool for clinical microbiology at Hôpital Principal de Dakar, Senegal (West Africa) PLoS ONE. 2015;10:e0145889. doi: 10.1371/journal.pone.0145889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambou M, Aubadie-Ladrix M, Fenollar F, Fall B, Bassene H, Almeras L, et al. Comparison of matrix-assisted laser desorption ionization–time of flight mass spectrometry and molecular biology techniques for identification of Culicoides (Diptera: Ceratopogonidae) biting midges in Senegal. J Clin Microbiol. 2015;53:410–418. doi: 10.1128/JCM.01855-14. [DOI] [PMC free article] [PubMed] [Google Scholar]


