Abstract
The MalaCards human disease database (http://www.malacards.org/) is an integrated compendium of annotated diseases mined from 68 data sources. MalaCards has a web card for each of ∼20 000 disease entries, in six global categories. It portrays a broad array of annotation topics in 15 sections, including Summaries, Symptoms, Anatomical Context, Drugs, Genetic Tests, Variations and Publications. The Aliases and Classifications section reflects an algorithm for disease name integration across often-conflicting sources, providing effective annotation consolidation. A central feature is a balanced Genes section, with scores reflecting the strength of disease-gene associations. This is accompanied by other gene-related disease information such as pathways, mouse phenotypes and GO-terms, stemming from MalaCards’ affiliation with the GeneCards Suite of databases. MalaCards’ capacity to inter-link information from complementary sources, along with its elaborate search function, relational database infrastructure and convenient data dumps, allows it to tackle its rich disease annotation landscape, and facilitates systems analyses and genome sequence interpretation. MalaCards adopts a ‘flat’ disease-card approach, but each card is mapped to popular hierarchical ontologies (e.g. International Classification of Diseases, Human Phenotype Ontology and Unified Medical Language System) and also contains information about multi-level relations among diseases, thereby providing an optimal tool for disease representation and scrutiny.
INTRODUCTION
With the advent of new high-throughput technologies in both research and clinical domains, new data across many fields pertaining to diseases are generated. While this presents opportunities for discovery, it also brings about new challenges in disease data acquisition, processing and unification. In 2013, we released MalaCards, an integrated compendium of diseases and their annotations (1). MalaCards tackles many of the problems that stem from the complexity of disease data and from the multiplicity of information sources. This is accomplished by employing sophisticated data-mining strategies modelled after the widely-used GeneCards database (2,3). The present report reviews these ongoing strategies, and highlights improvements and new implementations. One important change is an increase from 44 data sources in 2013 to 68 today.
One of the key issues in disease data integration is disease nomenclature, whereby very often a disease is named differently in different databases. MalaCards overcomes this difficulty by employing an elaborate aliases system, so that practically every name appears as a listed alias. This multifaceted approach is also reflected in MalaCards’ striving to portray complementary information, sometime at the price of a certain degree of redundancy, such as when showing multiple complete summaries from different sources. This approach optimizes the capacity of MalaCards to maximize the complete portrayal of disease attributes. This overview trait is strengthened by the free text search that allows users to present elaborate queries and effectively benefit from the wealth of stored information.
In recent years, new high-throughput technologies have greatly advanced the field of disease genetics and genomics. MalaCards continues to address this challenge with its comprehensive Genes section, in line with the systems approach that guides MalaCards. This section has undergone significant alterations, including score comparability among diseases and the introduction of the concept of Elite disease-gene association. In the same vein, the Drugs and Therapeutics section has been expanded, e.g. with clinical trials and FDA-approved drugs. With these and other improvements, MalaCards remains an invaluable tool for researchers and clinicians alike. We describe the database creation process, along with recent additions and improvements to the data and web interface. MalaCards data are available online at no cost, and through data dumps, upon request.
DISEASE DEFINITION
Disease unification
The MalaCards project constitutes an attempt to generate a complete lexicon of all human diseases. This is a daunting task for many reasons, and, therefore, we regard it as an effort to delineate a route toward attaining that goal. The main challenge of such a task is to overcome the lexical heterogeneity that prevails in the realm of diseases. We selected ten disease databases to serve as disease-name sources (Supplementary Table S1). In Version 1.11, these primary sources include a total of 83 923 unique name and alias strings, which underwent a textual unification process (1), resulting in almost 20 000 disease name groups. An inherent part of the process is that in each group, one of the names is defined as a ‘main name’ and the rest are defined as ‘aliases’. The main names constitute the basis for the MalaCards database, and define the titles of the ∼20 000 annotated disease web cards; each of them is called ‘MalaCard’ – a card for a disease/malady. The remaining 50 560 terms populate the Aliases and Classifications section of the cards.
In addition, there are 11 other data sources, defined as secondary, whose names and aliases are used to supply additional MalaCards aliases to existing cards, largely using the same name mapping algorithm. One of these sources, Unified Medical Language System (UMLS), is associated with a different mapping algorithm, the MetaMap program (4). Each MalaCards term (names and aliases) obtained in the first round is submitted to the MetaMap program with results restricted to UMLS concepts with semantic assignments of Pathologic Function, Cell or Molecular Dysfunction, Experimental Model of Disease, Disease or Syndrome, Mental or Behavioral Dysfunction and Neoplastic Process. A term that generates a maximal MetaMap Indexing ranking function score of 1000 (details available at http://skr.nlm.nih.gov/papers/references/ranking.pdf) to a UMLS concept is accepted as a legitimate alias for MalaCards. In total, 13 425 unique UMLS concepts were identified and mapped onto 12 817 unique maladies in MalaCards.
The relational database behind MalaCards allows us to perform extensive comparative analyses that help rationalize the relations among different disease compendia, including MalaCards. One important facet of our unification process is that MalaCards is fully inclusive: every disease entry in each of the data sources has a representation within MalaCards. While there are no accepted standards for objectively defining several textual strings as representing the same disease, which makes the MalaCards unification process difficult, it nonetheless guarantees that MalaCards has a remarkable capability to discover disease names from unrelated sources. We subjected MalaCards to an advanced search on 1000 randomly-selected disease names from DisGeNET (5). Searching for an exact match to MalaCards main names, main names and aliases, and all fields, respectively yielded results in 49%, 83% and 90% of the queries.
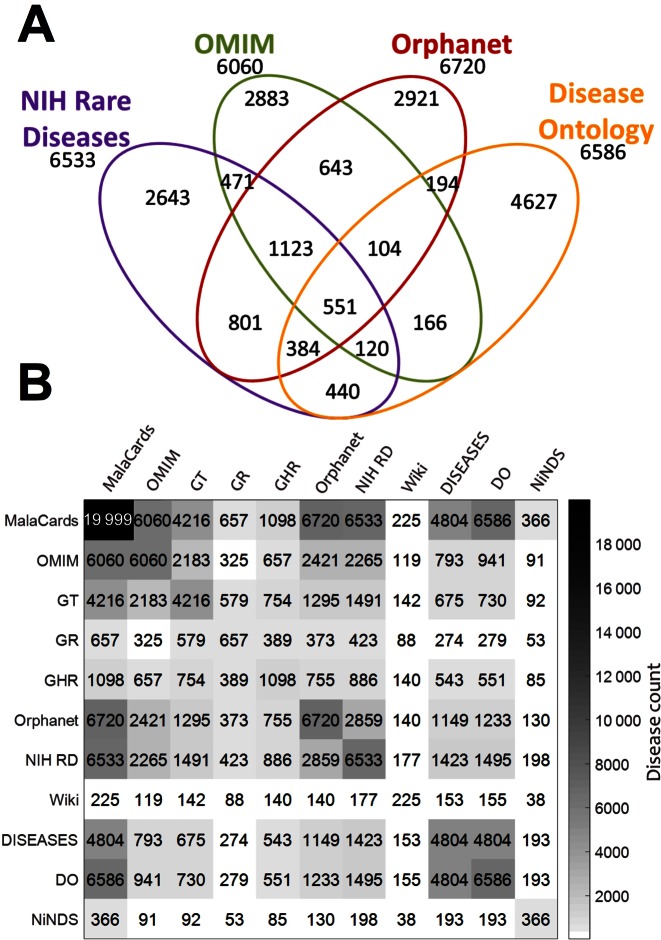
The MalaCards naming process provides a capacity to analytically compare disease coverage among different databases. One analysis involves four main data sources, with ∼6000 entries each (Figure 1A). It highlights the apparent name heterogeneity represented in these sources, whereby 40–70% of all entries are singletons, i.e. not unified with entries from other sources. Only 2282 entries are successfully unified among 3 or 4 of the sources. Another analysis (Figure 1B) portrays an all-against-all overlap in disease coverage among MalaCards and its ten primary naming data sources. It is evident that MalaCards successfully integrates partially overlapping sources, bringing forth the most comprehensive collection of disease entries. The different source sizes and varying patterns of overlap are likely related to differences in topical focus and granularity of disease definition.
Figure 1.
Overlaps among disease sources. (A) Venn diagram for the four major MalaCards name sources, according to MalaCards mapping. (B) A symmetric matrix showing the number of overlapping diseases between all pairs of primary name sources according to MalaCards mapping. Shading (as per color bar) and numerals represent degree of overlap in disease counts. Source abbreviations: DO– Disease Ontology, GR- Gene Reviews, NIH RD– NIH Rare Diseases, GT- GeneTests, GHR- Genetic Home Reference, NINDS– National Institute of Neurological Disorders and Stroke.
In one example, the overlap between Orphanet and OMIM is 2421 diseases, and between MalaCards and OMIM it is 6060 diseases (the entire OMIM disease count), probably due to Orphanet's policy of unifying several OMIM disease subtypes into one entry. However, as the overlap between Orphanet and MalaCards comprises 6720 diseases, there may be as many as 660 diseases that appear in both of the latter databases, but not in OMIM. The non-OMIM diseases show a disease category distribution as shown in Supplementary Figure S1, suggesting that many such missing diseases are either cancers, or fetal or infectious diseases.
Disease relatedness
Each disease defined by the naming and unification process is subsequently assigned a hierarchy of disease relatedness layers as follows: (i) Disease aliases (synonyms), shown in the Aliases and Classifications section. This section also contains a list of external identifiers that facilitates mapping and navigation in additional data facilities. The aliases constitute a rich source of nomenclature stemming from disease names in sources other than the one selected to define the disease's main name, as well as declared aliases in all relevant sources. (ii) Disease families. These are molded after the concept of phenotypic series in OMIM (6), generated via an in-house text-mining algorithm that highlights cases in which several diseases bear the same name, but with modifiers such as serial numeral/letter, type indication or inheritance mode. MalaCards defines 1299 families with 4.8 ± 8.3 members (range 2–132), including a total of 6292 diseases. (iii) Related diseases, where B is defined to be a related disease for disease A if the disease name A is mentioned verbatim, and in any section, in the MalaCard for disease B. Additional relatedness links are discovered by seeking significant gene sharing between the two diseases as seen in GeneAnaltytics (7). A composite relatedness score is generated and defines the top 20 related diseases portrayed in each MalaCard's weighted network image (see (8) for details). (iv) Disease SuperPaths, currently en route to implementation. These are generated from the gene-disease matrix (see Gene-disease connections below). The basic concept is defining a ‘disease pathway’ to be the group of genes related to a disease. We have shown that such pathways contain new information with respect to standard biological pathways (Rappaport et al., submitted). Adapting an algorithm previously constructed for the unification of biological pathways from different data sources (9), we obtain optimal disease SuperPaths, which afford the discovery of novel gene-disease and disease-disease relationships across a network path that spans several disease-gene and gene-disease edges. This approach may be extended in the future to topological calculations such as the disease-gene network distance of two diseases or of two genes.
Another form of disease-disease connection shown in the Related Diseases section is co-morbidity. A set of disease co-morbidity relationships (with P < 0.01) was obtained from the Phenotypic Disease Network (PDN) (10). These diseases are identified with ICD9 codes in the PDN. The ICD9 codes were used to identify the corresponding UMLS concepts that were checked against the list of UMLS concepts mapped to maladies. A total of 4989 relationships modelling co-morbidity of unique 741 MalaCards diseases were mapped.
ANNOTATIONS
Following the unification process, a MalaCard is generated for each amalgamated disease. It displays a disease web-card with diverse annotative information and provides an entry point for disease information, integrating textual information, disease network graphics, keywords and links to other databases. The card is divided into 15 sections. The left sidebar includes the list of sources contributing to the specific section in the specific disease. A ‘Jump to section’ element exists in each section header for easy navigation to different sections.
MalaCards employs several different methods to annotate its disease cards: (i) Direct mining of relevant text from a ‘named’ target source, i.e. one for which the unification process has generated a relationship between a MalaCards name and the source's disease name. This is exemplified by summaries from Genetic Home Reference, or symptoms from Disease Ontology (DO). (ii) Text mining for the MalaCards name in a target source, followed by mining of the required information, e.g. publications from PubMed, whereby the MalaCards name is matched within the PubMed title to associate publications with a disease. (iii) Identifier links connecting a MalaCard to a record target source, followed by information mining, as exemplified by variations from ClinVar. (iv) Manual curation of specific sections in a target source followed by obtainment of specific annotations. This is done in the case of disease-related drugs obtained from FDA.gov. (v) Set enrichment analysis via GeneAnalytics (7), by probing the overlap between genes associated with an entity in GeneCards (e.g. pathways, GO terms and mouse phenotypes) and disease-related genes. Where possible, annotative elements in the different sections are scored and prioritized based on their relevance level, and deep links to the sources of information are given.
Quality assurance
Automatic data mining affords rapid extraction and annotation of large amounts of data from multiple sources. This is nearly unavoidable for a project of MalaCards’ magnitude. However, automated mining methods may lead to both false positive and false negative annotations, which could result, among others, in improper disease unification. The search results minicards mechanism (see below), which shows the exact hit context, enables crowd-sourced elimination of some of these errors. In addition, quality assurance is instituted on every MalaCards update and version (we currently aim for three major versions per year, with additional interim updates). This QA includes automatic checks on database integrity and comparisons to previous versions. In addition, a dedicated team member and part-time consultants perform sample manual curation.
Symptoms and phenotypes
Key disease annotations are symptoms and phenotypes which typically represent changes from normal function or appearance. MalaCards currently obtains such information from five sources, four in the Symptoms section and one in the Animal Models section (mouse phenotypes). MalaCards shows the following: (i) Human Phenotype Ontology (HPO) phenotype entries of aspect ‘O’ (Phenotypic abnormality) (11). These are displayed in a table with phenotype description, frequency among the diseased individuals and source accession. (ii) DO symptoms (12), using the ‘has_symptom’ relationship. (iii) Orphanet (13) clinical signs. Recently, Orphanet has added HPO terminology, and this will be shown in MalaCards 1.12, assisting future unification. (iv) UMLS symptoms using semantic type assignment of Sign or Symptom. A total of 1868 distinct UMLS symptoms were mapped to 4619 distinct maladies via 21 675 relationships. (v) OMIM symptoms, included as links to the Clinical Features and Clinical Synopsis section. (vi) Mouse phenotypes are brought in from Mouse Genome Informatics (14). We note that the distinction between diseases and symptoms is not always well defined, hence the same textual terms may appear as symptoms for disease A and a MalaCards name for disease B (e.g. Glaucoma for Marfan syndrome).
Drugs and therapeutics
The Drugs and Therapeutics section has recently undergone considerable improvement, increasing the number of subsections from 2 to 7. This provides the user with a multisource overview in a central information domain for diseases. Described below are the Drug and Therapeutics tables shown in MalaCards.
A table based on the ClinicalTrials.gov registry. To obtain drug-disease connection we first mapped the source's conditions to MalaCards disease names using our name unification algorithm. Since ClinicalTrials.gov shows a list of clinical trials for each condition, and, in turn, includes a list of drug interventions for every clinical trial, we were able to generate a unique list of drugs for every MalaCard. The drug expandable table is further enriched with drug integrated annotations drawn from GeneCards drugs data, including a comprehensive drug synonym list, as well as trial phase and status (by which the table is sorted), CAS Registry Number and PubChem ID. A total of 8005 distinct diseases were mapped to 3017 distinct drugs via 966 338 relationships.
A table based on combined information from the UMLS. Since UMLS concepts are mapped onto MalaCards names, UMLS drugs, based on the National Drug File-Reference Terminology (NDF-RT) (15) can also be mapped to MalaCards diseases. For each mapped UMLS concept, all relationships that have an attribute of ‘may treat’ and semantic type assignment of ‘pharmacologic substance’ were examined. There were 1772 unique drug concepts that were mapped to 3080 unique maladies in MalaCards.
A manually curated table for cancer and respiratory drugs from FDA labels (http://labels.fda.gov/). The connection between the label drug indication in this source and the MalaCards name was curated manually. The table includes drug name, active ingredient(s), pharmaceutical company and approval date. In an expanded view, users may view information and summaries on the FDA label, indication and usage, drug target(s) from DrugBank and mechanism of action. This effort is being extended to additional diseases categories.
A table showing interventional clinical trial records from the ClinicalTrials.gov registry, mapped onto the specific disease by applying the name unification process on the aforementioned condition list. The table includes the title, status, phase and ID of the clinical trial.
In addition, the section contains (i) a link to a search of the disease name within the NIH clinical center (http://clinicalcenter.nih.gov/), yielding additional information regarding clinical trials; (ii) Cell-based therapeutic approaches from LifeMap Discovery (16), which include stem-cell-based therapeutics, and Embryonic/adult cultured cells (candidate therapeutic approaches); and (iii) a link to Mesh lookup in the Cochrane library of evidence-based medicine (17), which provides evidence enabling informed healthcare decision-making.
Categories and classification
To enhance its navigation and analysis capacities, we have added categorization and classification features to MalaCards, most of which appear in the Aliases and Classifications section. The first feature is disease characteristics, which includes mortality, age of onset, age of death and mode of inheritance, taken from HPO (11) (aspects ‘I’, ‘C’ and ‘M’) and Orphanet (epidemiological data, available at http://www.orphadata.org/cgi-bin/index.php/). Secondly, we display disease classifications: an International Classifications of Diseases (ICD10) tree (18), mapped to the disease using naming as well as identifier matching through intermediary source identifiers; and Orphanet classification for the disease, based on MalaCards name mapping. Thirdly, we show in-house MalaCards categories, including 6 general disease categories (rare, genetic, cancer, metabolic, fetal and infectious), as well as 18 major organ/tissue categories, with some degree of inter-category overlap. These are generated by mapping to accepted classification sources (e.g. DO, Orphanet, ICD10) as well as by mining specialized keywords in disease names and descriptions. Special pages list all diseases for each category, sorted by relevance, and including the members of each family, all of which, by a MalaCards rule, share the same category affiliations. Finally, MalaCards shows in the Anatomical Context section several data entries related to more detailed disease-tissue relationships. These include the in-house MalaCards organs/tissues relations selected from a broader repertoire of 82 anatomical entities, obtained by extensive text mining of individual cards; Foundational Model of Anatomy (FMA) ontology data connected to the disease via DO; Assignment of the cells, compartments, and organs relevant to the disease, obtained from LifeMap Discovery (16).
Summaries and publications
These annotations provide textual information pertaining to each disease in MalaCards. We have ten sources for entries in the Summaries section, including definition of the disease, etiology and main symptoms. This section is a major strength of MalaCards, as it brings under one roof a diversity of viewpoints, with different degrees of resolution and different levels of detail, providing the user with a multisource overview of the disease. Summaries are mapped in several ways, depending on the information source. For example, the Wikipedia summary is linked by searching for the MalaCards main name, using the MediaWiki search API (https://www.mediawiki.org/wiki/API:Search), and the SwissProtKB summary (http://www.uniprot.org/docs/humdisease) is linked by cross-referencing to the OMIM ID. Also included is MalaCards’ in-house automatically generated summary that judiciously groups central annotations in the specific card into a descriptive text.
The Publications sections shows a total of ∼2 000 000 articles associated with MalaCards diseases, obtained by searching appearances of all of the disease main names within all PubMed paper titles using the PubMed API (19). Publications are sorted according to descending date. All summaries and publication titles are searchable, and article abstracts will soon be added to the searchable index.
GENE-DISEASE CONNECTIONS
Disease genes
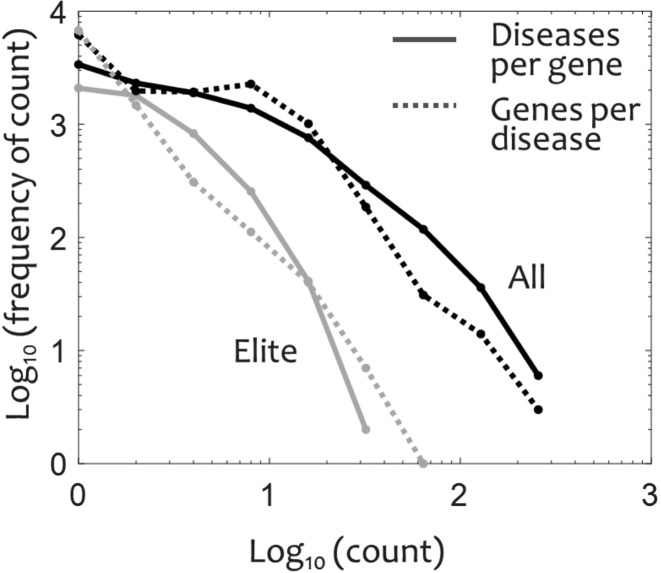
Each disease in MalaCards is associated with a prioritized list of genes, obtained from nine sources and shown in MalaCards’ Genes section. This basically defines the GeneCards Suite's gene-disease matrix, fully available via the ‘disease genes’ link on the in GeneCards home page, and is reflected in the GeneCards Disorders section of every relevant gene. The gene disease matrix currently connects 10 198 genes with 13 619 disease entries, spanning 1 to 318 genes per disease, and 1 to 386 diseases per gene (Figure 2). The sources for gene-disease relations are both manually-curated (e.g. OMIM, Orphanet, SwissProtKB) and text-mined resources (e.g. DISEASES (20), Novoseek (21)). For the latter type, MalaCards links to evidence-providing publications. The gene-disease priority scores are assigned both by the significance ascribed at the mined source, and by search scores strength for each association. This is accomplished by defining an overall disease-gene score SDG, computed as a weighted sum of individual scores derived from eight sources of information: OMIM, ClinVar, Orphanet, SwissProt-Humsavar, GeneTests, DISEASES, Novoseek and GeneCards, as described (Rappaport et al., Submitted). The individual score values depend on the level of manual curation of the information source, and on the confidence score assigned by the source to its different annotation classes. In MalaCards we further define an ‘elite’ gene for a disease as a gene with SDG > 2.5, and the overall score is designed to assure that gene-disease relations above this threshold come from at least one manually-curated source. Through a recent improvement, disease-gene connection scores are now comparable across different diseases. A total of 73.6% of the gene disease associations are supported by GeneCards through a text mining process in which the disease main name is mined from the cards of the genes in GeneCards by using a non-stemmed Solr index (http://lucene.apache.org/solr/) and Elasticsearch queries (https://www.elastic.co/). Data filtering heuristics at this stage reduce the noise level while retaining support for 75% of the elite associations. ‘Elite’ gene-disease associations are defined to be those from sources that are manually curated and contain strong and reliable associations. In parallel, for cancer diseases, census genes from COSMIC (22) are prominently tagged. We emphasize that the term ‘elite gene’ purely reflects the evidence for the strength of disease-gene association. It is not a gene annotation, as for or a given gene, elite status may prevail for some diseases and not others. Importantly, the implications column in the Genes section's table supplies evidence for the association, with links to respective sources/relevant GeneCards sections.
Figure 2.
Distributions for disease-gene relations. Solid black – diseases per gene, dashed black – genes per disease, solid grey – diseases per elite gene, dashed grey – elite genes per disease.
Gene-related disease annotations
Several annotation entries in MalaCards depend on the disease gene list, and are briefly described as follows:
Disease-related genetic testing information, for both inherited and other disorders, are displayed in the Genetic Tests section. This information is mined from two resources, GeneTests (23) and the recently added Genetic Testing Registry (24). These display a link to a disease page in the external source, where related laboratories and tests are listed. A future improvement will include the mining of the identity of the specific laboratory and the properties of the specific tests relevant to the disease. Notably, some of the tests in the linked disease pages constitute more broadly disposed gene panels, and some are more specific for the disease.
Disease-related Gene Ontology (GO) terms (25) of the three classes—cellular component, biological process and molecular function—are shown in a dedicated GO Terms section. As in the case of related diseases, we show here GO terms scored by relevance, based on GeneAnalytics gene-set analysis, which calculates significant enrichment of GO terms in the gene set associated with the disease (7,26).
Disease-related biological pathways. This is a recent improvement, which included the GeneCards SuperPaths (9) (clusters of individual pathways) enriched in the gene set associated with the disease. The table displays the SuperPaths along with their member pathways.
In a pilot, we show expression information for over 100 human diseases. The data includes the most differentially expressed genes (P-value threshold of 0.05, corrected for multiple testing) in the diseased tissue versus its matched normal tissue. The data are derived from the gene expression omnibus and/or manually curated from the scientific literature, extracted as described in (16).
Genetic variations
Causative variations for a given disease are gleaned from three sources, each shown in a separate table. Data from ClinVar (27) include variation name with a link to the information source, variation type, significance, dbSNP ID, genome assembly and location. Variations are taken from ClinVar only if most of its clinical significance attributes belong to the set of pathogenic, risk factor, drug response, protective, confers sensitivity or likely pathogenic. Data from UniProtKB/Swiss-Prot (http://www.uniprot.org/docs/humsavar) include nucleic acid and amino acid change, dbSNP ID and a link to further details at the source. For showing data from the catalogue of somatic mutations in cancer (COSMIC) (28) we algorithmically mapped the COSMIC cancer disease classification tags to MalaCards names. This was done by searching the classification terms in the following MC sections: Genes, Aliases and Classifications and Summaries, excluding non-specific terms like ‘mixed’, ‘NS’, etc. The variation score is a summation of the number of hits of each of the tags. In the future, we plan to unify the above three variations sources into one table.
SEARCH AND NAVIGATION
MalaCards aggregates textual annotations for each of its diseases from 68 data sources. Much of this information is included in the web card, and is indexed by the Lucene/Solr search engine. Therefore, searching MalaCards provides users with direct access to dozens of disease information sources in one go, akin to crowd wisdom (29). The MalaCards search provides a default set of section weights. For example, the Aliases and Classifications section is boosted, giving preference to keyword hits therein.
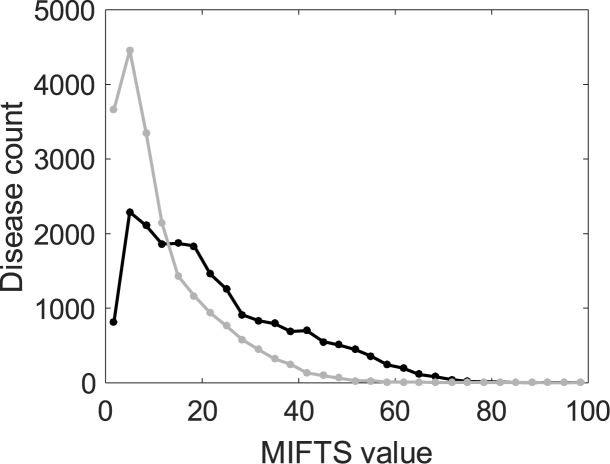
Conveniently, search results are initially shown in a format of one line per found-disease (microcards), showing disease name, family (parent/child) affiliation, relevance score (provided by Solr) and a disease's depth-of-annotation score (MalaCards information score – MIFTS (1)). MIFTS, reflects the amount of research devoted to a disease, hence, in parallel to the search score provides assessment of the importance of a disease among the search results. MIFTS also allows an evaluation of MalaCards’ progress over the years (Figure 3). This allows a quick view of the results, before proceeding. Clicking + in a microcard activates a recently installed feature, showing a minicard with textual context and section location for each of the found terms. Links are provided to the corresponding locations in the full MalaCard.
Figure 3.
Distribution of MalaCards Information Score (MIFTS). Grey – status in version 1.03 (2013), black – the current version 1.11. MIFTS values have almost doubled, from an average of 12.2 ± 10.3 to 22.5 ± 16, reflecting the progress made over three years in the knowledge recorded in MalaCards.
MalaCards search mechanism supports complex Boolean expressions, wild cards, stemming and exact match. A recently introduced advanced search enables further power and specificity. In this mode, users can limit the search to only match terms in one or more of MalaCards formal sections, taking advantage of the card's formal sectioning. For example, limiting a search to the ‘Symptoms’ section will only bring up diseases in which the keyword is formally defined as a symptom (‘symptomizer’ action). Finally, the ∼80 tables in the MalaCards SQL-based relational database allow, in addition, a plethora of sophisticated queries that enable the advanced user to discern unexpected trends and relationships in the realm of human disease.
COMPARISON WITH OTHER DATABASES
Ontologies
MalaCards has been constructed with the idea that each of its entries (disease cards) is devoted to the comprehensive coverage of a single topic – a defined disease. This is in contrast to the concept of ontology, an organizational system basically designed to portray the relationships between various concepts, as exemplified by GO, (25) DO (12) and HPO (11). Ontologies are often represented as graph or tree structures, whose navigation and utilization are often not straightforward. The ‘flat’, basic design of MalaCards is more akin to a glossary, whereby each record contains information on a single entry. This design provides a stable skeleton for annotation and for searches, but at the same time allows one to also define, when appropriate, relationships among entries. This is manifested, among others, in showing ontological relations as part of each card's annotations, and in MalaCards’ layers of relatedness.
Symptoms, phenotypes and diseases
A contributing factor to the complexity of the disease universe is that disease terminology and symptom/phenotype terms are often used interchangeably. This may lead to difficulties in databases in which searches are confined to either disease or symptom terms. MalaCards’ free text search format allows user flexibility via side-by-side inclusion of disease terms, as well as relevant symptom, phenotype and condition keywords in every disease card. At the same time, users wishing to focus on a specific keyword category may take advantage of MalaCards’ advanced search. Thus, the various search and exploration methods available in MalaCards enable users to query and identify a disease or condition based on disease, symptom, or phenotype information that are available in MalaCards, enabling users to examine the relationships and current integration of knowledge provided by MalaCards.
Disease genes comparison
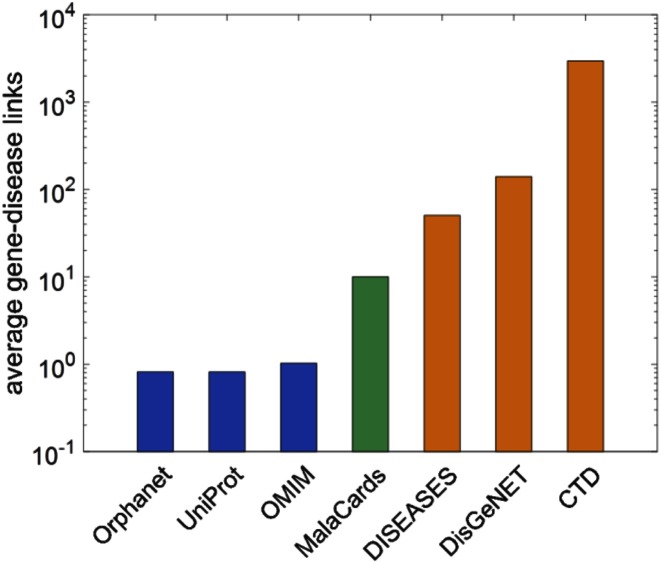
Given the importance of gene-disease associations, much research attention has been directed to improved algorithms for generating such connections (5,20,30,31). In order to evaluate MalaCards performance in this context, we performed a comparison to six other disease data sources. For the analysis we used a sample of 37 MalaCards diseases that have exactly 10 related genes in MalaCards, and are also mappable to all compared data sources. The average number of gene-disease associations mined from each source is shown in Figure 4. It is evident that three of the sources (OMIM, Orphanet and UniProtKB) have very few genes (about one gene per disease) and three other (CTD, DisGeNET and DISEASES) have a very large number of genes per disease, ranging from 1 to 24 745. MalaCards assumes a middle stand, representing a compromise between paucity and promiscuity, thus providing a balanced number of genes per disease. Our data mining mechanism assures that MalaCards never misses any of the genes in the stringent data sources, in fact, such genes are defined by MalaCards as Elite genes for a disease, reflecting the evidence for the strength of their association with such a disease.
Figure 4.
The average number of gene-disease associations across all compared sources for the set of 37 compared diseases.
The question at hand is whether MalaCards’ compromise between the economy and opulence portrays a good selection of additional genes, as derived from the promiscuous sources lists. We therefore examined, for each sample disease, the relationships between the MalaCards rank vector and the across-source consensus rank for the same genes, computed as an average rank across the 7 sources (including MalaCards). In two of the 37 diseases, very strong deviations from the consensus rank were found for some MalaCards genes. These were traced to gene symbol errors or data mining irregularities, and therefore these diseases were removed from the analysis.
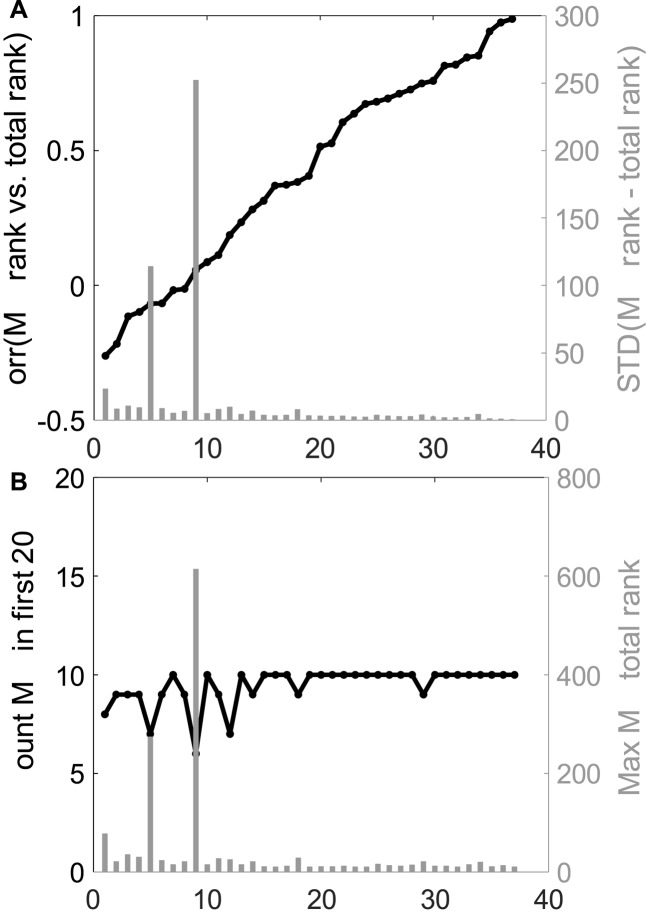
The analyses performed were as follows: (i) The average Pearson correlation between the MalaCards ranks and the consensus ranks was found to be 0.44 ± 0.37 (Figure 5A, left). (ii) The average standard deviation of the differences between the MalaCards rank and the consensus rank was 5.00 ± 4.16 (Figure 5A, right). (iii) The average number of MalaCards gene-disease association in the top 20 genes of the consensus rank is 9.6 ± 0.7 (Figure 5B, left). (iv) The average maximal consensus rank of a MalaCards gene was 18 ± 12.22 (Figure 5B, right, see also Supplementary Tables S2 and S3). All of these computed parameters attest to MalaCards’ capacity to effectively capture the consensus of all sources.
Figure 5.
Inter-source gene set comparison. (A) Left, black: Correlation between MalaCards gene rank vector and the consensus rank; Right, grey: Standard deviation of the difference between the MalaCards gene rank and the consensus rank. (B) Left, black: The number of MalaCards gene disease association in the top 20 consensus rank; Right, grey: The maximal consensus gene rank for a MalaCards gene. Raw data values for each disease are given in Supplementary Table S3.
The great discrepancy between very few genes per disease in the stringent sources and thousands of genes per disease in some automatically mined sources may be a cause of great concern. The stringent sources serve an important purpose in that they report only what is absolutely certain. This raises the questions of why add more gene-disease relations, and how many. We contend that the added disease genes are the basis of further research, whereby knowledge about less-certain gene-disease relations feeds new discoveries that make new relations more solid. This is done, among other things, via NGS interpretation tools. However, such tools suffer dearly from excess gene-disease promiscuity, and as MalaCards plays a central role in such tools (see next chapter), its compromise approach becomes a necessity. We further note that MalaCards does not utilize a simple-minded numerical cut-off values for deciding the mid-range gene count ((8), Rappaport et al., Submitted), and the number 10 mentioned in the foregoing analysis is for an example disease subgroup and intended to facilitate comparisons.
The two deviant diseases in Figure 5 portray interesting cases of artefacts incurring in the defining related genes for diseases. In one example, for van der Woude syndrome the gene CFAP57 ranked 3 in MalaCards, but was not present in any of the other sources. It turns out that CFAP57 is a new official HGNC symbol, with a previous symbol being WDR65. Under the older symbol a disease link is found in DISEASES (rank 4, Supplementary Table S2), but none of the other databases shows the gene in either notations. MalaCards benefits from GeneCards strict adherence to HGNC symbols, including immediate adoption of new symbols. In another example, for spinocerebellar ataxia 2 the gene ARID1B ranks 7 in MalaCards, but does not appear in any of the other sources. The disease implication arises from an early publication (32) which is mined by GeneCards, but not by any other source. We note that this gene has never since been reconfirmed as related to the disease, which may explain its absence from other sources for the disease.
The foregoing portrayals reflect the discrepancy of information between the resources integrated in MalaCards. The capacity to discover irregularities such as for van der Woude syndrome and spinocerebellar ataxia 2 is a powerful argument for developing tools like MalaCards, helping to alleviate domains of community confusion or poor use of terminology. In future work, a more comprehensive view of such discrepancies, with references to the different data sources provided by MalaCards, may be useful for further curation of the disease data by the experts and/or scientific community, including the development of tools for community curation.
USE CASES
Next generation sequencing
MalaCards data can be highly useful in analyzing clinical NGS cases. A crucial step in such analyses is gene-phenotype interpretation, which is performed subsequent to initial variant annotation and filtering, which, among other things, assigns variants to genes. NGS interpretation entails prioritizing the ensuing list of genes by seeking relationships of every gene to the patient's disease and phenotype terms (33). As this process is gene-centric, GeneCards is a natural candidate to assist in this goal. However, until recently, a considerable segment of crucially relevant information was present in MalaCards, but not in GeneCards. We therefore launched a GeneCards Suite modification, whereby information from three MalaCards sections, Aliases, Summaries and Symptoms, was fully integrated into the GeneCards search index (Rappaport et al., Submitted). This data enrichment is crucial for a comprehensive capacity to link genes with diseases and symptoms. Such a feature was then fully inherited by VarElect, the GeneCards-based NGS interpretation tool of the GeneCards Suite (34).
A relevant clinical example is the study of a patient afflicted with two seemingly disparate symptoms, distal motor neuropathy and ichthyosis, investigated in the laboratory of one of us (MG). A single affected individual, with suspected X-linked inheritance, underwent whole exome sequencing. After filtering for high protein impact and for control population frequency <0.01, about 1800 candidate variations in 1284 genes were identified. This relatively long gene list is typical of cases with only one sequenced individual and limited capacity for segregation analysis. The entire gene list was submitted to MalaCards-enriched VarElect, which can easily handle lists of this length. Only one gene, an ion pump, came out as jointly related to both phenotypes.
The decipherment of this disease highlighted the power of combining MalaCards and GeneCards information. Because the joint appearance of both phenotypes has never been reported in a single disease, a search in MalaCards alone for ‘distal motor neuropathy’ and ‘ichthyosis’ showed no results. However, VarElect provided a result because the embedded MalaCards information pointed to two different diseases, each with one of the phenotypes, but both related to the same gene. We note that this is in the vein of disease SuperPaths mentioned above, as we have here a case with one gene linked to two diseases, each of which related to more genes. Even a minute part of such a network is capable of shedding light on a previously undeciphered disease.
The applicability of MalaCards’ information to NGS interpretation has been demonstrated in additional projects (34). Currently, several hundred laboratories and clinical facilities worldwide utilize MalaCards-enriched VarElect to interpret sequenced genomes and identify culprit diseases.
A strong recent trend is a move in NGS analyses from exome sequencing to whole genome sequencing (WGS) (35). When an entire genome is sequenced, a great majority of the variations are found away from protein coding exons. MalaCards has a strong degree of readiness for WGS, as its Genes section includes all cases in which ncRNA genes have been implicated in a disease. In an example, an advanced search for MIR* in the Genes section of the current MalaCards version yields 25 diseases related to microRNA genes. A more elaborate query performed on the MalaCards MySQL database provides the result shown in Supplementary Figure S2, whereby on average, 3.5% of all genes in all disease categories are ncRNAs. This figure is expected to increase significantly as WGS becomes more prevalent. Finally, since with the advent of WGS, non-coding regulatory regions will be found to harbor variants, the MalaCards Genes section will have to be improved to accommodate enhancers and their target genes, utilizing tools such as GeneHancer (Fishilevich et al., In preparation), currently being embedded in the GeneCards Suite.
Transcriptomics
Transcriptome analyses are often used to tackle disease or condition mechanisms, where differential gene expression is registered by comparing disease with control, as a means of identifying disease-related genes. In one example (36), the authors asked whether a brain region (Nucleus accumbens, NAC), involved in natural rewards and addictions, was also related to the emotional reward of motherhood. Utilizing microarrays, NAC gene expression changes were monitored in postpartum female mice as compared to virgin controls. The MalaCards gene-disease matrix played a central role in identifying 100 addiction/reward related genes, several of which showing gene expression alterations. A second example was aimed at identifying potential therapeutic targets for papillary thyroid carcinoma (PTC) (37). Genes showing differential expression between PTC patients and normal individuals were identified and subjected to further bioinformatics analysis. MalaCards helped pinpoint six final candidate genes, all related to the disease.
Systems medicine
Due to its extremely broad knowledgebase, as well as sophisticated web search and database queries, MalaCards constitutes an effective Systems Medicine tool (8). In one example, MalaCards was used to assist transferring annotations from one ontology (the ICD code) to another (GO), aiming at integrating large-scale heterogeneous biomedical ontologies based on genomic relationships (38). The authors reconstructed a merged tree of GO and ICD9 codes and positively assessed it by comparing it with two disease-gene data sets (MalaCards and DO). A second example involves the construction of RNA Binding Protein (RBP) Expression and Disease Dynamics database (READ DB), a non-redundant, curated database of human RBPs. Adding to other RBP annotations such as RNA and protein expression levels, RNA recognition motifs and predicted binding targets, they included scored diseases associations from MalaCards, providing the disease dynamics aspects of RBPs in the context of post-transcriptional regulatory networks (39).
As mentioned under Disease relatedness layers above, it is possible to define the group of genes related to a disease as a ‘disease pathway’, and such pathways provide new information on gene mutual relations (Rappaport et al., Submitted). In this respect, a global view of disease pathways may bring forth new vistas on both genes and diseases. There are about 3000 biological pathways in GeneCards, obtained from 12 pathways sources, and these are unified into around 1000 SuperPaths. In comparison, the MalaCards gene-disease matrix, when its rows are viewed as disease pathways, has ∼5000 entries with 3 or more genes per disease, and ∼1000 entries with 10 or more genes. The total number of genes is equal, about 10 000. Thus, there is a comparable amount of informative gene groupings in each of the types.
An exciting future direction for MalaCards is taking such a comparison several steps further: one may ask how often it is possible to predict the validity of a disease gene candidate based on its being in the same biological pathway as a known gene for the same disease. Such ‘guilt by association’ logic prevails in NGS analyses, including in VarElect's indirect mode (34). In the inverse direction, perhaps there are numerous cases in which, having two genes in the same disease pathway attests to a yet undiscovered biological relationship between them. Finally, among systems analyses empowered by MalaCards, one could address questions such the degree of symptom sharing among diseases linked to the same gene (cf. (40)) using a broader network than hitherto available.
A relevant tool in this respect is GenesLikeMe (previously GeneDecks Partner Hunter (26)), another GeneCards Suite tools. Given a probe gene, GenesLikeMe provides a scored list of genes that bear similarity to the probe. This similarity is multi-dimensional, including similarities by shared sequence paralogs, protein domains, protein and RNA expression patterns across normal tissues, biological SuperPaths, GO terms and more. GeneLikeMe also provides similarity according to disease pathway sharing (disorder sharing). For any pair of genes, one can thus ascertain how many diseases include both genes in their gene list (disease pathway). This enables revealing disease-dependent, perhaps unexpected, gene-to-gene relationships.
Links from external databases
MalaCards’ extensive coverage of both the disease and gene universes makes it an effective target for incoming links. Current incoming links include: The UCSC genome browser (41), whereby relevant genes have disease track links to the appropriate MalaCards, based on the gene-disease matrix; Genetic Home Reference (GHR) (https://ghr.nlm.nih.gov/), and Diseasecard (42), both originating from their disease pages, employing MalaCards name unification scheme; PubMed LinkOuts (http://www.ncbi.nlm.nih.gov/projects/linkout/) from ∼100 000 highly relevant publications to the corresponding disease cards; UniProtKB, from the ‘pathology & biotech’ section of a protein card to relevant MalaCards, employing the gene-disease matrix. Such links provide users of external gene and disease databases access to considerable additional information.
Acknowledgments
The authors thank Raphael Zidovetzki, Gil Stelzer, Iris Bahir and Yaron Golan for assistance with data source allocation and database improvements. The authors also thank MalaCards users for their feedback and support and the anonymous reviewersfor their valuable comments and suggestions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
LifeMap Sciences Inc., CA, USA; Crown Human Genome Center at the Weizmann Institute of Science. Funding for open access charge: Weizmann Institute of Science.
Conflict of interest statement. None declared.
REFERENCES
- 1.Rappaport N., Nativ N., Stelzer G., Twik M., Guan-Golan Y., Stein T.I., Bahir I., Belinky F., Morrey C.P., Safran M., et al. MalaCards: an integrated compendium for diseases and their annotation. Database. 2013;2013:bat018. doi: 10.1093/database/bat018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Safran M., Dalah I., Alexander J., Rosen N., Iny Stein T., Shmoish M., Nativ N., Bahir I., Doniger T., Krug H., et al. GeneCards Version 3: the human gene integrator. Database. 2010;2010:baq020. doi: 10.1093/database/baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S., Stein T.I., Nudel R., Lieder I., Mazor Y., et al. The GeneCards Suite: from gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinformatics. 2016;54:1.30.1–1.30.33. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 4.Aronson A.R., Lang F.M. An overview of MetaMap: historical perspective and recent advances. J. Am. Med. Inform. Assoc. 2010;17:229–236. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pinero J., Queralt-Rosinach N., Bravo A., Deu-Pons J., Bauer-Mehren A., Baron M., Sanz F., Furlong L.I. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database. 2015;2010:bav028. doi: 10.1093/database/bav028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amberger J.S., Bocchini C.A., Schiettecatte F., Scott A.F., Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM(R)), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-Ari Fuchs S., Lieder I., Stelzer G., Mazor Y., Buzhor E., Kaplan S., Bogoch Y., Plaschkes I., Shitrit A., Rappaport N., et al. GeneAnalytics: an integrative gene set analysis tool for next generation sequencing, RNAseq and microarray data. OMICS. 2016;20:139–151. doi: 10.1089/omi.2015.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rappaport N., Twik M., Nativ N., Stelzer G., Bahir I., Stein T.I., Safran M., Lancet D. MalaCards: a comprehensive automatically-mined database of human diseases. Curr. Protoc. Bioinformatics. 2014;47 doi: 10.1002/0471250953.bi0124s47. doi:10.1002/0471250953.bi0124s47. [DOI] [PubMed] [Google Scholar]
- 9.Belinky F., Nativ N., Stelzer G., Zimmerman S., Iny Stein T., Safran M., Lancet D. PathCards: multi-source consolidation of human biological pathways. Database: J. Biol. Dtabases Curation. 2015;2015:bav006. doi: 10.1093/database/bav006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hidalgo C.A., Blumm N., Barabasi A.L., Christakis N.A. A dynamic network approach for the study of human phenotypes. PLoS Comput. Biol. 2009;5:e1000353. doi: 10.1371/journal.pcbi.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohler S., Doelken S.C., Mungall C.J., Bauer S., Firth H.V., Bailleul-Forestier I., Black G.C., Brown D.L., Brudno M., Campbell J., et al. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014;42:D966–D974. doi: 10.1093/nar/gkt1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kibbe W.A., Arze C., Felix V., Mitraka E., Bolton E., Fu G., Mungall C.J., Binder J.X., Malone J., Vasant D., et al. Disease Ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data. Nucleic Acids Res. 2015;43:D1071–D1078. doi: 10.1093/nar/gku1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maiella S., Rath A., Angin C., Mousson F., Kremp O. [Orphanet and its consortium: where to find expert-validated information on rare diseases] Rev Neurol (Paris) 2013;169(Suppl. 1):S3–S8. doi: 10.1016/S0035-3787(13)70052-3. [DOI] [PubMed] [Google Scholar]
- 14.Eppig J.T., Richardson J.E., Kadin J.A., Ringwald M., Blake J.A., Bult C.J. Mouse Genome Informatics (MGI): reflecting on 25 years. Mamm. Genome. 2015;26:272–284. doi: 10.1007/s00335-015-9589-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chute C.G., Carter J.S., Tuttle M.S., Haber M., Brown S.H. Integrating pharmacokinetics knowledge into a drug ontology: as an extension to support pharmacogenomics. AMIA Annu. Symp. Proc. 2003;2003:170–174. [PMC free article] [PubMed] [Google Scholar]
- 16.Edgar R., Mazor Y., Rinon A., Blumenthal J., Golan Y., Buzhor E., Livnat I., Ben-Ari S., Lieder I., Shitrit A., et al. LifeMap Discovery: the embryonic development, stem cells, and regenerative medicine research portal. PLoS One. 2013;8:e66629. doi: 10.1371/journal.pone.0066629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts I., Ker K. Cochrane: the unfinished symphony of research synthesis. Syst. Rev. 2016;5:115. doi: 10.1186/s13643-016-0290-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bramer G.R. International statistical classification of diseases and related health problems. Tenth revision. World Health Stat. Q. 1988;41:32–36. [PubMed] [Google Scholar]
- 19.McEntyre J. Linking up with Entrez. Trends Genet. 1998;14:39–40. doi: 10.1016/S0168-9525(97)01325-5. [DOI] [PubMed] [Google Scholar]
- 20.Pletscher-Frankild S., Palleja A., Tsafou K., Binder J.X., Jensen L.J. DISEASES: text mining and data integration of disease-gene associations. Methods. 2015;74:83–89. doi: 10.1016/j.ymeth.2014.11.020. [DOI] [PubMed] [Google Scholar]
- 21.Allende R.A. Accelerating searches of research grants and scientific literature with novo|seekSM. Nat. Methods. 2009;6 http://www.nature.com/nmeth/journal/v6/n5/full/nmeth.f.250.html [Google Scholar]
- 22.Futreal P.A., Coin L., Marshall M., Down T., Hubbard T., Wooster R., Rahman N., Stratton M.R. A census of human cancer genes. Nat. Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pagon R.A. GeneTests: integrating genetic services into patient care. Am. J. Hum. Genet. 2007;81:658–659. doi: 10.1086/521407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rubinstein W.S., Maglott D.R., Lee J.M., Kattman B.L., Malheiro A.J., Ovetsky M., Hem V., Gorelenkov V., Song G., Wallin C., et al. The NIH genetic testing registry: a new, centralized database of genetic tests to enable access to comprehensive information and improve transparency. Nucleic Acids Res. 2013;41:D925–D935. doi: 10.1093/nar/gks1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gene Ontology C. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stelzer G., Inger A., Olender T., Iny-Stein T., Dalah I., Harel A., Safran M., Lancet D. GeneDecks: paralog hunting and gene-set distillation with GeneCards annotation. OMICS. 2009;13:477–487. doi: 10.1089/omi.2009.0069. [DOI] [PubMed] [Google Scholar]
- 27.Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J., et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forbes S.A., Beare D., Gunasekaran P., Leung K., Bindal N., Boutselakis H., Ding M., Bamford S., Cole C., Ward S., et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sonabend A.M., Zacharia B.E., Cloney M.B., Sonabend A., Showers C., Ebiana V., Nazarian M., Swanson K.R., Baldock A., Brem H., et al. Defining Glioblastoma resectability through the wisdom of the crowd: a proof-of-principle study. Neurosurgery. 2016;2016 doi: 10.1227/NEU.0000000000001374. doi:10.1227/NEU.0000000000001374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis A.P., Grondin C.J., Lennon-Hopkins K., Saraceni-Richards C., Sciaky D., King B.L., Wiegers T.C., Mattingly C.J. The Comparative Toxicogenomics Database's 10th year anniversary: update 2015. Nucleic Acids Res. 2015;43:D914–D920. doi: 10.1093/nar/gku935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Famiglietti M.L., Estreicher A., Gos A., Bolleman J., Gehant S., Breuza L., Bridge A., Poux S., Redaschi N., Bougueleret L., et al. Genetic variations and diseases in UniProtKB/Swiss-Prot: the ins and outs of expert manual curation. Hum. Mutat. 2014;35:927–935. doi: 10.1002/humu.22594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imbert G., Saudou F., Yvert G., Devys D., Trottier Y., Garnier J.M., Weber C., Mandel J.L., Cancel G., Abbas N., et al. Cloning of the gene for spinocerebellar ataxia 2 reveals a locus with high sensitivity to expanded CAG/glutamine repeats. Nat. Genet. 1996;14:285–291. doi: 10.1038/ng1196-285. [DOI] [PubMed] [Google Scholar]
- 33.Lohmann K., Klein C. Next generation sequencing and the future of genetic diagnosis. Neurotherapeutics. 2014;11:699–707. doi: 10.1007/s13311-014-0288-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stelzer G., Plaschkes I., Oz-Levi D., Alkelai A., Olender T., Zimmerman S., Twik M., Belinky F., Fishilevich S., Nudel R., et al. VarElect: the phenotype-based variation prioritizer of the GeneCards Suite. BMC Genomics. 2016;17(Suppl. 2):444. doi: 10.1186/s12864-016-2722-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rabbani B., Nakaoka H., Akhondzadeh S., Tekin M., Mahdieh N. Next generation sequencing: implications in personalized medicine and pharmacogenomics. Mol. Biosyst. 2016;12:1818–1830. doi: 10.1039/c6mb00115g. [DOI] [PubMed] [Google Scholar]
- 36.Zhao C., Eisinger B.E., Driessen T.M., Gammie S.C. Addiction and reward-related genes show altered expression in the postpartum nucleus accumbens. Front. Behav. Neurosci. 2014;8:388. doi: 10.3389/fnbeh.2014.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao M., Wang K.J., Tan Z., Zheng C.M., Liang Z., Zhao J.Q. Identification of potential therapeutic targets for papillary thyroid carcinoma by bioinformatics analysis. Oncol. Lett. 2016;11:51–58. doi: 10.3892/ol.2015.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hashemikhabir S., Xia R., Xiang Y., Janga S. A framework for identifying genotypic information from clinical records: exploiting integrated ontology structures to transfer annotations between ICD codes and Gene Ontologies. IEEE/ACM Trans. Comput. Biol. Bioinform. 2015;2015 doi: 10.1109/TCBB.2015.2480056. doi:10.1109/TCBB.2015.2480056. [DOI] [PubMed] [Google Scholar]
- 39.Hashemikhabir S., Neelamraju Y., Janga S.C. Database of RNA binding protein expression and disease dynamics (READ DB) Database. 2015;2015:bav072. doi: 10.1093/database/bav072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou X., Menche J., Barabasi A.L., Sharma A. Human symptoms-disease network. Nat. Commun. 2014;5:4212. doi: 10.1038/ncomms5212. [DOI] [PubMed] [Google Scholar]
- 41.Speir M.L., Zweig A.S., Rosenbloom K.R., Raney B.J., Paten B., Nejad P., Lee B.T., Learned K., Karolchik D., Hinrichs A.S., et al. The UCSC Genome Browser database: 2016 update. Nucleic Acids Res. 2016;44:D717–D725. doi: 10.1093/nar/gkv1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lopes P., Oliveira J.L. An innovative portal for rare genetic diseases research: the semantic Diseasecard. J. Biomed. Inform. 2013;46:1108–1115. doi: 10.1016/j.jbi.2013.08.006. [DOI] [PubMed] [Google Scholar]