Abstract
Since 1992, FlyBase (flybase.org) has been an essential online resource for the Drosophila research community. Concentrating on the most extensively studied species, Drosophila melanogaster, FlyBase includes information on genes (molecular and genetic), transgenic constructs, phenotypes, genetic and physical interactions, and reagents such as stocks and cDNAs. Access to data is provided through a number of tools, reports, and bulk-data downloads. Looking to the future, FlyBase is expanding its focus to serve a broader scientific community. In this update, we describe new features, datasets, reagent collections, and data presentations that address this goal, including enhanced orthology data, Human Disease Model Reports, protein domain search and visualization, concise gene summaries, a portal for external resources, video tutorials and the FlyBase Community Advisory Group.
INTRODUCTION
Now in its 25th year, FlyBase continues to serve as the leading database and web portal for genetic, genomic, and functional information about the model organism Drosophila melanogaster (1,2). During this tenure, the primary literature has been and continues to be the major source of data in FlyBase (3). Over the last decade, there has been an additional focus on other types of data, with high-throughput data projects featuring prominently. Recent years have seen the completion of Release 6 of the Drosophila melanogaster reference genome (4,5); a complete reannotation of the reference genome on the basis of RNA-Seq coverage data, RNA-Seq junction data, and transcription start site profiles from the modENCODE project (6–10); and the generation of large public collections of insertion mutants by the Gene Disruption Project (11–14).
As biological knowledge continues to expand and new techniques are developed, FlyBase strives to provide our expanding user community with improved access to new data and data types. Recent years have seen the addition of Gene Group Reports (1), the design of multiple tools to access RNA-Seq data, improvements to FlyBase ontologies (15,16), the incorporation of the Disease Ontology (17,18), the addition of the user–curation tool Fast-Track Your Paper (19) and enhancements to our major search interface, QuickSearch.
In the past year, we have continued to add new types of data, features, tools and user-help options. Cooperation across the model organism communities has become an increasingly important issue, especially as it impacts research into human disease. In the interest of enhancing the accessibility of FlyBase to users outside our traditional community, we have incorporated new orthology data for Homo sapiens and eight model organisms, added a robust orthology search tool, and improved the display of orthology calls on FlyBase Gene Reports. We have also highlighted the utility of flies in human disease research through the addition of Human Disease Model Reports.
In addition, we have made improvements to existing data types and tools. Many FlyBase Gene Reports now include a Gene Snapshot, which is a short pithy overview of a gene's function, written by an expert researcher in language friendly to the non-specialist. Our genome browser GBrowse now includes a track for Pfam protein domains, with domain mappings to multiple protein isoforms for genes with multiple transcripts (20). We have incorporated several new high-throughput datasets and two major reagent collections, the P[acman] collection of BAC clones and the Vienna Tiles (VT) GAL4 collection of fly stocks into GBrowse (21,22).
There is a wealth of resources housed in external websites that are vitally important to FlyBase users. We have updated, expanded and reorganized the Resources page where we maintain lists of hundreds of third party websites, adding a portal page for the most popular resource categories, and a prominent new access button on the FlyBase front page.
FlyBase has also been working to improve engagement with our user community. We are now producing video tutorials, to help our users navigate FlyBase and FlyBase tools. Finally, we have recruited a FlyBase Community Advisory Group, with representatives from fly labs in over 40 countries, which we periodically survey about potential changes and improvements to FlyBase.
Enhanced orthology data
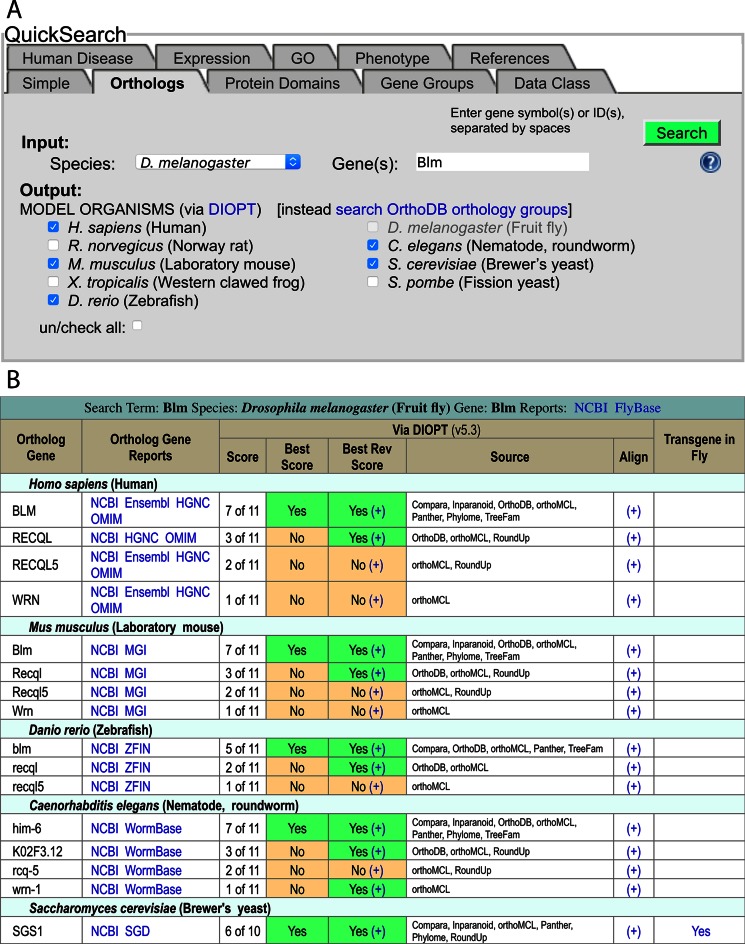
FlyBase has recently incorporated orthology data from the DRSC Integrative Ortholog Prediction Tool (DIOPT) (23). This dataset, added in the FB2016_02 release of FlyBase, integrates ortholog predictions between several species (currently H. sapiens, D. melanogaster and seven other model organisms) from multiple individual tools (currently Compara, HGNC, Homologene, Inparanoid, Isobase, OMA, OrthoDB, orthoMCL, Panther, Phylome, RoundUp, TreeFam and ZFIN). The DIOPT approach thereby provides a streamlined comparison of orthology predictions originating from different algorithms based on sequence homology, phylogenetic trees or functional similarity. FlyBase also includes a non-redundant orthology set derived specifically from OrthoDB (24). This set comprises ∼60 species, biased towards those organisms closely related to D. melanogaster, such as additional Drosophila species and other diptera. Both the DIOPT and OrthoDB datasets are searchable through the new ‘Orthologs’ tab of the QuickSearch tool on the FlyBase homepage (Figure 1) and are shown explicitly within the revised ‘Orthologs’ section of our Gene Reports.
Figure 1.
(A) The Orthologs tab of the QuickSearch tool, located on the FlyBase homepage. The D. melanogaster gene ‘Blm’ has been entered as the search term, and several species within the DIOPT set have been selected as the output. (B) The results page of that query—see text for details.
To use the new QuickSearch Orthologs tab, simply select the input species, enter one or more gene symbols or IDs, then select one or more output species (Figure 1A). Where the input species is D. melanogaster, there is a choice between searching the DIOPT or OrthoDB-derived datasets. For DIOPT-based searches, the choice of output species is identical to the input species; for OrthoDB-based searches, the output species are arranged into five distinct groups: ‘Drosophila species’, ‘non-Drosophila Dipterans’, ‘non-Dipteran Insects’, ‘non-Insect Arthropods’ and ‘non-Arthropod Metazoa’.
The results page shows the list of ortholog predictions arranged by species (Figure 1B). For each gene, the official gene symbol is shown alongside links to report pages at model organism databases, NCBI (National Center for Biotechnology Information), Ensembl and/or OMIM® (Online Mendelian Inheritance in Man). For DIOPT-based searches, the number of individual algorithms supporting a given orthologous gene-pair relationship, compared to the total number of relevant algorithms for that call, is expressed as the ‘Score’; the explicit list of supporting algorithms is also shown. The ‘Best score’ and ‘Best Rev Score’ columns indicate whether the given ortholog has the highest score for the query gene, and whether the reciprocal relationship is also true. Links are also provided to an alignment of amino acid sequences between predicted orthologs on the DIOPT site, and to FlyBase Gene Reports where a non-Drosophila gene has been expressed transgenically in flies. If desired, the results table may be downloaded in TSV format by clicking on the link at the top of the table. The results page for OrthoDB-based searches is similar, except that the DIOPT-specific columns are absent and the OrthoDB ‘orthology group' to which the species belongs is given.
A similar presentation is used to show DIOPT and OrthoDB-derived orthology data within the ‘Orthologs’ section of D. melanogaster Gene Reports (not shown). Orthology relationships between D. melanogaster and humans are highlighted in a separate ‘Human Orthologs’ subsection, which also includes links to the human gene reports and any associated diseases at the OMIM website (omim.org) (25). Furthermore, Gene Group Reports, which serve to describe and list sets of functionally related D. melanogaster genes or gene products (1), have been enhanced with a ‘View orthologs’ option—clicking this button automatically runs all listed genes through the QuickSearch Orthologs tool to generate the results table described above.
Human disease model reports
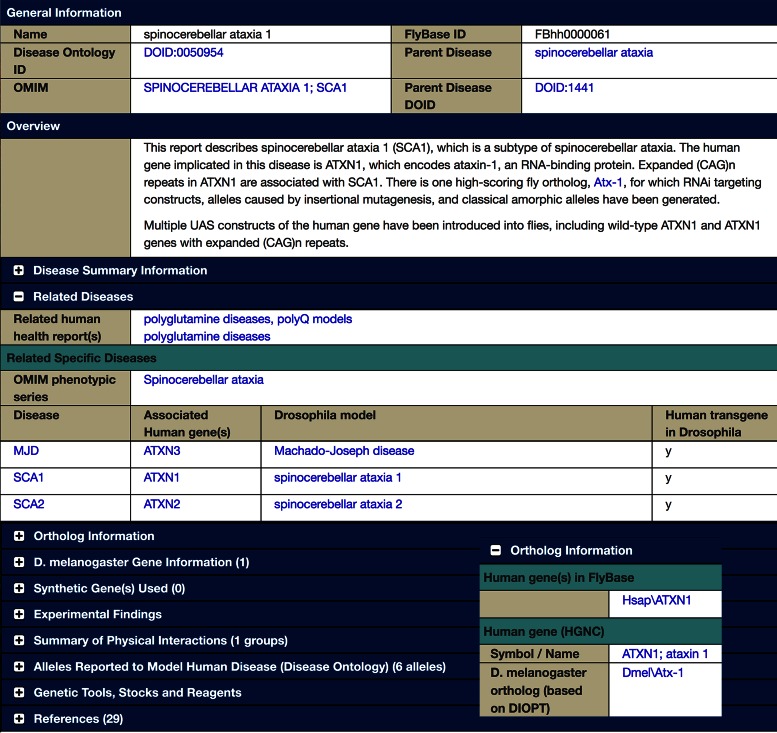
The Human Disease Model Report (Figure 2), first introduced in September 2015 (FB2015_04) is designed as an entry point for both non-Drosophila researchers unfamiliar with FlyBase, and for Drosophila researchers interested in human disease as modeled in flies (18). This new report integrates disease-related information from multiple locations across FlyBase, as well as from outside resources, and includes a bibliography of research papers and reviews. As of the October 2016 release (FB2016_05), FlyBase has produced 388 such reports.
Figure 2.
Human Disease Model Report for Spinocerebellar Ataxia 1. General Information, Overview, Related Diseases. Inset: Ortholog Information.
The top section of each report (Figure 2) includes links to the corresponding OMIM and/or FlyBase Term Reports for Disease Ontology terms, if available. This is immediately followed by an ‘Overview’ of key points from information deeper in the report. The next section, the ‘Disease Summary’, provides disease-specific information from resources outside of FlyBase, primarily from OMIM phenotype and genotype reports, organized as so to be useful to geneticists and molecular biologists. This section also includes links to informational resources such as GeneReviews® (26) and the MGI (Mouse Genome Informatics) Human-Mouse Disease Connection (27).
The ‘Related Diseases’ section (Figure 2) includes links to reports for diseases that have a relationship to the disease reported. ‘Related Specific Diseases’ are subtypes of a parent disease. For example, the disease spinocerebellar ataxia 1 is one of >40 genetically distinct autosomal dominant spinocerebellar ataxias. The ‘Related Specific Diseases’ table links to the OMIM phenotype and genotype report of each subtype that has been associated with a specific human gene, as well as to each disease subtype for which a FlyBase report has been made; this allows facile navigation between related reports. Other kinds of disease relationships, such as a common genetic or mechanistic basis, are displayed in the ‘Related human health report(s)’.
The ‘Ortholog Information’ section (Figure 2, inset), links to gene reports for the implicated human gene and the D. melanogaster ortholog(s). If the human gene has been transgenically introduced into flies, this section also links to a FlyBase Gene Report for the transgene. The ‘Synthetic Gene(s) Used’ section links to FlyBase Gene Report(s) for synthetic gene(s). These gene associations allow us to promote data from elsewhere in FlyBase into the Human Disease Model Report; promoted data types include Disease Ontology annotations of alleles of human, fly and synthetic genes that have been shown experimentally to model the given disease; physical interactions between proteins or transcripts encoded by the orthologous fly genes and other fly proteins or transcripts, and genetic tools, stocks, and reagents involving mutants and constructs of the human, fly, and synthetic genes.
The Human Disease Model Report format has allowed FlyBase to include multiple types of disease models. The majority of these are based on OMIM gene-disease associations, such as the subtypes of amyotrophic lateral sclerosis or xeroderma pigmentosum. Mechanistic models address diseases caused by a common underlying mechanism; many of these are modeled through use of a synthetic construct, such as ‘RNA repeat diseases’ or ‘polyglutamine diseases, polyQ models’. Induced models make use of an environmental or chemical perturbation, such as the model for ‘Parkinson-like disease, chemically-induced’, in which Parkinson disease-like phenotypes are induced by exposure to toxins such as rotenone or paraquat. Postulated disease models have not been definitively identified in human patients, but have been studied in flies. Postulated models can include potential diseases identified in genome-wide associated studies (GWAS), such as ‘Alzheimer disease, susceptibility to (postulated), SNRPN-related’. Other postulated models involve genes or proteins that appear to be involved in a human disease but have not yet been definitively implicated, such as ‘Parkinson disease (postulated), GPR37-related’.
A browsable list of all Human Disease Model Reports to date can be accessed via a link on the Human Disease QuickSearch tab. An update to this QuickSearch tab, available in the FB2016_06 release, supports searching for reports using a disease name, synonym, Disease Ontology term, or OMIM ID number; a human gene symbol or HGNC ID number; or a Drosophila gene symbol, synonym, or FlyBase ID number. Reports can also be accessed from a corresponding Disease Ontology Term Report page, and through links in the Human Disease Model Data section of FlyBase Gene Reports.
Protein domain display
Protein domain data in FlyBase are obtained from InterPro (28). InterPro combines signatures from several member databases (including Pfam, PROSITE and SMART) and provides protein domain predictions for the UniProtKB reference set of annotated polypeptides (29), which includes the Drosophila proteome. These domain data are currently shown in the ‘Protein Domains/Motifs’ and ‘Polypeptide Data’ sections of FlyBase Gene Reports. A ‘Protein Domains’ search option, recently added to the FlyBase QuickSearch tool, makes it easy to find genes with a given protein domain. Valid entries include InterPro signature names or identifier numbers, which include protein domains, families, repeats or sites. Searches are case-insensitive, and partial terms are supported (using * as a wildcard).
To better illustrate the organization of annotated polypeptides, protein domains encoded in the genome are shown in the new ‘Protein domains (PFAM)’ GBrowse track (listed in the ‘Aligned Evidence’ section). Pfam signatures (20) within the InterPro data were mapped by FlyBase to the reference D. melanogaster genome assembly. In all, ∼18 000 domain calls of ∼4000 domains for ∼10 000 genes were mapped to the genome, covering a total of ∼8 megabase pairs. Note that only Pfam domain signatures identified in the UniProtKB reference set are shown in Gbrowse; domains encoded by portions of the genome not represented by the UniProtKB reference set are not shown. Note also that these data are currently limited to the Pfam set of domain calls, which represents the largest set of domain calls available; additional domain calls available from the InterPro consortium may be incorporated at a future date.
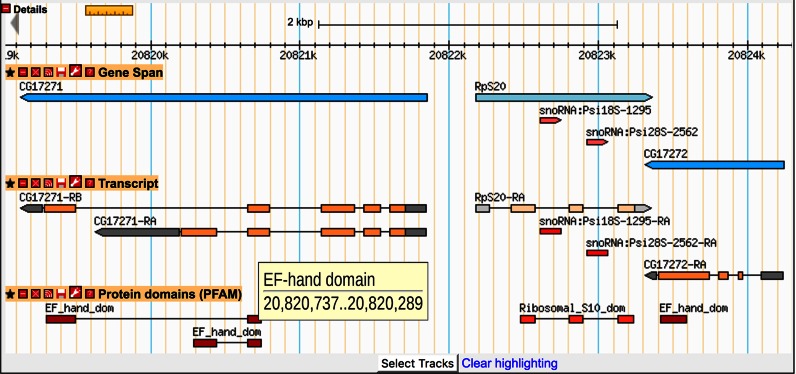
Domain mappings are depicted in GBrowse as orange (plus strand) or red (minus strand) blocks, with thin lines spanning exon junctions (Figure 3). Mousing over the block presents domain name and location information, while clicking on the block opens up a report at pfam.org with more data for the domain. This genomic view of protein domains offers several advantages, including a quick view of protein organization for all alternative isoforms, and the illumination of genomic regions encoding clusters of genes sharing similar sets of protein domains. For example, this track clearly illustrates how alternative C-terminal splicing of the CG17271 gene generates proteins with alternative EF-hand domains, as well as the presence of two EF-hand-encoding genes (CG17271 and CG17272) in this region. While the genomic view offers some advantages, it can be difficult to appreciate a protein's domain organization on the genome when splicing is complex or spans large introns. For this reason, FlyBase is also developing visualizations for our Gene and Polypeptide Reports that present Pfam domain data in the context of the uninterrupted polypeptide.
Figure 3.
Protein domain visualization in GBrowse. Screenshot of GBrowse in the region 3R:20 819 447..20 825 007. Annotated gene regions are shown in the ‘Gene Span’ track at the top as blue arrows, and transcript annotations are shown in the middle panel in the ‘Transcripts’ track (CDSs shown as orange blocks, UTRs shown as gray blocks and introns as thin lines). The ‘Protein domains (PFAM)’ track depicts the mapping of Pfam domain calls to the genome, with domains encoded by genes on the plus or minus strand shown as orange or red blocks, respectively. Many domain calls span multiple exons, in which case the different exonic portions of the domain call are linked by a thin line that bridges the exon junction. Mousing over a Pfam block brings up the domain name and genomic location in a pop-up window, while clicking on the Pfam block opens a report page at pfam.org.
New datasets and reagent collections
FlyBase regularly incorporates high-throughput datasets and large-scale reagent collections, and we highlight here some new additions. We now offer a GBrowse track that allows genome-wide browsing of the P[acman] collection of bacterial artificial chromosome (BAC) clones (21). These clones carry D. melanogaster genomic fragments in a BAC that permits conditional plasmid amplification and ΦC31 integrase-mediated genomic insertion to facilitate transgene construction. We also offer a GBrowse track that depicts the regulatory regions used to generate the Vienna Tiles GAL4 lines available at the VDRC stock center (22,30). As these regulatory regions are small, typically 2 kb in size, they each carry a distinct set of candidate cis-regulatory elements that may recapitulate only a subset of a gene's expression pattern. As such, these Vienna Tile GAL4 lines may be useful in studies of transcriptional regulation, or when a more restricted domain of GAL4 expression is desired. Both of these are listed in the ‘Other Reagents’ section of the GBrowse track selection menu. Clicking on the GBrowse glyph for a given reagent opens up a FlyBase report with additional information about the reagent, including links to the resource centers from which reagents can be obtained. In addition, for a given gene, overlapping BAC clones are listed in the ‘Stocks and Reagents’ section of the FlyBase Gene Report.
FlyBase has also recently incorporated several datasets which are offered as data tracks in GBrowse: RNA-Seq profiles of mRNA 5′-ends, RNA-Seq profiles of small RNA species, and ChIP studies of mesodermal transcription factor binding and histone modification enrichment. The RNA-Seq profiles of capped mRNA 5′-ends originate from the RAMPAGE and MachiBase studies (31,32), providing data for 43 different developmental stage ranges. The RNA-Seq profiles of small RNA (miRNA, piRNA and siRNA) were kindly provided by Eric Lai's group, who reanalyzed data from 274 small RNA-Seq experiments comprising 35 publications (33,34; full list at flybase.org/reports/FBrf0230987.html). These realigned RNA-Seq data were consolidated by sample type to give profiles for 26 cultured cell lines, 13 developmental stage ranges and 5 tissues. Finally, we highlight new ChIP data that were kindly provided by Eileen Furlong's group, who reanalyzed ChIP profiles from several studies to provide updated peak calls (35–39). These data include binding regions for 13 mesodermal transcription factors during embryogenesis, as well as regions enriched for six different histone modifications in purified embryonic mesodermal cells.
Short gene summaries: gene snapshots
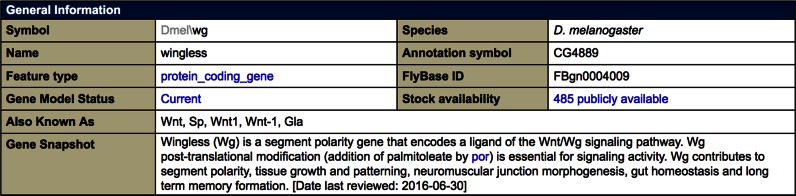
Gene Snapshots are short summaries designed to provide a quick overview of the function of a gene's products. They appear at the top of each Gene Report to directly provide a big picture of the gene's function (Figure 4) and are also downloadable in bulk via our Batch Download tool or a precomputed file (available from the ‘Downloads’ link of the navigation bar on any FlyBase page). Each Snapshot is associated with a review date, and cases that are in progress or are deemed to have insufficient data to summarize are stated as such. Gene Snapshots will be especially helpful for providing key information about the genes selected by genome-wide analyses and screens.
Figure 4.
Gene Snapshot for the gene wg.
In order to produce these, an automated algorithm was designed to select experts on the different genes, and these authors were contacted and asked to provide a couple of sentences/bullet points describing (i) how the protein functions, (ii) what pathway it is in (if relevant) and (iii) its main biological roles, preferably using terms suitable for a general, non-Drosophilist audience. These key points were then revised by FlyBase curators for consistency in length and format. As of FlyBase Release FB2016_05, we have added 1527 Gene Snapshots.
For those genes currently lacking a gene snapshot, FlyBase welcomes contributions through the online form accessible from the snapshot field of the gene. Feedback on existing Gene Snapshots can be made by clicking on the ‘Contact FlyBase’ link (accessible from the bottom of every FlyBase page) and selecting the ‘Gene Snapshots’ option. There is also a dedicated FlyBase Wiki page designed as a hub of information on the Gene Snapshots, which includes the list of contributors, the guidelines for writing a summary and the link to contribute.
The resources portal
FlyBase maintains lists of hundreds of external web sites that offer information and/or reagents of interest to Drosophila researchers. These have been updated and expanded and are all available from the new ‘Resources’ button located at the top of the FlyBase home page or from the ‘Resources’ link in the ‘Links’ drop down menu found on any FlyBase report or query page. FlyBase has taken two different approaches to organizing and displaying these links.
First, FlyBase has created a dedicated ‘Popular Resource Categories’ series of static pages, improving the accessibility of the most frequently visited resources. Links to these pages are located in a navigation bar at the top of the Resources Page. Currently, nine categories have been created: CRISPR, RNAi, Stocks, Antibodies, Model Organism Databases, Images, Neuroscience, Maps and Protocols (Table 1). Each of these pages has been populated with both links and information regarding its specific subject.
Table 1. Content of popular resources pages.
| Popular category | Resource Links Offered |
|---|---|
| Antibodies | Collections of Drosophila-specific antibodies, both commercial and non-profit. |
| CRISPR | gRNA design resources. Stocks and vectors. Selected method reviews and reports. |
| Images | Image-based resources. Tools for image analysis, visualization and annotation. |
| RNAi | Design/analysis tools. Stocks and cell-based reagents. Screening centers, phenotypes and result validation. |
| Neuroscience | Databases of expression, anatomy, and physiology. Tools for viewing, analyzing and querying images. Interactive maps. Nomenclature and protocols. |
| Stocks | Stock collections and links to collections by specific category (e.g. RNAi, Deficiencies, etc.). Species collections. |
| Maps | Genetic map position, cytological (polytene band) position, and genomic coordinates for Drosophila genes. Chromosome maps for D. melanogaster and various other non-melanogaster Drosophila species. |
| Model Organism Databases | Links to 16 different model organism databases. |
| Protocols | A linkout to flyrnai.org's collection of experimental protocols. |
In the second approach, FlyBase maintains extensive lists of resources organized by broad general area of interest. These have been divided into two main sections, ‘Network Resources’ and ‘Reagent Resources’. The first section, Network Resources, is subdivided into 23 subcategories of links centered around online tools and information, e.g. orthology prediction websites, general bioinformatic tools, protein analysis sites and transcription regulation databases and tools. The second section, Reagent Resources, contains 11 subgroups and is focused on providing access to physical reagents that are obtainable via online web portals, such as antibodies, primers, cDNAs and CRISPR vectors. FlyBase welcomes suggestions for new links to be added to these resource pages and updates to the current listings that can be made through the contact FlyBase links present on most FlyBase pages or at the bottom of every wiki page.
Video tutorials
FlyBase is producing video tutorials as a quick and efficient way for users to learn how to use FlyBase tools and discover new functionalities. All our tutorials can be found on our YouTube channel, FlyBase TV, which can be accessed from the ‘Help’ menu on the FlyBase navigation bar, from the YouTube icon button at the bottom of every FlyBase page, or directly from the YouTube website. The videos span a range of topics and target different audiences. They are categorized by themes into series, and users can play all the videos of one series by clicking on the playlist of interest (Table 2). The ‘Basic Navigation’ series is helpful for new users and describes basic operations in FlyBase. The ‘RNA-Seq’ series targets users interested in gene expression data and has a video for each of the RNA-Seq tools.
Table 2. FlyBase video tutorials.
| Basic Navigation Series | How to find all data related to a gene |
| How to generate an Excel file of all alleles of a gene | |
| Finding related genes in FlyBase: gene groups | |
| RNA-Seq Series | RNA-Seq Part I: using GBrowse |
| RNA-Seq Part II: using RNA-Seq profile search | |
| RNA-Seq Part III: searching for similarly expressed genes | |
| FlyBase Guidelines | How to cite FlyBase |
| Author guidelines |
We aim to produce a video for each tool in FlyBase, and others in response to frequent queries from the community. To be notified of newly released videos you can subscribe to FlyBase TV, by clicking the red button ‘Subscribe’ on our YouTube page.
The FlyBase community advisory group
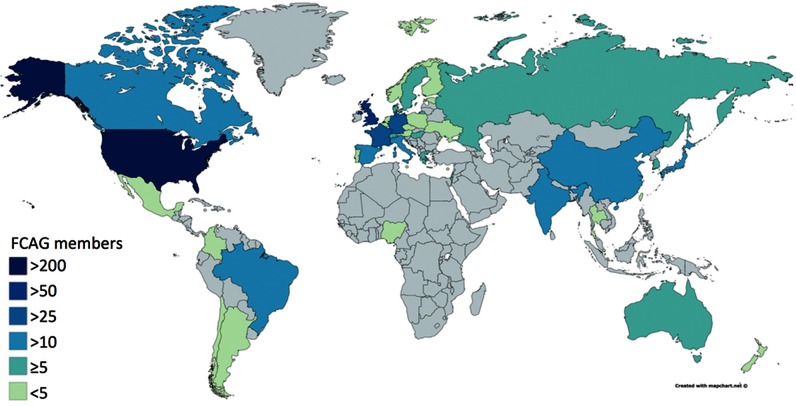
As the members of the FlyBase team work to improve the amount and usefulness of data in FlyBase, numerous questions arise regarding which data are the most important for Drosophila researchers and how these data can be most usefully presented on the website. The FlyBase Community Advisory Group (FCAG) was launched in September 2014 with the aim of gaining greater feedback from the community about changes in our database. The group consists of representatives from any Drosophila lab worldwide that uses FlyBase as part of its research. FCAG is currently comprised of 541 members representing 529 Drosophila labs from 41 countries (Figure 5).
Figure 5.
International distribution of the FlyBase Community Advisory Group. Map created using https://mapchart.net/.
Members of the group are sent up to six surveys a year on a variety of different subjects. To date, we have completed seven surveys, with an average response rate of 54%. Issues covered have ranged from a general survey on improvements people would like to see in FlyBase, to more targeted surveys on specific data types or feedback about new features. Several of the updates described above were made as a direct result of FCAG responses, including better representation of protein domains, the launch of video tutorials, the addition of the Gene Snapshot section of Gene Reports, and updating the external resources page with popular categories.
FlyBase aims to have a representative from every lab that uses FlyBase in their research. Interested researchers are encouraged to register by following the FCAG link under the Community header of the FlyBase navigation bar.
FlyBase, community and communication
The role of FlyBase extends beyond the distribution of curated data; we are not only a resource to the community but also a resource of community, facilitating communication from FlyBase to our users, users to FlyBase and users with one another. All our community resources can be found under the Community header in the navigation bar, or through the ‘Community’ button located on the left side of the FlyBase homepage. Users can receive news and information from FlyBase by subscribing to the FlyBase newsletter, the FlyBaseDotOrg Twitter feed, as well as by reading updates via the News button and rotating commentaries on the homepage. Researchers can send feedback or questions to FlyBase through the ‘Contact FlyBase’ link at the bottom of every FlyBase page, or can take a more active role by joining the FlyBase Community Advisory Group, described above. Finally, FlyBase facilitates communication between researchers through an opt-in People database of contact information, and through the FlyBase Forum discussion group.
Over the last 25 years, FlyBase has been a vital informational resource to the Drosophila research community. We continue this role today, while also striving to serve a more diverse user base, which includes human geneticists, clinicians, and biologists working with other model organisms. Engaging with this expanding community will be key as FlyBase looks to the future.
Acknowledgments
We would like to thanks the PIs, curators, and developers of FlyBase for their comments on the manuscript. At the time of writing, the members of the FlyBase Consortium included: Norbert Perrimon, Susan Russo Gelbart, Cassandra Extavour, Kris Broll, Madeline A. Crosby, Gilberto do Santos, David B. Emmert, L. Sian Gramates, Kathleen Falls, Beverley B. Matthews, Christopher J. Tabone, Pinglei Zhou, Mark Zytkovicz, Nicholas H. Brown, Giulia Antonazzo, Helen Attrill, Marta Costa, Silvie Fexova, Tamsin Jones, Aoife Larkin, Steven J. Marygold, Gillian H. Millburn, Alix J. Rey, Nicole Staudt, Jose-Maria Urbano, Thomas Kaufman, Joshua L. Goodman, Gary B. Grumbling, Victor B. Strelets, Jim Thurmond, Richard Cripps, Maggie Werner-Washburne, Phillip Baker.
Footnotes
Present addresses:
Laura Ponting, Wellcome Trust Sanger Institute, Wellcome Genome Campus, Hinxton CB10 1SA, UK.
Andrew J. Schroeder, Department of Biomedical Informatics, Harvard Medical School, Boston, MA 02115, USA.
FUNDING
National Human Genome Research Institute at the National Institutes of Health [U41 HG00739 to N.P.]; Medical Research Council (UK) [G1000968 to N.B.]. Funding for open access charge: National Human Genome Research Institute at the National Institutes of Health [U41 HG00739 to N.P.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Attrill H., Falls K., Goodman J.L., Millburn G.H., Antonazzo G., Rey A.J., Marygold S.J., FlyBase Consortium FlyBase: establishing a gene group resource for Drosophila melanogaster. Nucleic Acids Res. 2016;44:D786–D792. doi: 10.1093/nar/gkv1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marygold S.J., Crosby M.A., Goodman J.L., FlyBase Consortium . Using FlyBase, a database of Drosophila Genes & Genomes. In: Dahmann C, editor. Drosophila: Methods and Protocols. 2nd edn. Vol. 1478. NY: Springer; 2016. pp. 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marygold S.J., Leyland P.C., Seal R.L., Goodman J.L., Thurmond J., Strelets V.B., Wilson R.J., FlyBase consortium FlyBase: improvements to the bibliography. Nucleic Acids Res. 2013;41:D751–D757. doi: 10.1093/nar/gks1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoskins R.A., Carlson J.W., Wan K.H., Park S., Mendez I., Galle S.E., Booth B.W., Pfeiffer B.D., George R.A., Svirskas R., et al. The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res. 2015;25:445–458. doi: 10.1101/gr.185579.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.dos Santos G., Schroeder A.J., Goodman J.L., Strelets V.B., Crosby M.A., Thurmond J., Emmert D.B., Gelbart W.M., FlyBase Consortium FlyBase: introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res. 2015;43:D690–D697. doi: 10.1093/nar/gku1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.modENCODE Consortium. Roy S., Ernst J., Kharchenko P.V., Kheradpour P., Negre N., Eaton M.L., Landolin J.M., Bristow C.A., Ma L., Lin M.F., et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graveley B.R., Brooks A.N., Carlson J.W., Duff M.O., Landolin J.M., Yang L., Artieri C.G., van Baren M.J., Boley N., Booth B.W., et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoskins R.A., Landolin J.M., Brown J.B., Sandler J.E., Takahashi H., Lassmann T., Yu C., Booth B.W., Zhang D., Wan K.H., et al. Genome-wide analysis of promoter architecture in Drosophila melanogaster. Genome Res. 2011;21:182–192. doi: 10.1101/gr.112466.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matthews B.B., Dos Santos G., Crosby M.A., Emmert D.B., St Pierre S.E., Gramates L.S., Zhou P., Schroeder A.J., Falls K., Strelets V., Russo S.M., Gelbart W.M., FlyBase Consortium Gene model annotations for Drosophila melanogaster: Impact of high-throughput data. G3 (Bethesda) 2015;5:1721–1736. doi: 10.1534/g3.115.018929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crosby M.A., Gramates L.S., Dos Santos G., Matthews B.B., St Pierre S.E., Zhou P., Schroeder A.J., Falls K., Emmert D.B., Russo S.M., Gelbart W.M., FlyBase Consortium Gene model annotations for Drosophila melanogaster: The Rule-Benders. G3 (Bethesda) 2015;5:1737–1749. doi: 10.1534/g3.115.018937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagarkar-Jaiswal S., DeLuca S.Z., Lee P.T., Lin W.W., Pan H., Zuo Z., Lv J., Spradling A.C., Bellen H.J. A genetic toolkit for tagging intronic MiMIC containing genes. eLife. 2015;4:e08469. doi: 10.7554/eLife.08469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagarkar-Jaiswal S., Lee P.T., Campbell M.E., Chen K., Anguiano-Zarate S., Cantu Gutierrez M., Busby T., Lin W.W., He Y., Schulze K.L., et al. A library of MiMICs allows tagging of genes and reversible, spatial and temporal knockdown of proteins in Drosophila. eLife. 2015;4:e05338. doi: 10.7554/eLife.05338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spradling A.C., Bellen H.J., Hoskins R.A. Drosophila P elements preferentially transpose to replication origins. Proc. Natl. Acad. Sci. U.S.A. 2011;108:15948–15953. doi: 10.1073/pnas.1112960108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venken K.J.T., Schulze K.L., Haelterman N.A., Pan H., He Y., Evans-Holm M., Carlson J.W., Levis RW., Spradling A.C., Hoskins R.A., et al. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods. 2011;8:737–743. doi: 10.1038/nmeth.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costa M., Reeve S., Grumbling G., Osumi-Sutherland D. The Drosophila anatomy ontology. J. Biomed Semantics. 2013;4:32. doi: 10.1186/2041-1480-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osumi-Sutherland D., Marygold S.J., Millburn G.H., McQuilton P.A., Ponting L., Stefancsik R., Falls K., Brown N.H., Gkoutos G.V. The Drosophila phenotype ontology. J. Biomed Semantics. 2013;4:30. doi: 10.1186/2041-1480-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kibbe W.A., Arze C., Felix V., Mitraka E., Bolton E., Fu G., Mungall C.J., Binder J.X., Malone J., Vasant D., et al. Disease ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data. Nucleic Acids Res. 2015;43:D1071–D1078. doi: 10.1093/nar/gku1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Millburn G.H., Crosby M.A., Gramates L.S., Tweedie S., FlyBase Consortium FlyBase portals to human disease research using Drosophila models. Dis. Model Mech. 2016;9:245–252. doi: 10.1242/dmm.023317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bunt S.M., Grumbling G.B., Field H.I., Marygold S.J., Brown N.H., Millburn G.H., FlyBase Consortium Directly e-mailing authors of newly published papers encourages community curation. Database (Oxford) 2012;2012:bas024. doi: 10.1093/database/bas024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A., et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venken K.J., Carlson J.W., Schulze K.L., Pan H., He Y., Spokony R., Wan K.H., Koriabine M., de Jong P.J., White K.P., et al. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat. Methods. 2009;6:431–434. doi: 10.1038/nmeth.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kvon E.Z., Kazmar T., Stampfel G., Yáñez-Cuna O., Pagani M., Schernhuber K., Dickson B.J., Stark A. Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature. 2014;512:91–95. doi: 10.1038/nature13395. [DOI] [PubMed] [Google Scholar]
- 23.Hu Y., Flockhart I., Vinayagam A., Bergwitz C., Berger B., Perrimon N., Mohr S.E. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics. 2011;12:357. doi: 10.1186/1471-2105-12-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kriventseva E.V., Tegenfeldt F., Petty T.J., Waterhouse R.M., Simão F.A., Pozdnyakov I.A., Ioannidis P., Zdobnov E.M. OrthoDB v8: update of the hierarchical catalog of orthologs and the underlying free software. Nucleic Acids Res. 2015;43:D250–D256. doi: 10.1093/nar/gku1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amberger J.S., Bocchini C.A., Schiettecatte F., Scott A.F., Hamosh A. OMIM.org: Online mendelian inheritance in man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pagon R.A., Adam M.P., Ardinger H.H., Wallace S.E., Amemiya A., Bean L.J.H, Bird T.D., Ledbetter N., Mefford H.C., Smith R.J.H., Stephens K., editors. GeneReviews® [Internet] Seattle: University of Washington; 1993. 2016. [Google Scholar]
- 27.Eppig J.T., Richardson J.E., Kadin J.A., Smith C.L., Blake J.A., Bult C.J., MGD Team Mouse genome database: From sequence to phenotypes and disease models. Genesis. 2015;53:458–473. doi: 10.1002/dvg.22874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitchell A., Chang H.Y., Daugherty L., Fraser M., Hunter S., Lopez R., McAnulla C., McMenamin C., Nuka G., Pesseat S., et al. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 2015;43:D213–D221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dietzl G., Chen D., Schnorrer F., Su K.C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S., et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 31.Batut P., Dobin A., Plessy C., Carninci P., Gingeras T.R. High-fidelity promoter profiling reveals widespread alternative promoter usage and transposon-driven developmental gene expression. Genome Res. 2013;23:169–180. doi: 10.1101/gr.139618.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahsan B., Saito T.L., Hashimoto S., Muramatsu K., Tsuda M., Sasaki A., Matsushima K., Aigaki T., Morishita S. Nucleic Acids Res. 2009;37:D49–D53. doi: 10.1093/nar/gkn694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berezikov E., Robine N., Samsonova A., Westholm J.O., Naqvi A., Hung J.H., Okamura K., Dai Q., Bortolamiol-Becet D., Martin R., et al. Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res. 2011;21:201–215. doi: 10.1101/gr.116657.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wen J., Mohammed J., Bortolamiol-Becet D., Tsai H., Robine N., Westholm J.O., Ladewig E., Dai Q., Okamura K., Flynt A.S., et al. Diversity of miRNAs, siRNAs, and piRNAs across 25 Drosophila cell lines. Genome Res. 2014;24:1236–1250. doi: 10.1101/gr.161554.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonn S., Zinzen R.P., Girardot C., Gustafson E.H., Perez-Gonzalez A., Delhomme N., Ghavi-Helm Y., Wilczyński B., Riddell A., Furlong E.E. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat. Genet. 2012;44:148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- 36.Ciglar L., Girardot C., Wilczyński B., Braun M., Furlong E.E. Coordinated repression and activation of two transcriptional programs stabilizes cell fate during myogenesis. Development. 2014;141:2633–2643. doi: 10.1242/dev.101956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Junion G., Spivakov M., Girardot C., Braun M., Gustafson E.H., Birney E., Furlong E.E. A transcription factor collective defines cardiac cell fate and reflects lineage history. Cell. 2012;148:473–486. doi: 10.1016/j.cell.2012.01.030. [DOI] [PubMed] [Google Scholar]
- 38.Rembold M., Ciglar L., Yáñez-Cuna J.O., Zinzen R.P., Girardot C., Jain A., Welte M.A., Stark A., Leptin M., Furlong E.E. A conserved role for Snail as a potentiator of active transcription. Genes Dev. 2014;28:167–181. doi: 10.1101/gad.230953.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zinzen R.P., Girardot C., Gagneur J., Braun M., Furlong E.E. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature. 2009;463:65–70. doi: 10.1038/nature08531. [DOI] [PubMed] [Google Scholar]