Abstract
Viruses represent the most abundant life forms on the planet. Recent experimental and computational improvements have led to a dramatic increase in the number of viral genome sequences identified primarily from metagenomic samples. As a result of the expanding catalog of metagenomic viral sequences, there exists a need for a comprehensive computational platform integrating all these sequences with associated metadata and analytical tools. Here we present IMG/VR (https://img.jgi.doe.gov/vr/), the largest publicly available database of 3908 isolate reference DNA viruses with 264 413 computationally identified viral contigs from >6000 ecologically diverse metagenomic samples. Approximately half of the viral contigs are grouped into genetically distinct quasi-species clusters. Microbial hosts are predicted for 20 000 viral sequences, revealing nine microbial phyla previously unreported to be infected by viruses. Viral sequences can be queried using a variety of associated metadata, including habitat type and geographic location of the samples, or taxonomic classification according to hallmark viral genes. IMG/VR has a user-friendly interface that allows users to interrogate all integrated data and interact by comparing with external sequences, thus serving as an essential resource in the viral genomics community.
INTRODUCTION
Viruses are key players in nature able to infect organisms from the three domains of life and found across all known ecological niches (1) therefore affecting biogeochemical cycles and ecosystem dynamics (1–5). However, due to limitations primarily related to identifying and culturing them, the detection of environmental viruses remained very limited until the advent of metagenomic approaches (6). Since then, a number of environmental viromes have been scrutinized providing a broader view of the diversity and distribution of viruses (7–13). Unfortunately, this information usually remains scattered across different repositories -such as general data repository databases (e.g. GenBank (14) or EMBL (15)), or virus-specific databases (e.g. virus pathogen resource (16)), recombinant virus database (17), and hepatitis B database (18)). Furthermore, metadata such as isolation source or habitat where the virus was originally identified, or information about its putative host, often remains elusive or not available in several of these databases. More recent works are making a great progress towards an effort to provide a centralized resource for viral data and associated tools (19). However, despite the excellent existing resources, we still lack a data management and visualization environment integrating viral genes, genomes, clusters, functions, associated host and habitat with analytical tools that would enable large-scale comparative analysis of the global virome.
In order to alleviate some of the existing resource limitations, and enable the community to access and analyze an expanded version of the recently emerging viral genomics data we have developed IMG/VR, an integrated viral analysis system, within the Integrated Microbial Genomes with Microbiome samples (IMG/M) data management system (20).
IMG/VR provides the largest integration of viral sequences with associated metadata and allows users to explore these data to decipher biogeographical and habitat distribution patterns of viral species as well as traveling across all the identified hosts putatively infected with viral sequences. In addition, users can compare and analyze their sequences against IMG/VR's data (including viral protein family models, viral cluster and singleton information, distribution patterns of similar viral sequences across the globe, percent of known and unknown genes per sequence, and information regarding viral taxonomy and putative viral-host(s)), integrated with a variety of analytical tools.
We anticipate that IMG/VR will become a reference resource for sequence analysis of viral genomes and viral contigs derived from metagenomic samples.
RESULTS
IMG/VR is a data management resource for visualization and analysis of viral sequences integrated with associated metadata within the IMG/M system (20). IMG/VR provides a unique integration of viral sequences with associated metadata including connection to putative hosts, and habitat types.
Data integrated into IMG/VR
Viral sequences
The IMG/VR system is an integrated resource for viral data management and associated metadata within the IMG/M system (20). In its first public release, IMG/VR contains a total of 268 320 viral sequences from both isolate viral genomes (iVGs) and metagenomic viral contigs (mVCs). The 264 413 mVCs currently provided by the system were obtained from 2981 metagenomic samples (out of a list of over 6000 total samples screened) from geographically and ecologically diverse habitats according to the Genomes OnLine Database (GOLD) classification system (21,22).
mVCs were identified using a computational approach described in Paez-Espino et al. (11). Briefly, a set of over 25 thousand viral protein families (VPFs) was constructed from manually identified mVCs and isolate viral genomes of dsDNA viruses and retroviruses available at NCBI (as of April 2015). This set of VPFs (accession link in Supplementary data) was used as bait for identifying viral sequences from assembled metagenomic contigs longer than 5 kb. In approximately a quarter of all mVCs the total gene coverage per contig by VPFs was very high (at least 70%) although, interestingly, in another quarter (representing ∼60 000 mVCs) the coverage was under 35%, indicating that a great volume of the viral gene content still remains unknown. In total, the 264 413 mVCs encode 6.1 million proteins, most of which (94.9%) had no hits to genes of known function at the time of the annotation.
Viral sequence grouping
All viral sequences in IMG/VR are grouped into clusters of related sequences, ranging from 1 to 349 members per group. 122 665 sequences (46% of total) belong to single member clusters or singletons (represented with a ‘sg_’ prefix and a numeric identifier), while the remaining 145 655 sequences (143 532 mVCs and 2123 iVGs) were grouped into 39 701 viral clusters (represented with a ‘vc_’ prefix and a numeric identifier) of two members or more. From those, most groups (52%) have only two members while 4.5% have 10 or more members.
This clustering approach employed in IMG/VR has been modified from the method previously used in Paez-Espino et al. (11), which relied on both amino acid identity and total alignment fraction for pairwise comparison of viral sequences, by a more scalable method based on nucleotide sequence identity (23,24). The stringent thresholds used (90% nucleotide sequence identity over 75% of the sequence length) made it possible to recreate the viral groups generated in Paez-Espino et al., recapitulating the species-level grouping for 87% of viral clusters, and with the remainder grouping at genus level.
Host–virus identification
Traditionally, viruses infecting Bacteria or Archaea (i.e. phages) have been isolated from the host they have been infecting, and therefore the host–virus relation was delineated upfront (25). With the advent of metagenomics however, there is an increasing number of identification of viral sequences from environmental samples, for which the identification of a putative host is not as straight as it was for the isolate viruses. A number of computational methods have been proposed to bypass this limitation (11,26).
IMG/VR provides putative host information for 20 073 viral sequences (7.5% of all the viral sequences) using two computational approaches as previously described (11). The first approach is looking for viral clusters that contain isolate viral genomes with host information. Projecting the isolate viral-host information onto the cluster results in host assignment for 862 mVCs. The second approach depends on the CRISPR-Cas prokaryotic immune system, which retains viral fragments (proto-spacers) within microbial CRISPR arrays (27,28,29). Using this approach, 13 474 mVCs were assigned to putative hosts.
In total, genomes from 36 bacterial and archaeal phyla were linked to viral sequences (Table 1). A large number of these connections were previously unknown, including the identification of nine phyla (Atribacteria, Fervidibacteria, Armatimonadetes, Deferribacteres, Parcubacteria, Gemmatimonadetes, Ignavibacteria, Aminicenantes and Saccharibacteria) which were not previously reported to be infected by viruses in the NCBI RefSeq database or as prophages (30).
Table 1. Bacterial and archaeal host phyla with corresponding number of mVCs.
| Host phylum | Viral contig count | Host phylum | Viral contig count |
|---|---|---|---|
| Euryarchaeota | 91 | Armatimonadetes | 6 |
| Crenarchaeota | 41 | Caldiserica | 6 |
| Aigarchaeota | 1 | Tenericutes | 5 |
| Firmicutes | 2715 | c. Marinimicrobia | 4 |
| Proteobacteria | 2540 | c. Parcubacteria | 4 |
| Bacteroidetes | 1338 | Deferribacteres | 4 |
| Actinobacteria | 739 | Fibrobacteres | 4 |
| Fusobacteria | 435 | c. Fervidibacteria | 3 |
| Chloroflexi | 37 | Cloacimonetes | 3 |
| Cyanobacteria | 27 | Gemmatimonadetes | 3 |
| Deinococcus-Thermus | 23 | Synergistetes | 3 |
| Verrucomicrobia | 23 | Marinimicrobia | 2 |
| Nitrospirae | 18 | Acidobacteria | 1 |
| Chlorobi | 16 | c. Aminicenantes | 1 |
| Aquificae | 15 | c. Omnitrophica | 1 |
| Thermotogae | 15 | c. Saccharibacteria | 1 |
| Spirochaetes | 11 | Ignavibacteriae | 1 |
| c. Atribacteria | 9 | Planctomycetes | 1 |
Browsing iVGs and mVCs via viral datasets
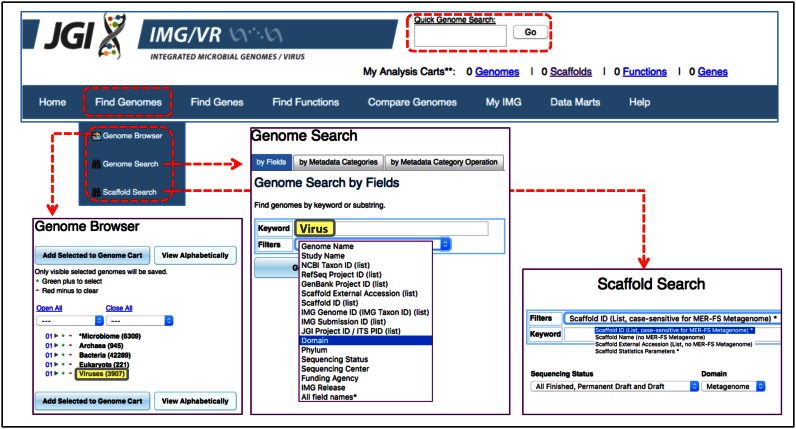
The search functionality in IMG/VR is similar to that in the IMG/M system (20). All isolate viral genomes (iVGs) can be accessed via ‘Quick Genome Search’ (by typing the virus name or taxon identifier (‘Taxon OID’)) or ‘Find Genomes’ tab (selecting viruses in ‘Genome Browser’ or ‘Genome Search’ tools) (Figure 1).
Figure 1.
General IMG/VR search functionality. Basic search tools from IMG/M's top menu bar (dashed red boxes) can be used to access the viral content of IMG/VR. ‘Quick Genome Search’ at the top menu can be used to query specific viral names, taxon identifiers or keywords. Alternatively, all isolate viral content can be retrieved from the ‘Find Genome’ tab, either selecting Viruses (boxed in grey with yellow background) from the ‘Genome browser’ display (bottom left panel) or searching for Virus (boxed in grey with yellow background) in the Domain filter of the ‘Genome Search’ tool (bottom central panel). To search for metagenomic viral contigs users need to access first the metagenomic sample (using any of the above tools). Then the ‘Scaffold Search’ tool can be used to select specific scaffolds (bottom right panel).
The predicted mVCs are stored as metagenome scaffolds and they remain under their corresponding metagenome datasets (i.e. metagenome ‘Taxon OID’). Thus, metagenome ‘Taxon OIDs’ can also be accessed the same way that any iVG and specific mVCs can be retrieved from the ‘Scaffold Search’ tool of the ‘Find Genomes’ tab (Figure 1).
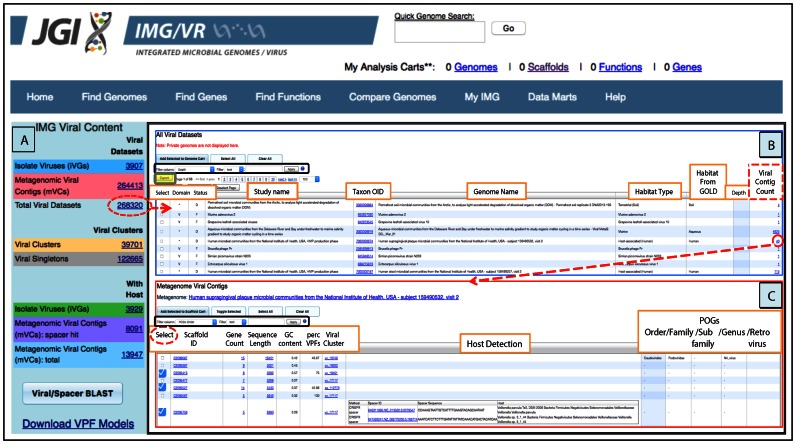
In order to further facilitate the identification and selection of viral sequences in IMG/VR, all iVGs and mVCs can be accessed from the left panel table (IMG Viral Content) available from the entry page (Home tab) (Figure 2A). This entry point enables browsing all viral datasets in the context of their associated samples and corresponding metadata, e.g. habitat type or depth of the metagenome sample from which a viral sequence was identified (Figure 2B). This table provides information about the total number of viral contigs per sample in IMG, allowing a quick identification of the samples with the largest number of viruses. Similar to other tables in IMG, the results can be exported in a tab-delimited text format compatible with a number of other tools for metagenomic analysis, as well as R and Microsoft Excel (Figure 2B). By clicking on the ‘Viral Contig Count’ number from the previous table, users can examine the list of viral contigs from individual samples (Figure 2C). The information displayed for a selected contig or group of contigs includes: scaffold identifier (Scaffold ID), gene count per contig (Gene Count), contig length (Sequence Length bp), guanine and cytosine content (GC Content), percent of genes per contig covered with viral protein families (Perc VPFs), viral species name identifier (Viral Cluster; detailed in ‘Sequence grouping’ section and Supplementary data), predicted host and method of prediction (Host Detection; detailed in ‘Host–virus identification’ section), taxonomic assignment at different levels based on clusters of orthologous genes of phages (POGs) (Supplementary data), and the putative retrovirus sequences (Supplementary data).
Figure 2.
Browsing IMG/VR viral datasets. (A) Total counts and access to the list of viral sequences from isolate viruses, metagenomic viral contigs, or their combination (dashed red oval). (B) Detailed table from ‘Total Viral Datasets’ link displaying study and sample name, Taxon OID, habitat information and number of metagenomic viral contigs. (C) List of viral metagenomic contigs found in a single sample. Columns in (B) and (C) can be sorted by clicking on the column header, and different filters can be used for specific searches (black boxes). Tables can be also exported in a tab-delimited text format by using the Export button (grey box with yellow background).
Browsing mVCs via environmental metadata
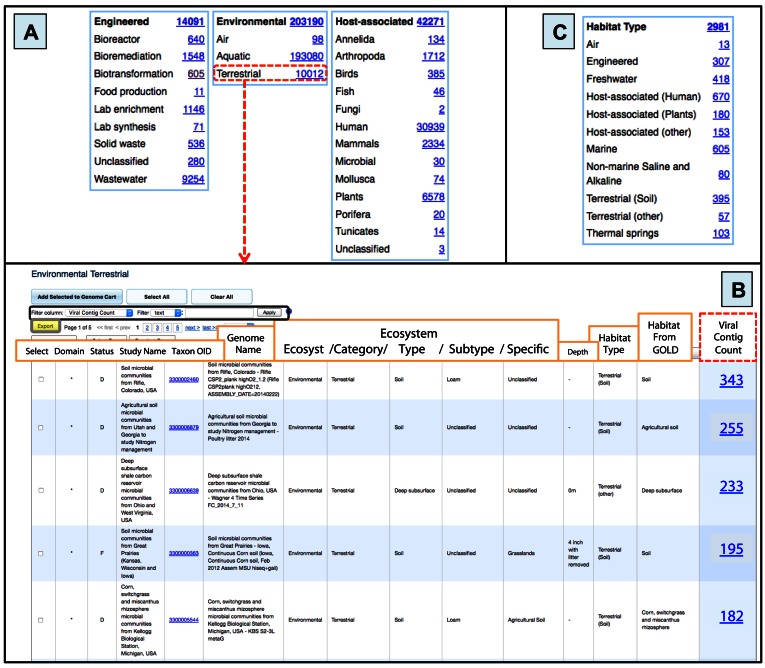
Metagenomic viral contigs can be viewed in relation to different environmental metadata associated with each sample. Two distinct curated environmental classifications systems are displayed at the bottom of the IMG/VR landing page, the ecosystem and the habitat type classification (11,21,22) (Figure 3).
Figure 3.
Accessing metagenomic viral contigs via associated environmental metadata. (A) Distribution of metagenomic viral contigs per ecosystem and ecosystem category information of samples according to GOLD classification. When a category is selected (e.g. Terrestrial samples—boxed in dashed red) a new table is displayed. (B) Detailed information of the selected Terrestrial samples. The total number of metagenomic viral contigs per sample (boxed in dashed red) can be viewed. Columns can be sorted by clicking on the column header, and different filters can be used for specific searches (black box). The table can be also exported in a tab-delimited text format by using the Export button (gray box with yellow background). (C) Number of mVCs per Habitat Type category of the sample where the mVCs were found.
The ecosystem classification is based on a previously developed five-tier hierarchical classification system (21). All metagenome data sets are organized in three main classes of the top ecosystem tier: engineered, environmental and host-associated; and then further divided into sub-tiers called ecosystem category, ecosystem type, ecosystem subtype and specific ecosystem (31) (Figure 3A). Currently, 78.3% of the mVCs belong to environmental samples, while 16.3% and 5.4% correspond to host-associated and engineered, respectively. Users can navigate through all samples at once or just reduce the search to any specific ecosystem class or category (i.e ‘Environmental Terrestrial’ Figure 3B), and from there, select particular types, subtypes or specific ecosystems.
The habitat type classification is based on 11 distinct manually curated habitat terms (e.g. air, freshwater, marine, host-associated human, host-associated plants, terrestrial soil) previously described (11). This classification allows the selection of mVCs from samples that belong to a single habitat type (Figure 3C).
Browsing mVCs via geographic location or human body site metadata
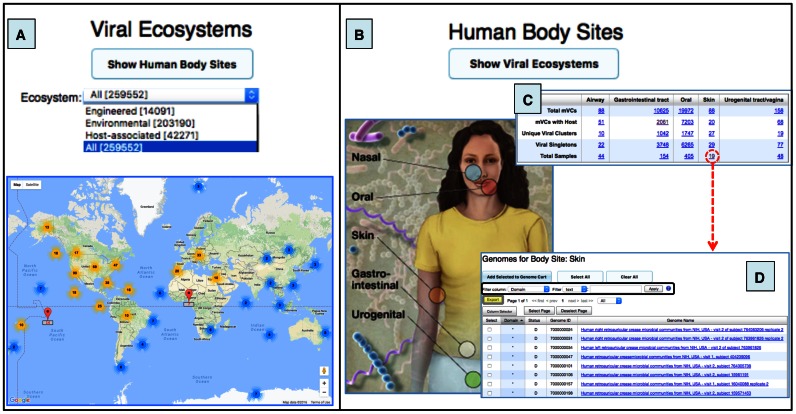
Viral contigs can be viewed based on the geographic coordinates of a corresponding sample. This functionality is available primarily for environmental metagenomes and allows the selection of samples with specific location via ‘Marker Clusterer for Google Maps’, a javascript API utility library that creates and manages per-zoom-level clusters for large amounts of markers. Ultimately, as users zoom in the map, a list of viral contigs that belong to a sample(s) can be retrieved -by clicking on a map pin- and selecting the count next to the metagenome of interest for that location (Figure 4A).
Figure 4.
Maps of samples containing viral contigs from environmental and human-associated metagenomes. (A) World interactive Google Map with a geographic location of metagenomic samples. All samples can be selected together or only those from any of the three major ecosystems. Map pins (in red) represent location counts of viral contigs and may contain multiple samples. Map pins are grouped into clusters and clusters themselves into larger clusters (bold number with a coloured halo based on number of members within the cluster) according to the Google Map javascript API utility library. As you zoom into any of the cluster locations, the number on the cluster decreases, and you begin to see the individual markers on the map. Zooming out of the map consolidates the markers into clusters again. (B) Human Body image showing the five body sites with available samples. All the metagenomic viral contigs identified in each body site can be accessed from the circles in the image. (C) Table provides information about mVC clusters/singletons, number of samples, and viral contigs with a host. (D) List of human skin samples with viral contigs. Columns can be sorted by clicking on the column header. Different filters can be used for specific searches (black box). This table can be exported by using the Export button (grey box with yellow background). Human body image credit: NIH Medical Arts and Printing.
Additionally, all viral contigs identified in samples from the human body can be displayed by clicking on the ‘Show Human Body Sites’ button (Figure 4B). This option allows access to viral contigs derived from samples of any of the five main human body sites (nose, mouth, skin, intestine, and vagina), together with general statistics of these viruses per body site (Figure 4C). From the default Human Body Sites summary table users can select all mVCs from a particular sample site or only those with a putative host (Figure 4D).
Browsing viral clusters and viral singletons
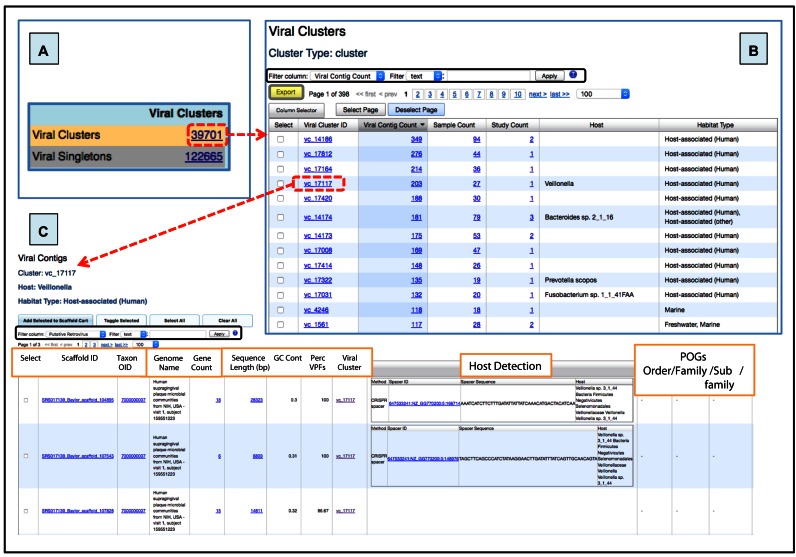
Viral clusters and singletons together represent the entire viral diversity within IMG/VR. A total of 39 701 viral clusters and 122 665 singletons are available from the left panel on IMG/VR's entry page (Figure 5A). Together, these represent 162 366 viral quasi-species identified numerically with the prefix ‘vc_’ or ‘sg_’ depending if they belong to a viral cluster or remain as a singleton.
Figure 5.
Viral diversity in IMG/VR. (A) Accession link with the number of viral clusters or singletons available in the system. (B) Detailed table from ‘Viral Clusters’ (boxed in dashed red from panel a) showing the number of metagenomic viral contigs per cluster. The number of distinct samples, and unique projects (‘Study Count’) is shown, besides information regarding host and habitat. (C) Viral cluster details table with host (via microbial CRISPR-spacer sequence matches) and taxonomic (via hallmark genes) information when available. Columns in (B) and (C) can be sorted by clicking on the column header, and different filters can be used for specific searches (black boxes). Tables can be also exported in a tab-delimited text format by using the Export button (grey box with yellow background).
By clicking on the viral cluster or singleton identifiers the users can obtain information about the number of members in the cluster (‘Viral Contig Count’), the number of samples in which they were found (‘Sample Count’), the number of independent projects these samples belong to (‘Study Count’), the proposed host (when detected, ‘Host’), and the sample's habitat (‘Habitat Type’) (Figure 5B).
By clicking on a single viral cluster, all members of the cluster are displayed with several related metadata, including the number of genes per viral contig, contig length, GC content, host assignment, and taxonomic information (Figure 5C).
Browsing viral clusters based on their host assignment
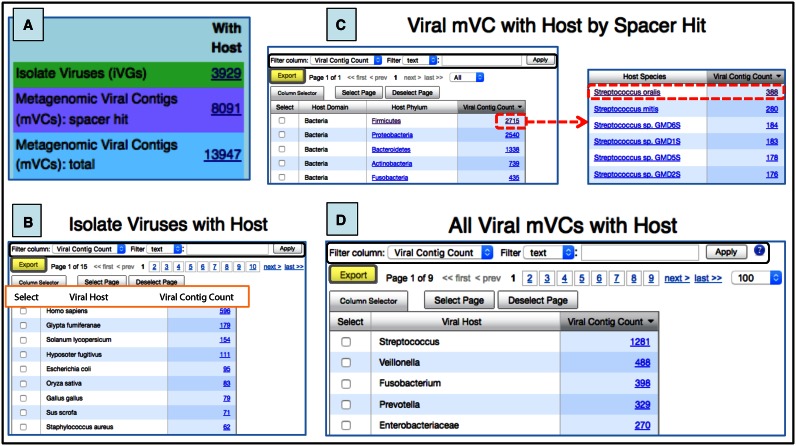
The third section of the left panel in the IMG/VR entry page shows the number of viral contigs associated with a host. Three different categories of host-linked contigs are provided (Figure 6A). First, the number of isolate viruses experimentally assigned to a host is reported. There are currently 3929 such viruses, which when accessed, are listed together with their corresponding host (Figure 6B).
Figure 6.
Viral data sets with host assignment. (A) Number of isolate viruses or metagenomic viral contigs with a predicted host. (B) ‘Isolate viruses with host’ table sorted by the hosts infected by the highest number of viral genomes. (C) Top microbial host species containing metagenomic viral contigs. (D) Microbial hosts (at different taxonomic levels) with the highest number of metagenomic viral contigs assigned. Columns in (B), (C), and (D) can be sorted by clicking on the column header and different filters can be used for specific searches (black boxes). Tables can be also exported in a tab-delimited text format by using the Export button (grey box with yellow background).
Second, the metagenomic viral contigs that bear a proto-spacer sequence match to a spacer from a microbial isolate genome (allowing a direct association virus–host at the species level) are reported. There are currently 8084 mVCs which can be listed in a table grouped with their associated hosts. As an example, there are 131 different viral species (representing a total of 388 mVCs) putatively infecting Streptococcus oralis (Figure 6C).
Finally, the total number of metagenomic viral sequences that can be assigned to a host (at the lowest possible taxonomic level) by projecting the host–virus information onto a viral cluster, is also presented. There are 13 947 in this category, whereby in the majority of the cases the virus-host link is at genus or species level. The microbial genera infected with the highest number of viral contigs are Streptococcus, Veillonella, Fusobacterium and Prevotella s (Figure 6D). In ∼9% of all assignments, the host connection is at a higher taxonomy range (ranging from family to phylum).
All the information from all the tables can be independently accessed by clicking on their corresponding links or could be exported in a tab-delimited text format by using the ‘Export’ button.
Sequence search
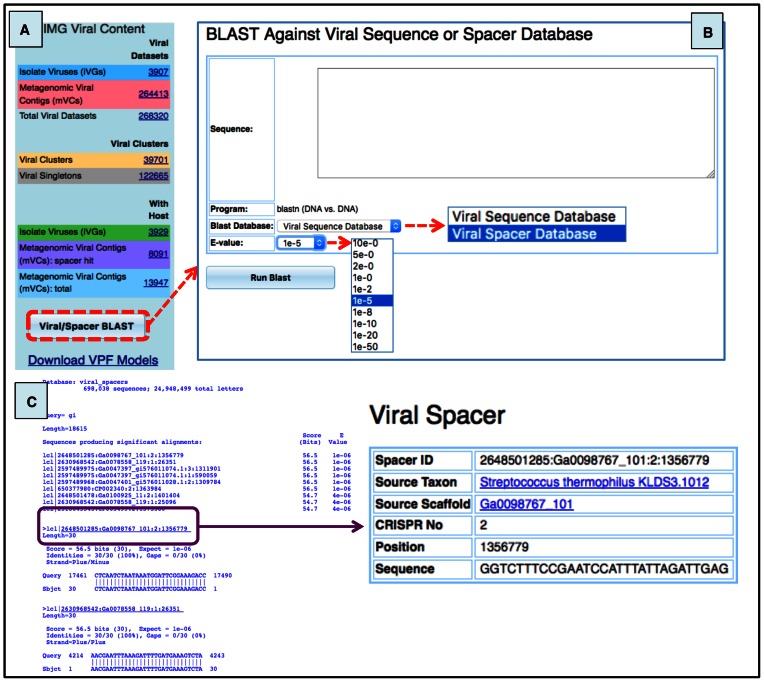
Users can compare their sequences against the sequence data integrated into IMG/VR. Specifically, the sequences of all the viral contigs and all the spacer sequences from the isolate genomes can be queried by using the ‘Viral/Spacer Blast’ option at the bottom of the home page (Figure 7A). Both queries can be selected from ‘Blast Database’ and rely on nucleotide BLAST searches (32) with customizable e-value cutoffs (Figure 7B).
Figure 7.
Viral searches against IMG/VR databases. (A) Location of blast tool in IMG/VR (dashed red box). (B) User interface to blast sequences. Exclusively nucleotide sequences can be queried currently in the system. Sequence(s) must be added into the blank area. Users can blast their sequence(s) against any of the two databases integrated into IMG/VR: ‘Viral Sequence’ or ‘Viral Spacer’ and customize the e-value cutoff. (C) Example of a blast output of an external partial viral sequence (Streptococcus phage 858) against the spacer database. When a sequence hit (purple box) is selected, a new panel (‘Viral Spacer’) is displayed showing details of the sequence spacer and the putative corresponding microbial host.
Matches against the viral database generate a list of viral sequences with a significant alignment based on the selected thresholds. These subject sequences can be directly accessed or selected to-be-added to the Scaffold Cart, where their associated metadata are also provided. Similarly, matches of external viral sequences against the spacer database generate a list of host(s) containing a CRISPR-spacer sequence with a significant alignment based on the selected cutoffs. These putative host(s) can be further explored by clicking on the host identifier. This redirects the user to detailed information of the spacer: source taxon name, location of the spacer within the CRISPR array, and spacer sequence (Figure 7C).
Future developments
We present the first version of a viral specific system within the IMG database. Almost 6000 metagenome datasets publicly available in IMG/M were mined in search of viral contigs at the time of the study (June 2016). Since IMG/M is continuously growing in number and size of metagenome studies, we anticipate that the number of viral sequences included in IMG/VR will continue to grow rapidly. Future versions of IMG/VR will complement the isolate and metagenomic viral contigs detected with prophage sequences identified from microbial genomes. This is expected to drive the identification of a larger number of virus-host connections and the viral clusters expansion connected to hosts. In addition, we are developing an RNA virus discovery pipeline from metatrascriptomic datasets that will complement the global DNA virome. We also plan to expand the current host–virus assignment with other prediction approaches (e.g. based on viral tRNA matches (11), specific lysozymes, or other computational approaches (12,26)) and to refine viral taxonomy in accordance with community standards that should be derived from the gene sharing networks emerging as a way to organize the viral sequence space, expanding the current information about eukaryotic and archaeal viruses as well as putative giant viruses and virophages.
Overall, the growing number of metagenomic datasets and the continuous detection of new viral contigs together with the ongoing development of analysis and search capabilities within the IMG system will render IMG/VR a critical community resource for the study of viruses.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the US Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, under contract number DE-AC02-05CH11231 and used resources of the National Energy Research Scientific Computing Center, supported by the Office of Science of the US Department of Energy.
Conflict of interest statement. None declared.
REFERENCES
- 1.Suttle C.A. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 2.Gomez P., Buckling A. Bacteria-phage antagonistic coevolution in soil. Science. 2011;332:106–109. doi: 10.1126/science.1198767. [DOI] [PubMed] [Google Scholar]
- 3.Pal C., Macia M.D., Oliver A., Schachar I., Buckling A. Coevolution with viruses drives the evolution of bacterial mutation rates. Nature. 2007;450:1079–1081. doi: 10.1038/nature06350. [DOI] [PubMed] [Google Scholar]
- 4.Brum J.R., Sullivan M.B. Rising to the challenge: accelerated pace of discovery transforms marine virology. Nat. Rev. Microbiol. 2015;13:147–159. doi: 10.1038/nrmicro3404. [DOI] [PubMed] [Google Scholar]
- 5.Fuhrman J.A. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399:541–548. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- 6.Breitbart M., Salamon P., Andresen B., Mahaffy J.M., Segall A.M., Mead D., Azam F., Rohwer F. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. U.S.A. 2002;99:14250–14255. doi: 10.1073/pnas.202488399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breitbart M., Miyake J.H., Rohwer F. Global distribution of nearly identical phage-encoded DNA sequences. FEMS Microbiol. Lett. 2004;236:249–256. doi: 10.1016/j.femsle.2004.05.042. [DOI] [PubMed] [Google Scholar]
- 8.Breitbart M., Rohwer F. Here a virus, there a virus, everywhere the same virus. Trends Microbiol. 2005;13:278–284. doi: 10.1016/j.tim.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Brum J.R., Ignacio-Espinoza J.C., Roux S., Doulcier G., Acinas S.G., Alberti A., Chaffron S., Cruaud C., de Vargas C., Gasol J.M., et al. Ocean plankton. Patterns and ecological drivers of ocean viral communities. Science. 2015;348:1261498. doi: 10.1126/science.1261498. [DOI] [PubMed] [Google Scholar]
- 10.Dinsdale E.A., Edwards R.A., Hall D., Angly F., Breitbart M., Brulc J.M., Furlan M., Desnues C., Haynes M., Li L., et al. Functional metagenomic profiling of nine biomes. Nature. 2008;452:629–632. doi: 10.1038/nature06810. [DOI] [PubMed] [Google Scholar]
- 11.Paez-Espino D., Eloe-Fadrosh E.A., Pavlopoulos G.A., Thomas A.D., Huntemann M., Mikhailova N., Rubin E., Ivanova N.N., Kyrpides N.C. Uncovering Earth's virome. Nature. 2016;536:425–430. doi: 10.1038/nature19094. [DOI] [PubMed] [Google Scholar]
- 12.Mizuno C.M., Rodriguez-Valera F., Kimes N.E., Ghai R. Expanding the marine virosphere using metagenomics. PLoS Genet. 2013;9:e1003987. doi: 10.1371/journal.pgen.1003987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roux S., Brum J.R., Dutilh B.E., Sunagawa S., Duhaime M.B., Loy A., Poulos B.T., Solonenko N., Lara E., Poulain J., et al. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature. 2016;537:689–693. doi: 10.1038/nature19366. [DOI] [PubMed] [Google Scholar]
- 14.Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W. GenBank. Nucleic Acids Res. 2016;44:D67–D72. doi: 10.1093/nar/gkv1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cook C.E., Bergman M.T., Finn R.D., Cochrane G., Birney E., Apweiler R. The European Bioinformatics Institute in 2016: Data growth and integration. Nucleic Acids Res. 2016;44:D20–D26. doi: 10.1093/nar/gkv1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pickett B.E., Greer D.S., Zhang Y., Stewart L., Zhou L., Sun G., Gu Z., Kumar S., Zaremba S., Larsen C.N., et al. Virus pathogen database and analysis resource (ViPR): a comprehensive bioinformatics database and analysis resource for the coronavirus research community. Viruses. 2012;4:3209–3226. doi: 10.3390/v4113209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ugai H., Murata T., Nagamura Y., Ugawa Y., Suzuki E., Nakata H., Kujime Y., Inamoto S., Hirose M., Inabe K., et al. A database of recombinant viruses and recombinant viral vectors available from the RIKEN DNA bank. J. Gene Med. 2005;7:1148–1157. doi: 10.1002/jgm.786. [DOI] [PubMed] [Google Scholar]
- 18.Hayer J., Jadeau F., Deleage G., Kay A., Zoulim F., Combet C. HBVdb: a knowledge database for Hepatitis B Virus. Nucleic Acids Res. 2013;41:D566–D570. doi: 10.1093/nar/gks1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bolduc B., Youens-Clark K., Roux S., Hurwitz B.L., Sullivan M.B. iVirus: facilitating new insights in viral ecology with software and community data sets imbedded in a cyberinfrastructure. ISME J. 2016 doi: 10.1038/ismej.2016.89. doi:10.1038/ismej.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Markowitz V.M., Chen I.M., Chu K., Szeto E., Palaniappan K., Pillay M., Ratner A., Huang J., Pagani I., Tringe S., et al. IMG/M 4 version of the integrated metagenome comparative analysis system. Nucleic Acids Res. 2014;42:D568–D573. doi: 10.1093/nar/gkt919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ivanova N., Tringe S.G., Liolios K., Liu W.T., Morrison N., Hugenholtz P., Kyrpides N.C. A call for standardized classification of metagenome projects. Environ. Microbiol. 2010;12:1803–1805. doi: 10.1111/j.1462-2920.2010.02270.x. [DOI] [PubMed] [Google Scholar]
- 22.Reddy T.B., Thomas A.D., Stamatis D., Bertsch J., Isbandi M., Jansson J., Mallajosyula J., Pagani I., Lobos E.A., Kyrpides N.C. The Genomes OnLine Database (GOLD) v.5: a metadata management system based on a four level (meta)genome project classification. Nucleic Acids Res. 2015;43:D1099–D1106. doi: 10.1093/nar/gku950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng L., Ignacio-Espinoza J.C., Gregory A.C., Poulos B.T., Weitz J.S., Hugenholtz P., Sullivan M.B. Viral tagging reveals discrete populations in Synechococcus viral genome sequence space. Nature. 2014;513:242–245. doi: 10.1038/nature13459. [DOI] [PubMed] [Google Scholar]
- 24.Konstantinidis K.T., Tiedje J.M. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. U.S.A. 2005;102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lederberg E.M., Lederberg J. Genetic studies of lysogenicity in Escherichia coli. Genetics. 1953;38:51–64. doi: 10.1093/genetics/38.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edwards R.A., McNair K., Faust K., Raes J., Dutilh B.E. Computational approaches to predict bacteriophage-host relationships. FEMS Microbiol. Rev. 2016;40:258–272. doi: 10.1093/femsre/fuv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paez-Espino D., Sharon I., Morovic W., Stahl B., Thomas B.C., Barrangou R., Banfield J.F. CRISPR immunity drives rapid phage genome evolution in Streptococcus thermophilus. MBio. 2015;6 doi: 10.1128/mBio.00262-15. doi:10.1128/mBio.00262-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 29.Paez-Espino D., Morovic W., Sun C.L., Thomas B.C., Ueda K., Stahl B., Barrangou R., Banfield J.F. Strong bias in the bacterial CRISPR elements that confer immunity to phage. Nat. Commun. 2013;4:1430. doi: 10.1038/ncomms2440. [DOI] [PubMed] [Google Scholar]
- 30.Roux S., Hallam S.J., Woyke T., Sullivan M.B. Viral dark matter and virus-host interactions resolved from publicly available microbial genomes. Elife. 2015;4:e08490. doi: 10.7554/eLife.08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pagani I., Liolios K., Jansson J., Chen I.M., Smirnova T., Nosrat B., Markowitz V.M., Kyrpides N.C. The Genomes OnLine Database (GOLD) v.4: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2012;40:D571–D579. doi: 10.1093/nar/gkr1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]