Abstract
DNA Customization of nutraceutical products is here. In the truest sense, “Gene Guided Precision Nutrition™” and KB220 variants (a complex mixture of amino–acids, trace metals, and herbals) are the pioneers and standard-bearers for a state of the art DNA customization. Findings by both, Kenneth Blum, Ph.D. and Ernest Noble, Ph.D. concerning the role of genes in shaping cravings and pleasure- seeking, opened the doors to comprehension of how genetics control our actions and effect our mental and physical health. Moreover, technology that is related to KB220 variants in order to reduce or eradicate excessive cravings by influencing gene expression is a cornerstone in the pioneering of the practical applications of nutrigenomics. Continuing discoveries have been an important catalyst for the evolution, expansion, and scientific recognition of the significance of nutrigenomics and its remarkable contributions to human health. Neuro-Nutrigenomics is now a very important field of scientific investigation that offers great promise to improving the human condition. In the forefront is the development of the Genetic Addiction Risk Score (GARS™), which unlike 23andMe, has predictive value for the severity of drug and alcohol abuse as well as other non-substance related addictive behaviors. While customization of neuronutrients has not yet been commercialized, there is emerging evidence that in the future, the concept will be developed and could have a significant impact in addiction medicine.
Keywords: DNA Customization, Nutraceuticals, Pro-Dopamine regulation (KB220 variants) Reward Deficiency Solution Sustem, GARS
INTRODUCTION
We are currently in a genomics era that promotes future medical advancements, particularly in the field of psychiatry. The comprehension of DNA and polymorphic associations with the brain reward circuitry has paved an innovative way in how we approach and think of addictive behaviors. As authors the basis of this article presents, a review of both neurogenetics and nutrigenomics related to addiction medicine. The goal is to provide the foundations of more accurate genetic diagnoses and the use of Dopamine agonist therapy (pro-dopamine regulation) to balance dopaminergic activation. As a result of several experiments on the nature of addiction, we are proposing a novel approach, challenging the recovery field to utilize such tools, and introduce them into inpatient/outpatient treatment programs in addiction clinics. The following are significant tools in changing the recovery landscape: The Genetic Addiction Risk Score (GARS™) for Reward Deficiency Syndrome (RDS) diagnoses; the Comprehensive Analysis Of Reported Drugs (CARD™) to establish agreement and abstinence during treatment; natural Dopamine agonistic therapy (KB220™); and ultimately, mRNA (patent pending) to resolve pre- and post-candidate gene expressions in Reward Deficiency Syndrome (RDS). Therefore, the proposal of this paradigm shift is appropriately named: “Reward Deficiency Solutions System (RDSS™)” [1].
Have you ever wondered why so many Americans and individuals across the globe are falling victim to the chain of addictive behaviors and becoming part of the worst epidemic in the history of the word?
The answer is, in part, both genetic and environmental (epigenetic). Recently, researchers found that epigenetic effects on the chromatin structure of our DNA are a legacy that passes on from generation to generation.
Scientists such as Stephen Hawkins suggest that we are made up of self–assembled molecules generated over 14 billion years. More interesting, is that, we as Homo sapiens, differ in our DNA by only 0.5% [2]. Recent evidence shows that each human has an average of 60 new mutations when compared to their parents. Even more remarkable, is that the human brain contains billions of neurons working in concert to provide us the gift of “well- being:” free of mental disease and stress. The number of neurons in the brain varies dramatically from species to species. One estimate (published in 2012) puts the human brain at about 85 billion neurons and approximately 85 trillion synapses. It turns out that 20% of our entire body’s energy is budgeted to keep our brain working normally. The differences between individual humans are 4.25 billion neurons and 4.25 trillion synapses that make us unique.
This known difference affects the 7.4 billion humans that roam earth, working and living together to achieve some degree of productivity and happiness. However, as the world turns, 21st century humans are faced daily with reminders of terrorism and horrific diseases that arise because these genetic and epigenetic differences lead to fatalities, not just from cancer, but from mental impairments that influence billions of neurons and trillions of synapses. This molecular rearrangement of our genome makes each of us unique. For example, how dopamine functions in our reward system may also be unique. One example, among other gene variations involved in brain reward, is that genetic differences account for the presence of Attention Deficit/Hyperactivity Disorder (ADHD), a subtype of Reward Deficiency Syndrome (RDS), in approximately 8–12 % of children in the United States and 4% of adults worldwide [3]. You may also be surprised to know that, at birth, an estimated 100,000 million people in the United States carry a gene form (allele) of just one genetic variation that involves brain dopamine D2 receptors. It is known that carriers of the allele DRD2 Taq A1 have 30–40% lower D2 receptors in the brain [4]. So what does this mean regarding our romance with getting high - “turning on” and “turning off” with potent psychoactive drugs (e.g., alcohol, cocaine, and opiates) and resultant addiction and fatalities seen in children?
In 1990, the first association of a variant (A1) on the dopamine D2 receptor gene (DRD2) and severe alcoholism was discovered and published by Blum et al. in JAMA. Later experiments showed that individuals who carry this variant have 30–40 % lower dopamine receptors than DRD2 A2 carriers [4]. Simply, being born with this single gene variation (DRD2 A1 form) known to cause low dopamine D2 receptors, sets an individual up to have a high addiction risk (vulnerability) to any substance or behavior that stimulates the neuronal release of dopamine. In fact, in 1996, Blum’s laboratory used a mathematical model (Bayesian Theorem) to predict that an individual born with the A1 allele (variant) has a 74.4% risk of developing a RDS behavior: addiction [5]. Individuals with this allele will have an initial acute response to using a psychoactive drug or experiencing pathological gambling, or whatever behavior stimulates enough neuronal dopamine for them to feel normal, possibly for the first time. Unfortunately, chronic consumption/experiences lead to epigenetic changes that further reduce dopamine receptor numbers, and a stronger need to abuse can lead to unwanted uncontrollable behaviors, and even narcotic overdose followed by death.
How were the genes involved in reward found? The chemical messengers (neurotransmitters) in the brain are like keys that turn on various functions of genes. The neurotransmitters that participate in evoking pleasurable feelings in the reward circuitry, work in a cascading fashion throughout the brain. These interactions (Brain Reward Cascade) may be viewed as activities of subsystems within a greater system, occurring simultaneously or in sequence, merging in cascade fashion toward a particular effect [6]. The goal is the generation of feelings of well being by the eventual release of just the right amount of dopamine at the reward site. In this scenario, there are at least seven major neurotransmitters and their pathways that are involved: serotonin, cannabis, endorphin (enkephalin), GABA, glutamine, acetylcholine, and dopamine. There are thousands of published studies about these reward genes and pathways that influence the function of these named neurotransmitters [7]. This research involved the identification of gene (DNA) variations or alleles that individuals are born with and epigenetic (environmental RNA) changes that may alter the healthy, intended function of DNA.
Dysfunctional DNA is due to what is referred to as single nucleotide polymorphisms, frequently called SNPs (pronounced “snips”). SNPs are the most frequent kind of genetic variation among individuals. Each SNP signifies a variance in a single DNA building block, or nucleotide. For example, a SNP may substitute the nucleotide cytosine (C) with the nucleotide thymine (T) in a specific strand of DNA. SNPs typically occur throughout one’s DNA. They occur once in every 300 nucleotides on average, which translates to approximately 10 million SNPs in the entire human genome [8]. Usually, these variations can be located in the DNA between genes. They can behave as biological markers, assisting scientists to find genes that are linked to disease. When SNPs occur within a gene or near a gene (in a regulatory region), they alter the gene’s function. If these SNPs are found in the Brain Reward Cascade’s [BRC] set of genes [6], the neurotransmission will be dysfunctional, resulting in a loss of dopamine regulation or balance (homeostasis). Too little dopamine will, at birth, predispose individuals to “want” or “like” psychoactive drugs or even, behaviors like hypersexuality and gambling. Compromised DNA with risk variations (alleles) can predispose them to become victim to the chain of addictive behaviors [9].
Following 25 years of extensive research from many scientists worldwide, a panel of ten reward gene risk variants called the Genetic Addiction Risk Score (GARS) has been developed. In unpublished work, when GARS was compared to Addiction Severity Index (ASI) used in many clinical settings, it was found to significantly predict the severity of both alcohol and drug dependency [10].
In support of early testing for addiction and other RDS subtypes, parents caught up in today’s horrific demographic of 127 people, both young and old, dying from opiate/opioid overdose every day in America, need help. Families in the past would have never guessed that their loved ones would die or could be in real danger due to opiate addiction. Author Bill Moyers published an article in Parade Magazine, where he reported that as he traveled around the United States, he found too many children with ADHD, and that many of those children had subsequent issues like substance abuse. He emphatically called for better ways to identify these children and treat them with approaches other than addictive pharmaceuticals.
When the GARS test becomes available and is approved by the FDA, especially for parents to test their children, and without such approval for clinicians to test their patients, the field will be able to assess the vulnerability of patient chemical dependency and more importantly, assess their children of RDS behaviors like addiction, ADHD, and autism spectrum disorders. The common thread across all these risk gene variants is that they lead to a low dopamine (hypodopaminergic) function or deficit. There are arguments against testing, such as the fear of labeling and knowing the risk. The real issue or challenge, however, is what can be done if risk alleles are found? It is understandable that when there is one gene–one disease (OGOD) involved such as in Huntington’s disease, when treatment is unavailable, and prevention remains a problem, why even know the risk? [11].
Have we found a safe non-addictive solution that will provide the brain with a means to balance the neurotransmitters involved in the BRC, culminating in true dopamine homeostasis? In spite of variant genes and epigenetic, environmental insults, holistic approaches like mindfulness, exercise, spirituality, and particularly, amino acid therapy (KB220 formulations), have been shown to reduce relapse and increase brain dopamine homeostasis [12]. We are suggesting that not only should clinicians in the near future be able to genetically test our children for unwanted reward gene risk variants that predispose them to dopamine deficiency and the lack of reward and risk for drug and non-drug addiction, but also possibly even prevent RDS behaviors. It is possible that genetic risk for substance abuse and other RDS behaviors can be identified by the GARS test and explain why some individuals are vulnerable and others not. With continued research, genetic and epigenetic dopamine deficiencies can be treated, relapse reduced, and addicts can free themselves from the clutches of powerful addictive behaviors and bring balance and happiness to their lives. This laudable goal requires intensive research.
There are hundreds of mutations and several thousand permutations and combinations of these mutations which results in for example, diabetes is often remembered and treated for the ‘raise in blood glucose level’ beyond certain arbitrary values accepted by one or the other professional bodies. Raise in blood sugar is in fact only one of the several ‘signs’ of DMT2. This is the way we manage most of the diseases in ‘modern medicine’ with certain notable exceptions, for example the use of antibiotics against certain infections. Another notable example from this decade is the use of’ imatinib’® an example of a designer drug used in the treatment multiple cancers, most notably Philadelphia chromosome-positive (Ph+) chronic myelogenous leukaemia. This is a drug we humans designed against a disease with an insight into the molecular pathology with certain degree of precision. Certainly there is strong evidence that alcohol causes cancer at seven sites in the body and probably others. Current estimates suggest that alcohol-attributable cancers at these sites make up 5.8% of all cancer deaths world-wide.
DNA CUSTOMIZATION IS A REALITY
The advent of DNA customization breaks through the traditional paradigm of a “one size fits all” dietary supplementation (and even drug therapy) and can present a promising new, more accurate and beneficial paradigm of nutrition. Individuals will in the near future, receive supplementation of nutraceutical products that are more precise for the customized to meet their unique, genetically determined needs. No more trial-and-error guesswork is necessary. It is possible that this new standard provides nutritional supplements that offer significantly greater value to the individual, than the conventional “one size fits all” formulations.
The introduction of DNA customized technology represents a fundamental change in dietary supplementation. As history demonstrates, in addition to availing hope and opportunity, change can also be met with skepticism, doubt, and even resistance. For various reasons, many refuse to change the status quo, especially those with a heavily vested interest in conventional dogma. On the contrary, there are others, who seeing the possibilities, want to partake in the new paradigm and market inferior versions of the technology. They offer the promise of DNA customization with woefully inadequate technology capabilities and tools. They claim personal care through genetics, while not measuring genotypes.
PRACTICAL APPLICATIONS
The field of science, out of which DNA customization has emerged, is called Nutrigenomics. Nutrigenomics is the study of the relationships between nutrition, the response of genes, and their combined effects on overall health and behavior [13]. However, not all nutrigenomic technologies are the same. To better evaluate the benefit and significance of DNA customization, a few questions need to be explored and answered:
What is the importance and value of DNA Customized Nutrition (Why use it)?
What exactly is DNA Customized Nutrition?
What criteria need to be satisfied for DNA Customized Nutrition to be worthwhile?
WHAT IS THE IMPORTANCE AND VALUE OF DNA CUSTOMIZED NUTRITION (WHY USE IT)?
As mentioned above, while we all have basic similarities that may appear to make us pretty much the same, or at least make us need the same things in general, the fact is that there are many distinct differences within humans that can alter individual basic needs. Some of our most obvious differences include: gender; age; race; body types; body sizes; tastes and appetites; physical and mental capabilities and activities; talents and skills; eye and hair colour; blood types; etc. Other less obvious differences can influence our bodies and behaviors just as profoundly. Some of these differences include, among others, variations in:
Metabolic processes (the ability to produce and regulate energy)
Ability to tolerate stress
Emotions
Behaviors
Lifestyle (i.e., food, drink and activity choices)
Immune system
The wide range of variations in these differences creates a variety of differences in our individual nutritional needs. These differences result from genetic differences: that is, polymorphisms in genotypes and their gene expressions. An example of this is that alcohol affects Asians and Native Americans quite differently than Caucasians, and that the alcohol dehydrogenase gene polymorphism has predictive value in alcohol addiction [14]. Genotypes represent specific gene variants. An individual is not and does NOT have only one genotype, but a vast number of genotypes. In order to effectively utilize DNA customized formulations, it is necessary to know the many genotypes of an individual across multiple biological and metabolic pathways.
In the earliest years of dietary supplementation, vitamin and mineral preparations were based on the lowest common denominator – what minimum amount of nutrients seemed to best address the most basic and common needs of the ‘masses.’ Many supplement companies followed the RDA guidelines to meet the basement of biological necessity. The inadequacies of that minimalist model from those early days through to the present day became obvious. This realization has led to elementary types of ‘category customization’ to address the different nutritional needs caused by those factors cited above, like gender, age, athletics, blood type, etc. Even with this elementary type of customization, a relative ‘sameness’ of formulas is still maintained within those groups (e.g., all women get the same type of women’s formula, etc.).
While addressing some ‘general’ differences within the categories, these conventional formulas were, and still are, “one size fits all” (i.e., young women need more iron and young men need more zinc, etc.). “One size fits all” formulas do not meet an individual’s specific needs, and can inadvertently create some nutritional imbalances.
Furthermore, the flaws and inadequacies of common lifestyles combined with “one size fits all” supplement formulas did not, and still do not, curb the epidemic increase in a wide range of chronic physical and mental disorders like cardiovascular disease, hypertension, osteoporosis, diabetes, obesity, inflammatory disorders, Alzheimer’s, Parkinson’s (and all of the neurodegenerative disorders), depression, conduct disorders, ADHD, OCD, and the current opiate/opioid epidemic. Importantly, certain groups of people were, and are, more prone (predisposed) to suffer a higher incidence and severity of certain types of conditions. Examples include: Caucasian and Asian women being more prone to osteoporosis; and Black people being prone to more severe cardiovascular disease (CVD), diabetes, and obesity. There is also evidence of high risk of the DRD2 as a susceptibility gene with heroin dependence in Chinese patients and the same gene polymorphism was associated with low risk of heroin dependence in Germans [15]. This fact is further punctuated by the enormous quantity and range of pharmaceutical drugs created to address these disorders in different ways, and the plethora of adverse side effects that can occur due to differences in how patients react to the drugs. Attempts to find natural remedies to address these ill health conditions gave rise to an ever-increasing selection of the “one size fits all” ‘condition-specific’ dietary supplement formulas as well.
The patient’s obsession with finding better ways to improve their health has resulted in a meteoric rise in the quantity and range of dietary choices and alternatives, especially in dietary supplements. This fact is punctuated by the availability of a mind-numbing array of dietary supplements from which to choose the “one that’s best for me”: clear evidence of an in-born desire for customization. The search for the best supplement to achieve a health objective (i.e., weight loss or increased athletic performance) or address an ill health condition (i.e., lower cholesterol, resolve digestive disorders, and address depression, etc.) is almost endless.
The issue is that we are inclined to pay attention to ‘what worked for someone else’ and then, think that product will therefore work for me. Moreover, sometimes the product effect backfires, promoting negative results or reactions confirming evidence that we have genetic differences. Nevertheless, even dietary supplement companies try to get in on the popular trends and formulate products very similar to popular selling versions on the market, perpetuating the “one size fits all” and “works one way” formula fads. Innovation seems to be confined to the next best miracle fruit extract or the like.
With the discovery of DNA, and the advent of the human genome project, an entirely new dimension of opportunity for nutritional customization was born in the field of nutrigenomics. An important fact must now be highlighted: “one size” does not fit all for your clothes or your health needs. Another important factor crucial to the success of a DNA customized nutraceutical product, as previously mentioned, is whether the genes being targeted for nutraceutical customization are the most appropriate to provide the greatest benefit? In fact, DNA customization eliminates the trial and error guesswork of finding the best formulation for the individual, ensuring the greatest and most sustainable benefit. This is easily said than done, but remains a laudable goal for the future of generable medicine and in this context, addiction medicine. Blum’s group has already published on this possibility with regard to KB220 variants in obesity [16–18].
WHAT EXACTLY IS A DNA CUSTOMIZED NUTRACEUTICAL?
DNA analysis and nutraceutical customization is a real possibility following additional required research. The following is an overview of the nutrigenomic DNA customization process:
Find the right science-based ingredients that influence therapeutically important pathways - a crucial factor.
-
Existing research must validate the dose-dependent mechanisms of action (MOA) and beneficial effects of those ingredients.
-
Determine if, and by how much, those ingredients influence gene expression.
Dose-dependent results are reviewed to establish the baseline dose needed to achieve beneficial gene expression effects in a reasonable portion of the population.
Research is then either conducted, or if available, reviewed to determine a range of dose-dependent gene expression results.
Dose-dependent gene expression results are then compared to results in the MOA and efficacy research.
Ingredients selected through this process become “DNA Qualified” and are used in DNA customized formulations.
Research is then reviewed to determine which genotypes exhibit the kind of gene expressions noted in the gene expression studies.
Algorithms are then established for DNA customized dose-dependent applications for each DNA qualified ingredient based on the genotypes for each pathway.
Then research is reviewed or conducted on select populations of genotype-qualified candidates to confirm, and/or better define and catalogue the efficacy parameters.
Once this process is completed, genuine DNA qualified ingredients can be customized for the gene polymorphisms of an individual.
Specific genes of an individual are analyzed, via analysis of a buccal swab.
Genotypes for crucial pathways are determined and ingredient potencies adjusted to accommodate gene expression variations of the individual.
Risk alleles of a number of important physiological pathways are selected on the basis of a common thread (i.e., hypodopaminergic trait/state). Each risk allele must have been associated with a particular risk compared to controls in many published studies.
WHAT CRITERIA NEED TO BE SATISFIED FOR DNA CUSTOMIZED NUTRITION TO BE WORTHWHILE?
“Any one part of your body cannot be well, unless you care for the whole body.”
– Plato, Classical Greek Philosopher
It is clearly beneficial to address foundational health factors that affect the whole body in conjunction with other more focused targets (synergistic symbiosis). For example, to address weight loss without first considering genetic factors and the controlling involvement of the brain is a formula for failure, repeatedly demonstrated with conventional methods. The shortcoming of most conventional therapeutic tactics, even nutraceutical ones, is that they rely solely on narrow, compartmentalized, and/or downstream symptomatic targets [19]. Some examples of these types of benefit targets include:
Anti-inflammatory
Appetite suppression
Blocking carbohydrate absorption
Blocking fat absorption
Diuretics
Stimulants
Calming and Soothing
Libido enhancement
Cholesterol lowering
Blood sugar lowering
Exercise and nutritional needs-
Conventional nutraceutical formulations that address these types of ‘benefit’ targets, routinely do not address the gene expression-based mechanisms of underlying ‘upstream’ causes, promoting their ‘downstream’ conditions. They are generally targeted at mechanisms of action that achieve superficial and/or temporary symptomatic relief at best. One very important model in terms of Precision Medicine has been developed by Body/Sync genetic testing. Developed by genetic scientists, genomic nutritionists and fitness professionals, Body/Sync model and test has been designed to help people better understand and manage their genetic nutrition and fitness goals targeting well-researched genes.
There could be many ‘causes’ for a supposed malady and its symptoms. For instance, a depressed individual might try taking St. John’s Wort [20] to feel better (trial and error). Importantly, there are many causes and types of depression. What works for one cause and type, might not work for another, or even make other types worse. Knowing the genotypes of genes in certain pathways in the brain would enable a DNA customized formula to promote healthy brain function and reduce guesswork.
Another example of this premise is the use of fat blockers to reduce fat calorie absorption and promote ‘weight’ loss. The fact is that, in the end, calorie blocking and deprivation work against the body’s genetic survival instructions. The initial effect could be to reduce fat calorie absorption, resulting in some initial weight loss. But such deprivation tactics create a survival crisis, triggering a genetically mandated protection response. The long-term result of this effect is to lower the basal metabolic rate (rate of calorie burning), increase fat storage, and increase cravings and food consumption to ensure survival, which promotes greater weight re-gain (also known as the ‘Yo-Yo’ rebound weight gain) [21].
Many conventional nutraceutical products, especially condition-specific formulas, target symptomatic relief. In contrast, by exerting a nutraceutical influence on gene expressions, in terms of precision medicine, DNA customized nutraceutical products must address the whole body, and importantly, the brain’s control of all bodily functions (neuro-nutrigenomics). The result corrects the effects of specific gene variants (polymorphisms), normalizing target tissue function. Moreover, DNA customized formulas should be symbiotic, improving the overall health and function of the body in a synergistic manner, and satisfying the criteria required to provide effective and sustainable outcomes. Those criteria are:
Criteria 1
The biological effect should be “body friendly” (i.e., biologically and immunologically compatible).
DNA customized nutrition is satisfying a need of the body. Therefore, the body should not have to protect, reject, or retaliate against the effects the formula’s ingredients have on the body. For example, pharmaceutical drugs, stimulants, fat blockers, and/or mega-dose nutrient overloading (pharmacologic-like) effects, including all stimulants, suppressants, and calorie deprivation tactics, could fight against what has been termed “body friendly” [22].
Criteria 2
Gene polymorphisms identified and targeted for nutrigenomic effects need to exert a significant influence on the origin of a problem and the source of the targeted outcome. In simple terms, get to the genetic source. For example, in contrast to single-effect stimulants and/or calorie deprivation tactics, weight loss needs to be the result of correcting bio-system inadequacies. At least 5 major pathways need to be simultaneously addressed:
Energy metabolism (production and regulation)
Reward and craving management in the brain
Neuro-endocrine management
Stress management
Immune system function
Criteria 3
The nutraceutical benefit should not only correct the target tissue structure and/or function, it should make a significant contribution to overall health.
In terms of developing DNA customized nutrient specific formulae for treatment, and possible prevention of Reward Deficiency Syndrome (RDS) [23] and all substance and non-substance seeking addictive behaviors, it is the entire ingredient base that works in concert through, for example, balancing the Brain Reward Cascade (BRC) that will provide an effect on the overall health of an individual.
Criteria 4
The beneficial effects should be obvious, different, transformational, and sustainable for most of the patients.
In-vitro studies can simulate a powerful mechanism of action in cell cultures. However, when human studies are conducted on the same product to find a specific benefit, like blocking fat absorption to induce weight loss, the outcomes are usually skewed across a range of benefits. The results could look like:
30% of the people received strong benefits (also a possible placebo-effect)
30% received minor benefits
15% received no benefits
15% actually experienced reverse effects (i.e., increased weight gain)
According to a conventional perspective, this outcome would still be considered positive and have significant medical value. However, in this example, these results suggest that as much as 70% of the patients will experience some level of disappointment. This is the primary reason that many people stop taking a weight loss product within 3 to 6 months. This also does not consider the “Yo-Yo” rebound weight gain effect (the failure effect), which appears to happen in at least 95% of the cases with time. The need for precision medicine will come of age, especially when genetic testing could provide meaningful results and medical benefits.
CONVENTIONAL METHODS DO NOT COMPLY WITH ALL OF THE 4 IMPORTANT CRITERIA
First, conventional methods may not be “body friendly”
In many cases, products impose a reduced fat and calorie intake (blocking) on the body. The initial effect could be weight loss (especially, in the 30% of people). However, the long-term effect will be the result of the body’s attempt to protect itself from the ‘famine’ as cited in the “Thrifty Gene Hypothesis” [24] that has been created. This effect might be significantly greater in certain individuals in the 70% group. The body will ultimately try to protect against reduced nutrient absorption with increased fat storage, a natural survival mechanism.
Second, ‘fat-blocking’ methods do not address multiple metabolic or genetic causes of the problem
Just blocking fat absorption is not addressing the many other issues related to obesity, whereby many genes and subsequent polymorphisms play important roles in the induction of aberrant cravings [25], enhancing brown fat synthesis [26], other addictions [27], and inducing metabolic anomalies [28].
Third, conventional methods do not make a significant contribution to overall health
It is indisputable that weight loss is associated with many health benefits. However, owing to the facts cited in first two criteria, and to the fact that absorption of very important lipids (i.e., Omega 3’s) and fat soluble vitamins (i.e., vitamins A, D, E, and K) are being blocked, the long-term effect will be reduced health that is further amplified by “Yo-Yo” rebound weight gain.
Fourth, conventional methods do not provide obvious, different, and positive transformational effects that are sustainable for most of patients
We must keep in mind that most dietary supplement research, especially for weight loss products, evaluates results over an 8–12 week period. This is a very short-term window in a long-term process. Considering the reality of the weight gain rebound effect, this is an inadequate time to evaluate more telling long-term effects. In fact, history reveals that conventional weight loss programs, almost without fail, have failed. If the products work at all, conventional calorie deprivation weight loss tactics (including fat blockers), without exception, provide results for only a temporary period. Ultimately, conventional methods, result in the dreaded weight re-gain. Given this, and due to the facts cited in the first three criteria, for the vast majority of people, the results are not obvious, not different from other similar-type products, not transformational, and not sustainable.
A major reason that human research results are skewed is due to the wide variation in genetic factors. Identifying these factors enables the use of the right therapeutic modalities in the appropriate amounts to achieve the greatest benefits in the most people for the longest time. When done correctly, the incredible value of DNA customization will be indisputable.
GENETIC TESTING: FACTS OR FICTION
While some physicians may think that by using 23andMe genomic testing, they could adequately determine a person’s risk for RDS behaviors, they are mistaken. In the first place, the FDA did not allow 23andMe to provide any interpretive data to an individual related to medical health issues and in fact, threatened removal of a misleading test. Secondly, there are a number of SNPs that can influence addictive behaviors that are not obtained in the 23andMe test. In fact, there is not one study published on the 23andMe test concerning accuracy as revealed in PubMed (07-19-16). The following information will help inform the medical community about genetic testing cautions in general.
In terms of molecular genetic testing, there are three types that are of current interest. These include: Pharmacogenetics (primarily evaluating metabolizing enzymes for high and low metabolizers with, for example, opiates); Genetic Addiction Risk Score (to determine, through a panel of reward gene polymorphisms, stratification risk or vulnerability to all RDS behaviors including pain tolerance); and Pharmacogenomics (personalized addiction medicine based on genotyping an individual and targeting specific gene loci).
Pharmacogenetic Testing
Various alleles in the P450 system are currently utilized in pain medicine clinics to evaluate metabolic concerns to help identify high and low metabolizers. For the most part, this has not translated to significant clinical utility, but may have some relevance in terms of buprenorphine/naloxone treatments. When used in conjunction with GARS, the alleles could provide valuable information [29].
GENETIC ADDICTION RISK SCORE
Genetic Addiction Risk Score
It is now known that in terms of nature (genes) and nurture (environment) and behavioral outcome in Homo sapiens, the contribution is 50% genes and 50% epigenetics. Thus, molecular genetics or DNA testing is very important, especially linking aberrant behaviors to any individual.
Blum’s laboratory proposed that any disturbance along the brain reward cascade due to either gene variations (polymorphisms) or environment (epigenetics) will result in aberrant addictive behaviors or RDS. In spite of a global search to uncover specific or candidate genes, or even clusters of genes characterized from high-density SNP arrays, it is well known that many attempts have not replicated or have been inconclusive. However, Palmer et al. [30] recently showed that between 25–36% of the variance in the generalized vulnerability to substance dependence is attributable to common single nucleotide polymorphisms. Moreover, the additive effect of common single nucleotide polymorphisms is shared across principal indicators of comorbid drug problems. Furthermore, as a result of these studies, more recent evidence has revealed that specific candidate gene variants account for risk prediction.
Adopting a Bayesian approach, earlier studies from Blum’s laboratory [5] determined a Positive Predictive Value (PPV) for the DRD2 A1 variant (low number of D2 receptors) of 74%, indicating that if a child is born with this polymorphism, they have a very high risk of becoming addicted to either drugs, food, or aberrant behaviors at some point in their lives [31]. Over the many years to come since the 1990 finding on the DRD2 gene association of the Taq A1 allele and severe alcoholism [32], laboratories all across the globe including NIDA and NIAAA not only confirmed this early work [33], but also extended the magnitude of many other candidate genes, especially genes and second messengers located in the reward circuitry of the brain [1].
An example includes: Moeller et al. [34] suggested that drug cues contribute to relapse, and their neurogenetic results have identified the DAT1R 9R allele as a vulnerability allele for relapse, especially during early abstinence (e.g., detoxification). The DAT1R 9R allele influences the fast acting transport of dopamine, sequestered from the synapse, leading to a hypodopaminergic trait.
It is important to be cautious to accept such genetic testing that uncovers reward circuitry gene polymorphisms, particularly those linked to dopaminergic pathways as well as opioid receptor(s) as a method of obtaining better treatment results. Comprehending the relationship between the reward circuitry’s participation in buprenorphine outcomes and its corresponding genotypes deliver an innovative model to enhance a patient’s clinical experience and improvements throughout opioid replacement therapy [35]. In fact, Blum’s group’s genetic risk score represents a panel of known reward genes and associated risk polymorphisms providing genetic risk for addiction and other behaviors, including medical monitoring and clinical outcome response [35].
Pharmacogenomics – Customized Addiction Medicine
As mentioned earlier, along these lines and in conjunction with one of us (KB), and Gerald Kozlowski, developed the “Brain Reward Cascade” (BRC) [6]. This concept served as a blue print for how neurotransmitters interact in the reward system of the brain. In addition, it has been firmly established that respective reward genes that regulate these chemical messengers ultimately control the amount of dopamine released into not only the reward site, but also other regions of the brain.
Moreover, it is well established that resting state functional connectivity integrity is important for normal homeostatic functioning. Zhang et al. [36] recently showed that in heroin addicts, there is reduced connectivity between the dorsal anterior cingulate cortex (dACC) and the rostral anterior cingulate cortex (rACC), as well as reduced connectivity between the subcallosal anterior cingulate cortex (sACC) and the dACC. Their findings of variations of functional connectivity in the three sub-regions of the ACC in heroin addicts implied that these sub-regions of the ACC, together with other key brain areas (i.e., dorsal striatum, putamen, orbital frontal cortex, dorsal striatum, cerebellum, amygdala, etc.), potentially play important roles in heroin addiction. Most recently, Blum’s laboratory, along with Zhang’s group [37], in abstinent heroin addicts that KB220Z™, a complex putative dopamine D2 agonist, was shown to induce an increase in blood-oxygen-level dependent (BOLD) activation in the caudate-accumbens-dopaminergic pathways when compared to a placebo following one-hour of acute administration. Furthermore, KB220Z™ also reduced resting state activity in the putamen of abstinent heroin addicts. In the second phase of this pilot study, of all ten abstinent heroin-dependent subjects, three brain regions of interest (ROIs) were observed to be appreciably triggered from resting state by KB220Z™ when compared to the placebo group (P < 0.05).
Augmented functional connectivity was observed in a putative system that was comprised of the dorsal anterior cingulate, medial frontal gyrus, nucleus accumbens, posterior cingulate, occipital cortical regions, and cerebellum. In utilizing DNA-based testing, there is a successful development of polymorphic gene testing, which enables customized (personalized) anti-obesity compounds. This serves as the basis of futuristic personalized addiction medicine utilizing GARS.
Fiction
While there is a plethora of very positive experiments involving thousands of studies for many candidate gene associations with all RDS behaviors including pain, there are also negative results [38–41]. Currently, a number of companies have entered the genetic testing arena in the addiction and pain industrial space claiming “personalized care.” However, we believe these companies have not exhibited proper scientific research in these arenas. These issues include exaggerated claims such as using Blum’s original work [5, 42], stating that their genetic test is 74% predictive. This is indeed false because they use one gene (DRD2) to back their claim and commercialize a full panel of other candidate genes and have never carried out any outcome studies with their panel. Additionally, they make other false claims suggesting that patient’s results are compared to population controls. Review of their “disease free” controls reveal significant flaws, especially in light of not controlling for a remarkable list of RDS behaviors [43]. They would have to utilize what has been termed “Super-Controls.” Simply stated, population controls may carry many invisible RDS behaviors that must be identified, so that the control group is RDS-free. Otherwise, utilization will lead to spurious and false results [44].
Another issue is that these companies have selected genes that may be involved in risky behavior, but they do not utilize the correct variant in their tests or use very rare variants that do not truly prove addiction risk. Specifically, Mayer and Höllt [45] correctly proposed that the “vast number of non-coding, intronic or promoter polymorphisms in the opioid receptors may influence addictive behavior, but these polymorphisms are far less studied, and their physiological significance remains to be demonstrated.” Most importantly, these companies have never performed research to show whether or not their genetic full panel test significantly predicts anything, let alone addiction risk or any associated behaviors.
Facts
Though we, the authors, may have a particular bias due to several years of work dedicated to cultivate a precise genetic test to forecast accurate liability/risk for RDS and its related behaviors, we will try to elucidate the reasons why our present laboratory testing site has successfully developed the first Genetic Addiction Risk Score (GARS™) in conjunction with the Institute of Behavioral Genetics, at the University of Colorado, Boulder.
To develop GARS™, we first selected ten reward candidate genes (DRD1, 2, 3, 4; DAT1; serotonin transporter; COMT; MAO; GABA; Mu opiate receptor) and several SNPs and point mutations that affect the net discharge of dopamine at the brain reward site. The variants or SNPs, incorporating point mutations, were selected to reveal a hypodopaminergic trait. In considering validation, we joined the creators of the Addiction Severity Index-Media Version (ASI-MV), a test required in 18 states, for both alcohol and drug severity risk scores [45].
In unpublished work, we contacted eight very diverse treatment centers across the United States, resulting in a total of 393 subjects that were genotyped using the selected GARS™ panel. All data was genotyped and statistically analyzed at the Institute for Behavioral Genetics (IBG) at the University of Colorado, Boulder. We discovered a noteworthy relationship between a summed score of all GARS panel risk alleles (variant types) and both the ASI-MV alcohol (p<0.004) and drug (P<0.05) severity indices in a total of 273 subjects. We did find that age was an important variable for both alcohol and drug severity.
In fact, we found that the greater the quantity (number) of risk alleles, the greater the likelihood of alcohol or drug use severity. It was also observed that family difficulties, psychological concerns, and medicalization, suggestively correlated. One critical caveat was that if we altered any particular SNP, the significance was absent. This indicates how vital the selected GARS™ panel is, and any deviation will produce false results that may occur with other commercial testing technology that have no research to validate their tests. It was also found that if we try weighting each allele by increasing its score power, we also lose significance, suggesting the importance of the clustering of genes implicated in the BRC, rather than any single gene polymorphism by itself.
Benefits of GARS
We found that a ten gene panel, driven by risk alleles for a hypodopaminergic state, consisting of 22 (mother & father) SNPs, was significantly associated with the level of ASI-MV alcohol and drug risk severity score and thus, has important clinical relevance and utility.
To our knowledge, this is the first and only correlation of a panel of genes with established polymorphisms that reflect the BRC [6] with the ASI–MV alcohol and drug risk severity score ever accomplished to date. While other studies are required to further confirm and extend the GARS test to include other genes and polymorphisms that associate with a hypodopaminergic trait, these results provide clinicians with a non-invasive genetic test.
Unlike “23andMe,” other than finding ancestral heritage, genomic testing such as GARS can be used to improve clinical interactions and decision-making with the following precise polymorphic associations:
Attenuation of guilt and denial;
Corroboration of family gene-o-grams;
Risk severity-based decisions about appropriate, therapies including pain medications and risk for addiction liability;
Appropriate level of care placement (i.e., inpatient, outpatient, intensive outpatient, residential);
Length of stay in treatment;
Genetic severity-based relapse and recovery liability and vulnerability;
Pharmacogenetic medical monitoring for better clinical outcomes (i.e., the A1 allele of the DRD2 gene reduces the binding to delta receptors in the brain, reducing Naltrexone’s clinical effectiveness); and
Medical necessity for insurance scrutiny.
HAVE WE HATCHED DNA CUSTOMIZED NUTRITION?
In consideration of these initial hypothesis generating studies, and a lack of research, we were determined to create a study that assessed the practice of DNA customization of a nutritional solution for both wellness and weight management. We have reviewed several study outcomes [16–18], whereby Blum’s laboratory genotyped 1,058 subjects, and these subjects were administered KB220Z [formerly LG9939, Recomposize, Genotrim] (a complex Neuroadaptagen nutraceutical-dl-phenylalanine, chromium, l-tyrosine, and other select amino-acids and adaptogens) based on polymorphic outcomes. The resultant customized formulae involved a minimum of 175 Single Nucleotide Polymorphisms (SNPs) covering 16 genes important to the BRC and most importantly, involved in “dopamine homeostasis.” In a small subset, simple t-tests relating various parameters before and 80 days on the nutraceutical were done.
The meaningful clinical results are the following: weight loss (p < 0.008); decrease in sugar craving (p < 0.008); appetite suppression (p < 0.004); decrease in snacking (p < 0.005); decrease in late night binging (p < 0.007); increased awareness of over-eating (p < 0.02); improved energy (p < 0.004); improvement of sleep quality (p < 0.02); and increase in happy emotions (p < 0.02). Polymorphic correlates were acquired for several genes (PPAR gamma 2, MTHFR, 5-HT2a, and DRD2 genes) with positive clinical factors tested in this study. Notably, of all the results and gene polymorphisms, only the DRD2 gene polymorphism (Al allele) had a significant Pearson correlation with treatment days (r = 0.42, p = 0.045). This 2-fold increase is a significant genotype for treatment compliance [16,18].
Additionally, Blum’s team methodically assessed the effect of the polymorphisms of these five candidate genes as critical targets for the design of a DNA customized nutraceutical, KB220Z, to fight obesity with distinctive stress on body re-composition as measured by Body Mass Index (BMI) [17]. A total of 21 individuals were evaluated in a preliminary investigational study of KB220Z.
The investigation was grounded on the outcomes of buccal swab genotyping for every subject. An individualized customized nutraceutical formula was specified as a utility of measured gene polymorphisms of the five gene candidates evaluated. At the start of the investigation, and every two weeks successively, every subject completed an adapted Blum-Downs OPAQuE Scale™ (Overweight Patient Assessment Questionnaire). The alleles involved were the DRD2 Al; MTHFR C 677T; 5HT2a 1438G/A; PPAR-γProl2Ala; and Leptin Ob1875 < 208bp. Pre- and post-hoc analyses discovered a significant difference between the initial BMI and the BMI following an average of 41 days (28–70 d) of KB220Z ingestion in the 21 subjects. The pre-BMI was 31.2 (weight/Ht2) as compared to the post-BMI of 30.4 (weight/Ht2) with a significance value of P < 0.034 (one tailed). Comparably, the pre-weight in pounds (lb) was 183.52 as compared to the post-weight of 179 lb with a significance value of P < (0.047). They also observed trends in the decrease of late night snacking, carbohydrate craving, stress, and waist circumference. Furthermore, in the 41-day time frame, they observed a trend in weight loss, whereby 71.4% of the subjects lost weight. Therefore, 15 out of 21 subjects lost weight with a z score of 2.4 and a significance value of P < (0.02). In this particular group, 53% lost an average of over 2.5% of their initial weight.
Further preliminary findings demanding additional experiments, using a Path Analysis (non-customized KB220Z), also discovered significant links to anti-obesity related behaviors. In a one-year cross-sectional open trial study of 24 unscreened subjects using an oral KB220Z variant, the following beneficial outcomes occurred: decreased stress; sleep improvement; increased energy; universal well-being; decreased cravings (sugar/carbohydrates); enhancement in mental focus/memory; enhancement in blood sugar levels; decreased food intake; decreased waist circumference; weight loss; decreased blood pressure; enhanced exercise performance; decreased drug seeking; decreased hyperactivity; and decreased cholesterol levels [46].
GENETIC TESTING FOR CUSTOMIZATION
We hereby suggest that the outcome of using this natural dopaminergic stimulating (potentially DNA customized) method over time should lead to neuronal DA discharge at the NAc, potentiating an abundance of D2 receptors [47]. We are confident since earlier outcomes using KB220Z, in terms of neuroimaging and qEEG studies [37, 48,49], presented both improved resting state functional connectivity in abstinent heroin addicts and control of extensive theta activity in the cingulate gyrus of abstinent psychostimulant abusers, similarly to the effects as seen in alcoholics.
Additional evidence in humans is derivative of anti-craving effects as seen in several peer-reviewed published clinical trials, involving randomized double blind placebo-controlled studies [12]. In fact, animal gene therapy utilizing cDNA vectors of the DRD2 gene implanted into the NAc results in decreased alcohol and cocaine craving behavior [50–53].
We, the authors, are aware that dopaminergic activation in the long run, rather than blocking dopamine [54], should be used to treat not only alcohol, cocaine, and nicotine cravings, but also glucose cravings and other known behavioral addictions (e.g. gambling [55], hypersexuality [56], etc.). Thus, the coupling of genetic antecedents, such as the GARS and nutrition, may be a very viable alternative approach for the treatment of all RDS behaviors.
Further research from Blum’s laboratory designed a hypothetical modeling study, in which they sought to assess health and economic consequences of a nutrigenomic product for weight loss. Blum’s team [57] assembled a nutrigenomic economic model by relating (1) published study data correlated with the value of a product and/or ingredients; (2) confirmed clinical evaluations that have been previously linked to health economic data; and (3) data including condition frequency and inclusive cost of illness. In this hypothetical model, we validate that a DNA customized nutraceutical positively decreases the cost of illness at the macroeconomic and microeconomic level based upon cost efficiency and cost-benefit analyses. From this suggested model, they have predicted the significant prognostic health economic consequences of a nutrigenomic intervention to establish a hypothetical model of nutrigenomic economics as it relates to obesity.
CONCLUSION
Numerous replications amounting to 15 variant formulae, permitted Blum’s team [16–18] to utilize polymorphic targets of a number of reward genes (Serotonergic, Opioidergic, GABAergic, and Dopaminergic) to customize KB220 [Neuroadaptogen-amino-acid therapy (NAAT)] by specific algorithms. To reiterate, identifying over 1,000 obese subjects in the Netherlands, a subsequent small subset was administered various KB220 formulae, tailored according to personal DNA polymorphisms personalized, that decoded to substantial reduction in both Body Mass Index (BMI) and weight (lb).
Recently, Blum’s laboratory alongside Brett Haberstick, Andrew Smolen and others, have created an unpublished panel of genes called the “Genetic Addiction Risk Score” (GARSTM). When they chose 10 genes with suitable variants, a statistically significant connection between the ASI-Media Version alcohol and drug severity scores and GARS was discovered. This particular observation was seen in 273 patients appearing at seven distinct treatment centers. This significant link could possibly set the stage for initial clinical identification of susceptibility, and associate personalized medicine as a nutrigenomic key for all RDS behaviors.
Most notably, as mentioned earlier, Blum et al. [37] reported that a well- researched variant of KB220Z, in abstinent heroin addicts, remarkably increased resting state functional connectivity. It was observed that this enhanced activation of resting state functional connectivity was observed in a putative network that included the dorsal anterior cingulate, medial frontal gyrus, nucleus accumbens, posterior cingulate, occipital cortical areas, and cerebellum.
In further unpublished rat studies performed at the University of Florida, Febo, Blum, and others, found that KB220Z considerably stimulates, above placebo, seed ROIs, comprising the left nucleus accumbens, cingulate gyrus, anterior thalamic nuclei, hippocampus, pre-limbic, and infra-limbic loci. This response induced by KB220Z demonstrates significant functional connectivity, increased brain volume recruitment, and enhanced dopaminergic functionality across the brain reward circuitry. This robust, yet, selective response implies clinical relevance.
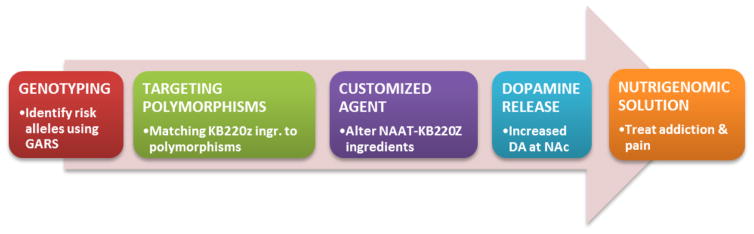
Finally, we, the authors, are now paused to propose a Reward Deficiency System Solution™ that promotes early identification and stratification of risk alleles by utilizing GARS, allowing for customized nutrigenomic targeting of these risk alleles by altering neuronutrient amino–acid therapy ingredients as an algorithmic function of carrying these polymorphic DNA SNPs. By doing so, following required research, this novel approach could potentially yield the first ever nutrigenomic solution for addiction and pain and other RDS addictive behaviors [Figure 1]. This introduces the long-awaited innovative era of genomic addiction medicine [59].
Fig: 1.
Schematic of DNA customization application for a Nutrigenomic solution to RDS (with permission: Blum et al. [58]).
HIGHLIGHTS.
Precision Medicine in terms of not only genetic testing for a predisposition of Reward Deficiency (Hypodopaminergic ) risk for all addictive behaviors, but for identification of specific SNPS and the potential of DNA customization is now plausible. The field of Nutrigenomics is emerging and will be most beneficial in the treatment and prevention of abarrant seeking behavior with the promise of curtailing the global epidemic of unwanted addictions.
Acknowledgments
The authors appreciate the expert edits of Margaret A. Madigan. We are also appreciative of Mary Hauser and staff of Dominion Diagnostics and Pauline Gee of BodySync. We also appreciate the important work by Drs. Pauline Gee of BodySync and Andrew Smolen and Bret Haberstick of Colorado University, Institute of Behavioral Genetics.
Footnotes
CONFLICT OF INTEREST
Kenneth Blum, PhD is the holder of a number of US and Foreign patents issued and pending related to Nutrigenomics and Nutraceuticals. Together with IGENE LLC., a developmental agreement is being established with Body/Sync Inc. for the commercialization of the Genetic Addiction Risk Score (GARS)™ with a sales license to Dominion Diagnostics, LLC. Kenneth Blum is a paid consultant (Chief Scientific Advisor) of Dominion Diagnostics, LLC, and is President& CEO of IGENE, LLC. Dr. Blum is a member of the scientific advisory board of Dominion Diagnostics, LLC and Rivermend Health. Dr. Blum is Chief Scientific Officer of LaVitaRDS. Dr. Blum serves as Neuroscience advisor to Summit Estates Recovery Center and The Shores Treatment & Recovery Center. Drs. Blum (Chairman), Badgaiyan, Thanos and Febo are members of LaVitaRDS Scientific Advisory Board. Dr. Blum is also the Scientific Director of Path Foundation NY. The authors state that there are no other conflicts of interest.
FINANCIAL DISCLOSURE
This work was supported by National Institutes of Neurological Disorder and Stroke, Grant Number:1R01NS073884; National Institutes of Mental Health, Grant Number: 1R21MH073624 awarded to Dr. Badgaiyan. Kenneth Blum and Eric R. Braverman are the recipients of a grant from Life Extension Foundation, Ft. Lauderdale, Florida, awarded to PATH Foundation NY. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Blum K, Febo M, McLaughlin T, et al. Hatching the behavioral addiction egg: Reward Deficiency Solution System (RDSS)™ as a function of dopaminergic neurogenetics and brain functional connectivity linking all addictions under a common rubric. J Behav Addict. 2014 Sep;3(3):149–56. doi: 10.1556/JBA.3.2014.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rena G, Begg F, Ross A, et al. Molecular cloning, genomic positioning, promoter identification, and characterization of the novel cyclic amp-specific phosphodiesterase PDE4A10. Mol Pharmacol. 2001 May;59(5):996–1011. doi: 10.1124/mol.59.5.996. [DOI] [PubMed] [Google Scholar]
- 3.Blum K, Chen AL, Braverman ER. Attention-deficit-hyperactivity disorder and reward deficiency syndrome. Neuropsychiatr Dis Treat. 2008 Oct;4(5):893–918. doi: 10.2147/ndt.s2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noble EP, Blum K, Ritchie T, et al. Allelic association of the D2 dopamine receptor gene with receptor-binding characteristics in alcoholism. JAMA Psychiatry. 1991 Jul;48(7):648–54. doi: 10.1001/archpsyc.1991.01810310066012. [DOI] [PubMed] [Google Scholar]
- 5.Blum K, Wood RC, Braverman ER, et al. The D2 dopamine receptor gene as a predictor of compulsive disease: Bayes’ theorem. Funct Neurol. 1995 Jan-Feb;10(1):37–44. [PubMed] [Google Scholar]
- 6.Blum K, Kozlowski GP. Ethanol and Neuromodulator influences. A cascade model of reward. In: Ollat H, Parvez S, Parvez H, editors. Alcohol and Behaviour: Basic and Clinical Aspects Progress in Alcohol Research. Utrecht, Netherlands: VSP International Science Publishers; 1990. pp. 131–150. [Google Scholar]
- 7.Blum K, Oscar-Berman M, Demetrovics Z, et al. Genetic Addiction Risk Score (GARS): molecular neurogenetic evidence for predisposition to Reward Deficiency Syndrome (RDS) Mol Neurobiol. 2014;50(3):765–796. doi: 10.1007/s12035-014-8726-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim D, Kim WY, Lee SY, Lee SY, Yun H, Shin SY, Lee J, Hong Y, Won Y, Kim SJ, Lee YS, Ahn SM. Revising a personal genome by comparing and combining data from two different sequencing platforms. PLoS One. 2013 Apr 8;8(4):e60585. doi: 10.1371/journal.pone.0060585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blum K, Gardner E, Oscar-Berman M, et al. “Liking” and “wanting” linked to Reward Deficiency Syndrome (RDS): hypothesizing differential responsivity in brain reward circuitry. Curr Pharm Des. 2012;18(1):113–8. doi: 10.2174/138161212798919110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blum K, Giordano J, Hauser M, et al. Coupling Genetic Addiction Risk Score (GARS™), Comprehensive Analysis of Reported Drugs (CARD™) dopamine agonist therapy (KB220z™): Reward Deficiency Solution System (RDSS). Abstract accepted to European Psychiatry Congress; Viena, Austria. March 18–20th, 2014. [Google Scholar]
- 11.Plomin R, Owen MJ, McGuffin P. The genetic basis of complex human behaviors. Science. 1994 Jun 17;264(5166):1733–9. doi: 10.1126/science.8209254. [DOI] [PubMed] [Google Scholar]
- 12.Blum K, Oscar-Berman M, Stuller E, et al. Neurogenetics and Nutrigenomics of Neuro-Nutrient Therapy for Reward Deficiency Syndrome (RDS): Clinical Ramifications as a Function of Molecular Neurobiological Mechanisms. J Addict Res Ther. 2012 Nov 27;3(5):139. doi: 10.4172/2155-6105.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rana S, Kumar S, Rathore N, et al. Nutrigenomics and its Impact on Life Style Associated Metabolic Diseases. Curr Genomics. 2016 Jun;17(3):261–78. doi: 10.2174/1389202917666160202220422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gitik M, Srivastava V, Hodgkinson CA, et al. Association of Superoxide Dismutase 2 (SOD2) Genotype with Gray Matter Volume Shrinkage in Chronic Alcohol Users: Replication and Further Evaluation of an Addiction Gene Panel. Int J Neuropsychopharmacol. 2016 May 9; doi: 10.1093/ijnp/pyw033. pii: pyw033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu K, Lichtermann D, Lipsky RH, et al. Association of specific haplotypes of D2 dopamine receptor gene with vulnerability to heroin dependence in 2 distinct populations. JAMA Psychiatry. 2004 Jun;61(6):597–606. doi: 10.1001/archpsyc.61.6.597. [DOI] [PubMed] [Google Scholar]
- 16.Blum K, Chen TJH, Williams L, et al. A short term pilot open label study to evaluate efficacy and safety of LG839, a customized DNA directed nutraceutical in obesity: exploring nutrigenomics. Gene Ther Mol Biol. 2008;12(2):371–382. [Google Scholar]
- 17.Blum K, Chen AL, Chen TJ, Rhoades P, Prihoda TJ, et al. LG839: anti-obesity effects and polymorphic gene correlates of reward deficiency syndrome. Adv Ther. 2008;25(9):894–913. doi: 10.1007/s12325-008-0093-z. [DOI] [PubMed] [Google Scholar]
- 18.Blum K, Chen ALC, Chen TLC, Rhoades P, Prihoda P, et al. Dopamine D2 Receptor Taq A1 allele predicts treatment compliance of LG839 in a subset analysis of a pilot study in The Netherlands. Gene Therapy Mol Biol. 2008;12(1):129–140. [Google Scholar]
- 19.Ríos-Hoyo A, Gutiérrez-Salmeán G. New Dietary Supplements for Obesity: What We Currently Know. Curr Obes Rep. 2016 Jun;5(2):262–70. doi: 10.1007/s13679-016-0214-y. [DOI] [PubMed] [Google Scholar]
- 20.Yildiz M, Batmaz S, Songur E, et al. State of the art psychopharmacological treatment options in seasonal affective disorder. Psychiatr Danub. 2016 Mar;28(1):25–9. [PubMed] [Google Scholar]
- 21.German AJ. Obesity Prevention and Weight Maintenance After Loss. Vet Clin North Am Small Anim Pract. 2016 May 31; doi: 10.1016/j.cvsm.2016.04.011. pii: S0195-5616(16)30027-4. [DOI] [PubMed] [Google Scholar]
- 22.Downs BW, Chen AL, Chen TJ, et al. Nutrigenomic targeting of carbohydrate craving behavior: can we manage obesity and aberrant craving behaviors with neurochemical pathway manipulation by Immunological Compatible Substances (nutrients) using a Genetic Positioning System (GPS) Map? Med Hypotheses. 2009 Sep;73(3):427–34. doi: 10.1016/j.mehy.2009.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Downs B, Oscar-Berman M, Waite R, et al. Have We Hatched the Addiction Egg: Reward Deficiency Syndrome Solution System™. J Genet Syndr Gene Ther. 2013 Jun 3;4(136):14318. doi: 10.4172/2157-7412.1000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higginson AD, McNamara JM, Houston AI. Fatness and fitness: exposing the logic of evolutionary explanations for obesity. Proc Biol Sci. 2016 Jan 13;283(1822) doi: 10.1098/rspb.2015.2443. pii: 20152443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blum K, Chen AL, Oscar-Berman M, et al. Generational association studies of dopaminergic genes in reward deficiency syndrome (RDS) subjects: selecting appropriate phenotypes for reward dependence behaviors. Int J Environ Res Public Health. 2011 Dec;8(12):4425–59. doi: 10.3390/ijerph8124425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polyák Á, Winkler Z, Kuti D, et al. Brown adipose tissue in obesity: Fractalkine-receptor dependent immune cell recruitment affects metabolic-related gene expression. Biochim Biophys Acta. 2016 Jul 12; doi: 10.1016/j.bbalip.2016.07.002. pii: S1388-1981(16)30183-4. [DOI] [PubMed] [Google Scholar]
- 27.Leménager T, Dieter J, Hill H. Exploring the Neural Basis of Avatar Identification in Pathological Internet Gamers and of Self-Reflection in Pathological Social Network Users. J Behav Addict. 2016 Jul 14;:1–15. doi: 10.1556/2006.5.2016.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo P, Li Y, Eslamfam S, Ding W, et al. Discovery of Novel Genes Mediating Glucose and Lipid Metabolisms. Curr Protein Pept Sci. 2016 Jun 26; doi: 10.2174/1389203717666160627084304. [DOI] [PubMed] [Google Scholar]
- 29.Stamer UM, Stüber F. Genetic factors in pain and its treatment. Curr Opin Anaesthesiol. 2007 Oct;20(5):478–84. doi: 10.1097/ACO.0b013e3282ef6b2c. [DOI] [PubMed] [Google Scholar]
- 30.Palmer RH, Brick L, Nugent NR, et al. Examining the role of common genetic variants on alcohol, tobacco, cannabis and illicit drug dependence: genetics of vulnerability to drug dependence. Addiction. 2015 Mar;110(3):530–7. doi: 10.1111/add.12815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ducci F, Goldman D. The genetic basis of addictive disorders. Psychiatr Clin North Am. 2012;35(2):495–519. doi: 10.1016/j.psc.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blum K, Noble EP, Sheridan PJ, Montgomery A, Ritchie T, Jagadeeswaran P, Nogami H, Briggs AH, Cohn JB. Allelic association of human dopamine D2 receptor gene in alcoholism. JAMA. 1990;263(15):2055–2060. [PubMed] [Google Scholar]
- 33.Grandy DK, Litt M, Allen L. The human dopamine D2 receptor gene is located on chromosome 11 at q22-q23 and identifies a TaqI RFLP. Am J Hum Genet. 1989;45(5):778–785. [PMC free article] [PubMed] [Google Scholar]
- 34.Moeller SJ, Parvaz MA, Shumay E, et al. Gene x abstinence effects on drug cue reactivity in addiction: multimodal evidence. J Neurosci. 2013;33(24):10027–10036. doi: 10.1523/JNEUROSCI.0695-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blum K, Oscar-Berman M, Jacobs W, et al. Buprenorphine Response as a Function of Neurogenetic Polymorphic Antecedents: Can Dopamine Genes Affect Clinical Outcomes in Reward Deficiency Syndrome (RDS)? J Addict Res Ther. 2014;5 doi: 10.4172/2155-6105.1000185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Gong J, Xie C, et al. Alterations in brain connectivity in three sub-regions of the anterior cingulate cortex in heroin-dependent individuals: Evidence from resting state fMRI. Neuroscience. 2015;284:998–1010. doi: 10.1016/j.neuroscience.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 37.Blum K, Liu Y, Wang W, et al. rsfMRI effects of KB220ZTM on neural pathways in reward circuitry of abstinent genotyped heroin addicts. Postgrad Med. 2015;127(2):232–241. doi: 10.1080/00325481.2015.994879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levey DF, Le-Niculescu H, Frank J, et al. Genetic risk prediction and neurobiological understanding of alcoholism. Transl Psychiatry. 2014;4:e391. doi: 10.1038/tp.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farris SP, Arasappan D, Hunicke-Smith S, et al. Transcriptome organization for chronic alcohol abuse in human brain. Mol Psychiatry. 2014 doi: 10.1038/mp.2014.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan J, Aliev F, Webb BT, et al. Using genetic information from candidate gene and genome-wide association studies in risk prediction for alcohol dependence. Addict Biol. 2014;19(4):708–721. doi: 10.1111/adb.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goldman D. Candidate genes in alcoholism. Clin Neurosci. 1995;3(2):174–181. [PubMed] [Google Scholar]
- 42.Blum K, Sheridan PJ, Wood RC, et al. The D2 dopamine receptor gene as a determinant of reward deficiency syndrome. J R Soc Med. 1996;89(7):396–400. doi: 10.1177/014107689608900711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen TJ, Blum K, Mathews D, et al. Are dopaminergic genes involved in a predisposition to pathological aggression? Hypothesizing the importance of “super normal controls” in psychiatric genetic research of complex behavioral disorders. Med Hypotheses. 2005;65(4):703–707. doi: 10.1016/j.mehy.2005.04.037. [DOI] [PubMed] [Google Scholar]
- 44.Mayer P, Höllt V. Pharmacogenetics of opioid receptors and addiction. Pharmacogenet Genomics. 2006;16(1):1–7. doi: 10.1097/01.fpc.0000182781.87932.0d. [DOI] [PubMed] [Google Scholar]
- 45.Butler SF, McNaughton EC, Black RA. Tapentadol abuse potential: a post marketing evaluation using a sample of individuals evaluated for substance abuse treatment. Pain Med. 2015;16(1):119–130. doi: 10.1111/pme.12524. [DOI] [PubMed] [Google Scholar]
- 46.Blum K, Chen TJ, Meshkin B, et al. Reward deficiency syndrome in obesity: a preliminary cross-sectional trial with a Genotrim variant. Adv Ther. 2006 Nov-Dec;23(6):1040–51. doi: 10.1007/BF02850224. [DOI] [PubMed] [Google Scholar]
- 47.Blum K, Febo M, Badgaiyan RD, et al. Common Neurogenetic Diagnosis and Meso-Limbic Manipulation of Hypodopaminergic Function in Reward Deficiency Syndrome (RDS): Changing the Recovery Landscape. Curr Neuropharmacol. 2016 May 12; doi: 10.2174/1570159X13666160512150918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blum K, Chen TJ, Morse S, et al. Overcoming qEEG abnormalities and reward gene deficits during protracted abstinence in male psychostimulant and polydrug abusers utilizing putative dopamine D2 agonist therapy: part 2. Postgrad Med. 2010 Nov;122(6):214–26. doi: 10.3810/pgm.2010.11.2237. [DOI] [PubMed] [Google Scholar]
- 49.Miller DK, Bowirrat A, Manka M, et al. Acute intravenous synaptamine complex variant KB220™ “normalizes” neurological dysregulation in patients during protracted abstinence from alcohol and opiates as observed using quantitative electroencephalographic and genetic analysis for reward polymorphisms: part 1, pilot study with 2 case reports. Postgrad Med. 2010 Nov;122(6):188–213. doi: 10.3810/pgm.2010.11.2236. [DOI] [PubMed] [Google Scholar]
- 50.Myers RD, Robinson DE. Mmu and D2 receptor antisense oligonucleotides injected in nucleus accumbens suppress high alcohol intake in genetic drinking HEP rats. Alcohol. 1999;18(2–3):225–33. doi: 10.1016/s0741-8329(99)00015-4. [DOI] [PubMed] [Google Scholar]
- 51.Thanos PK, Volkow ND, Freimuth P, Umegaki H, Ikari H, Roth G, Ingram DK, Hitzemann R. Overexpression of dopamine D2 receptors reduces alcohol self-administration. J Neurochem. 2001;78(5):1094–103. doi: 10.1046/j.1471-4159.2001.00492.x. [DOI] [PubMed] [Google Scholar]
- 52.Thanos PK, Rivera SN, Weaver K, Grandy DK, Rubinstein M, Umegaki H, Wang GJ, Hitzemann R, Volkow ND. Dopamine D2R DNA transfer in dopamine D2 receptor-deficient mice: effects on ethanol drinking. Life Sci. 2005;77(2):130–9. doi: 10.1016/j.lfs.2004.10.061. [DOI] [PubMed] [Google Scholar]
- 53.Thanos PK, Michaelides M, Umegaki H, Volkow ND. D2R DNA transfer into the nucleus accumbens attenuates cocaine self-administration in rats. Synapse. 2008;62(7):481–6. doi: 10.1002/syn.20523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blum K, Chen AL, Chen TJ, et al. Activation instead of blocking mesolimbic dopaminergic reward circuitry is a preferred modality in the long term treatment of reward deficiency syndrome (RDS): a commentary. Theor Biol Med Model. 2008 Nov 12;5:24. doi: 10.1186/1742-4682-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gyollai A, Griffiths MD, Barta C, et al. The genetics of problem and pathological gambling: a systematic review. Curr Pharm Des. 2014;20(25):3993–9. doi: 10.2174/13816128113199990626. [DOI] [PubMed] [Google Scholar]
- 56.Blum K, Badgaiyan RD, Gold MS. Hypersexuality Addiction and Withdrawal: Phenomenology, Neurogenetics and Epigenetics. Cureus. 2015 Oct 12;7(10):e348. doi: 10.7759/cureus.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meshkin B, Blum K. Folate nutrigenetics: a convergence of dietary folate metabolism, folic acid supplementation, and folate antagonist pharmacogenetics. Drug Metab Lett. 2007 Jan;1(1):55–60. doi: 10.2174/187231207779814319. [DOI] [PubMed] [Google Scholar]
- 58.Blum K, Simpatico T, Badgaiyan RD, Demetrovics Z, Fratantonio J, et al. Coupling Neurogenetics (GARS™) and a Nutrigenomic Based Dopaminergic Agonist to Treat Reward Deficiency Syndrome (RDS): Targeting Polymorphic Reward Genes for Carbohydrate Addiction Algorithms. J Reward Defic Syndr. 2015;1(2):75–80. doi: 10.17756/jrds.2015-012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blum K, Badgaiyan RD. Addiction Research and Therapy in the 21st Century: Providing a Forum for Evidence -Based Addiction Medicine. J Addict Res Ther. 2013;4 doi: 10.4172/2155-6105.1000e117. pii: 1000e117. [DOI] [PMC free article] [PubMed] [Google Scholar]