Abstract
Recent discoveries indicate that the foamy virus (FV) (Spumavirus) ancestor may have been among the first retroviruses to appear during the evolution of vertebrates, demonstrated by foamy endogenous retroviruses present within deeply divergent hosts including mammals, coelacanth, and ray-finned fish. If they indeed existed in ancient marine environments hundreds of millions of years ago, significant undiscovered diversity of foamy-like endogenous retroviruses might be present in fish genomes. By screening published genomes and by applying PCR-based assays of preserved tissues, we discovered 23 novel foamy-like elements in teleost hosts. These viruses form a robust, reciprocally monophyletic sister clade with sarcopterygian host FV, with class III mammal endogenous retroviruses being the sister group to both clades. Some of these foamy-like retroviruses have larger genomes than any known retrovirus, exogenous or endogenous, due to unusually long gag-like genes and numerous accessory genes. The presence of genetic features conserved between mammalian FV and these novel retroviruses attests to a foamy-like replication biology conserved for hundreds of millions of years. We estimate that some of these viruses integrated recently into host genomes; exogenous forms of these viruses may still circulate.
Keywords: paleovirology, endogenous retrovirus, foamy virus, fish, phylogeny
1. Introduction
Foamy viruses (FV) constitute one of seven extant genera of Retroviridae (Linial 1999). The lone genus of the Spumaretrovirinae subfamily, FVs are complex retroviruses distinguished from Orthoretrovirinae primarily because of disparate replication strategies (Rethwilm 2010). FV infection is persistent and nonpathogenic in vivo, but causes characteristic fluid-filled syncytial vacuoles in vitro with a “foamy” appearance (Meiering and Linial 2001; Delelis et al. 2004).
Known infectious FVs are thus far confined to mammalian hosts (Katzourakis et al. 2014), including felines (Winkler et al. 1997), horses (Tobaly-Tapiero et al. 2000), cows (Renshaw and Casey 1994), bats (Wu et al. 2012), and wide variety of primates (Kupiec et al. 1991; Renne et al. 1992; Herchenröder et al. 1994; Bieniasz et al. 1995; Thümer et al. 2007; Pacheco et al. 2010) harboring species-specific FVs. Among primate FVs, host-virus coevolution appears to have been the norm for the last 30 million years (Cong et al. 2005). Despite attempts to link a cryptic FV infection to common human diseases (Meiering and Linial 2001), a circulating human-specific FV has never been described, though simian foamy virus (SFV) zoonoses do occur (Switzer et al. 2004; Betsem et al. 2011).
FVs, in common with all retroviruses (RV), integrate reverse-transcribed dsDNA into the genome of an infected cell. The integrated virus then exploits the host’s cellular machinery to generate proteins for virion assembly and egress (Coffin et al. 1997). Endogenous retroviruses (ERV) are the result of integration into germline cells and subsequent parent-offspring vertical transmission as genomic DNA (Weiss 2006). After endogenization, ERVs evolve at the host neutral evolutionary rate and can remain detectable tens of millions of years after integration as viral “fossils” (Patel et al. 2011). Newly discovered ERVs are traditionally classified by phylogenetic proximity to exoge nous RVs; class I ERVs are related to Gammaretrovirus isolates, class II are most related to Betaretrovirus, and class III to Spumaretrovirus, or FV. ERVs may represent extinct RVs that no longer exist in exogenous form. For example, though human-specific FVs are unknown, the human genome is rife with the relics of ancestral infection by FV-like class III elements (Bénit et al. 1999), some of which are widely retained among other primates (Greenwood et al. 2005).
The discovery of endogenous foamy viruses (EFV) in the genomes of the two-toed sloth (Katzourakis et al. 2009) and Cape golden mole (Han and Worobey 2014; Katzourakis et al. 2014), and an exogenous galago FV (Katzourakis et al. 2014), provide evidence for at least 100 million years of host-virus coevolution for Spumaretrovirinae. A coelacanth EFV suggests the possibility of a marine FV origin 400 million years ago (Ma) (Han and Worobey 2012). The presence of foamy-like ERVs in zebrafish (Llorens et al. 2009), platyfish, and cod (Schartl et al. 2013) lend credence to this hypothesis, though the possibility remains that these elements represent distinct and possibly extinct lineages that branched very early in the evolution of animal retroviruses (Katzourakis et al. 2014) . Whether these fish ERVs represent “true” EFVs or early diverging foamy-like class III retroelements, their existence is congruent with other lines of evidence that FVs are the most ancient genus of extant RVs. Uniquely nonexistent pathogenicity in living hosts has been cited as possible evidence for ancient origins (Linial 2000), along with a replication strategy that resembles orthoretroviruses in some respects, but pararetroviruses in others (Rethwilm and Bodem 2013). Whether FVs are truly ancient or a highly successful RV lineage with a very wide host range, we might expect a significant undiscovered diversity of foamy-like ERVs in host clades that diverged early in the evolution of vertebrates. Ray-finned fishes are the most species-rich clade of living vertebrates, with at least 26,000 taxa (Helfman et al. 2009), and represent an inadequately explored host lineage. We report here the discovery of 23 novel FV-like elements in teleost genomes by database and PCR screens.
2. Materials and Methods
2.1. Genome screening
We queried all sequences all actinopterygian species available in GB whole genome shotgun (WGS) databases using the tblastn algorithm and a set of exogenous and endogenous FV Pol sequences (prototype foamy virus, CAA68999.1; equine foamy virus, AF201902.1; feline foamy virus, CAA70075.1; coelacanth EFV —Han and Worobey 2012; sloth EFV— Katzourakis et al. 2009). We recursively screened each species, querying top hits with blastx against the GB non-redundant protein database. Contigs matching best with FVs were screened for open reading frames (ORFs) with UGENE (Okonechnikov et al. 2012). Pol coding DNA sequences (cds) ∼3,000 nt in length with N-terminal Pro D(T/S)GA, and downstream RT YXDD(I/V) and Int DDX35E motifs were favored. Conserved motifs were annotated with CD-search (Marchler-Bauer and Bryant 2004). Pol cds detected in multiple reading frames were conservatively corrected by adding a one or two position gap where the reading frames met.
WGS sequence contigs that grouped with exogenous FVs in a phylogenetic context were investigated further. We screened each species WGS database with a conspecific contig as a blastn query to identify all contigs with >2,000 kb of pol coding sequence at >70 percent nt identity, lower than the common arbitrary cut-off for ERV families (Jern et al. 2005). All-against-all BLAST searches identified ERVs with intact LTR and paralogs within these families. We used CD-HIT (Huang et al. 2010) and bootstrap neighbor-joining tree searches in ClustalX (Larkin et al. 2007) to redundantly search for orthologous ERVs between closely related species.
2.2. Quasi-consensus construction and annotation
ERVs with secondary integrations or large deletions indicated by alignment gaps were built into a quasi-consensus for annotation purposes; consensus sequences were not used for tests of selection or age estimation. RepeatMasker (Smit et al. 2015) was used to identify and remove probable secondary insertions. Each quasi-consensus is an intact cds (typically Pol) used to anchor an alignment of flanking proviral genomic sequences. For example: Amphilophus citrinellus ERV; the most intact provirus-bearing contig, AcERV-1 (acc: CCOE01001074.1), has a complete putative Pol ORF, but flanking ERV regions contain premature stop codons and small poly-N assembly artifacts. Using the region from the PBS to the Pol start codon, and the Pol stop codon to the 3′ PPT, as blastn queries, we aligned ERV fragments with ≥80 percent query coverage using the ClustalX algorithm (Larkin et al. 2007) in UGENE (Okonechnikov et al. 2012). The AcERV quasi-consensus is a strict majority consensus of these alignments. We repeated this procedure as necessary for ERVs in other hosts.
ORFs were screened against GB the non-redundant protein database using blastx, and against UniProtKB with HMMER (Finn et al. 2015). Identities were called based on conserved functional motifs for pol, position in genome for gag, and presence of TM-helix motifs predicted by TMHMM Server v2.0 (Krogh et al. 2001) for env genes. Protease cleavage sites were predicted with ProP 1.0 Server (Duckert et al. 2004). ERVs were scanned for conserved FV-like features with a custom Perl script or the regular expression search feature in UGENE (Okonechnikov et al. 2012). Accession numbers for sequences in this analysis are listed in Supplementary Table 1.
2.3. ERV age estimation
We aligned LTR with the ClustalW (Larkin et al. 2007) algorithm and calculated the Kimura (K80) corrected divergence (Kimura 1980) using MEGA 5.2.2 (Tamura et al. 2011). We applied the formula , where t is time, d is divergence, and µ is the host mutation rate. Host mutation rates were derived from several sources (Fu et al. 2010; Fraser et al. 2015; Kratochwil et al. 2015), and used the Poecilia formosa rate (Fraser et al. 2015) when species lacked a direct estimate of mutation rate. The zebrafish mutation rate was increased to 4.3e−8/site/year. Where ERVs shared a flank with another ERV, we aligned with ClustalW the longest paralogous flanking sequence possible, detected by blastn (2313–4661 positions). Flank-divergence estimates of ERV age were also calculated with the formula , where d is K80 divergence, and µ is the host mutation rate. For DrFV-3, we detected six contigs sharing a 5′ flank. After alignment, we selected Hasegawa–Kishino–Yano as the best-fitting substitution model using jmodelTest 2 (Guindon and Gascuel 2003; Darriba et al. 2012). We conducted a two-million-generation Bayesian MCMC analysis of time to most recent common ancestor (TMRCA) with BEAST v1.8.2 (Drummond et al. 2012), specifying 4.3e−8/site/year as the rate prior, a strict clock, and Yule model of speciation. Outputs were analyzed using Tracer to ensure convergence (Rambaut et al. 2014). Accession numbers for sequences in this analysis are listed in Supplementary Table 1.
2.4. De novo fish ERV sequencing
Using a pol alignment of fish ERVs from WGS contigs and mammal FVs, blocks of conserved positions were identified using a custom Perl script parsing a ClustalX q-score file (Sievers et al. 2011) with an 18-nt sliding window. The RT DNA-binding motif QYP(L/I)N and RT active site motif YIDD(I/V)F are suitable primer binding sites ∼450 nt apart and therefore able to detect fragmented ERVs. We designed a highly degenerate pair of PCR primers: FishFPol472F 5′-CARTAYCSIHTNAA and FishFPol919R 5′-GWRIANRTCRTCDATRTA, where I is inosine. Numbers in the primer name refer to the annealing site position in DrFV-2 pol.
Several institutions gifted ethanol-suspended tissues, which are listed by order and species in Supplementary Table 2. We extracted genomic DNA using a DNeasy Blood & Tissue kit (Qiagen) and quantified it with a Qubit 2.0 fluorometer (Invitrogen). We assayed DNA quality by amplifying a ∼1,500 nt fragment of RAG1 with PCR primers from previous studies (López et al. 2004; Li and Ortí 2007), modifying them when necessary (Supplementary Table 3).
PCR amplification of EFV sequences was performed on 5–20 ng template DNA in a 25-µl reaction mixture containing ThermoPol Buffer at 1X concentration (New England BioLabs), 2.0 mM MgSO4, 150 ng each of forward and reverse primer, 0.2 mM dNTPs, and 1 unit Taq DNA Polymerase (New England BioLabs). Reactions proceeded in a stepdown thermal cycler protocol with the following steps: 95°C—3 m, (95°C—30 s, 64°C—20 s, 68°C—45 s) × 4, (95°C—30 s, 60°C—20 s, 68°C—45 s) × 4, (95°C—30 s, 56°C—20 s, 68°C—45 s) × 4, (95°C—30 s, 52°C—20 s, 68°C—45 s) × 30, 68°C—5 m. PCR products were loaded into a specially prepared PCR-clean 2 percent agarose gel, followed by excision and purification of 400–500 nt gel bands with a Zymoclean Gel DNA Recovery Kit (Zymo Research). Recovered DNA was cloned with a pGEM-T Easy Vector System (Promega) and JM109 competent cells (Promega). Vector inserts were PCR amplified with m13 primers and reverse strands were sequenced with m13R primer at the University of Arizona Genetics Core.
2.5. Phylogenetic analysis
For the phylogeny of representative retroviruses, we retrieved Pol cds (Supplementary Table 4) and aligned them with fish ERVs using ClustalX (Larkin et al. 2007). In-frame stop codons were treated as gaps in the aa sequence. Ambiguously aligned positions were removed with trimAl using the gappyout option (Capella-Gutierrez et al. 2009), producing an alignment of 693 positions. We used MrBayes v3.2.5 for phylogenetic reconstruction, selecting a mixed-model amino acid model prior (Ronquist et al. 2012). MCMC chains ran for five million generations, sampling trees every 100 generations, and discarding the first 25 percent as burn-in. Convergence and adequate estimated sample size of parameters was confirmed using Tracer (Rambaut et al. 2014).
The smaller RV Pol phylogeny was created with a set of representative RVs, and translated sequences from PCR sequencing assay, aligned with ClustalX (Larkin et al. 2007). Ambiguously aligned and gap-ridden positions were eliminated with trimAl (Capella-Gutierrez et al. 2009), producing an alignment with 702 aa positions, including gap positions in short ERV fragments sequenced in this study and gap positions representing in-frame stop codons. Phylogenetic reconstruction was done in MrBayes v3.2.5 (Ronquist et al. 2012), setting a mixed model amino acid prior, running for three million generations, sampling every 100, and discarding the first 25 percent as burn-in.
3. Results
3.1. Foamy-like ERVs in fish genomes
We screened all 52 available actinopterygian WGS databases in GenBank (GB) with exogenous and endogenous FV Pol amino acid (aa) queries. Our search revealed FV-like pol sequences within 17 teleost species genomes (Table 1).
Table 1.
EFV-harboring fish genome contigs
| Order Species | Common name | Contig (acc) | BLASTx match (acc) | % ID | E-value | ERV components |
|---|---|---|---|---|---|---|
| Cypriniformes | ||||||
| D. rerioa | Zebrafish | CAAK05053864.1 | SFVspm (ABV59399) | 29 | 6E−83 | LTR-gag-pol-env-LTR |
| P. promelas | Fathead minnow | JNCD01029789.1 | SFVcpz (AKM21185) | 30 | 5E−87 | tLTR-gag-pol-env-LTR |
| Salmoniformes | ||||||
| O. mykiss | Rainbow trout | CCAF010042932.1 | EFV (NP_054716) | 30 | 5E−36 | pol |
| Gadiformes | ||||||
| G. morhuab | Cod | CAEA01131311.1 | PFV (AAA66556) | 34 | 9E−71 | pol |
| Pleuronectiformes | ||||||
| C. semilaevis | Tongue sole | AGRG01061695.1 | SFVspm (ABV59399) | 33 | 2E−64 | pol |
| Cichliformes | ||||||
| A. citrinellus | Midas cichlid | CCOE01001074.1 | EFV (NP_054716) | 28 | 7E−100 | LTR-gag-pol-env-LTR |
| Cyprinodontiformes | ||||||
| N. furzeri | Turquoise killifish | JNBZ01063262.1 | EFV (NP_054716) | 31 | 5E−69 | pol |
| F. heteroclitus | Mummichog | JXMV01054445.1 | SFVmac (AFA44809) | 33 | 1E−33 | pol |
| X. maculatusb | Platyfish | AGAJ01041163.1 | SFVorg (CAD67562) | 29 | 6E−92 | tLTR-gag-pol-env |
| P. formosa | Guppy | AYCK01027102.1 | SFVmac (AFA44809) | 29 | 2E−92 | LTR-gag-polt-env-LTR |
| P. reticulata | Amazon molly | AZHG01028727.1 | SFVspm (ABV59399) | 29 | 4E−86 | LTR-gag-pol-env-LTR |
| Perciformes | ||||||
| A. fimbria | Sablefish | AWGY01041462 | SFVcpz (AFX98084) | 37 | 4E−58 | pol |
| L. crocea | Yellow croaker | JRPU01012463.1 | SFVspm (ABV59399) | 26 | 3E−81 | LTR-gag-pol-envt |
| P. magnuspinnatus | Korean mudskipper | JACL01052273.1 | SFVspm (ABV59399) | 30 | 4E−74 | pol |
| S. rubrivinctus | Flag rockfish | AUPQ01030678.1 | SFVspm (ABV59399) | 32 | 3E−62 | pol |
| S. nigrocinctus | Tiger rockfish | AUPR01019601.1 | SFVspm (ABV59399) | 31 | 1E−60 | pol |
| Formerly perciformes | ||||||
| S. partitus* | Bicolor damselfish | JMKM01039980.1 | SFVspm (ABV59399) | 33 | 2E−72 | tLTR-gag-polt-env-LTR |
Host species higher taxonomic levels per Betancur-R et al. (2013) , except (*) incertae sedis member of Ovalentaria assigned by aforementioned. Common names per FishBase classification (Harel et al. 2015). (a) Related ERV described in Llorens et al. (2009). (b) Previously described in Schartl et al. (2013). ERV components marked (t) are truncated.
EFV, Equine foamy virus; PFV, Prototype foamy virus; SFVcpz, Simian foamy virus – chimpanzee; SFVorg, Simian foamy virus – orangutan, SFVmac, Simian foamy virus – macaque; SFVspm, Simian foamy virus – spider monkey.
Three of these ERVs have been described previously. The platyfish genome (Xiphophorus maculatus) was reported to contain two near-intact FV-like insertions (Schartl et al. 2013). We recovered one insert encoding nearly a full proviral genome, though it lacks long terminal repeats (LTR), designated XmERV. A second large insert, also lacking LTR, is missing 32 percent its length. These and the fifteen other fragments are highly similar with minimum 80 percent nucleotide (nt) identity. From the zebrafish genome (Danio rerio), in addition to the EFV-like element designated DrFV-1 (Llorens et al. 2009), we recovered two additional full length ERV sharing 96 percent global nucleotide identity along the aligned length excluding the 5′ LTR. We designate these DrFV-2 and DrFV-3 following the naming precedent set by Llorens et al. (2009). The EFV fragment detected in the cod genome (Gadus morhua) contained only the reverse transcriptase (RT) region of pol (Schartl et al. 2013) . However, we detected a second FV-like sequence. A conserved domain search (Marchler-Bauer and Bryant 2004) revealed the presence of RNase H, and integrase (Int) (E-value = 5.67 × 10−17, 6.36 × 10−23, respectively). Visually inspecting the conceptual translations of this sequence revealed a retroviral protease (Pro) motif within a reading frame lacking RT. This same contig also encoded two trans-membrane (TM) motifs among broken reading frames within 3.5 kb downstream, evidencing its origin as an infectious RV. Infectious mammal FVs encode Env with two TM regions that associate the protein with host endoplasmic reticulum membrane before being processed by host proteases for assembly into virions (Rethwilm 2010). Concatenating then translating these sequences produced a 1,069-aa protein with one premature stop codon that shares 66 percent aa identity when aligned with either platyfish ERV Pol sequence.
Among novel fish foamy-like ERVs, eight were characterized by a pol gene alone, as neither canonical gag, env, nor potential ORFs corresponding to FV bel or tas accessory genes were detected among WGS contigs. Rainbow trout (Oncorhynchus mykiss), Korean mudskipper (Periophthalmus magnuspinnatus), tongue sole (Cynoglossus semilaevis), flag rockfish (Sebastes rubrivinctus), tiger rockfish (Sebastes nigrocinctus), and sablefish (Anoplopoma fimbria) genomes each contain a single fragment, none of them comprising a complete ORF, for which we extracted >1,700 nt of sequence homologous to pol. Turquoise killifish (Nothobranchius furzeri) FV-like sequences were also characterized by pol sequence alone, but there are numerous copies of complete FV-like pol genes in this host genome. These killifish pol sequences are highly similar (>80 percent identity), and possibly paralogous. Similarly, the mummichog genome (Fundulus heteroclitus) has four non-functional copies that encode Pol after conservative frameshift corrections, but with premature stop codons. Aside from a TM motif in a fragmented reading frame downstream of Pol in one contig, there are no other discernable ERV features.
Five species retain viral genomic insertions of better quality. The yellow croaker genome (Larimichthys crocea) contains numerous ERV integrations, some with intact LTR. The most intact fragment featured both 5′ and 3′ LTR, a gag-like ORF, pol, and a partial env gene encoding a TM motif (LcERV). The ten pol-containing EFV-like fragments in this genome share >92 percent nt identity with the single intact ERV. The bicolor damselfish genome (Stegastes partitus) contains one LTR-complete ERV (SpERV), and numerous smaller fragments with largely intact gag-pol-env or some derivation, but no LTR. The single intact ERV was degraded with numerous stop codons and frameshift indels.
The Midas cichlid genome (Amphilophus citrinellus) contains four LTR-complete EFV-like elements (AcERV-1 through AcERV-4) in addition to five fragments lacking either the 5′ or 3′ LTR. None of these shared flanking host genomic sequences, thus are likely not the result of segmental duplication. Each shares ∼85 percent global nt identity with other A. citrinellus EFV-like elements.
Guppy (Poecilia reticulata) and Amazon molly (P. formosa) genomes both contain two full ERVs. In guppies, both LTR are truncated in one insert, the other has inverted LTR. In Amazon mollies, one viral genome is complete, but its putative accessory genes are also found upstream of the 5′ LTR. The other complete insert has a truncated 3′ LTR. FV-like ERVs in these species are not orthologous. There are no detectable shared ERV flanking sequences between these species. A clustering analysis of all Poecilia FV-like pol fragments >1,000 nt displayed greater within-species similarity than between-species similarity (%ID mean: P. reticulata, 94.1 percent; P. formosa, 94.0 percent).
The fathead minnow genome (Pimephales promelas), the nearest relative to zebrafish among these results, contains two FV-like insertions. The first, which we designate PpERV-1, lacks LTR but otherwise encodes nonfunctional RV core and accessory genes. PpERV-1 shares 77 percent global identity with several smaller fragments in the genome, which may be the product of a single infection. The second intact ERV is highly divergent, yet remarkably intact, as it retains partial LTRs and encodes nonfunctional RV core genes. Designated PpERV-2, It shares only 40 percent aa identity with PpERV-1. Among fish ERVs in this study, it is the only example of integration by multiple distinct foamy-like viruses.
Finally, the Nile tilapia (Oreochromis niloticus) and the giant mudskipper (Periophthalmodon schlosseri) genomes may contain an FV-related pol fragment, demonstrated by a blastx search with exogenous FVs being the best hits (Nile tilapia, GB accession number (acc): AERX01018483.2, E-value = 4 × 10−36; giant mudskipper, acc: JACM01045479.1, E-value = 2 × 10−19). Both contained numerous indels, with Nile tilapia ERV missing RT motifs. We were unable to assign either ERV to an RV clade with confidence; therefore we excluded these from further analysis and did not include them in the total number of novel fish ERVs.
3.2. Sister Clade to Known FV comprised of fish ERVs
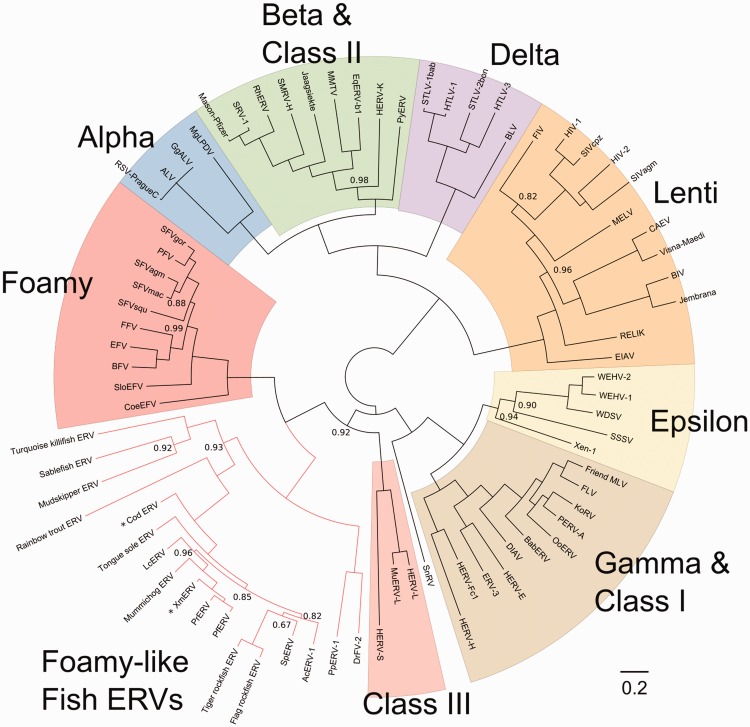
Fish ERV Pol sequences discovered in this study were aligned with a set of exogenous and endogenous retroviruses (Supplementary Table 4), and with previously discovered platyfish, cod, and zebrafish EFVs. A Bayesian phylogeny of 693 aa aligned positions (Fig. 1) shows that Fish FV-like elements group robustly (posterior probability = 1.0) as the sister group to all other FV. Similarly, all major clades of exogenous and endogenous RVs group robustly in branching orders reflecting known relationships. We could not discern certain finer relationships among fish ERVs with precision, indicated by several nodes with low posterior probability. Critically, the novel fish ERV clade shares a closer phylogenetic relationship with mammal FVs than class III ERVs in mammals, including HERV-L and MuERV-L.
Figure 1.
Retrovirus Pol phylogeny. MrBayes consensus tree estimated from 692 aligned Pol protein positions and rooted at its midpoint for illustrative purposes. Branch lengths estimated as expected substitutions per site. Nodes with posterior probability <1.0 are labeled. Seven genera of exogenous and endogenous retroviruses in colored blocks; Snakehead retrovirus, genus yet unclassified, is uncolored; foamy-like teleost endogenous retroviruses represented by salmon-colored branches. Asterisks (*) mark foamy-like fish ERVs discovered in previous studies. Class I ERVs are grouped with gammaretroviruses and class II with betaretroviruses for simplicity. RV taxa and source citations are available in Supplementary Table 4.
On a coarse scale, fish ERVs group into recognizable fish host clades. ERVs from the four Fundulidae (mummichog, platyfish, guppy, and Amazon molly) form a clade that branches in an order matching host species divergence. The two Cypriniformes species ERVs (zebrafish and fathead minnow) are also grouped. Cod and rainbow trout ERVs both branch before the large percomorph ERV crown group in a pattern that resembles the relationships of those hosts.
3.3. Genomic organization
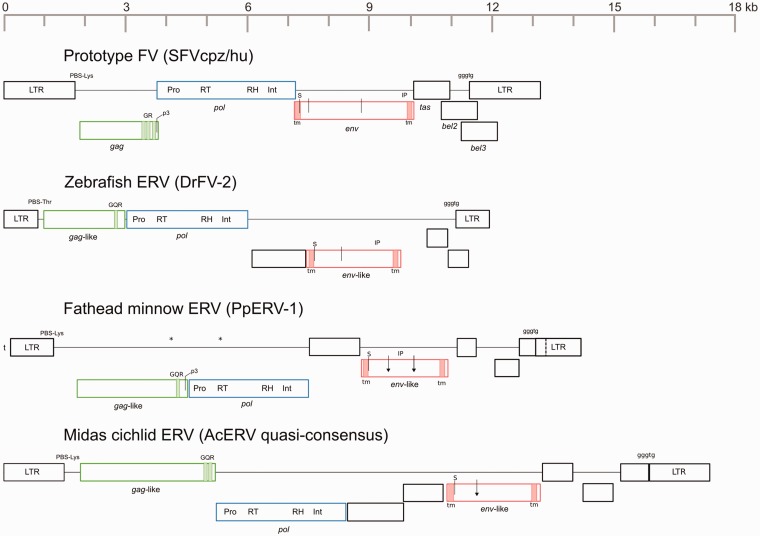
To determine whether the molecular characteristics of these viruses resemble known FV characteristics, we annotated intact ERVs and compared them with the prototype FV (SFVcpz/hu), acc: Y07725.1 (Fig. 2).
Figure 2.
Fish ERV genetic features. SFVcpz/hu (acc: Y07725.1) is the prototype virus for the FV clade. Open boxes represent ORFs. Premature stop codons marked with an asterisk (*). Reading frames represented by vertical orientation. Downward arrow represents furin cleavage site; arrows surmounted by ‘S’ are signal peptidase cleavage sites; p3-cleavage site marked by ‘p3’ arrow; dotted line represents ORF boundary in LTR. PPT:LTR boundary ‘gggtg’ motif marked as such. Hypothetical gag genes are green, pol are blue, and hypothetical env are red. Black boxes are unknown accessory genes unless labeled. Boxes and genome length are to scale (kb). GQR, glycine-glutamine-arginine rich box; GR, glycine-arginine rich box; IP, internal promoter; Int, integrase; LTR, long terminal repeat; PBS, primer binding site; Pro, protease; RH, ribonuclease H; RT, reverse transcriptase; TM, trans-membrane motif.
Zebrafish (DrFV-2, acc: CAAK05053864.1) and fathead minnow (PpERV-1, acc: JNCD01029789.1) ERVs contain ORFs (two premature stop codons noted in PpERV-1, Fig. 2) corresponding to RV core genes and several putative accessory genes. Possible DrFV gag and env genes possess no detectable sequence homology with any other known RV, FV or otherwise, as determined by BLAST searches, CD-HIT (Huang et al. 2010), and a final protein homology screen using HMMER (Finn et al. 2015). The same is true for PpERV-1 and the remaining annotated ERVs.
DrFV-2 and PpERV-1 encode two or three accessory genes downstream from env, much like mammalian FVs universally encode bel1/tas transactivator and bel2/bet regulatory proteins in the same region (Rethwilm 1995; Omoto et al. 2004). Whether the DrFV and PpERV 3′ accessory genes maintain similar functions is unknown. Neither share functional domains with FV counterparts. Both viruses also contain an ORF, 1335 and 1263 nt, respectively, between pol and env coding sequences. Mammal FVs do not possess this, nor do known class III ERVs (Ribet et al. 2008). This feature is also unique among infectious fish retroviruses (Hart et al. 1996; Paul et al. 2006; Rovnak and Quackenbush 2010). The closest comparison to be made is among lentiviruses, which often encode a viral infectivity factor (Vif) or transactivator (Tat) in this region (Tang et al. 1999).
The Midas cichlid ERV, AcERV-1, retains intact LTR and pol. We constructed a quasi-consensus anchored with the pol gene to obviate premature stop codons and frameshifting indels in regions flanking pol. To test whether this consensus resembles the genetic organization of the virus pre-integration, we calculated the pairwise ratio of non-synonymous to synonymous substitutions (dN/dS) in pol and env between the four intact and unmodified AcERV sequences that constitute the consensus. A group of closely related, highly-intact ERVs originating recently from subsequent germ-line infections will retain a signal of purifying selection (dN/dS < 1) in both pol and env. In contrast, a replication-competent yet non-infectious ERV can replicate then reintegrate into the host genome while accumulating deleterious mutations in its env gene. In the latter case, because sequence divergence occurred post-endogenization, pol will still retain a signal of purifying selection, but env will have evolved under a neutral selection regime (dN/dS ∼ 1). Pairwise pol (dN/dS = 0.086–0.120) and pairwise env comparisons (dN/dS = 0.149–0.228) were consistent with a history of purifying selection. A codon-based Z-test rejected the hypothesis of neutral evolution for both genes (pol, p = 0; env, p = 0). A similar test of unmodified pre-consensus DrFV-2 and DrFV-3 pol and env coding regions demonstrated similar results (pol, dN/dS = 0.136; env, dN/dS = 0.123) and we likewise rejected the hypothesis of neutral evolution (P = 0). Apparent purifying selection could be explained also by ERV domestication, which we are unable to rule out. Co-opted retroviral gene families can retain a strong signal of selection after millions of years, perhaps falsely implying RV infectivity. Primate syncytin genes, for example, originated as orthoretroviral env in an early primate ancestor, yet are well conserved among distant lineages (Blaise et al. 2003).
We annotated coding sequences in five more intact ERVs: Platyfish (XmERV), amazon molly (PrERV), guppy (PfERV), bicolor damselfish (SpERV), and yellow croaker (LcERV) (see Supplementary Fig. 1). PrERV and LcERV necessitated construction of a quasi-consensus due to numerous nonsense or frameshift mutations. Among these fish ERVs, increased complexity is the common characteristic and genetic organization can be sorted into two types. Comprising the first type, DrFV-2 and PpERV-1, both integrated into Cypriniformes hosts, contain ERVs of similar length and organization. At 11,967, DrFV-2 is large but still within the spectrum of RV genome size. PpERV-1 is larger at 14,122 nt after taking into account the truncated 5′ LTR. ERVs from each species possess a comparably arranged set of putative accessory genes. We again calculated pairwise dN/dS between DrFV-2 and DrFV-3 and tested for a signal of purifying selection to determine if these are post-endogenization artifacts. We rejected the null hypothesis of neutral evolution for ORF-1 (dN/dS = 0.189, P = 0.002), located between pol and env, and ORF-3 (dN/dS = 0.246, P = 0.007), downstream of env. We were unable to do so for ORF-2 (dN/dS = 0.55, P = 0.379). Though PpERV-1 and DrFV-2 ORFs are similarly placed and sized, they share no tangible sequence homology.
The second type, those most like AcERV, all share multiple similarly arranged genes which contribute to a very large RV genome size >17,000 nt. These ERVs are larger than any previously described RV genome, and very likely more complex in terms of cell interactions. The function of these numerous large putative accessory genes is unknown. Pairwise dN/dS calculations between the four intact AcERV sequences of ORF-1 (dN/dS = 0.132–0.182) and ORF-2 (dN/dS = 0.476–0.818), between pol and env, and ORF-3 (dN/dS = 0.106–0.153), ORF-4 (dN/dS = 0.125–0.262), and ORF-5 (dN/dS = 0.151–0.191), downstream of env, demonstrated that these genes probably had a functional role before endogenization. A codon-based Z-test rejected the hypothesis of neutral evolution for all five genes (ORF-1, P = 0; ORF-2, P = 0.026; ORF-3, P = 0; ORF-4, P = 0; ORF-5, P = 0).
For ERVs with AcERV-like organization, we aligned the aa sequences of putative accessory genes. The first three ORFs are homologous among AcERV, XmERV, PrERV, PfERV, SpERV, and LcERV (ORF-1, 29–74 per cent ID; ORF-2, 20–65 percent ID; ORF-3, 23–75 percent ID). AcERV ORF-4 is unique among ERVs described here, sharing no sequence homology with the rest. AcERV-5 is somewhat unique, sharing some aa sites with ORF-4 in LcERV (79 pos., 34 percent ID, E-value = 3e−12) and SpERV (224 pos., 28 percent ID, E-value = 2e−10), as determined by a blastp query. ORF-4 aa sequences in PrERV, PfERV, and XmERV are still more similar to each other, likely reflecting host relatedness (37–60 percent ID). Identity matrices derived from these alignments are available as Supplementary Table 5a–d.
A conserved domain search produced no matches with known retroviruses or otherwise. We were unable to detect TM motifs in accessory genes among these ERVs, indicating that the products of these genes are not directly involved in virus budding and maturation. Gag-like genes are very large in these ERVs, 3,342–3,411 nt for those of AcERV type. For comparison, FV gag ranges from 1,413 nt for coelacanth endogenous FV (CoeEFV) (Han and Worobey 2012), to 1,947 nt for SFVcpz/hu (Maurer et al. 1988). Based on host phylogeny, these very large and accordingly complex RVs could have arisen at some stage within percomorph evolution. This relationship may be transient, as there are no examples of complete FV-like ERVs outside of Percomorphaceae and Cyprinodontiformes.
3.4. Fish ERV foamy-like features
Shared genetic architecture and functional motifs between these fish ERVs and mammalian FVs would be evidence for a FV common ancestor between these clades. Though not necessarily a phylogenetically conserved trait (Blomberg et al. 2009), nearly all class III ERVs use the 3′ end of a partially unwound Lysine tRNA to prime reverse transcription at a primer binding site (PBS) near the 5′ end of the RNA genome. Exceptions to this strategy include the recently discovered galago prosimian FV (Katzourakis et al. 2014), with a PBS corresponding to asparagine tRNA, and HERV-S, which utilizes serine tRNA (Yi et al. 2004). The majority of fish ERVs in this study with intact genomes utilize tRNA-Lys. PBS sequences correspond exactly, except for XmERV, SpERV, AcERV-1, and AcERV-4, which contain a single mismatch for vertebrate tRNALys. Zebrafish, both DrFV-2 and DrFV-3, utilize a threonine tRNA PBS, while guppy and Amazon molly ERVs use asparagine tRNA.
FVs have two transcription promoters, one shared by all RVs in the 5′LTR, and an internal transcription promoter (IP) located in env (Campbell et al. 1994). The IP encourages production of Tas, which acts in trans to further increase the activity of the IP. We detected a strong IP (TATAAATA) toward the 3′ terminus of the env-like region in both DrFV-2 and DrFV-3 (Fig. 2), and near the midpoint for PpERV-1 and PpERV-2. Notably, each of these contain up to three more TATA-boxes, but none of them are located near the 3′ end of env-like ORFs. Because of these IP signatures are generally >1,000 bp upstream of putative accessory genes, it is unclear whether they have a functional cis-regulatory effect. PfERV, LcERV, and SpERV also possess a detectable env-like IP. Most of them also include a strong TATA-box (TATATAAG) in the second ORF after pol. Again, whether this IP is functional is unknown.
All fish ERVs detected in this study share a GGGTG nt motif on the border between the polypurine tract (PPT) and 3′LTR (Fig. 2) where these features exist. This is a peculiar feature also shared by all FVs (Delelis et al. 2004), and this structure may be responsible for the unique process of FV integration. In contrast to Orthoretrovirinae integration, FV Int only cleaves one end of proviral dsDNA of its terminal dinucleotide (Juretzek et al. 2004). The reigning explanation models FV dsDNA as a 2-LTR circle that must be cleaved at the palindromic site joining the LTR ends, rather than a blunt-ended linear molecule (Delelis et al. 2005). The strict conservation of the GGGTG PPT:LTR motif points to its probable role in the aforementioned FV pre-integration process. This feature alone suggests a FV last common ancestor (LCA) of these fish ERVs and FVs in mammals.
A side effect of proviral DNA integration is a target site duplication (TSD). After engaging host genomic DNA, RV Int cleaves both strands at positions 4–6 nt apart, then reforms phosophodiester between the ragged genomic DNA ends and proviral dsDNA stands, leaving a 4–6 nt direct repeat abutting both ends of the newly integrated ERV (Brown 1997). Because RV Int mediates this process, the precise number of duplicated nucleotides tends to be conserved among RV genera. Both FVs and Gammaretrovirus form a 4-nt TSD with little exception (Serrao et al. 2015). Among fish ERVs in this study, with one exception (AcERV-3) all complete FV-like integrations also have 4 nt TSDs.
Fish foamy-like ERV env-like products possibly share typical FV function. Furin-like cellular proteases cleave Env into three products: a short leader peptide (LP), a surface unit (SU), and TM unit, requiring at minimum an RXXR recognition site (Nakayama 1997) in two places (Fig. 2: PFV). Compare this with orthoretroviral Env, cleaved once at the SU-TM junction by furin-like proteases, and cleaved of a quickly-degrading LP unit by signal peptidase (SPase) (Coffin et al. 1997; Rethwilm 2010). DrFV-2, PpERV-1, AcERV, and SpERV all contain a putative SU-TM cleavage site, and a likely N-terminal signal peptide is found in all intact fish ERV Env-like protein except LcERV and SpERV. FV-characteristic tripartite Env processing is an apparent feature in one intact ERV: PpERV-1. This 718 aa protein features an SPase cleavage site at aa position (pos.) 48: VLG|LI, abutting a TM helix at pos. 30–52; a strong furin-like cleavage site at pos. 209: RKAR|SL, and pos. 420: RLKR|VG, and a C-terminal TM helix at pos. 647–669 (Fig. 2). Though lacking a Gag-interacting WXXW motif, this protein is hypothetically processed in a very FV-like fashion. Whether these sites are missing elsewhere because of alternate Env processing or because of sequence degradation is unknown. We favor the latter explanation because the lack of furin-like cleavage sites in XmERV, PfERV, PrERV, and LcERV is difficult to explain otherwise when this mechanism of Env glycoprotein processing is ubiquitous among infectious RVs.
A Gag trait conserved among infectious FVs and certain EFVs (Katzourakis et al. 2014), including PpERV-1 (Fig. 2), is the p3 cleavage site: a viral Pro cleavage site motif VX|XV (Kehl et al. 2013) very near the C-terminus. In PpERV-1, this motif is VG|RV, -34 aa from the C-terminus. Though not cleaved in all virions (Rethwilm 2010), removal of this short terminal peptide is necessary for both replication and infectivity (Enssle et al. 1997; Zemba et al. 1998). Where Orthoretrovirinae Gag are typically cleaved completely into matrix, capsid, and nucleocapsid domains by Pro (Coffin et al. 1997), Spumaretrovirinae are not (Müllers 2013). Secondary Pro cleavage sites are found in Gag, but efficiency at these sites is low, and the large unprocessed Gag protein (minus the p3 cleavage) is not cleaved again until the virus docks with a host cell. The p3 cleavage site was not found in any other Fish ERVs, though potential secondary Pro cleavage sites were evident.
Conspicuously absent from Gag-like proteins in FV-like Fish ERVs are Cys-His boxes. Orthoretrovirinae encode at least one zinc finger domain, bearing the motif CX2CX4HX4C (Llorens et al. 2009), toward the C-terminus of Gag which are indispensable to the retroviral life cycle and maintenance of infectivity(Gorelick et al. 1999; Lee and Linial 2006). FV Gag alternatively contains glycine-arginine rich motifs (GR box) that fulfill the much same function as Cys-His zinc fingers (Müllers 2013). One is critical for viral packaging of Pol, which complexes with Gag to package viral RNA (Lee and Linial 2008), another serves as a nuclear localization signal (Tobaly-Tapiero et al. 2008). While fish ERVs are generally enriched for glycine/arginine toward the C-terminus of the likely gag gene, there are no clearly definable GR boxes. Rather, the presence of at least one glycine-glutamine-arginine (GQR) domain is conserved; DrFV-2: RRGRQQR at −101 aa from C-terminus; PpERV-1: GDRQRDQ −88 aa from C-terminus; AcERV: GQRTQQ −90 aa and QGQQR at −73 aa from C-terminus. PrERV (RQPRGG; −39 aa), XmERV (QGRPFQQG; −90 aa), SpERV (RGNQQRTQ; −78 aa), and LcERV (GGDQRR; −57 aa) also contain GQR boxes in similar C-terminal positions. Given the absence of a Cys-His boxes, and the well known DNA/RNA binding affinity of glutamine (Tan and Frankel 1998; Luscombe et al. 2001), we hypothesize that FV-like Fish RVs utilize a GQR box, fulfilling the function of conventional GR boxes. This notion is reminiscent of feline FV, which carries the aspartic protease catalytic motif DTQA (Winkler et al. 1997), a unique variant of the typical D(T/S)GA active site motifs thought to be immutable for a properly functioning RV Pro domain.
3.5. Estimated age of ERV integration
Provirus LTRs are identical upon integration; due to the mechanics of reverse transcription (Coffin et al. 1997), any mismatches must have occurred since. Where possible, we coupled pairwise divergence of LTR within intact ERVs with an estimated per generation host mutation rate (Johnson and Coffin 1999) to estimate time since integration. We also estimated coalescent age of paralogous fragments that arose through segmental duplication, identified by LTR sharing a common flanking sequence where LTR-complete ERVs are absent.
Unmodified ERV sequences in six species proved amenable to age estimation (Table 2). Midas cichlid mutation rate, measured empirically with breeding experiments, has been calculated at 6.6e−8/site/generation (Recknagel et al. 2013), with a generation time of ∼1 year (Kratochwil et al. 2015). Amazon molly mutation rate has been estimated to be 4.89e−8/sites/year (Fraser et al. 2015). We took the Amazon molly rate for the guppy rate, as they are closely related, and for yellow croaker and bicolor damselfish, as no mutation rate estimates exist for these species. The zebrafish rate of synonymous substitution has been estimated at 4.3e−9/site/year based on comparisons of conserved non-coding regions (Fu et al. 2010). Assuming the LTR and immediate flanking region are evolving neutrally, we should use non-synonymous substitutions in the rate estimate. To be conservative, we therefore increased this rate estimate by an order of magnitude, to 4.3e−8/site/year, bringing it in line with Midas cichlid and Amazon molly mutation rates.
Table 2.
Estimates of ERV integration age
| ERV | Methods | Divergence: d | Mutation rate: µ | Estimated age: t |
|---|---|---|---|---|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Divergence calculated with K80 nucleotide substitution model (Krogh et al. 2001). Mutation rates drawn from previous studies (Luscombe et al. 2001; Tamura et al. 2011; Kratochwil et al. 2015). DrFV-3 divergence was calculated from multiple sequence alignment using BEAST (Guindon and Gascuel 2003), and the 95 percent highest posterior density interval is included. Confidence intervals could not be calculated for other estimates due to our estimation methods, and these should not be interpreted as precise estimates lacking uncertainty. Where more than one estimation was possible, these are presented as upper and lower estimates of minimum age
HPD, highest posterior density; pos, nucleotide positions; TMRCA, time to most recent common ancestor; y, years. Accession numbers listed in Supplementary Table 1.
DrFV-2 is the highest quality ERV discovered in this study. Its LTR nucleotide sequences are identical, and barring LTR homogenization mediated by gene conversion, this virus infected zebrafish very recently. DrFV-3 shares 96 percent nt identity with DrFV-2 from gag to 3′LTR, but only 85 percent identity between 5′LTR. DrFV-3 is clearly recombinant, as it shares a 5′ flanking sequence with a fourth fragmented copy. DrFV-3 LTR divergence is not likely to give an accurate estimate. We detected six ERV fragments, several of them solo LTR, which share a long 5′ flanking sequence with DrFV-3, which we used to estimate TMRCA. That DrFV-3 integrated ∼284,000 years ago (TMRCA: 2.84e5, 95 percent highest posterior density: 2.47e5–3.24e5 years ago), while DrFV-2 likely integrated much more recently, is an interesting observation. Similarly, PpERV-1 and PpERV-2 are highly divergent and either represent superinfection by distantly related foamy-like RVs, or asynchronous infection and integration. Only PpERV-1 is nearly LTR-complete, but has identical LTR and no shared flanks. SpERV is also recombinant, having 3′ coding region upstream of its 5′ LTR. Despite this, we estimated its minimum age through LTR-divergence because no EFV-like fragments share flanking regions. The estimate of ∼71,000 years is probably less than the time since integration.
The four complete AcERV integrations retain LTR, and we dated them with LTR-divergence. We estimated their ages to range from 15,000 to 83,000 years since integration. None of them appear to be recombinant. This disparity between integration ages has two possible explanations; either LTR at different loci evolve at different rates, or four integration events happened over a span of 60,000 years. We favor the latter explanation, if only because FV superinfection is virally regulated and probably rare (Nethe et al. 2005).
PfERV is represented in the Amazon molly genome by two sequences; PfERV-1 has short LTR, probably having been truncated on either end of the ERV. PfERV-2 has intact LTR differing in length by one nt pos., and we favor the upper estimate of ∼150,000 years for this reason. Within guppies, PrERV-1 has an inverted 5′ LTR. Low-level divergence from the 3′ LTR might indicate that this occurred through recombination, but this is unclear. PrERV-2 has incomplete LTR and no shared flanking region to calculate flank-divergence. However, a ∼7,000-nt PrERV fragment shares a 3′ flank with a solo LTR. Again we favor the upper age estimate: ∼160,000 years since the segmental duplication that produced these shared flanks. The similarity between these Poecilia ERV estimates is probably coincidental, as we found no evidence of orthology and these species diverged well before these minimum integration dates (Meredith et al. 2010).
A single LTR-intact ERV in the yellow croaker genome represents LcERV, and we estimated ∼102,000 years since integration. It also shares a 5′LTR and upstream flank with a fragment elsewhere in the genome for which we estimated ∼31,000 years since integration. The lower age estimate is probably accurate, but only reflects years passed since the segmental duplication that created the fragment of LcERV. The upper age is closer to the true age of insertion and is a more accurate minimum age for LcERV.
3.6. De novo sequencing of FV-like sequences in diverse teleosts
We designed a pair of highly degenerate PCR primers to amplify a 450–500 nt region of FV RT and screened ninety-one preserved ray-finned fish tissues from taxa spanning the actinopterygian tree of life (Supplementary Table 2). We detected FV-like endogenous pol in nine of these fish (Table 3). Four of these species belong to the Ovalentaria crown group within the percomorph “bush”: houndfish (Beloniformes: Tylosus crocodilus), Bennet’s flying fish (Beloniformes: Cheilopogon pinnatibarbatus), and doubleline clingfish (Blenniformes [Betancur et al. 2013]: Lepadichtys lineatus) I belong to orders with no WGS representation, while freshwater angelfish (Cichliformes: Pterophyllum scalare) is a South American cichlid related to the Midas cichlid.
Table 3.
Hosts with FV-like fragments; de novo sequencing
| Order species | Common name | Source catalog no. | BLASTx hit (acc) | % ID | E-value |
|---|---|---|---|---|---|
| Anguilliformes | |||||
| G. miliaris | Goldentail moray | KU 145 | SFVspm (ABV59399) | 27 | 9E−15 |
| Cypriniformes | |||||
| H. placitus | Plains minnow | MSB 57953 | SFVmac (AFA44809) | 47 | 1E−19 |
| Gadiformes | |||||
| A. pectoralis | Giant grenadier | KU 2298 | EFV (NP_054716) | 35 | 2E−21 |
| Sygnathiformes | |||||
| D. volitans | Flying gurnard | KU 237 | SFVspm (ABV59399) | 35 | 3E−25 |
| Cichliformes | |||||
| P. scalare | Freshwater angelfish | KU 2846 | SFVspm (ADE05995) | 39 | 7E−27 |
| Blenniformes | |||||
| L. lineatus | Doubleline clingfish | KU 7020 | SFVspm (ABV59399) | 38 | 3E−29 |
| Beloniformes | |||||
| C. pinnabarbatus | Bennet’s flying fish | KU 2780 | SFVspm (ABV59399) | 36 | 4E−21 |
| T. crocodilus | Houndfish | KU 5842 | SFVspm (ADE05995) | 40 | 1E−28 |
| Perciformes | |||||
| S. aequidens | Deepwater serrano | SIO 08-90 | SFVspm (ABV59399) | 38 | 2E−27 |
Listed are the best hits for a representative cloned sequence successfully amplified from genomic DNA. Orders after Betancur-R et al. (2013). Common names from FishBase (Harel et al. 2015)
KU, University of Kansas; MSB, Museum of Southwest Biology; SIO, Scripps Institute of Oceanography. Blastx hit abbreviations: EFV, equine foamy virus; SFVmac, simian foamy virus – macaque; SFVspm, simian foamy virus – spider monkey.
The deepwater serrano (Serranus aequidens) is a Perciformes like the yellow croaker. The flying gurnard (Dactylopterus volitans) is a Sygnathiformes, more closely related to Scombriformes, such as tuna and mackerel, than to the other percomorph lineages identified in this study. ERVs in this species suggest that foamy-like retroviruses might be widespread across Percomorphaceae, a massive grouping of fish representing approximately half of all ray-finned fish (Alfaro et al. 2009). The plains minnow (Hybognathus placitus) belongs to Cypriniformes, in the same family as zebrafish and fathead minnow. Giant grenadier (Albatrossia pectoralis) belongs to Gadiformes along with cod. Last, the goldentail moray eel (Gymnothorax miliaris) is an Anguilliformes of the basal-teleost Elopomorpha lineage. These species share a more basal LCA than the host species identified by genomic screens. With the detection of EFV-like sequences in goldentail moray, the host range of these viruses includes all teleosts.
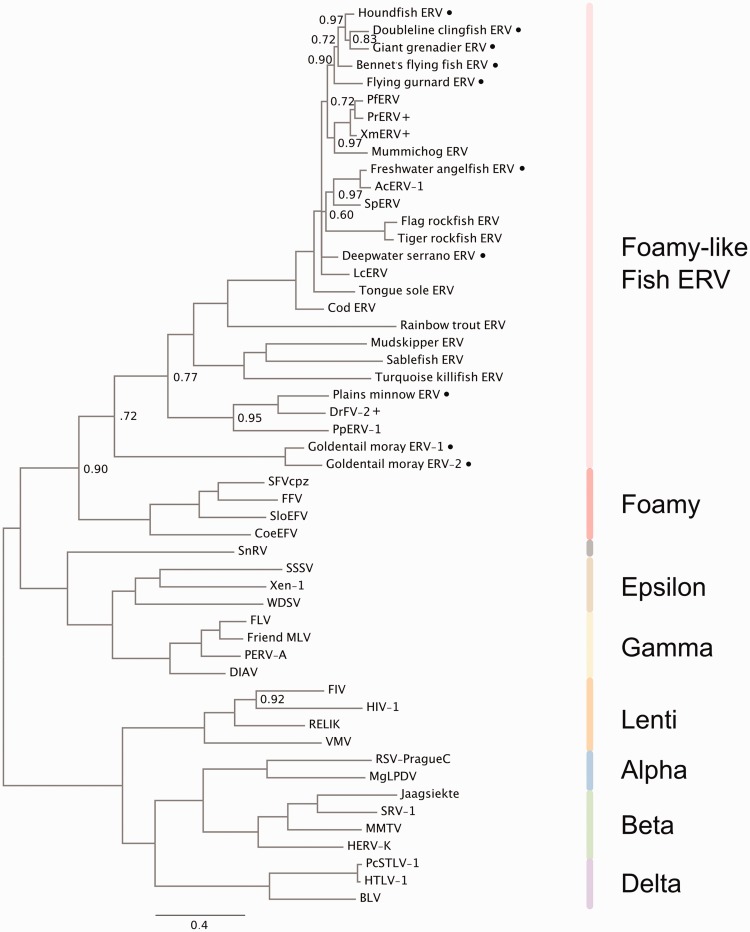
A phylogeny that includes representatives of these sequences and a smaller set of distantly related RVs show that these ERVs also belong to an FV sister clade (Fig. 3). Both goldentail moray ERVs are found at the root of this tree, in the same position of the host species among teleosts in the fish tree of life (Betancur et al. 2013).
Figure 3.
Phylogeny of RVs and twenty-seven foamy-like fish ERVs. MrBayes consensus tree estimated from 702 Pol aa positions, including gap positions introduced with short fragments sequenced in this study. Tree is midpoint rooted for clarity. Nodes with posterior probability <1.0 are marked. Branch lengths estimated as expected substitutions per site. RV clades are marked with colored bars to right. Included are ERVs discovered and sequenced by PCR-based assay in this study (•), and ERV sequences from WGS data alone. Also labeled are taxa from WGS databases that we PCR amplified and sequenced as positive controls (+).
4. Discussion
The discovery of these foamy-like ERVs in fish hosts means that there are now more described teleost EFVs than there are mammal EFVs ( Katzourakis et al. 2009, 2014; Han and Worobey 2012; Wu et al. 2012). Fish EFVs are not a novel discovery for paleovirology (Llorens et al. 2009; Schartl et al. 2013), and though undiscovered fish ERV diversity was predicted, it was not expected that 34 percent of species with WGS data (eighteen of fifty-two spp.) would contain evidence of past foamy-like RV infection. Compare this fraction to the 5 percent of sequenced mammal genomes known to contain EFVs (4 of 113 spp.). This mammalian percentage is also smaller than EFV-like sequences discovered through de novo PCR screens in this study: 9 percent (nine of ninety-one spp.). Interestingly, we were unable to detect any orthologous fish EFVs, even within congeneric Poecilia species pairs or within congeneric Sebastes species pairs. This is somewhat surprising given the estimates of relatively recent divergence of within these pairs (25 Ma [Meredith et al. 2010]; and 3–4 Ma [Hyde and Vetter 2007], respectively). Considering this point, and the high proportion of EFV-positive fish genomes, it is readily apparent that teleost fish have been frequent hosts to foamy-like RV infection.
Neither our PCR-based nor our in silico assays exhausted the possibility of undetected foamy-like RV diversity within fish species producing negative results. After endogenization, RV DNA sequence conservation is expected to decay, though this process may be halted if its host domesticates it. A practical limit exists for the detection of ERVs by PCR. Our assay was constrained by the necessary specificity of PCR primer sequences and relied on the conservation of complementary annealing sites. Our in silico assay of WGS databases was limited by a theoretical age-of-integration maximum, outside which ERV sequences will have decayed beyond recognition. Furthermore, the absence of detectable foamy-like ERVs in a lineage does not demonstrate a historical absence of exogenous counterparts because germ-line endogenization is not a guaranteed outcome of RV infection.
The fish EFV clade at large is only loosely grouped into recognizable fish clades, implying a history of frequent cross-species transmission. This is not surprising if it is presumed that virus transmission is more likely in an aquatic environment than a terrestrial environment. Little is actually known about how animal viruses spread in aquatic ecosystems (Suttle 2007), but there have been documented cases of a virus rapidly spreading among multiple geographically distant species in a short amount of time among fish hosts (Einer-Jensen 2004).
Fish EFVs possess the largest genomes of any known retrovirus to date. Most of this size increase is caused by a much larger than normal gag-like gene and the acquisition of several accessory genes. The functions of these genes are unknown but it may be assumed that replication pathways for these viruses are more complex than mammal FVs. Aside from the increase in genetic coding potential, these fish ERVs are foamy-like in organization. Long LTR, independently transcribed gag and pol, 4 nt TSD, and the presence of internal transcription promoters are all shared by FVs, although not exclusively (Coffin et al. 1997; Rethwilm 2010).
We detected discrete features unique to FV biology among known RVs. The tripartite Env processing regime found in PpERV-1 is foamy-like, as is the putative p3 cleavage site next to the C-terminus PpERV-1 Gag-like protein. These features have presumably been obscured by post-endogenous neutral substitutions in other fish ERVs annotated in this study. Most fish ERVs examined in this study use tRNALys to prime reverse transcription, and those that do not still mostly use known FV alternatives. All fish ERV examined in this study possess the GGGTG PPT:LTR junction motif that characterizes FV asymmetrical provirus processing (Delelis et al. 2004). Likewise is the absence of Cys-His zinc finger Gag motifs unique to FVs (Müllers 2013) and similarly absent in fish foamy ERVs with an intact gag-like ORF.
We did not detect the CTRS signature (Eastman and Linial 2001) used by FVs to localize virus assembly in the cytoplasm. Nor did we locate a WXXW Gag–Env interaction motif used by FVs to direct virus budding to the ER (Kehl et al. 2013). Conservation of the latter would necessitate conservation of the Gag substrate it interacts with. Gag is the fastest-evolving core gene in known exogenous FVs, followed by env (Rethwilm and Bodem 2013); therefore, it is difficult to treat absence of evidence as evidence of absence for this supposedly requisite feature FV replication. A CTRS functional domain is found in all exogenous FVs, but not exclusively. RV cytoplasmic targeting and encapsidation was first noted in Betaretrovirus mouse mammary tumor virus and Mason-Pfizer monkey virus (Choi et al. 1999). Its active site consists of a single arginine, an abundant residue in fish EFVs. Though necessary for FV-like replication, the sequence conservation among even closely related mammal FVs is suspect (Kehl et al. 2013). Despite our inability to characterize certain apparently indispensible FV-like domains among these fish ERVs, the specific motifs and features that were detected, when taken together, point to a basic replication strategy conserved between these fish ERVs and nearest phylogenetic relations—CoeEFV and mammal FVs.
There is a strong indication that these viruses are still circulating in some infectious form, though there is no real expectation that exogenous descendants of ERVs remain among hosts. In contrast to other early-diverging ERV lineages recently discovered (Cui et al. 2012; Tarlinton et al. 2013; Wang et al. 2013; Han 2015), the six fish ERV integrations that could be dated in this study are all very young, even when compared with the relatively recent integration of fish endogenous Epsilonretrovirus in zebrafish and other species (Basta et al. 2009). The oldest, zebrafish DrFV-3, is estimated to have integrated ∼284,000 years ago (Table 1). DrFV-2 in the same species is the most recent integration and has identical LTR sequences, indicating host infection over a considerable period of time. The same may be true for fathead minnow ERVs, of which PpERV-1 probably integrated very recently and PpERV-2 is highly divergent. One explanation for this is contemporaneous integration of two EFV types into the fathead minnow genome; the other is a long but not necessarily continuous period of infection. AcERV in Midas cichlids is very young, is present in many copies with estimated integrations ∼15,000–83,000 years ago, and closely related to other South American freshwater fish foamy-like ERVs (Fig. 1). That distantly related ERVs are so young and possibly accompanied by exogenous counterparts for so long points to the likelihood of present day persistence.
Evidence for an ancient origin of these viruses is more elusive. The quality of ERVs extracted from fish genomes is highly variable. Some integrated relatively recently, while others are degraded to the limit of recognition. ERV fragments from three host species in particular; P. magnuspinnatus, N. furzeri, and A. fimbria, are percomorph species closely related (in relative terms) to the fish group circumscribing guppies and yellow croaker, for example. In addition, they are at present geographically and ecologically disparate. Turquoise killifish occupy temporary freshwater pools in Zimbabwe and Mozambique, growing from egg to adult in a mere month (Harel et al. 2015). Sablefish are benthic deep-sea dwellers with a trans-Pacific distribution (Orlov and Biryukov 2005), while mudskippers occupy mudflats in Korea and mainland China (Baeck et al. 2008). Yet ERVs from these fish occupy a basal position within fish foamy ERVs. This may be attributable to phylogenetic signal degradation over millions of years of accumulated neutral mutations since integration.
With no overwhelming pattern of host–virus codivergence and no clear evidence of geographic clustering, it is difficult to generate hypotheses that better explain the proposed ancient marine origin proposed for FV (Han and Worobey 2012). Accepting that fish foamy-like RVs are widely distributed across teleost hosts, if the LCA of these fish carried the LCA of these viruses, then we can surmise their emergence in the Permian Period 283 Ma at minimum (Betancur et al. 2013). This estimate is rather easy to accept given the deep divergence between the mammal-coelacanth FV clade and the fish ERV clade (Fig. 1).
This widespread diversity of foamy-like ERVs in fish hosts is consistent with a hypothetical ancient marine origin of FVs (Han and Worobey 2012). The infectious progenitors of these ERVs were clearly capable of invading phylogenetically disparate hosts. They form a deeply diverging monophyletic sister clade with known FVs, and are more closely related to such than known class III FV-like ERVs in mammals (Fig. 1). Further sampling of fish taxa, and the possible eventual discovery of an extant strain, could further elucidate FV origins. These results, however, are nonetheless consistent with the idea that FVs arose in the ocean hundreds of millions of years ago.
Data Availability
All DNA sequences produced during this study have been made available in GenBank under accession numbers KX354941-KX354950.
Supplementary Material
Acknowledgements
We thank Tom Watts and Guanzhu Han for their technical advice and helpful discussions. We thank Brendan Larsen for reading our manuscript and providing comments. We also thank Andrew Bentley, Lex Snyder, H. J. Walker, Kevin Fitzsimmons, and the Rainbow Trout Farm at Oak Creek Canyon in Sedona, AZ for kindly providing us with fish tissue samples.
Supplementary data
Supplementary data are available at Virus Evolution online.
Funding
This study was supported by NIH/NIAID R01AI084691 and the David and Lucile Packard Foundation.
Conflict of interest: None declared.
References
- Alfaro M. E., et al. (2009) ‘Nine Exceptional Radiations Plus High Turnover Explain Species Diversity in Jawed Vertebrates’, Proceedings of the National Academy of Sciences of the United States of America, 106/32: 13410–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeck G. W., Takita T., Yoon Y. H. (2008) ‘Lifestyle of Korean Mudskipper Periophthalmus magnuspinnatus with Reference to a Congeneric Species Periophthalmus modestus’, Ichthyological Research, 55: 43–52. [Google Scholar]
- Basta H. A., et al. (2009) ‘Evolution of Teleost Fish Retroviruses: Characterization of New Retroviruses with Cellular Genes’, Journal of Virology, 83/19: 10152–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bénit L., et al. (1999) ‘ERV-L Elements: A Family of Endogenous Retrovirus-Like Elements Active Throughout the Evolution of Mammals’, Journal of Virology, 73/4: 3301–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur-R R., et al. (2013) ‘The Tree of Life and a New Classification of Bony Fishes’, PLoS Currents Tree of Life, 1–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betsem E., et al. (2011) ‘Frequent and Recent Human Acquisition of Simian Foamy Viruses through Apes’ Bites in Central Africa’, PLoS Pathogen, 7/10: e1002306.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniasz P. D., et al. (1995) ‘A Comparative Study of Higher Primate Foamy Viruses, Including a New Virus from a Gorilla’, Virology, 207/1: 217–28. [DOI] [PubMed] [Google Scholar]
- Blaise S., et al. (2003) ‘Genomewide Screening for Fusogenic Human Endogenous Retrovirus Envelopes Identifies Syncytin 2, a Gene Conserved on Primate Evolution’, Proceedings of the National Academy of Sciences of the United States of America, 100/22: 13013–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomberg J., et al. (2009) ‘Classification and Nomenclature of Endogenous Retroviral Sequences (ERVs)’, Gene, 448/2: 115–23., [DOI] [PubMed] [Google Scholar]
- Brown P. O. (1997) Integration In: JM Coffin, SH Hughes, HE Varmus. eds. Retroviruses. Cold Spring Harbor: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Campbell M., et al. (1994) ‘Characterization of the Internal Promoter of Simian Foamy Viruses’, Journal of Virology, 68/8: 4811–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez S., Silla-Martinez J. M., Gabaldon T. (2009) ‘trimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses’, Bioinformatics, 25/15: 1972–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi G., et al. (1999) ‘Identification of a Cytoplasmic Targeting/Retention Signal in a Retroviral Gag Polyprotein’, Journal of Virology, 73/7: 5431–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin JM, Hughes SH, Varmus HE, ed. (1997) Overview of Reverse Transcription In: Retroviruses. Cold Spring Harbor: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Coffin JM, Hughes SH, Varmus HE, ed. (1997) Synthesis and Organization of Env Glycoproteins In: Retroviruses. Cold Spring Harbor: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Coffin JM, Hughes SH, Varmus HE, ed. (1997) Synthesis, Assembly, and Processing of Viral Proteins In: Retroviruses. Cold Spring Harbor: Cold Spring Harbor Laboratory Press. [PubMed] [Google Scholar]
- Coffin JM, Hughes SH, Varmus HE, ed.(1997). The Place of Retroviruses in Biology Retroviruses. Cold Spring Harbor: Cold Spring Harbor Laboratory Press. [PubMed] [Google Scholar]
- Cong M., et al. (2005) ‘Ancient Co-speciation of Simian Foamy Viruses and Primates’, Nature, 434: 376–80. [DOI] [PubMed] [Google Scholar]
- Cui J., et al. (2012) ‘Identification of Diverse Groups of Endogenous Gammaretroviruses in Mega- and Microbats’, Journal of General Virology, 93: 2037–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D., et al. (2012) ‘jModelTest 2: More Models, New Heuristics and Parallel Computing’, Nature Methods, 9/8: 772.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delelis O., et al. (2005) ‘A Novel Function for Spumaretrovirus Integrase: An Early Requirement for Integrase-Mediated Cleavage of 2 LTR Circles’, Retrovirology, 2: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delelis O., Lehmann-Che J., Saïb A. (2004) ‘Foamy Viruses—A World Apart’, Current Opinion in Microbiology, 7/4: 400–6. [DOI] [PubMed] [Google Scholar]
- Drummond A. J., et al. (2012) ‘Bayesian Phylogenetics with BEAUti and the BEAST 1.7’, Molecular Biology and Evolution, 29/8: 1969–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckert P., Brunak S., Blom N. (2004) ‘Prediction of Proprotein Convertase Cleavage Sites’, Protein Engineering Design & Selection, 17/1: 107–12. [DOI] [PubMed] [Google Scholar]
- Eastman S. W., Linial M. L. (2001) ‘Identification of a Conserved Residue of Foamy Virus Gag Required for Intracellular Capsid Assembly’, Journal of Virology, 75/15: 6857–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einer-Jensen K. (2004) ‘Evolution of the Fish Rhabdovirus Viral Haemorrhagic Septicaemia Virus’, Journal of General Virology, 85/5: 1167–79. [DOI] [PubMed] [Google Scholar]
- Enssle J., et al. (1997) ‘Carboxy-Terminal Cleavage of the Human Foamy Virus Gag Precursor Molecule Is an Essential Step in the Viral Life Cycle’, Journal of Virology, 71/10: 7312–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., et al. (2015) ‘HMMER Web Server: 2015 Update’, Nucleic Acids Research, 43/W1: W30–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser B. A., et al. (2015) ‘Population Genomics of Natural and Experimental Populations of Guppies (Poecilia reticulata)’, Molecular Ecology, 24/2: 389–408. [DOI] [PubMed] [Google Scholar]
- Fu B., et al. (2010) ‘The Rapid Generation of Chimerical Genes Expanding Protein Diversity in zebrafish’, BMC Genomics 11/1: 657.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelick R. J., et al. (1999) ‘Strict Conservation of the Retroviral Nucleocapsid Protein Zinc Finger Is strongly influenced by its role in viral infection processes: characterization of HIV-1 particles Containing Mutant Nucleocapsid Zinc-Coordinating Sequences’, Virology, 256: 92–104. [DOI] [PubMed] [Google Scholar]
- Greenwood A. D., et al. (2005) ‘The Distribution of Pol Containing Human Endogenous Retroviruses in Non-human Primates’, Virology, 334/2: 203–13. [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. (2003) ‘A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood’, Systematic Biology, 52/5: 696–704. [DOI] [PubMed] [Google Scholar]
- Han G.-Z. (2015) ‘Extensive Retroviral Diversity in Shark’, Retrovirology, 12/1: 10–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G.-Z., Worobey M. (2012) ‘An Endogenous Foamy Virus in the Aye-Aye (Daubentonia madagascariensis)’, Journal of Virology, 86/14: 7696–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G.-Z., Worobey M. (2012) ‘An Endogenous Foamy-Like Viral Element in the Coelacanth Genome’, PLoS Pathogen, 83/6: e1002790.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G.-Z., Worobey M. (2014) ‘Endogenous Viral Sequences from the Cape Golden Mole (Chrysochloris asiatica) Reveal the Presence of Foamy Viruses in All Major Placental Mammal Clades’, PLoS One, 9/5: 3–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel I., et al. (2015) ‘A Platform for Rapid Exploration of Aging and Diseases in a Naturally Short-Lived Vertebrate’, Cell, 160/5: 1013–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart D., et al. (1996) ‘Complete Nucleotide Sequence and Transcriptional Analysis of Snakehead Fish Retrovirus’, Journal of Virology, 70/6: 3606–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfman G. S., Collette B. B., Facey D. E., Bowen B. W. (2009) The Diversity of Fishes: Biology, Evolution, and Ecology, vol. 2 New York: Wiley; … 736 p. [Google Scholar]
- Herchenröder O., et al. (1994) ‘Isolation, Cloning, and Sequencing of Simian Foamy Viruses from Chimpanzees (SFVcpz): High Homology to Human Foamy Virus (HFV)’, Virology, 201/2: 187–99. [DOI] [PubMed] [Google Scholar]
- Huang Y., et al. (2010) ‘CD-HIT Suite: A Web Server for Clustering and Comparing Biological Sequences’, Bioinformatics, 26/5: 680–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde J. R., Vetter R. D. (2007) ‘The Origin, Evolution, and Diversification of Rockfishes of the Genus Sebastes (Cuvier)’, Molecular Phylogenetics and Evolution, 44/2: 790–811. [DOI] [PubMed] [Google Scholar]
- Jern P., Sperber G. O., Blomberg J. (2005) ‘Use of Endogenous Retroviral Sequences (ERVs) and Structural Markers for Retroviral Phylogenetic Inference and Taxonomy’, Retrovirology, 2: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W. E., Coffin J. M. (1999) ‘Constructing Primate Phylogenies from Ancient Retrovirus Sequences’, Proceedings of the National Academy of Sciences of the United States of America, 96/18: 10254–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juretzek T., et al. (2004) ‘Foamy Virus Integration’, Journal of Virology, 78/5: 2472–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzourakis A., et al. (2009) ‘Macroevolution of Complex Retroviruses’, Science, 325/5947: 1512.. [DOI] [PubMed] [Google Scholar]
- Katzourakis A., et al. (2014) ‘Discovery of Prosimian and Afrotherian Foamy Viruses and Potential Cross Species Transmissions Amidst Stable and Ancient Mammalian Co-evolution’, Retrovirology, 11: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehl T., Tan J., Materniak M. (2013) ‘Non-simian Foamy Viruses: Molecular Virology, Tropism and Prevalence and Zoonotic/Interspecies Transmission’, Viruses, 5/9: 2169–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. (1980) ‘A Simple Method for Estimating Evolutionary Rates of Base Substitutions through Comparative Studies of Nucleotide Sequences’, Journal of Molecular Evolution, 16/2: 111–20. [DOI] [PubMed] [Google Scholar]
- Kratochwil C. F., Sefton M. M., Meyer A. (2015) ‘Embryonic and Larval Development in the Midas Cichlid Fish Species Flock (Amphilophus spp.): A New Evo-devo Model for the Investigation of Adaptive Novelties and Species Differences’, BMC Developmental Biology, 15/12: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., et al. (2001) ‘Predicting Transmembrane Protein Topology with a Hidden Markov model: Application to Complete Genomes’, Journal of Molecular Biology, 305/3: 567–80. [DOI] [PubMed] [Google Scholar]
- Kupiec J. J., et al. (1991) ‘Sequence Analysis of the Simian Foamy Virus Type 1 Genome’, Gene, 101: 185–94. [DOI] [PubMed] [Google Scholar]
- Larkin M. A., et al. (2007) ‘Clustal W and Clustal X version 2.0’, Bioinformatics, 23/21: 2947–8. [DOI] [PubMed] [Google Scholar]
- Lee E.-G., Linial M. L. (2006) ‘Deletion of a Cys-His Motif from the Alpharetrovirus Nucleocapsid Domain Reveals Late Domain Mutant-like Budding Defects’, Virology, 347/1: 226–33. [DOI] [PubMed] [Google Scholar]
- Lee E.-G., Linial M. L. (2008) ‘The C Terminus of Foamy Retrovirus Gag Contains Determinants for Encapsidation of Pol Protein into Virions’, Journal of Virology, 82/21: 10803–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Ortí G. (2007) ‘Molecular Phylogeny of Clupeiformes (Actinopterygii) Inferred from Nuclear and Mitochondrial DNA Sequences’, Molecular Phylogenetics and Evolution, 44: 386–98. [DOI] [PubMed] [Google Scholar]
- Linial M. L. (1999) ‘Foamy Viruses Are Unconventional Retroviruses’, Journal of Virology, 73/3: 1747–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linial M. L. (2000) ‘Why Aren’t Foamy Viruses Pathogenic?’, Trends in Microbiology, 8: 284–9. [DOI] [PubMed] [Google Scholar]
- Llorens C., et al. (2009) ‘Network Dynamics of Eukaryotic LTR Retroelements beyond Phylogenetic Trees’, Biology Direct, 4/1: 41.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López J. A., Chen W.-J., Ortí G. (2004) ‘Esociform Phylogeny’, Copeia, 2004/3: 449–64. [Google Scholar]
- Luscombe N. M., Laskowski R. A., Thornton J. M. (2001) ‘Amino Acid-Base Interactions: A Three-Dimensional Analysis of Protein-DNA Interactions at an Atomic Level’, Nucleic Acids Research, 29/13: 2860–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Bryant S. H. (2004) ‘CD-Search: Protein Domain Annotations on the Fly’, Nucleic Acids Research, 32: W327–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer B., et al. (1988) ‘Analysis of the Primary Structure of the Long Terminal Repeat and the Gag and Pol Genes of the Human Spumaretrovirus’, Journal of Virology, 62/5: 1590–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiering C. D., Linial M. L. (2001) ‘Historical Perspective of Foamy Virus Epidemiology and Infection’, Clinical Microbiology Reviews, 14/1: 165–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith R. W., et al. (2010) ‘Molecular Phylogenetic Relationships and the Evolution of the Placenta in Poecilia (Micropoecilia) (Poeciliidae: Cyprinodontiformes)’, Molecular Phylogenetics and Evolution, 55/2: 631–9. [DOI] [PubMed] [Google Scholar]
- Müllers E. (2013) ‘The Foamy Virus Gag Proteins: What Makes Them Different?’, Viruses, 5/4: 1023–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K. (1997) ‘Furin: A Mammalian Subtilisin/Kex2p-like Endoprotease Involved in Processing of a Wide Variety of Precursor Proteins’, Biochemical Journal, 327: 625–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nethe M., Berkhout B., van der Kuyl A. C. (2005) ‘Retroviral Superinfection Resistance’, Retrovirology, 2: 52.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonechnikov K., Golosova O., Fursov M. (2012) ‘Unipro UGENE: A Unified Bioinformatics Toolkit’, Bioinformatics, 28/8: 1166–7. [DOI] [PubMed] [Google Scholar]
- Omoto S., et al. (2004) ‘Feline Foamy Virus Tas Protein Is a DNA-Binding Transactivator’, Journal of General Virology, 85/Pt 10: 2931–5. [DOI] [PubMed] [Google Scholar]
- Orlov A. M., Biryukov I. A. (2005) ‘First Report of Sablefish in Spawning Condition off the Coast of Kamchatka and the Kuril Islands’, ICES Journal of Marine Sciences, 62/5: 1016–20. [Google Scholar]
- Pacheco B., et al. (2010) ‘Species-Specific Inhibition of Foamy Viruses from South American Monkeys by New World Monkey TRIM5 Proteins’, Journal of Virology, 84/8: 4095–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M. R., Emerman M., Malik H. S. (2011) ‘Paleovirology—Ghosts and Gifts of Viruses Past’, Current Opinion in Virology, 1/4: 304–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul T. A., et al. (2006) ‘Identification and Characterization of an Exogenous Retrovirus from Atlantic Salmon Swim Bladder Sarcomas’, Journal of Virology, 80/6: 2941–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Suchard M., Xie D., Drummond A. (2014) Tracer v1.6. available from http://beast.bio.ed.ac.uk/Tracer.
- Recknagel H., Elmer K. R., Meyer A. (2013) ‘A Hybrid Genetic Linkage Map of Two Ecologically and Morphologically Divergent Midas Cichlid Fishes (Amphilophus spp.) Obtained by massively parallel DNA sequencing (ddRADSeq)’, G3, 3/1: 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renne R., et al. (1992) ‘Genomic Organization and Expression of Simian Foamy Virus Type 3 (SFV-3)’, Virology, 186/2: 597–608. [DOI] [PubMed] [Google Scholar]
- Renshaw R. W., Casey J. W. (1994) ‘Transcriptional Mapping of the 3’ End of the Bovine Syncytial Virus Genome’, Journal of Virology, 68/2: 1021–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rethwilm A. (1995) ‘Regulation of Foamy Virus Gene Expression’, Current Topics in Microbiology and Immunology, 193: 1–24. [DOI] [PubMed] [Google Scholar]
- Rethwilm A. (2010) ‘Molecular Biology of Foamy Viruses’, Medical Microbiology and Immunology, 199/3: 197–207 [DOI] [PubMed] [Google Scholar]
- Rethwilm A., Bodem J. (2013) ‘Evolution of Foamy Viruses: The Most Ancient of All Retroviruses’, Viruses, 5/10: 2349–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribet D., et al. (2008) ‘Murine Endogenous Retrovirus MuERV-L Is the Progenitor of the "Orphan" Epsilon Viruslike Particles of the Early Mouse Embryo’, Journal of Virology, 82/3: 1622–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., et al. (2012) ‘MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space’, Systematic Biology, 61/3: 539–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovnak J., Quackenbush S. L. (2010) ‘Walleye Dermal Sarcoma Virus: Molecular Biology and Oncogenesis’, Viruses, 2/9: 1984–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schartl M., et al. (2013) ‘The Genome of the Platyfish, Xiphophorus maculatus, Provides Insights into Evolutionary Adaptation and Several Complex Traits’, Nature Genetics , 45/5: 567–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrao E., et al. (2015) ‘Key Determinants of Target DNA Recognition by Retroviral Intasomes’, Retrovirology, 12/1: 39.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., et al. (2011) ‘Fast, Scalable Generation of High-Quality Protein Multiple Sequence Alignments Using Clustal Omega’, Molecular Systems Biology, 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A., Hubley R., Green P. (2015) RepeatMasker Open-4.0 [Internet]. Available from: http://www.repeatmasker.org
- Suttle C. A. (2007) ‘Marine Viruses—Major Players in the Global Ecosystem’, Nature Reviews Microbiology, 5/10: 801–12. [DOI] [PubMed] [Google Scholar]
- Switzer W. M., et al. (2004) ‘Frequent Simian Foamy Virus Infection in Persons Occupationally Exposed to Nonhuman Primates’, Journal of Virology, 78/6: 2780–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., et al. (2011) ‘MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutioanry Distance, and Maximum Parsimony Methods’, Molecular Biology and Evolution, 28/10: 2731–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan R., Frankel A. D. (1998) ‘A Novel Glutamine-RNA Interaction Identified by Screening Libraries in Mammalian Cells’, Proceedings of the National Academy of Sciences of the United States of America, 95/8: 4247–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Kuhen K. L., Wong-Staal F. (1999) ‘Lentivirus Replication and Regulation’, Annual Reviews of Genetics, 33/1: 133–70. [DOI] [PubMed] [Google Scholar]
- Tarlinton R. E., et al. (2013) ‘Characterisation of a Group of Endogenous Gammaretroviruses in the Canine Genome’, The Veterinary Journal, 196/1: 28–33. [DOI] [PubMed] [Google Scholar]
- Thümer L., et al. (2007) ‘The Complete Nucleotide Sequence of a New World Simian Foamy Virus’, Virology, 369/1: 191–7. [DOI] [PubMed] [Google Scholar]
- Tobaly-Tapiero J., et al. (2000) ‘Isolation and Characterization of an Equine Foamy Virus’, Journal of Virology, 74/9: 4064–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobaly-Tapiero J., et al. (2008) ‘Chromatin Tethering of Incoming Foamy Virus by the Structural Gag Protein’, Traffic, 9/2: 1717–27. [DOI] [PubMed] [Google Scholar]
- Wang L., et al. (2013) ‘Ancient Invasion of an Extinct Gammaretrovirus in Cetaceans’, Virology, 441/1: 66–9. [DOI] [PubMed] [Google Scholar]
- Weiss R. A. (2006) ‘The Discovery of Endogenous Retroviruses’, Retrovirology, 3: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler I., et al. (1997) ‘Characterization of the Genome of Feline Foamy Virus and Its Proteins Shows Distinct Features Different from Those of Primate Spumaviruses’, Journal of Virology, 71/9: 6727–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., et al. (2012) ‘Virome Analysis for Identification of Novel Mammalian Viruses in Bat Species from Chinese Provinces’, Journal of Virology, 86/20: 10999–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J.-M., et al. (2004) ‘Human Endogenous Retroviral Elements Belonging to the HERV-S Family from Human Tissues, Cancer Cells, and Primates: Expression, Structure, Phylogeny and Evolution’, Gene, 342/2: 283–92. [DOI] [PubMed] [Google Scholar]
- Zemba M., et al. (1998) ‘The Carboxy-Terminal p3Gag Domain of the Human Foamy Virus Gag Precursor Is Required for Efficient Virus Infectivity’, Virology, 247/1: 7–13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All DNA sequences produced during this study have been made available in GenBank under accession numbers KX354941-KX354950.