Abstract
The Pathosystems Resource Integration Center (PATRIC) is the bacterial Bioinformatics Resource Center (https://www.patricbrc.org). Recent changes to PATRIC include a redesign of the web interface and some new services that provide users with a platform that takes them from raw reads to an integrated analysis experience. The redesigned interface allows researchers direct access to tools and data, and the emphasis has changed to user-created genome-groups, with detailed summaries and views of the data that researchers have selected. Perhaps the biggest change has been the enhanced capability for researchers to analyze their private data and compare it to the available public data. Researchers can assemble their raw sequence reads and annotate the contigs using RASTtk. PATRIC also provides services for RNA-Seq, variation, model reconstruction and differential expression analysis, all delivered through an updated private workspace. Private data can be compared by ‘virtual integration’ to any of PATRIC's public data. The number of genomes available for comparison in PATRIC has expanded to over 80 000, with a special emphasis on genomes with antimicrobial resistance data. PATRIC uses this data to improve both subsystem annotation and k-mer classification, and tags new genomes as having signatures that indicate susceptibility or resistance to specific antibiotics.
INTRODUCTION
The Pathosystems Resource Integration Center (PATRIC) is the all-bacterial Bioinformatics Resource Center (BRC) (https://www.patricbrc.org) (1). Established by the National Institute of Allergy and Infectious Diseases (NIAID), PATRIC provides researchers with an online resource that stores and integrates a variety of data types—e.g. genomics, transcriptomics, protein–protein interactions, 3D protein structures and sequence typing data, and associated metadata. The PATRIC website is primarily organism-centric, with various levels of genomic data and associated information related to each included organism. While the PATRIC homepage highlights the 22 NIAID Category A–C genera for easy access to data associated with many pathogenic species, PATRIC's compilation and organization of all relevant data for all available sequenced bacteria are standardized according to bacterial (NCBI) taxonomy (2), with options for viewing sets of genomes within the hierarchical bacterial tree.
With an emphasis on consistency in comparative genomic analysis, PATRIC has standardized annotations for over 80 000 bacterial genomes (as of August 2016) using the Rapid Annotation using Subsystems Technology toolkit (RASTtk) (3,4). This number includes more than 6000 genomes from antibiotic resistance studies that whose reads were obtained from the Sequence Read Archive (SRA) (5) and then assembled, annotated and made public in PATRIC. It also includes nearly 500 genomes submitted to PATRIC by the United States Agricultural Department (USDA) that were part of their pathogen monitoring process. In addition to the RASTtk-based annotations, PATRIC preserves and provides the legacy annotations from GenBank (6) and RefSeq (7), allowing researchers to contrast differences between the two. Summaries of the different data types (e.g. genes, RNAs, etc.) from both PATRIC and RefSeq annotations are available across different taxonomic levels, and are also provided for both genomes and for individual genes.
WHAT'S NEW IN PATRIC?
PATRIC has made significant improvements since it was last described in Nucleic Acids Research (NAR) (1). The web interface has been redesigned to: accommodate the increase in genomic data volume; improve the search capability; enhance the workspace; and provide a ‘group-centric’ view of genomes where researchers can create collections of genomes and then see all the information and data on those groups summarized together, apart from the larger collection of public data. With the increased concern over antimicrobial resistance (AMR), annotation efforts have focused on AMR genes, including collecting and curating genomes with AMR metadata. Perhaps the most significant improvement to the resource is a series of new services that allow our users to import their raw data and analyze it within the privacy of their own workspace. A ‘virtual integration’ of the data is provided, which allows researchers to keep their data private, and compare it across any of the public data available in PATRIC. The improvements to the PATRIC resource are described below.
New website design
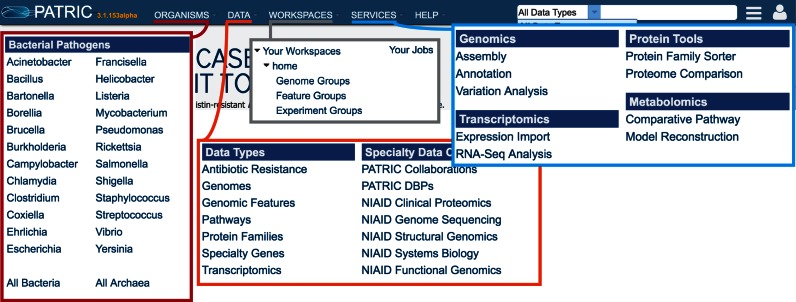
Since its original design, the PATRIC website has grown substantially in terms of complexity, and the amount of data has increased by more than two orders of magnitude. As a result, issues with data searches, information portrayal and website navigation have arisen. To meet this need, the website has been redesigned to enhance the user experience. All data types are now indexed with SOLR and interfaced with the PATRIC Data API in order to achieve the user interface goals of providing access to large, complex datasets, both public and private. This access allows for searching, displaying, filtering and downloading genomes, genomic feature, specialty genes, protein families, pathways and experiment data. Easier access to tools, services and the workspace are now provided on the PATRIC home page (Figure 1). In addition, integrated views of user-selected groups have been added, this being directly driven by feedback from researchers using the resource.
Figure 1.
New layout of the PATRIC homepage (https://www.patricbrc.org) with easy access to data, tools and the private user workspace.
New services
Since 2013, PATRIC has expanded and improved its research capabilities for users by building and incorporating a set of services that are designed to streamline and simplify common bioinformatic workflows. These services include genome assembly, genome annotation, RNA-Seq analysis, expression import, proteome comparison, metabolic model reconstruction and variation analysis. All of these services are available through a private workspace that allows users to compare their private data to the public data in the PATRIC database. Researchers can now upload their data files (e.g. sequence reads or assembled genomes), run the desired analysis services, integrate their private data with the data in PATRIC for comparative analysis, store the resulting output files and download the results.
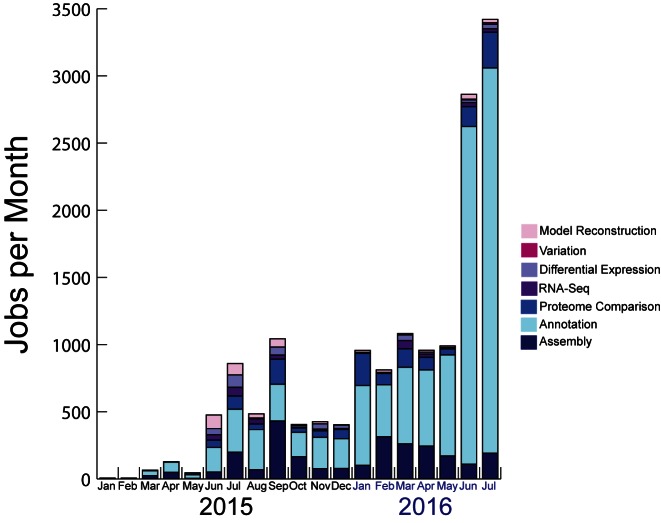
The PATRIC team has focused considerable effort on making the set of services rapid and easy to use, and as a result, they have been gaining in popularity (Figure 2). In 2015, a total of 4987 jobs were processed, and to date in 2016, 20 635 jobs have been submitted. Currently, the most popular services are annotation and assembly with a total of 10 372 and 2620 total jobs run to date respectively. Use of our available services, as measured by the number of registered accounts, is also increasing. By the end of 2015, there were 4023 registered PATRIC users, and to date in 2016 we have received 1318 new registered users. The PATRIC services are organized under the services tab on the PATRIC homepage, and the workspace is now accessed as a tab on the top of the page, and also at https://www.patricbrc.org/login.
Figure 2.
The growth in the number of jobs submitted and processed by registered users at PATRIC since the new assembly, annotation, metabolic modeling, proteome comparison, expression import and RNA-Seq services were implemented in 2015. The variation service was a recent addition in May 2016.
Genome assembly
A variety of command-line programs exist for assembling sequence reads into contigs, but these often require programming skills and considerable expertise to use effectively (8). To simplify this task, PATRIC integrates the Assembly RAST (ARAST) service to enable researchers to assemble short reads that are single or paired-end (typically from Illumina), and also long reads that include bax.h5 read files and filtered FASTQ files from PacBio (9) and Oxford Nanopore (10,11). PATRIC provides several assembly strategies (Figure 3) that include algorithms for base calling correction [BayesHammer (12) and Kmergenie (13)], and assemblers [Velvet (14), IDBA (15), Spades (16), MEGAHIT (17) and MiniASM (18)]. This is followed by scoring and two assembly evaluation frameworks, ALE (19) and a custom ARAST evaluation procedure. Finally, QUAST (20,21)—run at the end of all recipes to evaluate the contig sets on various metrics as well as give a summary comparing the resulting assemblies—is used for manual and visual inspection. The assembly results are downloadable from the workspace and, along with the contigs, they include a report that helps users choose which assembly they prefer for submission to the annotation service, or for further processing elsewhere. The Genome Assembly service can be through the Services tab, or directly at https://www.patricbrc.org/app/Assembly.
Figure 3.
The various assembly strategies now offered at PATRIC.
Genome annotation
In order to enable comparative analyses, all of the genomes in PATRIC are consistently annotated using a customized version of the RAST tool kit (RASTtk) (4). This allows users to compare proteins and other genomic features across the entire collection using a consistent vocabulary. Likewise, when a user submits a genome to the PATRIC Genome Annotation service, it is processed identically so that comparisons can be made between any private genome and all of the public genomes. In addition to the RASTtk analysis steps [described previously (4)] several custom annotation steps have been added to the PATRIC Annotation service. For instance, a BLAST-based comparison (22) is performed with several externally curated ‘Specialty Gene’ databases, including the Antibiotic Resistance Database (ARDB) (23) and the Comprehensive Antibiotic Resistance Database (CARD) (24) for antibiotic resistance proteins. VICTORS, the Virulence Factor Database (VFDB) (25) and a separate collection of PATRIC virulence factors (26) are used to identify virulence factors. The Therapeutic Drug Targets Database (27) and DrugBank (28) are used for drug targets, and the human genome for human homologs (Genome Reference Consortium Human Build 38 at https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.26). These results are displayed under the ‘Specialty Genes’ tab for a given genome, and users can filter the output by database, BLAST threshold and the taxonomy of the corresponding match. PATRIC also assigns protein family membership to protein encoding genes in order to drive its comparative analysis tools and metabolic pathway information, which can be viewed on the KEGG map (29) under the ‘Pathways’. Finally, for select species, a machine learning based prediction of antibiotic susceptibility or resistance (described below), which can be viewed on the landing page for the annotated genome (30), is performed. The Genome Annotation service can be accessed through the Services tab, or directly at https://www.patricbrc.org/app/Annotation.
Proteome comparison
PATRIC provides a Proteome Comparison service that can be used to compare up to nine genomes to a reference in a bidirectional best BLASTP analysis (22). Originally built for the RAST website (3), the improved service adds new functionality for PATRIC. Researchers can now use any public or private genome as a reference or a comparison genome. In addition, they can upload genomes with different annotations, or specific sets of proteins. This enables a search for homologs that is not limited to annotations, and also allows examination of specific groups. Other updates include a high-resolution downloadable circular diagram with direct links between it and the and underlying gene pages. Researchers can now filter on the BLAST E-value and the minimum percentage sequence coverage and identity, the results of all being available in the downloadable file for each genome in the comparison. The Proteome Comparison service can be accessed through the Services tab, or directly at https://www.patricbrc.org/app/SeqComparison.
RNA-Seq analysis
PATRIC has improved its RNA-Seq Analysis service, now giving researchers two ways to examine their data. The new service, which can analyze sets of single or paired-end reads, allows users to choose between two workflows for processing RNA-Seq data: Rockhopper (31) or Tuxedo (32). Tuxedo, based on the Tuxedo tool suite (32), is similar to Pathogen Portal's former service, the RNA Rocket (33). The new service provides SAM/BAM output for alignment, tab-delimited files profiling expression levels, and differential expression test results between conditions. The SAM/BAM files can be uploaded and displayed on the genome browser for the reference strain, and the differential expression file can be displayed in the heatmap and gene expression filtering tools that are part of PATRIC's transcriptomic analysis suite by using the Differential Expression Import tool. The RNA-Seq Analysis service can be accessed through the Services tab, or directly at https://www.patricbrc.org/app/Rnaseq.
Expression import
As part of the workspace upgrade, PATRIC has migrated the functionality to upload and transform expression data to an updated Expression Import service. This service allows users to upload differential expression data into their private workspace and then compare it with other expression data available in PATRIC. The service supports gene expression, protein expression, and phenotype array data in the form of log ratios generated by comparing samples, conditions or time points. The Expression Import service can be accessed through the Services tab, or directly at https://www.patricbrc.org/app/Expression.
Variation analysis
The new Variation Analysis service can be used to identify and annotate sequence variations. The service enables users to upload one or multiple short read files and align them with a closely related reference genome. For each sample, the service computes the variations against the reference and presents a detailed list of Single Nucleotide Polymorphisms (SNPs), Multiple Nucleotide Polymorphism (MNPs), insertions, and deletions with confidence scores and effects such as ‘synonymous mutation’ and ‘frameshift.’ It includes a choice of five different aligners and two different SNP callers. The aligners include BWA-mem and BWA-mem-strict (34), Bowtie2 (35,36), MOSAIK (37) and LAST (38). FreeBayes (39) and SAMtools (40) are the two SNP callers. High confidence variations are downloadable in the standard VCF (Variant Calling Format) augmented by SNP annotation. A summary table illustrating how the variations are shared across the samples is also available. The Variation service can be accessed through the Services tab, or directly at https://www.patricbrc.org/app/Variation.
Model reconstruction
With the release of the Model Reconstruction service, users can construct their own metabolic model for any genome in PATRIC. The service includes support for model gap-filling, Flux Balance Analysis (FBA), essential gene prediction and export of models in SBML format. The Model Reconstruction service leverages capabilities of the ModelSEED (41) and is available in PATRIC both from the Services tab at the top of any page and at https://www.patricbrc.org/app/Reconstruct.
Data enhancements
Genomes and antimicrobial resistance data (AMR)
PATRIC imports genomes from GenBank (6) on a monthly basis, and the collection of genomes has been growing at a near exponential rate for several years (Figure 4). As genomes have become plentiful, it has become increasingly apparent that the value of adding a new genome to the database is linked to what is known about that genome, or its metadata. With each genome that is brought into PATRIC, the metadata from the GenBank file is parsed and added to the PATRIC database, where it can be browsed on the website.
Figure 4.

The number of genomes available in PATRIC have an almost exponential growth since the resource began in 2004.
With sufficient genomes and metadata it is possible to use statistical and machine learning-based approaches to generate hypotheses. For instance, the genomes in PATRIC and their AMR metadata (susceptibility or resistance to a given antibiotic) have been used to build machine learning-based classifiers in order to predict the regions in the genome associated with AMR. When a genome is submitted to the annotation service, these classifiers are also used to predict if the organism is susceptible or resistant to an antibiotic. To date, this analysis has been performed on Acinetobacter, Mycobacterium, Streptococcus and Staphylococcus (30) genomes in PATRIC.
Since the submission of metadata with a genome is almost entirely voluntary, and many researches have been electing to submit their genomes to SRA (5) or the European Nucleotide Archive (ENA) (42) instead of assembling and annotating them, we have been searching the literature for publications with large-scale AMR studies and assembling those genomes in order to bolster the AMR prediction effort. To date we have assembled approximately 8500 genomes with AMR metadata from SRA and ENA, which have been added to PATRIC. The entire set of genomes with AMR metadata is shown in Table 1.
Table 1. Genomes with AMR metadata that are available in PATRIC.
| Genomes with AMR metadata | Genomes with MIC dataa | Genomes with SIR Datab | Antibiotics testedc | |
|---|---|---|---|---|
| Achromobacter | 1 | 1 | 0 | 18 |
| Acinetobacter | 506 | 95 | 505 | 31 |
| Aerococcus | 1 | 1 | 0 | 18 |
| Citrobacter | 6 | 6 | 2 | 25 |
| Corynebacterium | 1 | 1 | 0 | 14 |
| Enterobacter | 69 | 66 | 14 | 31 |
| Enterococcus | 9 | 0 | 9 | 8 |
| Escherichia | 51 | 47 | 26 | 29 |
| Hafnia | 1 | 1 | 0 | 9 |
| Klebsiella | 343 | 270 | 160 | 31 |
| Leclercia | 1 | 1 | 0 | 18 |
| Lelliottia | 1 | 0 | 1 | 16 |
| Mycobacterium | 5110 | 177 | 5110 | 19 |
| Neisseria | 491 | 491 | 450 | 8 |
| Pseudomonas | 519 | 394 | 516 | 38 |
| Serratia | 4 | 4 | 3 | 16 |
| Staphylococcus | 2150 | 85 | 2150 | 27 |
| Streptococcus | 4439 | 1245 | 3553 | 30 |
aGenomes with minimum inhibitory concentration (MIC) data include data from a variety of testing methods such as Kirby–Bauer disc diffusion, E-test, BD Phoenix Automated Microbiology System, etc.
bGenomes where phenotypes have been determined as being susceptible, intermediate or resistant (SIR) to a given antibiotic. In these cases, the determination was made by the authors of the study.
cThe list of antibiotics may include data for entire classes of antibiotics, such as beta-lactams or carbapenems.
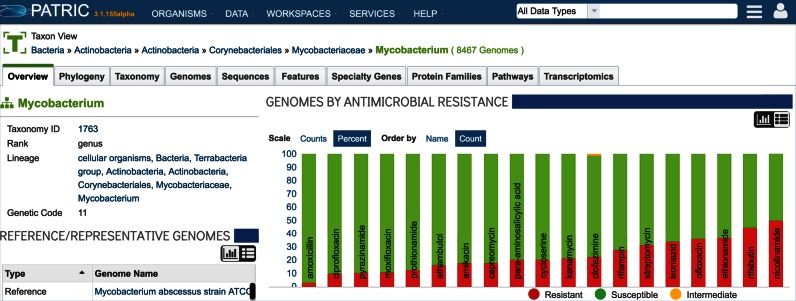
The antimicrobial resistance metadata is displayed on the PATRIC website on the Genome list pages (Figure 5). Currently researches can sort on the terms ‘Susceptible,’ ‘Resistant’ or ‘Intermediate’ to locate the genomes tagged with that information. In addition, researchers can filter on the source of the information via the ‘Antimicrobial Evidence’ field, which provides the ability, for example, to only include genomes that have AMR metadata from panel information versus metadata that has been parsed from the genome record. The entire collection of AMR metadata for the PATRIC genomes can be downloaded from the FTP site (ftp://ftp.patricbrc.org/patric2/current_release/RELEASE_NOTES/).
Figure 5.
Antimicrobial Resistance Data summaries are now available on organism landing pages at PATRIC. This example shows the current data available for Mycobacterium genomes.
In addition to seeking out published genomic studies that provide AMR information, PATRIC has undertaken a large AMR annotation effort with the goal of providing PATRIC users with the ability to rapidly recognize and project known AMR-related genome features to new genomes in an automated fashion, and to use comparative genomics approaches for the discovery of new resistance mechanisms. Curation thus far has focused on encoding the following mechanisms of drug resistance: outer membrane porins, multidrug efflux mechanisms, and capsular and extracellular polysaccharides. To date, 25 subsystems covering 265 unique functional roles relating to antimicrobial resistance have been encoded. These subsystems are used by the annotation system to guide the projection of these specific functional roles.
Protein families
In order to provide a rich comparative analysis environment, PATRIC maintains an up-to-date set of protein families covering the entire collection of genomes. These protein families provide the necessary data for many of the PATRIC tools, including the protein family heatmap analysis tool. Early in the project, when there were fewer genomes, we generated protein families using BLAST (22) and OrthoMCL (43). As the number of genomes increased and it became infeasible to compute alignment-based protein families, we began maintaining protein families based on FIGfams (44), which are used by the ‘classic’ version of the RAST annotation system (3,45). Using FIGfam-based protein families in PATRIC has two main drawbacks. First, they are generated from the PubSEED database, which contains ∼12 000 genomes (45,46). This meant that most of the proteins in PATRIC were being assigned to a family by projection, and many proteins lacked an assignment. Second, since FIGfams were designed for projecting annotations across broad phylogenetic distances, they tended to be more inclusive than families built from alignment-based algorithms.
Last year, we developed a new algorithm for generating protein families that could be applied to the entire collection of PATRIC genomes (47). The method works by using the annotation process to guide family formation (4). For each set of proteins within a genus that have the same annotation, the ‘signature’ amino acid k-mers that were used in the annotation process are used to create a distance matrix for the members of the set. The set is then clustered using a Markov Cluster Algorithm (MCL) (21). For proteins with no annotation, we cluster using BLAST similarity (22). Then in order to create global families that span multiple genera, a merger of local families is performed with the same annotation, using a more relaxed inclusion criterion. This new collection of protein families is called ‘PATtyFams’.
Overall, the PATtyFam algorithm is rapid and generates protein families resembling those created by alignment-based algorithms (47). When a genome is submitted to the PATRIC annotation service, protein families are automatically assigned to each protein by projection, thus enabling a user to compare their private genome with the PATRIC collection. Both the local genus-level families (prefixed ‘PLF’ for PATRIC local family) and the global families (prefixed ‘PGF’ for PATRIC global family) are displayed on the landing page for each protein. As the genome collection increases, we periodically regenerate the protein families for the entire PATRIC database. PATRIC's current set of PATtyFams consists of 281 million proteins, forming 11.4 million genus-level families and 8.3 million global families.
FTP site
As the number of genomes increases, the ability to download the associated data becomes more difficult. To meet the growing need to allow our users easy access to all the data that is associated with these large numbers of genomes, the PATRIC FTP site—available from ftp://ftp.patricbrc.org/patric2—has been completely reorganized and updated. It now contains downloadable, updated files for all genomes genome annotations, and AMR metadata, as well as protein-protein interaction and other data.
FUTURE DIRECTIONS
As part of the comparative analysis process, PATRIC provides legacy annotations from RefSeq and GenBank. In order to enhance this experience and keep the data current, PATRIC will update assembly versions from GenBank and incorporate them into PATRIC on a monthly basis. In addition, annotations for the RefSeq and GenBank genomes will be refreshed at least annually, providing researchers with the ability to compare these with annotations from RASTtk.
Future development and services at PATRIC will be directed on three fronts. First, PATRIC will increase efforts aimed at making data and services more useful for those performing clinical research. These efforts will move the PATRIC system closer to the translational research that occurs in a clinical setting by providing support that creates services and views on PATRIC data that would be useful to clinicians working in a research setting.
PATRIC will also increase its focus on modeling in the upcoming years. With genome sequencing now a common practice, more advanced computational approaches that make use of the increasing numbers of genomes, associated metadata, and multi-omics data integration become an important part of understanding complex correlation. Support for generating, hosting and comparing models will emerge as PATRIC continues to develop.
Finally, future activities will increase focus on support in the area of therapeutics. As the pathogen metabolic modeling community matures, opportunities to apply new computational concepts to target identification will develop. Advances in the construction of metabolic models and flux balance analysis, compiling essential genes and simulating the effects of double knockouts across a wide range of pathogens can be used to form new reference data sets, which will reveal new targets for experimental studies involving approaches from phage therapy to small molecule discovery, which can lead to novel therapeutics.
Footnotes
Present address: Alice R. Wattam, Biocomplexity Institute, Virginia Tech University, Blacksburg, VA 24061-0477, USA.
FUNDING
PATRIC has been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services [HHSN272201400027C]. Funding for open access charge: Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services [HHSN272201400027C].
Conflict of interest statement. None declared.
REFERENCES
- 1.Wattam A.R., Abraham D., Dalay O., Disz T.L., Driscoll T., Gabbard J.L., Gillespie J.J., Gough R., Hix D., Kenyon R., et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014;42:D581–D591. doi: 10.1093/nar/gkt1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Federhen S. Type material in the NCBI Taxonomy Database. Nucleic Acids Res. 2015;43:D1086–D1098. doi: 10.1093/nar/gku1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A., Formsma K., Gerdes S., Glass E.M., Kubal M. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:1. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brettin T., Davis J.J., Disz T., Edwards R.A., Gerdes S., Olsen G.J., Olson R., Overbeek R., Parrello B., Pusch G.D., et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015;5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kodama Y., Shumway M., Leinonen R. The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res. 2012;40:D54–D56. doi: 10.1093/nar/gkr854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W. GenBank. Nucleic Acids Res. 2016;44:D67–D72. doi: 10.1093/nar/gkv1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D., et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker M. De novo genome assembly: what every biologist should know. Nat. Methods. 2012;9:333. [Google Scholar]
- 9.Rhoads A., Au K.F. PacBio sequencing and its applications. Genomics Proteomics Bioinformatics. 2015;13:278–289. doi: 10.1016/j.gpb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Branton D., Deamer D.W., Marziali A., Bayley H., Benner S.A., Butler T., Di Ventra M., Garaj S., Hibbs A., Huang X. The potential and challenges of nanopore sequencing. Nat. Biotechnol. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laver T., Harrison J., O'Neill P., Moore K., Farbos A., Paszkiewicz K., Studholme D.J. Assessing the performance of the Oxford Nanopore Technologies MinION. Biomol. Detect. Quantif. 2015;3:1–8. doi: 10.1016/j.bdq.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nikolenko S.I., Korobeynikov A.I., Alekseyev M.A. BayesHammer: Bayesian clustering for error correction in single-cell sequencing. BMC Genomics. 2013;14:1. doi: 10.1186/1471-2164-14-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Namiki T., Hachiya T., Tanaka H., Sakakibara Y. MetaVelvet: an extension of Velvet assembler to de novo metagenome assembly from short sequence reads. Nucleic Acids Res. 2012;40:e155. doi: 10.1093/nar/gks678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng Y., Leung H.C., Yiu S.-M., Chin F.Y. Research in Computational Molecular Biology. Lisbon: Proceedings of the 14th Annual International Conference; 2010. IDBA-A practical iterative de Bruijn graph de novo assembler; pp. 426–444. [Google Scholar]
- 16.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li D., Liu C.-M., Luo R., Sadakane K., Lam T.-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 18.Li H. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics. 2015;15:2103–2110. doi: 10.1093/bioinformatics/btw152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark S., Egan R., Frazier P.I., Wang Z. ALE: a generic assembly likelihood evaluation framework for assessing the accuracy of genome and metagenome assemblies. Bioinformatics. 2013;29:435–443. doi: 10.1093/bioinformatics/bts723. [DOI] [PubMed] [Google Scholar]
- 20.Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Dongren S. Graph Clustering by Flow Simulation. Utrecht: University of Utrecht; 2001. [Google Scholar]
- 22.Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., Madden T.L. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu B., Pop M. ARDB—antibiotic resistance genes database. Nucleic Acids Res. 2009;37:D443–D447. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McArthur A.G., Waglechner N., Nizam F., Yan A., Azad M.A., Baylay A.J., Bhullar K., Canova M.J., De Pascale G., Ejim L. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013;57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen L, Zheng D, Liu B, Yang J, Jin Q. VFDB 2016: hierarchical and refined dataset for big data analysis–10 years on. Nucleic Acids Res. 2016;44:D694–D697. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mao C., Abraham D., Wattam A.R., Wilson M.J., Shukla M., Yoo H.S., Sobral B.W. Curation, integration and visualization of bacterial virulence factors in PATRIC. Bioinformatics. 2015;31:252–258. doi: 10.1093/bioinformatics/btu631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang H., Qin C., Li Y.H., Tao L., Zhou J., Yu C.Y., Xu F., Chen Z., Zhu F., Chen Y.Z. Therapeutic target database update 2016: enriched resource for bench to clinical drug target and targeted pathway information. Nucleic Acids Res. 2016;44:D1069–D1074. doi: 10.1093/nar/gkv1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Law V., Knox C., Djoumbou Y., Jewison T., Guo A.C., Liu Y., Maciejewski A., Arndt D., Wilson M., Neveu V., et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 2014;42:D1091–D1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis J.J., Boisvert S., Brettin T., Kenyon R.W., Mao C., Olson R., Overbeek R., Santerre J., Shukla M., Wattam A.R., et al. Antimicrobial resistance prediction in PATRIC and RAST. Sci. Rep. 2016;6:27930. doi: 10.1038/srep27930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McClure R., Balasubramanian D., Sun Y., Bobrovskyy M., Sumby P., Genco C.A., Vanderpool C.K., Tjaden B. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 2013;41:e140. doi: 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Warren A.S., Aurrecoechea C., Brunk B., Desai P., Emrich S., Giraldo-Calderon G.I., Harb O., Hix D., Lawson D., Machi D., et al. RNA-Rocket: an RNA-Seq analysis resource for infectious disease research. Bioinformatics. 2015;31:1496–1498. doi: 10.1093/bioinformatics/btv002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee W.P., Stromberg M.P., Ward A., Stewart C., Garrison E.P., Marth G.T. MOSAIK: a hash-based algorithm for accurate next-generation sequencing short-read mapping. PLoS One. 2014;9:e90581. doi: 10.1371/journal.pone.0090581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frith M.C., Wan R., Horton P. Incorporating sequence quality data into alignment improves DNA read mapping. Nucleic Acids Res. 2010;38:e100. doi: 10.1093/nar/gkq010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garrison E., Marth G. Haplotype-based variant detection from short-read sequencing. arXiv preprint. 2012 arXiv:1207.3907. [Google Scholar]
- 40.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henry C.S., DeJongh M., Best A.A., Frybarger P.M., Linsay B., Stevens R.L. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat. Biotechnol. 2010;28:977–982. doi: 10.1038/nbt.1672. [DOI] [PubMed] [Google Scholar]
- 42.Leinonen R., Akhtar R., Birney E., Bower L., Cerdeno-Tarraga A., Cheng Y., Cleland I., Faruque N., Goodgame N., Gibson R., et al. The European Nucleotide Archive. Nucleic Acids Res. 2011;39:D28–D31. doi: 10.1093/nar/gkq967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen F., Mackey A.J., Stoeckert C.J., Roos D.S. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meyer F., Overbeek R., Rodriguez A. FIGfams: yet another set of protein families. Nucleic Acids Res. 2009;37:6643–6654. doi: 10.1093/nar/gkp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Overbeek R., Olson R., Pusch G.D., Olsen G.J., Davis J.J., Disz T., Edwards R.A., Gerdes S., Parrello B., Shukla M., et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014;42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Overbeek R., Begley T., Butler R.M., Choudhuri J.V., Chuang H.-Y., Cohoon M., de Crécy-Lagard V., Diaz N., Disz T., Edwards R. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davis J.J., Gerdes S., Olsen G.J., Olson R., Pusch G.D., Shukla M., Vonstein V., Wattam A.R., Yoo H. PATtyFams: protein families for the microbial genomes in the PATRIC Database. Front. Microbiol. 2016;7:118. doi: 10.3389/fmicb.2016.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]