Abstract
Pseudouridylation is a common post‐transcriptional modification in RNA, but its functional consequences at the cellular level remain largely unknown. Using a proximity‐biotinylation assay, we identified a protein module in mitochondrial RNA granules, platforms for post‐transcriptional RNA modification and ribosome assembly, containing several proteins of unknown function including three uncharacterized pseudouridine synthases, TRUB2, RPUSD3, and RPUSD4. TRUB2 and RPUSD4 were previously identified as core essential genes in CRISPR/Cas9 screens. Depletion of the individual enzymes produced specific mitochondrial protein synthesis and oxidative phosphorylation assembly defects without affecting mitochondrial mRNA levels. Investigation of the molecular targets in mitochondrial RNA by pseudouridine‐Seq showed that RPUSD4 plays a role in the pseudouridylation of a single residue in the 16S rRNA, a modification that is essential for its stability and assembly into the mitochondrial ribosome, while TRUB2/RPUSD3 were similarly involved in pseudouridylating specific residues in mitochondrial mRNAs. These results establish essential roles for epitranscriptomic modification of mitochondrial RNA in mitochondrial protein synthesis, oxidative phosphorylation, and cell survival.
Keywords: epitranscriptomic modification, mitochondrial protein synthesis, oxidative phosphorylation, pseudouridine synthase, ribosome assembly
Subject Categories: Membrane & Intracellular Transport, Protein Biosynthesis & Quality Control, RNA Biology
Introduction
Genomewide mapping studies of the epitranscriptome in human cells have revealed widespread post‐transcriptional modifications in both non‐coding and coding RNA 1, 2. Pseudouridine (ψ), the most common RNA modification, has been suggested to play a role in RNA stability, RNA folding/secondary structure, and translation efficiency and fidelity; however, the functional consequences of this modification at the cellular level, especially in mRNA, generally remain obscure 3. Human cells contain 13 pseudouridine synthases, but the majority remain uncharacterized, and their substrates and subcellular distribution are unknown 3. The mitochondrial 16S rRNA contains a single known pseudouridine site 4, and mitochondrial tRNAs are extensively pseudouridylated by PUS1, an enzyme that also modifies cytosolic tRNAs, and when mutated causes mitochondrial myopathy and sideroblastic anemia 5, 6. Genomewide pseudouridine profiling suggested that in addition to these known pseudouridine sites in mitochondrial RNA, specific mRNAs, and the 12S rRNA might be pseudouridylated in human cells 7, but these results have not been confirmed, nor has the enzymatic machinery been identified.
Mammalian mitochondrial RNA (mtRNA) is transcribed as two polycistronic transcripts, which are further processed to produce 2 rRNAs, 11 mRNAs (including two bicistronic transcripts), and 22 tRNAs. Post‐transcriptional handling of mitochondrial transcripts, as well as the assembly of the mitochondrial ribosome, occurs in non‐membrane delimited structures called mitochondrial RNA granules 8, 9, 10, 11. These structures contain newly synthetized mtRNA and a number of RNA‐binding proteins responsible for stabilization, processing, modification, folding, and degradation of mtRNA. Here we have uncovered a protein module containing three uncharacterized pseudouridine synthases that localizes to these granules. We identify the pseudouridylated sites in mitochondrial RNA that are the targets of these enzymes and investigate the functional consequences of these epitranscriptomic modifications for mitochondrial ribosome biogenesis, mitochondrial translation, and assembly of the oxidative phosphorylation complexes.
Results and Discussion
Identification of a mitochondrial pseudouridine synthase protein module using BioID
Several RNA‐binding proteins that localize to the RNA granule have been linked to human disease 12, and loss‐of‐function variants in one such protein, FASTKD2, cause infantile mitochondrial encephalopathy 13. FASTKD2 binds the mitochondrial 16S rRNA 9, 14, and it is required for the assembly of the large 39S subunit (mt‐LSU) of the mitochondrial ribosome 9, but the molecular mechanism remains unknown. To further investigate FASTKD2 function, we searched for interacting protein partners using affinity purification–mass spectrometry and BioID, a proximity‐biotinylation assay that identifies interacting protein partners in vivo independent of the strength of the interaction 15. FASTKD2 specifically interacted with known RNA granule proteins, several proteins of the mitochondrial ribosome, other mitochondrial enzymes, as well as four proteins, MTERFD1, TRUB2, WBSCR16, NGRN (Fig 1A and Table EV1), whose presence in mitochondrial RNA granules has not been previously reported, and which we confirmed by immunofluorescence studies (Fig EV1). We observed only partial overlap with GRSF1, a bone fide RNA granule protein, and it is possible that many RNA granule proteins transit into and out of the granules in response to physiological signals, a hypothesis that requires further investigation. MTERFD1 (mitochondrial transcription termination factor MTERF3) is required for the assembly of the mt‐LSU 16, whereas TRUB2 (a pseudouridine synthase), WBSCR16 (Williams‐Beuren Syndrome Chromosome Region 16), and NGRN (Neugrin) are uncharacterized 17. Using BioID, we reciprocally confirmed the specific interaction of TRUB2 with all these proteins (Fig 1B) and identified two other pseudouridine synthases, RPUSD3 and RPUSD4, forming a plausible functional module for mitochondrial pseudouridylation. Both TRUB2 and RPUSD4 were identified as core essential genes in CRISPR/Cas9 screens of multiple human cell lines, and RPUSD3 was essential in two of the cell lines investigated 18. NGRN, WBSCR16, TRUB2, RPUSD3, and RPUSD4 were also identified in a Crispr/Cas9 death screen for essential OXPHOS genes, and subsequent immunoprecipitation and mass spectrometry studies suggested that they formed a module, independently confirming the results we obtained using the BioID assay 19. TRUB2 also interacted with METTL15 (Fig 1B), an orthologue of the E. coli protein RsmH, which methylates cytidine 1402 (at the N4 position) in bacterial 16S rRNA 20, a modification conserved in the mammalian mitochondrial 12S rRNA.
Figure 1. Mitochondrial pseudouridylation module.
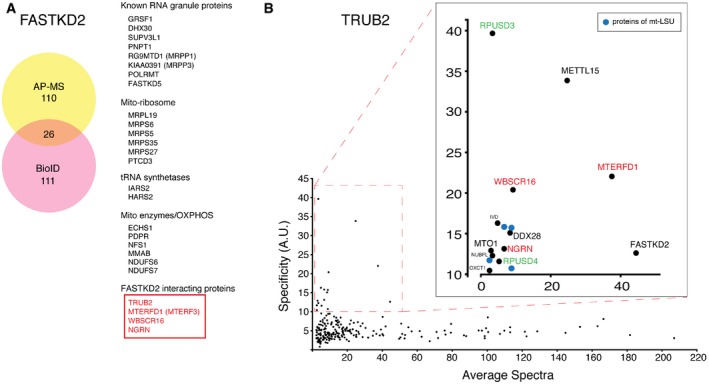
- Identification of FASTKD2 interacting proteins by AP–MS and BioID. 26 high‐confidence interactors detected by both techniques are listed on the right.
- Prey specificity graph for BioID interactome of TRUB2 protein. Prey specificity was calculated as the relative enrichment of interaction of individual preys and TRUB2, compared to their interaction with 138 other baits (42 mitochondrial baits, 96 baits from other cellular compartments).
Figure EV1. Co‐localization of the proteins in the pseudouridylation module with mitochondrial RNA granules.
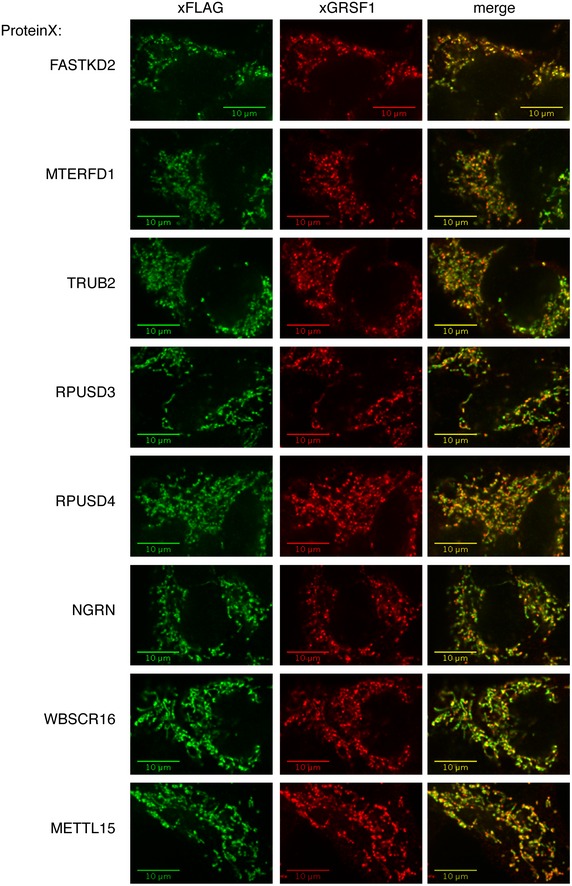
Flp‐In T‐REx 293 cells transduced with a ProteinX‐BirA*‐FLAG construct were treated with tetracycline for 24 h to induce expression of the protein, and the co‐localization of ProteinX (anti‐FLAG antibody) with a mtRNA granule protein (anti‐GRSF1) was visualized by immunofluorescence. All proteins in the pseudouridylation module co‐localized with GRSF1, albeit to various extents. Scale bars: 10 μm.
Depletion of the pseudouridine synthases produces specific defects in mitochondrial translation and assembly of the OXPHOS complexes
Mitochondrial tRNAs and rRNAs are modified at several positions, and these modifications include methylation, thiolation, formylation, taurine addition, and pseudouridylation 6, 21. Although some of the enzymes involved have been characterized, only one pseudouridine synthase, PUS1, which pseudouridylates both cytosolic and mitochondrial tRNAs 5, has been investigated to date; however, it was not detected in our studies as part of the module. To investigate the role of the three mitochondrial pseudouridine synthases identified in the pseudouridylation module, we used siRNA to deplete each of them individually in human 143B cells, a cell line that we used to characterize the composition of mitochondrial RNA granules 8 (Figs 2A and EV2A). The efficiency of suppression at the protein level was between 77 and 95%. No commercially available anti‐RPUSD3 antibody could detect a specific band on an immunoblot, so the efficiency of the knockdown (95%) was confirmed by qRT–PCR (Table EV2). Interestingly, depletion of MTERFD1, used as a positive control, led to a significant decrease in the levels of FASTKD2, TRUB2, RPUSD4, and NGRN (Fig EV2A), suggesting that it plays a role as a regulator of the stability of this protein module. Depletion of TRUB2 resulted in a 50% decrease in WBSCR16 isoform 1, and a small, but significant, decrease in FASTKD2. Depletion of each of the three pseudouridine synthases led to a decrease in the levels of MRPL11, a protein of the mt‐LSU, implicating the pseudouridine synthase module in mitochondrial translation and the biogenesis of the mitochondrial ribosome.
Figure 2. Mitochondrial pseudouridine synthases TRUB2, RPUSD3, and RPUSD4 are necessary for OXPHOS biogenesis.
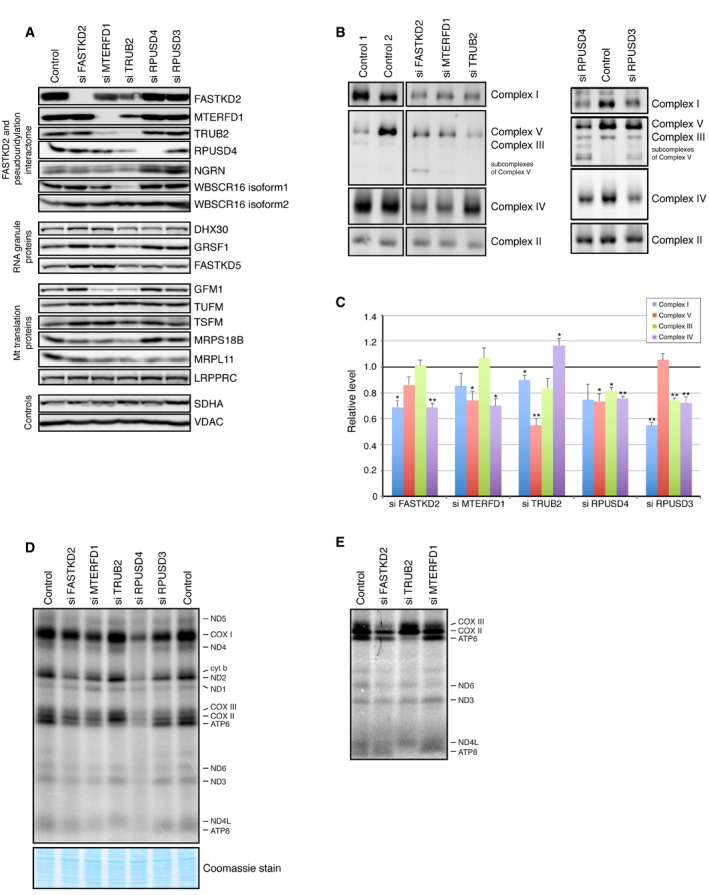
-
AImmunoblot analysis of indicated proteins in control and siRNA‐treated cells.
-
BBN–PAGE analysis of siRNA‐mediated depletion shows an OXPHOS defect as revealed by subunit‐specific antibodies against individual OXPHOS complexes.
-
CQuantification of the levels of individual OXPHOS complexes normalized to complex II levels. The graph represents the relative abundance of individual complexes in cells treated with the specified siRNA versus controls. The bars represent the mean ± SEM of 3–7 independent experiments. P‐values were calculated using paired two‐tailed t‐test (*P < 0.05; **P < 0.01).
-
D, EPulse‐labeling mitochondrial translation experiment of the 13 mitochondria‐encoded polypeptides (seven subunits of complex I [ND], three subunits of complex IV [COX], two subunits of complex V [ATP], and one subunit of complex III [cyt b]) in control and siRNA‐treated cells. (E) Detail of a mitochondrial pulse‐labeling experiment showing a severe decrease in the translation of ATP6 and ATP8 in TRUB2‐depleted cells.
Source data are available online for this figure.
Figure EV2. Quantification of the levels of pseudouridine synthase interacting proteins, the rate of mitochondrial translation, and the levels of 12S and 16S rRNA in siRNA‐treated cells.
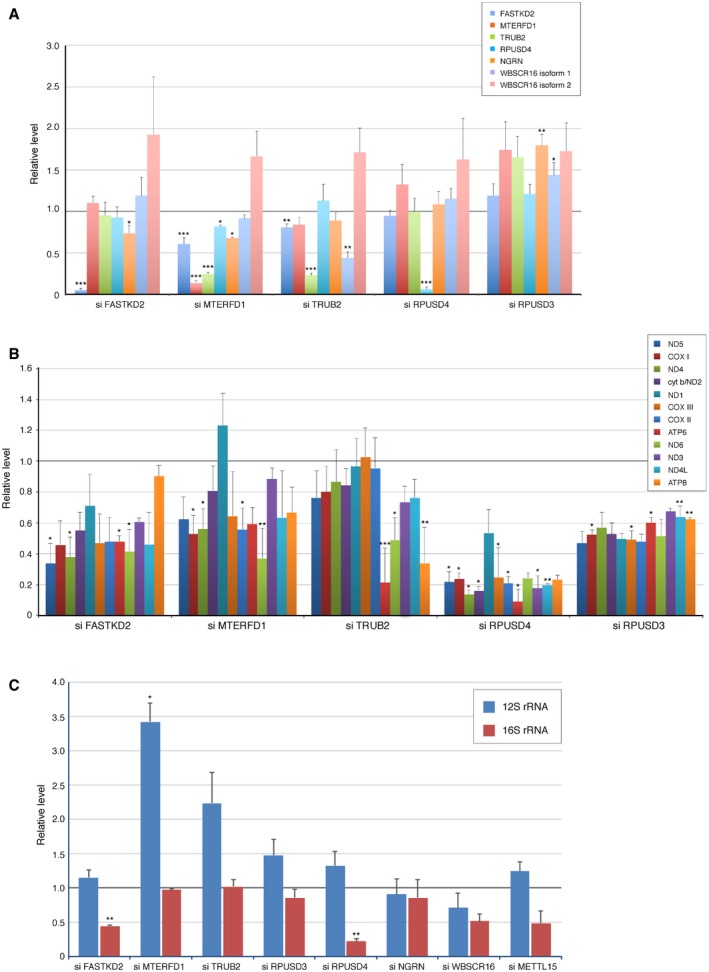
- Quantification of the levels of individual pseudouridine synthase interacting proteins normalized to SDHA in cells treated with siRNA. The graph represents the relative protein levels compared to controls. The bars represent mean ± SEM of 4–13 independent experiments.
- Quantification of the synthesis of individual mitochondria‐encoded polypeptides in siRNA‐treated cells. The graph represents the relative levels compared to controls. The bars represent mean ± SEM of 2–4 independent experiments.
- qRT–PCR analysis of 12S and 16S rRNA in siRNA‐treated cells. The graph represents the relative levels compared to controls. The bars represent mean ± SEM of 2–3 independent experiments.
Depletion of each of the pseudouridine synthases also resulted in combined oxidative phosphorylation (OXPHOS) assembly defects (Fig 2B and C) due to the decreased synthesis of the mtDNA‐encoded polypeptides (Figs 2D and EV2B). Mitochondrial protein synthesis, as measured by a pulse‐translation assay, was decreased to 76, 20, and 53% of control in cells in which TRUB2, RPUSD4, and RPUSD3 were depleted, but the nature of the defect differed in all three cases. The decrease in mitochondrial protein synthesis in TRUB2‐, RPUSD3‐, or RPUSD4‐depleted cells was not caused by a decrease in the levels of individual mRNAs (Table EV2); however, the level of the 16S rRNA was decreased to 22% of control in RPUSD4‐depleted cells (Table EV2), consistent with the severe global decrease in mitochondrial protein synthesis. In 143B cells, only depletion of RPUSD4 and FASTKD2 resulted in a significant decrease in 16S rRNA (Fig EV2C), in contrast to the results reported after Crispr/Cas9 knockdown in K562 cells 19. It is possible that the differences between the two studies simply reflect the fact that the studies were carried out in different cell types, but this will require further investigation. Remarkably, in TRUB2 knockdown cells, the synthesis of ATP6 and ATP8 proteins (encoded on a bicistronic transcript) was severely reduced (Figs 2E and EV2B), a pattern not seen previously in cells in which any component of the mitochondrial RNA metabolism or translation machinery was depleted or mutated.
Depletion of TRUB2 and RPUSD4 leads to defects in mitochondrial ribosome assembly
To investigate the effects of depleting the individual pseudouridine synthases on the assembly of the mitochondrial ribosome, we performed a sucrose gradient sedimentation analysis in both control and depleted cells. Nearly all immunodetectable TRUB2 and RPUSD4 co‐sedimented with the mt‐LSU (Fig 3A). Depletion of both TRUB2 and RPUSD4 led to a decrease in the assembly mt‐LSU, as well as the 55S monosome (Figs 3B and C, and EV3), with the most severe defect appearing in cells lacking RPUSD4. The assembly of the 28S small ribosomal subunit (mt‐SSU) was either not affected (siRPUSD4) or slightly increased (siTRUB2). In contrast, knockdown of RPUSD3 did not affect the assembly of the mitochondrial ribosome, suggesting that these enzymes have non‐overlapping functions in mitochondrial protein synthesis.
Figure 3. Interactions between the pseudouridine synthase module and the mitochondrial ribosome.
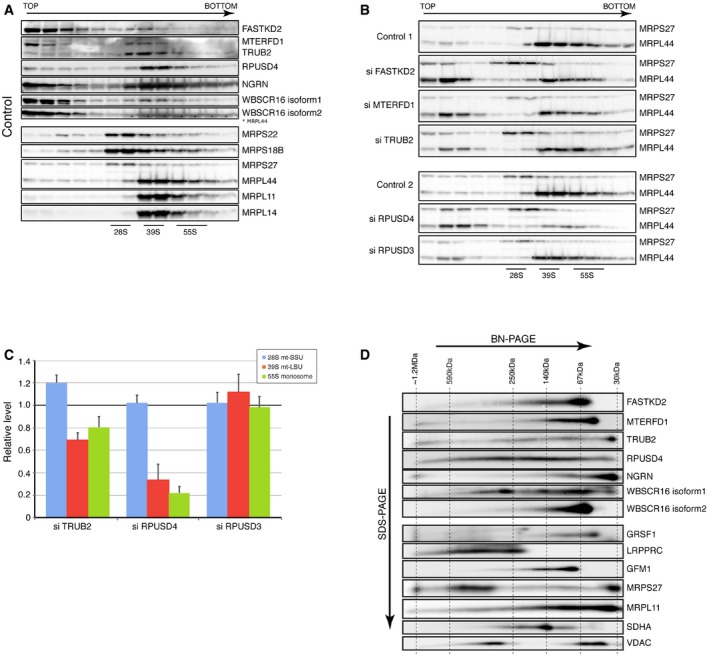
-
A, BIdentification of mitochondrial ribosomal proteins and pseudouridine synthase interacting proteins by sucrose gradient centrifugation in control cells (A) and cells treated with siRNA (B). Individual fractions were separated by SDS–PAGE and immunoblotted with the indicated antibodies. Panel control 2 in (B) is identical to the one shown in (A). The migration of the mt‐SSU (28S), the mt‐LSU (39S), and the mitochondrial monosome (55S) is shown.
-
CQuantification of the levels of the mt‐SSU (28S), the mt‐LSU (39S), and the mitochondrial monosome (55S) normalized to the SDHA levels. The graph represents the relative abundance of individual subunits in cells treated with specified siRNA versus controls. Quantification of the mt‐SSU, mt‐LSU, and monosome was done by averaging the intensity of the signals from three different antibodies directed against specific structural subunits of the ribosome. The bars represent the mean ± SEM.
-
D2D‐immunoblot analysis (BN–PAGE/SDS–PAGE) of pseudouridine synthase interacting proteins. The migration (sizes in kDa) of known protein complexes in the first dimension is indicated on the top of the blot.
Source data are available online for this figure.
Figure EV3. Assembly of the mitochondrial ribosome.
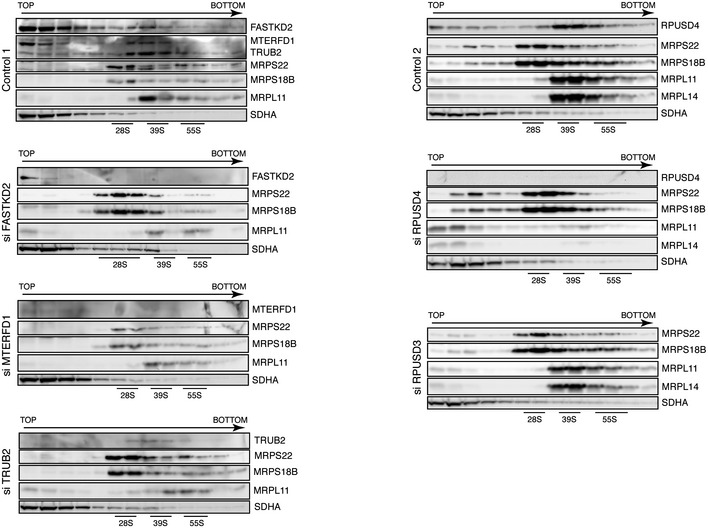
Depletion of the pseudouridine synthases TRUB2 and RPUSD4 leads to an impaired assembly of the mitochondrial ribosome. Individual fractions from a sucrose gradient centrifugation of mitochondria isolated from control and siRNA‐treated cells were separated by 12.5% SDS–PAGE and immunoblotted for the indicated mitochondrial ribosomal proteins and pseudouridine synthase interacting proteins. SDHA was used as a loading control. The migrations of the mt‐SSU (28S), the mt‐LSU (39S), and the mitochondrial monosome (55S) are indicated. Some of the panels in control 1 and control 2 are identical to those shown in Fig 3A in the main text.
To assess whether any of the pseudouridylation module proteins formed stable high molecular weight complexes, we performed a 2D‐gel analysis (Fig 3D). TRUB2, FASTKD2, MTERFD1, and NGRN migrated largely at their predicted monomeric molecular weights, while RPUSD4 and WBSCR16 were present predominantly in higher molecular weight complexes of different sizes. Consistent with our observation that UV cross‐linking was necessary to immunoprecipitate the proteins interacting with FASTKD2, these data demonstrate that the proteins in the pseudouridine synthase module do not form a stable multimeric complex, suggesting that their interactions are transient and facilitated by mitochondrial RNA.
Pseudouridine‐Seq (ψ‐Seq) identifies modified sites in mitochondrial RNA
To determine the specific sites modified by TRUB2, RPUSD3, and RPUSD4, we employed pseudouridine‐Seq (ψ‐Seq) on purified mitochondrial RNA (Fig 4A). This method utilizes an alkaline‐resistant modification of pseudouridine by N‐cyclohexyl‐N′‐(2‐morpholinoethyl)carbodiimide metho‐p‐toluenesulfonate (CMC), reported to cause a termination in reverse transcription at a position 3′ to ψ‐CMC 7, 22. In our experiments, CMC treatment caused pseudouridylated base skipping during reverse transcription, rather than a complete stop (Fig 4A), leading to a deletion of the base in the resulting sequencing reads (Fig EV4B). We computed the difference between the deletion rate in treated versus untreated samples (Δ) for all nucleotides in the mitochondrial genome. Pseudouridylated sites were determined using a cutoff of Δ > 2.5% in duplicate control samples (seven sites, Table EV3), which were further filtered to remove sequencing artifacts (Table EV3). We detected four high‐confidence sites (Figs 4B and EV4B): 16S rRNA (ψ3069), mt‐tRNA‐Leu (ψ3259), COXI (ψ6294), and COXIII (ψ9904–9906). The precise position for ψ in COXIII mRNA could not be determined due to the alignment ambiguity of three consecutive U's. Analysis of the motifs surrounding the ψ site in both the 16S rRNA and the mRNAs showed a similar consensus sequence (Fig EV5A), and in silico predictions of the secondary structure of the RNAs showed that all ψ sites are either in a loop, or in a stem adjacent to a loop, and thus likely to be accessible (Fig EV5B). We did not observe the presence of the previously suggested ψ in 12S rRNA or COXII mRNA 7. Neither the ψ11155 in ND4 mRNA (Δ = 1.9%) nor the ψ7658 in COXII mRNA (Δ = 2.2%) passed our cutoff; however, they remain potential, low‐level pseudouridine sites. We only detected one high‐confidence modification in mt‐tRNA, as our library preparation selected for products > 200 base pairs (mt‐tRNAs are mostly ~70 nt long). Nonetheless, another plausible ψ3286 site in mt‐tRNA‐Leu was detected, corresponding to a mt‐tRNA position 55, a site known to be pseudouridylated in mt‐tRNA‐Glu, mt‐tRNA‐Gln, mt‐tRNA‐Ser(UCN), and mt‐tRNA‐Tyr 6. Additional studies will be needed to verify the existence of this modification.
Figure 4. Mitochondrial ψ‐Seq analysis.
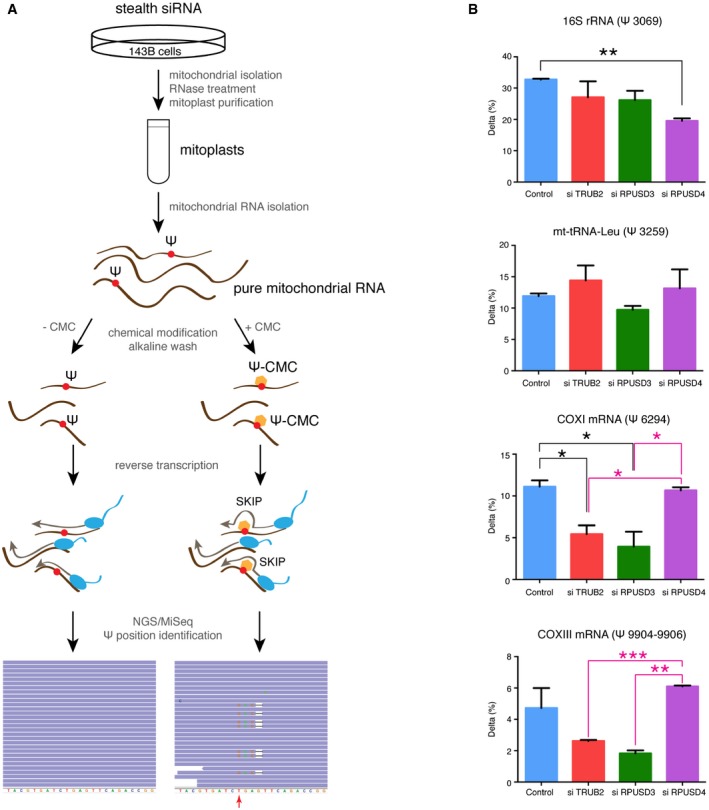
- Experimental approach used for the identification of pseudouridine sites in mitochondrial RNA. A combination of the CMC modification and the next‐generation sequencing on pure mitochondrial RNA from control and siRNA‐treated cells was used. CMC‐ψ modification causes skips in reverse transcription rather than stops, resulting in 1‐bp deletions in sequence alignments. The red arrow indicates a putative pseudouridine site.
- Quantification of Δ (the difference between the rate of deletions in CMC‐treated versus untreated cells) for individual ψ sites in control and siRNA‐treated cells. The bars represent the mean ± SEM of two independent experiments. P‐values were calculated using a one‐tailed t‐test (*P < 0.05; **P < 0.01; ***P < 0.001).
Figure EV4. Pseudouridine modification.
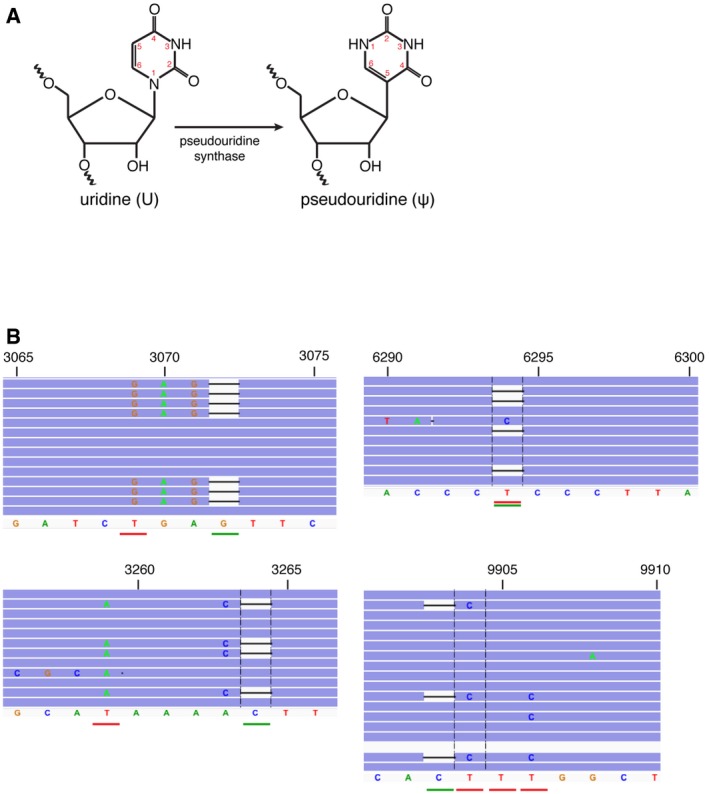
- A schematic of an enzymatic reaction of pseudouridine (ψ) synthesis by pseudouridine synthase.
- IGV screenshot of reads in CMC‐treated samples mapping to the top four pseudouridylated sites. The position of the deletion is underlined in green, and the putative pseudouridylated site is underlined in red.
Figure EV5. Primary and secondary structures of the pseudouridine sequence motif.
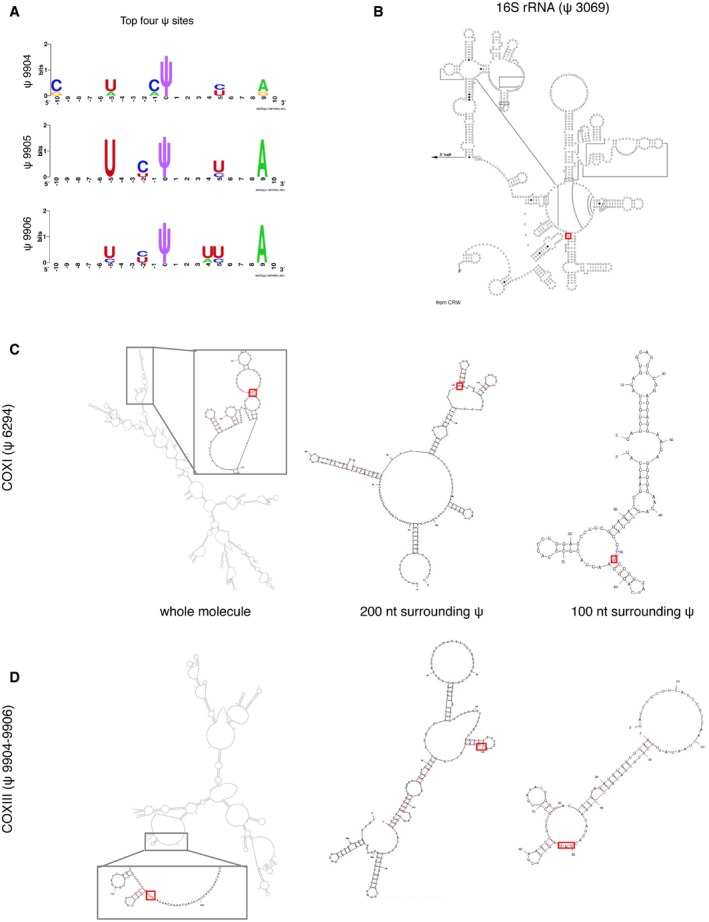
-
ASequence motifs surrounding the mitochondrial ψ sites generated by WebLogo software at http://weblogo.berkeley.edu. Three motifs are suggested for the top four ψ sites, because the COXIII (ψ 9904–9906) position is uncertain due to the ambiguity of the sequence alignments.
-
B–DPositions of identified ψ sites (red rectangle) within the secondary structures of individual RNAs. (B) Known secondary structure of the 3′‐terminus of 16S rRNA 37 and predicted structures for COXI (C) and COXIII mRNAs (D). Predictions were based either on sequence fragments of 100 nt and 200 nt surrounding each ψ site, or using the whole mRNA molecule as the input. Pseudouridine is predicted to be accessible in all cases, located at a loop or in the boundary between a stem and a loop/bulge structure.
ψ‐Seq analysis is not able to determine the absolute levels of pseudouridine in a single RNA. When a fully pseudouridylated probe was used in a ψ‐Seq experiment, only 43% of reads terminated 3′ to the ψ site 23. In our experiments, 32% of reads at the ψ3069 site in 16S rRNA contained a deletion, suggesting that a high percentage (likely all) of the mitochondrial 16S rRNA is pseudouridylated. The percentage of deletion‐containing reads was lower for the other identified ψ sites, implying the co‐existence of both the ψ‐ and U‐containing transcripts, and suggesting that the modification is likely to be dynamic.
Identification of the enzymes responsible for pseudouridylating mitochondrial RNAs
Pseudouridylation analysis in TRUB2‐, RPUSD3‐, and RPUSD4‐depleted cells showed the presence of pseudouridine modifications at the same positions as in control cells, but we detected clear differences in the level of pseudouridylation at some of these sites (Fig 4B). The levels of ψ3259 modification in mt‐tRNA‐Leu, which is modified by PUS1 5, were not different among the investigated samples, indicating that, as expected, neither TRUB2, RPUSD3, or RPUSD4 are involved in this modification. In RPUSD4‐depleted cells, the level of the 16S rRNA modification (ψ3069) was specifically reduced to 60% of control, while the levels of ψ modification at the other two sites in mRNAs were comparable to control. The ψ3069 modification was not affected in TRUB2 or RPUSD3 knockdown cells. These results strongly suggest that RPUSD4 is responsible for the pseudouridylation of the 16S rRNA, and that this modification is essential for the stability of the transcript, as the depletion of RPUSD4 causes a fourfold decrease in 16S rRNA levels, resulting in a severely reduced capacity to assemble the mt‐LSU, diminished synthesis of mtDNA‐polypeptides, and an OXPHOS assembly defect.
Depletion of both TRUB2 and RPUSD3 resulted in decreased pseudouridylation in the COXI (ψ6294) and COXIII (ψ9904–9906) mRNAs. Although the decrease in COXIII pseudouridylation was not significantly different from the control samples, the level of modification at this position was significantly reduced compared to RPUSD4‐depleted cells. We cannot exclude the possibility that both TRUB2 and RPUSD3 enzymes are modifying these mRNAs; however, three lines of evidence suggest that RPUSD3 is the major modifying enzyme for COXI and COXIII mRNA and that the effect of TRUB2 is secondary. First, depletion of RPUSD3 leads to a significant decrease in the synthesis of COXI and COXIII polypeptides (Figs 2D and EV2B), resulting in a substantial decrease in the assembly of complex IV (Fig 2B and C), demonstrating that mitochondrial mRNA pseudouridylation of these mRNAs is important for the efficient synthesis of these polypeptides (without affecting the stability of the mRNA). Second, depletion of TRUB2 resulted in a 50% decrease in RPUSD3 mRNA (Table EV2). Until an appropriate anti‐RPUSD3 antibody becomes available, RPUSD3 protein levels and the interdependence of these two proteins cannot be directly interrogated. Finally, depletion of TRUB2 leads primarily to a defect in the synthesis of ATP6 and ATP8 subunits, while complex IV is relatively spared (Fig 2D and E). The mechanism underlying translation suppression of the bicistronic ATP8/6 transcript by TRUB2 depletion will require further investigation.
In summary, our results show that the pseudouridine synthase module we identify is in fact more complex than the suggested 16S rRNA regulatory module 19. The BioID assay also identified additional components of the protein module (MTERFD1 and METTL15) that play a role in ribosome biogenesis and thus the control of mitochondrial protein synthesis. Although we have not uncovered the exact molecular function of FASTKD2, it is clearly involved in the post‐transcriptional handling of the 16S rRNA, and we have now placed this function in the much larger context of a post‐transcriptional regulatory module. Our data strongly suggest that RPUSD4, a core essential enzyme, pseudouridylates the mitochondrial 16S rRNA, and that this is required for its assembly into a functional mitochondrial ribosome. mRNA pseudouridine modifications, catalyzed by TRUB2/RPUSD3, modulate the efficiency of mitochondrial protein synthesis without apparent changes in transcript abundance or stability. These results establish clear roles for pseudouridine modifications in mitochondrial non‐coding and coding RNA that are essential for expression of the mitochondrial transcriptome and for cell viability.
Materials and Methods
Cell lines
Flp‐In T‐REx 293 and 143B cell lines were grown in high‐glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, at 37°C in an atmosphere of 5% CO2.
Antibodies
The following antibodies were used in this study: anti‐FASTKD2 (Proteintech Group 17464‐1‐AP), anti‐MTERFD1 (Sigma HPA002966), anti‐TRUB2 (Proteintech Group 19891‐1‐AP), anti‐RPUSD4 (Sigma HPA039689), anti‐NGRN (Proteintech Group 14885‐1‐AP), anti‐WBSCR16 (Proteintech Group 13796‐1‐AP), anti‐FLAG (Sigma F1804), anti‐GRSF1 (Sigma HPA036985), anti‐DHX30 (Abcam ab85687), anti‐FASTKD5 (Sigma SAB2700438), anti‐GFM1 (in house), anti‐TUFM/TSFM (a kind gift of Linda Spremulli), anti‐MRPS18B (Proteintech Group 16139‐1‐AP), anti‐MRPL11 (Cell Signalling #2066S), anti‐LRPPRC (in house), anti‐SDHA (Abcam ab14715), anti‐Porin (Abcam ab14734), anti‐NDUFA9 (Abcam ab14713), anti‐COX IV (Abcam ab110261), anti‐ATP5A1 (Abcam ab14748), anti‐UQCRC2 (Abcam ab14745), anti‐MRPS27 (Proteintech Group 17280‐1‐AP), anti‐MRPS22 (Proteintech Group 10984‐1‐AP), anti‐MRPL44 (Proteintech Group 16394‐1‐AP), anti‐MRPL14 (Sigma SAB4502786).
Affinity purification–mass spectroscopy (AP–MS) analysis
143B cells grown on plates (diameter 15 cm2) were washed in PBS and UV cross‐linked in a UV Stratalinker 1800 (Stratagene) for 10 min on ice. Mitochondria (400 μg) isolated from UV cross‐linked 143B cells were extracted in 200 μl of extraction buffer [50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1% taurodeoxycholate, and Complete protease inhibitors without EDTA (Roche)] on ice for 40 min, followed by a centrifugation at 9,000 g at 4°C for 20 min, and the supernatant was pre‐cleared overnight with non‐coated Dynabeads protein A (Invitrogen) to reduce non‐specific protein binding. Binding of FASTKD2 antibody to Dynabeads protein A (Invitrogen) was performed according to the manufacturer's instructions (version no. 004), with the exception that the incubation of the antibody with the beads was carried out overnight. The pre‐cleared extracts were used in the immunoprecipitation experiment with antibody cross‐linked beads and non‐coated beads (control‐IP). The immunoprecipitation reaction was performed overnight at 4°C. Samples were eluted using 0.1 M glycine (pH 2.5) supplemented with 1% dodecyl maltoside, TCA precipitated and analyzed on an Orbitrap (Thermo Scientific, Watlham, MA) at the Institute de Recherches Cliniques de Montreal.
Mitochondrial isolation
143B cells were resuspended in ice‐cold 250 mM sucrose/10 mM Tris–HCl (pH 7.4) and homogenized with seven passes in a pre‐chilled zero‐clearance homogenizer (Kimble/Kontes, Vineland, NJ). A post‐nuclear supernatant was obtained by centrifugation of the samples twice for 10 min at 600 g. Mitochondria were pelleted by centrifugation for 10 min at 10,000 g and washed once in the same buffer. Protein concentration was determined by Bradford assay.
BioID
BirA*‐FLAG constructs were generated using Gateway cloning into the pDEST5‐BirA*‐FLAG‐C‐ter vector. As starting clones, clones number BC001457 and BC001544 were used for cloning of TRUB2 and FASTKD2, respectively. Human cells [Flp‐In T‐REx 293 cells] were transfected using Lipofectamine 2000 (Invitrogen). Cells were seeded at 250,000 cells per well in a 6‐well plate in 2 ml DMEM supplemented with 10% FBS and 1% pen/strep (100 U/ml). The next day, cells were transfected with 200 ng pDEST5‐ProteinX‐BirA*‐FLAG, and 2 μg of pOG44 in 250 μl of 1× Opti‐MEM (Invitrogen) mixed with 5 μl of Lipofectamine 2000 reagent in 250 μl of 1× Opti‐Mem. The Opti‐MEM/Lipofectamine solution was added to the Opti‐MEM/plasmid solution and incubated 20 min before addition to the cells (in medium without antibiotics). The medium was changed 4 h after transfection. The next day, transfected cells were passaged into 10 cm2 plates, and the following day selected by the addition of hygromycin (Calbiochem) at a final concentration of 200 μg/ml. This selection media was changed every 2–3 days until clear visible colonies were present. Up to six colonies per construct were picked and expanded, and the localization and the expression level of the construct were assessed by immunofluorescence using an anti‐FLAG antibody. The selected clones were scaled up to six 15 cm2 plates for treatment and harvesting. Cells were grown to 70% confluency before induction of protein expression using 1 μg/ml tetracycline (Sigma), and media supplementation with 50 μM biotin for protein labeling. Cells were harvested 24 h later as follows: Cell medium was decanted, cells were washed twice with 5 ml of PBS per 15 cm2 plate and then harvested by scraping in 5 ml of PBS. Cells from 3 × 15 cm2 plates were pelleted at 800 rpm for 3 min, PBS aspirated and pellets transferred to a −80°C freezer. Purification of biotinylated proteins followed by their identification by mass spectrometry was performed as described previously 24, and data were compared to negative controls expressing the BirA* tag alone, the BirA* tag fused to GFP, and untransfected cells. The prey specificity module of the ProHits‐viz software (http://prohitsviz.lunenfeld.ca) was used to analyze the TRUB2 BioID data, using 42 mitochondrial baits and 96 baits from other cellular compartments as controls.
Immunocytochemistry
Flp‐In T‐REx 293 cells expressing ProteinX‐BirA‐FLAG constructs grown on coverslips were induced using 1 μg/ml tetracycline (Sigma) for 24 h, fixed with 4% formaldehyde solution, solubilized by Triton X‐100, and incubated with the anti‐FLAG and anti‐GRSF1 antibodies. The appropriate anti‐species secondary antibodies coupled with Alexa fluorochromes (Invitrogen) were subsequently used at a dilution of 1:1,000. Cells were imaged with an Olympus IX83 microscope connected to a Yokogawa CSU‐X confocal scanning unit. Images were analyzed using ImageJ software Fiji 25.
siRNA transfection
Stealth RNA interference duplex constructs (Invitrogen) were used for transient knockdown of TRUB2 (HSS120266), RPUSD3 (HSS138721+HSS138722), RPUSD4 (HSS131380), MTERFD1 (HSS121476), and FASTKD2 (HSS176985) in 143B cells. Stealth siRNA duplexes were transiently transfected into cells using Lipofectamine RNAiMAX (Invitrogen), according to the manufacturer's specifications. The transfection was repeated on day 3, and the cells were harvested on day 6 for analysis.
Denaturing, native, and two‐dimensional PAGE
SDS–PAGE was used to separate isolated mitochondrial extracts and fractions from sucrose gradient sedimentation. In general, isolated mitochondria were extracted with 1.5% dodecyl maltoside/PBS after which 20 μg of protein were run on either 10 or 12.5% polyacrylamide gels.
Blue‐native PAGE (BN–PAGE) was used to separate individual OXPHOS complexes. Isolated mitochondria were solubilized with 1% dodecyl maltoside and centrifuged for 20 min at 20,000 g. 10–20 μg of supernatant was run in the first dimension on 6–15% polyacrylamide gradient gels as previously described 26. For the second dimension analysis, BN–PAGE/SDS–PAGE was carried out as detailed previously 27. Separated proteins were transferred to a nitrocellulose membrane using a semi‐dry system (GE Healthcare Life Sciences), and immunoblot analysis was performed with the indicated antibodies.
Mitochondrial translation assay
Pulse labeling of mitochondrial translation products in 143B control and siRNA‐treated cells was performed with 200 μCi/ml of a [35S]‐methionine/cysteine mix (Perkin Elmer Health Sciences), in DMEM lacking methionine and cysteine and containing 100 μg/ml of the cytoplasmic translation inhibitor emetine, for 60 min as described in detail elsewhere 28. The gel was stained with Bio‐Safe Coomassie stain (Bio‐Rad), dried, and exposed to a phosphorimager cassette.
qRT–PCR
Total RNA was isolated from control and siRNA‐treated cells using miRNeasy kit (Qiagen). qRT–PCR analysis for TRUB2, RPUSD3, and RPUSD4 mRNAs, as well as all mitochondrial transcripts, was performed at the Institute for Research In Immunology and Cancer (IRIC).
Sucrose density gradient sedimentation
Mitochondria from control and siRNA‐treated cells (400 μg) were lysed in lysis buffer (260 mM sucrose, 100 mM KCl, 20 mM MgCl2, 10 mM Tris–Cl pH 7.5, 1% Triton X‐100, 5 mM β‐mercaptoethanol, protease inhibitor cocktail without EDTA (Roche)) on ice for 20 min, centrifuged at 9,400 g for 45 min at 4°C and subsequently loaded on a 1 ml 10–30% discontinuous sucrose gradient (50 mM Tris–Cl, 100 mM KCl, 10 mM MgCl2), and centrifuged at 32,000 rpm (130,000 g) for 130 min at 4°C in a Beckman SW60‐Ti rotor. After centrifugation, 13 fractions were collected from the top and used for SDS–PAGE analysis. Quantification of the mt‐SSU, mt‐LSU, and monosome was done by averaging the intensity of the signals from three different antibodies directed against specific structural subunits of the ribosome.
Mitoplast preparation and isolation of pure mitochondrial RNA
Pure mitochondrial RNA was prepared as described previously 29. Freshly prepared mitochondria from control and siRNA‐treated cells (from ten 15 cm2 plates per sample) were pelleted at 15,000 g for 10 min at 4°C and resuspended in 100 μl STE buffer (250 mM sucrose, 10 mM Tris–HCl (pH 7.4), 1 mM EDTA, 1× protein inhibitors (Roche)). RNase A (10 mg/ml) was added to a final concentration of 100 μg/ml and incubated on ice for 30 min to digest the contaminating cytoplasmic RNA. The mitochondrial suspension was layered on a discontinuous sucrose gradient (1.7 M and 1 M in 10 mM Tris–HCl (pH 7.6), 1 mM EDTA) and centrifuged at 70,000 g for 40 min at 4°C to isolate pure mitochondria. The interphase between the two sucrose cushions was collected by a Pasteur pipette (~400 μl) and washed twice in 1 ml STE and centrifuged at 15,000 g for 10 min at 4°C. To prepare mitoplasts, the pure mitochondrial fraction was resuspended in 25 μl of STE/BSA (STE supplemented with 0.1% (w/v) fat‐free BSA) and stirred on ice. 250 μl of digitonin (6.25 mg/ml in STE/BSA) was added dropwise over ~1–2 min, and the solution was stirred on ice for total 15 min. 2 ml STE/BSA was added, and the suspension was homogenized by four strokes of a zero‐clearance homogenizer, centrifuged at 15,000 g for 10 min at 4°C. Mitoplasts were washed in STE and centrifuged at 15,000 g for 10 min at 4°C. Pure mitochondrial RNA was isolated from mitoplasts using the miRNeasy kit (Qiagen). RNA quality and quantity were determined using a Bioanalyzer 2100 (Agilent) and NanoDrop (Thermoscientific) at the McGill University and Génome Québec Innovation Centre.
CMC modification of pure mitochondrial RNA
The CMC (N‐cyclohexyl‐N′‐(2‐morpholinoethyl)carbodiimide metho‐p‐toluenesulfonate; Santa Cruz Biotechnology) modification was performed essentially as described previously 30 with minor adjustments. 1.5 μg (in 30 μl) of pure mitochondrial RNA was divided into two samples: 18 μl (+CMC sample) and 12 μl (−CMC sample), and the volume of both samples was brought up to 20 μl. 2.9 μl of 40 mM EDTA (pH 8.0) was added, and the samples were denatured at 80°C for 3 min and placed on ice. Fresh CMC (0.4 M) was prepared in BEU buffer (50 mM bicine pH 8.5, 4 mM EDTA, 7 M urea). 100 μl of CMC or 100 μl BEU buffer was added to +CMC and −CMC samples, respectively. Samples were incubated at 40°C for 45 min in Thermomixer (Eppendorf) at 1,000 rpm. RNA was precipitated by the addition of 2 μl GlycoBlue (Invitrogen), 50 μl 3 M sodium acetate (pH 5.2), and 1 ml ice‐cold 100% ethanol at −80°C for a minimum of 30 min and centrifuged at 20,000 g for 30 min at 4°C. Precipitated RNA was washed twice with 500 μl ice‐cold 70% ethanol, centrifuged at 20,000 g for 10 min at 4°C, and air‐dried for ~2 min. To reverse the binding of CMC to U and G residues, RNA was resuspended in 30 μl sodium carbonate buffer (50 mM sodium carbonate (pH 10.4), 2 mM EDTA) and incubated at 50°C for 2 h in a Thermomixer at 1,000 rpm. RNA was precipitated by addition of 2 μl GlycoBlue (Invitrogen), 3.5 μl 3 M sodium acetate (pH 5.2), and 90 μl ice‐cold 100% ethanol at −80°C for minimum 30 min and centrifuged at 20,000 g for 30 min at 4°C. Precipitated RNA was washed twice with 500 μl ice‐cold 70% ethanol, centrifuged at 20,000 g for 10 min at 4°C, air‐dried for ~2 min, and resuspended in 12 μl of water. RNA quality and quantity were determined using the Bioanalyzer 2100 and NanoDrop.
RNASeq
RNA quality assessment, RNASeq library preparation (Kapa Stranded RNA‐Seq; Illumina), and MiSeq 150 nt pair‐end sequencing were performed at the McGill University and Génome Québec Innovation Centre.
RNA‐sequencing analysis
Sequencing runs were processed with Illumina CASAVA software. Trimmomatic v0.32 31 was used to trim reads, including removal of low‐quality bases at the end of reads (phred33<30), clipping of the first three bases and clipping of Illumina adaptor sequences using the palindrome mode. We performed quality trimming with a sliding window, cutting once the average quality of a window of four bases fell below 30. We discarded reads shorter than 30 base pairs after trimming. Trimmed reads were aligned to the reference genome hg19 using STAR v2.3.0e 32. Quality control was performed using metrics obtained with FASTQC v0.11.4, SAMtools 33, BEDtools 34, and custom scripts. Bigwig tracks were produced with custom scripts, using BEDtools 34 and UCSC tools. Data were visualized using the Integrative Genomics Viewer (IGV) 35.
Pseudouridylation quantification
CMC treatment caused skips of pseudouridylated bases during reverse transcription (RT), leading to a deletion of the base in the resulting sequencing reads (Fig EV4B), rather than complete stops of the RT described in previous studies 7, 22. In order to identify putative pseudouridylation sites, we used SAMtools 33 to summarize base‐calls of aligned reads to each position of the mitochondrial genome. Number of deletions at each position normalized by total read depth was obtained for each site using custom scripts, and the final score computed was defined as
where and are the number of deletions and total number of reads observed at position i, respectively, with or without treatment. The score was computed for all sites in the genome (not limited to U's), since ambiguities in the alignment step can result in deletions observed a few base pairs away from the actual pseudouridylation site (Fig EV4B). A cutoff threshold of Δ > 2.5% in both control samples was used to identify pseudouridylated sites. Resulting potential candidate sites were individually examined using IGV to resolve alignment ambiguities.
Sequence motif and secondary structure analyses
Sequence motifs surrounding the pseudouridylated sites were obtained using WebLogo (weblogo.berkeley.edu). Secondary structures of the regions surrounding the ψ sites in COXI and COXIII mRNAs were predicted using the Mfold web server 36. Regions of 100, 200 nucleotides surrounding each of the ψ sites or the complete mRNA molecule were used for predictions.
Statistical analysis
Data are presented as the mean ± SEM. Statistical analysis was performed using either Graph Pad or Microsoft Excel software. P‐values were calculated using a t‐test as indicated in figure legends.
Author contributions
HA designed and performed the experiments and co‐wrote the manuscript, KC and CLK analyzed and interpreted the pseudouridine‐Seq data, and Z‐YL and A‐CG performed the BioID experiments and helped analyze the data. EAS helped with experimental design, supervised the project, and co‐wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
Table EV2
Table EV3
Review Process File
Source Data for Figure 2
Source Data for Figure 3
Acknowledgements
This work was supported by grants from the CIHR (MT‐15460) and UMDF to EAS, and a grant from the CIHR (FDN 143301) to A‐CG. CLK is a Chercheur Boursier of the FRQS (Fonds de Québec‐Santé), and KC is supported by a doctoral fellowship from the FRQS. We thank Kathleen Daigneault for technical assistance, James D.R. Knight and Christopher Go for assistance with analysis of the BioID data.
EMBO Reports (2017) 18: 28–38
References
- 1. Gilbert WV, Bell TA, Schaening C (2016) Messenger RNA modifications: form, distribution, and function. Science 352: 1408–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liu N, Pan T (2016) N6‐methyladenosine‐encoded epitranscriptomics. Nat Struct Mol Biol 23: 98–102 [DOI] [PubMed] [Google Scholar]
- 3. Li X, Ma S, Yi C (2016) Pseudouridine: the fifth RNA nucleotide with renewed interests. Curr Opin Chem Biol 33: 108–116 [DOI] [PubMed] [Google Scholar]
- 4. Ofengand J, Bakin A (1997) Mapping to nucleotide resolution of pseudouridine residues in large subunit ribosomal RNAs from representative eukaryotes, prokaryotes, archaebacteria, mitochondria and chloroplasts. J Mol Biol 266: 246–268 [DOI] [PubMed] [Google Scholar]
- 5. Patton JR, Bykhovskaya Y, Mengesha E, Bertolotto C, Fischel‐Ghodsian N (2005) Mitochondrial myopathy and sideroblastic anemia (MLASA): missense mutation in the pseudouridine synthase 1 (PUS1) gene is associated with the loss of tRNA pseudouridylation. J Biol Chem 280: 19823–19828 [DOI] [PubMed] [Google Scholar]
- 6. Suzuki T, Suzuki T (2014) A complete landscape of post‐transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res 42: 7346–7357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carlile TM, Rojas‐Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV (2014) Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515: 143–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Antonicka H, Sasarman F, Nishimura T, Paupe V, Shoubridge EA (2013) The mitochondrial RNA‐binding protein GRSF1 localizes to RNA granules and is required for posttranscriptional mitochondrial gene expression. Cell Metab 17: 386–398 [DOI] [PubMed] [Google Scholar]
- 9. Antonicka H, Shoubridge EA (2015) Mitochondrial RNA granules are centers for posttranscriptional RNA processing and ribosome biogenesis. Cell Rep 10: 920–932 [DOI] [PubMed] [Google Scholar]
- 10. Jourdain AA, Koppen M, Wydro M, Rodley CD, Lightowlers RN, Chrzanowska‐Lightowlers ZM, Martinou JC (2013) GRSF1 regulates RNA processing in mitochondrial RNA granules. Cell Metab 17: 399–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tu YT, Barrientos A (2015) The human mitochondrial DEAD‐Box protein DDX28 resides in RNA granules and functions in mitoribosome assembly. Cell Rep 10: 854–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Powell CA, Nicholls TJ, Minczuk M (2015) Nuclear‐encoded factors involved in post‐transcriptional processing and modification of mitochondrial tRNAs in human disease. Front Genet 6: 79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ghezzi D, Saada A, D'Adamo P, Fernandez‐Vizarra E, Gasparini P, Tiranti V, Elpeleg O, Zeviani M (2008) FASTKD2 nonsense mutation in an infantile mitochondrial encephalomyopathy associated with cytochrome c oxidase deficiency. Am J Hum Genet 83: 415–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Popow J, Alleaume AM, Curk T, Schwarzl T, Sauer S, Hentze MW (2015) FASTKD2 is an RNA‐binding protein required for mitochondrial RNA processing and translation. RNA 21: 1873–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Roux KJ, Kim DI, Raida M, Burke B (2012) A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 196: 801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wredenberg A, Lagouge M, Bratic A, Metodiev MD, Spahr H, Mourier A, Freyer C, Ruzzenente B, Tain L, Gronke S et al (2012) MTERF3 regulates mitochondrial ribosome biogenesis in invertebrates and mammals. PLoS Genet 9: e1003178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Calvo SE, Clauser KR, Mootha VK (2016) MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res 44: D1251–D1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, Mis M, Zimmermann M, Fradet‐Turcotte A, Sun S et al (2015) High‐resolution CRISPR screens reveal fitness genes and genotype‐specific cancer liabilities. Cell 163: 1515–1526 [DOI] [PubMed] [Google Scholar]
- 19. Arroyo JD, Jourdain AA, Calvo SE, Ballarano CA, Doench JG, Root DE, Mootha VK (2016) A genome‐wide CRISPR death screen identifies genes essential for oxidative phosphorylation. Cell Metab 24: 875–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kimura S, Suzuki T (2010) Fine‐tuning of the ribosomal decoding center by conserved methyl‐modifications in the Escherichia coli 16S rRNA. Nucleic Acids Res 38: 1341–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rorbach J, Minczuk M (2012) The post‐transcriptional life of mammalian mitochondrial RNA. Biochem J 444: 357–373 [DOI] [PubMed] [Google Scholar]
- 22. Bakin A, Ofengand J (1993) Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry 32: 9754–9762 [DOI] [PubMed] [Google Scholar]
- 23. Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, Leon‐Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES et al (2014) Transcriptome‐wide mapping reveals widespread dynamic‐regulated pseudouridylation of ncRNA and mRNA. Cell 159: 148–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Couzens AL, Knight JD, Kean MJ, Teo G, Weiss A, Dunham WH, Lin ZY, Bagshaw RD, Sicheri F, Pawson T et al (2013) Protein interaction network of the mammalian Hippo pathway reveals mechanisms of kinase‐phosphatase interactions. Sci Signal 6: rs15 [DOI] [PubMed] [Google Scholar]
- 25. Schindelin J, Arganda‐Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al (2012) Fiji: an open‐source platform for biological‐image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leary SC, Sasarman F (2009) Oxidative phosphorylation: synthesis of mitochondrially encoded proteins and assembly of individual structural subunits into functional holoenzyme complexes. Methods Mol Biol 554: 143–162 [DOI] [PubMed] [Google Scholar]
- 27. Antonicka H, Ogilvie I, Taivassalo T, Anitori RP, Haller RG, Vissing J, Kennaway NG, Shoubridge EA (2003) Identification and characterization of a common set of complex I assembly intermediates in mitochondria from patients with complex I deficiency. J Biol Chem 278: 43081–43088 [DOI] [PubMed] [Google Scholar]
- 28. Sasarman F, Shoubridge EA (2012) Radioactive labeling of mitochondrial translation products in cultured cells. Methods Mol Biol 837: 207–217 [DOI] [PubMed] [Google Scholar]
- 29. Rackham O, Filipovska A (2014) Analysis of the human mitochondrial transcriptome using directional deep sequencing and parallel analysis of RNA ends. Methods Mol Biol 1125: 263–275 [DOI] [PubMed] [Google Scholar]
- 30. Carlile TM, Rojas‐Duran MF, Gilbert WV (2015) Pseudo‐seq: genome‐wide detection of pseudouridine modifications in RNA. Methods Enzymol 560: 219–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA‐seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thorvaldsdottir H, Robinson JT, Mesirov JP (2013) Integrative Genomics Viewer (IGV): high‐performance genomics data visualization and exploration. Brief Bioinform 14: 178–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM et al (2002) The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3: 2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Table EV1
Table EV2
Table EV3
Review Process File
Source Data for Figure 2
Source Data for Figure 3


