Abstract
RNAcentral is a database of non-coding RNA (ncRNA) sequences that aggregates data from specialised ncRNA resources and provides a single entry point for accessing ncRNA sequences of all ncRNA types from all organisms. Since its launch in 2014, RNAcentral has integrated twelve new resources, taking the total number of collaborating database to 22, and began importing new types of data, such as modified nucleotides from MODOMICS and PDB. We created new species-specific identifiers that refer to unique RNA sequences within a context of single species. The website has been subject to continuous improvements focusing on text and sequence similarity searches as well as genome browsing functionality. All RNAcentral data is provided for free and is available for browsing, bulk downloads, and programmatic access at http://rnacentral.org/.
INTRODUCTION
Non-coding RNAs (ncRNAs) are a critical component of cellular machinery of all organisms. For example, the ribosomal RNA has been shown to be a ribozyme responsible for peptide bond synthesis (1), and the activity of the eukaryotic spliceosome is mediated by ncRNAs (2). Apart from being the main player in those central processes, ncRNAs provide additional layers of subtle regulation of gene expression. MicroRNAs have been shown to regulate the expression of the majority of mRNAs in animals and plants (3), and the range of regulatory roles of lncRNAs, including by genomic scaffolding and chromatin remodelling and modification (4), is becoming clearer. There is an intense scientific interest in ncRNAs resulting in a large number of ncRNA databases, but until recently searching and comparing them was challenging, and there was no uniform way to access or reference ncRNA sequences. To this end, we developed RNAcentral, a database of ncRNA sequences that serves as a single entry point to the data from a large collection of collaborating ncRNA resources that cover ncRNA sequences of all types and from all organisms. First conceived in 2011 (5), RNAcentral was made public in 2014 (6). This paper gives an update on the status of the database and related activities.
DATA OVERVIEW
New Expert Databases
RNAcentral aggregates ncRNA sequence data from an international consortium of RNA resources that we call Expert Databases. In the past two years, 12 additional Expert Databases have been integrated into RNAcentral (see Table 1). Among the newly imported resources were two major ribosomal RNA databases, SILVA (7) and Greengenes (8), which complement rRNAs from ENA and Rfam, as well as a high quality subset of rRNA sequences from RDP (9). Ribosomal RNAs represent the majority of sequences in RNAcentral due to their use in environmental sampling.
Table 1. Expert Databases imported into RNAcentral since release 1.
| Database name | Description | URL |
|---|---|---|
| DictyBase | A model organism database for the social amoeba Dictyostelium discoideum | http://dictybase.org |
| Greengenes | A full-length 16S rRNA gene database that provides a curated taxonomy based on de novo tree inference | http://greengenes.secondgenome.com/downloads |
| LNCipedia | An integrated database of human lncRNAs | http://www.lncipedia.org |
| MODOMICS | A comprehensive database of RNA modifications | http://modomics.genesilico.pl |
| NONCODE | An integrated knowledge database dedicated to ncRNAs (excluding tRNAs and rRNAs) | http://www.noncode.org |
| PDB | A repository of information about the 3D structures of large biological molecules | http://www.wwpdb.org/ |
| PomBase | A comprehensive database for the fission yeast Schizosaccharomyces pombe | http://www.pombase.org |
| SGD | An integrated database for the budding yeast | http://yeastgenome.org |
| SILVA | A resource for quality checked and aligned ribosomal RNA sequence data | http://www.arb-silva.de/ |
| snoPY | A database of snoRNAs, snoRNA gene loci, and target RNAs as well as snoRNA orthologues | http://snoopy.med.miyazaki-u.ac.jp/ |
| TAIR | A database of genetic and molecular biology data for the model higher plant Arabidopsis thaliana | http://www.arabidopsis.org |
| WormBase | A resource for genomic and genetic data about nematodes with primary emphasis on Caenorhabditis elegans | http://www.wormbase.org |
Sequences from five Model Organism Databases (DictyBase (10), PomBase (11), SGD (12), TAIR (13) and WormBase (14)) have been imported into RNAcentral. Long non-coding RNA (lncRNA) coverage was extended by the addition of NONCODE (15) and LNCipedia (16) datasets. The inclusion of PDB (17) as an Expert Database helps to map between the worlds of ncRNA sequence and structure. Small nucleolar RNAs (snoRNAs) play an important role in guiding the modification of other ncRNA and mRNAs (18), and snoPY (19) provides to RNAcentral a dataset of snoRNAs found in human, fly, worm, yeast, and thale cress. An up-to-date list of all RNAcentral Expert Databases is available at http://rnacentral.org/expert-databases.
Database growth
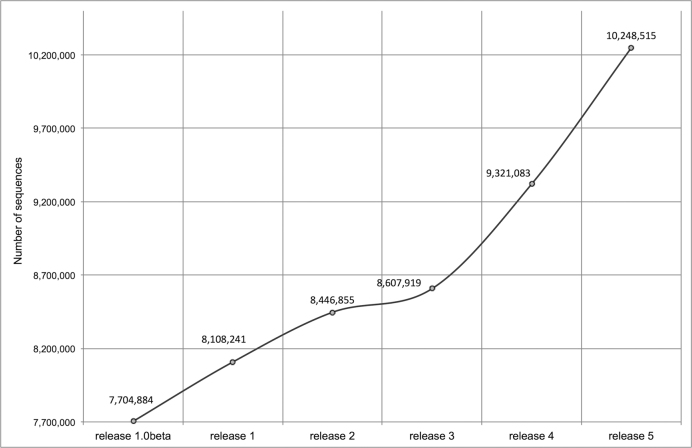
RNAcentral currently holds 10.2 million distinct ncRNA sequences (an increase of 2.1 million since release 1) with about 28 million cross-references (up 11 million since release 1) to 22 Expert Databases (Figure 1). The sequences come from over 720 000 organisms from all domains of life, with half of all sequences deriving from bacteria and ∼40% from eukaryotes.
Figure 1.
Growth in the number of unique RNA sequences since release 1. An up-to-date version of the chart is available at http://rnacentral.org/about-us.
Species-specific identifiers
Since its inception, RNAcentral has provided unique identifiers for each distinct RNA sequence. For example, URS00004C905 is the identifier for the sequence UUUGGUCCCCUUCAACCAGCUG, which is the miRNA miR-133a-3p. The exact same sequence is observed in more than 10 species. These unique and stable identifiers are useful for a number of tasks, such as unambiguously referring to an RNA sequence, reducing redundancy in sequence datasets, and keeping track of cross-references. However, it is essential to be able to refer uniquely to a ncRNA sequence annotation in a particular species. In order to address this issue, we have introduced species-specific RNA sequence identifiers. The new identifiers are composed of a unique RNA sequence identifier and a NCBI taxonomic identifier separated by underscore. For example, URS00004C9052_9606 is the human copy of the miRNA hsa-miR-133a-3p (9606 being the taxonomic identifier for Homo sapiens) and URS00004C9052_10090 corresponds to the mmu-miR-133a-3p sequence from mouse. RNAcentral text searches now return species-specific entries by default.
One of the use cases for the species-specific RNA sequence identifiers is curation of literature references to assign ontology terms to an RNA sequence from a specific organism, for example to annotate the molecule's function. The biocuration community has already begun using RNAcentral identifiers for assigning Gene Ontology terms to human miRNAs (20). We are investigating assigning identifiers that differentiate between multiple occurrences of the same sequence in a genome.
Modified nucleotides
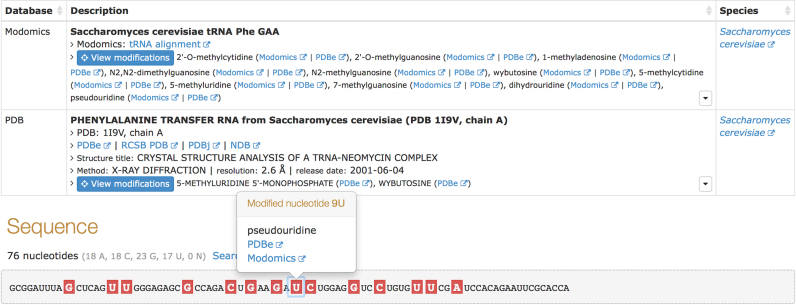
Modified nucleotide residues play important roles in functions of many ncRNAs. For example, modifications of ribosomal RNAs are essential for the assembly and stability of ribosomes (21), and tRNA modifications can influence protein gene expression (22). In order to begin capturing information about modified residues in RNA molecules and enable comparison of different datasets, we imported modified rRNA and tRNA sequences from MODOMICS (23), a database of RNA modification pathways, as well as all ncRNA modifications from Protein Data Bank. So far RNAcentral contains over 170 different chemical modifications found at over 8000 positions in about 600 unique sequences. Figure 2 shows a web interface that provides a unified view of the modifications from different databases. For each modified residue, cross-references to the MODOMICS and PDB databases are provided to enable easy access to more detailed information about each modification. In future releases we will continue importing information about modified nucleotides from MODOMICS and other resources as more data become available thanks to new developments in sequencing technology (24,25).
Figure 2.
Web interface displaying modified nucleotides for a Saccharomyces cerevisiae tRNA(Phe) sequence (RNAcentral entry URS000011107D_4932).
WEBSITE UPDATES
The RNAcentral website has been subject to continuous improvement based on user feedback and several interactive workshops held during annual RNAcentral consortium meetings. The homepage was redesigned to reflect the three main ways to access the data: text search, sequence similarity search and genome browser, each of which will be discussed further below.
Text search
RNAcentral text search provides a flexible way for exploring RNAcentral data using a faceted interface powered by EBI search (26). In the past two years, the search functionality was improved both in terms of user interface and searchable data. For example, publication metadata (such as paper titles, PubMed identifiers or author names) associated with ncRNA sequences can now be searched, which makes it possible to look up ncRNA sequences submitted to sequence archives when new papers are published. For example, a recent paper describes TRM10 (27), a mRNA-derived small RNA that acts as ribosome inhibitor. By searching RNAcentral with PubMed identifier 24685157 the sequence is easily found (see entry URS00007E15D1) and can be used for further analysis, such as sequence similarity search. Moreover, it is possible to compare sequences reported in different papers. For example, one can identify mitochondrial rRNA sequences shared by a Danish (28) and an Iranian (29) population by searching in RNAcentral with both publication titles. These search results can be exported in multiple formats, thereby facilitating more detailed investigation by the user.
Sequence similarity search
RNAcentral sequence search is the first online tool that enables sequence similarity searches against a comprehensive set of ncRNAs. The service is powered by the nhmmer software, which has a comparable speed to BLAST but is more sensitive (30). The web interface supports searching using an RNA or DNA sequence as a query and displays pairwise sequence alignments for each match. The results can be sorted by E-value, sequence identity and other criteria. If an exact match for a query sequence already exists in the database, its RNAcentral identifier is retrieved using the RNAcentral API without having to wait for the full search results to become available.
Genome browser
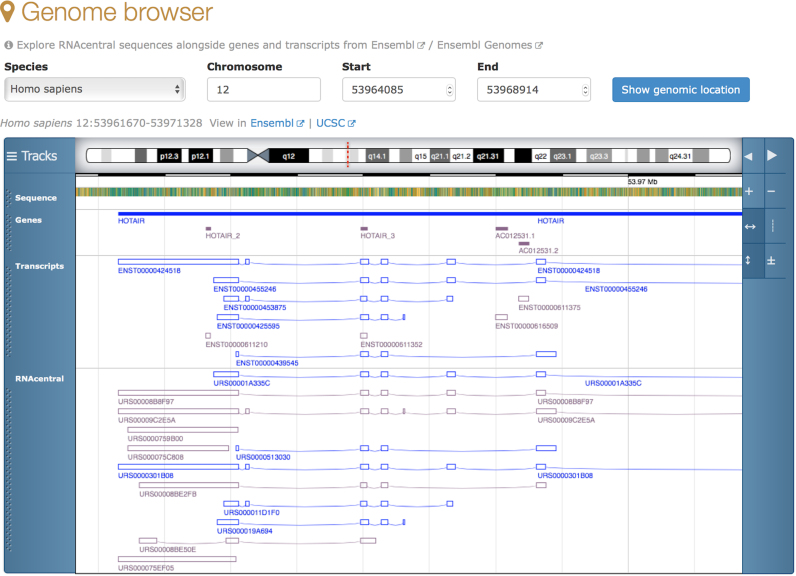
Viewing sequences in their genomic context can provide important biological insights. For example, one can visualise snoRNAs found in introns of lncRNA GAS5 (31) using a built-in genome browser (see RNAcentral entry URS00008B3C85). In a recent update, we extended this functionality to enable browsing RNAcentral starting with a genomic location. The embedded genome browser, powered by Genoverse (http://genoverse.org), currently supports 13 key species, including human, mouse, fly, worm, and yeast (Figure 3). RNAcentral sequences are displayed alongside genes and transcripts from Ensembl (32) and Ensembl Genomes (33) with links to fully featured genome browsers, such as UCSC (34) and Ensembl. The genomic data are also available via a programmatic interface and downloadable files in BED/GFF formats.
Figure 3.
RNAcentral genome browser showing HOTAIR lncRNA in human chromosome 12.
RNACENTRAL USE CASES
Citations to RNAcentral are beginning to appear in the scientific literature, and currently fall into three main categories of use: (i) RNAcentral is used as a comprehensive source of ncRNA annotations and a reference for identification of novel ncRNAs in species like rainbow trout or cow (35,36). (ii) RNAcentral identifiers are used for literature curation, for example, human miRNAs are annotated with GO terms using RNAcentral species-specific identifiers to refer to RNA sequences (20). (iii) The RNAcentral API is used for sequence or identifier retrieval (37–39). For example, given an RNAcentral sequence identifier the Forna tool can predict and visualise its secondary structure. Over the past two years, the RNAcentral website has been accessed by over 33 000 unique visitors from 156 countries who performed over 100 000 text and 12 000 sequence similarity searches.
TRAINING AND OUTREACH
We continuously engage in outreach activities and provide user support by email and on GitHub. We have delivered over 20 presentations to date at scientific conferences and research institutes worldwide and organised a training event at the Wellcome Genome Campus. We also developed an online training course and recorded a live webinar (available on YouTube). All training materials can be accessed at http://rnacentral.org/training. We are open to suggestions from our user community by email, on GitHub and on Twitter. The contact information can be found at http://rnacentral.org/contact.
FUTURE DIRECTIONS
The main goal of RNAcentral is to provide a comprehensive set of ncRNA sequences, so integrating new Expert Databases and importing more data will remain a priority. For example, our goal is to integrate the remaining Model Organism Databases, such as FlyBase (40) and RGD (41), in order to provide uniform access to high-quality ncRNA sequence and annotations from key species. More than 20 participating ncRNA resources still need to be integrated and new Expert Databases are continually joining the consortium. We welcome relevant databases to contact us about membership.
In the second phase of development, we will begin to integrate new types of annotations that can provide insight into the functions of ncRNA sequences found in RNAcentral. We will work on importing secondary structure information from Comparative RNA Website (42), GtRNAdb (43) and Rfam (44) databases. Work is underway on integrating miRNA-mRNA interactions from TarBase (45) and miRNA–lncRNA interactions from LncBase (46) into RNAcentral. We will enrich existing annotations by importing ontology terms from external resources and assigning ontology terms automatically where possible. We also plan to use sequence alignment-based mapping to connect more RNAcentral sequences to reference genomes. RNAcentral is a young and fast-growing resource, but it has already proved useful for many applications, and its utility will be increased as more data are integrated and the associated services mature.
ACKNOWLEDGEMENTS
RNAcentral has been prepared by Anton I. Petrov, Simon J.E. Kay, Ioanna Kalvari, Kevin L. Howe, Kristian A. Gray, Elspeth A. Bruford, Paul J. Kersey, Guy Cochrane, Robert D. Finn, Alex Bateman at the European Bioinformatics Institute (EMBL-EBI); Ana Kozomara, Sam Griffiths-Jones (University of Manchester); Adam Frankish (Wellcome Trust Sanger Institute); Christian W. Zwieb (University of Texas), Britney Y. Lau, Kelly P. Williams (Sandia National Laboratories); Patricia P. Chan, Todd M. Lowe (University of California Santa Cruz); Jamie J. Cannone, Robin R. Gutell (University of Texas at Austin); Magdalena A. Machnicka, Janusz M. Bujnicki (International Institute of Molecular and Cell Biology and Adam Mickiewicz University); Maki Yoshihama, Naoya Kenmochi (University of Miyazaki); Benli Chai, James R. Cole (Michigan State University); Maciej Szymanski, Wojciech M. Karlowski (Adam Mickiewicz University); Valerie Wood (University of Cambridge); Eva Huala, Tanya Z. Berardini (The Arabidopsis Information Resource and Phoenix Bioinformatics); Yi Zhao, Runsheng Chen (Chinese Academy of Sciences); Weimin Zhu (Data Science, National Center for Protein Science); Maria D. Paraskevopoulou, Ioannis S. Vlachos, Artemis G Hatzigeorgiou (University of Thessaly and Hellenic Pasteur Institute); SILVA team (Jacobs University Bremen and Max Planck Institute for Marine Microbiology); Lina Ma, Zhang Zhang (Beijing Institute of Genomics, Chinese Academy of Sciences); Joern Puetz (University of Strasbourg); Peter F. Stadler (University of Leipzig); Daniel McDonald (University of California San Diego); Siddhartha Basu, Petra Fey (Northwestern University); Stacia R. Engel, J. Michael Cherry (Stanford University); Pieter-Jan Volders, Pieter Mestdagh (Ghent University and Cancer Research Institute Ghent); Jacek Wower (Auburn University); Michael Clark (University of Oxford and Garvan Institute of Medical Research); Xiu Cheng Quek, Marcel E. Dinger (Garvan Institute of Medical Research).
Footnotes
Present address: Magdalena A. Machnicka, Faculty of Mathematics, Informatics and Mechanics (MIM), University of Warsaw, Banacha 2, 02-097 Warsaw, Poland.
Contributor Information
The RNAcentral Consortium:
Anton I Petrov, Simon J E Kay, Ioanna Kalvari, Kevin L Howe, Kristian A Gray, Elspeth A Bruford, Paul J Kersey, Guy Cochrane, Robert D Finn, Alex Bateman, Ana Kozomara, Sam Griffiths-Jones, Adam Frankish, Christian W Zwieb, Britney Y Lau, Kelly P Williams, Patricia P Chan, Todd M Lowe, Jamie J Cannone, Robin Gutell, Magdalena A Machnicka, Janusz M Bujnicki, Maki Yoshihama, Naoya Kenmochi, Benli Chai, James R Cole, Maciej Szymanski, Wojciech M Karlowski, Valerie Wood, Eva Huala, Tanya Z Berardini, Yi Zhao, Runsheng Chen, Weimin Zhu, Maria D Paraskevopoulou, Ioannis S Vlachos, Artemis G Hatzigeorgiou, Lina Ma, Zhang Zhang, Joern Puetz, Peter F Stadler, Daniel McDonald, Siddhartha Basu, Petra Fey, Stacia R Engel, J Michael Cherry, Pieter-Jan Volders, Pieter Mestdagh, Jacek Wower, Michael B Clark, Xiu Cheng Quek, and Marcel E Dinger
FUNDING
Biotechnology and Biological Sciences Research Council (BBSRC) [BB/J019232/1]. Funding for open access charge: Research Councils UK (RCUK).
Conflict of interest statement. Janusz M. Bujnicki is an Executive Editor of Nucleic Acids Research.
REFERENCES
- 1.Beringer M., Rodnina M.V. The ribosomal peptidyl transferase. Mol. Cell. 2007;26:311–321. doi: 10.1016/j.molcel.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 2.Hang J., Wan R., Yan C., Shi Y. Structural basis of pre-mRNA splicing. Science. 2015;349:1191–1198. doi: 10.1126/science.aac8159. [DOI] [PubMed] [Google Scholar]
- 3.Axtell M.J., Westholm J.O., Lai E.C. Vive la différence: biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 2011;12:221. doi: 10.1186/gb-2011-12-4-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomita S., Abdalla M.O.A., Fujiwara S., Yamamoto T., Iwase H., Nakao M., Saitoh N. Roles of long noncoding RNAs in chromosome domains. Wiley Interdiscip. Rev. RNA. 2016 doi: 10.1002/wrna.1384. doi:10.1002/wrna.1384. [DOI] [PubMed] [Google Scholar]
- 5.Bateman A., Agrawal S., Birney E., Bruford E.A., Bujnicki J.M., Cochrane G., Cole J.R., Dinger M.E., Enright A.J., Gardner P.P., et al. RNAcentral: A vision for an international database of RNA sequences. RNA. 2011;17:1941–1946. doi: 10.1261/rna.2750811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Consortium, RNAcentral. RNAcentral: an international database of ncRNA sequences. Nucleic Acids Res. 2015;43:D123–D129. doi: 10.1093/nar/gku991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDonald D., Price M.N., Goodrich J., Nawrocki E.P., DeSantis T.Z., Probst A., Andersen G.L., Knight R., Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole J.R., Wang Q., Fish J.A., Chai B., McGarrell D.M., Sun Y., Brown C.T., Porras-Alfaro A., Kuske C.R., Tiedje J.M. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basu S., Fey P., Pandit Y., Dodson R., Kibbe W.A., Chisholm R.L. DictyBase 2013: integrating multiple Dictyostelid species. Nucleic Acids Res. 2013;41:D676–D683. doi: 10.1093/nar/gks1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDowall M.D., Harris M.A., Lock A., Rutherford K., Staines D.M., Bähler J., Kersey P.J., Oliver S.G., Wood V. PomBase 2015: updates to the fission yeast database. Nucleic Acids Res. 2015;43:D656–D661. doi: 10.1093/nar/gku1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cherry J.M., Hong E.L., Amundsen C., Balakrishnan R., Binkley G., Chan E.T., Christie K.R., Costanzo M.C., Dwight S.S., Engel S.R., et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berardini T.Z., Reiser L., Li D., Mezheritsky Y., Muller R., Strait E., Huala E. The Arabidopsis information resource: Making and mining the ‘gold standard’ annotated reference plant genome. Genesis. 2015;53:474–485. doi: 10.1002/dvg.22877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yook K., Harris T.W., Bieri T., Cabunoc A., Chan J., Chen W.J., Davis P., de la Cruz N., Duong A., Fang R., et al. WormBase 2012: more genomes, more data, new website. Nucleic Acids Res. 2012;40:D735–D741. doi: 10.1093/nar/gkr954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y., Li H., Fang S., Kang Y., Wu W., Hao Y., Li Z., Bu D., Sun N., Zhang M.Q., et al. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2016;44:D203–D208. doi: 10.1093/nar/gkv1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Volders P.-J., Verheggen K., Menschaert G., Vandepoele K., Martens L., Vandesompele J., Mestdagh P. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 2015;43:D174–D180. doi: 10.1093/nar/gku1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Velankar S., van Ginkel G., Alhroub Y., Battle G.M., Berrisford J.M., Conroy M.J., Dana J.M., Gore S.P., Gutmanas A., Haslam P., et al. PDBe: improved accessibility of macromolecular structure data from PDB and EMDB. Nucleic Acids Res. 2016;44:D385–D395. doi: 10.1093/nar/gkv1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dupuis-Sandoval F., Poirier M., Scott M.S. The emerging landscape of small nucleolar RNAs in cell biology. Wiley Interdiscip. Rev. RNA. 2015;6:381–397. doi: 10.1002/wrna.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshihama M., Nakao A., Kenmochi N. snOPY: a small nucleolar RNA orthological gene database. BMC Res. Notes. 2013;6:426. doi: 10.1186/1756-0500-6-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huntley R.P., Sitnikov D., Orlic-Milacic M., Balakrishnan R., D'Eustachio P., Gillespie M.E., Howe D., Kalea A.Z., Maegdefessel L., Osumi-Sutherland D., et al. Guidelines for the functional annotation of microRNAs using the Gene Ontology. RNA. 2016;22:667–676. doi: 10.1261/rna.055301.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osterman I.A., Sergiev P.V., Tsvetkov P.O., Makarov A.A., Bogdanov A.A., Dontsova O.A. Methylated 23S rRNA nucleotide m2G1835 of Escherichia coli ribosome facilitates subunit association. Biochimie. 2011;93:725–729. doi: 10.1016/j.biochi.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 22.Duechler M., Leszczyńska G., Sochacka E., Nawrot B. Nucleoside modifications in the regulation of gene expression: focus on tRNA. Cell. Mol. Life Sci. 2016;73:3075–3095. doi: 10.1007/s00018-016-2217-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Machnicka M.A., Milanowska K., Osman Oglou O., Purta E., Kurkowska M., Olchowik A., Januszewski W., Kalinowski S., Dunin-Horkawicz S., Rother K.M., et al. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res. 2013;41:D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cozen A.E., Quartley E., Holmes A.D., Hrabeta-Robinson E., Phizicky E.M., Lowe T.M. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat. Methods. 2015;12:879–884. doi: 10.1038/nmeth.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krogh N., Jansson M.D., Häfner S.J., Tehler D., Birkedal U., Christensen-Dalsgaard M., Lund A.H., Nielsen H. Profiling of 2’-O-Me in human rRNA reveals a subset of fractionally modified positions and provides evidence for ribosome heterogeneity. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw482. doi:10.1093/nar/gkw482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Squizzato S., Park Y.M., Buso N., Gur T., Cowley A., Li W., Uludag M., Pundir S., Cham J.A., McWilliam H., et al. The EBI Search engine: providing search and retrieval functionality for biological data from EMBL-EBI. Nucleic Acids Res. 2015;43:W585–W588. doi: 10.1093/nar/gkv316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pircher A., Bakowska-Zywicka K., Schneider L., Zywicki M., Polacek N. An mRNA-derived noncoding RNA targets and regulates the ribosome. Mol. Cell. 2014;54:147–155. doi: 10.1016/j.molcel.2014.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li S., Besenbacher S., Li Y., Kristiansen K., Grarup N., Albrechtsen A., Sparsø T., Korneliussen T., Hansen T., Wang J., et al. Variation and association to diabetes in 2000 full mtDNA sequences mined from an exome study in a Danish population. Eur. J. Hum. Genet. 2014;22:1040–1045. doi: 10.1038/ejhg.2013.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Derenko M., Malyarchuk B., Bahmanimehr A., Denisova G., Perkova M., Farjadian S., Yepiskoposyan L. Complete mitochondrial DNA diversity in Iranians. PLoS One. 2013;8:e80673. doi: 10.1371/journal.pone.0080673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wheeler T.J., Eddy S.R. nhmmer: DNA homology search with profile HMMs. Bioinformatics. 2013;29:2487–2489. doi: 10.1093/bioinformatics/btt403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith C.M., Steitz J.A. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5’-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol. Cell. Biol. 1998;18:6897–6909. doi: 10.1128/mcb.18.12.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yates A., Akanni W., Amode M.R., Barrell D., Billis K., Carvalho-Silva D., Cummins C., Clapham P., Fitzgerald S., Gil L., et al. Ensembl 2016. Nucleic Acids Res. 2016;44:D710–D716. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kersey P.J., Allen J.E., Armean I., Boddu S., Bolt B.J., Carvalho-Silva D., Christensen M., Davis P., Falin L.J., Grabmueller C., et al. Ensembl Genomes 2016: more genomes, more complexity. Nucleic Acids Res. 2016;44:D574–D580. doi: 10.1093/nar/gkv1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Speir M.L., Zweig A.S., Rosenbloom K.R., Raney B.J., Paten B., Nejad P., Lee B.T., Learned K., Karolchik D., Hinrichs A.S., et al. The UCSC Genome Browser database: 2016 update. Nucleic Acids Res. 2016;44:D717–D725. doi: 10.1093/nar/gkv1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Al-Tobasei R., Paneru B., Salem M. Genome-wide discovery of long non-coding RNAs in rainbow trout. PLoS One. 2016;11:e0148940. doi: 10.1371/journal.pone.0148940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Durkin K., Rosewick N., Artesi M., Hahaut V., Griebel P., Arsic N., Burny A., Georges M., Van den Broeke A. Characterization of novel Bovine Leukemia Virus (BLV) antisense transcripts by deep sequencing reveals constitutive expression in tumors and transcriptional interaction with viral microRNAs. Retrovirology. 2016;13:33. doi: 10.1186/s12977-016-0267-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kerpedjiev P., Hammer S., Hofacker I.L. Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams. Bioinformatics. 2015;31:3377–3379. doi: 10.1093/bioinformatics/btv372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jossinet F., Ludwig T.E., Westhof E. Assemble: an interactive graphical tool to analyze and build RNA architectures at the 2D and 3D levels. Bioinformatics. 2010;26:2057–2059. doi: 10.1093/bioinformatics/btq321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eggenhofer F., Hofacker I.L., Höner Zu Siederdissen C. RNAlien - unsupervised RNA family model construction. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw558. doi:10.1093/nar/gkw558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Attrill H., Falls K., Goodman J.L., Millburn G.H., Antonazzo G., Rey A.J., Marygold S.J., FlyBase Consortium FlyBase: establishing a Gene Group resource for Drosophila melanogaster. Nucleic Acids Res. 2016;44:D786–D792. doi: 10.1093/nar/gkv1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimoyama M., De Pons J., Hayman G.T., Laulederkind S.J.F., Liu W., Nigam R., Petri V., Smith J.R., Tutaj M., Wang S.-J., et al. The Rat Genome Database 2015: genomic, phenotypic and environmental variations and disease. Nucleic Acids Res. 2015;43:D743–D750. doi: 10.1093/nar/gku1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cannone J.J., Subramanian S., Schnare M.N., Collett J.R., D'Souza L.M., Du Y., Feng B., Lin N., Madabusi L.V., Müller K.M., et al. The comparative RNA Web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:1–31. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan P.P., Lowe T.M. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016;44:D184–D189. doi: 10.1093/nar/gkv1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nawrocki E.P., Burge S.W., Bateman A., Daub J., Eberhardt R.Y., Eddy S.R., Floden E.W., Gardner P.P., Jones T.A., Tate J., et al. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 2015;43:D130–D137. doi: 10.1093/nar/gku1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vlachos I.S., Paraskevopoulou M.D., Karagkouni D., Georgakilas G., Vergoulis T., Kanellos I., Anastasopoulos I.-L., Maniou S., Karathanou K., Kalfakakou D., et al. DIANA-TarBase v7.0: indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 2015;43:D153–D159. doi: 10.1093/nar/gku1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paraskevopoulou M.D., Vlachos I.S., Karagkouni D., Georgakilas G., Kanellos I., Vergoulis T., Zagganas K., Tsanakas P., Floros E., Dalamagas T., et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016;44:D231–D238. doi: 10.1093/nar/gkv1270. [DOI] [PMC free article] [PubMed] [Google Scholar]