Abstract
Deep phenotyping has been defined as the precise and comprehensive analysis of phenotypic abnormalities in which the individual components of the phenotype are observed and described. The three components of the Human Phenotype Ontology (HPO; www.human-phenotype-ontology.org) project are the phenotype vocabulary, disease-phenotype annotations and the algorithms that operate on these. These components are being used for computational deep phenotyping and precision medicine as well as integration of clinical data into translational research. The HPO is being increasingly adopted as a standard for phenotypic abnormalities by diverse groups such as international rare disease organizations, registries, clinical labs, biomedical resources, and clinical software tools and will thereby contribute toward nascent efforts at global data exchange for identifying disease etiologies. This update article reviews the progress of the HPO project since the debut Nucleic Acids Research database article in 2014, including specific areas of expansion such as common (complex) disease, new algorithms for phenotype driven genomic discovery and diagnostics, integration of cross-species mapping efforts with the Mammalian Phenotype Ontology, an improved quality control pipeline, and the addition of patient-friendly terminology.
INTRODUCTION
The Human Phenotype Ontology (HPO) provides comprehensive bioinformatic resources for the analysis of human diseases and phenotypes, offering a computational bridge between genome biology and clinical medicine. The HPO was initially published in 2008 (1) with the goal of enabling the integration of phenotype information across scientific fields and databases. Since then, the project has grown in terms of coverage, scope and sophistication, and has become a core component of the Monarch Initiative, allowing computational cross-species analysis (2).
HPO has also become part of the core Orphanet (3) rare disease database content. The Orphanet nomenclature of rare diseases, whose adoption has been recommended by the European Commission expert group of rare diseases for codification of rare-disease (RD) patients in health information systems (recommendation on ways to improve codification for rare diseases in health information systems: http://ec.europa.eu/health/rare_diseases/docs/recommendation_coding_cegrd_en.pdf), is being annotated with HPO terms in order to allow for deep phenotyping of RD in health records and registries.
The description of phenotypic variation has become a central topic for translational research and genomic medicine (4–7), and ‘computable’ descriptions of human disease using HPO phenotypic profiles (also known as ‘annotations’) have become a key element in a number of algorithms being used to support genomic discovery and diagnostics. Here, we describe the latest improvements to the tools and resources being developed by the HPO Consortium and the Monarch Initiative, and provide an overview of external tools and databases that are using the HPO for translational research and diagnostic decision support.
HPO: NEW TERMS, ANNOTATIONS AND ONTOLOGY INTEGRATION
The HPO is organized as independent subontologies that cover different categories. The largest category is Phenotypic abnormality. The Mode of inheritance subontology allows disease models to be defined according to Mendelian or non-Mendelian inheritance modes. The Mortality/Aging subontology similarly allows the age of death typically associated with a disease or observed in a specific individual to be annotated. Finally, the clinical modifier subontology is designed to provide terms to characterize and specify the phenotypic abnormalities defined in the Phenotypic abnormality subontology, with respect to severity, laterality, age of onset, and other aspects.
Ontology
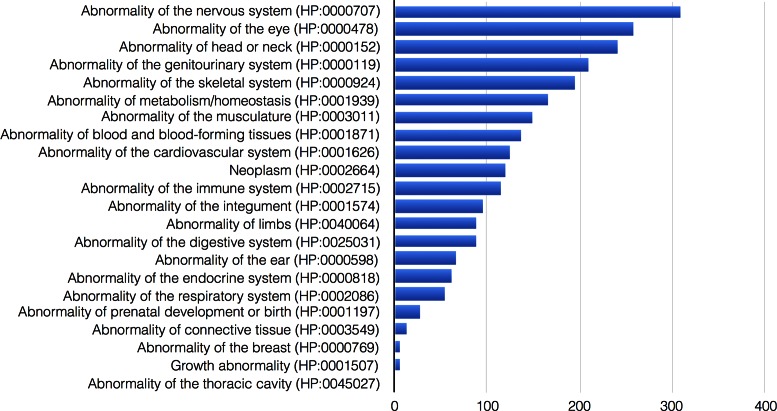
The HPO has grown substantially since the first Nucleic Acids Research database article in 2014 (Version: 30 July 2013) (8) to the September 2016 release (Version: 3 September 2016). There are 1725 additional terms (10 088 in 30 July 2013 versus 11 813 in 3 September 2016, see Figure 1) and 2269 additional subclass relationships (13 326 versus 15 595). We obsoleted 82 HPO classes (44 versus 126). We have added 2024 textual definitions (6603 versus 8627) and 8063 synonyms (6265 versus 14 328). Logical definitions were constructed for an additional 1126 HPO classes, bringing the total number to 5717. These definitions refer to ontologies for biochemistry, gene function, anatomy, and others, and allow cross-species mapping by means of automated semantic reasoning. There are now 123 724 annotations of HPO terms to rare diseases and 132 620 to common diseases.
Figure 1.
Distribution of HPO class additions per general category of phenotypic abnormalities. The figure shows the number of terms added per category since the previous Nucleic Acids Research database article in 2014 (8).
Annotations
The main domain application of the HPO has, to date, been on rare disorders, and we have in the past provided a large corpus of disease-HPO annotation profiles using OMIM, Orphanet and DECIPHER for disease entities (8). With recent advances in personalized medicine, it is becoming increasingly important to provide a computational foundation for phenotype-driven analysis of genomes and other translational research in other fields of medicine. Consequently, we have extended our work to common human disease phenotypes by means of a text-mining approach (9) toward analyzing the 2014 PubMed corpus, which allowed us to infer 132 620 HPO annotations for 3145 common diseases (10). These annotations were validated against a manually curated subset of disorders and experimental results showed an overall precision of 67%. We showed statistically significant phenotypic overlap between common diseases that share one or more associated genetic variants (‘Genome-wide association study [GWAS] hit’), as well as phenotypic overlaps between rare and common disease that are linked to the same genes (10). The HPO has also been adopted by several resources for genotype-phenotype data in the field of complex disease and genome-wide association study (GWAS) analysis, including GWAS Central (11) and GWASdb (12), and is likely to be adopted for phenome-wide association studies with electronic health records in the future (13).
Precision annotation of deep phenotyping data
The performance of computational search algorithms within and across species can improve if a comprehensive list of phenotypic features is recorded. It is helpful if the person annotating thinks of the set of annotations as a query against all known phenotype profiles. Therefore, the set of phenotypes chosen for the annotation must be as specific as possible, and represent the most salient and important observable phenotypes. The Monarch Initiative has developed an annotation sufficiency meter that assesses the breadth and depth of the phenotype annotation profile using a five-star rating system for a given patient in the context of all curated human and model organism phenotypes, with the goal of helping the annotator to generate an annotation profile specific enough to exclude similar diseases and to identify model organisms with similar phenotypes that may have mutations in relevant genes or pathways (14). The Monarch annotation sufficiency meter is displayed within PhenoTips (15) and PhenomeCentral (16).
Integration
The scope and specificity of phenotypes useful for diagnosis and clinical decisions support differ considerably from phenotypes useful for medical billing and quality-of-care assessment. What sets HPO apart from other ontologies is that it is purpose built for the diagnosis and care use case and that it is designed to facilitate cross-species comparisons so that non-human data can be brought to bear as well. Moreover, to accomplish this task the HPO must also have extremely broad coverage of concepts. In an evaluation of HPO content versus the numerous vocabularies integrated within the Unified Medical Language System (UMLS), Winnenburg and Bodenreider showed that the coverage of HPO phenotype concepts in the UMLS is 54% and only 30% in SNOMED CT (17). The UMLS is a terminology integration system developed by the U.S. National Library of Medicine that integrates many standard biomedical terminologies (18). In order to improve the coverage of phenotype data, the UMLS has now integrated the entire HPO starting with the 2015AB release. This enables an easy process to map HPO-encoded data to standard health-care terminologies such as SNOMED CT (19). HPO has contributed to the establishment of the International Consortium of Human Phenotype Terminologies (ICHPT; http://www.ichpt.org) to provide the community with standards that achieve interoperability among databases incorporating human phenotypic features. The outcome is a set of over 2300 terms which should be incorporated in any terminology and which is fully cross-referenced with HPO terms. These terms are not arranged in a hierarchy and so can be mapped to or incorporated into any ontology.
The HPO project data are available at http://www.human-phenotype-ontology.org. Requests for new terms or other amendments can be made using the GitHub issues tracker https://github.com/obophenotype/human-phenotype-ontology/issues. Further information on HPO-related publications and general announcements can be found on the HPO website at http://www.human-phenotype-ontology.org and on the HPO twitter feed @hp_ontology.
CLINICAL UTILITY
Although exome sequencing and other forms of genomic diagnostics have greatly accelerated the pace of discovery of novel disease-associated genes and have begun to be implemented in diagnostic settings in medical genetics, the overall diagnostic yield can still be low. It has been estimated that the genetic cause of only about half of the currently named ∼7000 rare diseases has been identified (20,21); in order to confidently assert that pathogenic variants in a given gene are associated with a given Mendelian disease, the community norm is to require the identification of at least two unrelated cases. The HPO team therefore continues to collaborate with clinical groups to refine and extend current terms and annotations to support efforts to match patient phenotype and genotype data. Table 1 provides an overview of public-facing clinical databases that use HPO to annotate patient data.
Table 1. A selection of public-facing clinical databases using HPO to annotate patient data for disease-gene discovery projects.
| Name | URL | Ref |
|---|---|---|
| PhenomeCentral | phenomecentral.org | (16) |
| DDD (Deciphering Developmental Disorders) | www.ddduk.org | (61,62) |
| DECIPHER (DatabasE of genomiC varIation and Phenotype in Humans using Ensembl Resources) | decipher.sanger.ac.uk | (63) |
| ECARUCA (European Cytogeneticists Association Register of Unbalanced Chromosome Aberrations) | http://umcecaruca01.extern.umcn.nl:8080/ecaruca/ecaruca.jsp | (64) |
| The 100 000 Genomes Project | https://www.genomicsengland.co.uk/ | (65) |
| Geno2MP (Exome sequencing data linked to phenotypic information from a wide variety of Mendelian gene discovery projects) | http://geno2mp.gs.washington.edu | (21) |
| NIH UDP (Undiagnosed Diseases Program) | available via phenomecentral.org | (66) |
| NIH UDN (Undiagnosed Diseases Network) | available via phenomecentral.org | (16) |
| HDG (Human Disease Gene Website series) | www.humandiseasegenes.com | |
| Phenopolis (An open platform for harmonization and analysis of sequencing and phenotype data) | https://phenopolis.github.io | |
| GenomeConnect (Patient portal developed by ClinGen (67) | www.genomeconnect.org | (68) |
| FORGE Canada & Care4Rare Consortium | available via phenomecentral.org | (69) |
| RD-Connect | platform.rd-connect.eu | (28) |
| Genesis | thegenesisprojectfoundation.org | |
The HPO has been extensively applied to the phenotypic characterization of bone dysplasias (rare genetic bone disorders). The Bone Dysplasia Ontology (BDO) (22) is an ontological representation of the International Skeletal Dysplasia Society's Nosology of Genetic Skeletal Disorders, the de facto standard classification for human bone dysplasias. The BDO uses HPO terms for the phenotypic description of each disorder. Using the BDO and HPO, decision support methods were developed to predict the correct bone dysplasia diagnosis from a set of HPO terms, and their methods outperformed many clinicians (23).
DECIPHER (https://decipher.sanger.ac.uk) was established in 2004 as a web-based system for interpretation and sharing of genomic variants and their associated phenotypes. DECIPHER now supports sequence variation and copy number variation in the nuclear and mitochondrial genomes. DECIPHER was an early adopter of HPO and is the platform through which data from the Deciphering Developmental Disorders study (DDD study) is shared (24). At the outset of the project, the DDD study (www.ddduk.org) funded a week-long workshop to improve the HPO ontology by reducing redundancy of terms and improving coverage in the rare disease space. DECIPHER currently has 21,689 open-access patient records annotated with 60,521 HPO-encoded phenotype observations.
PhenoTips (15) is an open-source clinical phenotype and genotype data collection tool. It provides simple user interfaces to select and explore HPO annotations and suggest diagnoses from OMIM. Records within PhenoTips can be de-identified and pushed to PhenomeCentral (16) to participate in phenotypic and genotypic matching with other cases in PhenomeCentral and in connected databases through the Matchmaker Exchange. PhenomeCentral makes use of HPO terms to measure semantic similarity between patient phenotypes and prioritize exome data using the Exomiser. At the time of this writing, PhenomeCentral contains 2640 matchable cases, of which 2059 have at least one HPO term, 172 are from the NIH UDP and 28 from the NIH UDN.
Patient Archive (PA) (2) is a clinical-grade phenotype-oriented platform for managing patient data; PA combines the richness of the HPO with highly intuitive user interfaces to aid the discovery and decision-making process in the context of clinical genomics. PA enables clinicians to use free text clinical notes as the starting point for structured HPO-centric patient phenotyping to support clinical diagnostics and care. To this end, an instance has been installed in the Western Australian Department of Health for clinical genetic use, both within and outside of, the Undiagnosed Diseases Program (UDP)—Western Australia; a clinical public health service. It has also been nominated as the platform of choice for the UDP Australia which participates in the Undiagnosed Diseases Network International (25). Relatedly, and building on the principles of founding work (26), the integration of automated annotation of HPO terms to 3D facial images as part of a suite of approaches in the clinical workflow continues to be developed through the Rare and Undiagnosed Diseases Diagnostic Service at Genetic Services of Western Australia (27).
Phenopolis is an interactive platform built on genomic and phenotypic data from over 4000 patients. With the help of phenotype quantification using HPO, Phenopolis is able to prioritize causative genes using prior knowledge from OMIM, Pubmed publications and existing tools such as Exomiser. Additionally, it helps novel gene discovery by looking for potential gene-HPO relationships among the patients without using any prior knowledge. This unbiased approach may provide valuable information for hospitals and researchers to optimize their resources on diagnosis and functional studies for the relevant genetic diseases.
Numerous rare-disease research consortia are using HPO for patient annotation and analysis. In order to review and expand the HPO to better represent specific disease areas, the HPO consortium has conducted workshops with consortia including the European FP7 projects RD-Connect (28), EURenOmics and NeurOmics. Using advanced omics technologies, NeurOmics, an EU-funded translational research project, aims to characterize the causes, pathomechanisms and clinical features across ten major neurodegenerative and neuromuscular disease groups affecting the brain and spinal cord, peripheral nerves and muscle. EURenOmics is using high-throughput technologies to characterize new genes causing or predisposing to kidney diseases, concentrating on five groups of renal disease.
RD-Connect is an integrated platform connecting databases, registries, biobanks and clinical bioinformatics for rare disease research that brings together multiple datasets on patients with rare diseases at a per-patient level. Deep phenotyping of affected individuals is an essential component of these projects, and is being addressed by using the HPO as a mechanism for linking a computationally accessible phenotypic record with a genomic dataset. The projects performed a review of available ontologies at an early stage and concluded that the HPO was the most appropriate ontology for their gene discovery focus (28).
Both NeurOmics and EURenOmics performed mapping exercises in order to transform data items suggested by clinicians as essential items to record for each patient presenting with a particular clinical profile into HPO terms, and in most cases this mapping was able to produce exact matches to an already existing HPO term. Missing areas were then addressed in the expert workshops described above. Several of these projects make use of PhenoTips (15) in order to capture clinical data to a highly granular level through an interface that is user-friendly for clinicians. Independently of the data entry mechanism, the use of the HPO means that the data generated by these consortia is fully interoperable with other datasets internationally. Currently, the RD-Connect platform contains ∼2000 exome, genome and panel sequencing datasets linked with HPO-coded phenotypic profiles from a range of rare diseases.
The HPO was used within the EuroEPINOMICS-Rare Epilepsy Syndrome (RES) project to systematically assess phenotypes in patients with epileptic encephalopathies (29,30). A first analysis of clustering of epilepsy phenotypes was presented as a poster at the 2012 European Congress of Epileptology (31), while a more comprehensive analysis of the obtained HPO terms including exome sequencing data is currently underway. Clustering of patient phenotypes in 171 patients with epileptic encephalopathies identified a subgroup of eight patients with closely related phenotypes. A review of manually curated phenotype data suggested these patients had a subset of Infantile Spasms with a good outcome. This preliminary analysis suggested that the use of HPO terms in patients with epilepsy is worthwhile, given that the identified epilepsy phenotype was both homogeneous and clinically meaningful.
The use of HPO terms for patients with epilepsy is challenging. In contrast to many other genetic disorders, the phenotypic features in epilepsy patients are dynamic and specific features such as a complex seizure semiology are often difficult to fully include in systematic phenotype ontologies. For example, a patient with simple febrile seizures may have self-limiting febrile seizures (FS), may have recurrent febrile seizures past the age of six years (FS+), or may develop the intractable, fever-related epilepsy of Dravet Syndrome over time. All three entities are distinct, but depending on the age of the patients, may be coded identically in the HPO if modifiers coding the patient's age are not used. The dilemma of fully representing dynamic neurological phenotypes emphasizes the need for the ongoing use of HPO modifiers to achieve dimensionality in phenotype data.
The HPO has been used to incorporate clinical data into the analysis of a diagnostic next-generation sequencing panel with nearly all known Mendelian disease-associated genes; the algorithm, Phenotypic Interpretation of Exomes (PhenIX) contributed to a diagnostic rate of 28% in children in whom previous extensive workups had failed to reveal a diagnosis (32). Using HPO to generate individualized phenotype-driven gene panels for diagnostics led to an increase in the diagnostic yield (33).The ThromboGenomics Consortium reported that computational prioritization of candidate rare variants identified in patients with bleeding, thrombotic or platelet disorders using HPO-coded phenotypes assigned the highest scores to pathogenic or likely pathogenic variants in 85% of cases, demonstrating that HPO-based algorithms can make multidisciplinary diagnostic meetings more efficient (34).
Once such a causative link between rare pathogenic variants in a given gene has been established, it is essential to assess the clinical variability attributed to other mutations in that gene. For this, several novel approaches have currently been developed, such as the Human Disease Gene Website series (HDG). HDG is an international library of websites (www.humandiseasegenes.com) for professional information about genes and copy number variants and their clinical consequences using HPO to annotate the phenotype. Here, professionals will find relevant information that helps with interpretation of variants and counseling of their patient/families with such a rare genetic disorder and also have the opportunity to share clinical data. Moreover, patients, parents, and caregivers will find useful information on the rare genetic disease in their family.
Sanford Health, one of the largest non-profit rural health care systems in the United States, has embarked on clinical genotyping of a substantial portion of its patient population to provide precision prevention and pharmacogenetics. As part of this process, it has incorporated tools within the patient portal of the electronic medical record (EMR) to enable patients to characterize themselves in HPO. Similarly, it has incorporated Phenotips within the EMR to enable clinical staff to characterize in HPO all patients prescribed diagnostic molecular testing. For both the patient self-characterization and the clinician characterization, the Monarch Initiative sufficiency score is used to guide depth of characterization. The HPO terms, data within the EMR and molecular test results are integrated to define diagnoses and best practice guidelines entered into the EMR.
The 100 000 Genomes Project (www.genomicsengland.co.uk) is sequencing 100 000 whole genomes from NHS patients in England with rare diseases or cancer. Recruitment to the Rare Disease Programme currently occurs across approximately 200 diseases. A vital aspect of the project is to link rare disease participants’ genomes with their phenotype profile to enable genome diagnostics and in-depth genotype-phenotype analyses. The phenotype profiles need to be detailed, specific, consistently applied, computationally accessible and concordant with existing standards. The project has developed HPO-based models for each rare disease. These comprise, typically, 20–40 HPO terms that describe the key features of the disease. These are presented to recruiting clinicians as a questionnaire—additional HPO terms can also be entered. This approach requires less prior knowledge of HPO to achieve in-depth phenotyping than simple ‘free entry’, and encourages recording of the absence of phenotypes as well as their presence. The models are typically developed by mapping HPO terms to an existing case report form, published review, registry schema or through interaction with clinical experts. Models are analysed to ensure practicality, consistency and specificity using the Monarch annotation sufficiency score described above. Where clinical terms that are not contained in HPO are identified during model development they are submitted for inclusion. The collected phenotypes for each program participant are used extensively in analysis pipelines, and for manual clinical interpretation and automated prioritization using algorithms such as Exomiser (35) and Phevor (36).
USE OF HPO IN GENE IDENTIFICATION RESEARCH
The HPO has been used in many ways in research on disease pathophysiology, diagnostics and gene-discovery projects. It has been used to provide lists of genes associated with one or more HPO terms in order to filter lists of candidate genes (37–39), to prioritize candidate genes in Exome-sequencing studies via PhenIX, Phevor or Exomiser (40–43), and to identify known or novel disease genes or to analyze structural variation in large cohorts (44–46). The Deciphering Developmental Disorders (DDD) study analyzed 4125 families with diverse developmental disorders and identified four novel disease-gene associations by combined analysis of the genotypes and the phenotypic similarity of patients with recessive variants in the same candidate gene (47). The BRIDGE-BPD Consortium (48) used genome sequencing combined with HPO coding to identify a gain-of-function variant in DIAPH1 in two unrelated pedigrees with deafness and macrothrombocytopenia (49). This finding was supported by Phenotype Similarity Regression (SimReg), an algorithm for identifying composite phenotypes associated with rare variation in specific genes (50). HPO-based phenotype analysis also allowed matching of human phenotypes to mouse phenotypes by cross-species analysis and thereby aided the discovery of a dominant gain-of-function mutation in SRC that causes thrombocytopenia, myelofibrosis, bleeding and bone abnormalities (51).
The Matchmaker exchange (MME) platform provides a systematic approach to rare disease-gene discovery with a federated network of phenotype-genotype databases that enable data sharing and discovery of relevant data (52,53) over a secure API (54). The HPO is the standard vocabulary for communicating phenotype data. The MME currently connects over 30 000 rare disease cases across six different patient databases.
TRANSLATIONAL RESEARCH AND DIAGNOSTICS WITH HPO: ALGORITHMS AND TOOLS
The HPO is a computational resource that allows algorithms to ‘compute over’ clinical phenotype data in an increasing number of contexts through a growing number of tools from the HPO Consortium and other groups (Table 2). The tools use the ontological structure of the HPO that allows individual terms to be associated with an information content, a measure of specificity (55), or with the underlying logical definitions of the terms, such that HPO terms can be linked to other resources such as model organisms (56,57).
Table 2. Tools and applications using HPO.
| Tool | Reference |
|---|---|
| Phenotype-driven differential diagnosis | |
| Phenomizer | (70) |
| BOQA | (71) |
| FACE2GENE | (72) |
| Phenolyzer | (73) |
| Phenotype-driven exome/genome analysis | |
| Exomiser | (35,74) |
| PhenIX | (32) |
| Phevor | (36) |
| PhenoVar | (75) |
| eXtasy | (76) |
| OMIMExplorer | (77) |
| Phen-Gen | (78) |
| Geno2MP | (21) |
| Genomiser | (79) |
| SimReg | (50) |
| ontologySimilarity | * |
| Functional and network analysis | |
| TopGene/ToppFunn | (80) |
| WebGestalt | (81) |
| SUPERFAMILY | (82) |
| GREAT | (83) |
| Random walk on heterogeneous network | (84) |
| PANDA | (85) |
| PREDICT | (86) |
| Clinical data management and analysis | |
| Phenotips | (15) |
| Patient Archive | (2) |
| GENESIS (GEM.app) | (87) |
| Cross-species phenotype analysis | |
| PhenoDigm | (88) |
| MouseFinder | (89) |
| Monarch | (2,53) |
| PhenomeNet | (90) |
| UberPheno | (56) |
| MORPHIN | (91) |
| PhenogramViz | (92) |
| Phenotype knowledge resources and databases | |
| Orphanet | (3) |
| MalaCards | (93) |
| NIH genetic testing registry | (94) |
| OMIM | (95) |
| dcGO | (96) |
| ClinVar | (97) |
| GeneSetDB | (98) |
| MSeqDR | (99) |
| DIDA (digenic diseases database) | (100) |
| Genetic and Rare Diseases (GARD) Information Center | (101) |
| Visualization | |
| PhenoStacks | (102) |
| PhenoBlocks | (103) |
| DECIPHER (phenogram) | (63) |
| phenogrid | (2) |
| ontologyPlot | * |
*Greene, D., Richardson, S. and Turro, E. OntologyX: a suite of R packages for working with ontological data, under review.
PUBLISHING PROCESSES AND DATA EXCHANGE
It is non-trivial to collect patient phenotypes reliably, whether retrospectively from existing medical data or prospectively. The overwhelming majority of clinical descriptions in the medical literature are available only as natural language text, meaning that searching, analysis and integration of medically relevant information is challenging. An important step to increase the amount and quality of phenotype data in databases is to obtain the relevant information from authors upon submission of articles. The journal Cold Spring Harbor Molecular Case Studies requires authors to select HPO terms for research papers that are displayed alongside the manuscript and that can be used to search journal content for other cases with overlapping HPO terms (58). Short Reports in Clinical Genetics require authors to submit HPO-coded phenotype data to PhenomeCentral (16). An important goal of the HPO and the Monarch Initiative is to provide computational standards that will allow for exchange of detailed genotype and phenotype data by means of the emerging PhenoPackets standard (http://phenopackets.org).
PATIENT PHENOTYPING
Patient-reported phenotype data in patient registries such as J-RARE for rare diseases has been increasingly exploited in scientific research; for instance, indicating symptoms still unknown to physicians. A barrier to the use of patient-reported data for understanding the natural history and phenotypic spectrum of diseases lies in the fact that clinical terminology is often unfamiliar to patients. The HPO consortium has therefore increased the usability of the HPO by patients, as well as scientists and clinicians, by systematically adding new, ‘plain language’ terms, either as synonyms to existing classes or by tagging existing HPO class labels as "layperson". These layperson terms provide increased access to the HPO—for example, a patient may know they are ‘color-blind’, but may not be familiar with the clinical term ‘Dyschromatopsia’. As a result of this effort, the HPO now contains over 6000 layperson terms that can be integrated into patient registries, making the terminology useful for data interoperability across clinicians and patients. Future work will include validation studies using data from patient registries to demonstrate the utility of the HPO layperson synonyms in informing rare disease diagnosis (59).
HPO: an assessment by the NIHR rare disease initiatives
HPO is used as the system to capture of phenotypic information for the UK's National Institute for Health Research (NIHR) Rare Disease initiatives on projects such as NIHR RD-TRC (Rare Disease—Translational Research Collaboration, http://rd.trc.nihr.ac.uk) and the NIHR BioResource Rare Disease NIHR BR-RD. HPO is employed in all of these broad wide-ranging studies and includes data integration from a variety of sources such as multiple EHR systems, in a variety of locations and specialities. In some disease areas, for example, bleeding and platelet disorders, HPO has been the platform for new gene discovery and innovative research findings (44); the advantage of HPO its support for statistical power associations across phenotypes across different diseases and in different branches of the HPO ontology.
The NIHR Rare Disease initiatives use a common infrastructure and clinical coding for the RD-TRC (56 studies), BR-RD (14 studies) and also in our contribution to the 100 000 Genome project (160+ targeted diseases, as mentioned above). This produces a large and diverse dataset with a growing ‘data dictionary’ containing terms mapped across different systems and coding schemes and includes clinically relevant signs outside of HPO—for example, lab test results or exercise questionnaires.
In a short update, it is difficult to present the breadth of the contribution HPO makes to NIHR-RD research, which indeed is growing as more diseases are characterized and encoded using HPO. The HPO is now being employed in numerous NIHR-RD studies and it is anticipated that its use will be extended into all studies in which phenotype data are captured.
The NIHR-RD-TRC has developed a qualitative scale for the maturity of HPO across different disease areas which adopts a four stage assessment (Table 3). The current, subjective, assessment of HPO maturity by the NIHR RD-TCR is shown in Table 4. The assessment will be used to prioritize areas requiring most attention in our future work.
Table 3. NIHR-RD-TRC assessment scale.
| Stage | Description | Example |
|---|---|---|
| Foundation | The basis of characterizing the disease in HPO needs to be developed | HPO is good for describing dysmorphologies especially across species: how do you model and use dyslexia? |
| Formulation | The theory is defined but key details need to be defined and handled in the ontology computations | HPO models biology, where diseases are caused by environmental factors, e.g. cancers — how can an environment ontology be included? |
| Refinement | The key data sets and definitions for the disease are identified and available but require ‘translation’ | Theme based registry systems hold collections of data in other coding systems (registry-specific or ICD) — how can these be mapped onto HPO? |
| Maturity | The HPO framework is in place and productive results are being obtained, the HPO term set continues to evolve | The HPO basics are in place and a set of Phenotypes in place — do we need more terms or do existing terms need modification? |
Table 4. NIHR-RD-TRC assessment of HPO maturity.
| Theme | Foundation | Formulation | Refinement | Maturity |
|---|---|---|---|---|
| Cancer | ✓✓✓✓ | ✓✓ | ||
| Cardiovascular | ✓✓✓✓✓ | ✓✓✓ | ✓✓ | |
| Central Nervous System | ✓✓✓ | |||
| Eye Diseases | ✓✓✓✓✓ | ✓✓✓✓✓ | ✓✓✓✓✓ | ✓✓ |
| Gastrointestinal | ✓✓✓✓ | ✓✓✓ | ||
| Immunological Disorders | ✓✓✓✓✓ | ✓✓✓ | ✓✓ | |
| Paediatric (cross-cutting) | ✓✓✓✓✓ | ✓✓✓ | ✓✓✓ | ✓ |
| Metabolic & Endocrine Diseases | ✓✓✓✓✓ | ✓✓ | ||
| Musculoskeletal Disorders | ✓✓✓✓✓ | ✓✓✓✓✓ | ✓✓✓ | |
| Muscle & Nerve Diseases | ✓✓✓✓✓ | ✓✓✓ | ✓ | |
| Non-malignant Haematology | ✓✓✓✓✓ | ✓✓✓✓✓ | ✓✓✓✓✓ | ✓✓✓ |
| Renal | ✓✓✓✓✓ | ✓✓✓✓✓ | ✓✓✓ | ✓ |
| Respiratory Diseases | ✓✓✓ | ✓ | ||
| Skin Diseases | ✓✓✓✓✓ | ✓✓✓✓✓ | ✓✓✓ |
FUTURE DEVELOPMENTS AND OUTLOOK
Development of the HPO has continued steadily since its initial publication in 2008 (1), and has focused on providing a well defined, comprehensive, and interoperable resource for computational analysis of human disease phenotypes, and has been used as a basis for a wide panoply of tools to perform analysis in clinical and in research settings. The HPO has been adopted by a growing number of groups internationally, and efforts are underway to translate the HPO into six languages, as we will report on in the future.
Orphanet serves as a reference portal for rare diseases populated by literature curation and validated by international experts (3). The HPO project and Orphanet are working on the creation of an integrated RD-specific informatics ecosystem that will build on the HPO as well as the Orphanet Rare Disease Ontology (ORDO), an open-access ontology developed from the Orphanet information system (60).
While the initial focus of the HPO was placed on rare, mainly Mendelian diseases, HPO annotations are now available also for 3145 common diseases (10). Current work will involve the extension of HPO resources for precision medicine, cancer, and disorders such as congenital heart malformations that are characterized by non-Mendelian inheritance.
Acknowledgments
The authors are grateful for the work of Miranda Jarnot and Tammy Powell at the National Library of Medicine for leading the work to import the HPO into the UMLS. The views expressed in this publication are those of the authors and not necessarily those of the funding agencies involved.
FUNDING
National Institutes of Health (NIH) Monarch Initiative [NIH OD #5R24OD011883]; E-RARE 2015 program, Hipbi-RD (harmonizing phenomics information for a better interoperability in the RD field); Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under [DE-AC02-05CH11231]; Bundesministerium für Bildung und Forschung (BMBF) [0313911]; Raine Clinician Research Fellowship (to G.B.); Stanley Institute for Cognitive Genomics at Cold Spring Harbor Laboratory (CSHL to G.J.L.); European Union Seventh Framework Programme [FP7/2007-2013] supported RD-Connect [305444], EURenOmics [2012-305608] and NeurOmics [2012-305121]; Fight for Sight and Retinitis Pigmentosa Fighting Blindness (to N.P.); National Institute for Health Research Biomedical Research Centre at Moorfields Eye Hospital National Health Service Foundation Trust and UCL Institute of Ophthalmology (UK) (to V.C.); University of Kiel, by a grant from the German Research Foundation [HE5415/3-1 to I.H.] within the EuroEPINOMICS framework of the European Science Foundation and grants of the German Research Foundation [DFG, HE5415/5-1, HE5415/6-1], German Ministry for Education and Research [01DH12033, MAR 10/012] and by the German chapter of the International League against Epilepsy (DGfE); International League Against Epilepsy (ILAE to I.H.) within the Epilepsiome initiative of the ILAE Genetics Commission (www.channelopathist.net); National Library of Medicine [R44 LM011585-02 to M.S.]. BBAdV is funded by the Dutch Organisation for Health Research and Development (ZON-MW grants 912-12-109). Funding for open access charge: NIH [R24-OD011883].
Conflict of interest statement. None declared.
REFERENCES
- 1.Robinson P.N., Köhler S., Bauer S., Seelow D., Horn D., Mundlos S. The Human Phenotype Ontology: a tool for annotating and analyzing human hereditary disease. Am. J. Hum. Genet. 2008;83:610–615. doi: 10.1016/j.ajhg.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McMurry J.A., Köhler S., Washington N.L., Balhoff J.P., Borromeo C., Brush M., Carbon S., Conlin T., Dunn N., Engelstad M., et al. Navigating the phenotype frontier: the monarch initiative. Genetics. 2016;203:1491–1495. doi: 10.1534/genetics.116.188870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rath A., Olry A., Dhombres F., Brandt M.M.C., Urbero B., Ayme S. Representation of rare diseases in health information systems: the Orphanet approach to serve a wide range of end users. Hum. Mutat. 2012;33:803–808. doi: 10.1002/humu.22078. [DOI] [PubMed] [Google Scholar]
- 4.Biesecker L.G. Phenotype matters. Nat. Genet. 2004;36:323–324. doi: 10.1038/ng0404-323. [DOI] [PubMed] [Google Scholar]
- 5.Robinson P.N., Webber C. Phenotype ontologies and cross-species analysis for translational research. PLoS Genet. 2014;10:e1004268. doi: 10.1371/journal.pgen.1004268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson P.N. Deep phenotyping for precision medicine. Hum. Mutat. 2012;33:777–780. doi: 10.1002/humu.22080. [DOI] [PubMed] [Google Scholar]
- 7.Deans A.R., Lewis S.E., Huala E., Anzaldo S.S., Ashburner M., Balhoff J.P., Blackburn D.C., Blake J.A., Burleigh J.G., Chanet B., et al. Finding our way through phenotypes. PLoS Biol. 2015;13:e1002033. doi: 10.1371/journal.pbio.1002033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Köhler S., Doelken S.C., Mungall C.J., Bauer S., Firth H.V., Bailleul-Forestier I., Black G.C.M., Brown D.L., Brudno M., Campbell J., et al. The Human phenotype ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014;42:D966–D974. doi: 10.1093/nar/gkt1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groza T., Köhler S., Doelken S., Collier N., Oellrich A., Smedley D., Couto F.M., Baynam G., Zankl A., Robinson P.N. Automatic concept recognition using the human phenotype ontology reference and test suite corpora. Database. 2015;2015:bav005. doi: 10.1093/database/bav005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groza T., Köhler S., Moldenhauer D., Vasilevsky N., Baynam G., Zemojtel T., Schriml L.M., Kibbe W.A., Schofield P.N., Beck T., et al. The human phenotype ontology: Semantic Unification of Common and Rare Disease. Am. J. Hum. Genet. 2015;97:111–124. doi: 10.1016/j.ajhg.2015.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beck T., Hastings R.K., Gollapudi S., Free R.C., Brookes A.J. GWAS Central: a comprehensive resource for the comparison and interrogation of genome-wide association studies. Eur. J. Hum. Genet. 2014;22:949–952. doi: 10.1038/ejhg.2013.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li M.J., Liu Z., Wang P., Wong M.P., Nelson M.R., Kocher J.-P.A., Yeager M., Sham P.C., Chanock S.J., Xia Z., et al. GWASdb v2: an update database for human genetic variants identified by genome-wide association studies. Nucleic Acids Res. 2016;44:D869–D876. doi: 10.1093/nar/gkv1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bush W.S., Oetjens M.T., Crawford D.C. Unravelling the human genome-phenome relationship using phenome-wide association studies. Nat. Rev. Genet. 2016;17:129–145. doi: 10.1038/nrg.2015.36. [DOI] [PubMed] [Google Scholar]
- 14.Washington N.L., Haendel M.A., Köhler S., Lewis S.E., Robinson P., Smedley D., Mungall C.J. How good is your phenotyping? Methods for quality assessment. 2014. http://phenoday2014.bio-lark.org/ Phenotype Day @ ISMB2014.
- 15.Girdea M., Dumitriu S., Fiume M., Bowdin S., Boycott K.M., Chénier S., Chitayat D., Faghfoury H., Meyn M.S., Ray P.N., et al. PhenoTips: patient phenotyping software for clinical and research use. Hum. Mutat. 2013;34:1057–1065. doi: 10.1002/humu.22347. [DOI] [PubMed] [Google Scholar]
- 16.Buske O.J., Girdea M., Dumitriu S., Gallinger B., Hartley T., Trang H., Misyura A., Friedman T., Beaulieu C., Bone W.P., et al. PhenomeCentral: a portal for phenotypic and genotypic matchmaking of patients with rare genetic diseases. Hum. Mutat. 2015;36:931–940. doi: 10.1002/humu.22851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rainer W., Bodenreider O. Proceedings of the Joint BioOntologies and BioLINK ISMB’2014 SIG session ‘Phenotype Day’. 2014. Coverage of phenotypes in standard terminologies; pp. 41–44. [Google Scholar]
- 18.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32:D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dhombres F., Bodenreider O. Interoperability between phenotypes in research and healthcare terminologies—investigating partial mappings between HPO and SNOMED CT. J. Biomed. Semantics. 2016;7:3. doi: 10.1186/s13326-016-0047-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boycott K.M., Vanstone M.R., Bulman D.E., MacKenzie A.E. Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nat. Rev. Genet. 2013;14:681–691. doi: 10.1038/nrg3555. [DOI] [PubMed] [Google Scholar]
- 21.Chong J.X., Buckingham K.J., Jhangiani S.N., Boehm C., Sobreira N., Smith J.D., Harrell T.M., McMillin M.J., Wiszniewski W., Gambin T., et al. The genetic basis of mendelian phenotypes: discoveries, challenges, and opportunities. Am. J. Hum. Genet. 2015;97:199–215. doi: 10.1016/j.ajhg.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groza T., Hunter J., Zankl A. The bone dysplasia ontology: integrating genotype and phenotype information in the skeletal dysplasia domain. BMC Bioinformatics. 2012;13:50. doi: 10.1186/1471-2105-13-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paul R., Groza T., Hunter J., Zankl A. Decision support methods for finding phenotype—disorder associations in the bone dysplasia domain. PLoS One. 2012;7:e50614. doi: 10.1371/journal.pone.0050614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bragin E., Chatzimichali E.A., Wright C.F., Hurles M.E., Firth H.V., Bevan A.P., Swaminathan G.J. DECIPHER: database for the interpretation of phenotype-linked plausibly pathogenic sequence and copy-number variation. Nucleic Acids Res. 2014;42:D993–D1000. doi: 10.1093/nar/gkt937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taruscio D., Groft S.C., Cederroth H., Melegh B., Lasko P., Kosaki K., Baynam G., McCray A., Gahl W.A. Undiagnosed diseases network international (UDNI): white paper for global actions to meet patient needs. Mol. Genet. Metab. 2015;116:223–225. doi: 10.1016/j.ymgme.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 26.Claes P., Liberton D.K., Daniels K., Rosana K.M., Quillen E.E., Pearson L.N., McEvoy B., Bauchet M., Zaidi A.A., Yao W., et al. Modeling 3D facial shape from DNA. PLoS Genet. 2014;10:e1004224. doi: 10.1371/journal.pgen.1004224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baynam G., Pachter N., McKenzie F., Townshend S., Slee J., Kiraly-Borri C., Vasudevan A., Hawkins A., Broley S., Schofield L., et al. The rare and undiagnosed diseases diagnostic service - application of massively parallel sequencing in a state-wide clinical service. Orphanet. J. Rare Dis. 2016;11:77. doi: 10.1186/s13023-016-0462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson R., Johnston L., Taruscio D., Monaco L., Béroud C., Gut I.G., Hansson M.G., ’t Hoen P.-B.A., Patrinos G.P., Dawkins H., et al. RD-Connect: an integrated platform connecting databases, registries, biobanks and clinical bioinformatics for rare disease research. J. Gen. Intern. Med. 2014;29(Suppl. 3):S780–S787. doi: 10.1007/s11606-014-2908-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suls A., Jaehn J.A., Kecskés A., Weber Y., Weckhuysen S., Craiu D.C., Siekierska A., Djémié T., Afrikanova T., Gormley P., et al. De novo loss-of-function mutations in CHD2 cause a fever-sensitive myoclonic epileptic encephalopathy sharing features with Dravet syndrome. Am. J. Hum. Genet. 2013;93:967–975. doi: 10.1016/j.ajhg.2013.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nava C., Dalle C., Rastetter A., Striano P., de Kovel C.G.F., Nabbout R., Cancès C., Ville D., Brilstra E.H., Gobbi G., et al. De novo mutations in HCN1 cause early infantile epileptic encephalopathy. Nat. Genet. 2014;46:640–645. doi: 10.1038/ng.2952. [DOI] [PubMed] [Google Scholar]
- 31.Albers J.A., Weckhuysen S., Suls A., Coessens B., Robinson P.N., De Jonghe P., Euroepinomics RES, Helbig I. Data-driven phenomic analysis of epileptic encephalopathy phenotypes using an ontology-based phenotype database. Euroepinomics Res. 2012;53:1–245. [Google Scholar]
- 32.Zemojtel T., Köhler S., Mackenroth L., Jäger M., Hecht J., Krawitz P., Graul-Neumann L., Doelken S., Ehmke N., Spielmann M., et al. Effective diagnosis of genetic disease by computational phenotype analysis of the disease-associated genome. Sci. Transl. Med. 2014;6:252ra123. doi: 10.1126/scitranslmed.3009262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ales M., Luca L., Marija V., Gorazd R., Karin W., Ana B., Alenka H., Peterlin B. Phenotype-driven gene target definition in clinical genome-wide sequencing data interpretation. Genet. Med. 2016 doi: 10.1038/gim.2016.22. doi:10.1038/gim.2016.22. [DOI] [PubMed] [Google Scholar]
- 34.Simeoni I., Stephens J.C., Hu F., Deevi S.V., Megy K., Bariana T.K., Lentaigne C., Schulman S., Sivapalaratnam S., Vries M.J., et al. A high-throughput sequencing test for diagnosing inherited bleeding, thrombotic, and platelet disorders. Blood. 2016;127:2791–2803. doi: 10.1182/blood-2015-12-688267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson P.N., Köhler S., Oellrich A., Sanger Mouse Genetics Project Sanger Mouse Genetics Project. Wang K., Mungall C.J., Lewis S.E., Washington N., Bauer S., Seelow D., et al. Improved exome prioritization of disease genes through cross-species phenotype comparison. Genome Res. 2014;24:340–348. doi: 10.1101/gr.160325.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singleton M.V., Guthery S.L., Voelkerding K.V., Chen K., Kennedy B., Margraf R.L., Durtschi J., Eilbeck K., Reese M.G., Jorde L.B., et al. Phevor combines multiple biomedical ontologies for accurate identification of disease-causing alleles in single individuals and small nuclear families. Am. J. Hum. Genet. 2014;94:599–610. doi: 10.1016/j.ajhg.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinto D., Delaby E., Merico D., Barbosa M., Merikangas A., Klei L., Thiruvahindrapuram B., Xu X., Ziman R., Wang Z., et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am. J. Hum. Genet. 2014;94:677–694. doi: 10.1016/j.ajhg.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merico D., Roifman M., Braunschweig U., Yuen R.K.C., Alexandrova R., Bates A., Reid B., Nalpathamkalam T., Wang Z., Thiruvahindrapuram B., et al. Compound heterozygous mutations in the noncoding RNU4ATAC cause Roifman Syndrome by disrupting minor intron splicing. Nat. Commun. 2015;6:8718. doi: 10.1038/ncomms9718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sulem P., Helgason H., Oddson A., Stefansson H., Gudjonsson S.A., Zink F., Hjartarson E., Sigurdsson G.T., Jonasdottir A., Jonasdottir A., et al. Identification of a large set of rare complete human knockouts. Nat. Genet. 2015;47:448–452. doi: 10.1038/ng.3243. [DOI] [PubMed] [Google Scholar]
- 40.Bowles N.E., Jou C.J., Arrington C.B., Kennedy B.J., Earl A., Matsunami N., Meyers L.L., Etheridge S.P., Saarel E.V., Bleyl S.B., et al. Exome analysis of a family with Wolff-Parkinson-White syndrome identifies a novel disease locus. Am. J. Med. Genet. A. 2015;167:2975–2984. doi: 10.1002/ajmg.a.37297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pippucci T., Parmeggiani A., Palombo F., Maresca A., Angius A., Crisponi L., Cucca F., Liguori R., Valentino M.L., Seri M., et al. A novel null homozygous mutation confirms CACNA2D2 as a gene mutated in epileptic encephalopathy. PLoS One. 2013;8:e82154. doi: 10.1371/journal.pone.0082154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Requena T., Cabrera S., Martín-Sierra C., Price S.D., Lysakowski A., Lopez-Escamez J.A. Identification of two novel mutations in FAM136A and DTNA genes in autosomal-dominant familial Meniere's disease. Hum. Mol. Genet. 2015;24:1119–1126. doi: 10.1093/hmg/ddu524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Covone A.E., Fiorillo C., Acquaviva M., Trucco F., Morana G., Ravazzolo R., Minetti C. WES in a family trio suggests involvement of TECPR2 in a complex form of progressive motor neuron disease. Clin. Genet. 2016;90:182–185. doi: 10.1111/cge.12730. [DOI] [PubMed] [Google Scholar]
- 44.Westbury S.K., Turro E., Greene D., Lentaigne C., Kelly A.M., Bariana T.K., Simeoni I., Pillois X., Attwood A., Austin S., et al. Human phenotype ontology annotation and cluster analysis to unravel genetic defects in 707 cases with unexplained bleeding and platelet disorders. Genome Med. 2015;7:36. doi: 10.1186/s13073-015-0151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh T., Kurki M.I., Curtis D., Purcell S.M., Crooks L., McRae J., Suvisaari J., Chheda H., Blackwood D., Breen G., et al. Rare loss-of-function variants in SETD1A are associated with schizophrenia and developmental disorders. Nat. Neurosci. 2016;19:571–577. doi: 10.1038/nn.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuen R.K.C., Thiruvahindrapuram B., Merico D., Walker S., Tammimies K., Hoang N., Chrysler C., Nalpathamkalam T., Pellecchia G., Liu Y., et al. Whole-genome sequencing of quartet families with autism spectrum disorder. Nat. Med. 2015;21:185–191. doi: 10.1038/nm.3792. [DOI] [PubMed] [Google Scholar]
- 47.Akawi N., McRae J., Ansari M., Balasubramanian M., Blyth M., Brady A.F., Clayton S., Cole T., Deshpande C., Fitzgerald T.W., et al. Discovery of four recessive developmental disorders using probabilistic genotype and phenotype matching among 4,125 families. Nat. Genet. 2015;47:1363–1369. doi: 10.1038/ng.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lentaigne C., Freson K., Laffan M.A., Turro E., Ouwehand W.H., BRIDGE-BPD Consortium and the ThromboGenomics Consortium Inherited platelet disorders: toward DNA-based diagnosis. Blood. 2016;127:2814–2823. doi: 10.1182/blood-2016-03-378588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stritt S., Nurden P., Turro E., Greene D., Jansen S.B., Westbury S.K., Petersen R., Astle W.J., Marlin S., Bariana T.K., et al. A gain-of-function variant in DIAPH1 causes dominant macrothrombocytopenia and hearing loss. Blood. 2016;127:2903–2914. doi: 10.1182/blood-2015-10-675629. [DOI] [PubMed] [Google Scholar]
- 50.Greene D., NIHR BioResource NIHR BioResource. Richardson S., Turro E. Phenotype similarity regression for identifying the genetic determinants of rare diseases. Am. J. Hum. Genet. 2016;98:490–499. doi: 10.1016/j.ajhg.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Turro E., Greene D., Wijgaerts A., Thys C., Lentaigne C., Bariana T.K., Westbury S.K., Kelly A.M., Selleslag D., Stephens J.C., et al. A dominant gain-of-function mutation in universal tyrosine kinase SRC causes thrombocytopenia, myelofibrosis, bleeding, and bone pathologies. Sci. Transl. Med. 2016;8:328ra30. doi: 10.1126/scitranslmed.aad7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Philippakis A.A., Azzariti D.R., Beltran S., Brookes A.J., Brownstein C.A., Brudno M., Brunner H.G., Buske O.J., Carey K., Doll C., et al. The Matchmaker Exchange: a platform for rare disease gene discovery. Hum. Mutat. 2015;36:915–921. doi: 10.1002/humu.22858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mungall C.J., Washington N.L., Nguyen-Xuan J., Condit C., Smedley D., Köhler S., Groza T., Shefchek K., Hochheiser H., Robinson P.N., et al. Use of model organism and disease databases to support matchmaking for human disease gene discovery. Hum. Mutat. 2015;36:979–984. doi: 10.1002/humu.22857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buske O.J., Schiettecatte F., Hutton B., Dumitriu S., Misyura A., Huang L., Hartley T., Girdea M., Sobreira N., Mungall C., et al. The Matchmaker Exchange API: automating patient matching through the exchange of structured phenotypic and genotypic profiles. Hum. Mutat. 2015;36:922–927. doi: 10.1002/humu.22850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pesquita C., Faria D., Falcão A.O., Lord P., Couto F.M., Bourne P.E. Semantic similarity in biomedical ontologies. PLoS Comput. Biol. 2009;5:e1000443. doi: 10.1371/journal.pcbi.1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Köhler S., Dölken S., Ruef B., Washington S.B.N., Westerfield M., Gkoutos G., Schofield P., Smedley D., Robinson P.N., Mungall C.J. Construction and accessibility of a cross-species phenotype ontology along with gene annotations for biomedical research. F1000 Res. 2013;2:30. doi: 10.12688/f1000research.2-30.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Washington N.L., Haendel M.A., Mungall C.J., Ashburner M., Westerfield M., Lewis S.E. Linking human diseases to animal models using ontology-based phenotype annotation. PLoS Biol. 2009;7:e1000247. doi: 10.1371/journal.pbio.1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robinson P.N., Mungall C.J., Haendel M. Capturing phenotypes for precision medicine. Cold Spring Harb. Mol. Case Stud. 2015;1:a000372. doi: 10.1101/mcs.a000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vasilevsky N., Engelstad M., Foster E., Mungall C., Robinson P., Köhler S., Haendel M. International Conference on Biological Ontology & BioCreative. 2016. Enhancing the human phenotype ontology for use by the layperson; p. IT402. [Google Scholar]
- 60.Vasant D., Chanas L., Malone J., Hanauer M., Olry A., Jupp S., Robinson P.N., Parkinson H., Rath A. ORDO: An ontology connecting rare disease, epidemiology and genetic data. 2014 http://phenoday2014.bio-lark.org/. Phenoday @ ISMB2014. [Google Scholar]
- 61.Firth H.V., Wright C.F., Study D. The Deciphering Developmental Disorders (DDD) study. Dev. Med. Child Neurol. 2011;53:702–703. doi: 10.1111/j.1469-8749.2011.04032.x. [DOI] [PubMed] [Google Scholar]
- 62.Wright C.F., Fitzgerald T.W., Jones W.D., Clayton S., McRae J.F., van Kogelenberg M., King D.A., Ambridge K., Barrett D.M., Bayzetinova T., et al. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet. 2015;385:1305–1314. doi: 10.1016/S0140-6736(14)61705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chatzimichali E.A., Brent S., Hutton B., Perrett D., Wright C.F., Bevan A.P., Hurles M.E., Firth H.V., Swaminathan G.J. Facilitating collaboration in rare genetic disorders through effective matchmaking in DECIPHER. Hum. Mutat. 2015;36:941–949. doi: 10.1002/humu.22842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Silfhout A.T.V., van Ravenswaaij C.M.A., Hehir-Kwa J.Y., Verwiel E.T.P., Dirks R., van Vooren S., Schinzel A., de Vries B.B.A., de Leeuw N. An update on ECARUCA, the European cytogeneticists association register of unbalanced chromosome aberrations. Eur. J. Med. Genet. 2013 doi: 10.1016/j.ejmg.2013.06.010. doi:10.1016/j.ejmg.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 65.Marx V. The DNA of a nation. Nature. 2015;524:503–505. doi: 10.1038/524503a. [DOI] [PubMed] [Google Scholar]
- 66.Bone W.P., Washington N.L., Buske O.J., Adams D.R., Davis J., Draper D., Flynn E.D., Girdea M., Godfrey R., Golas G., et al. Computational evaluation of exome sequence data using human and model organism phenotypes improves diagnostic efficiency. Genet. Med. 2016;18:608–617. doi: 10.1038/gim.2015.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rehm H.L., Berg J.S., Brooks L.D., Bustamante C.D., Evans J.P., Landrum M.J., Ledbetter D.H., Maglott D.R., Martin C.L., Nussbaum R.L., et al. ClinGen—the clinical genome resource. N. Engl. J. Med. 2015;372:2235–2242. doi: 10.1056/NEJMsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kirkpatrick B.E., Riggs E.R., Azzariti D.R., Miller V.R., Ledbetter D.H., Miller D.T., Rehm H., Martin C.L., Faucett W.A., ClinGen Resource GenomeConnect: matchmaking between patients, clinical laboratories, and researchers to improve genomic knowledge. Hum. Mutat. 2015;36:974–978. doi: 10.1002/humu.22838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beaulieu C.L., Majewski J., Schwartzentruber J., Samuels M.E., Fernandez B.A., Bernier F.P., Brudno M., Knoppers B., Marcadier J., Dyment D., et al. FORGE Canada consortium: outcomes of a 2-year national rare-disease gene-discovery project. Am. J. Hum. Genet. 2014;94:809–817. doi: 10.1016/j.ajhg.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Köhler S., Schulz M.H., Krawitz P., Bauer S., Dölken S., Ott C.E., Mundlos C., Horn D., Mundlos S., Robinson P.N. Clinical diagnostics in human genetics with semantic similarity searches in ontologies. Am. J. Hum. Genet. 2009;85:457–464. doi: 10.1016/j.ajhg.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bauer S., Köhler S., Schulz M.H., Robinson P.N. Bayesian ontology querying for accurate and noise-tolerant semantic searches. Bioinformatics. 2012;28:2502–2508. doi: 10.1093/bioinformatics/bts471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Basel-Vanagaite L., Wolf L., Orin M., Larizza L., Gervasini C., Krantz I.D., Deardoff M.A. Recognition of the cornelia de lange syndrome phenotype with facial dysmorphology novel analysis. Clin. Genet. 2016;89:557–563. doi: 10.1111/cge.12716. [DOI] [PubMed] [Google Scholar]
- 73.Yang H., Robinson P.N., Wang K. Phenolyzer: phenotype-based prioritization of candidate genes for human diseases. Nat Methods. 2015;12:841–843. doi: 10.1038/nmeth.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smedley D., Jacobsen J.O.B., Jäger M., Köhler S., Holtgrewe M., Schubach M., Siragusa E., Zemojtel T., Buske O.J., Washington N.L., et al. Next-generation diagnostics and disease-gene discovery with the Exomiser. Nat. Protoc. 2015;10:2004–2015. doi: 10.1038/nprot.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Trakadis Y.J., Buote C., Therriault J.-F., Jacques P.-É., Larochelle H., Lévesque S. PhenoVar: a phenotype-driven approach in clinical genomics for the diagnosis of polymalformative syndromes. BMC Med. Genomics. 2014;7:22. doi: 10.1186/1755-8794-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sifrim A., Popovic D., Tranchevent L.-C., Ardeshirdavani A., Sakai R., Konings P., Vermeesch J.R., Aerts J., De Moor B., Moreau Y. eXtasy: variant prioritization by genomic data fusion. Nat. Methods. 2013;10:1083–1084. doi: 10.1038/nmeth.2656. [DOI] [PubMed] [Google Scholar]
- 77.James R.A., Campbell I.M., Chen E.S., Boone P.M., Rao M.A., Bainbridge M.N., Lupski J.R., Yang Y., Eng C.M., Posey J.E., et al. A visual and curatorial approach to clinical variant prioritization and disease gene discovery in genome-wide diagnostics. Genome Med. 2016;8:13. doi: 10.1186/s13073-016-0261-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Javed A., Agrawal S., Ng P.C. Phen-Gen: combining phenotype and genotype to analyze rare disorders. Nat. Methods. 2014;11:935–937. doi: 10.1038/nmeth.3046. [DOI] [PubMed] [Google Scholar]
- 79.Smedley D., Schubach M., Jacobsen J.O.B., Köhler S., Zemojtel T., Spielmann M., Jäger M., Hochheiser H., Washington N.L., McMurry J.A., et al. A whole-genome analysis framework for effective identification of pathogenic regulatory variants in mendelian disease. Am. J. Hum. Genet. 2016;99:595–606. doi: 10.1016/j.ajhg.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen J., Bardes E.E., Aronow B.J., Jegga A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang J., Duncan D., Shi Z., Zhang B. WEB-based GEne SeT ANALYSIS toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41:W77–W83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wilson D., Pethica R., Zhou Y., Talbot C., Vogel C., Madera M., Chothia C., Gough J. SUPERFAMILY—sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic Acids Res. 2009;37:D380–D386. doi: 10.1093/nar/gkn762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jiang R. Walking on multiple disease-gene networks to prioritize candidate genes. J. Mol. Cell Biol. 2015;7:214–230. doi: 10.1093/jmcb/mjv008. [DOI] [PubMed] [Google Scholar]
- 85.Hart S.N., Moore R.M., Zimmermann M.T., Oliver G.R., Egan J.B., Bryce A.H., Kocher J.-P.A. PANDA: pathway and annotation explorer for visualizing and interpreting gene-centric data. PeerJ. 2015;3:e970. doi: 10.7717/peerj.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gottlieb A., Stein G.Y., Ruppin E., Sharan R. PREDICT: a method for inferring novel drug indications with application to personalized medicine. Mol. Syst. Biol. 2011;7:496. doi: 10.1038/msb.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gonzalez M., Falk M.J., Gai X., Postrel R., Schüle R., Zuchner S. Innovative genomic collaboration using the GENESIS (GEM.app) platform. Hum. Mutat. 2015;36:950–956. doi: 10.1002/humu.22836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Smedley D., Oellrich A., Köhler S., Ruef B., Project S.M.G., Westerfield M., Robinson P., Lewis S., Mungall C. PhenoDigm: analyzing curated annotations to associate animal models with human diseases. Database. 2013;2013:bat025. doi: 10.1093/database/bat025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen C.-K., Mungall C.J., Gkoutos G.V., Doelken S.C., Köhler S., Ruef B.J., Smith C., Westerfield M., Robinson P.N., Lewis S.E., et al. MouseFinder: candidate disease genes from mouse phenotype data. Hum. Mutat. 2012;33:858–866. doi: 10.1002/humu.22051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hoehndorf R., Schofield P.N., Gkoutos G.V. PhenomeNET: a whole-phenome approach to disease gene discovery. Nucleic Acids Res. 2011;39:e119. doi: 10.1093/nar/gkr538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hwang S., Kim E., Yang S., Marcotte E.M., Lee I. MORPHIN: a web tool for human disease research by projecting model organism biology onto a human integrated gene network. Nucleic Acids Res. 2014;42:W147–W153. doi: 10.1093/nar/gku434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Köhler S., Schoeneberg U., Czeschik J.C., Doelken S.C., Hehir-Kwa J.Y., Ibn-Salem J., Mungall C.J., Smedley D., Haendel M.A., Robinson P.N. Clinical interpretation of CNVs with cross-species phenotype data. J. Med. Genet. 2014;51:766–772. doi: 10.1136/jmedgenet-2014-102633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rappaport N., Nativ N., Stelzer G., Twik M., Guan-Golan Y., Stein T.I., Bahir I., Belinky F., Morrey C.P., Safran M., et al. MalaCards: an integrated compendium for diseases and their annotation. Database. 2013;2013:bat018. doi: 10.1093/database/bat018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rubinstein W.S., Maglott D.R., Lee J.M., Kattman B.L., Malheiro A.J., Ovetsky M., Hem V., Gorelenkov V., Song G., Wallin C., et al. The NIH genetic testing registry: a new, centralized database of genetic tests to enable access to comprehensive information and improve transparency. Nucleic Acids Res. 2013;41:D925–D935. doi: 10.1093/nar/gks1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Amberger J.S., Bocchini C.A., Schiettecatte F., Scott A.F., Hamosh A. OMIM.org: Online mendelian inheritance in man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fang H., Gough J. DcGO: database of domain-centric ontologies on functions, phenotypes, diseases and more. Nucleic Acids Res. 2013;41:D536–D544. doi: 10.1093/nar/gks1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Landrum M.J., Lee J.M., Riley G.R., Jang W., Rubinstein W.S., Church D.M., Maglott D.R. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Araki H., Knapp C., Tsai P., Print C. GeneSetDB: A comprehensive meta-database, statistical and visualisation framework for gene set analysis. FEBS Open Bio. 2012;2:76–82. doi: 10.1016/j.fob.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shen L., Diroma M.A., Gonzalez M., Navarro-Gomez D., Leipzig J., Lott M.T., van Oven M., Wallace D.C., Muraresku C.C., Zolkipli-Cunningham Z., et al. MSeqDR: A centralized knowledge repository and bioinformatics web resource to facilitate genomic investigations in mitochondrial disease. Hum. Mutat. 2016;37:540–548. doi: 10.1002/humu.22974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gazzo A.M., Daneels D., Cilia E., Bonduelle M., Abramowicz M., Van Dooren S., Smits G., Lenaerts T. DIDA: A curated and annotated digenic diseases database. Nucleic Acids Res. 2016;44:D900–D907. doi: 10.1093/nar/gkv1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Morgan T., Schmidt J., Haakonsen C., Lewis J., Della Rocca M., Morrison S., Biesecker B., Kaphingst K.A. Using the internet to seek information about genetic and rare diseases: a case study comparing data from 2006 and 2011. JMIR Res. Protoc. 2014;3:e10. doi: 10.2196/resprot.2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Glueck M., Gvozdik A., Chevalier F., Khan A., Brudno M., Wigdor D. PhenoStacks: cross-sectional cohort phenotype comparison visualizations. IEEE Trans. Vis. Comput. Graph. 2016 doi: 10.1109/TVCG.2016.2598469. doi:10.1109/TVCG.2016.2598469. [DOI] [PubMed] [Google Scholar]
- 103.Glueck M., Hamilton P., Chevalier F., Breslav S., Khan A., Wigdor D., Brudno M. PhenoBlocks: phenotype comparison visualizations. IEEE Trans. Vis. Comput. Graph. 2016;22:101–110. doi: 10.1109/TVCG.2015.2467733. [DOI] [PubMed] [Google Scholar]