Abstract
Antimicrobial resistance has become an imminent concern for public health. As methods for detection and characterization of antimicrobial resistance move from targeted culture and polymerase chain reaction to high throughput metagenomics, appropriate resources for the analysis of large-scale data are required. Currently, antimicrobial resistance databases are tailored to smaller-scale, functional profiling of genes using highly descriptive annotations. Such characteristics do not facilitate the analysis of large-scale, ecological sequence datasets such as those produced with the use of metagenomics for surveillance. In order to overcome these limitations, we present MEGARes (https://megares.meglab.org), a hand-curated antimicrobial resistance database and annotation structure that provides a foundation for the development of high throughput acyclical classifiers and hierarchical statistical analysis of big data. MEGARes can be browsed as a stand-alone resource through the website or can be easily integrated into sequence analysis pipelines through download. Also via the website, we provide documentation for AmrPlusPlus, a user-friendly Galaxy pipeline for the analysis of high throughput sequencing data that is pre-packaged for use with the MEGARes database.
INTRODUCTION
In recent years, antimicrobial resistance (AMR) has gained notoriety as a global threat to public health. Surveillance efforts aimed at the characterization of AMR have received increasing attention at the international level, as evidenced by the recent United Nations General Assembly high-level meeting on antimicrobial resistance, among other calls-to-arms from groups such as the United Nations, FAO, WHO, the White House, CDC, FDA, USDA, Public Health Agency of Canada, and the European Commission (1–8). Country-specific efforts have been important for monitoring trends in the prevalence of AMR so as to inform policy aimed at limiting the spread of resistance genes and the bacteria that harbor them (6,7). These surveillance programs have predominately used bacterial culture or polymerase chain reaction (PCR) to characterize select indicator bacteria (e.g. Escherichia coli, Salmonella) or specific gene targets related to AMR (6–8). While culture- and PCR-based methods have provided important insights into the prevalence of resistance, these techniques are limited to one, or at most a few, organisms/genes and therefore limit our ability to study the ecology of antimicrobial resistance within entire bacterial communities at the population level. This limitation is especially impactful to the study of AMR, which involves complex interactions among bacteria, horizontal transfer of genes, regulation by a variety of molecular pathways, and influences from the host and environment under study (9,10).
More recently, increasing accessibility to high-throughput quantitative PCR, microarrays, and high throughput sequencing (HTS) technologies have enabled identification of hundreds to thousands of AMR determinants in single genome or metagenomic samples (11–13). Metagenomic high-throughput sequencing specifically is currently being explored for expanded use in public health surveillance efforts related to AMR (1,4,9,14–17). However, where initiatives like the Human Microbiome Project (18) have developed standard pipelines for analysis of the microbiome (19,20), high-throughput analysis of AMR metagenomics suffers from a lack of tools specifically designed for this purpose. This dearth has led to the suboptimal use of bioinformatics tools that were designed for whole-genome sequencing data as well as a lack of consensus around a standardized metagenomics workflow (21,22). A central component of analytical pipelines used to characterize microbiome and metagenomic data are sequence classifiers, which utilize statistical or empirical information within the (meta)-genomic sequence data to assign a taxonomic label to a given DNA fragment. For classification of 16S rRNA gene sequence data, the mothur and Qiime microbiome pipelines make use of the Ribosomal Database Project (RDP), SILVA and Greengenes databases (23–25). In part, these 16S databases have enabled the success of pipelines including mothur and Qiime, as sequence classification is a critical step of the bioinformatics characterization of unknown DNA.
Several databases currently exist that thoroughly describe AMR genes and offer tools for analysis. However, these resources are primarily designed for for screening of a single genome or a few assembled contigs. These resources contain several limitations that hinder their utility for count-based analyses and classification of microbial community data using automated pipelines (26–28). Such limitations include the use of annotation headers with lengthy descriptions and non-conforming text symbols that do not interface well with UNIX- and Linux-based programs; errors in sequences and their annotations that are a result of semi-automated retrieval from public repositories; accessions with multiple AMR genes within a single sequence (e.g. plasmids, in the case of the NCBI Lahey beta-lactamase archive); and cyclical annotation structures with a large number of labels that do not lend themselves to certain statistical methods (e.g. naive Bayes) and the analysis of count data. While the contribution of the Antibiotic Resistance Ontology (ARO) developed by the Comprehensive Antibiotic Resistance Database (CARD) (28) is a notable improvement in AMR biocuration, and such a classification scheme is very useful for functional annotation, the ARO's highly descriptive and large annotation graph is not an optimal structure for other genomic efforts like ecological profiling and the construction of sequence classifiers. The use of databases with cyclical annotation graphs like the ARO can result in falsely inflated counts or the conflation of assignments in sequence classification. Additionally, while several existing databases offer BLAST functionality for data analysis, they do not provide a start-to-finish, GUI-based pipeline for easy integration into available bioinformatics tools. Researchers can currently use these existing databases to characterize AMR determinants within microbial communities with metagenomic sequencing data, but to do so they must substantially modify existing classification schemes, or they must verify each classification to ensure that it is assigned correctly and not double counted.
Thus, in order to further facilitate the characterization of AMR determinants in the context of large metagenomic studies, we present MEGARes, a hand-curated AMR database that has been specifically annotated for use in HTS data processing, the construction of sequence classifiers, and statistical analysis. Accessions in MEGARes have been manually verified for accuracy at the nucleotide and protein levels, and every annotation header has a standard format without non-conforming symbols, such that the database can be seamlessly integrated into custom scripting. To encourage the use of HTS of metagenomic samples for investigation of the ecology of AMR, we also provide an accessible, user friendly pipeline (named AmrPlusPlus) that is designed for metagenomic analysis and is fully integrated with the MEGARes database. Software used in this bioinformatic pipeline can be installed locally on Mac- and Linux-based operating systems using Docker, a software containerization platform, which aids in the portability and installation of complex pipelines like AmrPlusPlus (29).
DATABASE MOTIVATION AND DESCRIPTION
In sequence data analysis, the manner in which data are labeled can have a drastic impact on the results obtained, and the structure of annotation systems are therefore critical to analysis and correct interpretation. For example, the standard phylogenetic taxonomic classification scheme has been useful in microbiome analyses due in part to key characteristics of its annotation structure. Firstly, the standard phylogenetic taxonomy is hierarchical in nature. By drawing an edge between each taxonomic node, we can create a graph, linking the nodes together through their respective ranks, i.e. Bacteria is linked with Bacteroidetes, which is linked with Bacteroidales, and so on. One valuable aspect of the standard phylogenetic taxonomic structure is that its annotation graph is a tree; no two higher level ranks are linked to the same lower level rank, i.e. the graph has no cycles (Figure 1). For example, one bacterial genus can contain multiple species, but one species cannot belong to two different genera. Additionally, each organism is classified to exactly one location in the tree, such that an organism like Listeria monocytogenes has a unique classification path through the annotation graph. Because of this, we can assume independence between groups within the same level, which permits the use of faster methods such as the analytical calculation of probabilities using methods like naive Bayes. For large, complex data sets such as those that result from deep sequencing of metagenomic samples, having fast and robust statistical methods available is important, as the size of the data does not allow the use of computational methods that are substantively slower, such as BLAST.
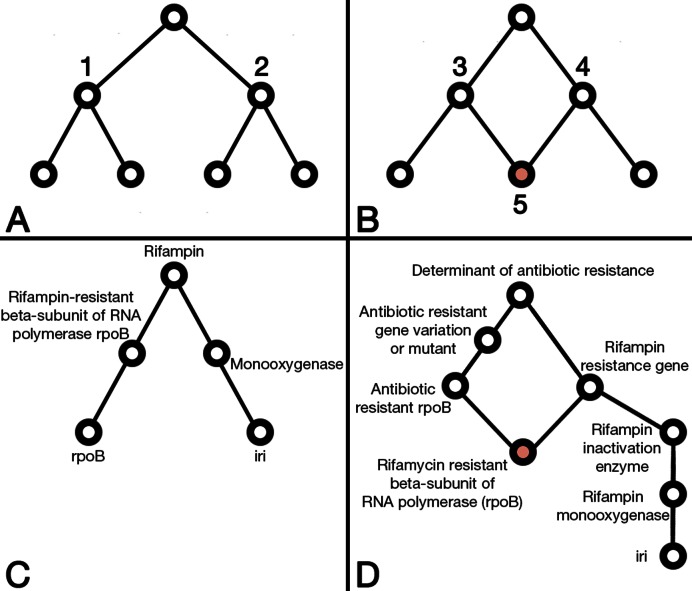
Figure 1.
(A) This annotation graph contains no cycles (is a tree), as nodes 1 and 2 do not share children and are therefore independent. (B) In contrast, node 3 and node 4 share node 5 as a child, which creates a cycle in the annotation graph and statistical dependencies between nodes 3 and 4. (C) The MEGARes annotation structure for two gene groups (rpoB and iri) that confer resistance to the rifampin class of antimicrobials. This annotation scheme contains no cycles. (D) The CARD annotation structure for the same two gene groups (rpoB and iri).
Additionally, the use of an acyclical annotation structure to label a reference database is critical for ensuring the veracity of output from count-based analyses (i.e. the number of reads or contigs that align to specific genes in the reference database). A cyclical graph structure can result in artificial count inflation when a single sequence (i.e. read or contig) is assigned to multiple categories at the same annotation level (i.e. if a gene is classified under two classes of resistance, such as rpoB-daptomycin and rpoB-rifampin). Such cycles also create uncertainty when training sequence classifiers on different annotation labels that share an identical sequence, as the classifier has difficulty in assigning the shared sequence to one category or the other. Therefore, an acyclical annotation structure, such as is used in the microbiome classification, is better suited to count-based analysis and classification within the context of an ecological- or community-level investigations.
With MEGARes, we have created an annotation structure that shares properties with the standard phylogenetic taxonomic annotations: each AMR sequence has a unique path through the annotation graph, and the graph contains no cycles. In order to facilitate hierarchical statistical analysis and the creation of robust classifiers, we have minimized the number of annotation levels and nodes such that each group has as many sequences as possible without creating nonsensical annotations.
We compare our database primarily to CARD, which has been recently updated and thoroughly curated (28). In contrast to the MEGARes annotation scheme, CARD's ARO has many more nodes and five additional classification levels, which results in sparse sequence membership within each node (Supplementary Table S1). Additionally, the CARD ARO contains 2966 cycles, which is a result of CARD's comprehensive annotation structure; thus, child nodes can have more than one parent node at any given level. Therefore, while the CARD ARO is comprehensive and serves the purpose of providing a descriptive labeling of each AMR sequence, its structure is not as conducive to statistical applications in high throughput analysis. Because of this, we believe MEGARes is a valuable, novel resource for enabling the high throughput analysis of AMR, particularly in a metagenomic, large data context.
Hierarchical annotation
The creation of an optimized annotation scheme for high throughput analysis involves a balance between preserving nucleotide identity and preserving functional grouping within gene clusters: sequence classifiers work best when classes are grouped by nucleotide similarity, however this can segregate biologically relevant categories into multiple groups, which in turn can result in model output that is difficult to interpret. Likewise, when biological category is prioritized over nucleotide identity, sequence classifiers that rely on sequence similarity cannot be accurately built. To this end, we have annotated MEGARes in such a way that identity is preserved and balanced with molecular function within sequence groups.
The MEGARes annotation scheme consists of three hierarchical levels regarding AMR genes: Drug Class, Mechanism, and Group. The Class level represents the major molecular category of resistance to different classes of antimicrobial drugs, e.g. tetracyclines, beta-lactams, glycopeptides. The Mechanism level is a child of the Class level and corresponds to the molecular mechanism that confers resistance to antibiotics. For example, tetracycline ribosomal protection proteins are a resistance mechanism within the tetracycline class; their function is to utilize GTP to free the tetracycline molecule from its binding site on the ribosome, resulting in resistance to tetracycline (30). Because molecular function often corresponds to conserved protein domains (as noted below in the description of clustering analysis), the Mechanism level of MEGARes preserves local regions of sequence similarity while allowing for variation in non-conserved regions. This differential sequence variation across the length of genes within Mechanism-level nodes is the natural result of the clustering procedure used during the verification and development of the MEGARes annotation structure, which is described in detail below. In addition, this underlying clustering method is ideal for models that aim to detect conserved regions of biological function, for example with profile Hidden Markov Models. The Group level is a child of the Mechanism level and is categorized based on information contained within the gene or operon. The Group annotation is meant to provide information on the major gene category for a given class of antimicrobials and varies depending on the class in question. For instance, the beta-lactamase genes contain group annotations corresponding to the gene names with which they are associated (31), e.g. SHV or TEM beta-lactamase. However, for classes such as the vancomycin resistance genes, the Group annotations correspond to the functional category within the vancomycin gene cluster operons, e.g. VanD-type accessory protein, VanA-type resistance protein. Again, this is meant to preserve the nucleotide identity within groupings and maintain reasonable biological categories across the database.
Data sources and curation
The comprehensive core database content was obtained by non-redundant compilation of sequences contained in Resfinder (November 2015), ARG-ANNOT (November 2015), the Comprehensive Antibiotic Resistance Database (CARD, v1.0.7), and the National Center for Biotechnology Information (NCBI) Lahey Clinic beta-lactamase archive (December 2015) (26–28,32). All data were included in the curation workflow except for the CARD protein wild-type sequences, as these represent genes that do not carry a known resistance mutation. Prior to inspection, sequences were collapsed at 100% identity using BLAT (v36×1) (33) with a maximum gap allowance of 0 to produce a set of unique sequences. For entries that contained header information with an NCBI accession number, Coding Sequence (CDS) regions were obtained by querying NCBI using the BioPython (v1.66) (34) module and the NCBI Entrez eUtils interface. Several entries, notably from the Lahey beta-lactamase archive, contained multiple CDS regions within the same sequence. In order to preserve sequence identity within gene annotation groups, these multiple CDS accessions were split into separate sequences along CDS boundaries and re-annotated based on the NCBI CDS annotation. For example, entries corresponding to plasmids that contained multiple antimicrobial genes were broken into gene fragments according to their CDS start/stop locations and were re-annotated with their respective gene category.
Gene annotations were validated through a combination of translated BLAST at >90% identity and gene clustering with USEARCH (35) at >80% identity. Each annotation was then manually inspected for accuracy. Sequences were considered potentially misannotated if any of the following were not concordant: the BLAST annotation and cluster annotation, the cluster annotation and current annotation, or the BLAST annotation and current annotation. This subset of questionably annotated genes was manually approved or re-annotated based on results from additional nucleotide and protein BLAST against the NCBI non-redundant nucleotide and protein databases (36). Genes that were identified as not related to AMR were excluded from the database. The 3824 resulting genes comprise a comprehensive and manually curated database of AMR genes that are derived from all high quality data sources that are currently available.
Database schema and features
MEGARes is structured as a relational database where the FASTA header of the gene sequence is the primary key: a FASTA file contains the gene sequences, and the annotations are stored in an additional comma-separated file. The database schema is updated through several Python scripts that allow for reproducible amendment of database information and the addition of new sequences following verification and header formatting.
Interactive browsing of the database annotations and sequences is offered through the web interface at (http://megares.meglab.org). For quick and focused searches, we offer a keyword-based search on the homepage of the website; users can input partial-match keywords and receive a comprehensive listing of matches to both database accession headers and nodes within the annotation file. Results of search refinements are updated in real-time. Alternatively, users can explore the hierarchical annotation structure through the ‘Browse’ feature by clicking on the annotation terms, which automatically submits queries to a MySQL server where the sequence and annotation tables are stored. A brief description of each annotation term is provided along with reference to primary literature sources that further describe the annotation category. For each gene annotation, the browsing interface lists both its children terms as well as other terms within the same level, allowing the user to navigate through the database by node. During any search or browsing session, sequences labeled with a given term can be downloaded as a flat FASTA file by clicking on the ‘Download Sequences’ link. Alternatively, the full database flat files can be downloaded as a compressed archive through the ‘Download’ option in the navigation bar.
The gene content of MEGARes is summarized in Supplementary Figure S1 and is provided as a D3 (37), interactive graphic available on the MEGARes website. To provide a reproducible mapping of sequence data, a file linking each sequence in MEGARes to the database and gene ID of origin can be downloaded as a flat file from the website. For users who wish to use MEGARes for analysis of large HTS datasets, we also provide an integrated bioinformatics pipeline that scales to large data more effectively than BLAST, which is described below.
AmrPlusPlus pipeline
A bottleneck to the widespread adoption of metagenomic methods for AMR analysis is the lack of accessible start-to-finish pipelines for HTS data processing. Furthermore, even if pipelines were defined, researchers would face substantial roadblocks when attempting to install dozens of different tools, many with multiple levels of dependencies (38). To overcome these barriers and provide accessibility to the MEGARes database, we offer AmrPlusPlus, a pipeline for resistome analysis of metagenomic datasets. The pipeline is fully integrated with Galaxy (39), an accessible, web-based platform that wraps command-line tools in an easy-to-use graphical interface. As such, AmrPlusPlus is an easy-install pipeline for local use; all tools required to run the workflow are pre-packaged and ready for use once downloaded onto the researcher's computer with a Mac- or Linux-based operating system. This ensures that users do not have to find, install, and configure the correct version of every tool required to run the workflow. Once downloaded, only four inputs are required: a resistance database in the form of a FASTA file, a host/background genome in FASTA format for the removal of host/background contamination, and a single or pair of FASTQ datasets. Each component in the workflow is configured to use default settings but can be modified by the user if desired. Installation and documentation can be found in the pipeline documentation (http://megares.meglab.org/amrplusplus). We have made the source files publicly available on GitHub (https://github.com/cdeanj/galaxytools).
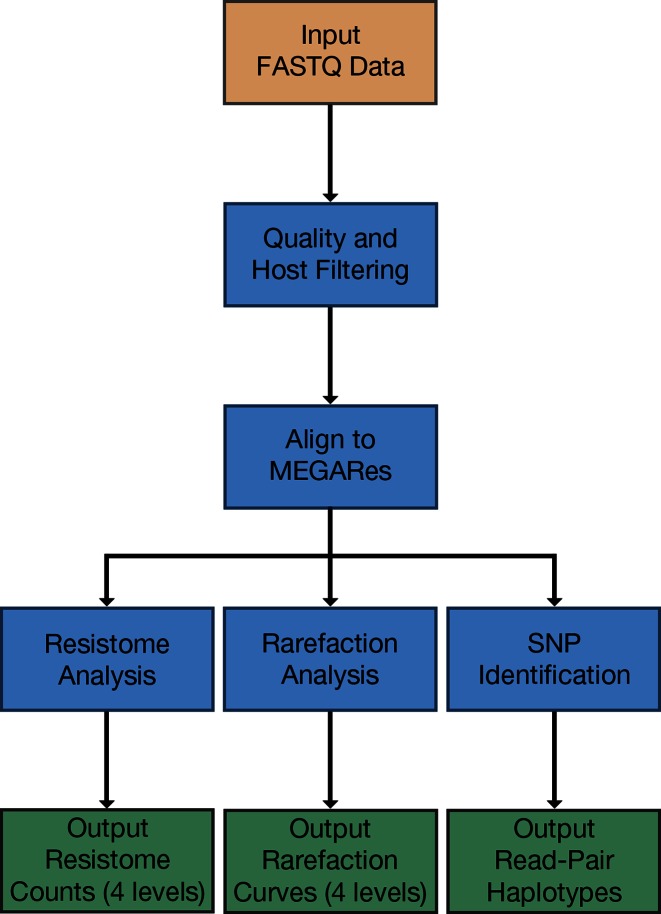
The AmrPlusPlus pipeline consists of five steps that allow for identification, quantification and read-pair haplotyping of AMR genes within metagenomic sequence data (Figure 2). Here, we provide a brief overview of the workflow, which is described in detail in the online documentation.
Figure 2.
A pipeline workflow diagram describing the steps involved in the AmrPlusPlus pipeline. The tan box denotes input files, blue boxes represent steps in the pipeline, green boxes denote outputs and arrows show the directionality of the workflow.
Read trimming and filtering
Trimmomatic (40) is used to remove adapter contamination and low quality reads. Additionally, for the analysis of host-associated metagenomic samples, the removal of contaminating host DNA can improve results. Therefore, the Burrows-Wheeler-Aligner (BWA) (41) is used to align sequence reads to a user selected host genome, and subsequent removal of aligned reads is performed with Samtools (42).
Align remaining reads to AMR database
Non-host reads from the previous step are then aligned to the user-specified resistance database using BWA; if users wish to change default BWA alignment criteria in order to relax or tighten identity matching requirements, they are able to do so through the Galaxy interface. By default, we offer MEGARes as the reference AMR database to provide a pipeline that is integrated with a database designed specifically for HTS analysis; however, users can also provide their own reference database. Output from this step is a BAM file that is then sorted and converted to a SAM file using Samtools. This SAM file is provided as input to Steps (3), (4) and (5), which utilize custom C++ tools developed specifically for resistome analysis.
Resistome identification
This program uses the SAM file from the alignment step to identify all AMR genes within the sequence data, using a user-specified gene fraction threshold. We define gene fraction as the proportion of nucleotides in a reference sequence to which at least one read from the sequence data is aligned. Given that short-read metagenomic data can result in false positive AMR gene identifications (43), the AmrPlusPlus pipeline can filter out genes with a gene fraction below a user-defined threshold using a custom C++ program called ResistomeAnalyzer (source code available at https://github.com/cdeanj/resistomeanalyzer). Counts of aligned reads are recorded at the gene, group, mechanism and class levels and provided as output to the user in tab-delimited format, which can then be used for statistical analysis.
Rarefaction analysis
The RarefactionAnalyzer program can perform rarefaction analysis of metagenomic data (source code available at https://github.com/cdeanj/rarefactionanalyzer); for more information, see the online documentation for AmrPlusPlus (http://megares.meglab.org/amrplusplus).
SNP detection
There are many programs available for identifying Single Nucleotide Polymorphisms (SNPs) in genomic data (44). However, because such programs were developed for single organism genomic sequencing, they utilize statistical assumptions that are not appropriate for metagenomic data. For instance, the copy number of genes can vary within organisms, between organisms, and also with the plasmid copy number. Few assumptions, if any, can be made about the mutation rate, population structure, or presence of bi- and tri-allelic minor alleles, as these metrics vary widely depending on the environment and population under study. Furthermore, the definition of an AMR ‘gene’ varies between classes and is currently debated within the scientific community, leading to confusion about whether a SNP denotes a gene haplotype or a new gene (45). This is particularly true for standards within the beta-lactamase class, which define a new ‘gene’ as any sequence with at least 1 amino acid difference from any known sequence. For MEGARes, we have adhered to formal and informal conventions for defining AMR genes, and therefore each accession within the database is considered the reference ‘gene’ as defined within existing databases. By extension, any SNP identified with respect to a reference sequence is defined as a SNP within a gene. In addition, while the traditional goals of SNP calling have been functional and population genetic analyses, the use of metagenomic data in surveillance efforts could portend a new use for SNPs; namely, as a type of DNA ‘fingerprint’ for tracking AMR genes over time and geography. Therefore, we provide a tool called SNPFinder that reports all possible SNPs and read pair haplotypes identified within a metagenomic sample and allows the user to determine which SNPs and read-pair haplotypes are significant based on their knowledge of the research question and population under study (source code available at https://github.com/cdeanj/snpfinder). The SNPs detected by SNPFinder are not intended to predict whether genetic changes will affect the resistance phenotype, but instead will allow genetic comparisons for ecological tracking, evolutionary studies, or other investigations of SNP profiles. SNPFinder takes each read (or read pair) that aligned to the resistance database and compares the read sequence to the reference sequence on a nucleotide-by-nucleotide basis. SNPs that fall on the same read pair are recorded as a read-pair haplotype within the corresponding resistance gene, and read-pair haplotypes within the same resistance gene are tallied to provide a count of occurrence of each haplotype by gene. The output of this program is a tab-delimited file with fields for gene, read-pair haplotype pattern, and count.
DISCUSSION
With MEGARes, we have presented a resource tailored specifically for the high throughput analysis of AMR genes within metagenomic data. MEGARes does not purport to replace the role of databases such as CARD and Resfinder, which provide users with rich gene descriptions and functional SNP annotation tools. Instead, MEGARes focuses specifically on resistome analysis as a natural extension of metagenomics data. As such, it forgoes detailed gene descriptions and multi-category annotations in favor of a simpler, hierarchical and acylic annotation scheme and short, script-friendly gene headers. We have deliberately maintained a relatively sparse annotation hierarchy in the hopes that MEGARes can serve as a solid foundation for further development of resistome-centered analytical methods such as sequence classifiers and hierarchical statistical models. In order to facilitate increased use of metagenomic datasets in resistome analyses, we have included MEGARes in AmrPlusPlus; however, the database is also readily downloadable for use in additional programmatic workflows of the user's choosing. AmrPlusPlus not only increases the accessibility of resistome analysis, but also provides users with 3 new integrated tools (ResistomeAnalyzer, RarefactionAnalyzer and SNPFinder) which we hope will help to bridge the gap between the bioinformatics and the statistical analysis of metagenomics data. As the scientific and regulatory communities explore the use of metagenomics in AMR surveillance and public health epidemiology (14,46), we hope that MEGARes proves to be a useful and usable tool for many researchers in the area of AMR.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Infectious Disease Supercluster and CSU Ventures at Colorado State University; Beef Checkoff; United States Department of Agriculture [2014-05257, 2015-03538]; National Institutes of Health [5T32OD012201]. Funding for open access charge: United States Department of Agriculture [2014-05257].
Conflict of interest statement. None declared.
REFERENCES
- 1.The White House. National Action Plan for Combating Antibiotic Resistant Bacteria. 2015. https://www.whitehouse.gov/sites/default/files/docs/national_action_plan_for_combating_antibotic-resistant_bacteria.pdf (25 October 2016, date last accessed)
- 2.United NationsNews Centre. At Resistance UN, global leaders commit to act on antimicrobial resistance: New York UN, global leaders commit to act on antimicrobial resistance: New York. 2016. http://www.un.org/apps/news/story.asp?NewsID=55011#.WBDSdeErLTo (25 October 2016, date last accessed)
- 3.Food and Agricultural Organization of the United Nations. The FAO action plan on antimicrobial resistance 2016-2020: Rome, Italy. 2016. http://www.fao.org/3/a-i5996e.pdf (25 October 2016, date last accessed)
- 4.United States Department of Agriculture, Office of Inspector General. 2016. Resistance USDA's Response to Antibiotic Resistance: Audit Report 50601-0004-31 : Washington, D.C.https://www.usda.gov/oig/webdocs/50601-0004-31.pdf (25 October 2016, date last accessed) [Google Scholar]
- 5.European Commission. Evaluation of the Action Plan against the rising threats from antimicrobial resistance. 2016. http://ec.europa.eu/dgs/health_food-safety/amr/docs/amr_evaluation_2011-16_evaluation-action-plan.pdf (25 October 2016, date last accessed)
- 6.United States Food and Drug Administration. 2013. Report National Antimicrobial Resistance Monitoring System—Enteric Bacteria. NARMS Integrated Report: 2012-2013: Silver Spring, MD.http://www.fda.gov/downloads/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/UCM453398.pdf (25 October 2016, date last accessed) [Google Scholar]
- 7.Public Health Agency of Canada. 2016. Canadian Antimicrobial Resistance Surveillance System – Report 2016: Guelph, Canada.http://healthycanadians.gc.ca/publications/drugs-products-medicaments-produits/antibiotic-resistance-antibiotique/alt/pub-eng.pdf (25 October 2016, date last accessed) [Google Scholar]
- 8.World Health Organization. 2015. Implementation Global Antimicrobial Surveillance System: Manual for Early Implementation: Geneva, Switzerland.http://apps.who.int/iris/bitstream/10665/188783/1/9789241549400_eng.pdf (25 October 2016, date last accessed) [Google Scholar]
- 9.Baquero F., Tedim A.-S.P., Coque T.M. Antibiotic resistance shaping multi-level population biology of bacteria. Front. Microbiol. 2013;4:15. doi: 10.3389/fmicb.2013.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacLean R.C., Hall A.R., Perron G.G., Buckling A. The population genetics of antibiotic resistance: integrating molecular mechanisms and treatment contexts. Nat. Rev. Genet. 2010;11:405–414. doi: 10.1038/nrg2778. [DOI] [PubMed] [Google Scholar]
- 11.Zhu Y.-G., Johnson T.A., Su J.-Q., Qiao M., Guo G.-X., Stedtfeld R.D., Hashsham S.A., Tiedje J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. PNAS. 2013;110:3435–3440. doi: 10.1073/pnas.1222743110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li B., Yang Y., Ma L., Ju F., Guo F., Tiedje J.M., Zhang T. Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME J. 2015 doi: 10.1038/ismej.2015.59. doi:10.1038/ismej.2015.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noyes N.R., Yang X., Linke L.M., Magnuson R.J., Dettenwanger A., Cook S., Geornaras I., Woerner D.E., Gow S.P., McAllister T.A., et al. Resistome diversity in cattle and the environment decreases during beef production. eLife. 2016;5:e13195. doi: 10.7554/eLife.13195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.EMBL-EBI Metagenomics. Local surveillance of infectious diseases and antimicrobial resistance from sewage: Project (ERP015410) 2016. https://www.ebi.ac.uk/metagenomics/projects/ERP015410 (25 October 2016, date last accessed)
- 15.Miller R.R., Montoya V., Gardy J.L., Patrick D.M., Tang P. Metagenomics for pathogen detection in public health. Genome Med. 2013;5:81. doi: 10.1186/gm485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Centers for Disease Control and Prevention. AMD Projects: Combatting Healthcare-associated Infections: Washington, D.C. 2016. http://www.cdc.gov/amd/pdf/factsheets/amd-projects-hai-2016.pdf (25 October 2016, date last accessed)
- 17.Interagency Task Force on Antimicrobial Resistance. 2012. Update a public health action plan to combat antimicrobial resistance - 2012 Update: Washington, D.C.http://www.cdc.gov/drugresistance/pdf/action-plan-2012.pdf (25 October 2016, date last accessed) [Google Scholar]
- 18.NIH HMP Working Group. Peterson J, Garges S., Giovanni M., McInnes P., Wang L., Schloss J.A., Bonazzi V., McEwen J.E., Wetterstrand K.A., et al. The NIH Human Microbiome Project. Genome Res. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I., et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J., et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oulas A., Pavloudi C., Polymenakou P., Pavlopoulos G.A., Papanikolaou N., Kotoulas G., Arvanitidis C., Iliopoulos I. Metagenomics: tools and insights for analyzing next-generation sequencing data derived from biodiversity studies. Bioinform Biol. Insights. 2015;9:75–88. doi: 10.4137/BBI.S12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ju F., Zhang T. Experimental design and bioinformatics analysis for the application of metagenomics in environmental sciences and biotechnology. Environ. Sci. Technol. 2015;49:12628–12640. doi: 10.1021/acs.est.5b03719. [DOI] [PubMed] [Google Scholar]
- 23.Cole J.R., Wang Q., Fish J.A., Chai B., McGarrell D.M., Sun Y., Brown C.T., Porras-Alfaro A., Kuske C.R., Tiedje J.M. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., Andersen G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta S.K., Padmanabhan B.R., Diene S.M., Lopez-Rojas R., Kempf M., Landraud L., Rolain J.-M. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014;58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., Aarestrup F.M., Larsen M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McArthur A.G., Waglechner N., Nizam F., Yan A., Azad M.A., Baylay A.J., Bhullar K., Canova M.J., Pascale G.D., Ejim L., et al. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013;57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merkel D. Docker: lightweight Linux containers for consistent development and deployment. Linux J. 2014;239:2. [Google Scholar]
- 30.Connell S.R., Tracz D.M., Nierhaus K.H., Taylor D.E. Ribosomal Protection Proteins and Their Mechanism of Tetracycline Resistance. Antimicrob. Agents Chemother. 2003;47:3675–3681. doi: 10.1128/AAC.47.12.3675-3681.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kong K.-F., Schneper L., Mathee K. Beta-lactam antibiotics: from antibiosis to resistance and bacteriology. APMIS. 2010;118:1–36. doi: 10.1111/j.1600-0463.2009.02563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bush K., Jacoby G.A. Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother. 2010;54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kent W.J. BLAT—the BLAST-Like Alignment Tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cock P.J.A., Antao T., Chang J.T., Chapman B.A., Cox C.J., Dalke A., Friedberg I., Hamelryck T., Kauff F., Wilczynski B., et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 36.NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2013;41:D8–D20. doi: 10.1093/nar/gks1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teller S. Data Visualization with d3.Js. Birmingham: Packt Publishing Ltd; 2013. pp. 1–180. [Google Scholar]
- 38.Morrison-Smith S., Boucher C., Bunt A., Ruiz J. Elucidating the role and use of bioinformatics software in life science research. Proceedings of the 2015 British HCI Conference. 2015 doi:10.1145/2783446.2783581. [Google Scholar]
- 39.Giardine B., Riemer C., Hardison R.C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina Sequence Data. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu170. doi:10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H., Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gibson M.K., Forsberg K.J., Dantas G. Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J. 2015;9:207–216. doi: 10.1038/ismej.2014.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mielczarek M., Szyda J. Review of alignment and SNP calling algorithms for next-generation sequencing data. J. Appl. Genet. 2016;57:71–79. doi: 10.1007/s13353-015-0292-7. [DOI] [PubMed] [Google Scholar]
- 45.Martínez J.L., Coque T.M., Baquero F. What is a resistance gene? Ranking risk in resistomes. Nat. Rev. Micro. 2015;13:116–123. doi: 10.1038/nrmicro3399. [DOI] [PubMed] [Google Scholar]
- 46.Baquero F. Metagenomic epidemiology: a public health need for the control of antimicrobial resistance. Clin. Microbiol. Infect. 2012;18(Suppl. 4):67–73. doi: 10.1111/j.1469-0691.2012.03860.x. [DOI] [PubMed] [Google Scholar]