Abstract
Philadelphia-like B-cell precursor acute lymphoblastic leukemia (Ph-like ALL) is characterized by distinct genetic alterations and inferior prognosis in children and younger adults. The purpose of this study was a genetic and clinical characterization of Ph-like ALL in adults. Twenty-six (13%) of 207 adult patients (median age: 42 years) with B-cell precursor ALL (BCP-ALL) were classified as having Ph-like ALL using gene expression profiling. The frequency of Ph-like ALL was 27% among 95 BCP-ALL patients negative for BCR-ABL1 and KMT2A-rearrangements. IGH-CRLF2 rearrangements (6/16; P=0.002) and mutations in JAK2 (7/16; P<0.001) were found exclusively in the Ph-like ALL subgroup. Clinical and outcome analyses were restricted to patients treated in German Multicenter Study Group for Adult ALL (GMALL) trials 06/99 and 07/03 (n=107). The complete remission rate was 100% among both Ph-like ALL patients (n=19) and the “remaining BCP-ALL” cases (n=40), i.e. patients negative for BCR-ABL1 and KMT2A-rearrangements and the Ph-like subtype. Significantly fewer Ph-like ALL patients reached molecular complete remission (33% versus 79%; P=0.02) and had a lower probability of continuous complete remission (26% versus 60%; P=0.03) and overall survival (22% versus 64%; P=0.006) at 5 years compared to the remaining BCP-ALL patients. The profile of genetic lesions in adults with Ph-like ALL, including older adults, resembles that of pediatric Ph-like ALL and differs from the profile in the remaining BCP-ALL. Our study is the first to demonstrate that Ph-like ALL is associated with inferior outcomes in intensively treated older adult patients. Ph-like adult ALL should be recognized as a distinct, high-risk entity and further research on improved diagnostic and therapeutic approaches is needed. (NCT00199056, NCT00198991)
Introduction
Acute lymphoblastic leukemia (ALL) is a heterogeneous disease with a complex pattern of molecular changes including fusion proteins, copy number alterations, and gene mutations.1 Considerable research is being dedicated to achieving better molecular characterization of ALL patients without known prognostic factors and to identifying patients with molecular aberrations that can be targeted by specific compounds (e.g. tyrosine kinase inhibitors in the case of the BCR-ABL1 fusion). In children, a subgroup of B-cell precursor ALL (BCP-ALL) with a gene expression profile similar to BCR-ABL1 (Philadelphia chromosome; Ph)-positive ALL, but lacking the BCR-ABL1 fusion gene, has been described and found to be associated with inferior outcomes compared to those of other subtypes of BCP-ALL.2,3 In pediatric patients this subgroup of ALL, named Ph-like or BCR-ABL1-like ALL, is associated with a number of genetic lesions that are potential candidates for targeted treatment.4 One study identified rearrangements of CRLF2 (IGH-CRLF2 or P2RY8-CRLF2) in approximately 50% of Ph-like ALL patients, and concomitant JAK1/JAK2 mutations in 50% of the CRLF2-rearranged patients.5 The remaining Ph-like ALL patients harbor a variety of kinase alterations, including translocations involving ABL class kinases or JAK2.4,5 Alterations (sequence mutations or deletions) of IKZF1 are also frequently observed in patients with Ph-like ALL.4,5 Several groups have verified these findings in children, adolescents and younger adults and demonstrated an increasing incidence of Ph-like ALL in adolescents and younger adults compared to children.5–9 We recently showed that the incidence of the Ph-like ALL subtype is highest in adolescents and younger adults (19–27%) and then decreases significantly with increasing age.10 So far, the data on the prognostic impact and molecular characteristics of Ph-like ALL in adults are limited and inconsistent.5,11
We set out to study the genetic characteristics of Ph-like ALL in adults, in comparison to other BCR-ABL1- and KMT2A-negative BCP-ALL, using gene expression profiling, mutational profiling by targeted amplicon sequencing and assessment of common copy number alterations. Furthermore, we analyzed the clinical characteristics and prognostic impact of the Ph-like ALL subtype in patients treated in prospective trials of the German Multicenter Study Group for Adult ALL (GMALL) for adult patients aged 15–65 years.
Methods
Patients
This analysis included 207 consecutive patients with newly diagnosed BCP-ALL studied between 1999 and 2005 at two reference laboratories.12,13 The routine diagnostic work-up included immunophenotyping, fluorescent in situ hybridization (FISH) for BCR-ABL1 and KMT2A (MLL) rearrangements (MLL-t), cytogenetics and molecular analyses of BCR-ABL1 translocations and MLL-t. Details of the patients’ characteristics are shown in Online Supplementary Table S1. The selection of patients for further molecular analysis was focused on two molecular BCP-ALL subgroups and based on available material: Ph-like ALL (n=16) and remaining BCP-ALL (Ph-negative, MLL-t-negative, non-Ph-like) (n=27). The study design is shown in Figure 1. Analyses of clinical characteristics and outcome were focused on patients treated in the GMALL trials 06/99 and 07/03. These trials incorporated intensive, pediatric-based therapy and risk-adapted allocation to allogeneic stem cell transplantation as previously described.14 Ph-negative BCP-ALL patients with high white blood cell count (>30,000/μL), pro-B-ALL, or failure to achieve complete remission after induction phase I were allocated to the high-risk group. Both studies were approved by the institutional review board of the University of Frankfurt, Germany and are registered at clinicaltrials.gov (NCT00199056, NCT00198991). All patients had given signed informed consent to participation in the trials.
Figure 1.
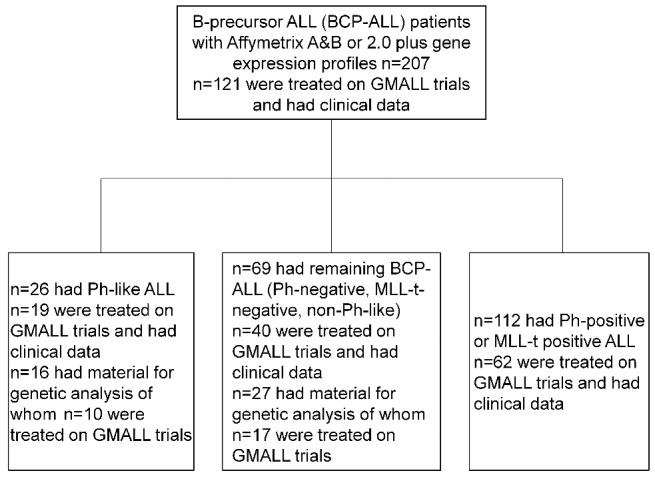
Study design. Flow chart showing the study design and distribution of patients.
Gene expression analysis
All patients were analyzed with the Affymetrix HGU-133 A, B Set (n=109) or with the Affymetrix HGU-133 Plus 2.0 chip (n=98). Details of sample preparation, hybridization and image acquisition have been described previously.15 No cell sorting was performed. The median percentage of leukemic cells in the samples analyzed was 90% (range, 30%–100%). The mean percentage was 87%. The HG-U133 A, B chips and HG-U133 Plus 2.0 chips were normalized separately by the robust multichip average method as described by Irizarry et al.16 Only the 44,754 probe sets present on both the Affymetrix HG-U133A, B chips and the HG-U133 Plus 2.0 chips were included in the analysis. To correct the batch effect resulting from the use of different chip designs, we applied an empirical Bayesian method as described previously.17 We used the published classification algorithm of Roberts et al. to identify Ph-like ALL patients by clustering.4 The analysis was performed based on 240 of the 257 probe sets used by Roberts et al. present in both chips (Online Supplementary Table S2). To validate this classification, microarray files were independently analyzed and classified using prediction analysis of microarrays.4 Patients with a predicted class 2 coefficient ≥0.5 were classified as having the Ph-like ALL gene expression signature. A direct comparison of the results from both algorithms is shown in the Online Supplement (Online Supplementary Table S3).
Patients with high CRLF2 expression we defined as those with CRLF2 expression in the highest quintile of the whole dataset (Online Supplementary Figure S1). The gene expression dataset is publicly available (Gene Expression Omnibus ID GSE66006).
Analysis of minimal residual disease
Minimal residual disease (MRD) was measured by quantitative polymerase chain reaction of individual IGH and TR rearrangements in a central laboratory as described previously.14 Molecular remission was defined as no MRD detection above the level of 10−4, confirmed with a minimum sensitivity of 10−4 according to published standards.18 The preferred time-point for evaluation of molecular response was before first consolidation. If not available, results of samples collected immediately after induction or after first consolidation were considered.
Analysis of IGH and CRLF2 rearrangements and P2RY8 deletions
FISH analyses were performed on pretreatment samples using standard techniques and commercial probes according to the manufacturers’ instructions. The presence of IGH translocations was determined by interphase FISH using the LSI IGH Dual Color, Break Apart Rearrangement Probe (Abbott). In addition, a CRLF2 break apart probe and a P2RY8 deletion probe (both Cytocell aquarius) were used.
Quantitative polymerase chain reaction for detection of the genomic P2RY8-CRLF2 fusion
Genomic DNA was amplified using primers designed to flank the fusion breakpoint (P2RY8_q_fw: 5′-AGCCACCCTTCCTTTAATAACTCAT-3′, CRLF2_q_rv: 5′-TCTGAGCTCCATGGTTCGTCA-3′).19 Quantitative polymerase chain reaction was performed on a LightCycler 480 (Roche) using a probe for real-time detection of the fusion amplicons (P-C_q_pr: FAM-TGGGCGGATCACGAGGTCAGGA-TAMRA).
Analysis of copy number alterations
Copy number alterations were analyzed using the SALSA multiplex ligation-dependent probe amplification kit P335-B1 (MRC Holland) according to the manufacturer’s protocol.20 The probe mix contains probes for IKZF1, PAX5, ETV6, RB1, BTG1 and the BTG1 downstream region, EBF1, CDKN2A-CDKN2B, ZFY, JAK2 and for the Xp22.33 region (PAR region, CRLF2, CSF2RA, IL3RA and P2RY8 genes). The multiplex ligation-dependent probe amplification data were analyzed using Coffalyser.Net analysis software version 131211.1524 provided by the manufacturer using default settings. Reference samples were used as recommended in the manufacturer’s protocol.
Targeted amplicon sequencing
A selection of 131 genes (Online Supplementary Table S4) known to be recurrently mutated in ALL were studied by targeted amplicon sequencing (Haloplex, Agilent) in 16 patients with Ph-like ALL and 23 with remaining BCP-ALL. The resulting libraries were sequenced in three runs on a MiSeq instrument. A quality metrics summary of sequencing data is shown in Online Supplementary Table S5. Sequence data were aligned to the human reference genome (version hg19) using BWA-MEM.21,22 Single nucleotide variants and short insertions or deletions were called using VarScan 2 and Pindel, respectively.23 Only genomic regions with a coverage of ≥30 fold were analyzed. Non-synonymous variants (single nucleotide polymorphisms and indels) in coding regions with ≥10 variant reads of specific leukemia-associated genes were reported (Online Supplementary Table S6). Details regarding the analysis algorithm have been published previously.17
Statistics
Statistical analyses were performed using R 3.0.1 software24 and routines from the biostatistics software repository Bioconductor and SPSS version 21.0 (SPSS Inc., Chicago, IL, USA). The two-sided Fisher exact test was used to compare categorical variables, while the Wilcoxon-Mann-Whitney test was applied for continuous variables. All patients whose clinical characteristics and outcome were analyzed were registered at the GMALL Study Center, where statistical analysis was performed with the SAS program (SAS-PC, version 8.02; SAS Institute, Cary, NC, USA). For statistical comparisons of the clinical variables, the two-sided Fisher exact test was applied for categorical variables and the Wilcoxon test for continuous variables. The survival analysis was based on the Kaplan-Meier method. Overall survival was calculated from date of diagnosis until death or last follow-up. Continuous complete remission (remission duration) was calculated from date of first complete remission to relapse or last follow-up in complete remission including patients with death in complete remission. Disease-free survival was calculated from date of first complete remission to relapse or death from any cause. Survival rates are given as probabilities of survival at 5 years, with a 95% confidence interval. The log-rank test was used to compare survival curves. A Cox model was used for multivariate analysis of the impact of different factors on remission duration and overall survival. For all analyses, P values ≤0.05 are considered statistically significant.
Results
Identification of patients with a Philadelphia-like gene expression profile
In total, 207 patients with BCP-ALL were analyzed (Figure 1), of whom 95 were negative for BCR-ABL1 and MLL-t. Of the 207 patients, we classified 26 patients (13%) as having Ph-like ALL based on their gene expression profile and the absence of the BCR-ABL1 fusion: this corresponds to a prevalence of 27% (26/95) in patients negative for BCR-ABL1 and MLL-t. The other 69 patients were then classified as having “remaining BCP-ALL” (Ph-negative, MLL-t-negative, non-Ph-like) and served as the reference group, similarly to another recent study in adults.11 Other studies, particularly in pediatric populations, compared Ph-like ALL to B-other patients defined as patients negative for the BCR-ABL1, MLL-t, ETV6-RUNX1 or TCF3-PBX1 fusions and hyperdiploidy or hypodiploidy. The incidence of Ph-like ALL in B-other patients in our cohort was 32% (26/82).
Since there is no generally accepted definition of high-risk features of adult ALL, it is unclear whether the B-other or remaining BCP-ALL group is a better control group for comparison with the Ph-like subtype. To account for this difficulty, we mention both comparisons in this report.
A detailed description of the age distribution of the patients and their immunophenotypic and molecular parameters is given in Online Supplementary Figure S2 and in Online Supplementary Table S1.
Philadelphia-like acute lymphoblastic leukemia is associated with specific genetic lesions and expression changes
Most patients with high CRLF2 expression clustered in the Ph-like ALL subgroup (15/26, 58% versus 7/69, 10% of remaining BCP-ALL; P<0.001). Online Supplementary Figure S3 shows the distribution of CRLF2 expression in BCP-ALL subtypes.
Further molecular analysis was focused on the 43 patients of the Ph-like and remaining BCP-ALL subgroups (Ph-like ALL: n=16; remaining BCP-ALL: n=27; Figure 2A/B). The selection was based on available material. Unfortunately, no RNA or cDNA for further molecular characterization, especially for ABL or JAK fusions, was available.
Figure 2.
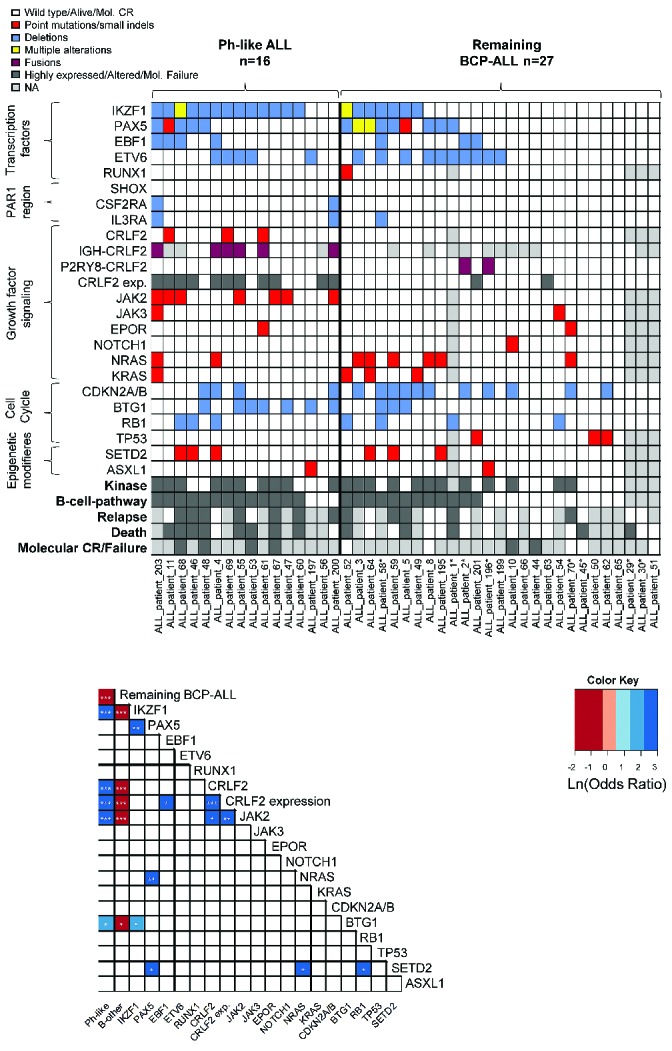
Distribution of genetic alterations in Ph-like and remaining BCP-ALL. (A) Panel A shows the distribution of common mutations, deletions and fusions and CRLF2 expression in Ph-like and remaining BCP-ALL (Ph-negative, MLL-t negative, non-Ph-like) patients. The definition of kinase and B-cell pathway reported by Roberts et al.5 was used. Patients represented by light gray boxes for Relapse and Death were not treated in GMALL trials 06/99 and 07/03 and no clinical data were available for them. Patients marked with “*” had either ETV6-RUNX1 or TCF3-PBX1 fusions or ALL with high hyperdiploidy. (B) Panel B shows the correlation of common mutations, deletions and fusions and CRLF2 expression in Ph-like and remaining BCP-ALL (Ph-negative, MLL-t negative, non-Ph-like).
All patients with an IGH-CRLF2 rearrangement (n=6) were in the Ph-like subgroup (only 29/43 patients could be analyzed), whereas P2RY8-CRLF2 fusions (n=2) were only found in the remaining BCP-ALL group (P=0.002 and P>0.05, respectively). Analysis of common copy number alterations by multiplex ligation-dependent probe amplification revealed that 77% of patients (33/43) had at least one deletion. Deletions of IKZF1 were significantly more common in Ph-like ALL than in remaining BCP-ALL (13/16, 81% versus 7/27, 30%; P<0.001). Mutational profiling of 131 genes recurrently mutated in ALL was performed by targeted amplicon sequencing (Online Supplementary Table S4). A total of 115 non-synonymous mutations affecting 50 genes was identified. At least one mutation could be identified in 92% (36/39) of patients. The distribution of the most common mutations is shown in Figure 2A and in more detail in Online Supplementary Table S6. Nine mutations in JAK2 were identified in seven patients. These mutations were found exclusively in the Ph-like ALL subgroup (7/16, 44% versus 0/23, 0%; P<0.001), and commonly associated with CRFL2 over-expression. The majority of JAK2 mutations (7/9) resulted in the change of amino acid R683 to G (n=5) or S (n=2). All mutations spared codon V617 and were located in the protein tyrosine kinase (PTK) domains (Online Supplementary Figure S4).
Rearrangements or sequence mutations of CRLF2 were found exclusively in the Ph-like subgroup (7/16 versus 0/23; P<0.001). Molecular alterations associated with ALL subtypes and co-occurrence of certain molecular alterations are shown in Figure 2B. The Ph-like ALL subgroup was associated with alterations of IKZF1 (P<0.001), CRLF2 (P<0.001), JAK2 (P<0.001), BTG1 (P=0.02) and high CRLF2 expression (P<0.001).
Clinical characteristics and prognosis of Philadelphia-like acute lymphoblastic leukemia
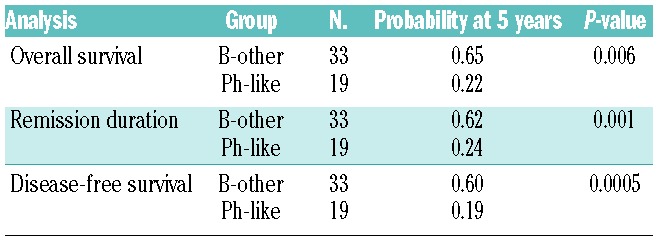
The clinical and outcome analyses were restricted to BCP-ALL patients intensively treated in GMALL trials 06/99 and 07/03 (n=107). This group contained Ph-positive (n=37), Ph-like (n=19), MLL-t (n=11) and remaining BCP-ALL (n=40) patients. The analysis focused on the comparison of Ph-like and remaining BCP-ALL patients. No significant differences in baseline characteristics, including age, sex, white-cell count, hemoglobin, platelet count and risk group, were observed between the Ph-like and remaining BCP-ALL subgroups (Table 1). Fifty-eight percent (11/19) of the Ph-like patients belonged to the standard-risk group, compared to 50% of the remaining BCP-ALL group. The complete remission rate after induction was 100% for both the Ph-like and the remaining BCP-ALL patients. However, significantly fewer patients reached molecular remission in the Ph-like ALL subgroup (4/12, 33% versus 15/19, 79%; P=0.02) (Table 2). Patients with Ph-like ALL had a significantly inferior probability of continuous complete remission at 5 years (26% versus 60%; P=0.03) (Figure 3). Overall, 12/19 Ph-like patients relapsed very rapidly after induction therapy with a median time to relapse of only 122 days (range, 25–844) compared to 555 days (range, 96–1248) in the remaining BCP-ALL group (P=0.03). Consequently, the realization of stem cell transplantation in first complete remission as stipulated in the protocol was significantly lower among high-risk patients in the Ph-like subgroup than among high-risk patients in the remaining BCP-ALL group (2/8, 25% versus 15/20, 75%; P=0.01). Overall, three patients with Ph-like ALL (2 high risk, 1 standard risk) received a stem cell transplant in first complete remission (1 relapsed after the transplant, 1 died in complete remission and 1 patient is in continuous complete remission). Overall survival (22% versus 64% at 5 years; P=0.006) and disease-free survival (19% versus 57% at 5 years, P<0.001) were significantly inferior in the Ph-like group than in the remaining BCP-ALL group (Figure 3). The outcome of Ph-like ALL patients was particularly poor within the high-risk group, mainly because of early relapse (Online Supplementary Figure S5AD). There was no significant difference in results when patients who underwent stem cell transplantation in first complete remission were excluded (Online Supplementary Figure S5E-F) and if Ph-like ALL patients were compared to the more stringently defined B-other ALL group (Table 3).
Table 1.
Characteristics of BCP-ALL patients treated in GMALL studies 06/99 and 07/03.
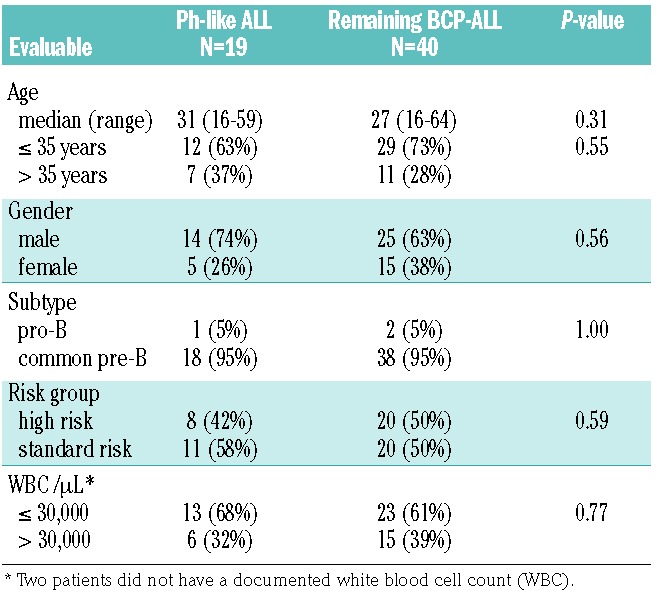
Table 2.
Clinical and molecular response of Ph-like and remaining BCP-ALL treated on GMALL studies 06/99 and 07/03.
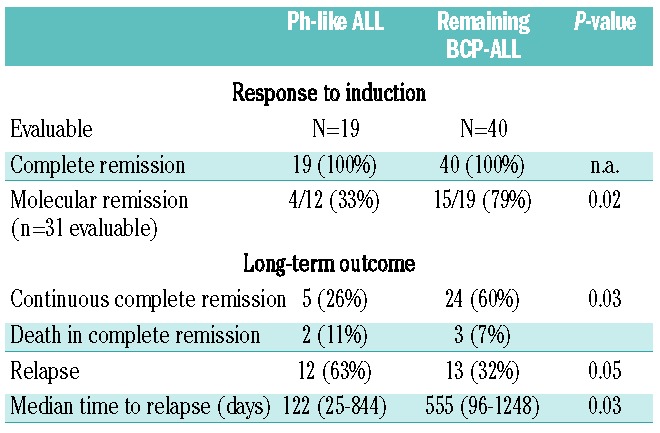
Figure 3.
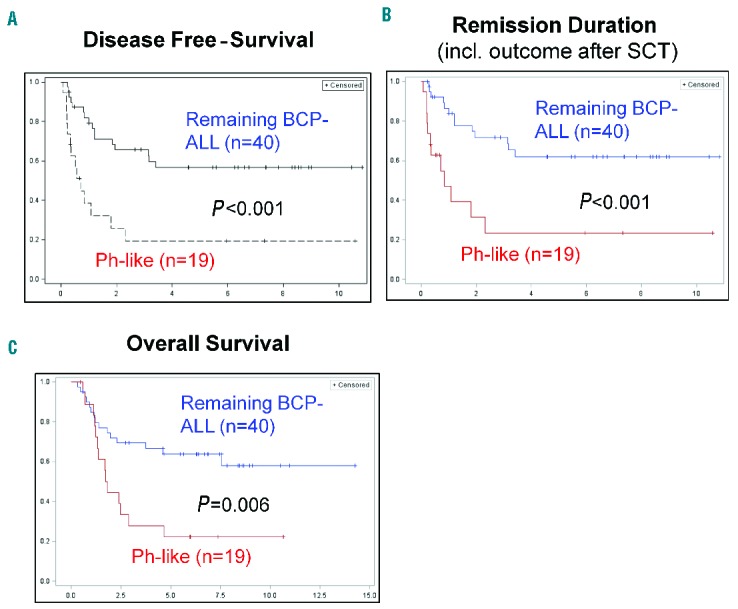
Disease-free survival, remission duration and overall survival of patients with Ph-like or remaining BCP-ALL. (A) Disease-free survival, (C) remission duration and (C) overall survival in ALL patients treated in GMALL trials 06/99 and 07/03, comparing the Ph-like ALL subgroup with remaining BCP-ALL patients (Ph-negative, MLL-t negative, non-Ph-like).
Table 3.
Comparison of outcomes of the Ph-like ALL subgroup with patients classified as having B-other ALL (negative for the BCR-ABL1, MLL-t, ETV6-RUNX1 or TCF3-PBX1 fusions and hyperdiploidy or hypodiploidy).
For Ph-like ALL patients with available MRD data (n=12), the molecular response was a significant predictor of relapse. One of four patients with molecular complete remission relapsed compared to seven of eight patients who did not achieve molecular complete remission. The remission duration was significantly different between patients with Ph-like ALL who did or did not achieve molecular complete remission (log-rank test, P=0.02).
Association of molecular alterations with survival
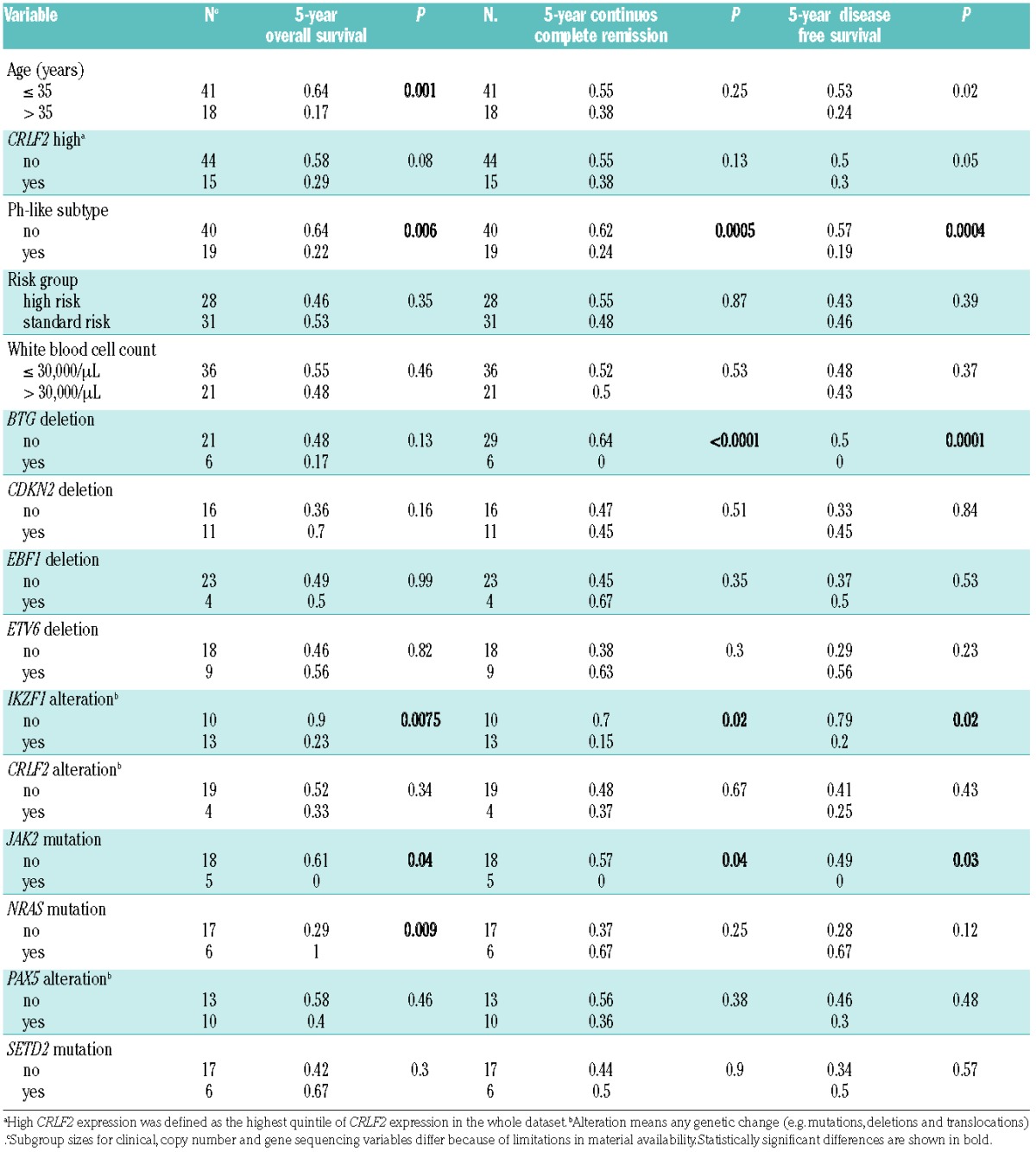
To further characterize the influence of different variables on survival in Ph-like and remaining BCP-ALL, univariate Cox analyses were used. The variables age (>35 years), JAK2 and NRAS mutation, IKZF1 alteration and the Ph-like subtype were significant predictors of inferior overall survival (Table 4), whereas Ph-like subtype, BTG deletion, IKZF1 alterations and JAK2 mutations were significant predictors of remission duration.
Table 4.
Univariate prognostic factors in Ph-like ALL and remaining BCP-ALL patients treated in GMALL studies 06/99 and 07/03.
Because of the limited overlap of patients with MRD analyses and further molecular characterization we calculated a multivariate model focusing the analysis on remission duration in 31 patients with information on Ph-like subtype and MRD. Both factors maintained significance with P=0.008 and a hazard ratio of 3.4 for molecular response and P=0.04 with a hazard ratio of 1.8 for Ph-like status. In a similar model for overall survival only molecular response maintained significance with P=0.002 and a hazard ratio of 4.7.
IGH-CRLF2 and/or JAK2 mutations identify a subset of Philadelphia-like acute lymphoblastic leukemia with high specificity
In our data set, FISH analysis for IGH-CRLF2 rearrangements and sequencing of JAK2 PTK was sufficient to identify Ph-like cases with a sensitivity and specificity of 63% and 100%, respectively. Using these parameters we were able to predict the Ph-like ALL subtype (Online Supplementary Figure S6) in a publicly available dataset (n=1,726)5 with a sensitivity of 33% and specificity of 100% regardless of age groups. If only patients aged ≥16 years were considered 41% of Ph-like cases could be identified (specificity 100%). The frequency of JAK2 mutations and IGH-CRLF2 rearrangements increased significantly with older age (<16 years versus ≥16 years; P=0.008).
Discussion
The aim of our study was to retrospectively identify Ph-like ALL in a cohort of adult patients up to the age of 64 years treated on prospective protocols of the GMALL study group. Several groups have demonstrated the adverse prognostic impact of the Ph-like ALL subtype in children, adolescents and younger adults and more recently in adults.2,3,5,11 However, data from adults with Ph-like ALL are still limited and inconsistent with regards to classification algorithms, the frequency of additional genetic alterations and the impact of this ALL subtype on survival.5,11
Our study used a simplified version of the St. Jude classifier2 to identify Ph-like ALL patients. The classification approach was independently validated using prediction analysis of microarrays and showed high concordance.4 There were three additional cases classified as Ph-like ALL by prediction analysis of microarrays, but they had weaker coefficients, and likely harbored sequence mutations rather than kinase rearrangements (Online Supplementary Table S3).
A recently published study comparing the St. Jude classifier with the DCOG/Erasmus MC signature3,9 has shown poor concordance, with the DCOG/Erasmus MC classifier identifying nearly twice as many patients as Ph-like compared to the St. Jude classifier.25
In our study, using the St. Jude classification algorithm, the overall frequency of the Ph-like subtype in adults with BCP-ALL was 13% and 32% in B-other ALL, which is in the range of published data.5,10,11
Beside our analysis, two other studies investigated Ph-like ALL in adult patients.5,11 When comparing these studies, it is important to note the difference in patient cohorts, classification algorithms and treatment protocols. A recently reported study by the HOVON group analyzed the clinical course of 21 Ph-like adolescents, younger adults and older adults (median age 25 years; range, 15–59) and compared it to that of 50 patients with remaining BCP-ALL (median age 34 years; range, 16–68).11 The landmark study by Roberts et al. included 710 adolescents and young adults with B-other ALL (n=169 Ph-like), with a median age of <20 years and an upper age limit of 39 years.5 The 19 Ph-like patients in our trial who were evaluable for outcome analysis had a median age of 31 years (range, 16–59) and were compared to 40 remaining BCP-ALL patients with a median age of 27 years (range, 16–64). Whereas the study by Roberts et al. focused on younger ALL patients, the HOVON trial and our study analyzed the clinical course of a cohort of older adults with BCP-ALL.
Regarding the treatment schedule, only three of the 21 Ph-like ALL patients in the HOVON cohort were treated according to pediatric-inspired, high-intensive protocols (HOVON-70 or HOVON-71). The adolescents and younger adults analyzed by Roberts et al. were treated in eight different clinical trials by the Children’s Oncology Group (COG), the Eastern Cooperative Oncology Group (ECOG), the MD Anderson Cancer Center (MDACC), Alliance – Cancer and Leukemia Group B protocols (CALGB) and St. Jude Hospital. These trials included various protocols and treatment schedules. Our own study included only patients intensively treated with GMALL pediatric-based protocols (06/99 and 07/03) and these patients therefore constitute a homogenously treated cohort with approaches comparable to current therapeutic strategies. This may explain the differences in complete remission rates (HOVON: 71%, St. Jude: not reported, GMALL: 100%) in Ph-like ALL patients between our analyses.5,11
In the HOVON study there were no significant differences in event-free survival and overall survival between adult patients with Ph-like ALL and those with remaining BCP-ALL, although there was a trend to higher relapse rates among the former.11 Roberts et al. reported that adolescents and younger adults with Ph-like ALL had inferior event-free survival and overall survival and a significant association with elevated MRD levels (analyzed only among patients included in COG trials).5 Our study demonstrated inferior disease-free survival, remission duration and overall survival, higher relapse rates and persistence of MRD in adults with Ph-like ALL. The most obvious reason for the discrepancies between the study by Roberts et al., our study and the HOVON analysis is the different classification algorithm used in the HOVON study which identified a different subset of patients. This is further accentuated by the differences in molecular findings. Deletions of IKZF1 were found in only 35% of cases of Ph-like ALL in the HOVON study (mutations not analyzed), whereas Roberts et al. found IKZF1 alterations (point mutations or deletions) in 68% of patients. We identified alterations of IKZF1 in 81% of the patients with Ph-like ALL. The higher incidence in our cohort compared to that in the St. Jude study could be due to an association of these alterations with older age. CRLF2 expression, JAK2 mutations or IGH-CRLF2 translocations were not reported in the HOVON study. We found that adult Ph-like ALL is also strongly associated with JAK2 mutations (44%), CRLF2 alterations (44%) and increased CRLF2 expression (58%), which is in accordance with the results in adolescents and younger adults reported by Roberts et al. Furthermore, we show that the frequency of JAK2 mutations and IGH-CRLF2 translocations increases significantly with age. Unfortunately, a full genetic characterization, for example by RNA-sequencing, was not possible because of limitations in available material, especially in cases with low CRLF2 expression.
The results of studies using the St. Jude classification algorithm demonstrate that some cases of Ph-like ALL are characterized by highly specific molecular alterations (especially JAK2 mutations and IGH-CRLF2). As gene expression analysis26 is not performed in routine diagnostics, testing for JAK2 PTK mutations and the IGH-CRLF2 translocation may represent a first and easily implementable option to identify at least a subgroup of patients with Ph-like ALL with high specificity until a consensus classification is available. It is, however, important to note that other studies showed that JAK2 mutations and IGH-CRLF2 fusions were not associated with the Ph-like ALL subtype in all cases.4 Furthermore, we were not able to demonstrate a prognostic impact of the combined analysis of these variables in our dataset (data not shown). This analysis is preliminary and has currently no clinical consequence but could help to promote further research. Further studies are warranted to evaluate the specificity and prognostic impact of these alterations for the classification of Ph-like ALL.
The optimal treatment of Ph-like ALL, and particularly of those patients with persistent MRD remains to be defined. Specifically, the potential impact of stem cell transplantation in Ph-like ALL needs to be established. In a recent pediatric trial, the negative prognostic impact of Ph-like ALL was almost completely eliminated by MRD-based treatment stratification including more intensive post-remission therapy and stem cell transplantation in Ph-like ALL patients with persistent MRD.8 Due to the limited number of patients in whom MRD was evaluated in our study, it remains unclear whether the Ph-like sub type would still have an independent prognostic impact, if MRD evaluation were to be performed in all adult patients. However, so far, our study is the first to report MRD data for older adults with Ph-like ALL. In our study only 4/12 (25%) patients with Ph-like ALL achieved a molecular complete remission and three of them remain in continuous complete remission. Whether the same would occur in larger cohorts of adult patients needs to be addressed by future studies. The relevance of molecular response in the general BCP-ALL subgroup has already been shown in far larger groups of patients who have undergone MRD assessment at distinct, well-defined time-points.14 The current strategy of the GMALL study group includes stem cell transplantation preceded by targeted therapies, such as antibody therapies,27 whenever available in patients with persistent MRD after first consolidation (including Ph-like ALL).
In summary, our study shows that the molecular and clinical features of adults with Ph-like ALL resemble those characteristic of pediatric Ph-like ALL patients. For the first time, our study demonstrates the negative prognostic impact on survival of the Ph-like ALL subtype in intensively treated older adults. Future studies should elucidate the role of thorough MRD monitoring and allogeneic transplantation, which are potentially effective in the Ph-like ALL subgroup.
Supplementary Material
Acknowledgments
The authors thank all participants and recruiting centers of the GMALL trials.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/102/1/130
Funding
This work was supported by a grant from the Friedrich-Baur Stiftung to TH, start-up funding from the Ludwig-Maximilians-Universität to KHM (FöFoLe 783) and by grant support from Deutsche Forschungsgemeinschaft [DFG SFB 1243, TP A06 (KHM, KS)]; Wilhelm Sander Stiftung [no. 2013.086.1 (TH, UM and KS)]; and the German Cancer Consortium (Deutsches Konsortium für Translationale Krebsforschung, Heidelberg, Germany). The CRLF2 and P2RY8 FISH probes were kindly provided by Cytocell aquarius. The GMALL studies were supported by Deutsche Krebshilfe (702657Ho2) and the José
References
- 1.Inaba H, Greaves M, Mullighan CG. Acute lymphoblastic leukaemia. Lancet. 2013;381 (9881):1943–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mullighan CG, Su X, Zhang J, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360 (5):470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Den Boer ML, van Slegtenhorst M, De Menezes RX, et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol. 2009;10(2):125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roberts KG, Morin RD, Zhang J, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22(2):153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts KG, Li Y, Payne-Turner D, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med. 2014;371(11):1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iijima K, Yoshihara H, Ohki K, et al. An analysis of Ph-like ALL in Japanese patients. Blood. 2013;122(21):352–352. [Google Scholar]
- 7.Silvestri D, Vendramini E, Fazio G, et al. Philadelphia-like signature in childhood acute lymphoblastic leukemia: the AIEOP Experience. Blood. 2013;122(21):353–353. [Google Scholar]
- 8.Roberts KG, Pei D, Campana D, et al. Outcomes of children with BCR-ABL1-like acute lymphoblastic leukemia treated with risk-directed therapy based on the levels of minimal residual disease. J Clin Oncol. 2014;32(27):3012–3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Veer A, Waanders E, Pieters R, et al. Independent prognostic value of BCR-ABL1-like signature and IKZF1 deletion, but not high CRLF2 expression, in children with B-cell precursor ALL. Blood. 2013;122(15): 2622–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herold T, Baldus CD, Gokbuget N. Ph-like acute lymphoblastic leukemia in older adults. N Engl J Med. 2014;371(23):2235. [DOI] [PubMed] [Google Scholar]
- 11.Boer JM, Koenders JE, van der Holt B, et al. Expression profiling of adult acute lymphoblastic leukemia identifies a BCR-ABL1-like subgroup characterized by high non-response and relapse rates. Haematologica. 2015;100(7):e261–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haferlach T, Kohlmann A, Wieczorek L, et al. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the International Microarray Innovations in Leukemia Study Group. J Clin Oncol. 2010;28(15):2529–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haferlach T, Kohlmann A, Schnittger S, et al. Global approach to the diagnosis of leukemia using gene expression profiling. Blood. 2005;106(4):1189–1198. [DOI] [PubMed] [Google Scholar]
- 14.Gokbuget N, Kneba M, Raff T, et al. Adult patients with acute lymphoblastic leukemia and molecular failure display a poor prognosis and are candidates for stem cell transplantation and targeted therapies. Blood. 2012;120(9):1868–1876. [DOI] [PubMed] [Google Scholar]
- 15.Herold T, Jurinovic V, Metzeler KH, et al. An eight-gene expression signature for the prediction of survival and time to treatment in chronic lymphocytic leukemia. Leukemia. 2011;25(10):1639–1645. [DOI] [PubMed] [Google Scholar]
- 16.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. [DOI] [PubMed] [Google Scholar]
- 17.Herold T, Metzeler KH, Vosberg S, et al. Isolated trisomy 13 defines a homogeneous AML subgroup with high frequency of mutations in spliceosome genes and poor prognosis. Blood. 2014;124(8):1304–1311. [DOI] [PubMed] [Google Scholar]
- 18.Bruggemann M, Schrauder A, Raff T, et al. Standardized MRD quantification in European ALL trials: proceedings of the Second International Symposium on MRD assessment in Kiel, Germany, 18–20 September 2008. Leukemia. 2010;24(3):521–535. [DOI] [PubMed] [Google Scholar]
- 19.Morak M, Attarbaschi A, Fischer S, et al. Small sizes and indolent evolutionary dynamics challenge the potential role of P2RY8-CRLF2-harboring clones as main relapse-driving force in childhood ALL. Blood. 2012;120(26):5134–5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwab CJ, Jones LR, Morrison H, et al. Evaluation of multiplex ligation-dependent probe amplification as a method for the detection of copy number abnormalities in B-cell precursor acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2010;49(12):1104–1113. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16): 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.R_Core_Team. R: A Language and Environment for Statistical Computing.
- 25.Boer JM, Marchante JR, Evans WE, et al. BCR-ABL1-like cases in pediatric acute lymphoblastic leukemia: a comparison between DCOG/Erasmus MC and COG/St. Jude signatures. Haematologica. 2015;100(9):e354–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang H, Roberts KG, Chen I-ML, et al. Development and validation of a highly sensitive and specific gene expression classifier to prospectively screen and identify B-precursor acute lymphoblastic leukemia (ALL) patients with a Philadelphia chromosome-like (“Ph-like” or “BCR-ABL1-Like”) Signature Blood. 2013;122(21):826–826. [Google Scholar]
- 27.Topp MS, Gokbuget N, Zugmaier G, et al. Long-term follow-up of hematologic relapse-free survival in a phase 2 study of blinatumomab in patients with MRD in B-lineage ALL. Blood. 2012;120(26):5185–5187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


